Abstract
Mix progenitors are short-lived multipotential cells formed as intestinal epithelial stem cells initiate a differentiation program. Clone dynamics indicates that various epithelial cell lineages arise from Mix via a sequence of progressively restricted progenitor states. Lateral inhibitory Notch signaling between the daughters of Mix (DOM) is thought to break their initial symmetry, thereby determining whether a DOM invokes a columnar (absorptive) or granulocytic (secretory) cell lineage program. This is supported by the absence of granulocytes following enforced Notch signaling or Atoh1 deletion. Conversely, granulocytes increase in frequency following inhibition of Notch signaling or Hes1 deletion. Thus reciprocal repression between Hes1 and Atoh1 is thought to implement a Notch signaling-driven cell-fate-determining binary switch in DOM. The brush (tuft) cells, a poorly understood chemosensory cell type, are not incorporated into this model. We report that brush cell numbers increase dramatically following conditional Atoh1-deletion, demonstrating that brush cell production, determination, differentiation and survival are Atoh1-independent. We also report that brush cells are derived from Gfi1b-expressing progenitors. These and related results suggest a model in which initially equivalent DOM progenitors have three metastable states defined by the transcription factors Hes1, Atoh1, and Gfi1b. Lateral inhibitory Notch signaling normally ensures that Hes1 dominates in one of the two DOMs, invoking a columnar lineage program, while either Atoh1 or Gfi1b dominates in the other DOM, invoking a granulocytic or brush cell lineage program, respectively, and thus implementing a cell fate-determining ternary switch.
Keywords: Brush cell, Tuft cell, Hes1, Atoh1, Gfi1b, Notch signaling, Intestinal stem cell
Introduction
The adult mammalian intestinal epithelium is a continuously renewed tissue comprised of four principal and several relatively minor cell lineages, all ultimately derived from a resident stem cell population. The highly structured and dynamic nature of the tissue makes it an excellent system in which to study stem cell biology and lineage determination. The stem cells (S) are self-renewing multipotent cells uncommitted to specific epithelial lineages. The scope of the set of intestinal epithelial cells with stem cell potential remains contentious, but clearly includes the crypt base columnar cells, which were initially thought to be distributed throughout cell positions 1–9 of the crypt base (Cheng and Leblond, 1974a,b), but were subsequently understood to reside in a stem-cell zone in cell positions 1–4 (Bjerknes and Cheng, 1979, 1981a,b, 1999). The crypt base columnar cells express Lgr5 (Barker et al., 2007), which enabled their isolation and clonal culture (Sato et al., 2009). Progeny of S that leave the stem cell zone and initiate differentiation give rise to short-lived Mix progenitors (Bjerknes and Cheng, 1999) that in turn divide to generate the daughters of Mix (DOM), likely in a region just above the stem cell zone referred to as the common origin of differentiation (COD; Bjerknes and Cheng, 2006a,b, 2010).
The four principal lineages are the columnar, mucous, Paneth and enteroendocrine cell lineages. Mature columnar lineage cells are the preponderant epithelial cell type, explaining their commonly used alias ‘enterocytes’, meaning ‘gut cells’. They participate in multiple aspects of mucosal defense, digestion, and nutrient uptake, the latter function motivating yet another alias, ‘absorptive cells’. The mucous, Paneth and enteroendocrine cell lineages share many features. Most obvious are the eponymic secretory granules characteristic of the mature cells of these lineages, hence they are collectively referred to as the secretory or granulocytic lineages. Less obvious is the fact that all granulocytes express the basic helix–loop–helix transcription factor Atoh1 (also known as Math1 and Hath1). Granulocytes are absent from Atoh1-deficient epithelium (Shroyer et al., 2007; Yang et al., 2001); conversely forced Atoh1 expression in fetal intestine results in increased expression of granulocytic markers (VanDussen and Samuelson, 2010), indicating that Atoh1 expression promotes granulocytic lineage programs. Hence the granulocytic or secretory lineages may also be usefully referred to as Atoh1-dependent lineages. However, it remains unclear whether the Atoh1-dependence of the granulocytic lineages is due to a shared origin from an Atoh1-dependent common progenitor or due to a shared dependence on Atoh1 for their formation, differentiation or survival (Bjerknes and Cheng, 2006a,b, 2010; Yang et al., 2001).
Columnar lineage cells do not normally express Atoh1 and columnar cells are produced in Atoh1-deficient epithelium, demonstrating that Atoh1 is not required for their formation (Shroyer et al., 2007; Yang et al., 2001). Instead of Atoh1, early columnar lineage cells express the Notch signaling target Hes1 (Jarriault et al., 1998; Jensen et al., 2000; Kayahara et al., 2003; Schroder and Gossler, 2002). Granulocytes are more numerous in Hes1-deficient epithelium (Jensen et al., 2000), suggesting that Hes1 acts to repress granulocyte generation, probably by repressing Atoh1 expression (Akazawa et al., 1995; Jensen et al., 2000; Yang et al., 2001).
These results indicate, by analogy with other systems, that lateral inhibitory Notch signaling (Fortini, 2009) is involved in lineage specification in the epithelium, in large part by modulating the expression of the opposing transcription factors Hes1 and Atoh1 (Jensen et al., 2000; Yang et al., 2001). Accordingly DOM progenitors are thought to display Notch family transmembrane receptor proteins and ligands on their cell surface. One of the sister DOMs receives increased Notch signaling and consequently increases Notch receptor expression, while its sister DOM increases expression of Notch ligand (collectively Delta). We will refer to DOM entering these states as DOMNotch and DOMDelta, respectively. Increased Hes1 expression in DOMNotch represses Atoh1 and invokes a columnar lineage program leading DOMNotch to become a columnar lineage progenitor. Its sister DOMDelta receives diminished Notch signaling, and as a consequence increases Atoh1 expression which represses Hes1 and invokes a granulocytic lineage program. Thus, the interaction between Notch signaling, Hes1, and Atoh1 in the initially equivalent DOMs is thought to break their symmetry, thereby implementing a lineage-determining binary switch specifying the columnar and granulocytic lineages (Bjerknes and Cheng, 2005; Jensen et al., 2000; Yang et al., 2001).
Evidence continues to accumulate that lateral inhibitory Notch signaling participates in intestinal epithelial lineage specification. Reduction of Notch signaling by application of gamma-secretase inhibitors (Milano et al., 2004; Wong et al., 2004) or of antibodies against Notch1 and Notch2 (Wu et al., 2010), or by partial Notch1 and Notch2 inducible knockout (Riccio et al., 2008), leads to increased granulocyte production. Conversely, activating the Notch signaling pathway by forced expression of a transgene encoding an active intracellular fragment of Notch1 (Notch-IC) inhibits granulocyte production (Fre et al., 2005, 2009; Stanger et al., 2005). Similar mechanisms are operative in the intestinal epithelium of zebrafish (Crosnier et al., 2005) and Drosophila (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2007).
The origin of the brush cell lineage, a minor cell lineage derived from the stem cells via committed brush cell lineage progenitors, is not explained by current lineage specification models and hence its elucidation is likely to reveal additional insights into the early stages of lineage commitment in the epithelium. During the preparation of this manuscript, Gerbe et al. (2011) reported that brush cells are absent in Atoh1-deficient epithelium and hence constitute a fourth Atoh1-dependent or secretory lineage in addition to the mucous, Paneth, enteroendocrine lineages. However, our results directly contradict this finding and conclusion.
Brush cells (also known as tuft, caveolated, multivesicular, peculiar, undifferentiated, fibrillovesicular, s-, agranular light, or solitary chemosensory cells) are relatively rare cells in the small intestine (Sato, 2007). They have a narrowed apical region topped by a prominent collection of long microvilli (the brush or tuft). In the electron microscope, the apical region is further characterized by numerous bundles of filaments, microtubules, vesicles, and tortuous invaginations that run from the apical surface deep into the cytoplasm (Nabeyama and Leblond, 1974; Sato, 2007). Similar cells are found in the epithelium of multiple structures derived ontogenically from the fore-, mid-, and hind-gut (Sato, 2007; Sbarbati et al., 2010).
Brush cells are continuously renewed, but brush cells are not labeled an hour after a pulse of 3H-thymidine and mitotic brush cells have not been reported indicating that brush cells are post-mitotic and hence must originate from other sources (Tsubouchi and Leblond, 1979). Immature brush cells are seen in small intestinal crypts indicating that the brush cells are likely derived from S (Cheng and Leblond, 1974b). This was formally demonstrated when brush cells were shown to be among the cell types produced by stem cell derived clones (Bjerknes and Cheng, 1999). They also described brush cell clones containing no other lineages, indicating that intermediate committed brush cell lineage progenitors exist.
Brush cells express multiple elements of the taste receptor signaling pathway cascade (Sbarbati et al., 2010) including α-gustducin (Höfer et al., 1996) and Trpm5 (Bezençon et al., 2007, 2008; Kaske et al., 2007; Kokrashvili et al., 2009), and hence brush cells are a type of solitary chemosensory cell in the gut. Comparative microarray analysis of the transcriptomes of Trpm5-expressing versus non-expressing cells revealed dozens of genes that are expressed at higher levels amongst the Trpm5-expressing cells, including the transcription factor Gfi1b (Bezençon et al., 2008).
Gfi1b is a Zinc-finger transcriptional repressor (Doan et al., 2004; Vassen et al., 2005) that has been shown to play a significant role in erythro- and megakarypoiesis, and in hematopoietic stem cells (Garçon et al., 2005; Hernandez et al., 2010; Huang et al., 2004; Khandanpour et al., 2010; Laurent et al., 2009; Osawa et al., 2002; Randrianarison-Huetz et al., 2010; Saleque et al., 2002; 2007; Vassen et al., 2007). Gfi1b is closely related to Gfi1, a transcriptional repressor known to stabilize the mucous and Paneth cell lineages by repressing the pro-enteroendocrine transcription factor Neurog3 (Bjerknes and Cheng, 2010). Accordingly, enteroendocrine cells are more frequent, and mucous and Paneth cells are less frequent in Gfi1-null mice (Bjerknes and Cheng, 2010; Shroyer et al., 2005). The potential role of Gfi1b in stem cell function or cell lineage determination in the intestinal epithelium has not been examined.
Here we report that Gfi1b is expressed by brush cells but not by other epithelial lineages, and we use Gfi1b expression as a marker to characterize the brush cell lineage progenitor. We then investigate whether the brush cell lineage arises from the DOMNotch or DOMDelta side of the Notch signaling divide and conclude that it arises from DOMDelta. However, Atoh1 is not expressed by brush lineage cells. Furthermore, we report that brush cell numbers increase dramatically following conditional Atoh1 deletion in adult intestinal epithelium, demonstrating that the brush cell lineage does not require Atoh1 for its determination, production, differentiation, or survival. Thus despite its DOMDelta origin the brush cell lineage is not an Atoh1-dependent granulocytic cell lineage. We conclude by proposing a model in which Hes1, Atoh1, and Gfi1b participate in a genetic network forming a Notch-signaling driven ternary switch regulating early cell lineage determination in the intestinal epithelium.
Materials and methods
Mice
CD-1 mice used in this study were purchased from Charles River Canada. Gfi1bEGFP/+ mice (Vassen et al., 2007) and intestines were kindly provided by Drs. C. Kandapour and T. Möröy. Atoh1-Cre; Rosa26-LacZ intestines (Yang et al., 2010) were kindly provided by Drs. Q. Ding and L. Gan. Atoh1-CreERT2; Rosa26-LacZ intestines (Fujiyama et al., 2009), from mice treated with a single gavage of Tamoxifen in corn oil (270 mg/kg) and killed after 5 days, and from control mice treated with oil only, were kindly provided by Drs. T. Fujiyama and M. Hoshino. Atoh1-EGFP (Rose et al., 2009) and Atoh1−/− (Ben-Arie et al., 1997) intestines were kindly provided by Drs. T. Klisch and H. Zoghbi.
For the Atoh1 conditional knockout study, a female homozygous Atoh1fl/fl mouse (Shroyer et al., 2007; The Jackson Laboratory, 008681) was mated with a male homozygous Rosa26CreERT2 mouse (Ventura et al., 2007; The Jackson Laboratory, 008463). The F1 offspring were mated to generate Atoh1fl/fl;Rosa26CreERT2/+ experimental and Atoh1fl/+;Rosa26CreERT2/+ control mice. Mice were given 3 daily gavages of Tamoxifen in oil (200 mg/kg) and killed 6 days after the initiation of treatment. The epithelium was isolated and processed for brush cell staining.
Rosa26 floxed-STOP-Notch-IC-IRES-EGFP mice have targeted into the Rosa26 allele a floxed STOP cassette controlling a Notch-IC transgene and an internal ribosome entry site (IRES) followed by EGFP modified to contain a nuclear localization signal. When the floxed STOP cassette is excised by Cre recombinase, both Notch-IC and the clone marker nuclear EGFP are transcribed from the transgene. For the Notch-IC clone study, a female homozygous Rosa26 floxed-STOP-Notch-IC-IRES-EGFP mouse (Murtaugh et al., 2003; The Jackson Laboratory, 008159), was mated with a male homozygous Rosa26CreERT2 mouse to generate F1 Rosa26CreERT2/floxed-STOP-Notch-IC-IRES-EGFP mice. F1 mice (9 weeks old) were given a single gavage of 5 mg Tamoxifen in oil and killed 72 days later. Epithelium was isolated and processed for Notch IC clones.
All mice were housed in specific pathogen free facilities. Tissues from at least 3 mice of each genotype were collected in compliance with protocols approved by the Animal Care Committees of the University of Toronto, the Institut de recherches cliniques de Montré-al, the Center of Comparative Medicine of Baylor College of Medicine, the National Institute of Neuroscience, Japan, and the University of Rochester. For embryo staging, noon on the day that the vaginal plug was observed was counted as embryonic day 0.5 (E0.5) and yolk sacs or tails were collected before fixation for genotyping
Recombination was induced by gavage of Tamoxifen in oil. A suspension of 20 mg/ml Tamoxifen (Sigma, T5648) in sterile canola oil containing 5% ethanol was dissolved at 37 °C for 4 h. The solution was aliquoted, stored at −20 °C, and melted at 37 °C immediately before use.
Tissue preparation and microscopy
Intestinal epithelium was isolated from proximal jejunum using 30 mM EDTA as previously described (Bjerknes and Cheng, 1981c) and fixed with 4% paraformaldehyde in PBS. In some preparations intact intestine was fixed with 4% paraformaldehyde in PBS, either by transcardiac perfusion followed by immersion in the same fixative, or by immersion. Immunofluorescence on either isolated cryptvillus units or microdissected crypts and villi from intact intestine was performed as previously described (Bjerknes and Cheng, 2006a, 2010). The antibodies and lectin used are listed in Table 1. Tissues were imaged with an AxioImager® Z1 microscope with a cooled CCD AxioCam®. Pseudocolor-multichannel-images were generated (including both fluorescence and differential interference contrast, DIC, images) and the background adjusted with Axiovision® 4.8 software (Carl Zeiss Canada).
Table 1.
Antibodies and markers.
| Antigen target of primary antibodies | Common aliases | Host | Source | |
|---|---|---|---|---|
| Dclk1 | Doublecortin-like kinase 1 | DCAMKL1 | Rabbit | Abcam |
| Krt18 | Keratin, type I cytoskeletal 18 | cytokeratin 18; CK18 | Rabbit | Epitomics |
| Ptgs1 | Prostaglandin-endoperoxide synthase 1 | Cyclooxygenase 1; Cox1 | Goat | Santa Cruz Biotechnology, Inc. |
| Trpm5 | Transient receptor potential cation channel, subfamily M, member 5 | Rabbit | gift from Dr. R. Margolskee (Kokrashvili et al., 2009) | |
| Atoh1 | Atonal homolog 1 (Drosophila) | Math1, Hath1 | Rabbit | Gift from Dr. J. Johnson (Helms and Johnson, 1998) |
| Neurog3 | Neurogenin 3 | Ngn3, Atoh5 | Rabbit | Gift from Dr. O. Madsen (Zahn et al., 2004) |
| Hes1 | Hairy and enhancer of split 1 (Drosophila) | Transcription factor HES-1 | Rabbit | Gift from Dr. T. Sudo (Ito et al., 2000) |
| Gfi1b | Growth factor independent 1B | Goat | Santa Cruz Biotechnology, Inc. | |
| Insm1 | Insulinoma-associated 1 | IA-1 | Guinea pig | Gift from Dr. C. Birchmeier (Jacob et al., 2009) |
| Mki67 | Antigen identified by monoclonal antibody Ki 67 | Ki67 | Rat (TEC-3) | DakoCytomation (Scholzen and Gerdes, 2000); |
| Chga | Chromogranin A | cgA | Rabbit | DakoCytomation |
| β-gal | β-galactosidase | lacZ | Rabbit | Cappel |
|
| ||||
| Secondary antibodies | ||||
|
| ||||
| rabbit IgG | Alexa Fluor® 488-, 555- and 647-conjugated anti-rabbit IgG | Donkey | Molecular Probes® | |
| Guinea pig IgG | DyLight™649-conjugated anti-Guinea pig IgG | Donkey | Jackson ImmunoResearch Laboratories, Inc. | |
|
| ||||
| Lectin | ||||
|
| ||||
| UEA-I | TRITC-conjugated Ulex europaeus agglutinin Type I | EY Laboratories, Inc. | ||
Cells of interest in whole mounts of crypts were assigned a cell position by first determining the point of intersection between the luminal axis and the crypt base. If this point was contained in a single cell, that cell was defined to be in position 1. Otherwise the cells flanking the intersection were considered to be in position 1. Cells immediately adjacent to these, as projected along the luminal axis, were assigned to position 2, and so forth up the crypt.
Fluorescence activated cell sorting (FACS), and quantitative PCR (qPCR)
Isolated jejunal epithelium was dissociated and fixed on ice in 4% paraformaldehyde in PBS (Bjerknes and Cheng, 2010). Following washing in PBS, brush cells were labeled for sorting using anti-Dclk1 followed by Alexa Fluor® 647-conjugated donkey anti-rabbit IgG secondary and DAPI. Brush and non-brush cells were sorted into cold PBS using a BD FACSAria®. At least 50,000 cells of each type were sorted each session. DNA was isolated using Fermentas GeneJETTM Genomic DNA purification Kit (#K0721) following manufacturer’s instructions. qPCR was performed using iQ SYBR Green® supermix (BioRad) on a CFX96 Real-Time PCR Detection System (BioRad).
Conditional Atoh1 deletion efficiency
Three primers were designed for use in quantifying the Atoh1fl, Atoh1Δ, and Atoh1+ alleles in tissues from experimental Atoh1fl/fl; Rosa26CreERT2/+ and control Atoh1fl/+;Rosa26CreERT2/+ mice before and after Tamoxifen induced recombination, as shown in Fig. 1A. The primer sequences were:
Fig. 1.
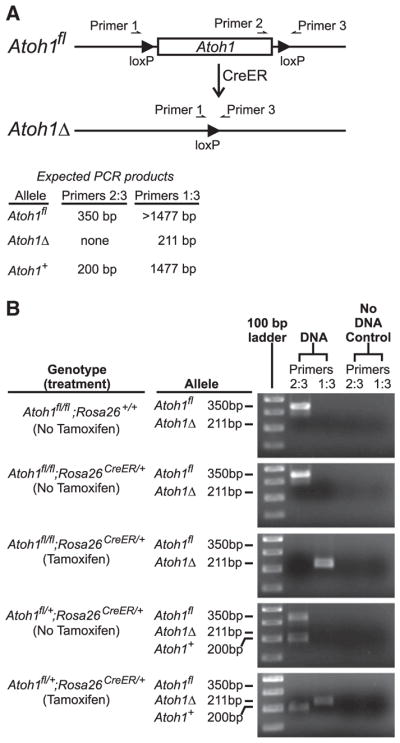
Design of the PCR scheme used to quantify floxed and recombined alleles of Atoh1 following Tamoxifen treatment of Atoh1fl/fl;Rosa26CreER/+ mice. (A) Schematic diagram showing the structure of the floxed Atoh1 allele before (Atoh1fl) and after (Atoh1Δ) Cre-induced recombination of the loxP sites that flank Atoh1. Note that primer 2 is excised following recombination, and the distance between primers 1 and 3 is decreased. The sizes of PCR products expected from the various alleles are indicated. (B) Images of agarose gels of the PCR products from the various genotypes before and after Tamoxifen treatment. Ear punch DNA was used for PCR reactions before Tamoxifen treatment. Isolated epithelial DNA was used after treatment. The floxed Atoh1 alleles were efficiently excised following Tamoxifen treatment.
primer 2—GACCTGTCGCCTTCGCTGCC, and
primer 3—GCGCGCTAGGAAGGGCATTTGG.
Two PCR primer pairs were used to probe the status of recombination of the Atoh1fl alleles. We denote these pairs Primers2:3 (qPCR efficiency 98.3%) and Primers1:3 (qPCR efficiency 99.4%). The PCR product expected from Primers1:3 spans the entire Atoh1 coding sequence and hence generates a long PCR product (1477 bp) from the wild type Atoh1+ allele, and a slightly longer product from the Atoh1fl allele (Fig. 1A). However, the PCR cycling program that we used incorporated a short extension time so that such long products were not efficiently amplified. Importantly, the recombined allele (denoted Atoh1Δ) yields a much shorter product that is efficiently amplified under the conditions used (Figs. 1A, B). The second primer pair, Primers2:3 spans the 3′ loxP site, and hence yields slightly differently sized products from the Atoh1+ and Atoh1fl alleles. Note that Primers2:3 generates no product following recombination induced excision of the Atoh1 coding region (Figs. 1A, B). Thus Primers2:3 provides a sensitive assay for any unrecombined Atoh1fl alleles following Tamoxifen activation of CreERT2 recombinase.
The fraction of Atoh1 alleles in a sample that were recombined, and thus transformed from Atoh1fl into Atoh1Δ alleles, was estimated from the qPCR CT determinations (primer pairs Primers1:3 and Primers2:3) using the formula 2ΔCT/(1+2ΔCT), where ΔCT = (CT(Primers2:3) −CT(Primers1:3)). The formula was derived as follows. The sample has R recombined (Atoh1Δ) and F floxed (Atoh1fl) alleles. The fraction of recombined alleles in the sample is therefore R/(F+R). The ratio R/F in the sample can be estimated from the qPCR results as R/F = 2ΔCT, assuming 100% PCR amplification efficiency for the primer pairs. Solving for R yieldsR = 2ΔCT F. Substituting this into R/(F+R) and simplifying yields the formula used.
We performed qPCR to determine the efficiency of recombination following 3 Tamoxifen doses and found that about 99.6% of Atoh1fl alleles in the epithelium underwent recombination (Table 2), demonstrating the nearly complete deletion of Atoh1 from the epithelium. The Atoh1fl allele in the control Atoh1fl/+;Rosa26CreERT2/+ mice was also efficiently recombined (Fig. 1B).
Table 2.
Recombination efficiency before and after Tamoxifen treatment.
| Genotype (treatment) | DNA source | ΔCT =(CT(Primers2:3) − CT(Primers1:3)) | Fraction of floxed Atoh1 alleles that are recombined = 2ΔCT/ (1 − 2ΔCT) |
|---|---|---|---|
| Atoh1fl/fl;Rosa26+/+ (none) | Ear punch | −∞a | 0±0 |
| Atoh1fl/fl;Rosa26CreER/+ (none) | Ear punch | −8.1±0.2 | 0.004±0.0004 |
| Atoh1fl/fl;Rosa26CreER/+ (Tamoxifen) | Intestinal epithelium | 7.9±0.3b | 0.996±0.0009b |
No product was detected with Primers1:3 under the PCR conditions used, hence by definition CT(Primers1 : 3) = −∞.
Significantly different from results obtained with DNA extracted from ear punches taken prior to Tamoxifen treatment.
It is also worth noting that this assay detected a low, but definite background recombination of Atoh1fl alleles in Atoh1fl/fl; Rosa26CreERT2/+ mice that had never been exposed to Tamoxifen (Table 2). No such background recombination was detected in Atoh1fl/fl;Rosa26+/+mice (i.e. no CreERT2). This low background recombination rate was of no consequence here but may be important in other studies such as lineage tracing experiments, especially if efficiently recombined targets are utilized. Appropriate controls need to be tested to determine the degree to which this is an issue in any specific case. We speculate that such background recombination may be due to exposure of genomic DNA to cytoplasmic CreERT2 during mitosis (following nuclear envelope disassembly), or to occasional degradation products of CreER that retain Cre recombinase activity but have lost the estrogen receptor module intended to restrict the fusion protein to the cytoplasm.
Data representation and statistics
Values are expressed as means±standard error of the mean (S.E.M.). Stated differences imply Student’s t-test returned P<0.05.
Results
Brush cell markers
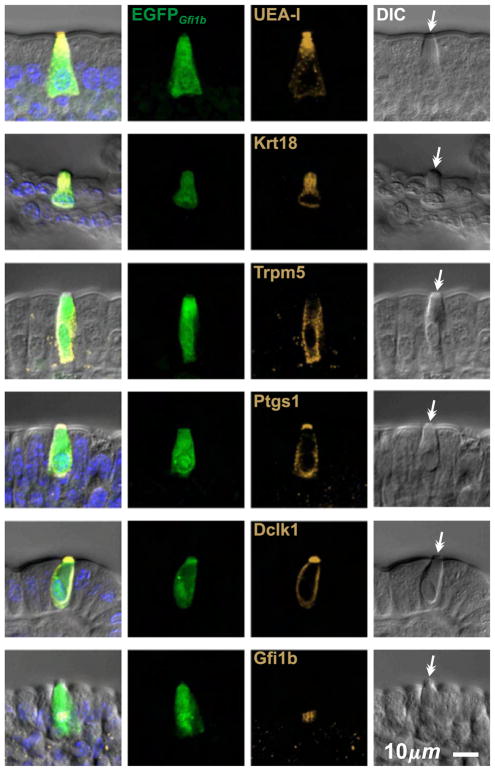
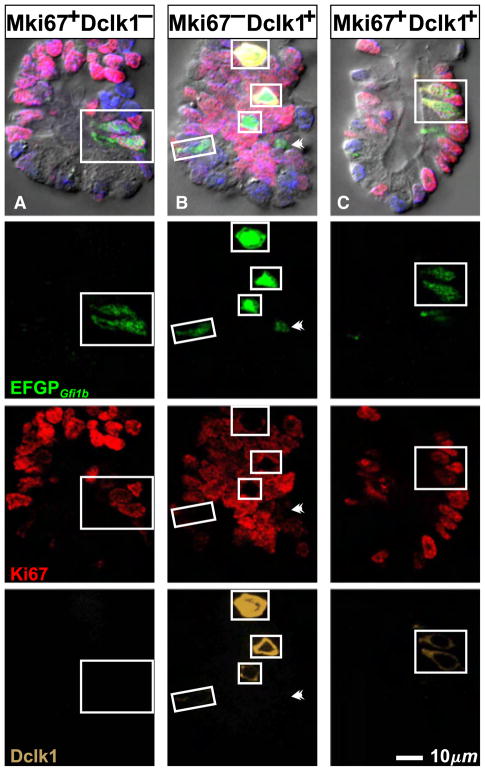
We confirmed that brush cells in epithelium isolated from CD-1 intestine are labeled by various reagents previously described to provide some selectivity for brush cells, including the lectin UEA-I (Gebhard and Gebert, 1999), and antibodies specific for Krt18 (Höfer and Drenckhahn, 1996), Trpm5 (Bezençon et al., 2007; Kaske et al., 2007; Kokrashvili et al., 2009), Ptgs1 (Bezençon et al., 2008), and Dclk1 (Bezençon et al., 2008; Gerbe et al., 2009) (Fig. 2). In making this assessment we relied on the distinctive appearance of brush cells in differential interference contrast (DIC) microscopy to confirm their identity and distinguish them from neighboring columnar or mucous cells. We found that UEA-I and anti-Dclk1 provided the best signal to noise ratio so we used them as primary brush cell markers in our studies.
Fig. 2.
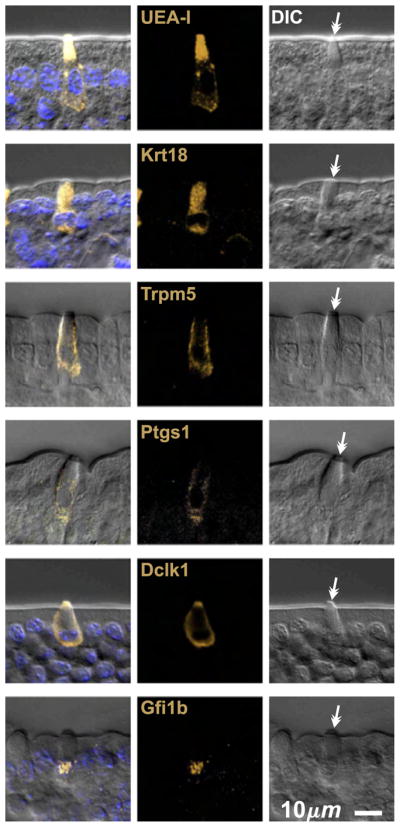
Optical sections of villus epithelium from CD-1 mice showing typical brush cells stained positively with the lectin UEA-I, and antibodies specific for Krt18, Trpm5, Ptgs1, Dclk1, and Gfi1b. Brush cell identity was confirmed in each case by differential interference contrast (DIC) microscopy, which heightens the distinctive appearance of the apical tip characteristic of brush cells (barbed arrows).
Established brush cell markers are not specific on their own, but in combination with anti-Insm1 and DIC can be used to reliably identify brush cells
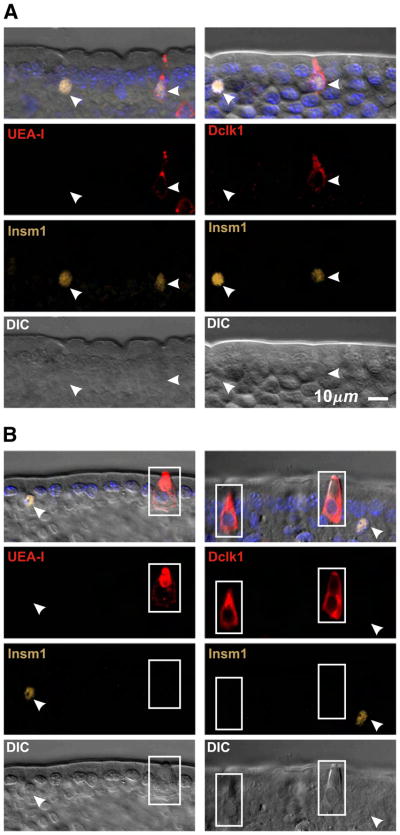
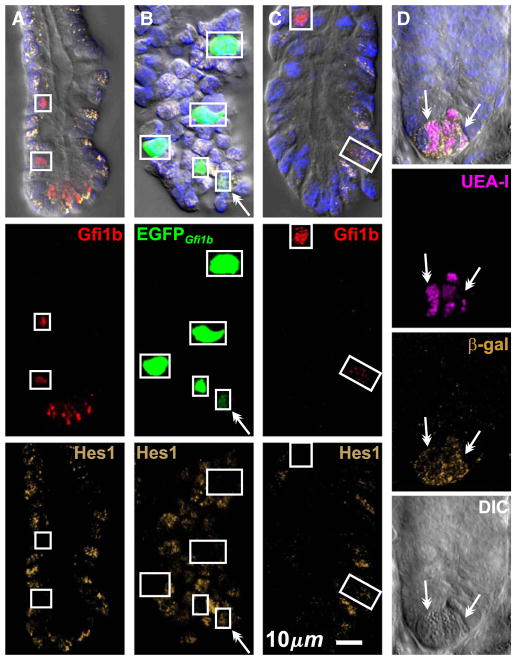
Previously described brush cell markers stain non-brush cell types to varying degrees, and hence on their own are not definitive brush cell markers (Table 3). Thus anti-Krt18 labels brush cells more strongly than other cell types, but other epithelial cell types are also stained confirming Höfer and Drenckhahn (1996). Anti-Trpm5 is more specific but may label some enteroendocrine cells (Bezençon et al., 2007; 2008), and the antibody preparation we used gave a high background that limited its usefulness. Anti-Ptgs1 is also fairly specific, but under the conditions of use here displayed a weak generalized background staining of most cells that limited its usefulness in the crypt. Anti-Dclk1 and UEA-I both stain brush cells robustly with an excellent signal to noise ratio. Dclk1 has been claimed to be a specific brush cell marker (Gerbe et al., 2009). However, we found that some Dclk1+ cells were also positive for the enteroendocrine cell lineage marker Insm1 (Fig. 3A), a transcription factor down stream of Neurog3, the enteroendocrine cell lineage determinant (Apelqvist et al., 1999; Bjerknes and Cheng, 2006a,b, 2010; Gierl et al., 2006; Jenny et al., 2002; Lee et al., 2002). Thus anti-Dclk1 labels a subset of enteroendocrine cells in addition to the brush cells. The Dclk1+Insm1+ enteroendocrine cells are weakly stained and usually have a volumetric flask-like shape, with a thinner neck and a smaller apical end than that of the more Eherlen-meyer flask-like shaped brush cells. The apical cytoplasm of the Dclk1+Insm1+ enteroendocrine cells is also not as refractile as that of brush cells in DIC, probably due to the brush cell’s extensive collection of apical fibrils (Nabeyama and Leblond, 1974). UEA-I similarly labels a subset of Insm1+ enteroendocrine cells (Fig. 3A). UEA-I also labels Paneth and to a lesser extent mucous cells.
Table 3.
Antibody staining patterns.
| Markers | Brush lineage progenitors | Mature brush cells | Enteroendocrine cells | Other epithelial cell types |
|---|---|---|---|---|
| Gfi1b | + | + | − | − |
| Dclk1 | −/+a | + | + (subset) | − |
| UEA-I | − | + | + (subset) | + (mucous and Paneth cells) |
| Krt18 | − | + | + (weak) | + (weak) |
| Trpm5 | − | + | + (subsetb) | − |
| Ptgs1 | − | + | Weak generalized staining | Weak generalized staining |
| Atoh1 | − | − | + | + (mucous and Paneth cells) |
| Hes1 | −/+c | − | − | −d |
| Insm1 | − | − | + | − |
| Neurog3 | − | − | −e,f | − |
| Chga | − | − | + (most) | − |
| Mki67 | + | − | −g | −g |
Brush cell lineage progenitors begin to express Dclk1 as they leave the cell cycle.
Expressed by rare immature EGFPGfi1b weak or Gfi1b+ cells in the COD, possibly DOM.
Crypt base columnar cells and columnar progenitors are Hes1 positive.
Progenitors in the enteroendocrine lineage are Neurog3 positive.
Immature cells in the enteroendocrine lineage are Neurog3 positive.
Actively cycling progenitors are Mki67 positive.
Fig. 3.
Optical sections of villus epithelium from CD-1 mice demonstrating the necessity of supplementing the existing brush cell markers with an enteroendocrine cell specific maker such as Insm1 to reliably distinguish enteroendocrine from brush lineage cells. (A) UEA-I and anti-Dclk1are not specific for brush cells because a subset of Insm1+ enter-oendocrine cells (arrow heads) also stain. (B) Brush cells (boxed) are UEA-I+ Dclk1+ but Insm1−, in contrast to Insm1+ enteroendocrine cells (arrow heads). DIC helps confirm brush cell identification.
We conclude that any of the brush cell markers can be used to confidently identify mature brush cells if they are used in combination with anti-Insm1 and DIC (i.e. marker+ Insm1− with stereotypical morphological features in DIC). This approach works particularly well with anti-Dclk1 or UEA-I (Fig. 3B).
Brush cells are Atoh1− and hence distinct from granulocytes, which are Atoh1+
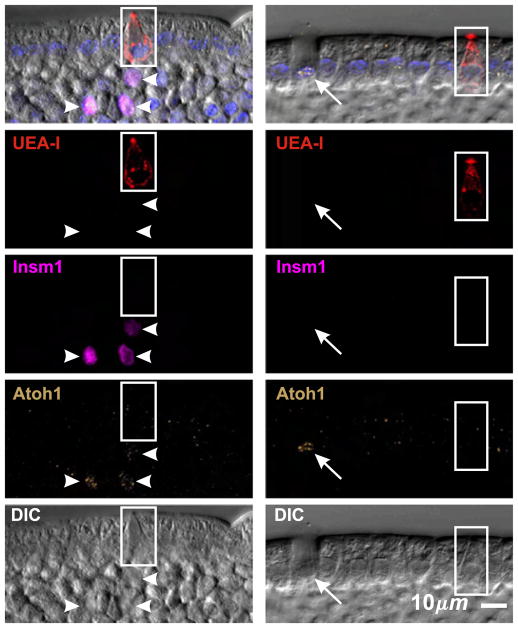
Granulocyte nuclei stained with anti-Atoh1. However, we were unable to find conditions under which brush cell nuclei were stained (Fig. 4), thereby distinguishing brush cells from the granulocytic cell lineages and indicating that brush cells constitute a distinct cell lineage (see below for additional evidence).
Fig. 4.
Brush cells (boxed) do not stain with an Atoh1-specific antibody, whereas enteroendocrine (arrow heads) and mucous (arrow) cell nuclei are Atoh1+.
Brush cells express the transcriptional repressor Gfi1b
Gfi1b is differentially expressed in sorted Trpm5+ cells (Bezençon et al., 2008). We found that anti-Gfi1b labels nuclei of brush cells (Fig. 2) but no other epithelial cell types. Therefore nuclear Gfi1b appears to be a specific brush cell marker. This finding prompted us to investigate an existing Gfi1bEGFP mouse model in which the sequence encoding Gfi1b was replaced by sequence encoding enhanced green fluorescent protein (EGFP) (Vassen et al., 2007). Accordingly, EGFP expression reflects Gfi1b promoter activity (but the subcellular localization of EGFP does not necessarily reflect that of Gfi1b protein). We will denote the EGFP expressed in these mice EGFPGfi1b. We found cells scattered throughout the intestinal epithelium of Gfi1bEGFP/+ mice. Closer examination by DIC revealed that the cells on the villus had typical brush cell morphology, and their identity was confirmed by staining with various brush cell markers, including anti-Gfi1b (Fig. 5). Importantly, EGFPGfi1b was seen only in brush cells and not in other epithelial cell types (Fig. 6). Furthermore, all cells were Atoh1− (Fig. 6). Thus brush cells express Gfi1b and the Gfi1bEGFP/+ mouse model reliably reflects that expression pattern.
Fig. 5.
Villus epithelium from Gfi1bEGFP/+ mice contains scattered, rare, cells exhibiting typical brush cell morphology. These cells stain positively with the lectin UEA-I and antibodies specific for Krt18, Trpm5, Ptgs1, Dclk1, and Gfi1b. Thus all cells are in the brush cell lineage. Importantly, the nuclei of all cells stained positively with anti-Gfi1b, confirming the correspondence between the promoter activity of the wild type and the Gfi1bEGFP alleles in the intestinal epithelium of this mouse model.
Fig. 6.
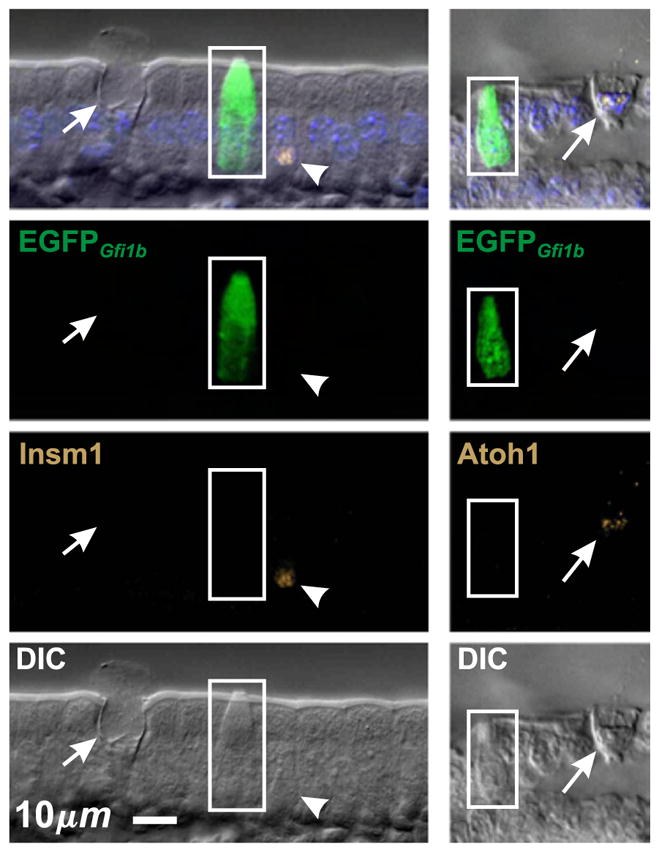
Villus epithelium from Gfi1bEGFP/+ mice confirming that EGFPGfi1b is expressed in brush cells (boxed), but not in other cell types such as mucous (arrow) or enteroendocrine (arrow head) cells. Accordingly, brush cells are Atoh1− and Insm1−.
Brush lineage cells originate in the crypt
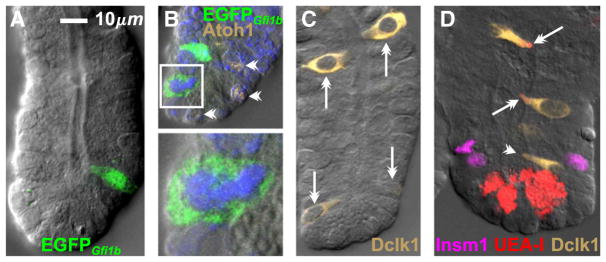
It was of particular interest to use the Gfi1bEGFP/+ mouse model to investigate the origin and differentiation of the brush cell lineage. We found scattered cells in Gfi1bEGFP/+ crypts, including small cells in the common origin of differentiation (COD, ~cell positions 5–8) just above the crypt base (Fig. 7A) and these were occasionally seen in mitosis (Fig. 7B) indicating that brush lineage progenitors are to be found amongst these cells (see below). The cells in the crypt were Atoh1−, like the mature brush cells on the villus. Dclk1+ cells were also seen in the crypt (Fig. 7C). In the upper crypt Dclk1+ cells were robustly stained and often had a mature brush cell form, while in the lower crypt they tended to be smaller and some, especially in the COD, were weakly stained. Maturing Dclk1+ cells in the upper crypt also tended to be UEA-I+, while the Dclk1+ cells around the COD were often UEA-I− (Fig. 7D). These observations indicate that brush lineage cells originate in the COD and then progressively differentiate as they move up the crypt. These qualitative observations are explored quantitatively in following sections.
Fig. 7.
The brush cell lineage originates in the lower crypt. (A) Gfi1bEGFP/+ crypt with an immature cell just above the crypt base. (B) Brush lineage progenitors likely exist in the lower crypt, as indicated by this cell in mitosis (boxed, enlarged 3× below; in mitosis nuclear EGFP disperses to the cytoplasm). Atoh1+ granulocytic lineage cells are also seen (barbed arrow heads). (C) Brush lineage cells in this CD-1 crypt (barbed arrows) exhibit an increasing differentiation gradient from the crypt base to the crypt top as demonstrated by increasing Dclk1 staining intensity and cell size. (D) Immature Dclk1+ cells near the crypt base do not express binding sites for UEA-I (barbed arrowhead), but accumulate UEA-I binding as they mature en route to the villus (barbed arrows). Note that UEA-I also stains Paneth cell granules in the crypt base.
Brush lineage cells display a gradient of differentiation along the crypt axis
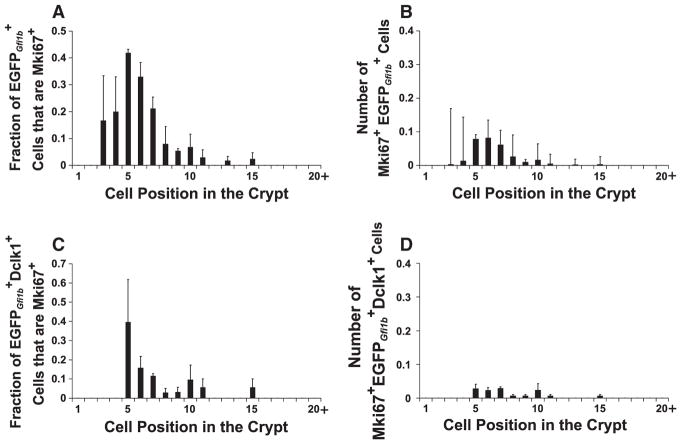
Brush lineage cells are relatively rare. The number of cells per crypt and their distribution along the crypt axis are shown in Fig. 8. Very few cells were observed in cell positions 1–4 of the crypt. cells were most frequent in the COD, cell positions 5–8, and were progressively less frequent in the higher positions, presumably due to their dilution and displacement by rapidly dividing columnar lineage cells in the mid-crypt (Fig. 8B).
Fig. 8.
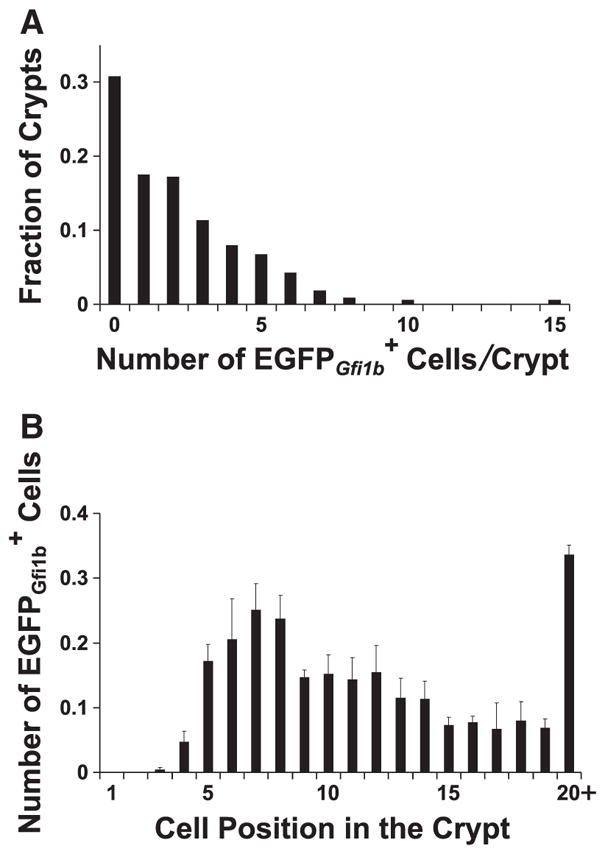
Graphs showing (A) the distribution of the number of cells contained in jejunal crypts from Gfi1bEGFP/+ mice, and (B) the distribution of cells along the crypt axis (mean±S.E.M.). A total of 694 cells were scored in 325 crypts from 3 mice.
The cells in the crypt were negative for the granulocytic lineage marker Atoh1, and were also negative for Neurog3, Insm1, and Chga (Figs. 7B, 9, 13). This indicates that Gfi1b expression in the crypt is normally restricted to the brush cell lineage and its immediate precursors. To investigate further we stained Gfi1bEGFP/+ crypts for various brush cell markers and observed that, depending on the marker, varying subsets of cells within the crypts were labeled (Figs. 9 and 10). There were also cells that were not labeled with any other brush lineage markers. These brush lineage marker negative cells tended to be located lower in the crypt and appeared to be smaller and less differentiated than the double labeled cells, indicating that Gfi1b appears earlier in the development of the brush cell lineage than the other brush cell lineage markers. Thus we used EGFPGfi1b labeling as a reference against which we quantified the appearance of the other markers. We recorded the cell position of every cell observed in a crypt and noted whether it was labeled or not for a particular brush cell marker (Fig. 10). The compiled data reflects the differentiation process of brush cells as they migrate up the crypt from their origin in the COD. Thus, early brush lineage cells express only Gfi1b, then Dclk1 becomes detectable, followed by Krt18, UEA-I binding sites, and later Ptgs1 (with the caveat that the observed nature of the order of appearance may also be a function of the sensitivity and quality of the detection reagents).
Fig. 9.
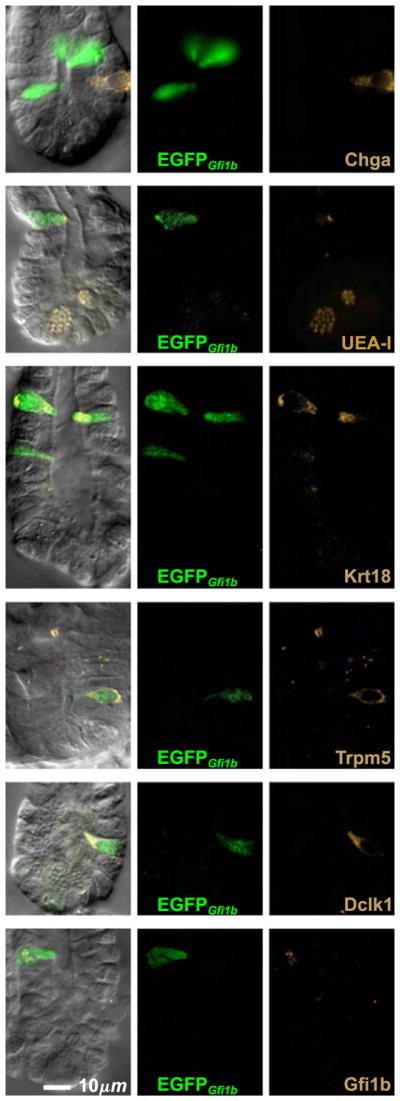
cells in crypts from Gfi1bEGFP/+ mice are brush lineage cells. None of these cells were stained with the enteroendocrine cell specific antibody Chga, but most were stained positively with brush cell markers such as the lectin UEA-I, and antibodies specific for Krt18, Trpm5, Dclk1, and Gfi1b. Note that anti-Insm1 was not necessary because all cells were Insm1−.
Fig. 13.
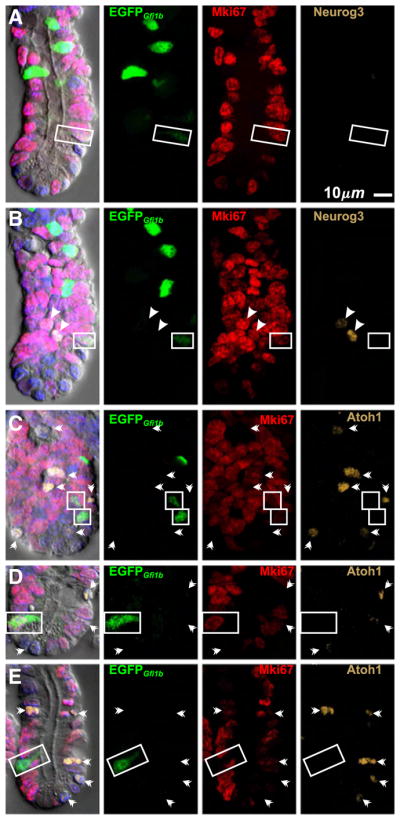
Brush lineage cells in crypts from Gfi1bEGFP/+ mice are Neurog3− and Atoh1−. (A) and (B) are different optical sections of the same crypt showing a proliferating (Mki67+) brush lineage cell (boxed) that is negative for Neurog3. The Neurog3+ cells (arrow heads) in (B) are . (C, D) Proliferating (Mki67+) and (E) post-mitotic (Mki67−) brush lineage cells (boxed) are Atoh1−. Atoh1+ cells (barbed arrow heads) do not express EGFPGfi1b.
Fig. 10.
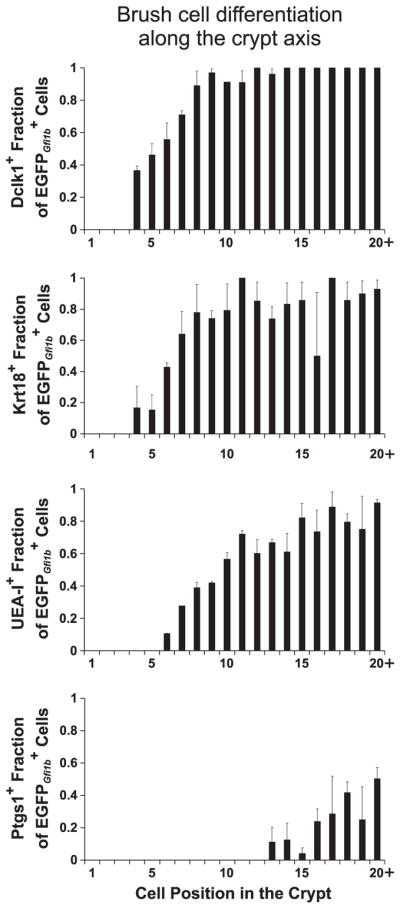
Graphs showing the accumulation of various brush cell differentiation markers as cells mature during their migration up the crypt towards the villus (mean±S.E.M.). The data was derived from crypts isolated from 3 Gfi1bEGFP/+ mice. The distributions were determined by scoring the following total numbers of cells: Dclk1+ (435), Krt18+ (386), UEA-I+ (626), and Ptgs1+ (283).
Characterization of brush cell lineage progenitors
Our observation of occasional mitotic figures in the lower crypt (Fig. 7B) indicated that the population contains a progenitor pool. Therefore we used the cell cycle marker Mki67 to characterize actively proliferating progenitors and determine their distribution within the crypt. When we stained Gfi1bEGFP/+ crypts with anti-Mki67 and anti-Dclk1, most cells in the crypt were Mki67− and Dclk1+ (Fig. 11B) and hence most of the brush cell lineage is post-mitotic. However, in the lower crypt we also observed Mki67+Dclk1− cells (Figs. 11A, 12A, B). These were small weakly cells, suggesting that they were recently formed, had just started expressing Gfi1b, and hence had accumulated only a small amount of EGFPGfi1b. The staining pattern suggests that some brush lineage cells emerge as Mki67+Dclk1− progenitors which then commence the brush cell differentiation program, including Dclk1 expression, as they exit the cell cycle.
Fig. 11.
Some of the cells in the lower crypt are proliferating, especially the immature cells that have not begun to accumulate Dclk1. (A) and (B) are different optical sections from the same Gfi1bEGFP/+ crypt. The boxed cells in (A) are Mki67+ but Dclk1−. (B) The boxed Dclk1+ brush lineage cells are Mki67−. The immature cell indicated by the barbed arrowhead is Dclk1−. (C) The boxed cells are rare examples of Mki67+ Dclk1+ brush lineage cells.
Fig. 12.
Graphs defining the distribution of proliferating brush lineage cells within the crypt (mean±S.E.M.). (A) The fraction of proliferating (Mki67+) brush lineage ( ) cells in each cell position. (B) The average number of proliferating brush lineage cells in each cell position per crypt. Most brush lineage progenitors were found in cell positions 5–8, corresponding to the common origin of differentiation for epithelial cells. The data was derived from 446 crypts from 3 Gfi1bEGFP/+ mice. The data set for (A) consists of 1020 cells and for (B) 133 Mki67+ cells. (C) The fraction of proliferating Dclk1-expressing brush lineage cells in each cell position. (D) The number of proliferating Dclk1-expressing brush lineage cells in each cell position per crypt. The data was derived from 178 crypts from 3 Gfi1bEGFP/+ mice, the data set for (C) consists of 358 Dclk1+ cells and for (D) 22 Mki67+ Dclk1+ cells. Comparing (D) with (B), it is clear that proliferating Dclk1-expressing brush cells constitute only a fraction of the proliferating brush lineage cells, indicating that the brush cell lineage progenitors exit the cell cycle as they begin to differentiate and accumulate Dclk1.
Occasional Mki67+Dclk1+ cells were also seen (Figs. 11C, 12C, D), contradicting previous claims that all Dclk1+ cells are Mki67− (Gerbe et al., 2009, 2011; Giannakis et al., 2006; May et al., 2008, 2009; Sureban et al., 2009). These cells tended to have stronger EGFPGfi1b, weak Dclk1 and usually weak Mki67 staining, which is consistent with the notion that they were recently derived from the Mki67+Dclk1− population and are leaving or have left the cell cycle.
The Mki67+ cells were most frequently seen around cell positions 5–8 (Fig. 12B). Thus the Mki67 staining pattern supports the view that committed brush lineage progenitors originate in the COD, commence the brush cell differentiation program, and exit the cell cycle.
Brush cells are Atoh1−, Insm1−, andChga− (Figs. 3B, 4, 6, 9), indicating that they are distinct from the granulocytic lineages. To determine whether brush cell lineage progenitors (Figs. 7B, 11A) are related to the granulocytic lineage progenitors we co-stained Gfi1bEGFP/+ crypts for Mki67 and Atoh1 or Neurog3. Although we observed many Atoh1+Mki67+ and Neurog3+Mki67+ cells, Mki67+ cells were Atoh1− and Neurog3− (Fig. 13). Thus in contrast to granulocytic progenitors, brush cell lineage progenitors do not express levels of Atoh1 detectable with antibodies, nor do they express detectable Neurog3.
Atoh1 and Hes1 are expressed by some brush lineage precursors
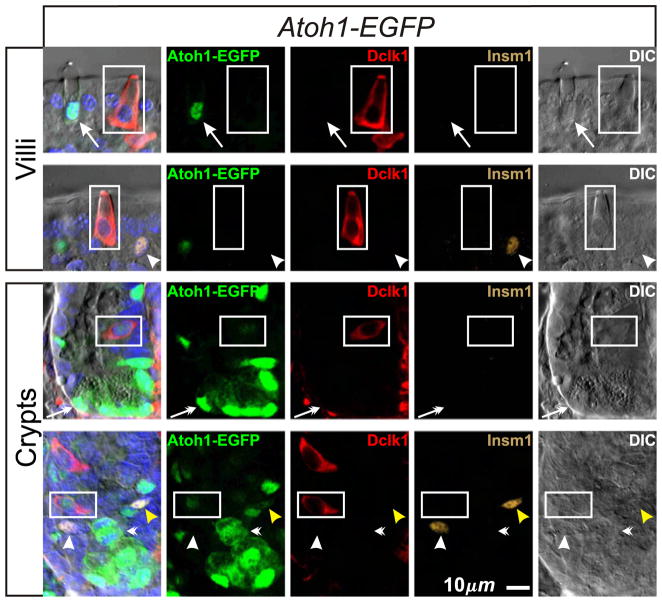
As an additional and potentially more sensitive check for Atoh1 expression in brush lineage cells or their precursors we used the Atoh1-EGFP mouse model in which EGFP sequence was inserted in-frame with Atoh1 sequence, thus encoding an Atoh1-EGFP fusion protein (Rose et al., 2009). Homozygous mice are viable and their intestinal epithelium contains granulocytes, implying that the fusion protein retains essential function. We observed Atoh1-EGFP in nuclei of all mucous and Paneth cells, and many Insm1+ enteroendocrine lineage cells in the lower crypt, but not in Insm1+ enteroendocrine lineage cells in the upper crypt and villi (Fig. 14), even though such cells stained with the anti-Atoh1 antibody.
Fig. 14.
Cells expressing Atoh1-EGFP in intestinal epithelium from Atoh1-EGFP mice. Atoh1-EGFP is expressed in all cells in the mucous (arrows) and Paneth (barbed-arrows) cell lineages. Immature Insm1+ enteroendocrine lineage cells in the lower crypt (yellow arrow head) are Atoh1-EGFP+, but surprisingly most mature enteroendocrine cells (arrow heads) did not exhibit detectable amounts of Atoh1-EGFP. Atoh1-EGFP+ mitotic figures are frequently seen in the crypt (barbed arrowheads). Most brush lineage cells (boxed; Dclk1+ Insm1−) do not express Atoh1-EGFP. However in the lower crypt, weak Atoh1-EGFP is occasionally observed in immature weakly Dclk1-expressing brush lineage cells.
This discrepancy may indicate that the manipulation of the Atoh1 gene has impacted the fidelity of Atoh1 promoter activity resulting in loss of Atoh1 expression in enteroendocrine cells as they mature. Interestingly, this result suggests that Atoh1 is not required for the final stages of enteroendocrine cell differentiation or survival because the Atoh1-EGFP fusion protein is the only source of Atoh1 in these mice; although it might be argued that functional EGFP-cleaved Atoh1 fragments may persist in these cells.
In the brush cell lineage, we found that most Dclk1+Insm1− cells in Atoh1-EGFP epithelium were EGFP−, confirming that mature brush cells are Atoh1− (Fig. 14). However, a few immature and weakly Dclk1+ cells in the lower crypt exhibited weak EGFP signal (see crypts in Fig. 14) indicating either a low level of Atoh1-EGFP expression in the cell, or that EGFP was carried over from an Atoh1-EGFP expressing precursor.
We used two Atoh1-Cre lineage tracing models to further investigate whether Atoh1 is expressed in the brush cell lineage or a precursor. In the first model, the Atoh1-Cre; ROSA26-LacZ reporter mouse, sequence encoding native Cre replaced one allele of the Atoh1 coding sequence and a lacZ floxed-stop reporter construct was incorporated into a Rosa26 allele. Therefore cells expressing Atoh1 also express Cre and hence have the potential to excise the floxed-STOP sequence resulting in the permanent and heritable expression of the lacZ gene, detectable by staining for its encoded product β-galactosidase. The second mouse model, the Atoh1-CreERT2; ROSA26-LacZ reporter mouse, was similar, except that sequence encoding Tamoxifen inducible CreERT2 was used rather than native Cre.
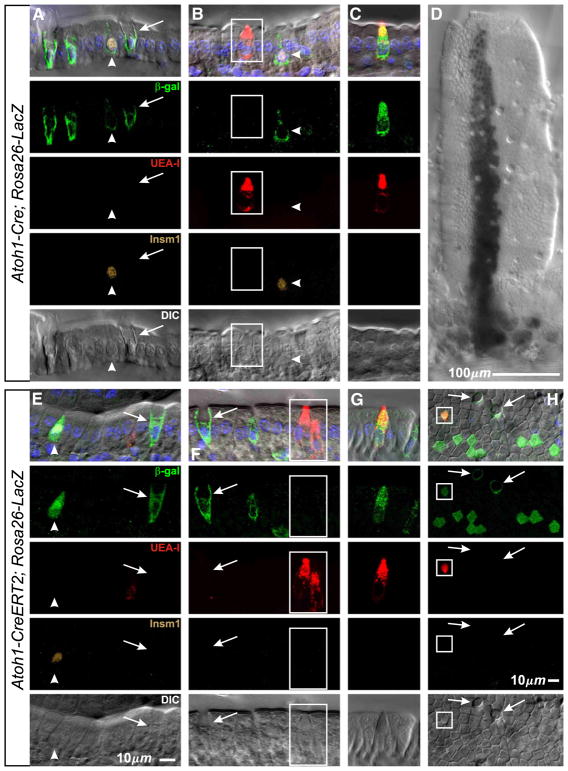
In Atoh1-Cre; ROSA26-LacZ reporter mouse intestine most mucous, enteroendocrine and Paneth cells were β-galactosidase+ (Figs. 15A, B, 16D; Table 4), but only 2% of brush cells were labeled (Figs. 15B, C; Table 4). Rare stem cell clones with large streams of cells emanating from labeled crypts and containing all epithelial cell types were also seen (Fig. 15D), presumably representing either genetic noise, a perturbed gene, or rare reversion of a downstream progenitor back into a stem cell and of no immediate relevance here. The results from inducible Atoh1-CreERT2; ROSA26-LacZ reporter mice, following a single dose of Tamoxifen, were similar except that more brush cells (14%) were β-galactosidase+ (Figs. 15E–G; Table 4). The CreERT2 recombinase, when bound to Tamoxifen, may be more efficiently localized to the nucleus relative to native Cre, and this may in part explain the different labeling efficiency of the brush cells in the 2 mouse models. This increased sensitivity in the CreERT2 model may also explain the regular observation of clusters of 8–32 β-galactosidase+ columnar cells (Fig. 15H). These probably represent clones derived from early columnar progenitors whose immediate precursor briefly expressed Atoh1-CreERT2 in the course of fate determination. β-galactosidase+ cells were not seen in control Atoh1- CreERT2 mice receiving only vehicle, nor in wild type mice without Cre.
Fig. 15.
Lineage tracing with Atoh1-Cre reporter mice indicates that brush lineage precursors transiently express Atoh1. (A–D) Optical sections of villi from Atoh1-Cre;Rosa26-LacZ lineage tracing mice showing, as expected, that Atoh1 expression has occurred in mucous (arrows) and enteroendocrine (arrow heads) lineage cells or their precursors. Evidence for the Paneth cell lineage is shown in Fig. 16D. In contrast, the vast majority of brush lineage cells (boxed) were negative, but a small number were β-gal+ (C) indicating Atoh1 expression occurred at some point in their past. (D) Low magnification image (X-gal stained) of a rare large stem cell derived clone on the villus. (E–H) Similar results were obtained from Atoh1-CreERT2; Rosa26-LacZ mice, except that β-gal+ brush cells (G) were more frequent, and importantly, that clones containing multiple columnar cells (H) were consistently seen indicating that a low level of transient Atoh1 expression occurs in early columnar lineage precursors, most likely the daughter of Mix (DOM) progenitors.
Fig. 16.
The nuclei of some immature Gfi1b expressing cells in the lower crypt stain weakly with Hes1 antibody. (A, B) The nuclei of most brush lineage cells (boxed) do not stain with Hes1 antibody. (A) A CD-1 mouse crypt with two Gfi1b+ Hes1− brush cell lineage nuclei (the Gfi1b staining of Paneth granules in the crypt base is nonspecific). (B) A tangential optical section of a Gfi1bEGFP/+ crypt containing 5 cells illustrating the increasing gradient of differentiation as brush lineage cells migrate up the crypt from their origin in the COD. The cells display an increasing gradient of cell size and EGFPGfi1b content. Only the small, immature cell in the crypt base is weakly positive for Hes1 (barbed arrow). (C) A weakly Gfi1b+ Hes1+ brush lineage cell in the base and a Gfi1b+ Hes1− brush lineage cell in the top of a crypt, isolated from an Atoh1 fl/fl; Rosa26 Cre/+ mouse after Tamoxifen treatment. (D) Optical section of a crypt from an Atoh1-Cre;Rosa26-LacZ mouse showing Atoh1 expression in Paneth cells (barbed arrows). Paneth cell granules are visualized by UEA-I staining.
Table 4.
Fraction of each cell type expressing the reporter following Atoh1-Cre lineage tracing.
| Genotype | Fraction of cells that were β-galactosidase+ (mean±S.E.M.) | |||
|---|---|---|---|---|
|
| ||||
| Mucous cells | Enteroendocrine cells | Paneth cells | Brush cells | |
| Atoh1Cre/+;Rosa26flox-STOP-lacZ/+ (total cells scored in 3 mice) | 0.66±0.04 (2317 cells) | 0.53±0.03 (685 cells) | 0.85±0.02 (978 cells) | 0.022±0.006 (1070 cells) |
| Atoh1CreERT2/+;Rosa26flox-STOP-lacZ/+ (total cells scored in 3 mice) | 0.71±0.04 (2761 cells) | 0.64±0.04 (587 cells) | 0.92±0.08 (591 cells) | 0.14±0.06 (485 cells) |
The infrequent labeling of brush cells in comparison to granulocytes in the Atoh1-Cre lineage tracing models (Table 4) indicates intermittent, weak, brief, or differential expression of Atoh1 in either brush lineage cells or more likely their precursors. The latter possibility is consistent with our failure to observe any evidence of Atoh1 labeling in the brush cell lineage with anti-Atoh1 antibody staining, and our observation of only faint EGFP in occasional very immature brush lineage cells in the Atoh1-EGFP mouse model. Thus it is likely that Atoh1 is expressed in a brush lineage precursor, with the caveat that Atoh1 regulation may have been perturbed by the genetic manipulations in these various mouse models.
Brush lineage precursors were further characterized by staining for the activated Notch target Hes1. While the nuclei of the vast majority of brush lineage cells in the crypt were Hes1− (Figs. 16A, B), occasional immature Gfi1b-expressing cells in the COD exhibited weak nuclear Hes1 staining (Figs. 16B, C).
Brush cells are present in proximal intestine by E18.5 in wild type, but not in Atoh1−/− embryos
It was of great interest to determine the effect of Gfi1b deletion on the brush cell lineage. Unfortunately erythropoiesis is dependent on Gfi1b, and hence homozygous deletion of Gfi1b is embryonic lethal by E15 (Garçon et al., 2005; Osawa et al., 2002; Saleque et al., 2002; Vassen et al., 2007). Therefore we investigated the time of appearance of brush lineage cells in the embryonic gut using the Gfi1bEGFP/+ mice. We did not observe brush lineage cells in E14.5 intestine, so it was not possible to directly determine the effect of Gfi1b deletion on brush cell lineage development.
We observed brush lineage cells in E18.5 Gfi1bEGFP/+ gastric epithelium and in the most proximal portion of the intestinal epithelium (Fig. 17), but only rarely in more distal regions. Most of these brush lineage cells were found in the troughs between villi (Fig. 17A), but some were occasionally seen on the villus (Figs. 17B, C). These results confirm Saqui-Salces et al. (2011), but differ from Gerbe et al. (2011) who reported that brush cells first appear on postnatal day 7. Brush cells have also been described in human embryos (Moxey and Trier, 1978), and in the rat gastric epithelium at birth (Iseki et al. (1991).
Fig. 17.
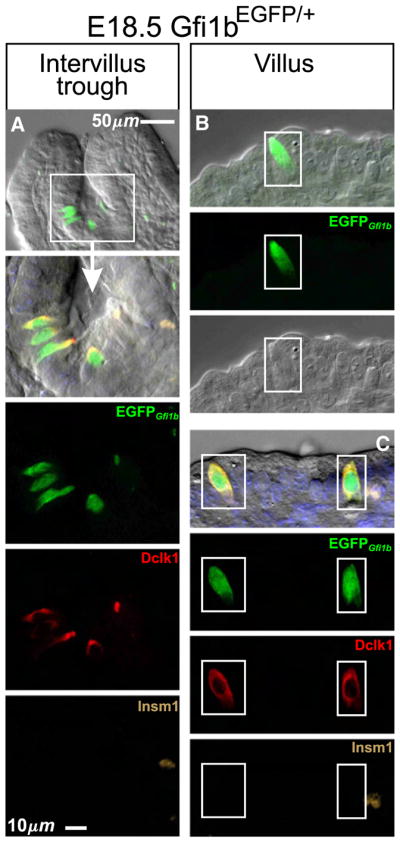
Brush lineage cells (boxed) are present in the proximal intestinal epithelium of E18.5 Gfi1bEGFP/+ embryos. (A) cells in the intervillus trough. These cells stained positively for Dclk1 but negatively for Insm1. (B, C) Occasionally, relatively mature brush cells were seen in the villus epithelium.
Atoh1 null mice fail to breath and hence die shortly after birth (Ben-Arie et al., 1997), so the presence of brush cells at E18.5 offered the opportunity to examine Atoh1 null intestine for any effects on brush cell lineage development. In E18.5 embryos with a functional Atoh1 allele (i.e. Atoh1+/− and Atoh1+/+ embryos) Dclk1+Insm1− brush cells were seen in the gastric epithelium and in the intervillus troughs of the most proximal intestinal epithelium (Fig. 18). In contrast, while the gastric epithelium of Atoh1-null littermates had brush cells at E18.5, they were absent from the intestinal epithelium (Fig. 18). Thus it appears that Atoh1 is required for normal embryonic development of the brush cell lineage in the intestinal epithelium, but not in the gastric epithelium. The effect could be a direct dependence of brush lineage formation on Atoh1, or more likely the time of appearance of the brush lineage in the proximal intestine could be perturbed as an epiphenomenon resulting from the multiple developmental abnormalities in these mice, including the complete absence of enteroendocrine cells (Ben-Arie et al., 1997; Yang et al., 2001). Results described below demonstrate that brush cell lineage formation does not depend on Atoh1, so the later is more likely.
Fig. 18.
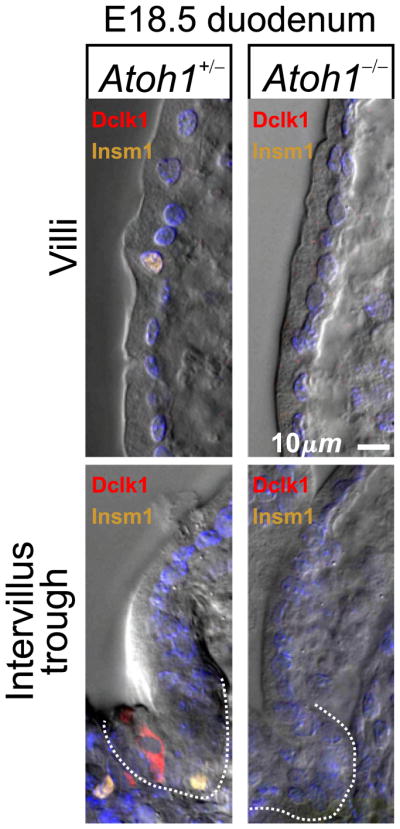
Dclk1+ Insm1− brush lineage cells in the intervillus trough (indicated by dotted lines) of E18.5 proximal intestine from an Atoh1+/− embryo. Brush, enteroendocrine and mucous lineage cells were not observed in the intestinal epithelium of E18.5 Atoh1−/− littermates.
The number of intestinal brush cells and their progenitors increases dramatically following conditional Atoh1 deletion in the adult
The effect of conditional Atoh1 deletion on the brush cell lineage in the adult intestinal epithelium was studied using homozygous Atoh1fl/fl;Rosa26CreERT2/+ mice. Heterozygous Atoh1fl/+;Rosa26CreERT2/+ mice served as controls because they retain a functional wild-type Atoh1 allele following Tamoxifen induced recombination. The Atoh1fl allele is efficiently recombined in the intestinal epithelium of both genotypes (Table 2; see Materials and methods for details).
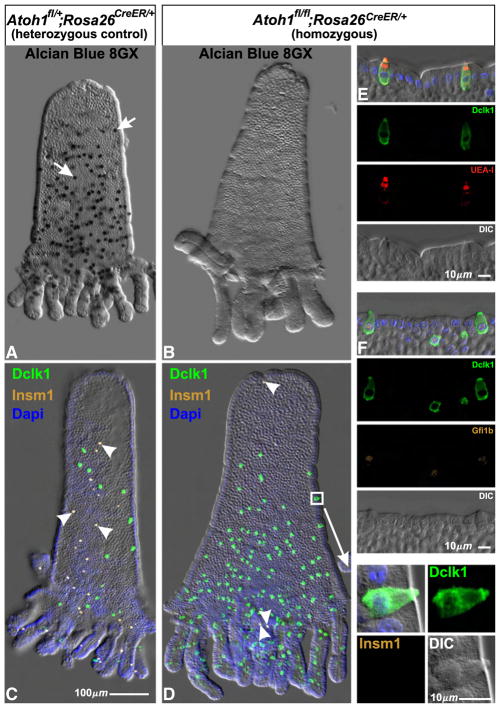
The impact of Atoh1 deletion was dramatic. We confirm previous reports (Shroyer et al., 2007; Yang et al., 2001) that mucous (Figs. 19A, B) and enteroendocrine cells (Figs. 19C, D) are largely absent following Atoh1 deletion (Table 5). In addition we observed a striking increase in the brush cell population of Tamoxifen treated Atoh1fl/fl;Rosa26CreERT2/+ mice in comparison to similarly treated control Atoh1fl/+;Rosa26CreERT2/+ mice (Figs. 19C, D; Table 5). These supernumerary brush cells appear normal morphologically and they express various brush cell markers, including Dclk1, UEA-I, and Gfi1b (Figs. 19D–F). There is a corresponding increase in the population of Gfi1b+ progenitors. Crypts from Tamoxifen treated Atoh1fl/fl; Rosa26CreERT2/+ and Atoh1fl/+;Rosa26CreERT2/+ mice were stained for Gfi1b, Dclk1, and the cell proliferation marker Mki67. Each Gfi1b+ nucleus in a crypt was scored and characterized for Dclk1 and Mki67 staining. Atoh1-deleted crypts contained dramatically more Gfi1b+ brush lineage cells and Gfi1b+ progenitors than did control crypts (Table 6). These results show that brush cells and their progenitors continue to be made following Atoh1-deletion. Therefore brush cell production is not Atoh1-dependent.
Fig. 19.
Isolated epithelium from adult heterozygous control Atoh1fl/+;Rosa26CreER/+ and homozygous Atoh1fl/fl;Rosa26CreER/+ mice 6 days after initiation of Tamoxifen treatment. (A) The numerous Alcian Blue 8GX positive mucous cells in control epithelium (arrows) were largely absent from Atoh1 deleted (Atoh1fl/fl;Rosa26CreER/+) epithelium (B). (C) Similarly, Insm1+ enteroendocrine cells found in control epithelium (arrowheads) were largely absent from Atoh1 deleted epithelium (D). In contrast, the Dclk1+ Insm1− brush cell population increased dramatically in homozygous epithelium (D) relative to heterozygous control (C). (E, F) The brush cells in Atoh1-deleted epithelium appear normal and stain with the standard brush cell markers including Dclk1, UEA-I, and Gfi1b (see also the enlargement of the region boxed in D).
Table 5.
Cell type counts after conditional Atoh1 deletion in Atoh1fl/+ control and Atoh1fl/fl experimental mice.
| Genotypea | Number of cells (mean±s.e.m.) | ||
|---|---|---|---|
|
| |||
| Mucous cells (Alcian blue+) | Enteroendocrine cells (Insm1+) | Brush cells (Dclk1+Insm1−) | |
| Per crypt | |||
|
|
|||
| Atoh1fl/+;Rosa26CreER/+ | 8.29±0.54 (138 crypts) | 4.24±0.16 (126 crypts) | 1.35±0.23 (126 crypts) |
| Atoh1fl/fl;Rosa26CreER/+ | 0.138±0.035b (537 crypts) | 0.205±0.080b (237 crypts) | 14.1±1.78b (237 crypts) |
| Per villus | |||
|
|
|||
| Atoh1fl/+;Rosa26CreER/+ | 210±23.7 (31 villi) | 31.2±3.58 (34 villi) | 14.3±3.45 (34 villi) |
| Atoh1fl/fl;Rosa26CreER/+ | 0.140±0.047b (98 villi) | 1.62±0.38b (50 villi) | 73.8±10.2b (50 villi) |
Data from 3 Tamoxifen treated mice of each genotype.
Significantly different from corresponding Tamoxifen treated Atoh1fl/+;Rosa26CreER/+ control results.
Table 6.
Counts of the number of Gfi1b expressing cells per crypt after conditional Atoh1 deletion in Atoh1fl/+ control and Atoh1fl/fl experimental mice.
| Genotypea | Number of cells per crypt (mean±s.e.m.) | |||
|---|---|---|---|---|
|
|
|
|||
| Gfi1b+ | Gfi1b+ Mki67+ | Gfi1b+ Dclk1+ | Gfi1b+ Dclk1+Mki67+ | |
| Atoh1fl/+;Rosa26CreER/+ (242 crypts) | 1.6±0.19 | 0.14±0.03 | 1.45±0.21 | 0.025±0.002 |
| Atoh1fl/fl;Rosa26CreER/+ (89 crypts) | 16.4±2.0b | 2.6±0.68b | 13.1±2.13b | 0.38±0.12b |
Data from 3 Tamoxifen treated mice of each genotype.
Significantly different from corresponding Tamoxifen treated Atoh1fl/+;Rosa26CreER/+ control results.
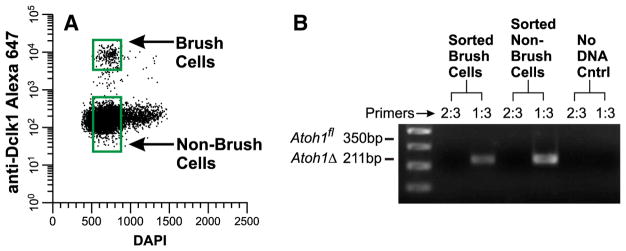
It can plausibly be argued that the persistence of the brush cell lineage following Atoh1 deletion might be due to a resistance to Cre recombinase in the brush cell lineage or its precursors, or that they fail to express Rosa26CreERT2, thus accounting for the 0.4% unrecombined alleles that were detected in the intestinal epithelium following Tamoxifen treatment (Table 2). Therefore it was important to directly determine the proportion of unrecombined Atoh1fl alleles in brush lineage cells. We purified brush cells from Tamoxifen-treated Atoh1fl/fl;Rosa26CreERT2/+ mice by fluorescence activated cell sorting (Fig. 20A). Only recombined alleles were observed in the sortpurified brush cells (Fig. 20B), and thus it is clear that Atoh1 is not required for the generation and maintenance of the brush cell lineage in adult intestinal epithelium. To the contrary, the brush cell lineage expands dramatically in the absence of Atoh1.
Fig. 20.
Brush cells isolated from Tamoxifen treated Atoh1fl/fl;Rosa26CreER/+ intestine have no functional Atoh1 alleles. (A) FACS dot plot illustrating the gating windows used to sort Brush cells and non-brush cells. Insm1 staining was not necessary because there were very few remaining enteroendocrine lineage cells in the Atoh1 deleted epithelium (see Table 5). (B) PCR products (generated from ~1500 cell equivalents of DNA isolated from sorted brush cells and ~4000 cell equivalents from non-brush cells) demonstrate predominantly recombined Atoh1Δ alleles.
Activated Notch signaling eliminates granulocyte production, but brush cell production continues at a reduced rate
The fact that the Notch signaling target Hes1 is weakly expressed in some brush lineage precursors and that brush cell production is Atoh1-independent despite evidence that Atoh1 is expressed in a brush lineage precursor means that under present models it is unclear from which side of the Notch signaling divide the brush cell lineage originates, or for that matter whether Notch signaling plays a role in the determination of the brush cell lineage.
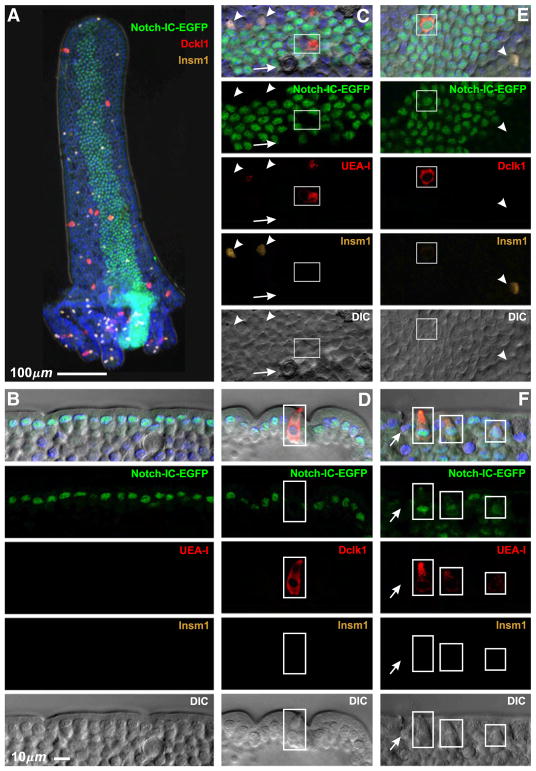
Expression of an intracellular fragment of the Notch receptor (Notch-IC) results in enforced Notch signaling and consequent increased expression of Hes1 and inhibition of formation of the granulocytic lineages (Fre et al., 2005, 2009; Stanger et al., 2005). Therefore, we investigated the effects of Notch-IC on the brush cell lineage using Rosa26CreERT2/floxed-STOP-Notch-IC-IRES-EGFP mice. Stem cells that successfully recombine the floxed STOP cassette generate long-lived clones expressing both Notch-IC and the clone marker nuclear EGFP. Notch-IC expressing long-lived clones induced by a single dose of Tamoxifen were readily identified in isolated crypt-villus units (Fig. 21A). Cells belonging to clones were easily distinguished by their EGFP+ nuclei. The vast majority of cells within clones were columnar cells (Fig. 21B). EGFP− granulocytes, columnar cells, and brush cells were often seen adjacent to and sometimes embedded within the clones (Figs. 21C, D), so single cell resolution was crucial in these studies to ensure that such cells were excluded from clonal consideration. Importantly, while we did not observe EGFP+ granulocytes, we did observe occasional EGFP+ brush cells (Figs. 21E, F). Out of a total of 1681 villi studied from 3 mice, 1602 clones were found and 7 of these contained at least one EGFP+ brush cell (a total of 14 EGFP+ brush cells were seen in the 7 clones). Thus brush cells are made in the clones, although at a greatly diminished rate. This indicates that, unlike granulocytes, generation and differentiation of brush cells still occur under conditions of diminished Atoh1 levels that result from persistent Notch signaling and up-regulation of Hes1. This confirms the Atoh1 conditional deletion result that brush cells are Atoh1-independent.
Fig. 21.
Effect of enforced Notch signaling seen in isolated epithelium from Rosa26CreERT2/floxed-STOP-Notch-IC-Ires-EGFP mice 72 days after induction by a single Tamoxifen treatment. (A) Low magnification view of an isolated villus with an attached Notch-IC-EGFP expressing crypt feeding a stripe of cells along the villus. Cells in the clone are identified by their nuclear EGFP. The other images are higher magnification views of various clones, some in sagittal section (B, D, F), others en face (C, E). (B–D) Notch-IC-EGFP clones consist mainly of columnar cells. Mucous (arrows), enteroendocrine (arrow heads), and brush (boxed) cells within or adjacent to the stripes are EGFP−. (E, F) However, infrequent Notch-IC-EGFP clones contained EGFP+ brush cells (boxed).
Discussion
Brush cells are continuously renewed post-mitotic cells (Nabeyama and Leblond, 1974; Tsubouchi and Leblond, 1979) ultimately derived from the intestinal epithelial stem cells, often via committed progenitors (Bjerknes and Cheng, 1999). Here we have explored the details of the origin of the brush cell lineage and have found additional insights into the mechanisms underlying cell lineage determination in the epithelium.
Contradictions with other reported results
Some of our central findings contradict conclusions reached in other recent reports. Further comment may be helpful.
Gerbe et al. (2011) reported that the intestinal epithelium of adult mice is ‘completely devoid’ of brush cells following conditional Atoh1 deletion. To the contrary, we found that the number of brush cells increases dramatically following Atoh1 deletion provided that care is taken to avoid Cre-induced intestinal toxicity. Watanabe et al. (1980) reported that the lethal Tamoxifen dose in mice is about 29 times less for intraperitoneal versus oral administration. We found in preliminary experiments that significant intestinal toxicity resulted from administering large Tamoxifen doses (for example, 5 daily doses by gavage of 333 mg/kg, about 5 mg per mouse). Under these toxic conditions (toxic in the context of Atoh1 deletion because heterozygous control mice appeared healthy at these doses) the villus epithelium was almost devoid of brush cells. Therefore a potential explanation of the discrepancy with Gerbe et al. is that they were misled by an artifact resulting from Cre-induced intestinal toxicity because they induced recombination by 4 daily intraperitoneal injections of 1 mg Tamoxifen per mouse, which may constitute a high effective dose due to the route of administration.
Dclk1 is used as a potential gastrointestinal epithelial stem cell marker (Dekaney et al., 2009; Giannakis et al., 2006; Jin et al., 2009; May et al., 2008; 2009; Sureban et al., 2009; von Furstenberg et al., 2011), despite reports that Dclk1 is expressed by brush cells (Bezençon et al., 2008) specifically (Gerbe et al, 2009; 2011). We find that Dclk1 is an excellent brush cell marker, but it also stains a subset of Insm1+ enteroendocrine cells. We also noticed in preliminary studies that at the 1:50 to 1:400 Dclk1 antibody dilutions used by others there was significant Dclk1 staining of the cell borders of most cells in the lower crypt, including all of the likely stem cell candidates. However, at the 1:40,000 dilution that we used here brush cell lineage staining was still robust but the cell surface staining of lower crypt cells was near background. Therefore antibody titer may help explain reported success using anti-Dclk1 to isolate cells with stem cell properties (May et al., 2008, 2009; Sureban et al., 2009).
The transcriptional repressor Gfi1b is expressed at all stages of the brush cell lineage but not in any of the other epithelial cell lineages
Our results demonstrate that nuclear Gfi1b is a specific marker for brush cells. All villus brush cells express Gfi1b, and conversely all Gfi1b-expressing villus epithelial cells are brush cells. In the crypt we observed a continuous progression from small immature cells in the COD, displaying barely perceptible EGFPGfi1b and no other brush cell markers, to large maturing cells in the upper crypt displaying robust EGFPGfi1b and all other brush cell markers. Thus Gfi1b is expressed at all stages of the brush cell lineage, and in particular may be used to identify the origin of the brush cell lineage deep in the crypt.
Brush cell lineage progenitors
EGFP protein has a half-life of ~26 h in mammalian cells (Corish and Tyler-Smith, 1999). This has two implications—cells accumulate EGFP if they continue to produce it, and EGFP serves as a tracer that can be used to follow cells for some time even if they cease EGFP production.
Occasional mitotic figures were seen, so we used Mki67 staining to characterize the progenitors among cells in Gfi1b+/EGFP intestinal epithelium. Most progenitors were located in the COD (cell positions 5–8; Fig. 12B), coincident with the location of the most immature brush lineage cells. We did not observe evidence of EGFPGfi1b in non-brush cell lineages in the crypt, indicating that the progenitors do not directly contribute to other lineages.
EGFPGfi1b also served as a tracer demonstrating continuity between progenitors and the brush cell lineage. Most progenitors were weakly , strongly Mki67+, and did not express other brush cell lineage markers. However a subset was also weakly Dclk1+. These weakly Dclk1+ cells tended to exhibit brighter EGFPGfi1b and weaker Mki67 staining, consistent with their derivation from the progenitors. This indicates that as progenitors leave the cell cycle they cease Mki67 production but continue to express Gfi1b and hence accumulate EGFPGfi1b. The cells also accumulate brush lineage cell markers, until in the crypt top they exhibit all brush cell markers and are robustly . Thus EGFPGfi1b expression and its accumulation links the progenitors in the COD to immature brush lineage cells as they commence differentiation, eventually forming maturing upper crypt brush cells. Additional evidence of the linkage between Gfi1b-expressing progenitors and the brush cell lineage comes from the conditional Atoh1-deletion experiment because the resulting increase in the brush cell population was accompanied by an increase in the number of Gfi1b+ progenitors in the crypts (Table 6). We conclude that the brush cell lineage, like the other epithelial cell lineages, originates from progenitors normally located in the common origin of differentiation (COD), just above the stem cell zone (Bjerknes and Cheng, 1981a,b,c, 2005, 2006a,b, 2010).
We were unable to detect anti-Atoh1 staining in any brush lineage cells including progenitors, distinguishing the brush cell lineage from the granulocytic lineages. We also did not detect Atoh1-EGFP in brush cells, and only a small fraction of brush cells were labeled in Atoh1-Cre and Atoh1-CreERT2 lineage tracing experiments. Early brush lineage cells also did not express key transcription factors known to play a role in granulocytic lineage determination. They don’t express the transcriptional repressor Gfi1 known to play a role in stabilizing the mucous and Paneth cell lineages (Bjerknes and Cheng, 2010). We also found no evidence of Neurog3 or Insm1 staining, confirming that brush cells are not a subset of the enteroendocrine cell lineage (Bjerknes and Cheng, 2006a,b, 2010; Gerbe et al., 2011; Gierl et al., 2006).
Kokrashvili et al. (2009) observed that brush cells express the signaling peptides uroguanylin, β-endorphin and Met-enkephalin leading them to suggest that they are a type of enteroendocrine cell. Yet brush cells and their progenitors don’t express the granulocytic lineage marker Atoh1 or the enteroendocrine lineage markers Neurog3 and Insm1 and hence brush cells are not in the enteroendocrine cell lineage as usually defined. Therefore to minimize potential confusion it would be best not to describe the brush cell lineage as enteroendocrine, despite the etymological and physiological reasonableness of the usage.
Brush lineage precursors
Only a small fraction of brush cells were labeled in Atoh1-Cre lineage tracing experiments and only occasional very immature brush lineage cells were weakly EGFP+ in mice expressing Atoh1- EGFP fusion protein. In light of the absence of detectable anti- Atoh1 staining in brush lineage cells, these genetic models indicate that Atoh1 is briefly, weakly, differentially or inconsistently expressed in a brush lineage precursor (assuming that the manipulation of the Atoh1 gene in these mice did not perturb their Atoh1 expression patterns). We also found that some brush cell precursors displayed nuclear Hes1 staining. Thus it appears that Hes1, Atoh1 and Gfi1b are all expressed in the earliest stages of brush cell lineage formation, possibly DOMDelta (see below and model), but only Gfi1b-expression persists in cells that engage the brush cell lineage program.
Brush cell lineage production, determination, differentiation and survival are Atoh1-independent; nonetheless the brush cell lineage originates from DOMDelta
Conditional Atoh1-deletion in adult Atoh1fl/fl;Rosa26CreERT2/+ mice yielded the expected Atoh1-dependence of granulocyte formation (Shroyer et al., 2007; Yang et al., 2001) because mucous and enteroendocrine cells were largely absent from the epithelium 6 days after initiation of Tamoxifen treatment. In contrast, brush cell numbers increased dramatically, and these brush cells were confirmed to lack functional Atoh1 alleles. Therefore, in contrast to granulocytic lineages, brush lineage cell production, determination, differentiation and survival do not require Atoh1 in the adult intestinal epithelium. The Atoh1-independence of brush cell lineage formation was also indicated by the continued formation of brush cells in Atoh1- repressed Notch-IC expressing clones.
The Atoh1-independence of the brush cell lineage suggests, under existing models, that the brush cell lineage originates from DOMNotch. If so, then the number of brush cells produced per crypt should double because in the absence of Atoh1 both sister DOMs should enter a DOMNotch-like state (i.e. both Atoh1-null DOMs should increase Hes1 expression). However, we observed a 10-fold increase, not a doubling of the number of brush cells per crypt. Thus we must consider a DOMDelta origin despite the Atoh1-independence of the brush cell lineage. In this scenario the 10-fold increase is due to all Atoh1-deficient DOMDelta engaging the brush cell lineage program rather than a granulocytic program. If so, then brush cell production per crypt should be roughly equivalent to that of the combined production of granulocytic and brush cell lineages in control crypts. This is what we observed. Atoh1-deleted crypts contained about 14 brush cells per crypt, while in control crypts brush, mucous, and enteroendocrine lineage cells together also totaled about 14 cells per crypt (Table 5). We ignored Paneth cells in this assessment because they are long-lived cells that are only rarely produced and hence make only a minor contribution to total daily cell production. The repression of brush cell production observed in the Notch-IC experiment is also consistent with a DOMDelta origin of the brush cell lineage.
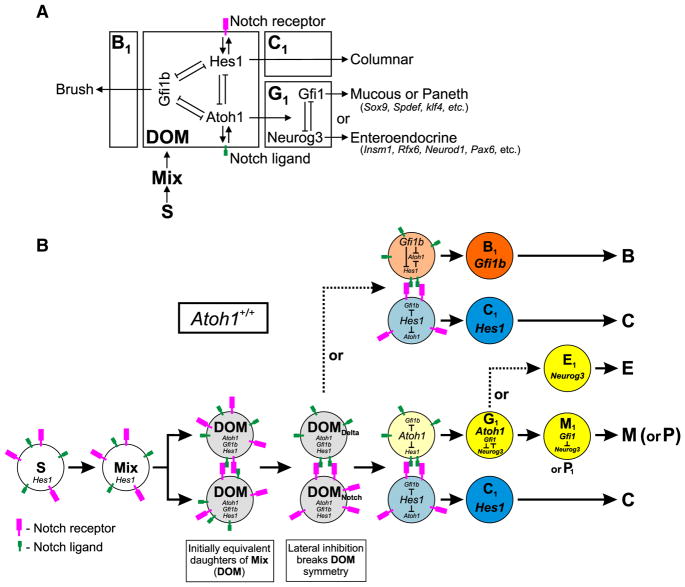
The origin of the brush cell lineage—a model
Current understanding is that the progeny of intestinal epithelial stem cells interact via Notch signaling to produce Hes1- and Atoh1- expressing progenitors that give rise, respectively, to the columnar and the granulocytic (mucous, enteroendocrine, and Paneth) cell lineages. The model needs to be reformulated to accommodate our findings regarding the brush cell lineage.
We propose a model based on the following definitions and observations (Figs. 22A, B):
Fig. 22.
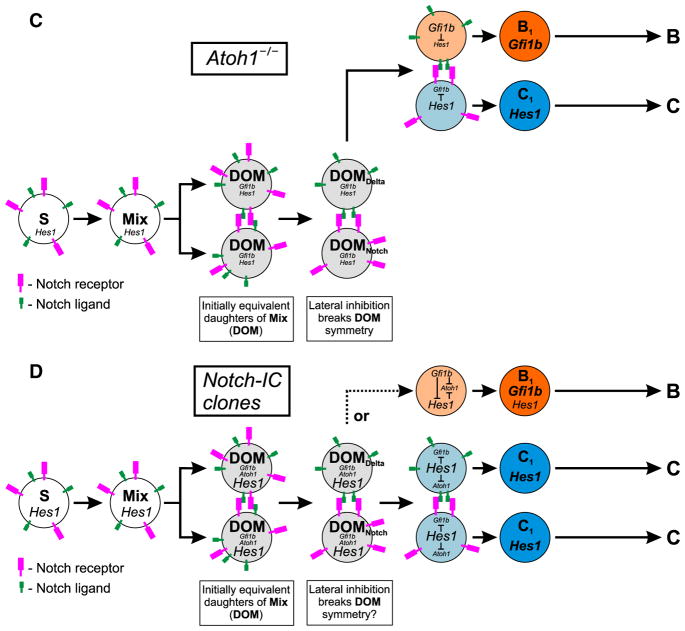
Proposed model of intestinal epithelial cell lineage derivation. (A) Schematic representation of the gene networks operating within the intestinal epithelium to define the various epithelial cell lineages. Stem cells, S, give rise to short-lived Mix progenitors as they leave the stem cell zone and initiate differentiation. The initially equivalent daughters of Mix (DOM) each express low levels of Hes1, Atoh1, and Gfi1b. Depending on whether Hes1, Atoh1, or Gfi1b dominates in a DOM the cell will invoke a columnar, granulocytic, or brush cell lineage program, respectively. (B) In wild type epithelium, lateral inhibitory Notch signaling between the two equivalent DOMs breaks their symmetry leading to DOMNotch and DOMDelta states which usually give rise to Hes1- and Atoh1-expressing cells, respectively. Hes1-expressing cells become the columnar cell lineage progenitors, C1. The Atoh1-expressing cells commence a granulocytic lineage program, G1, and immediately commit to one of the granulocytic lineages through the interaction of various downstream factors including Neurog3 and Gfi1, leading to formation of a mucous, enteroendocrine, or a Paneth progenitor (M1, E1, or P1). Alternatively, up regulation of Gfi1b in DOMDelta represses Atoh1 and invokes the brush cell lineage program. (C) In the absence of Atoh1, lateral inhibition between DOMs gives rise to Hes1- and Gfi1b-expressing cells. As a result Atoh1−/− epithelium lacks granulocytic cell lineages, and contains an expanded brush cell population in addition to the columnar cell lineage. (D) In Notch-IC clones from Tamoxifen-treated Rosa26CreERT2/floxed-STOP-Notch-IC-Ires-EGFP mice, Hes1 is over expressed due to the Notch-IC transgene, resulting in columnar cell lineage clones. Rarely, perhaps due to fluctuations in Hes1 levels allowing sufficient Gfi1b expression, a DOM invokes the brush cell lineage program.
We define Mix progenitors to be stem cell (S) progeny that have left the stem cell zone (Bjerknes and Cheng, 1981a,b) and initiated a differentiation program. These correspond to the Mix progenitors demonstrated by clone studies (Bjerknes and Cheng, 1999).
Mix mitosis results in daughters of Mix (DOM) progenitors, equivalent cells whose initial symmetry is broken by lateral inhibitory Notch signaling (Bjerknes and Cheng, 1999, 2005, 2006a,b, 2010; Jensen et al., 2000; Yang et al., 2001) resulting in DOMNotch and DOMDelta.
Hes1 staining is seen in non-granulocytic cells of the lower crypt (Jensen et al., 2000; Kayahara et al., 2003) indicating that Hes1 is likely expressed to some degree in S, Mix, and DOM. We observed immature Hes1+ Gfi1b+ cells in the crypt.
Atoh1 staining was not observed in brush cells and Atoh1 is not required for brush cell lineage determination and formation. However, some reporter-labeled brush cells were observed in two different Atoh1-Cre lineage tracing models. Similarly, weak Atoh1-EGFP was observed in occasional immature brush lineage cells in an Atoh1-EGFP model. Furthermore, in the Atoh1-CreERT2 lineage tracing model we regularly observed labeled columnar cell clones. Taken together these findings indicate that Atoh1 is transiently expressed in DOM prior to lineage commitment.
The greatly increased brush cell production following Atoh1- deletion, and the decreased production in the Notch-IC clones jointly indicate that the brush cell lineage is derived from the DOMDelta state, despite the fact that the brush cell lineage is Atoh1-independent.
Our results provide circumstantial evidence that Gfi1b regulates brush cell lineage determination, but we were unable to study Gfi1b-null intestine due to embryonic lethality. An important future experiment will be conditional Gfi1b deletion in adult intestinal epithelium.
Our results lead us to propose that the DOM express a low level of the relevant players (Hes1, Atoh1 and Gfi1b) enabling a gene network that interacts dynamically with Notch signaling to establish cell lineage commitment (Figs. 22A, B). Thus we envisage the DOM as having three metastable states defined respectively by Hes1, Atoh1, and Gfi1b. If Hes1 dominates then the DOM initiates the columnar lineage program and becomes a columnar lineage progenitor (C1). As in current models, lateral inhibitory Notch signaling ensures that this is the normal outcome for one of the two DOMs, DOMNotch, while Atoh1 or Gfi1b dominates in its sister DOMDelta. If Atoh1 dominates then the DOMDelta initiates a granulocytic lineage program and becomes an Atoh1-dependent granulocytic progenitor G1, which depending on downstream determinants, enters a mucous (M1), or an enteroendocrine (E1), or a Paneth (P1) cell progenitor state. If Gfi1b dominates, then the DOMDelta initiates a brush cell lineage program and becomes a brush cell lineage progenitor (B1).
Atoh1 deletion (Fig. 22C) leaves only two metastable states, defined by Hes1 and Gfi1b, respectively. Lateral inhibitory Notch signaling would then result in DOMNotch initiating a columnar lineage program while DOMDelta initiates a brush cell lineage program (Atoh1−/− DOM can’t engage a granulocytic lineage program). Thus in Atoh1-deficient epithelium the model predicts that brush cell production per crypt should be roughly equivalent to that of the combined production of granulocytic and brush cell lineages in control crypts, which is what we observed (Table 5).
Forced expression of Notch-IC (Fig. 22D) should result in most DOM initiating a columnar lineage program, and this is what we observed. Absence of granulocytes results from both the activation of a columnar lineage program and the consequent repression of Atoh1. The brush cell lineage, all else being equal, should have been similarly impacted. Activation of the columnar lineage program should repress Gfi1b and there should be a failure to initiate brush cell formation. However, while brush cell production was greatly reduced, occasional brush cells were still produced. One plausible explanation is that in comparison to Atoh1, Gfi1b is not as strongly repressed by Hes1. Furthermore, Hes1 expression is known to fluctuate greatly in many cells (Hirata et al., 2002; Kageyama et al., 2010). Perhaps fluctuations in Hes1 levels weakens the repression of Gfi1b expression in rare DOM, allowing Gfi1b to initiate the brush lineage program.
We have not explicitly incorporated the microfold cell into this model because its origin remains contentious. Some have concluded that they are derived from columnar cells under the influence of microorganisms or lymphocytes (Borghesi et al., 1999; Kerneis et al., 1997; Savidge, 1996; Savidge et al., 1991), while others conclude they are a distinct epithelial lineage (Gebert and Posselt, 1997; Lelouard et al., 2001; Miyazawa et al., 2006).
Acknowledgments
We thank Dr. H. Zoghbi for generously arranging access to Atoh1-EGFP and Atoh1-null tissues in her laboratory, Drs. C. Birchmeier, J. Johnson, O. Madsen, and R. Margolskee for kind gifts of antibodies, and Ms. D. White for assistance with the FACS experiments.
References
- Akazawa C, Ishibashi M, Shimizu C, Nakanishi S, Kageyama R. A mammalian helix–loop–helix factor structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. J Biol Chem. 1995;270:8730–8738. doi: 10.1074/jbc.270.15.8730. [DOI] [PubMed] [Google Scholar]
- Apelqvist Å, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, Hrabě de Angelis M, Lendahl U, Edlund H. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- Bezençon C, le Coutre J, Damak S. Taste-signaling proteins are coexpressed in solitary intestinal epithelial cells. Chem Senses. 2007;32:41–49. doi: 10.1093/chemse/bjl034. [DOI] [PubMed] [Google Scholar]
- Bezençon C, Fürholz A, Raymond F, Mansourian R, Métairon S, Le Coutre J, Damak S. Murine intestinal cells expressing Trpm5 are mostly brush cells and express markers of neuronal and inflammatory cells. J Comp Neurol. 2008;509:514–525. doi: 10.1002/cne.21768. [DOI] [PubMed] [Google Scholar]
- Bjerknes M, Cheng H. Stem-cell zone of the small intestinal epithelium. Anat Rec. 1979;193:484–485. [Google Scholar]
- Bjerknes M, Cheng H. The stem-cell zone of the small intestinal epithelium. I Evidence from Paneth cells in the adult mouse. Am J Anat. 1981a;160:51–63. doi: 10.1002/aja.1001600105. [DOI] [PubMed] [Google Scholar]
- Bjerknes M, Cheng H. The stem-cell zone of the small intestinal epithelium. III Evidence from columnar, enteroendocrine, and mucous cells in the adult mouse. Am J Anat. 1981b;160:77–91. doi: 10.1002/aja.1001600107. [DOI] [PubMed] [Google Scholar]
- Bjerknes M, Cheng H. Methods for the isolation of intact epithelium from the mouse intestine. Anat Rec. 1981c;199:565–574. doi: 10.1002/ar.1091990412. [DOI] [PubMed] [Google Scholar]
- Bjerknes M, Cheng H. Clonal analysis of mouse intestinal epithelial progenitors. Gastroenterology. 1999;116:7–14. doi: 10.1016/s0016-5085(99)70222-2. [DOI] [PubMed] [Google Scholar]
- Bjerknes M, Cheng H. Gastrointestinal stem cells. II Intestinal stem cells. Am J Physiol Gastrointest Liver Physiol. 2005;289:G381–G387. doi: 10.1152/ajpgi.00160.2005. [DOI] [PubMed] [Google Scholar]
- Bjerknes M, Cheng H. Neurogenin 3 and the enteroendocrine cell lineage in the adult mouse small intestinal epithelium. Dev Biol. 2006a;300:722–735. doi: 10.1016/j.ydbio.2006.07.040. [DOI] [PubMed] [Google Scholar]
- Bjerknes M, Cheng H. Intestinal epithelial stem cells and progenitors. Methods Enzymol. 2006b;419:337–383. doi: 10.1016/S0076-6879(06)19014-X. [DOI] [PubMed] [Google Scholar]
- Bjerknes M, Cheng H. Cell lineage metastability in Gfi1-deficient mouse intestinal epithelium. Dev Biol. 2010;345:40–63. doi: 10.1016/j.ydbio.2010.06.021. [DOI] [PubMed] [Google Scholar]
- Borghesi C, Taussig MJ, Nicoletti C. Rapid appearance of M cells after microbial challenge is restricted at the periphery of the follicle-associated epithelium of Peyer’s patch. Lab Invest. 1999;79:1393–1401. [PubMed] [Google Scholar]
- Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. I Columnar cell Am J Anat. 1974a;141:461–480. doi: 10.1002/aja.1001410403. [DOI] [PubMed] [Google Scholar]
- Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V Unitarian theory of the origin of the four epithelial cell types. Am J Anat. 1974b;141:537–562. doi: 10.1002/aja.1001410407. [DOI] [PubMed] [Google Scholar]
- Corish P, Tyler-Smith C. Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng. 1999;12(12):1035–1040. doi: 10.1093/protein/12.12.1035. [DOI] [PubMed] [Google Scholar]
- Crosnier C, Vargesson N, Gschmeissner S, Ariza-McNaughton L, Morrison A, Lewis J. Delta-Notch signalling controls commitment to a secretory fate in the zebrafish intestine. Development. 2005;132:1093–1104. doi: 10.1242/dev.01644. [DOI] [PubMed] [Google Scholar]
- Dekaney CM, Gulati AS, Garrison AP, Helmrath MA, Henning SJ. Regeneration of intestinal stem/progenitor cells following doxorubicin treatment of mice. Am J Physiol Gastrointest Liver Physiol. 2009;297:G461–G470. doi: 10.1152/ajpgi.90446.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan LL, Porter SD, Duan Z, Flubacher MM, Montoya D, Tsichlis PN, Horwitz M, Gilks CB, Grimes HL. Targeted transcriptional repression of GFI1 by GFI1 and GFI1B in lymphoid cells. Nucleic Acids Res. 2004;32:2508–2519. doi: 10.1093/nar/gkh570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini ME. Notch signaling: the core pathway and its posttranslational regulation. Dev Cell. 2009;16:633–647. doi: 10.1016/j.devcel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- Fre S, Pallavi SK, Huyghe M, Lae M, Janssen K-P, Robine S, Artavanis-Tsakonas S, Louvard D. Notch and Wnt signals cooperatively control cell proliferation and tumorigenesis in the intestine. Proc Natl Acad Sci USA. 2009:6309–6314. doi: 10.1073/pnas.0900427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyama T, Yamada M, Terao M, Terashima T, Hioki H, Inoue YU, Inoue T, Masuyama N, Obata K, Yanagawa Y, Kawaguchi Y, Nabeshima Y, Hoshino M. Inhibitory and excitatory subtypes of cochlear nucleus neurons are defined by distinct bHLH transcription factors, Ptf1a and Atoh1. Development. 2009;136:2049–2058. doi: 10.1242/dev.033480. [DOI] [PubMed] [Google Scholar]
- Garçon L, Lacout C, Svinartchouk F, Le Couédic JP, Villeval JL, Vainchenker W, Duménil D. Gfi-1B plays a critical role in terminal differentiation of normal and transformed erythroid progenitor cells. Blood. 2005;105:1448–1455. doi: 10.1182/blood-2003-11-4068. [DOI] [PubMed] [Google Scholar]
- Gebert A, Posselt W. Glycoconjugate expression defines the origin and differentiation pathway of intestinal M-cells. J Histochem Cytochem. 1997;45:1341–1350. doi: 10.1177/002215549704501003. [DOI] [PubMed] [Google Scholar]
- Gebhard A, Gebert A. Brush cells of the mouse intestine possess a specialized glycocalyx as revealed by quantitative lectin histochemistry. Further evidence for a sensory function. J Histochem Cytochem. 1999;47:799–808. doi: 10.1177/002215549904700609. [DOI] [PubMed] [Google Scholar]
- Gerbe F, Brulin B, Makrini L, Legraverend C, Jay P. DCAMKL-1 expression identifies tuft cells rather than stem cells in the adult mouse intestinal epithelium. Gastroenterology. 2009;137:2179–2180. doi: 10.1053/j.gastro.2009.06.072. [DOI] [PubMed] [Google Scholar]
- Gerbe F, van Es JH, Makrini L, Brulin B, Mellitzer G, Robine S, Romagnolo B, Shroyer NF, Bourgaux JF, Pignodel C, Clevers H, Jay P. Distinct Atoh1 and Neurog3 requirements define tuft Cells as a new secretory cell type in the intestinal epithelium. J Cell Biol. 2011;192:767–780. doi: 10.1083/jcb.201010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakis M, Stappenbeck TS, Mills JC, Leip DG, Lovett M, Clifton SW, Ippolito JE, Glasscock JI, Arumugam M, Brent MR, Gordon JI. Molecular properties of adult mouse gastric and intestinal epithelial progenitors in their niches. J Biol Chem. 2006;281:11292–11300. doi: 10.1074/jbc.M512118200. [DOI] [PubMed] [Google Scholar]
- Gierl MS, Karoulias N, Wende H, Strehle M, Birchmeier C. The zinc-finger factor Insm1 (IA-1) is essential for the development of pancreatic beta cells and intestinal endocrine cells. Genes Dev. 2006;20:2465–2478. doi: 10.1101/gad.381806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms AW, Johnson JE. Progenitors of dorsal commissural interneurons are defined by MATH1 expression. Development. 1998;125:919–928. doi: 10.1242/dev.125.5.919. [DOI] [PubMed] [Google Scholar]
- Hernandez A, Villegas A, Anguita E. Human promoter mutations unveil Oct-1 and GATA-1 opposite action on Gfi1b regulation. Ann Hematol. 2010;89:759–765. doi: 10.1007/s00277-009-0900-x. [DOI] [PubMed] [Google Scholar]
- Hirata H, Yoshiura S, Ohtsuka T, Bessho Y, Harada T, Yoshikawa K, Kageyama R. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science. 2002;298:840–843. doi: 10.1126/science.1074560. [DOI] [PubMed] [Google Scholar]
- Höfer D, Drenckhahn D. Cytoskeletal markers allowing discrimination between brush cells and other epithelial cells of the gut including enteroendocrine cells. Histochem Cell Biol. 1996;105:405–412. doi: 10.1007/BF01463662. [DOI] [PubMed] [Google Scholar]
- Höfer D, Puschel B, Drenckhahn D. Taste receptor-like cells in the rat gut identified by expression of alpha-gustducin. Proc Natl Acad Sci U S A. 1996;93:6631–6634. doi: 10.1073/pnas.93.13.6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DY, Kuo YY, Lai JS, Suzuki Y, Sugano S, Chang ZF. GATA-1 and NF-Y cooperate to mediate erythroid-specific transcription of gfi-1b gene. Nucleic Acids Res. 2004;32:3935–3946. doi: 10.1093/nar/gkh719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseki S, Kanda T, Hitomi M, Ono T. Ontogenic appearance of three fatty acid binding proteins in the rat stomach. Anat Rec. 1991;229:51–60. doi: 10.1002/ar.1092290107. [DOI] [PubMed] [Google Scholar]
- Ito T, Udaka N, Yazawa T, Okudela K, Hayashi H, Sudo T, Guillemot F, Kageyama R, Kitamura H. Basic helix-loop-helix transcription factors regulate the neu-roendocrine differentiation of fetal mouse pulmonary epithelium. Development. 2000;127:3913–3921. doi: 10.1242/dev.127.18.3913. [DOI] [PubMed] [Google Scholar]
- Jacob J, Storm R, Castro DS, Milton C, Pla P, Guillemot F, Birchmeier C, Briscoe J. Insm1 (IA-1) is an essential component of the regulatory network that specifies monoaminergic neuronal phenotypes in the vertebrate hindbrain. Development. 2009;136:2477–2485. doi: 10.1242/dev.034546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarriault S, Le Bail O, Hirsinger E, Pourquie O, Logeat F, Strong CF, Brou C, Seidah NG, Isral A. Delta-1 activation of notch-1 signaling results in HES-1 transactivation. Mol Cell Biol. 1998;18:7423–7431. doi: 10.1128/mcb.18.12.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny M, Uhl C, Roche C, Duluc I, Guillermin V, Guillemot F, Jensen J, Kedinger M, Gradwohl G. Neurogenin3 is differentially required for endocrine cell fate specification in the intestinal and gastric epithelium. EMBO J. 2002;21:6338–6347. doi: 10.1093/emboj/cdf649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen OD. Control of endodermal endocrine development by Hes-1. Nat Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- Jin G, Ramanathan V, Quante M, Baik GH, Yang X, Wang SS, Tu S, Gordon SA, Pritchard DM, Varro A, Shulkes A, Wang TC. Inactivating cholecystokinin-2 receptor inhibits progastrin-dependent colonic crypt fission, proliferation, and colorectal cancer in mice. J Clin Invest. 2009;119:2691–2701. doi: 10.1172/JCI38918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama R, Niwa Y, Shimojo H, Kobayashi T, Ohtsuka T. Ultradian oscillations in Notch signaling regulate dynamic biological events. Curr Top Dev Biol. 2010;92:311–331. doi: 10.1016/S0070-2153(10)92010-3. [DOI] [PubMed] [Google Scholar]
- Kaske S, Krasteva G, Konig P, Kummer W, Hofmann T, Gudermann T, Chubanov V. TRPM5, a taste-signaling transient receptor potential ion-channel, is a ubiquitous signaling component in chemosensory cells. BMC Neurosci. 2007;8:49. doi: 10.1186/1471-2202-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayahara T, Sawada M, Takaishi S, Fukui H, Seno H, Fukuzawa H, Suzuki K, Hiai H, Kageyama R, Okano H, Chiba T. Candidate markers for stem and early progenitor cells, Musashi-1 and Hes1, are expressed in crypt base columnar cells of mouse small intestine. FEBS Lett. 2003;535:131–135. doi: 10.1016/s0014-5793(02)03896-6. [DOI] [PubMed] [Google Scholar]
- Kerneis S, Bogdanova A, Kraehenbuhl JP, Pringault E. Conversion by Peyer’s patch lymphocytes of human enterocytes into M cells that transport bacteria. Science. 1997;277:949–952. doi: 10.1126/science.277.5328.949. [DOI] [PubMed] [Google Scholar]
- Khandanpour C, Sharif-Askari E, Vassen L, Gaudreau MC, Zhu J, Paul WE, Okayama T, Kosan C, Möröy T. Evidence that Growth Factor Independence 1b regulates dormancy and peripheral blood mobilization of hematopoietic stem cells. Blood. 2010;116:5149–5161. doi: 10.1182/blood-2010-04-280305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokrashvili Z, Rodriguez D, Yevshayeva V, Zhou H, Margolskee RF, Mosinger B. Release of endogenous opioids from duodenal enteroendocrine cells requires Trpm5. Gastroenterology. 2009;137:598–606. 606.e1–2. doi: 10.1053/j.gastro.2009.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent B, Randrianarison-Huetz V, Kadri Z, Romeo PH, Porteu F, Dumenil D. Gfi-1B promoter remains associated with active chromatin marks throughout erythroid differentiation of human primary progenitor cells. Stem Cells. 2009;27:2153–2162. doi: 10.1002/stem.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Perreault N, Brestelli JE, Kaestner KH. Neurogenin 3 is essential for the proper specification of gastric enteroendocrine cells and the maintenance of gastric epithelial cell identity. Genes Dev. 2002;16:1488–1497. doi: 10.1101/gad.985002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelouard H, Sahuquet A, Reggio H, Montcourrier P. Rabbit M cells and dome enterocytes are distinct cell lineages. J Cell Sci. 2001;114:2077–2083. doi: 10.1242/jcs.114.11.2077. [DOI] [PubMed] [Google Scholar]
- May R, Riehl TE, Hunt C, Sureban SM, Anant S, Houchen CW. Identifica-tion of a novel putative gastrointestinal stem cell and adenoma stem cell marker, doublecortin and CaM kinase-like-1, following radiation injury and in adenoma-tous polyposis coli/multiple intestinal neoplasia mice. Stem Cells. 2008;26:630–637. doi: 10.1634/stemcells.2007-0621. [DOI] [PubMed] [Google Scholar]
- May R, Sureban SM, Hoang N, Riehl TE, Lightfoot SA, Ramanujam R, Wyche JH, Anant S, Houchen CW. Doublecortin and CaM kinase-like-1 and leucine-rich-repeat-containing G-protein-coupled receptor mark quiescent and cycling intestinal stem cells, respectively. Stem Cells. 2009;27:2571–2579. doi: 10.1002/stem.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- Milano J, McKay J, Dagenais C, Foster-Brown L, Pognan F, Gadient R, Jacobs RT, Zacco A, Greenberg B, Ciaccio PJ. Modulation of notch processing by gamma-secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicol Sci. 2004;82:341–358. doi: 10.1093/toxsci/kfh254. [DOI] [PubMed] [Google Scholar]
- Miyazawa K, Aso H, Kanaya T, Kido T, Minashima T, Watanabe K, Ohwada S, Kitazawa H, Rose MT, Tahara K, Yamasaki T, Yamaguchi T. Apoptotic process of porcine intestinal M cells. Cell Tissue Res. 2006;323:425–432. doi: 10.1007/s00441-005-0086-z. [DOI] [PubMed] [Google Scholar]
- Moxey PC, Trier JS. Specialized cell types in the human fetal small intestine. Anat Rec. 1978;191:269–285. doi: 10.1002/ar.1091910302. [DOI] [PubMed] [Google Scholar]
- Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci U S A. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeyama A, Leblond CP. “Caveolated cells” characterized by deep surface invaginations and abundant filaments in mouse gastro-intestinal epithelia. Am J Anat. 1974;140:147–165. doi: 10.1002/aja.1001400203. [DOI] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science. 2007;315:988–992. doi: 10.1126/science.1136606. [DOI] [PubMed] [Google Scholar]
- Osawa M, Yamaguchi T, Nakamura Y, Kaneko S, Onodera M, Sawada K, Jegalian A, Wu H, Nakauchi H, Iwama A. Erythroid expansion mediated by the Gfi-1B zinc finger protein: role in normal hematopoiesis. Blood. 2002;100:2769–2777. doi: 10.1182/blood-2002-01-0182. [DOI] [PubMed] [Google Scholar]
- Randrianarison-Huetz V, Laurent B, Bardet V, Blobe GC, Huetz F, Dumenil D. Gfi-1B controls human erythroid and megakaryocytic differentiation by regulating TGF-beta signaling at the bipotent erythro-megakaryocytic progenitor stage. Blood. 2010;115:2784–2795. doi: 10.1182/blood-2009-09-241752. [DOI] [PubMed] [Google Scholar]
- Riccio O, van Gijn ME, Bezdek AC, Pellegrinet L, van Es JH, Zimber-Strobl U, Strobl LJ, Honjo T, Clevers H, Radtke F. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 2008;9:377–383. doi: 10.1038/embor.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MF, Ren J, Ahmad KA, Chao HT, Klisch TJ, Flora A, Greer JJ, Zoghbi HY. Math1 is essential for the development of hindbrain neurons critical for perinatal breathing. Neuron. 2009;64:341–354. doi: 10.1016/j.neuron.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleque S, Cameron S, Orkin SH. The zinc-finger proto-oncogene Gfi-1b is essential for development of the erythroid and megakaryocytic lineages. Genes Dev. 2002;16:301–306. doi: 10.1101/gad.959102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleque S, Kim J, Rooke HM, Orkin SH. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol Cell. 2007;27:562–572. doi: 10.1016/j.molcel.2007.06.039. [DOI] [PubMed] [Google Scholar]
- Saqui-Salces M, Keeley TM, Grosse AS, Qiao XT, El-Zaatari M, Gumucio DL, Samuelson LC, Merchant JL. Gastric tuft cells express DCLK1 and are expanded in hyperplasia. Histochem Cell Biol. 2011 doi: 10.1007/s00418-011-0831-1. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A. Tuft cells. Anat Sci Int. 2007;82:187–199. doi: 10.1111/j.1447-073X.2007.00188.x. [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build cryptvillus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Savidge TC. The life and times of an intestinal M cell. Trends Microbiol. 1996;4:301–306. doi: 10.1016/0966-842x(96)10052-4. [DOI] [PubMed] [Google Scholar]
- Savidge TC, Smith MW, James PS, Aldred P. Salmonella-induced M-cell formation in germ-free mouse Peyer’s patch tissue. Am J Pathol. 1991;139:177–184. [PMC free article] [PubMed] [Google Scholar]
- Sbarbati A, Bramanti P, Benati D, Merigo F. The diffuse chemosensory system: exploring the iceberg toward the definition of functional roles. Prog Neurobiol. 2010;91:77–89. doi: 10.1016/j.pneurobio.2010.01.010. [DOI] [PubMed] [Google Scholar]
- Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Schroder N, Gossler A. Expression of Notch pathway components in fetal and adult mouse small intestine. Gene Expr Patterns. 2002;2:247–250. doi: 10.1016/s1567-133x(02)00060-1. [DOI] [PubMed] [Google Scholar]
- Shroyer NF, Wallis D, Venken KJ, Bellen HJ, Zoghbi HY. Gfi1 functions downstream of Math1 to control intestinal secretory cell subtype allocation and differentiation. Genes Dev. 2005;19:2412–2417. doi: 10.1101/gad.1353905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroyer NF, Helmrath MA, Wang VY, Antalffy B, Henning SJ, Zoghbi HY. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology. 2007;132:2478–2488. doi: 10.1053/j.gastro.2007.03.047. [DOI] [PubMed] [Google Scholar]
- Stanger BZ, Datar R, Murtaugh LC, Melton DA. Direct regulation of intestinal fate by Notch. Proc Natl Acad Sci U S A. 2005;102:12443–12448. doi: 10.1073/pnas.0505690102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureban SM, May R, Ramalingam S, Subramaniam D, Natarajan G, Anant S, Houchen CW. Selective blockade of DCAMKL-1 results in tumor growth arrest by a Let-7a MicroRNA-dependent mechanism. Gastroenterology. 2009;137:649–659. doi: 10.1053/j.gastro.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi S, Leblond CP. Migration and turnover of enteroendocrine and caveo-lated cells in the epithelium of the descending colon, as shown by radioautography after continuous infusion of 3H-thymidine into mice. Am J Anat. 1979;156:431–451. doi: 10.1002/aja.1001560403. [DOI] [PubMed] [Google Scholar]
- VanDussen KL, Samuelson LC. Mouse Atonal homolog 1 directs intestinal progenitors to secretory cell rather than absorptive cell fate. Dev Biol. 2010;346:215–223. doi: 10.1016/j.ydbio.2010.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassen L, Fiolka K, Mahlmann S, Moroy T. Direct transcriptional repression of the genes encoding the zinc-finger proteins Gfi1b and Gfi1 by Gfi1b. Nucleic Acids Res. 2005;33:987–998. doi: 10.1093/nar/gki243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassen L, Okayama T, Moroy T. Gfi1b:green fluorescent protein knock-in mice reveal a dynamic expression pattern of Gfi1b during hematopoiesis that is largely complementary to Gfi1. Blood. 2007;109:2356–2364. doi: 10.1182/blood-2006-06-030031. [DOI] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumor regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- von Furstenberg RJ, Gulati AS, Baxi A, Doherty JM, Stappenbeck TS, Gracz AD, Magness ST, Henning SJ. Sorting mouse jejunal epithelial cells with CD24 yields a population with characteristics of intestinal stem cells. Am J Physiol Gastrointest Liver Physiol. 2011;300:G409–G417. doi: 10.1152/ajpgi.00453.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Tanaka H, Koizumi H, Tanimoto Y, Torii R, Yanagita T. General toxicity studies of tamoxifen in mice and rats. Jitchuken Zenrinsko Kenkyuko (CIEA Preclinical Reports) 1980;6:1–36. [Google Scholar]
- Wong GT, Manfra D, Poulet FM, Zhang Q, Josien H, Bara T, Engstrom L, Pinzon-Ortiz M, Fine JS, Lee HJ, Zhang L, Higgins GA, Parker EM. Chronic treatment with the gamma-secretase inhibitor LY-411,575 inhibits beta-amyloid peptide production and alters lymphopoiesis and intestinal cell differentiation. J Biol Chem. 2004 Mar;279:12876–12882. doi: 10.1074/jbc.M311652200. [DOI] [PubMed] [Google Scholar]
- Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, Finkle D, Venook R, Wu X, Ridgway J, Schahin-Reed D, Dow GJ, Shelton A, Stawicki S, Watts RJ, Zhang J, Choy R, Howard P, Kadyk L, Yan M, Zha J, Callahan CA, Hymowitz SG, Siebel CW. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- Yang H, Xie X, Deng M, Chen X, Gan L. Generation and characterization of Atoh1-Cre knock-in mouse line. Genesis. 2010;48:407–413. doi: 10.1002/dvg.20633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn S, Hecksher-Sørensen J, Pedersen IL, Serup P, Madsen O. Generation of monoclonal antibodies against mouse neurogenin 3: a new immunocytochemical tool to study the pancreatic endocrine progenitor cell. Hybrid Hybridomics. 2004;23:385–388. doi: 10.1089/hyb.2004.23.385. [DOI] [PubMed] [Google Scholar]