Abstract
Purpose
Homozygous deletions play important roles in carcinogenesis. The genome-wide screening for homozygously deleted genes in many different cancer types with a large number of patient specimens representing the tumor heterogeneity has not been done.
Experimental Design
We performed integrative analyses of the copy number profiles of 10,759 patients across 31 cancer types from The Cancer Genome Atlas project.
Results
We found that the Type-I interferon, α- and β-defensin genes were homozygously deleted in 19 cancer types with high frequencies (7%–31%, median = 12%; interquartile range = 10–16.5%). Patients with homozygous deletion of interferons exhibited significantly shortened overall or disease-free survival time in a number of cancer types, whereas patients with homozygous deletion of defensins did not significantly associate with worse overall or disease-free survival. Gene expression analyses suggested that homozygous deletion of interferon and defensin genes could activate genes involved in oncogenic and cell cycle pathways but repress other genes involved in immune response pathways, suggesting their roles in promoting tumorigenesis and helping cancer cells evade immune surveillance. Further analysis of the whole exomes of 109 melanoma patients demonstrated that the homozygous deletion of interferon (P = 0.0029, OR = 11.8) and defensin (P = 0.06, OR = 2.79) genes are significantly associated with resistance to anti-CTLA-4 (Cytotoxic T-Lymphocyte Associated protein 4) immunotherapy.
Conclusions
Our analysis reveals that the homozygous deletion of interferon and defensin genes are prevalent in human cancers, and importantly this feature can be used as a novel prognostic biomarker for immunotherapy resistance.
Keywords: Homozygous deletion, Type-I interferon, Defensin, immunotherapy resistance, cancer
Introduction
Somatic copy number alteration (SCNA) is one of the major sources of genomic instabilities that play critical roles in cancer development and progression. Previous studies estimate that 25% of the cancer genome is affected by arm-level SCNAs and 10% by focal SCNAs with 2% overlap (1). Mapping the focal SCNAs recurrent in different cancer types could potentially reveal the common molecular mechanisms of tumorigenesis and identify therapeutic targets. Through analyzing the copy number profiles of 4934 tumor genomes across 11 cancer types, The Cancer Genome Atlas (TCGA) project identified 140 recurrent focal SCNAs including 70 amplified regions affecting 959 genes and 70 deleted regions affecting 2084 genes (2). Another study exploring the copy number profiles of 3131 tumor genomes across 26 cancer types identified 158 recurrent focal SCNAs including 76 amplification affecting 1566 genes and 82 deletions affecting 2001 genes (1). However, distilling this genomic alteration information to enable cancer precise medicine is still limited.
Compared to genes with copy neutral, single-copy loss, copy gain or amplification, homozygously deleted genes provide a valuable resource to identify genes essential for cancer development, proliferation, and survival. Although similar work has been done in cell lines (3), genome-wide screening for homozygously deleted genes in many different cancer types with a large number of patient specimens representing the heterogeneity of tumors has not been done. In this study, we re-analyzed the copy number profiles of 10,759 patients across 31 cancer types with the aim to identify recurrent homozygously deleted genes implicated in carcinogenesis. Strikingly, we found 97% of identified homozygously deleted genes are located in only two loci including 9p21.3 and 8p23.3-21. In particular, we found type I interferon (IFN) gene cluster located in 9p21.3 and defensin (DEF) gene cluster located in 8p23.3-21.1 are homozygously deleted in at least 7% of patients in each of the 19 cancer types. Survival analyses of different tumor types indicated that patients with homozygous deletion of interferon or defensin genes exhibit much worse overall or disease-free survival. RNA-seq gene expression analyses between patients with and without IFN/DEF deletion in 19 cancer types indicate that homozygous deletions of IFN and DEF activate oncogenic and cell cycle pathways but repress immune response pathways. Reanalysis of the whole exome sequencing (WES) data generated from 109 metastatic melanoma patients suggests that homozygous deletion of IFN and DEF genes are significantly associated with resistance to anti-CTLA4 therapy (4). Since a large body of evidence suggest that interferons and defensins play major roles in tumor immunity by recognizing tumor cells and serve as a bridge to spontaneous adaptive T cell response (5–11), our findings suggest a common molecular mechanism mediated by the deletion of interferon and defensin genes, through which tumor cells escape immune detection and destruction. Given the pervasiveness of homozygous deletion of IFN and DEF genes in primary cancers, this study reveals universal and novel prognostic biomarkers to predict immunotherapy resistance, which has implications for personalized medicine.
Materials and Methods
The Cancer Genome Atlas Copy Number Variation Data and Analysis
The thresholded somatic copy number alteration data of 10,843 patients (33 cancer types) were downloaded from the Cancer Genome Hub at the University of California at Santa Cruz (https://genome-cancer.ucsc.edu/) (12). After removing CHOL (N = 36) and DLBC (N = 48) cohorts that have less than 50 patients, SCNA profiles of 10,759 patients were analyzed. These 10,759 patients compose 31 cancer types, which include 22 common cancers and 9 rare cancers. Common and rare tumor designation is defined by http://cancergenome.nih.gov/cancersselected/RareTumorCharacterizationProjects. Matched overall survival or disease-free survival data were downloaded from CBioPortal (http://www.cbioportal.org/) (13). In brief, Gistic2 generated gene level CNV estimates were thresholded into discrete values −2, −1, 0, 1, 2 representing homozygous deletion, single copy deletion, diploid neutral, low copy number gain and high copy number amplification, respectively. Homozygous deletion frequency is calculated as:
Gene expression analysis
For each tumor type, cancer patients were first divided into two groups according to the HDI (homozygous deletion of interferon) and/or HDD (homozygous deletion of defensin) status. Patients without RNA-seq data were removed. Gene level expression estimates (i.e. RSEM normalized read count) were downloaded from TCGA Data Portal (https://tcga-data.nci.nih.gov/). EdgeR was used to compare gene expression profiles between patients with HDI/HDD and those without HDI/HDD. Differentially expressed genes were determined by the FDR cutoff 0.01 (14).
Pathway and IPA analysis
Functional annotation of the homozygously deleted genes was performed by DAVID (https://david.ncifcrf.gov/) (15) and ConsensusPathDB gene set over-representation analysis (http://cpdb.molgen.mpg.de/CPDB) (16). Commonly up-regulated genes were defined as genes that significantly up-regulated in at least three cancer types but not down-regulated in any of the 17 cancer types. Commonly down-regulated genes were defined as genes that significantly down-regulated in at least three cancer types but not up-regulated in any of the 17 cancer types. Commonly up- and down-regulated genes were overlapped with hallmark gene sets collected in Molecular Signature Database (http://software.broadinstitute.org/gsea/msigdb/). Significant overlaps were determined by FDR q-value ≤ 0.05. Up- and down-regulated genes in each cancer types were uploaded into https://apps.ingenuity.com to do Ingenuity Pathway Analysis (IPA®).
Power analysis
We sought to determine if there is sufficient number of events available for overall and disease-free survival analyses for each cancer type. At given statistical significance criterion (α = 0.05, two-tailed) and statistical power 80% (i.e. β = 0.2), the minimum number of required events (K) is calculated as:
where Zα/2 = 1.96, Zβ = 0.84, π1 and π2 are the proportions to be allocated into two groups and were determined by the HDI/HDD frequencies in each cancer type. HR is the hazard ratio estimated using the logrank approach implemented in the survival package (https://cran.r-project.org/web/packages/survival/index.html).
where Oa and Ea are the numbers of observed and expected events in patients with HDI/HDD, and Ob and Eb are the numbers of observed and expected events in patients without HDI/HDD. HR > 1 indicates higher hazard of death or progression from the HDI/HDD group.
Survival and statistical analysis
To increase sample size, we combined COAD and READ as colorectal cancer, LUAD and LUSC as lung cancer (therefore 31 cancer types reduced to 29). We removed four rare tumors having no OS and DFS time available for survival analysis including adrenocortical carcinoma (ACC), mesothelioma (MESO), pheochromocytoma and paraganglioma (PCPG) and uterine carcinosarcoma (UCS). According to the power analysis described above, all cancer types except LGG were underpowered; we therefore required at least 10 events (i.e. death or progression) in HDI/HDD group. Based on this criterion, we further removed 10 cancer types having less than 10 events for both OS and DFS in HDI/HDD group including cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), acute myeloid leukemia (LAML), kidney chromophobe (KICH, rare tumor), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), testicular germ cell tumors (TGCT, rare tumor), thyroid carcinoma (THCA), thymoma (THYM, rare tumor), uterine corpus endometrial carcinoma (UCEC), uveal melanoma (UVM, rare tumor). Using the same criterion, PRAD (OS, Nevents = 2), ESCA (OS, Nevents = 6), SARC (DFS, Nevents = 8) were also removed. We performed either overall or disease-free survival analyses for the remaining 15 cancer types.
Survival analysis was performed using the “survival” R package available from http://cran.r-project.org. The Log-rank test was used to evaluate if the difference in overall survival or disease-free survival between the above two subgroups was statistically significant. Benjamini-Hochberg (BH) method was used to adjust log-rank test p-values for multiple comparisons. CoMEt exact test was used to evaluate the mutual exclusivity of HDIs and HDDs in each tumor type (17). Fisher’s exact test was used to evaluate the significance of association between HDI/HDD events and ipilimumab treatment responses in melanoma patients. Wilcoxon rank sum test (a.k.a. Mann-Whitney U test) was used to evaluate the significance the difference in diagnosis ages between LUSC patients with concurrent HDI/HDD and those without. Wilcoxon rank sum test was also used to compare HDI/HDD frequencies between common and rare cancer types.
Melanoma whole exome sequencing analysis
The whole exome sequencing data of tumors and matched normal tissues (i.e. adjacent normal tissues or peripheral blood mononuclear cells) for 109 melanoma patients were downloaded from dbGAP with accession number phs000452.v2.p1 (4). One patient (Pat121) was not included in the analysis because the file is larger than the maximum size allowed to download. Treatment responses to anti-CTLA-4 were retrieved from the supplementary data of the original publication. Raw reads alignments to the human reference genome (hg19/GRCh37) were extracted from the original SRA files using SRA-Toolkit (18). Somatic copy number variation (i.e. copy number change in tumor tissue versus normal tissues) for each patient was called by varscan-2 with the default parameters (19). The post-processing step of smoothing, segmentation and re-center were performed by DNACopy and in-house scripts (20). Genes with log2 (tumor/normal) smaller than −1 were considered as homozygous deletions. Patients were stratified into “responder” (N = 27), “non-responder” (N = 72) and “long-term survivor” (N = 10) following the original publication (4). Specifically, “responders” referred to patients who achieved clinical benefits (“complete response” or “partial response” or “stable disease” with overall survival (OS) > 1 year by RECIST criteria). “Non-responder” referred to those with minimal or no benefit from ipilimumab (“progressive disease” or “stable disease” with OS < 1 year by RECIST criteria). “Long-term survivors” refer to a cohort of patients who achieved long-term survival (OS > 2 years) after ipilimumab treatment but with early tumor progression (i.e. shorter progression-free survival (PFS) < 6 months).
Results
Homozygous deletions of interferon and defensin genes are pervasive in human cancers
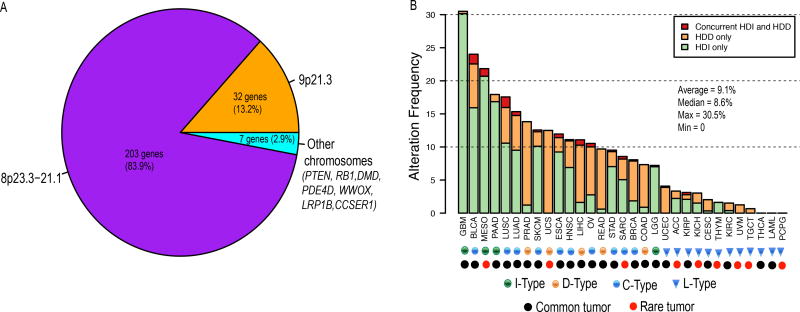
To identify the common, homozygously-deleted genes in human cancers, we analyzed the copy number profiles of 10,759 patients across 31 cancer types (Supplementary Table 1). We identified 242 genes whose average homozygous deletion frequencies across 31 cancer types are higher than 3% (Supplementary Table 2). Strikingly, we found 235/242 (97%) genes are clustered on either 9p21.3 or 8p23.3-21.1 with only seven (3%) genes (PTEN, RB1, DMD, PDE4D, WWOX, LRP1B, and CCSER1) residing on other chromosomes (Fig. 1A, Supplementary Table 2). As expected, all these seven genes except CCSER1 (alias FAM190A) are known tumor suppressors and have been identified as the potential drivers of focal deletions detected by previous studies (1,2). The molecular function CCSER1 is largely unknown, but its homozygous deletion has also been observed in various cancers (21–23). Functional classification of the 242 genes revealed that interferons (16 genes, q-value = 8.37×10−20) and defensins (24 genes, q-value = 1.65×10−30) are the most significantly enriched terms (Supplementary Table 3). The 16 type I interferon genes are located on 9p21.3, which include 13 IFN-α genes, 1 IFN-β, 1 IFN-ε and 1 IFN-ω gene (Supplementary Figure 1A). The 24 defensin genes are located on 8p23.3-21.1, which include 6 α-defensin and 18 β-defensin genes (Supplementary Figure 1B). Since both interferons and defensins are involved in innate immune response and play important roles in recognizing tumor cells and inducing an anti-tumor immune response, the widespread, homozygous deletion of these genes suggests a common molecular mechanism through which tumor cells escape immune destruction.
Figure 1. Genomic distribution of homozygously deleted genes and the deletion frequency of HDI/HDD in each tumor type.
(A) Genomic distribution of the 242 homozygously deleted genes identified. (B) Alteration frequency of HDI and HDD in 31 cancer types. Patients of each cancer type were divided into three groups: patients with HDI only (green), patients with HDD only (orange), and patients with concurrent HDI and HDD (red). The 31 cancers were classified into 4 types: L-type refers to cancer types with HDI/HDD frequencies less than 5% (blue triangle); I-type refers to cancer types in which HDI is the dominant alteration (green circle); D-type refers to cancer types in which HDD is the dominant alteration (orange circle); C-type refers to cancer types in which both HDI and HDD are prevalent (blue circle). Common and rare tumors are designated by TCGA and indicated using black and red circles, respectively. HDI, homozygous deletion of type-I interferons; HDD, homozygous deletion of defensin.
We observed the homozygous deletion of interferons (HDIs) and defensins (HDDs) in all 31 cancer types except PCPG (pheochromocytoma and paraganglioma, N = 162), THCA (thyroid carcinoma, N = 501) and LAML (acute myeloid leukemia, N = 191) (Fig. 1B, Supplementary Table 4). Interestingly, we found the frequencies of HDI/HDD are much higher in the 22 common cancer types (median frequency = 10%) than those in the nine rare cancer types (median frequency = 3%) (P = 0.1, two-tailed Wilcoxon rank sum test) (Supplementary Figure 2). This result suggests HDI/HDD as a common molecular mechanism involved primarily in the carcinogenesis of common cancers. Using 5% alternation frequency as the threshold, we defined 19 cancer types as the high HDI/HDD group, in which two brain tumor types LGG (brain lower grade glioma) and GBM (glioblastoma multiforme) exhibit the lowest (7%) and highest (31%) alternation frequencies, respectively (Fig. 1B, Supplementary Table 4). We further divided the 19 cancer types into three subtypes using the prevalence ratio (PR = {# of HDIs}/{# of HDDs}) of 5 as the threshold: The I-type is defined as cancer types with HDI at least 5 times more (PR ≥ 5) prevalent than HDD, including LGG, GBM, PAAD (Pancreatic adenocarcinoma) and MESO (Mesothelioma) (Fig. 2A–B, Supplementary Figure 3A–B). The D-type is defined as cancer types with HDD at least 5 times more (PR ≤ 0.2) prevalent than those of HDI, including PRAD (prostate adenocarcinoma), COAD (colon adenocarcinoma), READ (rectum adenocarcinoma), LIHC (liver hepatocellular carcinoma), UCS (uterine carcinosarcoma) (Fig. 2C–D, Supplementary Figure 3C–D). The C-type refers to those cancer types with both HDI and HDD (0.2 < PR < 5), including BLCA (bladder urothelial carcinoma), LUAD (lung adenocarcinoma), LUSC (lung squamous cell carcinoma), BRCA (breast invasive carcinoma), OV (ovarian serous cystadenocarcinoma), HNSC (head and neck squamous cell carcinoma), SKCM (skin cutaneous melanoma), ESCA (esophageal carcinoma), SARC (sarcoma), and STAD (stomach adenocarcinoma) (Fig. 2E–F, Supplementary Figure 3E–H).
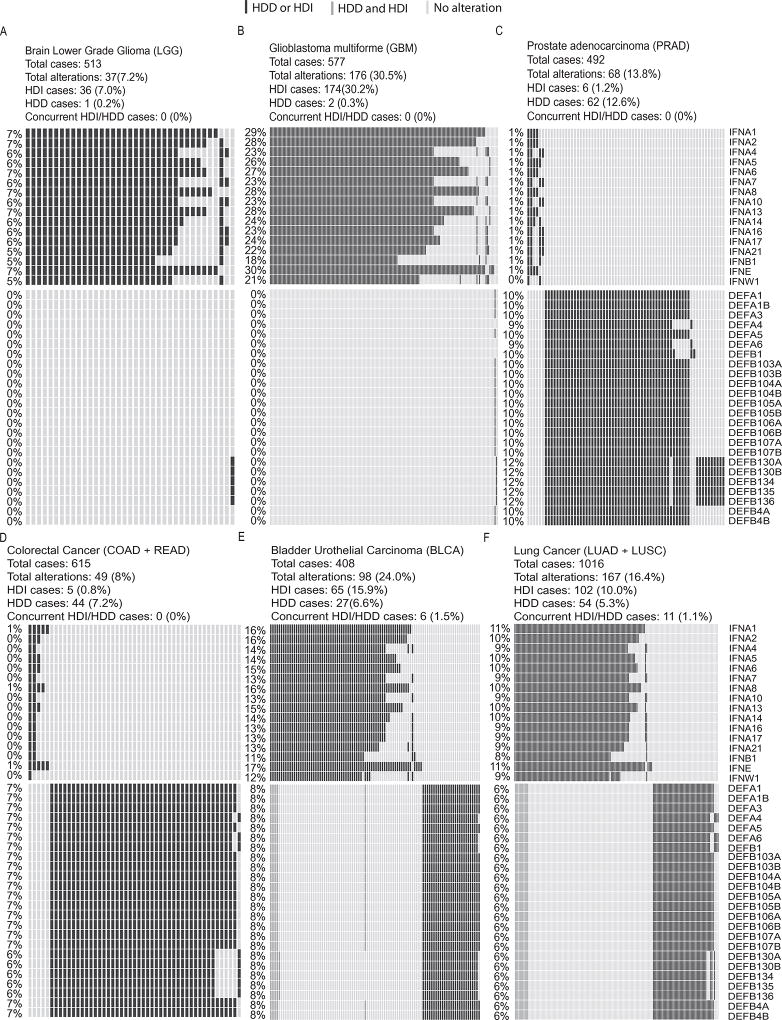
Figure 2. Homozygous deletions of interferon and defensin genes in six cancer types.
In each panel, rows represent genes and columns represent patients. Black bars indicate patients with HDD or HDI, grey bars indicate patients with concurrent HDD and HDI. Overall deletion frequency of each gene is listed on the left. (A–B) I-type cancers using brain lower grade glioma (LGG) and glioblastoma multiforme (GBM) as examples. (C–D) D-type cancers using prostate adenocarcinoma (PRAD) and colorectal cancer as examples. Colorectal cancer is the combined cohort of colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ). (E–F) C-type cancers using bladder urothelial carcinoma (BLCA) and lung cancer as examples. Lung cancer is the combined cohort of lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC). HDI, homozygous deletion of type-I interferons; HDD, homozygous deletion of defensin.
As shown in Fig. 2 and Supplementary Figure 3, I-type tumors almost exclusively have HDI but not HDD, and D-type tumors almost exclusively have HDDs but not HDIs. Even for C-type tumor such as BRCA (N = 1079), there were 20 (2%) patients having HDIs and 65 (6%) patients having HDDs but only 2 (0.2%) patients with concurrent HDI and HDD. These data suggested HDI and HDD had the tendency towards mutually exclusive both within and across cancer types, even though they did not reach statistical significance due to the limited number of cases (Supplementary Table 4). We expect patients with concurrent HDI and HDD would have the worse prognosis than those patients with only HDI or HDD, but the paucity of concurrent HDI and HDD preclude rigorous statistical analysis (Supplementary Table 4). However, we found in the LUSC cohort which has the most concurrent HDI and HDD cases (N = 8), the diagnosis age of patients with concurrent HDI and HDD (median = 60) is much smaller than other patients (median = 68) (P = 0.079, two-tailed Wilcoxon rank sum test) (Supplementary Figure 4). This result suggests the association between concurrent HDI and HDD and the early tumor onset in LUSC.
Homozygous deletions of interferon or defensin genes associate with worse overall or disease-free survival in human cancers
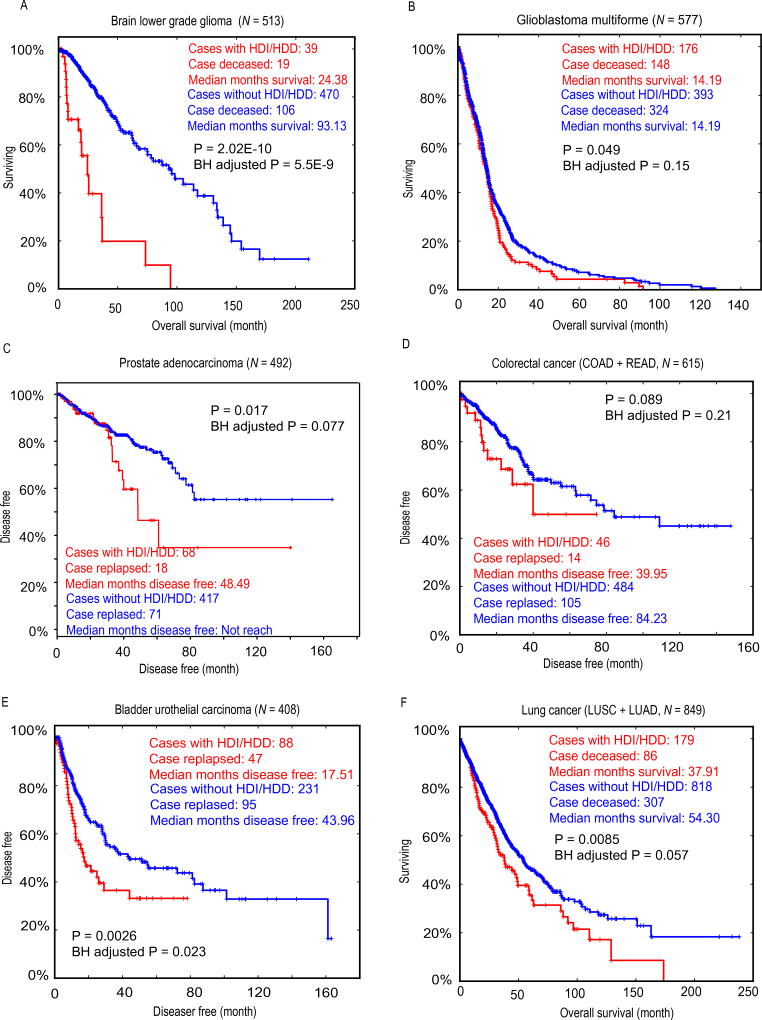
Since both interferons and defensins are implicated in anti-tumor immunity, we next investigated if patients with HDI or HDD lesions exhibit worse overall survival (OS) or disease-free survival (DFS). Indeed, we observed patients with I-type tumors (Fig. 3A–B) D-type tumors (Fig. 3C–D) and C-type tumors (Fig. 3E–F) had significantly worse OS or DFS. For examples, the median overall survival time of LGG patients with HDI or HDD (24 month) is only 26% of those patients without HDI or HDD (93 month) (P = 2.02×10−10, LogRank test) (Fig. 3A). Similarly, the median disease-free survival time of bladder cancer patients with HDI or HDD (17.5 month) is only 40% of those patients without HDI or HDD (44 month) (P = 0.0026) (Fig. 3E), and the median overall survival time of lung cancer patients with HDI or HDD is 70% of those patients without HDI or HDD (P = 0.0085) (Fig. 3F). We found that, in most cancer types, patients with HDI/HDD lesions consistently exhibit worse OS of DFS compared to those without HDI/HDD lesions, even though some cancer types did not reach statistical significance (Supplementary Figure 5). The associations of HDI/HDD status with worse clinical outcomes across cancer types highlight their prognostic values.
Figure 3. Comparing the overall or disease-free survival between patients with (red) and without (blue) HDIs/HDDs.
(A) Overall survival curves for brain lower grade glioma (LGG). (B) Overall survival curves for glioblastoma multiforme (GBM). (C) Disease-free survival curves for prostate adenocarcinoma (PRAD). (D) Disease-free survival curves for colorectal cancer. Colorectal cancer is the combined cohort of colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ). (E) Disease-free survival curves for bladder urothelial carcinoma (BLCA). (F) Overall survival curves for lung cancers. Lung cancer is the combined cohort of lung adenocarcinoma (LUAD) and lung quamous cell carcinoma (LUSC).
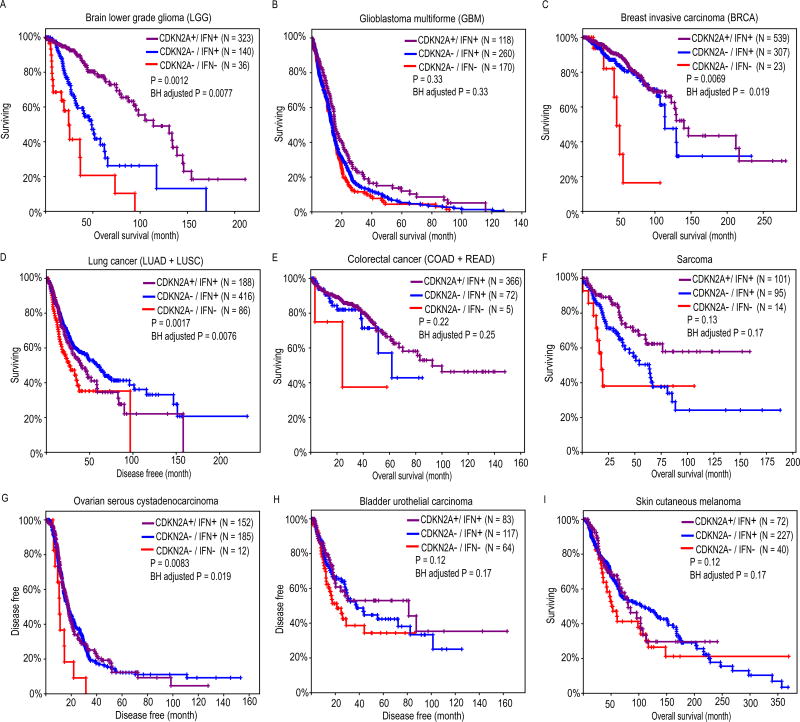
We found that IFN genes are located approximately 485 kb to 890 kb away from a well-known tumor suppressor CDKN2A (Cyclin Dependent Kinase Inhibitor 2A) (Supplementary Figure 1A). The homozygous deletion of this genes has been reported in mesothelioma and numerous cancer cell lines (3,24,25). We asked the question whether homozygous deletions of interferon genes are passive hitchhiking events due to the nearby CDKN2A deletion, or they play an active role in tumorigenesis and affect the patient survival. Therefore, we tested if the additional loss of IFN genes could further reduce patients’ overall or disease-free survival, comparing to patients with CDKN2A deletion only. We performed the survival analyses on three patient groups: (I) patients only have CDKN2A deletion (CDKN2A−/IFN+); (II) patients have CDKN2A deletion and additional IFN gene deletions (CDKN2A−/IFN−); and (III) patients have neither CDKN2A nor IFN gene deletions (CDKN2A+/IFN+). Very few patients have IFN deletion only (i.e. CDKN2A+/IFN−), so this group was excluded from our analysis. As expected, CDKN2A−/IFN− patients have the worst prognosis compared to CDKN2A−/IFN+ and CDKN2A+/IFN+ groups in a variety of cancer types including LGG, GBM, BRCA, lung cancer (LUAD + LUSC), colorectal cancer (COAD + READ), SARC, OV, BLCA, SKCM, ESCA and PAAD (Fig. 4, Supplementary Figure 6). After performing power analysis (see Materials and Methods), we found all tumor types except LGG are underpowered (Supplementary Table 5). Therefore, the insignificant P-values of OS/DFS analyses in some tumor types are most likely due to limited sample size. For instance; only 5 and 14 patients had both CDKN2A and IFN gene deletions for colorectal cancer and SARC, respectively. It could be also due to other confounding factors such as PTEN and RB1 deletions, which tend to be mutually exclusive with HDIs and HDDs (Supplementary Figure 7).
Figure 4. Comparing the overall survival between patients having neither CDKN2A nor interferon gene deletions (purple), patients having CDKN2A deletion only (blue), and patients having both CDKN2A and interferon gene deletions (red).
Survival curves for brain lower grade Glioma (LGG), glioblastoma multiforme (GBM), breast invasive carcinoma (BRCA), lung cancer, colorectal cancer, sarcoma (SARC), ovarian serous cystadenocarcinoma (OV), bladder urothelial carcinoma (BLCA), skin cutaneous melanoma (SKCM) are indicated in panel (A) – (I), respectively. Colorectal cancer is the combined cohort of colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ). Lung cancer is the combined cohort of lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC). To increase sample size, patients with single copy loss of CDKN2A, truncated mutations of CDKN2A/IFNs/DEFs were also included. Log rank test P-values were calculated by comparing patients having CDKN2A deletion only (blue) with patients having both CDKN2A and interferon gene deletions (red).
Homozygous deletions of interferon and defensin genes activate oncogenic and cell cycle pathways but repress immune response pathways
To identify gene expression signatures associated with the deletion of IFN and DEF genes, we performed gene expression analysis on each of the 19 cancer types (or 17 cancer types when COAD and READ are combined as colorectal cancer, LUSC and LUAD are combined as lung cancer) having high frequencies of HDI and HDD. Through differential gene expression analyses between patients with and without HDI/HDD lesions in 17 cancer types, we detected a total of 4599 genes that differentially expressed in at least one cancer type (Supplementary Table 6, Supplementary Figure 8A). On average, 204 (median = 145) and 254 (median = 232) genes are significantly (FDR < 0.01) up- and down-regulated in each cancer type, respectively (Supplementary Figure 8B).
To identify the implicated molecular pathway, we defined a set of genes commonly altered in multiple cancer types (see Materials and Methods) and overlapped them with the hallmark gene sets collected in Molecular Signature Database (http://software.broadinstitute.org/gsea/msigdb/index.jsp). We found “genes defining epithelial-mesenchymal transition (EMT)” (FDR = 4.6×10−18), “genes up-regulated by KRAS activation” (FDR = 6.7×10−9), and cell cycle genes (FDR = 1.3×10−6) are significantly enriched in genes up-regulated in patients with HDI/HDD (Supplementary Table 7). EMT is a biologic process involved in cancer progression and metastasis (26). KRAS is a proto-oncogene and its activation is implicated in various malignancies. Similarly, “genes regulated by NF-kB in response to tumor necrosis factor (TNF)” (FDR = 1.9×10−11), “genes up-regulated by STAT5 in response to IL2 stimulation” (FDR = 3.0×10−8) are significantly enriched in genes down-regulated in patients with HDI/HDD (Supplementary Table 7). It is known that NF-kB family members play key roles in regulating the transcription of cytokines and antimicrobial effectors and thereby controlling innate and adaptive immune responses (27) and that IL2 has essential roles in regulating immune tolerance through its direct effects on T cells (28).
We further performed Ingenuity Pathway Analysis (IPA®) analyses to identify common upstream regulators for genes differentially expressed between the two groups for each tumor type. Strikingly, we found TNF is detected as the top upstream regulator for all the 17 cancer types with extremely significant P-values (Supplementary Table 8). IFN-γ (IFNG) is detected as the second most common upstream regulator in 10 cancer types including MESO, PAAD, LGG, COAD, READ, LUSC, LUAD, BLCA, SARC and STAD (Supplementary Table 8). Both TNF and IFN-γ have been found to up-regulate immune checkpoint molecule PD-L1 (B7-H1) in a broad range of tumors (29), and have been linked to resistance to T-cell therapy and anti-CTLA-4 therapy in melanomas (30–32). In conclusion, HDD and HDI activate genes involved in oncogenic/cell cycle pathways and repress genes involved in immune response demonstrates their active role in facilitating tumorigenesis and immune escape.
Homozygous deletions of interferon/defensin genes associate with immunotherapy resistance in melanomas
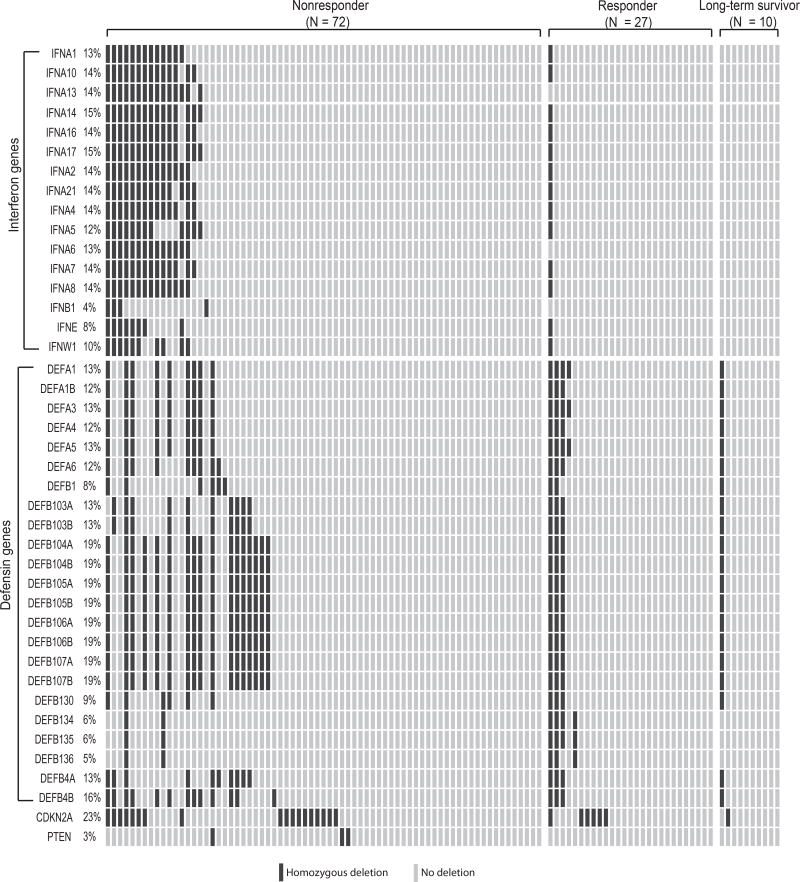
In a recent report, copy loss of type I interferon genes are found in 6 out of 12 melanoma patients that resist to anti-CTLA-4 treatment, but in none of the 4 responders (P = 0.23, two-tailed Fisher exact test) (31). To test if HDI and HDD are associated with immunotherapy resistance in an independent larger cohort, we analyzed the whole exome sequencing data of 109 metastatic melanoma patients treated with ipilimumab–a monoclonal antibody targeting CTLA-4 (4). The 109 melanoma patients are stratified into 72 non-responders (i.e. patients resistant to ipilimumab treatment), 27 responders and 10 long-term survivors according to RECIST criteria (see Materials and Methods) (33). HDIs are detected in 25% (18/72) of non-responders, 4% (1/27) responders, and none (0/10) of the long-term survivors (Fig. 5). Association analysis suggested that HDI is significantly associated with non-responders (P = 0.0029, odds ratio = 11.8) (Table 1). HDDs are also detected more frequently in non-responders, and the association is close to being statistically significant (P = 0.06, odds ratio = 2.79) (Table 2). As a comparison, deletions of two tumor suppressor genes CDKN2A (P = 0.63) and PTEN (P = 0.55) are not associated with responders or non-responders (Fig. 5). Taken together, our meta-analyses of two independent melanoma cohorts suggest the deletion of interferon and defensin genes is significantly associated anti-CTLA-4 treatment resistance, and might be a potential prognostic biomarker to predict resistance to other immunotherapies.
Figure 5. HDI/HDD alteration frequencies in 109 melanoma patients treated with ipilimumab.
Oncoprint plot showing the deletion landscape of interferon genes, defensin genes, CDKN2A and PTEN in 109 metastatic melanoma patients. Patients were divided into 3 groups including non-responder (N = 72), responder (N = 27) and long-term survival (N = 10), according the response outcomes to ipilimumab. HDI, homozygous deletion of type-I interferons; HDD, homozygous deletion of defensin.
Table 1.
Contingency table evaluating the association between HDI and non-responsiveness to ipilimumab treatment. Two-tailed Fisher Exact test, P = 0.0029, odds ratio (OR) = 11.8. HDI, homozygous deletion of type-I interferons.
| Type-I IFNs (Homozygous deletion) |
Type-I IFNs (1 or more copies) |
Total | |
|---|---|---|---|
| Non-responder | 18 | 54 | 72 |
| Responder + Long-term survivor | 1 | 36 | 37 |
| Total | 19 | 90 | 109 |
Table 2.
Contingency table evaluating the association between HDD and non-responsiveness to ipilimumab treatment. Two-tailed Fisher Exact test, P = 0.06, odds ratio (OR) = 2.79. HDD, homozygous deletion of defensin.
| α-, β-DEFs (Homozygous deletion) |
α-, β-DEFs (1 or more copies) |
Total | |
|---|---|---|---|
| Non-responder | 22 | 50 | 72 |
| Responder + Long-term survivor | 5 | 32 | 37 |
| Total | 27 | 82 | 109 |
Discussion
Immunotherapies have been demonstrated to be efficacious in many cancer types (34–36), however, therapeutic resistance is frequently observed and the mechanisms of both de novo and acquired immune-resistance are mostly unknown. In this study, we analyzed the copy number profiles of 10,759 primary cancer tissues and found the homozygous deletions of type I interferon and defensin genes are pervasive in 19 TCGA cancer types, and the alteration frequency is much higher than those of well-known tumor suppressors PTEN and RB1 in the same cancer type (Supplementary Figure 7). More importantly, homozygous deletions of interferon and defensin genes associate with worse overall or disease-free survival, and the resistance to anti-CTLA4 treatment in melanoma patients. Defects in the type I interferon signaling pathway have been proposed as a potential mechanism of cancer escape (insensitivity) to immunotherapy in mice and prostate cancer cell line (37,38). In mice, type I interferon signals are required to initiate the antitumor CD8+ response, and mice without IFN-α/β receptor cannot reject immunogenic tumor cells (9,10). Consistent with these preclinical observations, our observation that type I interferon genes being frequently deleted in many tumor types suggested a generic mechanism through which tumors develop immuno-resistance, and revealed new biomarkers to identify responsive patients. Homozygous deletions of interferon and defensin genes activate oncogenic pathways and repress immune response pathways further recapitulate their roles in promoting tumorigenesis and help tumor cells escape immunosurveillance.
Supplementary Material
Statement of translational relevance.
Immunotherapy has become a major treatment option for many cancers, however, therapeutic resistance is frequently observed and the genetic basis of lack of response is mostly unknown. By analyzing the genomes of 10,759 cancer patients across 31 cancer types, we found interferon and defensin genes are homozygously deleted with high frequencies in 19 cancer types, and the surviving time of patients with these deletions are significantly reduced. Further analysis of the genomes of 109 melanoma patients treated with ipilimumab – a monoclonal antibody targeting CTLA-4 demonstrated that the homozygous deletion of type-I interferon and defensin genes are significantly associated with resistance to immunotherapy. Given that interferon and defensin play important roles in tumor immunity, our study revealed a generic mechanism that might be utilized by tumor cells to escape immune destruction and identified that homozygous deletion of interferon/defensin as potential prognostic and predictive biomarker for immunotherapy resistance.
Acknowledgments
This study was partially supported by the Center for Individualized Medicine (Pharmacogenomics Program), Mayo Clinic; grants from the National Institutes of Health (CA134514, CA130908 and CA193239 to H.H.).
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–40. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox C, Bignell G, Greenman C, Stabenau A, Warren W, Stephens P, et al. A survey of homozygous deletions in human cancer genomes. Proc Natl Acad Sci USA. 2005;102:4542–7. doi: 10.1073/pnas.0408593102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–11. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gajewski TF, Schreiber H, Fu Y-X. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14:1014–22. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gajewski TF, Fuertes MB, Woo S-R. Innate immune sensing of cancer: clues from an identified role for type I IFNs. Cancer Immunol Immunother. 2012;61:1343–7. doi: 10.1007/s00262-012-1305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lichtenstein A, Ganz T, Selsted ME, Lehrer RI. In vitro tumor cell cytolysis mediated by peptide defensins of human and rabbit granulocytes. Blood. 1986;68:1407–10. [PubMed] [Google Scholar]
- 8.Droin N, Hendra J-B, Ducoroy P, Solary E. Human defensins as cancer biomarkers and antitumour molecules. Journal of proteomics. 2009;72:918–27. doi: 10.1016/j.jprot.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Fuertes MB, Kacha AK, Kline J, Woo S-R, Kranz DM, Murphy KM, et al. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8{alpha}+ dendritic cells. J Exp Med. 2011;208:2005–16. doi: 10.1084/jem.20101159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diamond MS, Kinder M, Matsushita H, Mashayekhi M, Dunn GP, Archambault JM, et al. Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J Exp Med. 2011;208:1989–2003. doi: 10.1084/jem.20101158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spaapen RM, Leung MYK, Fuertes MB, Kline JP, Zhang L, Zheng Y, et al. Therapeutic activity of high-dose intratumoral IFN-β requires direct effect on the tumor vasculature. J Immunol. 2014;193:4254–60. doi: 10.4049/jimmunol.1401109. [DOI] [PubMed] [Google Scholar]
- 12.Zhu J, Sanborn JZ, Benz S, Szeto C, Hsu F, Kuhn RM, et al. The UCSC Cancer Genomics Browser. Nat Methods. 2009;6:239–40. doi: 10.1038/nmeth0409-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012 doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 16.Kamburov A, Wierling C, Lehrach H, Herwig R. ConsensusPathDB--a database for integrating human functional interaction networks. Nucleic Acids Res. 2009;37:D623–8. doi: 10.1093/nar/gkn698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leiserson MDM, Wu H-T, Vandin F, Raphael BJ. CoMEt: a statistical approach to identify combinations of mutually exclusive alterations in cancer. Genome Biol. 2015;16:160. doi: 10.1186/s13059-015-0700-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kodama Y, Shumway M, Leinonen R International Nucleotide Sequence Database Collaboration. The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res. 2012;40:D54–6. doi: 10.1093/nar/gkr854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome research. 2012;22:568–76. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–72. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 21.Klorin G, Rozenblum E, Glebov O, Walker RL, Park Y, Meltzer PS, et al. Integrated high-resolution array CGH and SKY analysis of homozygous deletions and other genomic alterations present in malignant mesothelioma cell lines. Cancer Genet. 2013;206:191–205. doi: 10.1016/j.cancergen.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rentsch CA, Müller DC, Ruiz C, Bubendorf L. Comprehensive Molecular Characterization of Urothelial Bladder Carcinoma: A Step Closer to Clinical Translation? European urology. 2017 doi: 10.1016/j.eururo.2017.06.022. [DOI] [PubMed] [Google Scholar]
- 23.Calhoun ES, Hucl T, Gallmeier E, West KM, Arking DE, Maitra A, et al. Identifying allelic loss and homozygous deletions in pancreatic cancer without matched normals using high-density single-nucleotide polymorphism arrays. Cancer Res. 2006;66:7920–8. doi: 10.1158/0008-5472.CAN-06-0721. [DOI] [PubMed] [Google Scholar]
- 24.Illei PB, Rusch VW, Zakowski MF, Ladanyi M. Homozygous deletion of CDKN2A and codeletion of the methylthioadenosine phosphorylase gene in the majority of pleural mesotheliomas. Clin Cancer Res. 2003;9:2108–13. [PubMed] [Google Scholar]
- 25.Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, Andrews JM, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–8. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 27.Hayden MS, West AP, Ghosh S. NF-kappaB and the immune response. Oncogene. 2006;25:6758–80. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- 28.Malek TR, Castro I. Interleukin-2 receptor signaling: at the interface between tolerance and immunity. Immunity. 2010;33:153–65. doi: 10.1016/j.immuni.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8:793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 30.Landsberg J, Kohlmeyer J, Renn M, Bald T, Rogava M, Cron M, et al. Melanomas resist T-cell therapy through inflammation-induced reversible dedifferentiation. Nature. 2012;490:412–6. doi: 10.1038/nature11538. [DOI] [PubMed] [Google Scholar]
- 31.Gao J, Shi LZ, Zhao H, Chen J, Xiong L, He Q, et al. Loss of IFN-γ Pathway Genes in Tumor Cells as a Mechanism of Resistance to Anti-CTLA-4 Therapy. Cell. 2016;167:397–9. doi: 10.1016/j.cell.2016.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med. 2016;375:819–29. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eisenhauer EA, Therasse P, Bogaerts J. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) European Journal of …. 2009 doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 34.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–54. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brahmer JR, Tykodi SS, Chow LQM, Hwu W-J, Topalian SL, Hwu P, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–65. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Page DB, Postow MA, Callahan MK, Allison JP, Wolchok JD. Immune modulation in cancer with antibodies. Annu Rev Med. 2014;65:185–202. doi: 10.1146/annurev-med-092012-112807. [DOI] [PubMed] [Google Scholar]
- 37.Kaplan DH, Shankaran V, Dighe AS, Stockert E, Aguet M, Old LJ, et al. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci USA. 1998;95:7556–61. doi: 10.1073/pnas.95.13.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dunn GP, Sheehan KCF, Old LJ, Schreiber RD. IFN unresponsiveness in LNCaP cells due to the lack of JAK1 gene expression. Cancer Res. 2005;65:3447–53. doi: 10.1158/0008-5472.CAN-04-4316. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.