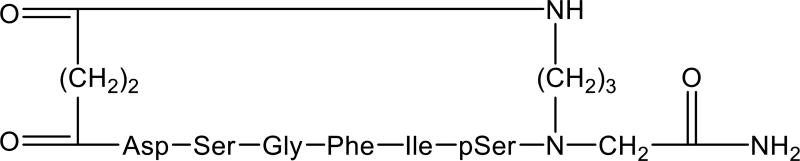Abstract
Protein-protein interactions (PPIs) are particularly important for controlling both physiologic and pathologic biological processes but are difficult to target due to their large and/or shallow interaction surfaces unsuitable for small molecules. Linear peptides found in nature interact with some PPIs, and protein active regions can be used to design synthetic peptide compounds for inhibition of PPIs. However, linear peptides are limited therapeutically by poor metabolic and conformational stability, which can compromise their bioactivity and half-life. Cyclic peptidomimetics (modified peptides) can be used to overcome these challenges because they are more resistant to metabolic degradation and can be engineered to adopt desired conformations. Backbone cyclization is a strategy that we developed to improve drug-like properties of linear peptide leads without jeopardizing the integrity of functionally relevant side-chains. Here, we provide the first description of an entire approach for developing backbone cyclized peptide compounds, based upon two straightforward ‘ABC’ and ‘DEF’ processes. We present practical examples throughout our discussion of revealing active regions important for PPIs and identifying critical pharmacophores, as well as developing backbone cyclized peptide libraries and screening them using cycloscan. Finally, we review the impact of these advances and provide a summary of current ongoing work in the field.
Keywords: Backbone Cyclization, Cyclization, Peptides, Peptidomimetics, Protein-Protein Interactions, Therapeutic
1.1. Introduction
Protein-protein interactions (PPIs) direct most of the processes in living cells. Signal transduction, cell cycle, apoptosis, metabolism, and proliferation are only a few examples of the fundamental functions regulated by PPIs. The importance of PPIs is demonstrated by their frequent dysregulation in disease [1] and is also reflected by the large number of studies dedicated to this subject, some of which are described in the following reviews [2–4]. Functional and structural studies of PPIs facilitate the characterization of these interactions at the molecular and cellular levels, and also provide sources for drug design.
Using computational prediction, rational design, genetic tools, structural data, and/or peptide libraries, peptides and truncated proteins can be derived from a specific protein binding site and can serve as competitive inhibitors of PPIs, which we focus on here. A significant advantage of peptides derived from protein binding surfaces as competitive inhibitors is that a small library developed from the protein interface can be generated without the need for a larger, untargeted library of heterogeneous molecules. In addition, modifying these peptides to optimize bioactivity (e.g. introduction of post-translational modifications), attaching labels (e.g. biotin or fluorescein isothiocyanate (FITC)), and improving stability (e.g. incorporation of non-natural amino acids and/or cyclization) is usually straightforward. Current methods for peptide synthesis and purification can be automated with high yield, purity and scale. As a result of these qualities, peptides derived from protein binding surfaces are ideal candidates for developing competitive inhibitors of PPIs.
The conversion of protein active regions into peptide-based drug-like molecules for PPI inhibition remains a significant challenge for medicinal chemists and the pharmaceutical industry. Although peptides have the desired characteristics needed to study and target PPIs, their unfavorable pharmacokinetic (PK) and pharmacodynamic (PD) properties, such as rapid metabolism, poor bioavailability and nonselective receptor binding, limit their broader use as drugs [5]. Thus, it is challenging to obtain a biologically active peptide with a desired stable structure based on a bioactive protein domain. One way to restrict the conformational space and stabilize the structure of a peptide is to use long peptide sequences, though this approach raises additional obstacles for synthesis, purification and solubility and it can be used only in cases where the structure is known.
Peptidomimetics (henceforth referred to interchangeably with peptides for simplicity) are modified peptides designed to maintain the biological function of the parent linear peptide, while simultaneously addressing the associated undesirable pharmacological properties [4, 6, 7]. Many types of modifications have been introduced to develop peptidomimetic compounds with improved pharmacological properties; these include local modifications, such as the incorporation of non-natural amino acids, as well as global modifications, such as, polypeptide chains that contain a circular sequence, or cyclization [7–11]. Cyclization is one of the most common strategies used to convert peptides into active pharmacological agents and drugs [12–16]. Along with improved pharmacological properties, cyclization also provides conformational stabilization and thus enables the formation of short structured formats that maintain the bioactive conformation [17, 18]. Several common cyclization strategies include metal cyclization [14, 19], metathesis-mediated cyclization [13, 20, 21], amide cyclization [12], and disulfide bridge cyclization [22, 23]. However, restricting the conformational space of peptides by the above-mentioned methods often results in reduced activity since bioactive residues are frequently used for cyclization or are entirely replaced by residues that allow cyclization. Since cyclization restricts the conformational space, cyclic peptides may lose their bioactivity because they are no longer capable of attaining the proper bioactive conformation. Thus, libraries must be screened to select cyclic peptide(s) that retain the desired biological activity [24, 25].
The backbone cyclization methodology that we introduced enables development of cyclic peptides without utilizing the residues that are part of the natural linear peptide. Specifically, backbone cyclization enables the preparation of cyclic peptide libraries without altering the functional groups of the side chain residues that are essential for bioactivity in the parent linear peptide [26]. This feature is extremely important when all the functional groups in a peptide sequence are essential for the biological activity and thus unavailable for cyclization [16]. In addition, backbone cyclization is an ideal strategy to explore the conformational space of cyclic peptides using the conformational library approach called cycloscan.
Here for the first time we describe an entire process for backbone cyclized peptidomimetic development, from identifying a protein active region to generating a backbone cyclic pharmacological agent, all with step-by-step considerations. The resultant molecules can be used as selective tools in basic research and for further study as drug leads.
2.1. Historical perspective
Peptides and proteins are key players in most diseases; hence, they are the basis for the development of many therapeutic compounds. In 1990, the cumulative number of approved peptide drugs was 28, most from natural sources. The pharmaceutical industry was reluctant to develop new peptide-based drugs due to the unfavorable PK and PD properties of peptides, such as rapid metabolism, poor bioavailability and nonselective receptor activation. In 1982, Kessler attributed some of these shortcomings (e.g., metabolic instability and lack of receptor selectivity) to their conformational flexibility [27]. He advocated that cyclization, which would lead to conformational stability, could overcome these shortcomings. Experimental evidence for the validity of Kessler’s theorem was demonstrated for enkephalins, somatostatin, GnRH, CCK, αMSH, and many other peptides in which cyclization resulted in receptor selectivity and metabolic stability, and in very few cases even allowed for conformational analysis of the bioactive conformation [26].
During this period, we performed structure activity relationship (SAR) studies on the tachykinin substance P. We noticed that contrary to other examples described on the success of cyclization, all attempts to prepare cyclic peptides (including end-to-end, end-to-side chain and side chain-to-side chain modes of cyclization) that involved the tachykinin hexapeptide active sequence X-Phe-X-Gly-Leu-Met-NH2 failed (Table 1 in [26]). Apparently, none of the amino acid side-chains and the terminal carboxamide in this active hexapeptide could be modified without destroying bioactivity. To overcome this limitation, we introduced backbone cyclization in which the cyclization is achieved by forming a bridge from the peptide bond nitrogen. We implemented the concept of backbone cyclization for the tachykinin active sequence to obtain a metabolically stable, highly selective and bioactive backbone cyclic analog [28].
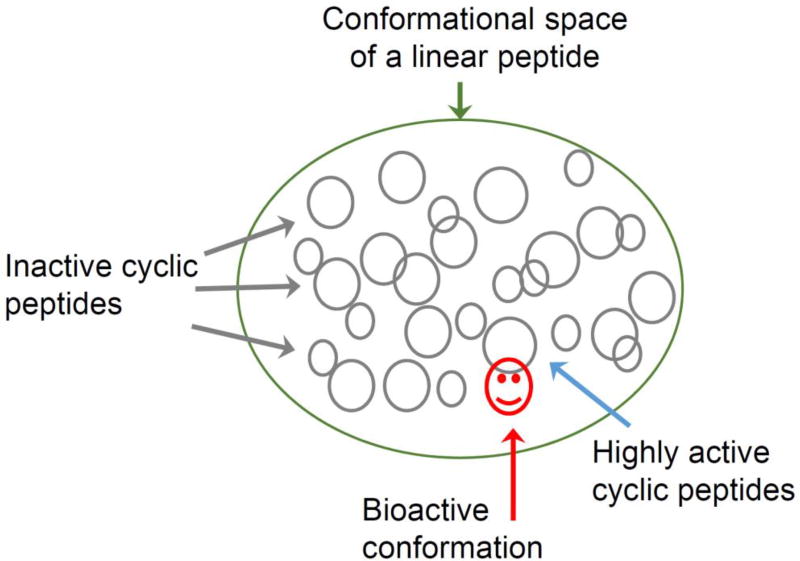
The introduction of combinatorial chemistry in the late 1980s and early 1990s [29] led to evaluation of the backbone cyclization concept in terms of libraries and diversity parameters. Restricting the conformational space of linear peptides by random cyclization may lead to many inactive cyclic peptides because they are unable to adopt the proper bioactive conformation. Since few cyclic peptides based on a given sequence will attain the bioactive conformation and will be active (Fig. 1), libraries must be screened to select cyclic peptide(s) that support the bioactive conformation together with desired “drug-like” properties.
Figure 1.
Restricting the conformational space of a linear peptide (green) by cyclization (gray) leads to many inactive cyclic peptides because they are unable to adopt the proper bioactive conformation (gray). Only very few peptides will attain the bioactive conformation and will be active (red).
Evidently, the cyclization strategies used at that time, namely end-to-end, end-to-side chain and side chain-to-side chain, had limited diversity, especially when Cys or Lys/Asp/Glu were used as side chains for cyclization. Thus, these compounds did not allow for proper screening of the conformational space of the linear parent peptide lead. However, the modes of backbone cyclization (backbone N-to-backbone N, backbone N-to-side chain and backbone N-to-ends) increase the diversity of cyclic peptide libraries. In addition, most backbone cyclization modes do not involve side chains. This feature is extremely important when all or most of the functional groups in the active linear parent peptide sequence are essential for its biological activity.
3.1. The ‘ABC’ and ‘DEF’ processes
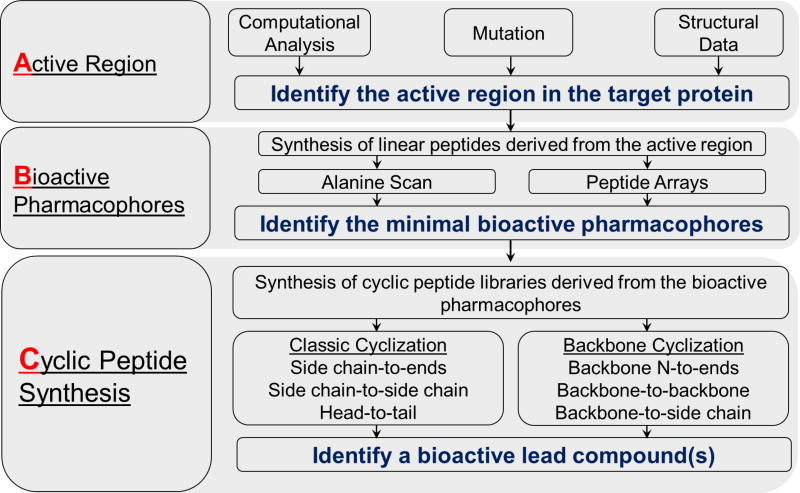
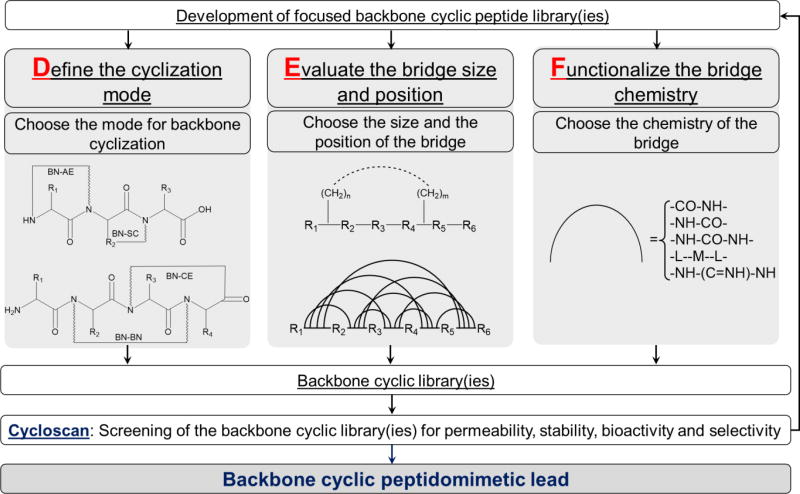
We divided the practice of developing backbone cyclized peptides into ‘ABC’ and ‘DEF’ processes, which guide the development of backbone cyclized peptidomimetic lead compounds. The ‘ABC’ process is a general procedure focused on identifying the protein interacting region, the bioactive pharmacophore in the interacting region of the target protein and synthesizing a peptide cyclic library. The initial step in the ‘ABC’ process is to identify the Active region that regulates or is involved in the target PPI. Next, a systematic approach is used to determine the Bioactive pharmacophore(s) in the active region. Finally, using Cyclic peptide synthesis, a library of cyclic peptides or peptidomimetics all derived from the native active pharmacophore is developed (Fig. 2). The ‘DEF’ process concerns construction of the backbone cyclic library, which is done based on the region and pharmacophore identified in the initial ‘ABC’ system. In the ‘DEF’ process, conformational libraries of variants with the same amino acid sequence are prepared by altering one or more of several parameters that determine the conformational space, in order to Define the cyclization mode, Evaluate the bridge size and position, and Functionalize the bridge chemistry (Fig. 3). These backbone cyclization and cycloscan techniques were applied to various bioactive peptides leading to the discovery of metabolically stable and receptor-selective drug leads (for example, somatostatin [30]). In addition, the above-described technologies have been successfully applied to target active regions in proteins for generating drug leads that inhibit specific PPIs.
Figure 2.
Flow chart describing the ‘ABC’ process for identifying and converting protein active regions into cyclic peptide drug leads.
Figure 3.
Flow chart describing the ‘DEF’ process. The flow chart shows the steps needed to convert a metabolically unstable, non-selective peptide or protein active region with poor absorption properties into a metabolically stable, selective, highly active, and orally available peptidomimetic drug lead with improved PK and PD properties [67]. (BN-AE - backbone-to-amino terminus, BN-SC - backbone-to-side chain, BN-CE - backbone-to-carboxy terminus, BN-BN - backbone-to-backbone).
3.2. Identifying an active region important for protein-protein interactions
Generally, a small number of residues comprising a protein’s active region are responsible for the vast majority of its interactions with binding partners [31, 32]. It is critical to define the active region responsible for a PPI in order to conduct an informed, targeted scan and characterization of the focused sequence (‘ABC’ step A, Fig. 2). This is especially worthwhile because PPI binding sites are often larger and far more complex than small ligand binding pockets. Depending on the target and level of prior knowledge, computational, genetic and/or structural approaches can be utilized to reveal the active region.
Computational and bioinformatics analyses can be used to predict active region interaction sites based on minimal sequence information. A variety of models that incorporate Monte Carlo simulations, support vector machines, or other approaches have been applied to identify residue-residue contacts in the surface of interacting proteins. For instance, Ofran and Rost employed a neural network to identify protein-protein interfaces from sequence information alone [33]. The authors trained the model on windows of nine residues for non-redundant proteins with solved structures from the Protein Data Bank (PDB) using feed-forward neural networks containing back-propagation and momentum terms. The algorithm identified 34 strongly predicted sites from 333 complexes found in the PDB, 94% of which were experimentally validated, indicating that prediction of some interaction sites based entirely on sequence is possible without consulting the solved PDB structures before prediction of PPIs. However, consideration of sequence homology and evolutionary conservation can enhance the accuracy and reliability of active region prediction. Amos-Binks and colleagues developed the protein-protein interaction prediction engine (PIPE)-Sites algorithm to predict more specific binding sites for PPI partners using re-occurring polypeptide sequences based only on the sequences for the proteins of interest and a database of known polypeptide sequence interactions [34]. This model performed superiorly to domain-domain interaction-based binding site prediction methods in a study of 265 yeast and 423 human interacting protein pairs with experimentally validated binding sites. Moreover, incorporation of information from predicted or solved protein structures can improve the accuracy of predicted binding site active regions further. Computational methods for protein docking to map interaction surfaces [35] and electrostatic desolvation profiling (where the average probe desolvation penalty is smaller for residues in binding sites) [36] are frequently used as the basis for rational design of PPI inhibitors as well as novel PPI surfaces.
Using tools for genetic manipulation and recombinant expression of protein variants or polypeptide chains, one can produce, display, screen, and reveal the active region of interacting protein partners. Genetic deletion can also be used to definitively demonstrate the necessity of a finite sequence to support a PPI. Yablonski and colleagues introduced deletions into the sequence of SLP-76, an adaptor protein required for T cell receptor (TCR) signaling via a PPI with phospholipase C-γ1 (PLC-γ1), to identify the specific binding site required for its constitutive interaction with PLC-γ1 [37]. The authors recombinantly produced sequential SLP-76 mutant constructs with either an N-terminal domain deletion, mutation of three functional tyrosine phosphorylation sites to phenylalanine, a deletion of the Gads-binding domain (previously known to be responsible for downstream signaling), an arginine to lysine mutation in the SH2 functional domain, an SH2 domain deletion, sequential deletions of P-1 through P-4 domains, or a C-terminal domain deletion. They demonstrated that each of the previously identified SLP-76 domains which contribute to the PLC-γ1 PPI (the N-terminal acidic domain with three tyrosine phosphorylation sites, the Gads-binding domain within the proline-rich region and the C-terminal SH2 domain) could be deleted individually without eliminating PLC-γ1-dependent SLP-76 function; however, the 67-amino acid P-1 domain within the proline-rich region of SLP-76 was essential for maintaining its activity downstream of the TCR by mediating a constitutive PPI with the PLC-γ1 SH3 domain. Moreover, additional genetic approaches for identifying active regions of protein interaction partners include mutation (e.g. to alanine), which we discuss further in the following section, and insertion, which has yet to be widely used.
When available, structural data can aid in efficiently defining active regions, especially when confronted with binding sites consisting of non-sequential residues. Combining computational methods with the growing number of 3D protein structures, Hugo et al. described an algorithm called SLiMDIet to detect short linear motifs (SLiMs) that frequently characterize transient PPIs by domain interface extraction and clustering based solely on 3D structural data [38]. This method involves extracting and clustering based on structural interaction interfaces belonging to the same domain from the protein of interest, followed by structural alignment of the extracted interaction interfaces to reveal the corresponding SLiM. Thus, SLiM active regions participating in PPIs of interest can be computationally identified for any protein with protein family database (Pfam) domain annotations and an available structure in the PDB (Fig. 2).
3.3. Identifying pharmacophores critical for bioactivity
Next, it is important to determine the pharmacophores within active regions that form the bioactive peptide and significantly contribute to the PPI (‘ABC’ step B, Fig. 2). Thermodynamic studies have demonstrated that despite the significant size difference between proteins and peptides, the deviation in average thermodynamics of protein-protein versus protein-peptide interactions is very small [39]. A probable explanation for these results is that in many PPIs only a small subset of amino acids within the overall interface contribute substantially to the binding affinity [31, 40–42].
Alanine scanning is a common approach to identify the critical amino acids in the PPI region. In the alanine scan technique, residues in the target protein are systematically substituted for alanine at selected positions and the resultant variants are assayed for function. This substitution eliminates side-chain interactions without significantly altering main-chain conformation or introducing steric or electrostatic effects. For example, Jin and coworkers studied the interaction between human growth hormone (hGH) and the monoclonal antibody 3 (MAb 3). They identified five key pharmacophores for this interaction: Arg 8, Asn 12, Arg 16, Asp 112, and Asp 116. Using an alanine scan in which they mutated up to 16 amino acids to alanine, the authors demonstrated that these modifications caused less than a 10-fold reduction in binding affinity, compared to mutating any of the five primary pharmacophores to alanine, which resulted in up to a 500-fold reduction in affinity. Interestingly, these pharmacophores were so critical that even replacing them with a homologous residue (i.e., Arg to Lys or Asp to Glu) caused nearly as large a reduction in the binding affinity as the alanine replacement [43]. Another study on the interaction between hGH and the extracellular domain of its bound receptor (hGHbp) using an alanine scan identified that although about 30 amino acid side chains from each protein played a role in the binding interaction, two tryptophan residues accounted for over 75% of the binding free energy [31]. This data was confirmed by solving the crystal structure of hGH and hGHbp, which indicated that the hydrophobic patch dominated by two tryptophans is a central pharmacophore assembled cooperatively and that several other residues contribute only marginally and indirectly to the binding [32].
Peptide arrays are an additional common technology used to identify pharmacophore(s) important for PPIs. In this approach, one synthesizes peptides derived from short sequential sequences of domains involved in the PPI [44]. For example, Colombo and coworkers identified the binding sequence of fibroblast growth factor-2 (FGF-2) to thrombospondin-1 (TSP-1) using peptide arrays, which they confirmed with binding assays using synthetic peptides and recombinant proteins. The authors designed peptide arrays based on the sequence of type III repeats in TSP-1. They synthesized 237 20-mer peptides with partially overlapping sequences and covalently linked these molecules to polypropylene cards so that binding of biotinylated FGF-2 to the peptides could be measured. Colombo et al. identified an FGF-2 binding sequence for TSP-1 in the 15-mer sequence, D739DDDDNDKIPDDRDN [45]. Taraboletti and coworkers used molecular dynamics analysis of the peptide and FGF-2 complex to confirm that the peptide adopted an extended conformation over the FGF-2 binding surface, occupying a heparin-binding site, with favorable electrostatic couplings between a Lys side chain on the FGF-2 surface and the negatively charged Asp groups of the peptide [46].
Above, we described the most common approaches to identify the minimal bioactive pharmacophores in a defined PPI region. This step is crucial for developing bioactive peptidomimetics, which are designed to overcome the pharmacological limitations associated with natural linear peptides, such as lack of proteolytic and conformational stability and low bioavailability. In order to address these challenges, it is critical to identify the minimum and most important residues for the PPI from which to develop the pharmacologically enhanced peptidomimetic lead. Further, defining which amino acids are unnecessary for PPI binding can provide opportunities for modifying positions or substituting residues with functional groups amenable to cyclization. By replacing only one or two unnecessary amino acids to perform cyclization, libraries of peptidomimetics in which all the primary residues required for binding are conserved can be used to screen the conformational space and identify the bioactive conformation, as we demonstrate in the following sections concerning backbone cyclization and cycloscan (‘ABC’ step C, Fig. 2 and Fig. 3).
4.1. Backbone cyclization
Backbone cyclization (‘ABC’ step C in Fig. 2, and detailed by the ‘DEF’ process in Fig. 3) is achieved through the covalent connection of two backbone amides, one backbone amide and a side chain functional group, or one backbone amide and the N- or C-terminus using an artificial spacer [26] (Fig. 3). The utilization of backbone amides for cyclization enables the conversion of linear peptides into cyclic peptides without the use or occupancy of essential bioactive pharmacophores. Furthermore, the use of artificial spacers and building blocks dramatically increases the diversity of possible ring structures and can thus be used as an efficient tool for conformational screening of a parent linear peptide.
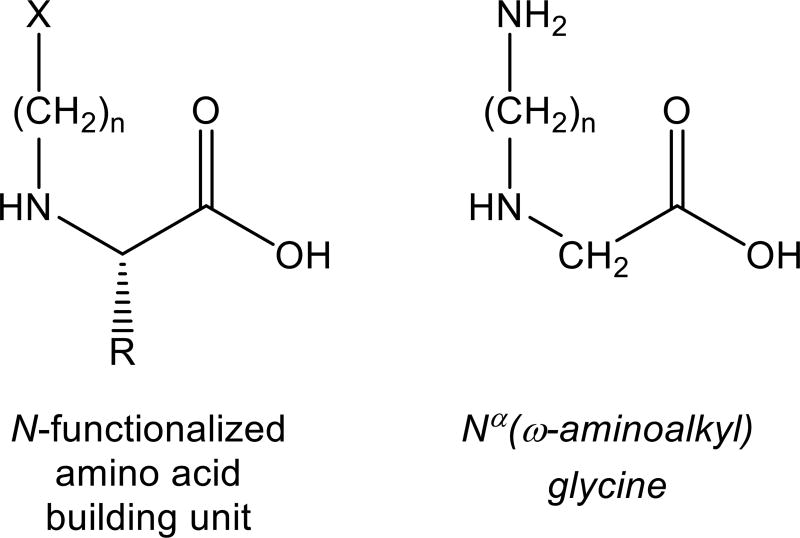
The synthesis of backbone cyclized peptides requires the use of non-natural N-functionalized amino acid building units. Although N-functionalized building units derived from all natural amino acids have been reported [47], by far the most commonly used units are derived from glycine and assume the Nα(ω-aminoalkyl) glycine format [48–53] (Fig. 4) due to their relative ease of preparation. These building units have been successfully used to prepare a variety of different backbone cyclic scaffolds including amide backbone cyclic peptides, where the building unit was covalently connected to the N-terminus via a dicarboxylic acid linker [28, 49, 54–56], as well as urea backbone cyclic peptides, where two building units were connected via a urea bridge [48].
Figure 4.
Structures of a general N-functionalized amino acid building unit and an Nα(ω-aminoalkyl) glycine building unit.
One of the main advantages of backbone cyclization is its suitability for conducting conformational screening using cycloscan. The use of artificial spacers and non-natural building blocks also allows for diversification. By changing one or more of the following four parameters, one can systematically scan the 3D peptide conformational space: (1) cyclization mode (i.e., backbone-to-backbone (BN-BN), backbone-to-amino terminus (BN-AE), backbone-to-carboxy terminus (BN-CE), or backbone-to-side chain (BN-SC)); (2) bridge size, which is comprised of the amino acid sequence within the macrocycle as well as the artificial bridge used for cyclization (i.e., the type of spacers, such as alkyl, poly(ethylene glycol) (PEG), etc.). In these bridge size libraries, the length of the linker attached to the backbone atom is varied (n and m); (3) bridge position, which determines the backbone atoms that will be part of the bridge and can be chosen arbitrarily based on SAR information and intuition. For each cyclization mode (e.g., BN-BN, BN-AE, BN-CE, or BN-SC), focused bridge size and/or bridge position libraries can be generated and screened; (4) bridge chemistry, defined by the type of chemical functions that exist on the bridge, namely the chemistry of the bond used for cyclization (e.g., amide, disulfide, urea, etc.). For each member of the bridge size and bridge position libraries, a bridge chemistry scan can be performed (Fig. 3).
4.2. Cycloscan: development of backbone cyclic peptides and their screening
As mentioned above in the ‘DEF’ process, the main considerations for developing backbone cyclic peptide libraries with conformational diversity are: defining the cyclization mode, evaluating bridge size and position, and functionalizing the bridge chemistry (Fig. 3). In this section we discuss examples of studies in which backbone cyclization and cycloscan methods have been used successfully, focusing in each example on a different parameter.
In order to construct a conformational library of backbone cyclized peptides, one first needs to develop the appropriate mode of cyclization (‘DEF’ step D, Fig. 3), which depends entirely on synthetic capabilities. The most convenient mode of cyclization, and the one routinely used by us, is the backbone-to-backbone cyclization mode (BN-BN, Fig. 3) since this does not involve side chains or termini that are essential for bioactivity. Moreover, this mode of cyclization allows for an exponential number of sub-libraries that enable scanning for the appropriate bridge size and/or position (Fig. 3).
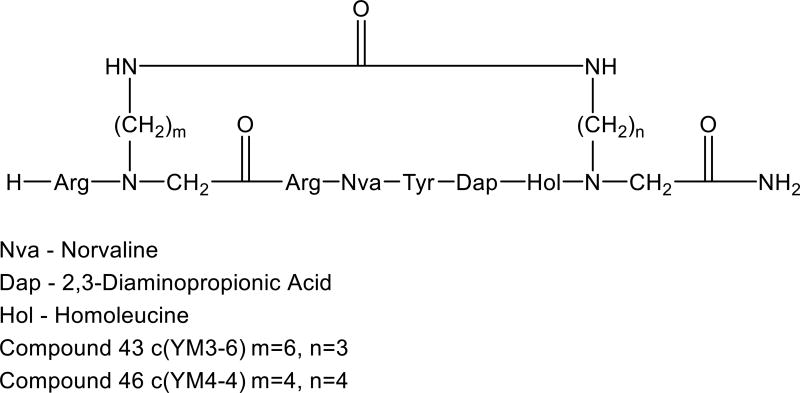
For example, Tal-Gan et al. designed a focused library of backbone-to-backbone (BN-BN) urea cyclic peptides aimed at targeting protein kinase B (PKB/Akt) that have been detected in many types of cancer. The authors developed backbone cyclic library based on a series of peptides derived from the protein glycogen synthase kinase 3 (GSK3, a PKB/Akt substrate), which demonstrated bioactivity and specificity as PKB/Akt inhibitors [57]. The authors positioned one building unit at the C-terminus and the second unit replaced the Pro residue located immediately prior to the N-terminal Arg. Screening members of this library in vitro led to characterization of two analogs, compound 43 and compound 46, that were ten-fold more active than the parent linear peptide (IC50 = 0.16 and 0.17 µM, respectively) (Fig. 5) [49].
Figure 5.
General structure of the backbone cyclic inhibitor library for PKB [49].
Another important factor for constructing a conformational library of backbone cyclized peptides is evaluating the bridge size (‘DEF’ step E, Fig. 3). Bridge size is a highly common parameter to vary and screen for in the conformational space of backbone cyclic peptides, using libraries assembled from building units with spacers of different lengths. Moreover, bridge size and position allow one to generate an exponential number of sub-libraries that enable scanning for the most bioactive and stable backbone cyclic peptide (Fig. 3). For example, Qvit et al. designed a focused library of backbone cyclic peptides to mimic the inhibitor kappa B protein, which binds nuclear factor kappa B (NF-κB). In this case, conformational screening was conducted by altering the bridge size through the use of building units of different lengths at the N- and C-terminal positions. One novel backbone-cyclized peptide, IκB31–37 (pBC-2,3), prevented the release of NF-κB in vitro, achieving approximately 90% inhibition of IκB ubiquitylation at a concentration of 3 µM. The peptide also decreased the translocation of NF-κB to the nucleus, which could be used to reduce its pathogenic effects in various diseases (Fig. 6) [55, 58].
Figure 6.
Structure of the backbone cyclic analog of NF-κB, IκB31–37(pBC-2,3) [55].
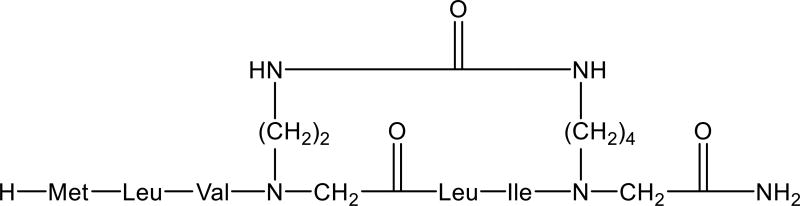
Bridge position scanning is more common when no information is available regarding the optimal position for cyclization (‘DEF’ step E, Fig. 3). In the following example, a “helix walk” bridge position scan involving repositioning of the helical region in the parent linear active sequence was a favored approach, enabling the discovery of a backbone cyclic peptide with drug like properties. Hurevich et al. developed a backbone cyclic peptide capable of inhibiting the dimerization of a specific G protein-coupled receptor (GPCR) involved in the pathogenesis of multiple sclerosis and other immune-mediated diseases [59]. The transmembrane helical bundle of GPCRs dimerize through helix-helix interactions in response to stimulation. Thus, the authors chose to use a backbone-to-backbone (BN-BN) urea cyclization where the two building units were located in i, i+3 positions relative to each other, with the first building unit containing a two-carbon linker and the second unit containing a four-carbon chain. Since no information was available regarding the optimal position for cyclization, a systematic “helix walk” scan was conducted where the two building blocks were systematically shifted across the peptide sequence while maintaining the i, i+3 distance. Five backbone cyclic peptides were constructed, and one of the analogs exhibited potent and selective inhibition of chemokine receptor 2 (CCR2)-mediated cell migration (Fig. 7). Furthermore, the peptide was also found to adopt an α-helix conformation, reaffirming that an α-helix is required for effective inhibition of dimerization [59].
Figure 7.
Structure of a backbone cyclic peptide analog based on the CCR2 dimerization region sequence [59].
Functional bridge chemistry is the last parameter to take into account when developing a backbone cyclic peptide library (‘DEF’ step F, Fig. 3). Several bridge chemistry approaches have been introduced for backbone cyclization, including backbone disulfide bridges [60, 61], backbone amide bridges [62], backbone metal bridges [63, 64], backbone urea bridges [48], and backbone azo bridges [65] (Fig. 3). In general, the selection of bridge chemistry should be based on biological and synthetic capabilities. Bridge chemistry can be chosen based upon available chemistries in the lab and/or intended application (i.e. specific metals for imaging purposes). As far as we know, to date there have been no comprehensive studies comparing different bridge chemistries for the same backbone cyclic peptides.
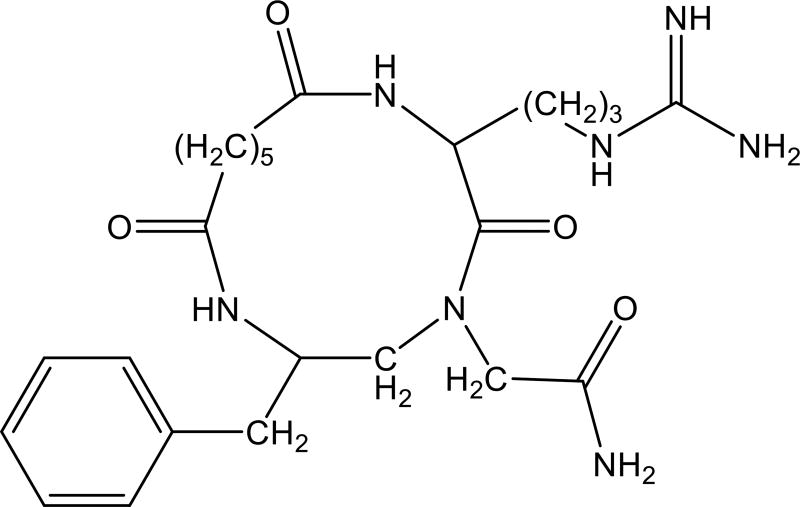
Most cyclization modes used currently to generate cyclic peptides from protein active regions (end-to-end, side-chain to-end and side-chain to side-chain) are limited to sequential active regions and cannot be used for non-continuous active regions. In contrast, backbone cyclization and cycloscan can be used to mimic both continuous and non-continuous protein active regions. Rational conversion of non-continuous topologies into a small orally bioavailable molecule is crucial for the discovery of many new drugs that inhibit PPIs. Hurevich et al. developed a method that utilizes backbone cyclization and cycloscan as intermediate steps for conversion of the CD4 non-continuous active region into small macrocyclic molecules. The authors demonstrated that this method is feasible by preparing a small inhibitor of human immunodeficiency virus (HIV) infection. The lead compound, CG-1, proved orally available in a rat model [66] (Fig. 8).
Figure 8.
Structure of the backbone cyclic peptide analog of the CD4 non-contiguous active region, CG-1, [66].
Although not in the scope of this review, optimization of the lead compound and scale up the synthesis should be considered after identifying a backbone cyclic lead compound. Ultimately, in vivo studies must also include PK, PD, toxicology, and efficacy.
5.1. Summary
The backbone cyclization methodology enables development of cyclic peptides without utilizing residues of the natural linear peptide, which may frequently be essential for bioactivity. Moreover, the technique facilitates synthesis of numerous peptides with the same primary sequence, allowing systematic screening using cycloscan of bioactive conformations for a given set of pharmacophores in search of drug leads. Backbone cyclization and cycloscan were applied to various bioactive peptides leading to the discovery of metabolically stable and receptor-selective drug leads. In this review for the first time we systemically define in detail the entire process of developing backbone cyclized peptidomimetic libraries. Starting from general steps for identifying a protein active region and the bioactive pharmacophore in this sequence, we described the use of the unique backbone cyclization and cycloscan methods for generating a backbone cyclic pharmacological agent, with detailed considerations and examples for each step.
Backbone cyclization and cycloscan techniques are optimal for design and development of selective compounds after identification of the protein-protein interface primary sequence, even without knowledge of the structure. While maintaining a constant primary sequence, one can screen the conformational space using various building blocks to synthesize numerous backbone cyclic peptidomimetics. To diversify bioactive conformations, one can manipulate the mode of cyclization, the size and the position of the bridge, as well as the bridge chemistry. The Achilles' heel of these techniques was previously the laborious preparation of building blocks that were usually constructed in solution [47]. Today, the variety of commercially available unnatural amino acids, in addition to our latest development of a synthetic procedure for generation of backbone cyclization building units on resin during solid phase peptide synthesis (SPPS) (unpublished results), allows one to buy or generate building units from almost every amino acid while avoiding many synthetic and purification challenges. We are in the process of applying these advances to therapeutically target additional PPIs, and we hope this will increase the popularity of the backbone cyclization and cycloscan approaches.
Acknowledgments
Y. T. thanks the Nevada INBRE (NIH GM103440) for the generous support of research in his laboratory.
Footnotes
CONFLICT OF INTEREST
The authors confirm that they have no conflicts of interest with the content of this article.
References
- 1.Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nature Reviews: Drug Discovery. 2004;3(4):301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 2.Jones S, Thornton JM. Principles of protein-protein interactions. Proceedings of the National Academy of Sciences. 1996;93(1):13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benyamini H, Friedler A. Using peptides to study protein–protein interactions. Future. 2010;2(6):989–1003. doi: 10.4155/fmc.10.196. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham AD, Qvit N, Mochly-Rosen D. Peptides and peptidomimetics as regulators of protein-protein interactions. Current Opinion in Structural Biology. 2017;44:59–66. doi: 10.1016/j.sbi.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fosgerau K, Hoffmann T. Peptide therapeutics: Current status and future directions. Drug Discovery Today. 2015;20(1):122–128. doi: 10.1016/j.drudis.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Adessi C, Soto C. Converting a peptide into a drug: strategies to improve stability and bioavailability. Current Medicinal Chemistry. 2002;9(9):963–978. doi: 10.2174/0929867024606731. [DOI] [PubMed] [Google Scholar]
- 7.Vagner J, Qu H, Hruby VJ. Peptidomimetics, a synthetic tool of drug discovery. Current Opinion in Chemical Biology. 2008;12(3):292–296. doi: 10.1016/j.cbpa.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toniolo C, Goodman M. Introduction to the Synthesis of Peptidomimetics. In Synthesis of Peptides and Peptidomimetics. Thieme; Stuttgart New York: 2002. [Google Scholar]
- 9.Naider F, Goodman M. Introduction to the Synthesis of Peptidomimetics. In Synthesis of Peptides and Peptidomimetics. Thieme; Stuttgart New York: 2002. pp. 1–16. [Google Scholar]
- 10.Gante J. Peptidomimetic-tailored enzyme inhibitors. Angewandte Chemie-International Edition in English. 1994;33(17):1699–1720. [Google Scholar]
- 11.Ahn JM, Boyle NA, MacDonald MT, Janda KD. Peptidomimetics and peptide backbone modifications. Mini Reviews in Medicinal Chemistry. 2002;2(5):463–473. doi: 10.2174/1389557023405828. [DOI] [PubMed] [Google Scholar]
- 12.Veber DF, Freidlinger RM, Perlow DS, Paleveda WJ, Jr, Holly FW, Strachan RG, Nutt RF, Arison BH, Homnick C, Randall WC, Glitzer MS, Saperstein R, Hirschmann R. A potent cyclic hexapeptide analogue of somatostatin. Nature. 1981;292(5818):55–58. doi: 10.1038/292055a0. [DOI] [PubMed] [Google Scholar]
- 13.Miller SJ, Blackwell HE, Grubbs RH. Application of ring-closing metathesis to the synthesis of rigidified amino acids and peptides. Journal of the American Chemical Society. 1996;118(40):9606–9614. [Google Scholar]
- 14.Giblin MF, Wang N, Hoffman TJ, Jurisson SS, Quinn TP. Design and characterization of alpha-melanotropin peptide analogs cyclized through rhenium and technetium metal coordination. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(22):12814–12818. doi: 10.1073/pnas.95.22.12814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dekker FJ, de Mol NJ, Fischer MJ, Kemmink J, Liskamp RM. Cyclic phosphopeptides for interference with Grb2 SH2 domain signal transduction prepared by ring-closing metathesis and phosphorylation. Organic & biomolecular chemistry. 2003;1(19):3297–3303. doi: 10.1039/b306681a. [DOI] [PubMed] [Google Scholar]
- 16.Demmer O, Frank AO, Kessler H. Peptide and Protein Design for Biopharmaceutical Applications. John Wiley & Sons, Ltd; 2009. pp. 133–176. [Google Scholar]
- 17.Garner J, Harding MM. Design and synthesis of alpha-helical peptides and mimetics. Organic & biomolecular chemistry. 2007;5(22):3577–3585. doi: 10.1039/b710425a. [DOI] [PubMed] [Google Scholar]
- 18.Haridas V. From peptides to non-peptide Alpha-helix inducers and mimetics. European Journal of Organic Chemistry. 2009;2009(30):5112–5128. [Google Scholar]
- 19.Ruan F, Chen Y, Itoh K, Sasaki T, Hopkins PB. Synthesis of peptides containing unnatural, metal-ligating residues: aminodiacetic acid as a peptide side chain. The Journal of Organic Chemistry. 1991;56(14):4347–4354. [Google Scholar]
- 20.Miller SJ, Grubbs RH. Synthesis of conformationally restricted amino acids and peptides employing olefin metathesis. Journal of the American Chemical Society. 1995;117(21):5855–5856. [Google Scholar]
- 21.Stewart ML, Fire E, Keating AE, Walensky LD. The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nature Chemical Biology. 2010;6(8):595–601. doi: 10.1038/nchembio.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gongora-Benitez M, Tulla-Puche J, Albericio F. Multifaceted roles of disulfide bonds. Peptides as therapeutics. Chemical Reviews. 2014;114(2):901–926. doi: 10.1021/cr400031z. [DOI] [PubMed] [Google Scholar]
- 23.Weckbecker G, Lewis I, Albert R, Schmid HA, Hoyer D, Bruns C. Opportunities in somatostatin research: biological, chemical and therapeutic aspects. Nature Reviews: Drug Discovery. 2003;2(12):999–1017. doi: 10.1038/nrd1255. [DOI] [PubMed] [Google Scholar]
- 24.Heavner GA, Audhya T, Doyle D, Tjoeng FS, Goldstein G. Biologically active conformations of thymopentin. Studies with conformationally restricted analogs. International Journal of Peptide and Protein Research. 1991;37(3):198–209. doi: 10.1111/j.1399-3011.1991.tb00271.x. [DOI] [PubMed] [Google Scholar]
- 25.Grdadolnik SG, Mierke DF, Byk G, Zeltser I, Gilon C, Kessler H. Comparison of the conformation of active and nonactive backbone cyclic analogs of substance P as a tool to elucidate features of the bioactive conformation: NMR and molecular dynamics in DMSO and water. Journal of Medicinal Chemistry. 1994;37(14):2145–2152. doi: 10.1021/jm00040a005. [DOI] [PubMed] [Google Scholar]
- 26.Gilon C, Halle D, Chorev M, Selinger Z, Byk G. Backbone cyclization - a new method for conferring conformational constraint on peptides. Biopolymers. 1991;31(6):745–750. doi: 10.1002/bip.360310619. [DOI] [PubMed] [Google Scholar]
- 27.Kessler H. Peptide conformations .19. Conformation and biological-activity of cyclic-peptides. Angewandte Chemie, International Edition in English. 1982;21(7):512–523. [Google Scholar]
- 28.Byk G, Halle D, Zeltser I, Bitan G, Selinger Z, Gilon C. Synthesis and biological activity of NK-1 selective, N-backbone cyclic analogs of the C-terminal hexapeptide of substance P. Journal of Medicinal Chemistry. 1996;39(16):3174–3178. doi: 10.1021/jm960154i. [DOI] [PubMed] [Google Scholar]
- 29.Furka A, Sebestyen F, Asgedom M, Dibo G. General method for rapid synthesis of multicomponent peptide mixtures. International Journal of Peptide and Protein Research. 1991;37(6):487–493. doi: 10.1111/j.1399-3011.1991.tb00765.x. [DOI] [PubMed] [Google Scholar]
- 30.Hornik V, Gellerman G, Afargan MEM. Google Patents. 2002 [Google Scholar]
- 31.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267(5196):383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 32.Clackson T, Ultsch MH, Wells JA, de Vos AM. Structural and functional analysis of the 1:1 growth hormone:receptor complex reveals the molecular basis for receptor affinity. Journal of Molecular Biology. 1998;277(5):1111–1128. doi: 10.1006/jmbi.1998.1669. [DOI] [PubMed] [Google Scholar]
- 33.Ofran Y, Rost B. Predicted protein-protein interaction sites from local sequence information. FEBS Letters. 2003;544(1–3):236–239. doi: 10.1016/s0014-5793(03)00456-3. [DOI] [PubMed] [Google Scholar]
- 34.Amos-Binks A, Patulea C, Pitre S, Schoenrock A, Gui Y, Green JR, Golshani A, Dehne F. Binding site prediction for protein-protein interactions and novel motif discovery using re-occurring polypeptide sequences. BMC Bioinformatics. 2011;12:225. doi: 10.1186/1471-2105-12-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith GR, Sternberg MJ. Prediction of protein-protein interactions by docking methods. Current Opinion in Structural Biology. 2002;12(1):28–35. doi: 10.1016/s0959-440x(02)00285-3. [DOI] [PubMed] [Google Scholar]
- 36.Fiorucci S, Zacharias M. Prediction of protein-protein interaction sites using electrostatic desolvation profiles. Biophysical Journal. 2010;98(9):1921–1930. doi: 10.1016/j.bpj.2009.12.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yablonski D, Kadlecek T, Weiss A. Identification of a phospholipase C-gamma1 (PLC-gamma1) SH3 domain-binding site in SLP-76 required for T-cell receptor-mediated activation of PLC-gamma1 and NFAT. Molecular and Cellular Biology. 2001;21(13):4208–4218. doi: 10.1128/MCB.21.13.4208-4218.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hugo W, Sung WK, Ng SK. Discovering interacting domains and motifs in protein-protein interactions. Methods in Molecular Biology. 2013;939:9–20. doi: 10.1007/978-1-62703-107-3_2. [DOI] [PubMed] [Google Scholar]
- 39.Stites WE. Protein–protein interactions: Interface structure, binding thermodynamics, and mutational analysis. Chemical Reviews. 1997;97(5):1233–1250. doi: 10.1021/cr960387h. [DOI] [PubMed] [Google Scholar]
- 40.Schreiber G, Fersht AR. Energetics of protein-protein interactions: analysis of the barnase-barstar interface by single mutations and double mutant cycles. Journal of Molecular Biology. 1995;248(2):478–486. doi: 10.1016/s0022-2836(95)80064-6. [DOI] [PubMed] [Google Scholar]
- 41.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. Journal of Molecular Biology. 1998;280(1):1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 42.Gonzalez-Ruiz D, Gohlke H. Targeting protein-protein interactions with small molecules: Challenges and perspectives for computational binding epitope detection and ligand finding. Current Medicinal Chemistry. 2006;13(22):2607–2625. doi: 10.2174/092986706778201530. [DOI] [PubMed] [Google Scholar]
- 43.Jin L, Wells JA. Dissecting the energetics of an antibody-antigen interface by alanine shaving and molecular grafting. Protein Science. 1994;3(12):2351–2357. doi: 10.1002/pro.5560031219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katz C, Levy-Beladev L, Rotem-Bamberger S, Rito T, Rudiger SG, Friedler A. Studying protein-protein interactions using peptide arrays. Chemical Society Reviews. 2011;40(5):2131–2145. doi: 10.1039/c0cs00029a. [DOI] [PubMed] [Google Scholar]
- 45.Colombo G, Margosio B, Ragona L, Neves M, Bonifacio S, Annis DS, Stravalaci M, Tomaselli S, Giavazzi R, Rusnati M, Presta M, Zetta L, Mosher DF, Ribatti D, Gobbi M, Taraboletti G. Non-peptidic thrombospondin-1 mimics as fibroblast growth factor-2 inhibitors: an integrated strategy for the development of new antiangiogenic compounds. Journal of Biological Chemistry. 2010;285(12):8733–8742. doi: 10.1074/jbc.M109.085605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taraboletti G, Rusnati M, Ragona L, Colombo G. Targeting tumor angiogenesis with TSP-1-based compounds: rational design of antiangiogenic mimetics of endogenous inhibitors. Oncotarget. 2010;1(7):662–673. doi: 10.18632/oncotarget.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gellerman G, Elgavi A, Salitra Y, Kramer M. Facile synthesis of orthogonally protected amino acid building blocks for combinatorial N-backbone cyclic peptide chemistry. Journal of Peptide Research. 2001;57(4):277–291. doi: 10.1046/j.1397-002x.2000.0780.x. [DOI] [PubMed] [Google Scholar]
- 48.Hurevich M, Tal-Gan Y, Klein S, Barda Y, Levitzki A, Gilon C. Novel method for the synthesis of urea backbone cyclic peptides using new Alloc-protected glycine building units. Journal of Peptide Science. 2010;16(4):178–185. doi: 10.1002/psc.1218. [DOI] [PubMed] [Google Scholar]
- 49.Tal-Gan Y, Gilon C, Hurevich M, Klein S, Ben-Shimon A, Rosenthal D, Hazan C, Shalev DE, Niv MY, Levitzki A. Backbone cyclic peptide inhibitors of protein kinase B (PKB/Akt) Journal of Medicinal Chemistry. 2011;54(14):5154–5164. doi: 10.1021/jm2003969. [DOI] [PubMed] [Google Scholar]
- 50.Qvit N, Monderer-Rothkoff G, Ido A, Shalev DE, Amster-Choder O, Gilon C. Development of bifunctional photoactivatable benzophenone probes and their application to glycoside substrates. Biopolymers. 2008;90(4):526–536. doi: 10.1002/bip.21010. [DOI] [PubMed] [Google Scholar]
- 51.Naveh S, Tal-Gan Y, Ling S, Hoffman A, Holoshitz J, Gilon C. Developing potent backbone cyclic peptides bearing the shared epitope sequence as rheumatoid arthritis drug-leads. Bioorganic and Medicinal Chemistry Letters. 2012;22(1):493–496. doi: 10.1016/j.bmcl.2011.10.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Linde Y, Ovadia O, Safrai E, Xiang Z, Portillo FP, Shalev DE, Haskell-Luevano C, Hoffman A, Gilon C. Structure-activity relationship and metabolic stability studies of backbone cyclization and N-methylation of melanocortin peptides. Biopolymers. 2008;90(5):671–682. doi: 10.1002/bip.21057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hess S, Linde Y, Ovadia O, Safrai E, Shalev DE, Swed A, Halbfinger E, Lapidot T, Winkler I, Gabinet Y, Faier A, Yarden D, Xiang Z, Portillo FP, Haskell-Luevano C, Gilon C, Hoffman A. Backbone cyclic peptidomimetic melanocortin-4 receptor agonist as a novel orally administrated drug lead for treating obesity. Journal of Medicinal Chemistry. 2008;51(4):1026–1034. doi: 10.1021/jm701093y. [DOI] [PubMed] [Google Scholar]
- 54.Friedler A, Friedler D, Luedtke NW, Tor Y, Loyter A, Gilon C. Development of a functional backbone cyclic mimetic of the HIV-1 Tat arginine-rich motif. Journal of Biological Chemistry. 2000;275(31):23783–23789. doi: 10.1074/jbc.M002200200. [DOI] [PubMed] [Google Scholar]
- 55.Qvit N, Hatzubai A, Shalev DE, Friedler A, Ben-Neriah Y, Gilon C. Design and synthesis of backbone cyclic phosphorylated peptides: The IκB model. Biopolymers. 2009;91(2):157–168. doi: 10.1002/bip.21098. [DOI] [PubMed] [Google Scholar]
- 56.Qvit N, Schechtman D, Peña DA, Berti DA, Soares CO, Miao Q, Liang L, Baron LA, Teh-Poot C, Martínez-Vega P, Ramirez-Sierra MJ, Churchill E, Cunningham AD, Malkovskiy AV, Federspiel NA, Gozzo FC, Torrecilhas AC, Manso Alves MJ, Jardim A, Momar N, Dumonteil E, Mochly-Rosen D. Scaffold proteins LACK and TRACK as potential drug targets in kinetoplastid parasites: Development of inhibitors. International Journal for Parasitology: Drugs and Drug Resistance. 2016;6:74–84. doi: 10.1016/j.ijpddr.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Litman P, Ohne O, Ben-Yaakov S, Shemesh-Darvish L, Yechezkel T, Salitra Y, Rubnov S, Cohen I, Senderowitz H, Kidron D, Livnah O, Levitzki A, Livnah N. A novel substrate mimetic inhibitor of PKB/Akt inhibits prostate cancer tumor growth in mice by blocking the PKB pathway. Biochemistry. 2007;46(16):4716–4724. doi: 10.1021/bi061928s. [DOI] [PubMed] [Google Scholar]
- 58.Qvit N, Hatzubai A, Shalev D, Ben-Neriah Y, Gilon C. 20th American Peptide Symposium. Vol. 611. Advances in Experimental Medicine and Biology; Montreal, Canada: 2009. Design and synthesis of backbone cyclic phosphopeptides: The IκB model; pp. 139–140. [DOI] [PubMed] [Google Scholar]
- 59.Hurevich M, Ratner-Hurevich M, Tal-Gan Y, Shalev DE, Ben-Sasson SZ, Gilon C. Backbone cyclic helix mimetic of chemokine (C-C motif) receptor 2: A rational approach for inhibiting dimerization of G protein-coupled receptors. Bioorganic and Medicinal Chemistry. 2013:3958–3966. doi: 10.1016/j.bmc.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 60.Gazal S, Gelerman G, Ziv O, Karpov O, Litman P, Bracha M, Afargan M, Gilon C. Human somatostatin receptor specificity of backbone-cyclic analogues containing novel sulfur building units. Journal of Medicinal Chemistry. 2002;45(8):1665–1671. doi: 10.1021/jm0100281. [DOI] [PubMed] [Google Scholar]
- 61.Qvit N, Reuveni H, Gazal S, Zundelevich A, Blum G, Niv MY, Feldstein A, Meushar S, Shalev DE, Friedler A, Gilon C. Synthesis of a novel macrocyclic library: Discovery of an IGF-1R inhibitor. Journal of Combinatorial Chemistry. 2008;10(2):256–266. doi: 10.1021/cc700113c. [DOI] [PubMed] [Google Scholar]
- 62.Altstein M, Ben-Aziz O, Daniel S, Schefler I, Zeltser I, Gilon C. Backbone cyclic peptide antagonists, derived from the insect pheromone biosynthesis activating neuropeptide, inhibit sex pheromone biosynthesis in moths. Journal of Biological Chemistry. 1999;274(25):17573–17579. doi: 10.1074/jbc.274.25.17573. [DOI] [PubMed] [Google Scholar]
- 63.Barda Y, Cohen N, Lev V, Ben-Aroya N, Koch Y, Mishani E, Fridkin M, Gilon C. Backbone metal cyclization: novel 99mTc labeled GnRH analog as potential SPECT molecular imaging agent in cancer. Nuclear Medicine and Biology. 2004;31(7):921–933. doi: 10.1016/j.nucmedbio.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 64.Fridkin G, Bonasera TA, Litman P, Gilon C. Backbone metal-cyclization: a novel approach for simultaneous peptide cyclization and radiolabeling. Application to the combinatorial synthesis of rhenium-cyclic somatostatin analogs. Nuclear Medicine and Biology. 2005;32(1):39–50. doi: 10.1016/j.nucmedbio.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 65.Fridkin G, Gilon C. Azo cyclization: peptide cyclization via azo bridge formation. Journal of Peptide Research. 2002;60(2):104–111. doi: 10.1034/j.1399-3011.2002.02993_1.x. [DOI] [PubMed] [Google Scholar]
- 66.Hurevich M, Swed A, Joubran S, Cohen S, Freeman NS, Britan-Rosich E, Briant-Longuet L, Bardy M, Devaux C, Kotler M, Hoffman A, Gilon C. Rational conversion of noncontinuous active region in proteins into a small orally bioavailable macrocyclic drug-like molecule: the HIV-1 CD4:gp120 paradigm. Bioorganic and Medicinal Chemistry. 2010;18(15):5754–5761. doi: 10.1016/j.bmc.2010.04.053. [DOI] [PubMed] [Google Scholar]
- 67.Ovadia O, Greenberg S, Laufer B, Gilon C, Hoffman A, Kessler H. Improvement of drug-like properties of peptides: the somatostatin paradigm. Expert opinion on drug discovery. 2010;5(7):655–671. doi: 10.1517/17460441.2010.493935. [DOI] [PubMed] [Google Scholar]