Abstract
Discriminative methods commonly produce models with relatively good generalization abilities. However, this advantage is challenged in real-world applications (e.g., medical image analysis problems), in which there often exist outlier data points (sample-outliers) and noises in the predictor values (feature-noises). Methods robust to both types of these deviations are somewhat overlooked in the literature. We further argue that denoising can be more effective, if we learn the model using all the available labeled and unlabeled samples, as the intrinsic geometry of the sample manifold can be better constructed using more data points. In this paper, we propose a semi-supervised robust discriminative classification method based on the least-squares formulation of linear discriminant analysis to detect sample-outliers and feature-noises simultaneously, using both labeled training and unlabeled testing data. We conduct several experiments on a synthetic, some benchmark semi-supervised learning, and two brain neurodegenerative disease diagnosis datasets (for Parkinson’s and Alzheimer’s diseases). Specifically for the application of neurodegenerative diseases diagnosis, incorporating robust machine learning methods can be of great benefit, due to the noisy nature of neuroimaging data. Our results show that our method outperforms the baseline and several state-of-the-art methods, in terms of both accuracy and the area under the ROC curve.
Index Terms: Linear discriminant analysis, semi-supervised learning, robust classification, feature selection, sample outlier detection, Alzheimer’s disease, Parkinson’s disease, biomarker identification, disease diagnosis, nuclear norm, regularization
1 Introduction
Discriminative methods learn a mapping from the input feature space to the output label space for a task of classification (or regression). Such methods usually achieve good classification (or regression) results, compared to the generative methods, when there is enough number of training samples. But they carry out limited abilities when there are a small number of labeled data [1]. On the other hand, when noise contaminates the data, discriminative models usually fail to find an optimal mapping. In many real-world applications, the data are usually contaminated by different levels of noise. In some cases, a whole bunch of samples are affected (e.g., deviations in neuroimaging data due to radiation or patient movements during the imaging process), and therefore not useful for the learning task. These types of deviations are often denoted as sample-outliers. On the other hand, sometimes only some specific predictor values or features are infected, known as intra-sample-outliers (or feature-noises).
Various efforts have been made to add robustness to different learning methods. For instance, Suzumaura et al. [2] and Xu et al. [3] introduced robustness to the conventional support vector machine formulation by proposing various regularization terms or suppressing the influence of the outliers. In other works, Kim et al. [4] and Croux et al. [5] proposed robust variations of Fisher/Linear Discriminant Analysis (LDA) method, and Li et al. [6] introduced a worst-case LDA, by minimizing the upper bound of the LDA cost function. These methods are all robust to sample-outliers. On the other hand, some methods were proposed to deal with the feature-noises, such as [7, 8]. Many previous methods use Robust Principal Component Analysis (RPCA) [9], to deal with feature-noises in an unsupervised manner. Furthermore, many robust approaches that denoise the data while training the model do not offer straightforward strategies to deal with the testing data. Often, the denoising procedure of the training and the testing data are conducted separately (e.g., in [10]), which might induce a bias to the whole learning process. One solution is to denoise the training and the testing data together, provided that the testing data are available. Therefore, we propose to take advantage of them as unlabeled data during the training phase. Under such semi-supervised setting, the constructed discriminative model can be more reliable, particularly for the cases with the small-sample-size problem. This could be attributed to the fact that more samples are being used to model the intrinsic geometry of the sample manifold.
The main application we are anticipating in this paper is the diagnosis of neurodegenerative diseases, based on neuroimaging data. This is a challenging problem, as the data is pretty much prone to noise and often there is a limited number of samples. Hence, there is a calling need for robust machine learning methods for such applications. Neurodegenrative diseases are debilitating and incurable conditions caused by progressive degeneration or death of the cells in the brain nervous system. These diseases affect millions of people around the world. Alzheimer’s Disease (AD) and Parkinson’s Disease (PD) are among the most common types. Although neurodegenerative diseases manifest with diverse pathological features, the cellular level processes resemble similar structures. For instance, in AD, deposits of tiny protein plaques result into brain damage and progressive loss of memory [11], while PD is mainly initiated by a selective loss of dopaminergic neurons in the Substantia Nigra (SN) brain region, leading to declining in the generation of a chemical messenger, dopamine. Lack of dopamine yields loss of ability to control body movements, along with several non-motor problems (e.g., depression, and anxiety) [12]. These diseases are often incurable; thus, early diagnosis and treatment are crucial to slow down their progression in the initial stages.
The challenges for building reliable diagnosis models include: (1) It is usually burdensome to acquire noise-free imaging data from the patients. Different sources of noise may affect the acquired data, including a wide variety of noises in the neuroimage acquisition procedure, the imposed artifacts due to preprocessing, and the large amount of inter-subject variabilities; (2) To build a good diagnosis model, through learning a classifier, we need a sufficiently large number of labeled subjects. However, acquiring reliably enough labeled data is costly and time-consuming. Therefore, models that can take advantage of unlabeled data (subjects that we are not certain about their disease) could be of great interest; (3) Different neurodegenerative diseases often affect different regions of the brain, i.e., only certain regions of the brain are associated with the disease. Thus, using all features can undermine the diagnosis performance, and we need to identify the imaging biomarkers for each specific disease while learning the diagnosis model.
To deal with the aforementioned challenges, we propose a semi-supervised discriminative classifier, to take advantage of the available unlabeled testing data. This leads to a more substantial number of samples, which can yield better modeling of the intrinsic geometry of the sample manifold. As a result, our model jointly estimates the noise model (both sample-outliers and feature-noises) on the whole labeled training and unlabeled testing data and simultaneously builds a discriminative model upon the denoised training data. Unlike many previous works on denoising medical images, we do not define the problem of denoising separately from the analysis part. In the sense that if a sample (or a feature value) does not act in accordance with others in building the model, it should be counted as a sample-outlier (or a feature-noise). This observation suggests that intertwining the denoising procedure with the learning framework will help to identify the sample-outliers and feature-noises more efficiently while learning a robust classification model. It is important to note that denoising and outlier detection has a long history in the area of medical image analysis and computing. The inter- and intra-subject variabilities, the noise sourced from the images devices, and the pre-processing errors emerge the study of robust methods for analyzing medical imaging data. For instance, in the recent years, several attempts have been made for denoising the medical images [13–16] or detecting outliers [17, 18], as a preprocessing step to any analysis on medical images.
1.1 Background and Overview of the Proposed Method
In this paper, we introduce a novel classification model based on LDA, which is robust against both sample-outliers and feature-noises, referred to as robust feature-sample linear discriminant analysis (RFS-LDA). The original LDA formulation finds the mapping between the sample space and the label space through a linear transformation matrix, maximizing a so-called Fisher discriminant ratio [4]. In practice, the major drawback of LDA is the small-sample-size problem, which arises when the number of available training samples is much less than the dimensionality of the feature space [19]. Original LDA finds the mapping by incorporating covariance matrices of the input feature matrices. In cases where the number of samples is much less than the number of features, these matrices are probably rank-deficient [20]. A reformulation of LDA based on the reduced-rank least-squares problem (known as LS-LDA) [20] tackles this problem. LS-LDA finds the mapping β ∈ ℝl×m by solving the following problem:
| (1) |
where Ytr ∈ ℝl×Ntr is a binary class label indicator matrix, for l different classes (or labels), and Xtr ∈ ℝm×Ntr is the matrix containing Ntr m-dimensional training samples. is a normalization factor that compensates for the different number of samples in each class [20]. As a result, the mapping β is a reduced rank transformation matrix [8, 20], which could be used to project a test data xtst ∈ ℝm×1 onto an l-dimensional space. Note that directly minimizing (1) avoids the small-sample-size problem by not using the covariance matrices. After it projects the samples to the output space, we need a simple step to infer the class labels. LDA maximizes inter-class variance, while minimizing the intra-class variance, in the mapped space. Thus, we expect that in the mapped space, same-class samples to be closer to each other. The class labels could, therefore, be simply determined using a k-NN strategy.
To make LDA robust against noisy data, Fidler et al. [7] estimate a robust basis, which consists all the discriminative information for classification or regression. In the testing phase, the estimated basis identifies the outliers in samples (images in their case) and then calculates the coefficients using a subsampling approach. On the other hand, Huang et al. [8] proposed a general formulation for Robust Regression (RR) and classification (i.e., Robust LDA or RLDA), where, they first denoise the training feature values using a strategy similar to RPCA [9], and then build the above LS-LDA model using the denoised data. In the testing stage, they denoise the testing samples using the denoised training data. This separate denoising procedure could not effectively form the underlying geometry of sample space to denoise the data. Furthermore, RR [8] only accounts for feature-noises by imposing a sparse noise model constraint on the features matrix, despite the fact that the least-squares data fitting term in (1) is vulnerable to large sample-outliers.
Recently, in robust statistics, it is found that ℓ1 functions are able to make more reliable estimations [21] than ℓ2 least-squares fitting functions. This has been previously adopted in many applications, including robust face recognition [22] and robust dictionary learning [23]. Reformulating the objective in (1) with ℓ1 loss entails the following problem:
| (2) |
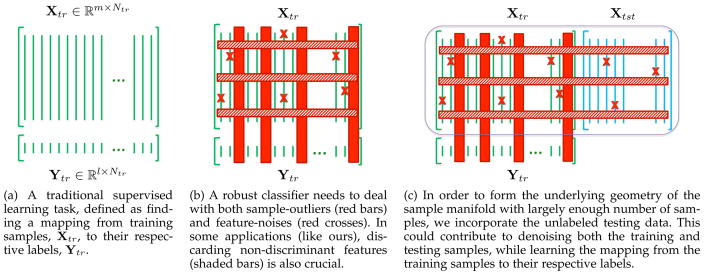
We incorporate this fitting function to deal with the sample-outliers, in this paper. We also adopt a strategy to simultaneously denoise the data from feature-noises. This is done through a semi-supervised setting to take advantage of all labeled and unlabeled data, and build the structure of the sample space more robustly. Figure 1 illustrates this idea, in which Fig. 1a shows a traditional learning problem. However, if the data contains sample-outliers or some samples suffer from noise in their feature values (Fig. 1b), traditional methods usually fail to build reliable models.
Fig. 1.
Overview of the proposed semi-supervised learning framework, robust to both sample-outliers and feature-noises.
Semi-supervised learning has long been of great interest in different fields, because it can make use of unlabeled or poorly labeled data to achieve better prediction models [24, 25]. For instance, Joulin and Bach [26] introduced a convex relaxation and used their model in different semi-supervised learning scenarios. In another work, Cai et al. [27] proposed a semi-supervised discriminant analysis, where the separation between different classes is maximized using the labeled data points, while the unlabeled data points estimate the structure of the data. Belkin et al. [28] similarly used the unlabeled data for regularization. In contrast, we incorporate the unlabeled testing data in our formulation to better estimate the intrinsic geometry of the sample manifold and denoise the data, while building the discriminative model upon the labeled training data. By incorporating the unlabeled testing data (Fig. 1c), we learn the classification model, while denoising both training and testing data and detecting sample-outliers.
We apply our method for the diagnosis of neurodegenerative brain disorders. Specifically, in this study, we use two popular databases: PPMI [29] and ADNI [30]. The former aims at investigating PD and its related disorders, while the latter is designed for diagnosing AD and its prodromal stage, known as Mild-Cognitive Impairment (MCI). In addition, to validate the proposed method, we further conduct experiments on synthetic data, as well as some benchmark datasets for semi-supervised learning.
1.2 Contributions
The contributions of this paper are multi-fold: (1) We propose an approach to dealing with the sample-outliers and feature-noises simultaneously and build a robust discriminative classifier. The sample-outliers are penalized through an ℓ1 fitting function. (2) Our proposed model operates under a semi-supervised setting, where the whole data (i.e., labeled training, and unlabeled testing samples) are incorporated to build the intrinsic geometry of the sample space, which leads to better data denoising. (3) We further select the most discriminative features for the learning process through regularizing the weights matrix with an ℓ1 norm. This is especially of great interest for the neurodegenerative disease diagnosis, where the features from different regions of the brain are extracted, but not all the regions are associated with a certain disease. Thus, the most discriminative regions associated with the disease would be identified, leading to a more reliable diagnosis model.
2 The Proposed Method: RFS-LDA
Suppose we have Ntr training and Ntst testing samples, each with a m dimensional feature vector, which leads to a set of N = Ntr + Ntst total samples. Let X ∈ ℝm×N denote the set of all samples (both training and testing), in which each column indicates a single sample, and also let yi ∈ ℝ1×N their corresponding ith labels. In general, with l different labels, we can define Y ∈ ℝl×N. Thus, X and Y are composed by stacking up the training and testing data as: and . Our goal is to determine the labels of the test samples, Ytst ∈ ℝl×Ntst.
Note that, throughout the paper, bold capital letters denote matrices (e.g., A), while bold lowercase letters denote vectors (e.g., a). All non-bold letters denote scalar variables. aij is the scalar in the row i and column j of A. 〈a1, a2〉 denotes the inner product between a1 and a2. and ||a||1 = Σi|ai| represent the squared Euclidean norm and the ℓ1 norm of a, respectively. , ||A||1,1 = Σj Σi |aij| and ||A||* designate the squared Frobenius norm, ℓ1,1 norm and the nuclear norm (sum of singular values) of A, respectively. IK ∈ ℝK×K denotes the identity matrix.
2.1 Formulation
All the available samples, both labeled and unlabeled, are arranged into a matrix, X ∈ ℝm×N, each of whose columns represents the feature vector of a sample. To achieve a robust classifier, we seek to denoise this matrix. Following [31, 32], this could be done by assuming that X can be spanned on a low-rank subspace and therefore should be rank-deficient. This assumption supports the fact that samples from same classes are more correlated [8, 32] and linearly-dependent. Accordingly, the original matrix X is decomposed into the summation of two counterparts, D ∈ ℝm×N and E ∈ ℝm×N. The former represents the denoised data matrix, while the latter is the error matrix. This is similar to RPCA [9], used in many computer vision applications. With this decomposition, we can assume that the denoised data matrix shall be rank-deficient and the error matrix sparse.
But as one can easily infer, this process of denoising does not incorporate the label information and is, therefore, unsupervised. Nevertheless, recall that we are also seeking a mapping between the denoised training samples and their respective labels. So, matrix D should be spanned on a low-rank subspace that would lead to a good classification model of its sub-matrix, Dtr. We incorporate the regression model in (2) as the fitting function to compute a mapping β. A schematic illustration of the proposed method is depicted in Fig. S1 of the supplementary material.
To ensure the rank-deficiency of the matrix D, like many previous works [9, 31, 32], we approximate the rank function using the nuclear norm (i.e., the sum of the singular values of the matrix). The noise is modeled using the ℓ1 norm of the matrix, which ensures a sparse noise model on the feature values. Accordingly, the objective function for RFS-LDA under a semi-supervised setting would be formed as:
| (3) |
where the first term is the ℓ1 regression model introduced in (2). This term only operates on the denoised training samples from matrix D with a row of all 1’s added to it (denoted as D̂), to counter for the bias in the linear model. The second and third terms, together with the first constraint, are similar to the RPCA formulation [9]. They denoise the labeled training and unlabeled testing data together, and in combination with the first term, we ensure that the denoised data also specifies a favorable regression. The last term is a regularization on the learned mapping coefficients, to avoid trivial or unexpectedly large values. The hyperparameters η, λ1 and λ2 are the scalar regularization hyperparameters, which will be discussed in detail later.
The regularization on the coefficients could be posed as a simple norm of the matrix, β. But, in many applications, like ours (disease diagnosis), many of the features in the feature vectors are redundant. This is because we extract features from different brain regions, but not all the regions contribute to a certain disease. Therefore, it is desirable to determine which features are the most relevant and the most discriminative for the task. Following [11, 22, 33], we are seeking a sparse set of weights that ensures incorporating the most discriminative features. Therefore, we propose a regularization on the weights matrix as a combination of the ℓ1 and Frobenius norms:
| (4) |
Evidently, the solution to the objective function in (3) is not easy to achieve. This is because it contains a quadratic term, and the minimization of the ℓ1 fitting function is not straightforward, due to its indifferentiability. To this end, we formalize the solution with a similar strategy as in Iteratively Re-weighted Least Squares (IRLS) [21]. The ℓ1 fitting term is approximated by a conventional ℓ2 least-squares, in which each of the samples in the D̂ matrix is weighted with the reverse of their regression residual. Additionally, since we regularize the weights β using a combination of ℓ1 and ℓ2 norms, the non-zero elements would represent the selected features by the algorithm. In order to reflect this to feature denoising scheme, we define a projection operator ℘β(.). This operator projects the values of the non-selected features (respective to zero values in β) to zero, to decrease their effect in minimizing the rank of the matrix D (in the second term). Therefore, the new problem would be:
| (5) |
where α̂ is a diagonal matrix, the ith diagonal element of which is the ith sample’s weight:
| (6) |
Hyperparameter δ is a small positive number (10−4 in our experiments), to prevent from any division by zeros in (6). In the next subsection, we introduce an algorithm to solve this optimization problem.
Our work is closely related to the RR formulations in [8], where the authors impose a low-rank assumption on the training data feature values and an ℓ1 assumption on the noise model. The discriminant model is learned similarly to LS-LDA, as described in (1). Whereas, we observed that to have a more robust regression model, we need to establish a strategy where we can weight the samples. This is because the ℓ1 noise model in [8] can only discard a controlled amount of sparse noise in the feature values, not the whole samples. On the other hand, our model operates under a semi-supervised setting, where both labeled training and unlabeled testing samples are denoised simultaneously, leading to a more robust denoising model. Also, our model further selects the most discriminative features to learn the regression model, by regularizing the learned weights and enforcing a sparsity condition on them.
To optimize the objective function in (5), we use the Alternating Direction Method of Multipliers (ADMM) [34]. The detailed optimization steps, along with the comprehensive analysis of the algorithm, its convergence properties and an upper bound for the time complexity of the proposed algorithm are provided in the supplementary material.
3 Experiments
To evaluate the proposed approach, we compare our method against several baselines and state-of-the-art methods in different scenarios. The first experiment evaluates our method on a synthetic set of data, which highlights how the proposed method is robust against sample-outliers or feature-noises separately, or when they occur at the same time. Then we employ some benchmark semi-supervised learning datasets and report results in comparisons with some baseline and state-of-the-art methods. The results of these two experiments (i.e., on synthetic and benchmark data) are reported in the supplementary material. We then apply the proposed RFS-LDA method to the problem of neurodegenerative brain disorder and disease diagnosis.
For the choice of hyperparameters, a set of possible values are first predefined, and the best hyperparameters are selected through 10-fold cross-validation, for all the competing methods. The RFS-LDA hyperparameters (as in Eq. (5)) are set with the same strategy as in [8]:
| (7) |
and ρ (controlling the {μ}s in the iterative optimization algorithm) is set to 1.01. We have set Λ1, Λ2, Λ3 and γ through inner-cross-validation grid-search in the range [10−4, 10].
3.1 Datasets
In this study, we use two real-world databases for two different brain neurodegenerative diseases, namely PD and AD. The first set of data is obtained from the Parkinson’s Progression Markers Initiative (PPMI) database [29], with the MRI data from 374 PD and 169 normal control (NC) subjects. The second dataset comes from the Alzheimer’s disease neuroimaging initiative (ADNI) database, which includes MRI and FDG-PET data. We used 93 AD patients, 202 MCI patients, and 101 NC subjects, each with complete MRI and FDG-PET data. The subjects’ brain images are preprocessed and regions of interest (ROI) features are extracted for each subject. For more detailed information about these two datasets and the preprocessing steps for feature extraction refer to the supplementary material.
3.2 Baseline Methods
We compare our proposed method with different baseline methods, including the conventional LS-LDA [20], RLDA [8], and linear Support Vector Machine (SVM). Another baseline method can be defined as running the same procedures as in the proposed method but disjointly. Therefore, we apply RPCA on the matrix X separately to first denoise, noted as RPCA+LS-LDA) [8]. To analyze the effectiveness of the feature selection strategy of the proposed method, we also include baseline methods which use sparse feature selection (SFS) together with SVM (SFS+SVM), and RLDA (SFS+RLDA). Except for RPCA+LDA, the other methods in comparison do not incorporate the testing data. In order to have a fair set of comparisons, we also compare against the transductive Matrix Completion (MC) approach [32] and the semi-supervised formulation of SVM (S3VM) [35]. These two methods incorporate the unlabeled testing data in the process of training their models. Additionally, in order to further evaluate the effect of the ℓ1 norm regularization on the weights matrix β, we also report results for RFS-LDA when regularized by only γ||β||F (denoted as RFS-LDA*), rather than the regularization term introduced in (4). Finally, we report results using the supervised version of our proposed method, which is denoted as supervised RFS-LDA (S-RFS-LDA). In S-RFS-LDA, we train our model using only the training data, where X in (5) is replaced with Xtr. In this way, we can examine the effect of using unlabeled testing data in the prediction model.
3.3 Disease Diagnosis
We evaluate our method with two popular datasets for neurodegenerative disease diagnosis, PPMI and ADNI, for diagnosis of PD and AD, respectively. These datasets, subject information, preprocessing steps, and feature extraction are explained in Section C of the supplementary material.
Results
The first row in Table 1 shows the diagnostic accuracy of the proposed technique (RFS-LDA) in comparisons with different baseline and state-of-the-art methods using 10-fold cross-validation. The results show that the proposed method outperforms all others. This can be attributed to the fact that our method better deals with feature-noises and sample-outliers. Recall that samples and their corresponding feature vectors extracted from the neuroimaging data are quite prone to noise, as discussed earlier. Therefore, some of the samples might not be useful, and some might be contaminated by a certain amount of noise. Our method can deal with both types of noises, as supported by the results. The second disease diagnosis experiment is conducted on ADNI, in which the goal is to discriminate normal controls (NC) from mild cognitive impairment (MCI) and AD subjects. Therefore, NC subjects form our negative class, while the positive class is defined as AD in one experiment, and MCI in the other. The diagnosis results of the AD vs. NC and MCI vs. NC are reported in the second and third rows in Table 1, respectively. As it can be seen, in comparison with the state-of-the-art, our method achieves better results in terms of both accuracy and the area under ROC curve.
TABLE 1.
Diagnosis accuracy of the proposed method (RFS-LDA) and the baseline methods on both PPMI and ADNI datasets. The † sign indicates a p-value > 0.05 in a Fisher exact test.
| RFS-LDA | RFS-LDA* | S-RFS-LDA | RLDA | SFS+RLDA | RPCA+LS-LDA | LS-LDA | SVM | SFS+SVM | S3VM | MC | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PD vs. NC | 79.8 | 76.1 | 74.1 | 68.3 | 70.5 | 61.0† | 58.5† | 66.1† | 69.2 | 71.5 | 70.6 |
| AD vs. NC | 92.1 | 89.8 | 88.3 | 87.8 | 90.0 | 87.6 | 82.7 | 85.4 | 87.1 | 90.5 | 88.7 |
| MCI vs. NC | 81.9 | 80.6 | 79.1 | 79.5 | 80.9 | 76.9 | 72.3† | 74.1 | 78.3 | 80.8 | 76.1 |
It is worth noting that running the model using a 10-fold cross-validation for the PD vs. NC (543 subjects), AD vs. NC (194 subjects), and MCI vs. NC (303 subjects) experiments on a PC (Intel® Core™ i7 @ 2.30 GHz and 8.00 GB of memory), with a parallel implementation in MATLAB® (i.e., using parfor for 4 workers) took approximately 6, 2 and 3.5 hours, respectively. Additionally, to test the statistical significance of the obtained results, we further conducted a Fisher exact test [36] on the accuracy score achieved by each of the methods. This test verifies that the method is significantly more accurate (with a p-value of p < 0.05) than randomly assigning the samples to the two classes. The results of this statistical test indicated that the proposed method achieves a p-value of even less than 0.001. This shows that there are no random associations with the obtained results. However, for some of the compared baseline methods, a p-value of p > 0.05 was observed, which is not appealing. These methods are marked with a † sign in Table 1. It is important to note that the comparisons between the supervised (S-RFS-LDA) and the semi-supervised (RFS-LDA) versions of the proposed algorithm in both Table 1 and Figure S5 of the supplementary material show that including the unlabeled testing data improves the results by a relatively notable margin. This can be because including more samples gives us a better representation of the sample manifold, leading to better denoising of simultaneous training and testing data, in a way that a better classifier is built.
Although the studies on Parkinson’s disease using modern machine learning techniques are scarce, there are quite a few studies in the literature for Alzheimer’s disease. State-of-the-art machine learning approaches for this purpose either aim at developing feature selection techniques or focus on designing delicate classifiers. The first type usually use sophisticated techniques for feature selection [37, 38], feature learning [39], or feature extraction[40–42] and then an straightforward classification technique (like SVM) is utilized. The second type develops task-specific classifiers to enhance the classification accuracies, e.g. [43–45]. In contrast, our method constructs the sample manifold using all labeled and unlabeled data to denoise the features and also selects the best features for classification, with a classification loss robust to sample-outliers. In Table 2, we compare our method with several state-of-the-art methods for Alzheimer’s disease diagnosis. The table includes all the information about the dataset and the methods they used for obtaining those results. This is only to show where our method stands among the previous works in the same field.
TABLE 2.
Comparisons of the proposed method with state-of-the-art methods for diagnosis of AD and MCI. N/A: indicates that the methods did not report results for that experiment; CSF: cerebrospinal fluid; Gen: categorical genetic information.
| Method | Subjects
|
Methodology | Modalities | AD vs. NC (%) | MCI vs. NC (%) | ||
|---|---|---|---|---|---|---|---|
| AD | MCI | NC | |||||
| Liu et al. [45] | 198 | N/A | 229 | Voxel GM+SVM Ensemble | MRI | 92.0 | N/A |
| Cuingnet et al. [42] | 137 | N/A | 162 | Voxel Direct D+SVM | MRI | 88.58 | N/A |
| Eskildsen et al. [41] | 194 | N/A | 226 | Cortical Thickness+SVM | MRI | 84.50 | N/A |
| Duche. et al. [40] | 75 | N/A | 75 | Tensor-based Morphometry+SVM | MRI | 92.0 | N/A |
| Min et al. [37] | 97 | N/A | 128 | Multi-Atlas ROI Features+SVM | MRI | 91.6 | N/A |
| Gary et al. [44] | 37 | 75 | 35 | Random Forest | MRI+PET+CSF+Gen | 89.0 | 74.6 |
| Tong et al. [43] | 35 | 75 | 77 | Graph Fusion | MRI+PET+CSF+Gen | 91.8 | 79.5 |
| Liu et al. [39] | 85 | 169 | 77 | Deep Feature Learning | MRI+PET | 91.4 | 82.1 |
| Ours | 93 | 202 | 101 | RFS-LDA | MRI+PET | 92.1 | 81.9 |
As discussed earlier, in medical imaging applications many sources of noise contribute to the acquired data, and therefore methods that can deal with noise and outliers are of great interest. Our method enjoys from a single optimization objective that can simultaneously suppress sample-outliers and feature-noises, which, compared to other methods, exhibits a good performance. One of the interesting functions of the proposed method is the regularization on the mapping coefficients with the ℓ1 norm, which would select a compact set of features to contribute to the learned mapping. The magnitude of the coefficients would show the relevance of the specific features for building the prediction model. In our application, the features from the whole brain regions are extracted, but not all the ROIs are associated with the disease (e.g., AD, MCI or PD). By exploring the learned coefficients by our method, we can determine which brain regions are highly associated with a certain disease.
Identification of Disease Biomarkers
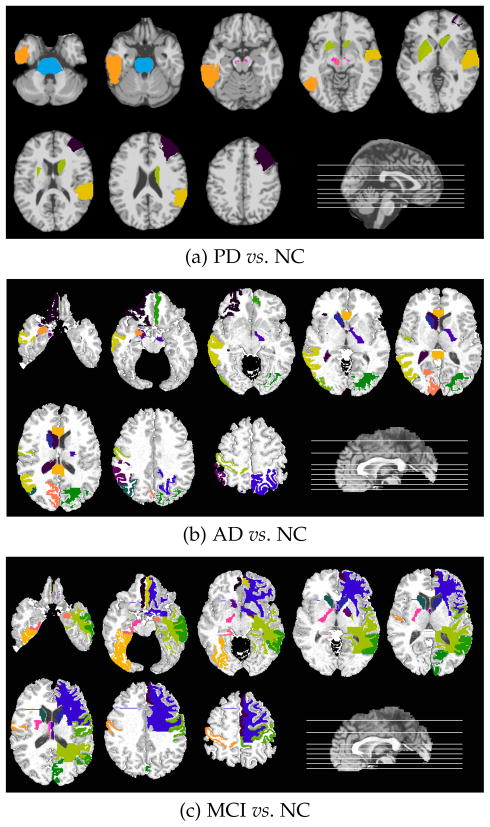
To extract these most relevant ROIs, we select the ROIs that were given larger weights in 50% of the ten repetitions of the 10-fold cross-validation tests. Fig. 2(a) visualizes the most relevant regions for PD on a raw brain template, including the middle frontal gyrus right, pons, substrata nigra left and right, red nucleus left, pallidum left, pautmen left, caudate right, inferior temporal left, and superior temporal gyrus right. As in the previous studies in the literature [46, 47], deep brain and striatum areas are known to play crucial roles for PD. Our study also confirms these clinical findings. Same experimental settings for AD and MCI identifies the top regions selected by our algorithm in AD vs. NC and MCI vs. NC classification scenarios (Figs. 2(b) and (c), respectively). These regions, including middle temporal gyrus, medial front-orbital gyrus, postcentral gyrus, caudate nucleus, cuneus, and amygdala, have also been reported to be associated with AD and MCI in the literature [11, 48]. The analysis of such selection of brain regions can be further incorporated for future clinical studies.
Fig. 2.
Top selected regions for each experiment. Selected regions are shown with different colors for clarity.
Method Discussions
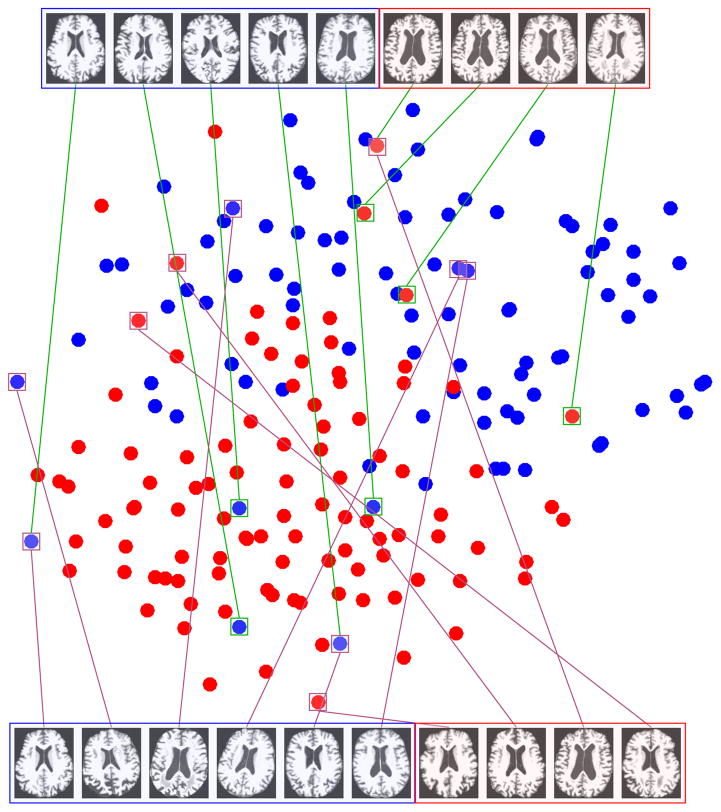
To analyze the effect of the sample-outlier detection in the proposed framework, we employ a dimensionality reduction technique to facilitate the visualization of the data points. We project the samples of the AD vs. NC experiment into the 2-D space using t-SNE [49]. The t-SNE projection technique visualizes high-dimensional data by giving each sample a location in a two-dimensional map. The map created by the t-SNE reveals the neighborhood structure of the sample manifold at many different scales [49]. This is particularly important for our application, in which the high-dimensional neuroimaging data lie on several different low-dimensional manifolds since the samples come from different subjects with or without the neurodegenerative disease. Fig. 3 shows the t-SNE projection in the 2-D space. In this figure, the samples, which received the smallest weights in their respective elements in the α̂ weight matrix (as in Equation (5)), are shown in the top part of the figure. We also depict the samples detected as outliers using the RANSAC [50] algorithm in the bottom part of the figure. Notably, as it is obvious in the figure, the samples detected as sample-outliers by our algorithm are those which are more controversial for the task of classification and lie outside the main neighborhood of each class. This is attributed to the fact that we detect them jointly with the classifier learning framework. On the other hand, the outliers detected by RANSAC are not always the best in terms of discriminability. This suggests that unsupervised outlier detection methods might not perform well when the aim is to learn a classifier or a regression model. In other words, in many learning tasks, the definition for sample-outliers might be different based on what the goal is.
Fig. 3.
t-SNE projection of AD vs. NC samples (better viewed in color). Top: Samples detected as outliers by our method. Bottom: Samples detected as outliers using RANSAC [50].
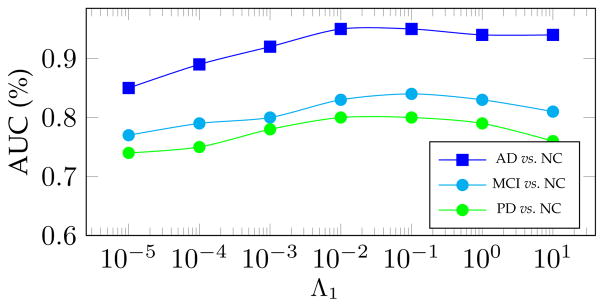
One of the important hyperparameters in the proposed RFS-LDA is λ1, as in Eq. (5), which controls the noise term. Modifying this hyperparameter leads to altered noise levels, detected by our algorithm. To analyze its effect on the learning performance, we fix all other hyperparameters and run the algorithm with different values of Λ1, and therefore λ1 (as discussed at the beginning of Section 3). The changes in the AUC for each of our experiments are illustrated in Fig. 4. As can be seen, the proposed method achieves reasonably good results with a wide range of the values of the hyperparameter.
Fig. 4.
Area under the ROC curve (AUC) as a function of the RFS-LDA hyperparameter Λ1, related to λ1 in (3).
It is worth noting that the proposed method works under a semi-supervised setting, which can be quite interesting for the application of disease diagnosis. When performing the diagnosis for new patients, all subjects whose clinical diagnosis has not been finalized (i.e., they are still in the process of evaluations and clinical monitoring) can yet be included in model building as unlabeled samples, to build a potentially more reliable classifier.
4 Conclusion
In this paper, we proposed a novel approach for discriminative classification, which is robust against both sample-outliers and feature-noises. Our method enjoys a semi-supervised setting, where all the labeled training and the unlabeled testing data are used to detect outliers and are denoised simultaneously. We have applied our method to several datasets, including synthetic, semi-supervised learning benchmark, and neurodegenerative brain disease diagnosis datasets, specifically for Parkinson’s disease and Alzheimer’s disease. The results showed that our method outperformed all competing techniques. As a direction for the future works, one can develop a multi-task learning reformulation of the proposed method to incorporate diagnosis from multiple modalities of neuroimaging data or extend the approach for the case of incomplete data.
Biographies

Ehsan Adeli is a postdoctoral research fellow at Stanford University. He received his Ph.D. from Iran University of Science and Technology, after which he worked as a postdoctoral researcher at the University of North Carolina at Chapel Hill. He has previously worked as a visiting research scholar at the Robotics Institute, Carnegie Mellon University in Pittsburgh, PA. Dr. Adeli’s research interests include machine learning, computer vision, medical image analysis and computational neuroscience.

Kim-Han Thung obtained his PhD degree from the University of Malaya, Malaysia, in 2012. He is currently a post-doctoral research associate at the University of North Carolina at Chapel Hill, USA. He was a research assistant at University of Malaya, Malaysia and a software engineer at Motorola Penang, Malaysia. His research interests include sparse learning, multi-task learning, deep learning, matrix completion, machine learning using incomplete data and medical image analysis.

Le An received the B.Eng. degree in telecommunications engineering from Zhejiang University in China in 2006, the M.Sc. degree in electrical engineering from Eindhoven University of Technology in Netherlands in 2008, and the Ph.D. degree in electrical engineering from University of California, Riverside in USA in 2014. His research interests include image processing, computer vision, pattern recognition, and machine learning.

Guorong Wu received the Ph.D. degree in computer science and engineering from Shanghai Jiao Tong University, Shanghai, China. He is currently with the Department of Radiology, The University of North Carolina, Chapel Hill, as an Assistant Professor. His research interests focus on fast and robust analysis of large population data, computer-assisted diagnosis, and image-guided radiation therapy.

Feng Shi received his Bachelors Degree in Electronics Engineering from Peking University, Beijing, China in 2002, and obtained his PhD degree in Computer Science from Institute of Automation, Chinese Academy of Sciences in 2008. He was then trained as a postdoctoral research associate in Medical Image Analysis at the University of North Carolina at Chapel Hill, NC, and later appointed as an Assistant Professor. Currently, he is an Assistant Professor in Cedars-Sinai Medical Center, Los Angeles, CA since 2016. His research interests include machine learning, neuro and cardiac imaging, multimodal image analysis and computer-aided early diagnosis.

Tao Wang graduated from the Shanghai Jiao-tong University School of Medicine with a doctoral degree in psychogeriatrics in 2012. He completed training as visiting scholar in UNC IDEA Group, Biomedical Research Imaging Center, UNC Chapel Hill, NC, USA in 2015–2016, and Banner Alzheimers Institute, Arizona, USA from June 2013 to July 2013. Since 2013 he has been an associate chief psychiatrist in the department of geriatric psychiatry, Shanghai Mental Health Center, Shanghai Jiao Tong University School of Medicine. Now he is the vice director of the department of geriatric psychiatry at the Shanghai Mental Health Center. His main research interest is neurocognitive disorder.

Dinggang Shen is Jeffrey Houpt Distinguished Investigator, and a Professor of Radiology, Biomedical Research Imaging Center (BRIC), Computer Science, and Biomedical Engineering in the University of North Carolina at Chapel Hill (UNC-CH). He is currently directing the Center for Image Analysis and Informatics, the Image Display, Enhancement, and Analysis (IDEA) Lab in the Department of Radiology, and also the medical image analysis core in the BRIC. He was a tenure-track assistant professor in the University of Pennsylvanian (UPenn), and a faculty member in the Johns Hopkins University. Dr. Shen’s research interests include medical image analysis, computer vision, and pattern recognition. He has published more than 800 papers in the international journals and conference proceedings. He serves as an editorial board member for eight international journals. He has also served in the Board of Directors, The Medical Image Computing and Computer Assisted Intervention (MICCAI) Society, in 2012–2015. He is Fellow of IEEE, and also Fellow of The American Institute for Medical and Biological Engineering (AIMBE).
Footnotes
Parts of the data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (http://adni.loni.ucla.edu), and Parkinson’s Progression Markers Initiative (PPMI) database (http://www.ppmi-info.org). The investigators within the ADNI or PPMI contributed to the design and implementation of the datasets and/or provided data but did not participate in analysis or writing of this paper. A complete listing of ADNI investigators can be found at: http://adni.loni.ucla.edu/wp-content/uploads/howtoapply/ADNIAcknowledgementList.pdf
Contributor Information
Ehsan Adeli, Stanford University, Stanford, CA 94305, USA. Biomedical Research Imaging Center (BRIC) and Department of Radiology, University of North Carolina at Chapel Hill, NC, 27599, USA.
Kim-Han Thung, Biomedical Research Imaging Center (BRIC) and Department of Radiology, University of North Carolina at Chapel Hill, NC, 27599, USA.
Le An, Biomedical Research Imaging Center (BRIC) and Department of Radiology, University of North Carolina at Chapel Hill, NC, 27599, USA.
Guorong Wu, Biomedical Research Imaging Center (BRIC) and Department of Radiology, University of North Carolina at Chapel Hill, NC, 27599, USA.
Feng Shi, Biomedical Imaging Research Institute, Cedars Sinai Medical Center, Los Angeles, CA 90048, USA.
Tao Wang, Department of Geriatric Psychiatry, Shanghai Mental Health Center and the Alzheimer’s Disease and Related Disorders Center, Shanghai Jiao Tong University, Shanghai, China.
Dinggang Shen, Biomedical Research Imaging Center (BRIC) and Department of Radiology, University of North Carolina at Chapel Hill, NC, 27599, USA. Department of Brain and Cognitive Engineering, Korea University, Seoul 02841, Republic of Korea.
References
- 1.Jordan A. On discriminative vs. generative classifiers: A comparison of logistic regression and naive bayes. NIPS. 2002;14:841. [Google Scholar]
- 2.Suzumura S, Ogawa K, Sugiyama M, Takeuchi I. Outlier path: A homotopy algorithm for robust svm; ICML; 2014. pp. 1098–1106. [Google Scholar]
- 3.Xu H, Caramanis C, Mannor S. Robustness and regularization of support vector machines. JMLR. 2009;10:1485–1510. [Google Scholar]
- 4.Kim S-J, Magnani A, Boyd S. Robust fisher discriminant analysis. NIPS; 2005. pp. 659–666. [Google Scholar]
- 5.Croux C, Dehon C. Robust linear discriminant analysis using s-estimators. Canadian J of Statistics. 2001;29(3):473–493. [Google Scholar]
- 6.Li H, Shen C, van den Hengel A, Shi Q. Worst-case linear discriminant analysis as scalable semidefinite feasibility problems. IEEE TIP. 2015;24(8) doi: 10.1109/TIP.2015.2401511. [DOI] [PubMed] [Google Scholar]
- 7.Fidler S, Skocaj D, Leonardis A. Combining reconstructive and discriminative subspace methods for robust classification and regression by subsampling. IEEE TPAMI. 2006;28(3):337–350. doi: 10.1109/TPAMI.2006.46. [DOI] [PubMed] [Google Scholar]
- 8.Huang D, Cabral R, De la Torre F. Robust regression. ECCV; 2012. pp. 616–630. [DOI] [PubMed] [Google Scholar]
- 9.Cand Eès, Li X, Ma Y, Wright J. Robust principal component analysis? Journal of the ACM. 2011;58(3):11:1–11:37. [Google Scholar]
- 10.Huang D, Cabral R, De la Torre F. Robust regression. IEEE TPAMI. 2015 doi: 10.1109/TPAMI.2015.2448091. [DOI] [PubMed] [Google Scholar]
- 11.Thung KH, Wee CY, Yap PT, Shen D. Neurodegenerative disease diagnosis using incomplete multi-modality data via matrix shrinkage and completion. NeuroImage. 2014;91:386–400. doi: 10.1016/j.neuroimage.2014.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ziegler D, Augustinack J. Harnessing advances in structural MRI to enhance research on Parkinson’s disease. Imag in med. 2013;5(2):91–94. doi: 10.2217/iim.13.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coup Pé, Yger P, Prima S, Hellier P, Kervrann C, Barillot C. An optimized blockwise nonlocal means denoising filter for 3-d magnetic resonance images. IEEE TMI. 2008;27(4):425–41. doi: 10.1109/TMI.2007.906087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodrigues I, Sanches J, Bioucas-Dias J. Denoising of medical images corrupted by poisson noise. ICIP; 2008. pp. 1756–1759. [Google Scholar]
- 15.Bhadauria H, Dewal M. Medical image denoising using adaptive fusion of curvelet transform and total variation. Computers and Electrical Engineering. 2013;39(5):1451–1460. [Google Scholar]
- 16.Manj Jón, Coupé P, Buades A, Louis Collins D, Robles M. New methods for mri denoising based on sparseness and self-similarity. Med Image Anal. 2012;16(1):18–27. doi: 10.1016/j.media.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 17.Fritsch V, Varoquaux G, Thyreau B, Poline J-B, Thirion B. Detecting outliers in high-dimensional neuroimaging datasets with robust covariance estimators. Med Image Anal. 2012;16(7):1359–1370. doi: 10.1016/j.media.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Mriaux S, Roche A, Thirion B, Dehaene-Lambertz G. Robust statistics for nonparametric group analysis in fmri. ISBI; 2006. pp. 936–939. [Google Scholar]
- 19.Li H, Jiang T, Zhang K. Efficient and robust feature extraction by maximum margin criterion. NIPS; 2003. pp. 97–104. [DOI] [PubMed] [Google Scholar]
- 20.De la Torre F. A least-squares framework for component analysis. IEEE TPAMI. 2012;34(6):1041–1055. doi: 10.1109/TPAMI.2011.184. [DOI] [PubMed] [Google Scholar]
- 21.Bissantz N, ümbgen LD, Munk A, Stratmann B. Convergence analysis of generalized iteratively reweighted least squares algorithms on convex function spaces. SIAM J on Optimization. 2009;19(4):1828–1845. [Google Scholar]
- 22.Wagner A, Wright J, Ganesh A, Zhou Z, Ma Y. Towards a practical face recognition system: Robust registration and illumination by sparse representation. CVPR; 2009. pp. 597–604. [DOI] [PubMed] [Google Scholar]
- 23.Lu C, Shi J, Jia J. Online robust dictionary learning. CVPR; June 2013.pp. 415–422. [Google Scholar]
- 24.Chapelle O, Schölkopf B, Zien A, editors. Semi-Supervised Learning. MIT Press; 2006. [Google Scholar]
- 25.Zhu X. Tech Rep 1530. Computer Sciences, University of Wisconsin-Madison; 2005. Semi-supervised learning literature survey. [Google Scholar]
- 26.Joulin A, Bach F. A convex relaxation for weakly supervised classifiers. ICML; 2012. [Google Scholar]
- 27.Cai D, He X, Han J. Semi-supervised discriminant analysis. CVPR; 2007. [Google Scholar]
- 28.Belkin M, Matveeva I, Niyogi P. Regularization and semi-supervised learning on large graphs. Learning Theory, vol. 3120 of LNCS. 2004:624–638. [Google Scholar]
- 29.Marek K, et al. The parkinson progression marker initiative (PPMI) Progress in Neurobiology. 2011;95(4):629–635. doi: 10.1016/j.pneurobio.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, Trojanowski JQ, Toga AW, Beckett L. Ways toward an early diagnosis in alzheimer’s disease: The alzheimer’s disease neuroimaging initiative (adni) Alzheimer’s & dementia: J of the Alzheimer’s Association. 2005;1:55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu G, Lin Z, Yan S, Sun J, Yu Y, Ma Y. Robust recovery of subspace structures by low-rank representation. IEEE TPAMI. 2013;35(1):171–184. doi: 10.1109/TPAMI.2012.88. [DOI] [PubMed] [Google Scholar]
- 32.Goldberg A, Zhu X, Recht B, Xu J-M, Nowak R. Transduction with matrix completion: Three birds with one stone. NIPS; 2010. pp. 757–765. [Google Scholar]
- 33.Elhamifar E, Vidal R. Robust classification using structured sparse representation. CVPR; 2011. [Google Scholar]
- 34.Boyd S, et al. Distributed optimization and statistical learning via the alternating direction method of multipliers. Found Trends Mach Learn. 2011;3(1):1–122. [Google Scholar]
- 35.Bennett KP, Demiriz A. Semi-supervised support vector machines. NIPS; 1998. pp. 368–374. [Google Scholar]
- 36.Fisher RA. The logic of inductive inference. Journal of the Royal Statistical Society. 1935;98(1):39–82. [Google Scholar]
- 37.Min R, Wu G, Cheng J, Wang Q, Shen D. Multi-atlas based representations for alzheimer’s disease diagnosis. Human brain mapping. 2014;35(10):5052–5070. doi: 10.1002/hbm.22531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu M, Zhang D, Adeli-Mosabbeb E, Shen D. Inherent structure based multi-view learning with multi-template feature representation for alzheimer’s disease diagnosis. IEEE TBME. 2016 doi: 10.1109/TBME.2015.2496233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu S, Liu S, Cai W, Che H, Pujol S, Kikinis R, Feng D, Fulham MJ. Multimodal neuroimaging feature learning for multiclass diagnosis of alzheimer’s disease. Biomedical Engineering, IEEE Transactions on. 2015;62(4):1132–1140. doi: 10.1109/TBME.2014.2372011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duchesne S, Caroli A, Geroldi C, Barillot C, Frisoni GB, Collins DL. Mri-based automated computer classification of probable ad versus normal controls. IEEE TMI. 2008;27(4):509–520. doi: 10.1109/TMI.2007.908685. [DOI] [PubMed] [Google Scholar]
- 41.Eskildsen SF, Coupé P, García-Lorenzo D, Fonov V, Pruessner JC, Collins DL, Initiative ADN, et al. Prediction of alzheimer’s disease in subjects with mild cognitive impairment from the adni cohort using patterns of cortical thinning. Neuroimage. 2013;65:511–521. doi: 10.1016/j.neuroimage.2012.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cuingnet R, Gerardin E, Tessieras J, Auzias G, Lehéricy S, Habert M-O, Chupin M, Benali H, Colliot O, et al. ADN Initiative. Automatic classification of patients with alzheimer’s disease from structural mri: a comparison of ten methods using the adni database. Neuroimage. 2011;56(2):766–781. doi: 10.1016/j.neuroimage.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 43.Tong T, Gray K, Gao Q, Chen L, Rueckert D. Machine Learning in Medical Imaging. Springer; 2015. Nonlinear graph fusion for multi-modal classification of alzheimer’s disease; pp. 77–84. [Google Scholar]
- 44.Gray KR, Aljabar P, Heckemann RA, Hammers A, Rueckert D, Initiative ADN, et al. Random forest-based similarity measures for multi-modal classification of alzheimer’s disease. NeuroImage. 2013;65:167–175. doi: 10.1016/j.neuroimage.2012.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu M, Zhang D, Shen D. Hierarchical fusion of features and classifier decisions for alzheimer’s disease diagnosis. Human brain mapping. 2014;35(4):1305–1319. doi: 10.1002/hbm.22254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Braak H, Tredici K, Rub U, de Vos R, Steur EJ, Braak E. Staging of brain pathology related to sporadic parkinson’s disease. Neurobio of Aging. 2003;24(2):197–211. doi: 10.1016/s0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- 47.Worker A, et al. Cortical thickness, surface area and volume measures in parkinson’s disease, multiple system atrophy and progressive supranuclear palsy. PLOS ONE. 2014;9(12) doi: 10.1371/journal.pone.0114167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pearce B, Palmer A, Bowen D, Wilcock G, Esiri M, Davison A. Neurotransmitter dysfunction and atrophy of the caudate nucleus in alzheimer’s disease. Neurochem Pathol. 1985;2(4):221–32. [PubMed] [Google Scholar]
- 49.Van der Maaten L, Hinton G. Visualizing data using t-SNE. JMLR. 2008;9(2579–2605):85. [Google Scholar]
- 50.Fischler MA, Bolles RC. Random sample consensus: A paradigm for model fitting with applications to image analysis and automated cartography. Commun ACM. 1981;24(6):381–395. [Google Scholar]