Abstract
Purpose
To describe the study protocol and baseline characteristics of the “African Descent and Glaucoma Evaluation Study (ADAGES) III: Contribution of Genotype to Glaucoma phenotype in African Americans”.
Design
Cross-sectional, case-control study
Participants
There were 3266 glaucoma patients and controls without glaucoma of African or European descent recruited from five study centers in different regions of the United States.
Methods
Individuals of African descent (AD) and European descent (ED) with primary open angle glaucoma (POAG) and control subjects completed a detailed demographic and medical history interview. Standardized height, weight and blood pressure measurements were obtained. Saliva and blood samples to provide serum, plasma, DNA and RNA were collected for standardized processing. Visual fields, stereoscopic disc photographs, and details of the ophthalmic examination were obtained and transferred to the University of California-San Diego Data Coordinating Center for standardized processing and quality review.
Main Outcome Measures
Participant gender, age, race, body mass index, blood pressure, history of smoking and alcohol use in POAG patients and controls are described. Ophthalmic measures included intraocular pressure, visual field mean deviation, central corneal thickness, glaucoma medication use or past glaucoma surgery. Ocular conditions including diabetic retinopathy, age-related macular degeneration and past cataract surgery were recorded.
Results
The 3266 ADAGES III study participants in this report include 2146 AD POAG patients, 695 ED POAG patients, 198 AD controls and 227 ED controls. AD POAG patients and controls were significantly younger (both 67.4 years) than ED POAG patients and controls (73.4 and 70.2 years, respectively). After adjusting for age, AD POAG patients had different phenotypic characteristics compared to ED POAG patients, including higher intraocular pressure, worse visual acuity and visual field mean deviation, and thinner corneas (all p<0.001). Family history of glaucoma did not differ between AD and ED POAG patients.
Conclusions
With its large sample size, extensive specimen collection and deep phenotyping of AD and ED glaucoma patients and controls from different regions in the United States, the ADAGES III Genomics Study will address gaps in our knowledge of the genetics of POAG in this high-risk population.
Glaucoma is the leading cause of irreversible blindness in individuals of African Descent (AD), and the second leading cause of irreversible blindness in all Americans1–3. As the population ages, the number of persons with primary open angle glaucoma (POAG) in the United States will increase by approximately 50% to over 3.3 million in 20204. Individuals of AD are more likely to become blind 5–7, and are fifteen times more likely to develop POAG-associated visual impairment8 than individuals of European Descent (ED).
There are well-established differences of the optic disc and visual field between AD and ED individuals. For example, it is well established in normal9–13, ocular hypertensive14, post-mortem15 and glaucomatous16 eyes, that individuals of AD have larger optic discs than individuals of ED. Further, healthy persons of AD reportedly have larger optic cup volumes 10,12,13, deeper optic cups12, and larger cup-to-disc ratios than healthy individuals of ED, although these differences should be interpreted with caution in light of the association between optic disc size and many other optic disc parameters17–20.
Although it is clear that AD individuals are more likely than ED individuals to have glaucoma and to go blind from the disease and that the glaucoma phenotype of AD individuals differ from ED individuals, the genetic characteristics that contribute to these differences are not understood. The African Descent and Glaucoma Evaluation Studies I and II studies provided detailed and extensive phenotype information on a cohort of healthy, glaucoma suspect and glaucoma patients of AD and ED. Specifically, the National Eye Institute funded ADAGES in 2003 to characterize structural and functional damage and change in AD patients and individuals with suspect disease in order to predict and eventually prevent POAG-related disability and blindness in this high risk population.21–23 Despite similar access to treatment provided by study clinicians, ADAGES demonstrated that AD individuals tend to have worse central and peripheral visual field (VF) damage,23 more variable VF results, and are more likely to be fast progressors.24 Most importantly, it was shown that the risk of developing VF damage as a function of IOP varies by race.25 Among glaucoma suspects at similar levels of elevated IOP during follow-up (≥ 21 mmHg), individuals of AD were 5.2 times more likely to develop VF damage than those of ED; however, no racial differences were found at lower IOP.25 Moreover, ADAGES results suggested that there are significant age- and race-related structural variations within the load-bearing connective tissue components of the optic nerve head and peripapillary sclera that significantly alter the biomechanical response of these tissues and may at least in part be responsible for racial differences in the pathophysiology of POAG.26–31 Thus, given these many racial differences, ADAGES III was designed to delineate the genetics of POAG in individuals of AD and to elucidate the genetic basis for the racial phenotypic differences found in ADAGES I and II.
This report describes the ADAGES III study design, methods and specimens collected, enrollment, demographics, and initial baseline clinical findings. More complete analyses of the genomics of POAG in individuals of AD will be reported separately.
Methods
Study Aims
The overall goal of ADAGES III is to identify the genetic factors associated with primary open angle glaucoma (POAG) in individuals of African descent (AD). To meet this goal, the following 4 aims are being addressed.
Collect samples and analyze phenotype and genotype: 1a) Obtain DNA samples from 3000+ subjects for genotyping; over two thirds that are AD. Cases and controls will be ascertained from both existing studies (African Descent and Glaucoma Evaluation Study (ADAGES), the Diagnostic Innovations in Glaucoma Study (DIGS)) and from new recruitment at the study centers. 1b) Establish the UCSD Shiley Eye Institute Data Coordinating Center database management system and sample repository to receive ocular phenotypic data, and maintain samples for this and future studies, as well establish a parallel genomics repository at the Genomics Institute of the Los Angeles Biomedical Research Institute at Harbor-UCLA (LABioMed).
Analyses of glaucoma as a qualitative trait, of glaucoma-related biometric traits, and of glaucoma severity: Analyze data for glaucoma as a qualitative trait and also perform a quantitative association analysis within the glaucoma cases for biometric traits (intraocular pressure, central cornea thickness, visual field severity and pattern of visual field loss, etc) as appropriate for both common and rare genetic variants. This analysis is conducted primarily within the ADAGES glaucoma cohort.
Confirmation and comparison of SNPs associated with glaucoma: Test the effect of known genes associated with POAG identified in individuals of ED descent in order to identify whether results in subjects with ED are generalizable to the African descent population: and test any novel findings from AD subjects in aims 1 and 2 in ED subjects with POAG.
Fine-mapping and functional analyses: Select regions and genes from Aims 1, 2, and 3 for further study in order to identify potential mechanisms for POAG pathophysiology. Identical and contemporaneous phenotyping (along with genotyping) will facilitate the identification of genotype-phenotype similarlities and differences between the AD and ED glaucoma participants. Better understanding of the genetic susceptibility and pathophysiology of progressive glaucoma will lead to improved means of treatment and prevention.
Synopsis of ADAGES III Study Design and Management
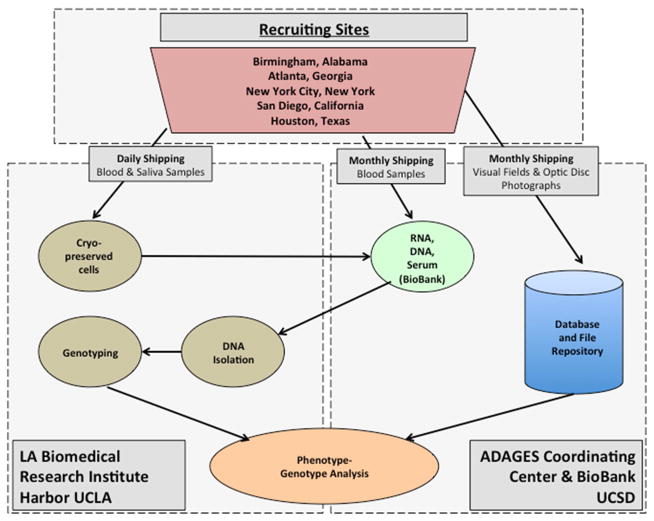
The UCSD ADAGES III Coordinating Center, ADAGES BioBank and the Genomics Institute of LABioMed (Figure 1) are responsible for the design, implementation and quality control of data and specimens collected in ADAGES III. Specifically, the ADAGES BioBank (Directors Drs. Ayyagari and Zangwill) and LABioMed genomics biobank (Directors Drs. Chen, Taylor and Rotter) are responsible for protocol development of the specimen collection, processing, shipping, and quality control of participant blood and saliva samples. The ADAGES BioBank staff certifies each technician involved in the specimen collection and/or processing to ensure that the all aspects of the ADAGES III specimen protocol are understood and followed. The ADAGES Coordinating Center (Director Dr. Zangwill) staff are responsible for technician certification of the overall study protocol including participant eligibility, anthropometric measurements, standardized interview, and transmission of existing visual fields, stereophotographs and optical coherence tomography images. In addition, the ADAGES Coordinating Center coordinates data entry, and completes quality control of the anthropometric measurements, the participant interview and ocular and non-ocular information. The ADAGES Coordinating Center, through its reading centers, the Imaging Data Evaluation and Analysis (IDEA) Center, and Visual Field Assessment Center (VisFACT) also is responsible for quality control of visual fields exams, optic disc stereophotographs and optical coherence tomography images.
Figure 1.
The African Descent and Glaucoma Evaluation Study (ADAGES) Genomics Study Study Design Flow Chart. Outline of specimen and data flow among recruiting sites, the UCSD ADAGES Coordinating Center & BioBank and the UCLA LA Biomedical Research Institute Harbor UCLA.
The following five recruiting sites participated in ADAGES III: University of California at San Diego (Shiley Eye Institute, Hamilton Glaucoma Center, University of California, La Jolla (SD, PI Dr. Weinreb), Edward S. Harkness Eye Institute, Columbia University Medical Center, New York, (NY, PI Dr. Liebmann), University of Alabama at Birmingham (AL, PI Dr. Girkin), University of Texas at Houston (TX, PI Dr. Feldman) and the clinical private practice of Dr. Dubiner, Atlanta, Georgia (GA). The study centers are responsible for participant recruitment, anthropometric measurements, participant interview and EYEPAD data entry, specimen collection, processing, and shipping as outlined below.
Study Population
POAG patients and controls without glaucoma who were of African or European descent by self-report were recruited from the study center clinics and from existing ADAGES and DIGS glaucoma participants. All participants provided written informed consent including widespread data sharing and dbGaP (NIH database of genotypes and phenotypes) deposition, and all procedures complied with the Health Information Portability and Accountability Act, and study centers received approvals by Institution Review Boards utilized by their institutions.
Patients with glaucoma, glaucoma progressors and controls without glaucoma of AD and ED were recruited as outlined in Table 1. Eligibility for inclusion as a POAG patient required glaucomatous visual field damage compatible with glaucomatous optic disc damage with no other ocular or non-ocular disease responsible for the visual field loss. Visual field damage was defined as a pattern standard deviation (PSD) or glaucoma hemifield test (GHT) outside normal limits. If good quality visual fields were not available due to advanced disease, clear documentation of glaucomatous optic disc damage by clinical examination or optic disc photography was required. Glaucomatous optic disc damage was defined as evidence of excavation, neuroretinal rim thinning or notching, localized or diffuse retinal nerve fiber layer (RNFL) defect, or an inter-eye asymmetry of the vertical cup disc ratio > 0.2. Classification as POAG required confirmation by the ADAGES Coordinating Center of the visual field and/or optic disc damage. In order to recruit a representative sample of POAG patients, exclusion criteria were limited to 1) ocular pathology that makes it difficult to determine whether there is characteristic visual field damage, 2) closed or occluded angles, 3) secondary glaucoma, 4) history of human immunodeficiency virus or hepatitis C infection, and 5) Non-African or European descent.
Table 1.
Inclusion and Exclusion Criteria
| Definitions | Glaucoma | ADAGES Glaucoma Progressors | ADAGES/DIGS Glaucoma Progressors | ADAGES Controls without glaucoma | ADAGES/DIGS Controls without glaucoma |
|---|---|---|---|---|---|
| Race | African Descent | African Descent | European Descent | African European | Descent Descent |
| Clinical Exam (chart review) |
|
Open Angles | Open Angles | ||
| Ocular Pathology (chart review) | Some other pathology allowable if there is still clear characteristic visual field damage | Other ocular pathology allowable if there is still clear characteristic glaucomatous visual field damage Other Ocular pathology is NOT allowed if:
|
|
||
| POAG Structural Damage (chart review) | 1 photograph, image, or clinical drawing clearly indicating OAG (no time requirement) |
|
|
||
| POAG visual field (chart review) | 1 reliable glaucomatous visual field required |
|
|
||
Progressing POAG patients as documented by structural changes, functional changes or disc hemorrhage were also recruited. To ensure that progressive changes were not due to other ocular conditions, participants with other ocular pathologies were excluded if 1) VF damage or progression was due to a cause other than POAG, 2) made interpretation of visual fields or photographs uncertain or difficult, or 3) visual acuity was worse than 20/50 due to a cause other than POAG. Glaucomatous progression is determined centrally by the ADAGES Coordinating Center using event-based and rate-based criteria.
African and European descent controls without glaucoma were also recruited. Eligibility for inclusion as a control without glaucoma was based on 1) no evidence of glaucomatous optic neuropathy based on a dilated clinical examination, and 2) an IOP < 21 mmHg. Family history of glaucoma was an exclusion criteria for a control if the individual reported 1) more than 1st degree relative with glaucoma, or 2) a first degree relative that was blind from glaucoma.
Study Visit
The study visit included obtaining informed consent, standardized blood pressure, height and weight measurements, specimen collection, a detailed interview and recording of information from the ophthalmic medical record, and ocular examination. All data were recorded into the Shiley Eye Institute secure custom IPAD-based data entry system, EYEChart, by certified personnel at each study center. Details of each component are outlined below.
Standardized blood pressure, height and weight measurements
Standardized protocols and instruments were used at each study center to obtain blood pressure, height, weight and heart rate measurements. Measurements for blood pressure were performed before any other measurements or blood drawing procedure. Blood pressure and pulse rate were measured using the Omron Automatic Blood Pressure (Model BP791IT) after participants were in a seated position for 5 minutes. Two measurements were taken, 5 minutes apart. If the two recordings were not within 10 mm Hg of each other, a 3rd measurement was completed. Height was measured to the 8th of an inch using the Seca Stadiometer (Model 213). Weight was measured in pounds using the Health-o-Meter Professional Remote Digital Scale after removal of jackets and/or bulky sweaters.
Interview
The standardized interview included a detailed assessment of the ancestry of the participants and their parents, marital status, education level, socioeconomic status, participant and parent’s history of non-ocular conditions and their treatment, participant’s, spouse’s, and first degree relatives’ history of ocular conditions, history of medical and surgical treatment for ocular conditions, history of over the counter medication and vitamin use, and past and current physical activity, smoking and alcohol use.
Ocular conditions and their treatment: ocular exam
Detailed information on the participants’ current and past ocular conditions and treatment was extracted from the participant’s ophthalmic medical record and entered into the EYEChart database entry system. Specifically, information on the following ocular tests was recorded: intraocular pressure (current and highest), visual acuity, central corneal thickness, refractive status, axial length, and gonioscopy. Information on the ophthalmic clinical examination of the anterior segment included detailed assessment of the cornea and conjunctiva, lens and iris. Information on the dilated examination of the optic nerve head included assessment of the optic nerve head and whether there was evidence of glaucomatous optic neuropathy (i.e. neuroretinal rim thinning, retinal nerve fiber layer thinning). Evidence of disc hemorrhages, peripapillary atrophy, large cup disc ratio, asymmetry between eyes, ophthalmic features that were not considered clear glaucomatous optic neuropathy was also recorded. Information on age-related macular degeneration, diabetic retinopathy, epiretinal membrane, drusen, macular edema, hemorrhages, retinal pigment epithelial changes, proliferative vitreoretinopathy and retinal dystrophies was also recorded from the dilated fundus examination.
Specimen Collection
The blood and saliva specimen collection protocol was designed to provide serum, plasma, DNA and RNA for multiple-omics and biochemistry analyses. Blood samples were collected in the following tubes 1) CPT tubes, 2) EDTA tubes, 3) paxgene RNA tube and 4) a saliva collection tube (Oragene, city and state). When possible, fasting blood samples were obtained.
The custom ADAGES III barcode labeling system, designed by the ADAGES Coordinating Center and Biobank, was used to label the study logs and forms, specimen collection tubes, transfer and storage tubes for ADAGES III specimens and documents. After certification by the ADAGES Biobank, technicians at each study center receive a secure log-in for entering participant information and printing unique bar-code labels for placement on specimen tubes, and acquisition and transfer logs. The ADAGES BioBank and LABioMed also access and print the bar-coded label system to label the processed specimens, storage boxes, and for use in their data management systems.
Blood specimen collections were processed into aliquots of whole blood, plasma, and serum. All aliquots were stored at −80C. DNA was isolated from the buffy coat layer of samples collected in EDTA tubes using standard isolation procedures. Saliva samples were collected in Oragene ORG-500 kits for DNA isolation. Peripheral blood mononuclear cells (PBMC) were isolated from the samples collected in CPT tubes and cryopreserved. Whole blood was collected in PAXgene RNA tubes and stored at −80C for future RNA isolation.
Genotyping Data
Genotyping of ADAGES cases is being performed at the Institute for Translational Genomics and Population Sciences at Los Angeles Biomedical (LABioMed) Research Institute, at Harbor-UCLAMedical Center. DNA is isolated from either whole blood or saliva using Qiagen kits in an automated system. Genotyping will be conducted with a GWAS panel (the Illumina Multi-Ethnic Genotyping Array (MEGA), Illumina, San Diego, CA); this chip contains SNPs specific for the study of African-American admixture (AIMS) and for exons of genes.
Genotyping data from the MEGA array for 1870 (exclude DM)/2048(include DM) “African-American convenience controls” will be provided in the form of raw intensity scores genotyped at the Genomics Core at Wake Forest School of Medicine. The studies include 1076 controls from the Wake Forest Diabetic and Non-Diabetic Kidney Disease studies32–34, 489 controls from the Insulin Resistance Atherosclerosis Family Study (IRASFS)35, 157 controls from the Insulin Resistance Atherosclerosis Study (IRAS)36 and 148 controls from the Arkansas African American Metabolic Cohort 37. The raw intensity scores (“idat files”) from ADAGES, generated by the LA Biomedical Institute and from the controls, generated by the Wake Forest School of Medicine team, will be combined into a single analysis for combined calling of alleles using Illumina software (Genome Studio). For additional quality control, the minor allele frequencies of the SNPS in the ADAGES controls will be compared with the minor allele frequencies in the Wake Forest controls. Thus both classical single variant as well as gene-based tests (also known as burden tests for rare variants) will be possible for both case-control (glaucoma presence/absence) as well as within case analyses (quantitative phenotypes).38–41
To expand ED controls, we have established collaborations with the Multi-Ethnic Study of Atherosclerosis (MESA) study which had GWAS data on 2623 ED controls. Given that the MESA GWAS was conducted on a different platform, we genotyped 86 MESA sample together with the ADAGES sample on the same MEGA array. Using the 86 samples with both GWAS chip data, we may filter out SNPs with poor imputation quality score.42
MESA (Multi-Ethnic Study of Atherosclerosis)
The Multi-Ethnic Study of Atherosclerosis (MESA) is a study of the characteristics of subclinical cardiovascular disease and the risk factors that predict progression to clinically overt cardiovascular disease or progression of the subclinical disease. MESA consisted of a diverse, population-based sample of an initial 6,814 asymptomatic men and women aged 45–84. 38 percent of the recruited participants were white, 28 percent African American, 22 percent Hispanic, and 12 percent Asian, predominantly of Chinese descent. Participants were recruited from six field centers across the United States: Wake Forest University, Columbia University, Johns Hopkins University, University of Minnesota, Northwestern University and University of California - Los Angeles. The first examination took place over two years, from July 2000 – July 2002. It was followed by four examination periods that were 17–20 months in length. Participants have been contacted every 9 to 12 months throughout the study to assess clinical morbidity and mortality. 43
Statistical Analysis
All analyses herein were completed using the patient as the unit of analysis. For eye specific measurements, the mean of both eyes was calculated for continuous variables, and the worse eye for categorical variables. Statistical comparisons were completed to identify racial differences and to identify differences between cases and controls. Specifically, we analyzed racial differences between AD and ED glaucoma patients, AD and ED progressing glaucoma patients, and AD and ED controls without glaucoma. In addition, we completed analyses stratified by race to compare AD cases and controls, and ED cases and controls. T-tests or Wilcoxon (Mann-Whitney) tests were used to evaluate continuous variables, and a chi-square test of association for categorical variables. Adjusted analyses were performed through using linear and generalized linear regression models including age as a covariate. In cases in linear modelling where the response was not normally distributed (e.g. skewness), a suitable transformation was utilized (Box Cox). A p-value <.05 was considered statistically significant. All statistical analyses were conducted using STATA 12.1 for Windows (StataCorp LP, College Station, TX) and R 3.1.0 (http://www.r-project.org).
Using the gwas data and AIMs on the genotyping chip, we will first carry our principal component analysis using smartPCA program.44 Top PCAs will be derived and adjusted in the following association analyses to account for potential population structures. Association between genetic variants and glaucoma as well as with quantitative measures will be carried out using linear mixed models while adjusting for age and top principle components. These analyses will be carried out using the program EPACT 45 and GENESIS 46–48, which have implemented statistical methods such as EMMAX 45 to efficiently carry-out the proposed tests. Quantile-quantile plots of results will be examined and genomic control will be applied as needed.
Power Considerations
We estimated power for both AD and ED studies. In AD, we have 2146 cases and a total of 2237 controls. For ED, we have 695 cases and 2850 controls. Power was calculated for candidate gene/SNP hypothesis (significance level of 0.05 for a single SNP, 0.005 for 10 SNPs), and for the multiple testing that occurs with a genome wide association analysis (p<5×10−8). Thus, for single SNP at genome wide significance level in the AD population, our study will have ≥ 86% power to identify novel SNPs with relative genetic risk of 1.4 and MAF of 0.2 (Table 2a). For the confirmation study of known genes associated with POAG, we calculated power at a significance level of 0.005 (=0.05/10 (POAG genes). Table 2a shows that we will have ≥88% power in AD population when the relative genetic risk is 1.2 and MAF is 0.3. The goal of the GWAS study is to identify novel loci. We understand that we can’t identify all risk loci in the current study. Power was therefore estimated power to identify at least one locus, assuming there are a total of 25, or a total of 50 loci, that are associated with the underlying risk of POAG. Our study will have 83% power to identify at least one SNP with genetic relaticve risk of 1.2 and MAF of 0.3 when there are a total of 25 risk SNPs, and a power of 97% if there are a total of 50 risk SNPs (Table 2b).
Table 2.
Power Calculations for 2146 African descent cases and 2237 African descent controls
| Table 2a. Power to detect known and novel SNPs | Table 2b. Power to identify at least one SNP | |||||
|---|---|---|---|---|---|---|
| MAF | Genetic Relative Risk | At a significance level of | Assuming existence of | |||
| 0.05 | 0.005* | 5.00E-08** | 25 SNPs | 50 SNPs | ||
| 0.1 | 1.1 | 0.28 | 0.07 | 0.0001 | 0.002 | 0.005 |
| 1.2 | 0.75 | 0.43 | 0.002 | 0.06 | 0.12 | |
| 1.4 | 0.99 | 0.98 | 0.33 | 0.99 | 0.99 | |
| 0.2 | 1.1 | 0.44 | 0.16 | 0.0001 | 0.002 | 0.005 |
| 1.2 | 0.94 | 0.75 | 0.02 | 0.47 | 0.72 | |
| 1.4 | 0.99 | 0.99 | 0.86 | 0.99 | 0.99 | |
| 0.3 | 1.1 | 0.54 | 0.23 | 0.0003 | 0.008 | 0.008 |
| 1.2 | 0.98 | 0.88 | 0.069 | 0.83 | 0.97 | |
| 1.4 | 0.99 | 0.99 | 0.97 | 0.99 | 0.99 | |
P-value for confirming known SNPs
P-value for identifying a novel SNP
For the ED population, we will have ≥80% power to identify novel SNPs with genetic relative risk of 1.5 and MAF 0.3 at genome wide significance level; and ≥80% power for the confirmation study of known genes when the relative genetic risk is 1.3 and MAF of 0.2. The power to identify at least one SNP is greater than 91% when there are a total of 25 risk SNPs and the SNP has genetic relative risk of 1.3 and MAF of 0.3. Assuming a total of 50 risk SNPs, we will have 84% power to identify a SNP with relative genetic risk of 1.3 and MAF of 0.2.
For quantitative phenotypes, we calculated power for ADAGES samples: 2146 AD and 695 ED. At genome-side significance level (p=5×10−8), we will have sufficient power (>=80%) to identify novel genes when it explains 1.9% variance in AD, and 5.5% in ED. The confirmation study will have >85% power to identify association when SNPs it explains 0.7% variance in AD sample, or >82% power when the SNP explains 2% of variance in ED sample.
Results
Study Population and Baseline Characteristics
Study recruitment began on August 1, 2014 and ended on August 31 2016. This report summarizes results on the 3266 participants who met eligibility criteria. After centralized review of the patient information, visual fields and stereophotographs by the ADAGES III Coordinating Center, 2146 AD POAG patients, 695 ED POAG patients, 198 AD controls, and 227 ED controls are included in this report. Recruitment by site, race and study group is presented in Table 3. Demographic information and ocular characteristics are presented in Tables 4 and 5. Although blood samples are not available on 222 (6.8%) of the 3266 participants, we have saliva for DNA extraction on 183/222 (82.4%) of these participants, leaving only 39 (1.1%) without a source of DNA for analysis. All eligible participants are included in the demographic analyses.
Table 3.
Recruitment by Site, Race and Study Group
| Site | Glaucoma Patients | Controls Without Glaucoma | Total | ||
|---|---|---|---|---|---|
| African Descent | European Descent | African Descent | European Descent | ||
| Alabama | 450 (21.0%) | 128 (18.4%) | 28 (14.1%) | 43 (18.9%) | 649 (19.9%) |
| Georgia | 331 (15.4%) | 83 (11.9%) | 95 (48.0%) | 74 (32.6%) | 583 (17.9%) |
| New York | 1093 (50.9%) | 63 (9.1%) | 35 (17.7%) | 0 (0.0%) | 1191 (36.5%) |
| San Diego | 106 (4.9%) | 343 (49.4%) | 21 (10.6%) | 12 (5.3%) | 482 (14.8%) |
| Texas | 166 (7.7%) | 78 (11.2%) | 19 (9.6%) | 98 (43.2%) | 361 (11.0%) |
| Total | 2146 (100.0%) | 695 (100.0%) | 198 (100.0%) | 227 (100.0%) | 3266 (100.0%) |
Table 4.
Demographic Information on Glaucoma Patients and Controls
| - | - | - | - | - | - | p-values (Mann Whitney for categorical) * age adjusted | ||
|---|---|---|---|---|---|---|---|---|
| Glaucoma Patients | Glaucoma-free Controls | Glaucoma Patients AD vs ED | AD Glaucoma vs normals | ED Glaucoma vs normals | ||||
| AD | ED | AD | ED | |||||
| N | 2147 | 695 | 198 | 227 | ||||
| mean (95% CI) | mean (95% CI) | |||||||
| Demographic information | Age at Blood Draw (mean, years) |
(n=2146) 67.4 (67.0, 67.9) |
(n=695) 73.4 (72.6, 74.1) |
(n=198) 67.4 (66.1, 68.6) |
(n=227) 70.2 (69.4, 71.0) |
<0.001 | 0.603 0.001 |
<0.001 |
| BMI* (mean) | (n=1908) | (n=658 | (n=194) | (n=219) | ||||
| 29.6 (29.3,29.9) | 26.5 (26.2,26.9) | 31.3 (30.3, 32.2) | 29.2 (28.4, 30.0) | <0.001 | <0.001 | |||
| n (%) | n (%) | |||||||
| Gender (frequency) | (n=2147) | (n=695) | (n=198) | (n=227) | 0.759 | <0.001 | 0.064 | |
| female | 1202 (56.0%) | 384 (55.2%) | 139 (70.2%) | 142 (62.6%) | ||||
| male | 944 (44.0%) | 311 (44.7%) | 59 (29.8%) | 85 (37.4%) | ||||
| Education (frequency) | (n=2089) | (n=691) | (n=196) | (n=226) | <0.001 | 0.008 | 0.026 | |
| Did Not Complete HS | 320 (15.3%) | 17 (2.5%) | 22 (11.2%) | 9 (4.0%) | ||||
| Completed HS | 590 (28.2%) | 100 (14.5%) | 55 (28.1%) | 47 (20.8%) | ||||
| 1+ Years of College | 932 (44.6%) | 358 (51.8%) | 107 (54.6%) | 116 (51.3%) | ||||
| 1+ Years of Grad School | 247 (11.8%) | 216 (31.3%) | 12 (6.1%) | 54 (23.9%) | ||||
| Marital Status (frequency) | (n=1972) | (n=661) | (n=196) | (n=225) | <0.001 | 0.704 | 0.526 | |
| Divorced/Separated | 399 (20.2%) | 66 (10.0%) | 40 (20.4%) | 29 (12.9%) | ||||
| Domestic Partner | 7 (0.4%) | 10 (1.5%) | 1 (0.5%) | 4 (1.8%) | ||||
| Married | 811 (41.1%) | 434 (65.7%) | 88 (44.9%) | 152 (67.6%) | ||||
| Single | 415 (21.0%) | 47 (7.1%) | 40 (20.4%) | 12 (5.3%) | ||||
| Widowed | 340 (17.2%) | 104 (15.7%) | 27 (13.8%) | 28 (12.4%) | ||||
| Lifestyle | Smoking (frequency) | (n=2074) | (n=683) | (n=195) | (n=225) | <0.001 | 0.035 | 0.754 |
| Never | 1260 (60.8%) | 367 (53.7%) | 106 (54.4%) | 125 (55.3%) | ||||
| Former Smoker | 650 (31.3%) | 288 (42.2%) | 63 (32.3%) | 93 (41.2%) | ||||
| Current Smoker | 164 (7.9%) | 28 (4.1%) | 26 (13.3%) | 8 (3.5%) | ||||
| Alcohol Consumption (frequency) | (n=2070) | (n=683) | (n=196) | (n=225) | <0.001 | 0.016 | <0.001 | |
| Current Consumer | 612 (29.6%) | 404 (59.1%) | 74 (37.8%) | 103 (45.8%) | ||||
| Systemic Conditions* | Diabetes (frequency) | (n=2110) | (n=695) | (n=193) | (n=226) | |||
| Diabetes Type 1 | 14 (0.7%) | 1 (0.1%) | 2 (1.0%) | 4 (1.8%) | 0.183 | 0.554 | 0.027 | |
| Diabetes Type 2 | 658 (31.2%) | 77 (11.1%) | 60 (31.1%) | 42 (18.6%) | <0.001 | 0.991 | 0.002 | |
| Cardiovascular Disease (frequency) | (n=2110) | (n=693) | (n=193) | (n=226) | 0.001 | 0.088 | 0.889 | |
| Yes | 265 (12.6%) | 146 (21.0%) | 32 (16.6%) | 40 (17.7%) | ||||
| Systemic Hypertension (frequency) | (n=2110) | (n=693) | (n=193) | (n=226) | <0.001 | 0.126 | 0.254 | |
| Yes | 1454 (68.9%) | 356 (51.4%) | 144 (74.6%) | 117 (51.8%) | ||||
| Treatment for Systemic Conditions | Taking Diabetes Medications (frequency) | (n=645) | (n=77) | (n=60) | (n=42) | <0.001 | 0.360 | <0.001 |
| Yes | 509 (78.9%) | 56 (72.7%) | 39 (65.0%) | 34 (81.0%) | ||||
| Taking Systemic Hypertension Medications (frequency) | (n=1427) | (n=342) | (n=144) | (n=117) | <0.001 | 0.830 | 0.011 | |
| Yes | 1036 (72.5%) | 258 (72.7%) | 85 (59.0%) | 89 (76.1%) | ||||
Table 5.
ADAGESIII Ocular Information by Study Group
| - | - | - | - | - | - | p-values (Mann Whitney for categorical)* age-adjusted | ||
|---|---|---|---|---|---|---|---|---|
| Glaucoma Patients | Controls without glaucoma | Glaucoma Patients AD vs ED | AD Glaucoma vs controls | ED Glaucoma vs controls | ||||
| AD | ED | AD | ED | |||||
| N | 2146 | 695 | 198 | 227 | ||||
| mean (95% CI) | mean (95% CI) | |||||||
| Visual Field Mean Deviation (dB) | n=1855 | n=673 | ||||||
| Visual Field Mean Deviation (dB) | −9.54 (−9.94, −9.14) | −8.17 (−8.65, −7.69) | - | - | < 0.001 | NA | NA | |
| IOP (mm Hg) | 16.9 (16.6 17.1) | 14.5 (14.1, 14.9) | 15.6 (15.2, 16.0) | 14.7 (14.4, 15.1) | <.001 | 0.016 | 0.185 | |
| Central Corneal Thickness | 533.7 (531.9, 535.5) | 544.1 (540.9 547.2) | NA | NA | <.001 | NA | NA | |
| Spherical Equivalent | −0.57 (−0.68, −0.46) | −0.76 (−0.93, −0.59) | −0.16 (−0.49, −0.18) | −0.28 (−0.57, 0.00) | <.001 | 0.001 | <0.001 | |
| n (%) | n (%) | |||||||
| Visual Acuity, worse eye (frequency) | 2138 | 694 | 198 | 225 | <0.001 | <0.001 | 0.072 | |
| equal or better than 20/20 | 264 (12.3%) | 119 (17.1%) | 44 (22.2%) | 47 (20.9%) | ||||
| 20/25 to 20/40 | 937 (43.8%) | 351 (50.5%) | 106 (53.5%) | 123 (54.7%) | ||||
| 20/50 to 20/160 | 482 (22.5%) | 142 (20.5%) | 35 (17.7%) | 43 (19.1%) | ||||
| equal or worse than 20/200 | 457 (21.4%) | 83 (12.0%) | 13 (6.6%) | 12 (5.3%) | ||||
| Glaucoma Treatment | Glaucoma Medication | 1975 (92.7%) | 592 (85.7%) | NA | NA | <.001 | ||
| Past Glaucoma Surgery | 765 (37.9%) | 458 (66.5%) | NA | NA | <.001 | |||
| Other Ocular Conditions | Pseudophakia | 834 (40.1%) | 389 (62.1%) | 37 (20.1%) | 65 (29.1%) | <.001 | <.001 | <.001 |
| Diabetic Retinopathy | 85 (4.2%) | 0 (0%) | 11 (6.0%) | 2 (0.9%) | NA | 0.248 | NA | |
| Macular degeneration | 25 (1.2%) | 17 (2.4%) | 3 (1.5%) | 5 (2.2%) | 0.351 | 0.539 | 0.466 | |
| Cataract Surgery | 1005 (48.3%) | 429 (68.59%) | 67 (36.4%) | 96 (43.0%) | <.001 | 333333 | <.001 | |
| Family History of Glaucoma | 1st degree relative | 1013 (47.6%) | 280 (40.9%) | 32 (16.4%) | 22 (9.9%) | 0.121 | <0.001 | <0.001 |
One hundred thirty two participants recruited for participation were excluded due to 1) Non-African or European descent (POAG: n=9, Controls: none), 2) absence of documented glaucomatous structural or functional damage (n=20 AD, n=11 ED), 3) secondary causes of glaucoma: n=22 AD, n=12 ED, Controls: 0), 4) ) tested positive for human immunodeficiency virus (HIV) or reported a history of hepatitis B or hepatitis C infection (POAG: n=33 AD, n=7 ED, Controls: n=2 AD, n=2 ED), 5) self-withdrawal from the study (POAG: n= 2 AD, n= 3 ED, Controls: n=1 AD, 0 ED), 6) did not meet inclusion criteria for a control (n=7 AD, n=1 ED). These excluded participants are not included in the analyses.
Demographic information and clinical characteristics of the AD and ED glaucoma patients and controls are presented in Table 4. The mean (95% CI) age of the AD glaucoma patients was similar to the AD control subjects, 67.4 (67.0, 67.9) years and 67.4 (66.1, 68.6) years, respectively. The ED glaucoma patients were significantly (p< 0.001) older than the ED controls; the mean (95% CI) age was 73.4 (72.6, 74.1) years and 70.2 (69.4, 71.0) years, respectively. In addition, AD POAG patients were significantly (p< 0.001) younger than ED POAG patients. For these reasons, adjustment for age was completed for relevant analyses. The majority of participants were female, with both the AD and ED control groups having a significantly higher proportion of female participants than the AD and ED glaucoma patient groups. Compared to ED POAG patients, fewer AD POAG patients attended college, were currently married, had ever smoked, and currently consume alcohol (all p <0.001).
Systemic conditions
Compared with POAG patients of ED, a significantly (p<0.001) larger proportion of AD POAG patients reported having a diagnosis of systemic hypertension (51.4% and 68.9%, respectively) and type 2 diabetes (11.1% and 31.2%, respectively). However, a significantly (p=0.001) smaller proportion of AD glaucoma patients reported having a diagnosis of cardiovascular disease (12.6%) compared to ED glaucoma patients (21.0%). AD glaucoma patients were similar to AD controls with respect to self-reported history of type II diabetes, cardiovascular disease and systemic hypertension. A similar proportion of ED glaucoma patients and controls reported a diagnosis of cardiovascular disease and systemic hypertension, but ED controls reported more type II diabetes (18.6%) than ED glaucoma patients (11.1%) (p=0.002).
Ocular characteristics
The ocular characteristics of the study population are presented in Table 5. Glaucoma patients of AD had higher mean IOP, lower mean CCT, and were more likely to be taking glaucoma medications, but less likely to have a history of glaucoma surgery than patients of ED (all p<0.001). The visual acuity of AD glaucoma patients was significantly worse than those of ED patients; the proportion of patients with a visual acuity 20/50 or worse was 43.9% and 32.5% (p<0.001), respectively. Glaucoma patients of AD were also more likely to have a first degree relative with glaucoma compared to patients of ED (47.6% and 40.9%, respectively), but the difference did not reach statistical significance after adjusting for age (unadjusted p =0.002, age adjusted p-value=0.121). Finally, AD glaucoma patients had significantly worse mean visual field MD (−9.54 db) compared to ED glaucoma patients (−8.17 dB) after adjusting for age (p<0.001). Historical visual field information is available on 1426 glaucoma patients. These patients with past VF testing data available have a mean of 7.6 visits and 5.6 years of follow-up.
Discussion
ADAGES III is designed to identify glaucoma genes in individuals of African descent. Identification of genes associated with POAG in this high-risk, minority population may lead to improved predictive models for diagnosis and progression detection, to the identification of pathways in the pathophysiology of POAG, and to the discovery of new drug targets for therapies to reduce the impact of glaucoma blindness for both the AD population, as well as all glaucoma patients. Through its deep phenotyping, and extensive biobanking of cell lines, serum, plasma and RNA, ADAGES III will provide important data for improving our understanding of the pathophysiology of POAG in this high-risk population.
Several lines of evidence support the paradigm that there is a genetic contribution to primary open angle glaucoma (POAG). First, there is a high concordance rate of POAG in monozygotic twins9,49 and up to 50% of POAG patients have a positive family history. 50 First-degree relatives of glaucoma patients have a 22% lifetime risk of developing POAG, compared to 2.3% risk in family members of non-glaucoma controls.51 The overall risk of developing POAG in 1st degree relatives is 7 to 10 times higher than the general population.50–52 Second, several rare Mendelian autosomal dominant or recessive diseases with glaucoma are well-known in pediatrics; subsequent work among these has identified mutations in the MYOC gene that contribute to juvenile open-angle glaucoma (Online Mendelian Inheritance in Man (OMIM) #137750); in PITX2 (OMIM#137600), FOXC1 (OMIM#601631), and PAX6 (OMIM#604229) that contribute to anterior segment dysgenesis; in CYP1B1 (OMIM#617315 & #231300) and LTBP2 (OMIM#613086) to congenital glaucoma; and in OPTN (OMIM#137760 & #606657) to familial normal-tension glaucoma.53 Based on this evidence, genome-wide association studies have been conducted more recently to find common variants contributing to adult-onset glaucoma with success, beginning with the observation of a strong association with the 9p21 region.54,55 Strikingly, this same locus also has a strong association with many adult-onset, common diseases, such as coronary artery disease.56,57 A recent meta-analysis of 3,853 cases and 33,480 controls from 8 datasets has demonstrated definitive, genome-wide significance, for the 9p21 region, and genes TMCO1, AFAP1, FOXC1, ABCA1, ATXN2, SIX6, GAS7, TXRND2.58
In addition, a number of novel associations have recently been identified by GWAS including variants near the genes CAV1 and CAV259,60, SIX1 and SIX661, SRBD162, CDKN2B54, TMCO154,61, and ELOVL5.62 In addition, ONH structural features (cup disc ratio, rim area, cup area, and total disc area) and other risk factors (IOP, CCT) have been analyzed extensively as POAG endophenotypes. 60,63–67 Specifically, CDKN2B 67, SIX1/665, ATOH763, CDC763, and CHEK268 have been found to be associated with ONH structural measurements while IOP has been linked to genetic factors near the genes TMCO169, CAV169, CAV270, FNDC3B69, and GAS7.71 Finally, CCT has been associated with FOXO172, ZNF46973, COL5A74, and FNDC3B.74 Recent meta-analyses of large cohorts have replicated many of these results and provided some evidence for additional genetic associations.68,75 Of note, these results are from populations of European descent and there are few studies on the genetics of POAG in populations of African descent. The Barbados Family Study of Open-Angle Glaucoma and a study of West Africans have shown regions on chromosomes 2p, 2q, and 10p to be linked to POAG76,77 and regions on 5q and 14q to elevated IOP78 in populations with African descent. Genes for these loci have not yet been clearly characterized. Candidate gene studies that have included Africans or African-Americans also confirm the association of 9p21 and perhaps SIX6. 79 Since up to now genetic studies of subjects with African descent have been much smaller, additional and larger genetic studies of POAG in African-Americans are warranted.
ADAGES III will test top results from large established GWAS cohorts of glaucoma in individuals of ED, the NEIGHBOR and GLAUGEN NEIGHBORHOOD (n=4500 cases) consortium,80,81 a unique collaborative effort involving investigators located throughout the United States. The goal of the consortium is to identify genetic variants associated with POAG in ED using an initial approach of GWAS. The eventual outcome of this work is to elucidate the molecular pathogenesis of POAG making it possible to implement effective screening and prevention strategies and to develop improved and novel therapies. We plan to identify whether and how the top results in subjects with AD are generalizable to individuals of ED through collaboration with the NEIGHBORHOOD consortium. More recently, the Primary Open-Angle African American Glaucoma Genetics (POAAGG) study,82 a large single-site study in Pennsylvania was initiated to investigate the genetics of African Americans. Other smaller studies probably have limited power to identify genes associated with POAG in AD individuals.83 ADAGES contribution will come both from its sample size and its detailed phenotyping, allowing assessment of the genetic contribution of glaucoma related quantitative traits, as well as comparing progression to non-progression. This paper describes the composition and types of data of the ADAGES III study. The study initially plans individual analyses utilizing the data described herein. In the longer term, the investigators of the study fully realize that the greatest success in the genomics field has come from consortium efforts that combine from a number of studies.58,84 We have initiated such efforts at establishing an African origin POAG GWAS consortium, specifically to maximize the power of several different investigative teams.
An advantage of this study is that participants are recruited from different regions in the United States. While the largest single recruitment of AD subjects were ascertained from New York, the study included many additional subjects; in order, from Alabama, Georgia, Texas and San Diego. We know that the ethnic background of the AD community in New York City population consists of a diverse group of immigrants including persons from the United States, the Caribbean, South America, and Africa, while the AD community in Alabama and Georgia consists mostly of individuals born in the South of the United States.85,86 The history of the AD population in the United States suggests that the genetic composition of this population may be modeled as an admixed population with genetic contribution from the continents of West Africa and Europe. While European and West African populations diverged as the result of different selective pressures over millennia, these populations were mixed as the result of the slave trade that brought West Africans to the Americas beginning in the 16th Century. This admixture creates two characteristics of the genomes of AD individuals: (1) mosaic chromosomes containing long segments from distinct continents87,88, but yet (2) shorter linkage disequilibrium within segments from African populations89–91. We have shown that in the ADAGESII cohort, the median African admixture among 244 self-reported AD was 92.0% (IQR: 75.4–97.5%), and among 245 self-report ED subjects was 0.54% (IQR: 0.39–0.98%). We found92 that a higher African admixture proportion was significantly correlated to a thinner cornea (ρ =−0.27; p<0.0001) and a larger disc area (ρ= 0.15; p<0.0001), and marginally associated with RNFL thickness (ρ = 0.20; p-value =0.092) in models adjusting for age, gender and diagnostic category. These characteristics present both the challenge to properly account for the population structure and to lower the false positive rate of the association study and, at the same time, the opportunity to perform joint ancestry and association testing.93 Therefore, this study will reflect the diverse genetic make-up of the individuals of AD in the United States, and evaluate whether biogeographic ancestry plays a role in the genetics of POAG in this population. Another advantage of this study is that a subset of participants are ADAGESII and DIGS participants with over 10 years of extensive ocular measurements and structural and functional testing. These data present a unique opportunity to analyze quantitative traits of progressing glaucoma patients at the highest risk of visual impairment and blindness from the disease.
The 3266 individuals recruited to participate in ADAGES III are included in this report. Consistent with other studies,94,95 the ADAGES III AD glaucoma patients are younger, have more self-reported diabetes and systemic hypertension, and have a higher BMI than ED glaucoma patients. For this reason, we controlled for the influence of age in our analyses. In addition, despite being younger, AD glaucoma patients have worse severity of disease as characterized by visual field MD, and worse visual acuity than the ED glaucoma patients enrolled in the study. AD glaucoma patients are also significantly less likely to have had cataract and glaucoma surgery than ED glaucoma patients, but are more likely to be on glaucoma medications (age-adjusted p<0.001 for all comparisons).
There are several possible limitations to this study. First, ADAGES recruited participants from 4 academic centers and one private practice, and therefore the enrollees may not be fully generalizable to the population of African and European descent individuals with POAG. However, its large sample size and inclusion of patients from 5 geographic locations provides a diverse cohort of AD and ED participants21 and therefore enhances its likelihood of generalizability to the U.S. AD population. Moreover, although the recruitment in New York, Alabama, San Diego and Texas are based at academic centers, satellite clinics of general ophthalmic practices in NY were utilized as recruitment centers and the GA study center was based in a large private ophthalmic practice. It is still possible, however, that there may be biases in the referral to academic centers that affects the generalizability of the results. Second, there are a small number of AD and ED controls without glaucoma participating in the study. A variety of convenience controls will be utilized to increase the size of the control group and improve the power to detect both common and rare variants in the case control analysis. However, as this is a descriptive report comparing characteristics of the various diagnostic groups and does not include hypothesis testing, we did not formally adjust the p-values for multiple comparisons.
In summary, with its large sample size, extensive specimen collection and deep phenotyping of glaucoma patients and controls of AD and ED from different regions in the United States, the ADAGES III Genomics Study will address gaps in our knowledge of the genetics of POAG in this high-risk population. The planned genotyping will improve our understanding of the genetics of POAG in individuals of AD and set the stage for developing methods for individualized assessment of the risk of developing progressive POAG, and personalized treatment options.
Supplementary Material
Acknowledgments
Finanicial Support
National Eye Institute: EY023704, P30EY022589, EY110008, EY019869, EY021818
National Institutes of Health: R01 DK087914, R01 DK066358, R01 DK053591, U01 DK105556, R01 HL56266, R01 DK070941, DRC DK063491, and CTSI UL1TR001881
Appendix: ADAGES III Genomics Study Group
Investigators: Robert N. Weinreb MD (Principal Investigator), Jerome I Rotter MD (Principal Investigator), Linda M. Zangwill PhD, Radha Ayyagari PhD, Jeffrey M Liebmann MD, Christopher A Girkin MD, Robert Feldman MD, Harvey Dubiner MD, Yii-der I Chen PhD, Xiuqing Guo PhD, Kent D Taylor PhD
Executive Committee: Robert N. Weinreb MD, Jerome I Rotter MD, Linda M. Zangwill PhD, Radha Ayyagari PhD, Jeffrey M Liebmann MD, Christopher A Girkin MD, Robert Feldman MD, Harvey Dubiner
ADAGESIII Resource Centers: (Principal Investigators are marked in italics)
ADAGESIII Data Coordinating Center
Linda M. Zangwill, PhD, Keri Dirkes MPH1, Eunice Williams-Steppe1, Suzanne Vega MPH, Maria Hunsicker, Naama Hammel MD1, Luke J Saunders PhD, Dan Auerbach, MS, Ken Newell,
ADAGESIII University of California San Diego Biobank:
Radha Ayyagari PhD, Matthew Holman, Natalie Kline, Heidi Amundson
ADAGESIII LaBioMed Genomics Biobank:
Jerome I Rotter MD2 Yii-der I Chen PhD2, Kent D Taylor PhD2, Xiuqing Guo PhD2, Kevin Sandow2, Kathryn Roll,2 Kelvin Lam Ubaydah Nasri2, Zorayr Arzumanyan2, Yeheng Liu2,
ADAGESIII Reading Centers
Visual Field Assessment Center (VisFACT): Hamilton Glaucoma Center:
Christopher Bowd, PhD, Linda M. Zangwill, Keri Dirkes, MPH, Suzanne Vega, MPH, Maria Hunsicker,
Imaging Data Evaluation and Assessment (IDEA) Reading Center: Hamilton Glaucoma Center:
Linda M. Zangwill, PhD, Christopher Bowd, PhD, Keri Dirkes, MPH, Suzanne Vega, MPH Maria Hunsicker,
ADAGESIII Clinical Centers: (Principal Investigators are marked in italics)
Hamilton Glaucoma Center, Department of Ophthalmology, University of California, San Diego,
Investigators: Robert N. Weinreb, MD, Felipe A. Medeiros, MD, PhD, Rigby Slight, MD;
Clinical Staff and Coordinators: Eunice Williams-Steppe, Tess Acera, Eric Cabezas, Joy McDonald, Veronica Rubio, Ana Marquez
New York Eye and Ear Infirmary and Columbia University
Investigators: Jeffrey M. Liebmann, MD, Joseph Panarelli MD, Lama A. Al-Aswad, Sung Chul Park, MD, Celso Tello MD, Jeremy Cotliar, MD, Rajendra Bansal, MD, Paul Sidoti MD, George A. Cioffi MD, Dana Blumberg MD, Robert Ritch MD, Elena Ilitchev, MD; Carlos Gustavo de Moraes, MD, MPH
Clinical Staff and Coordinators: Jeremy Reimann, MSPH, Marzhan Atakulova, William Elias, Shiming Luo, Maya Petashnick, Elizabeth Berg, Michael Deng, Anelya Buss, MD, Lam Lu
Department of Ophthalmology, University of Alabama, Birmingham:
Investigators: Christopher A. Girkin, MD, Brian Samuels MD
Clinical Staff and Coordinators: Demond Wiley, Cecily Range, Tracy Thomas, Alexis Delbridge
University of Texas Health Science Center, Houston, Texas
Investigators: Robert Feldman, MD, Nicholas Bell, MD Lauren Blieden, MD Garvin Davis, MD
Clinical Staff and Coordinators: Laura Baker, Ephrem Melese, Theodore Baker
Eye Care Center Management, Inc., Marrow, Georgia
Investigators:Harvey Dubiner MD
Clinical Staff and Coordinators: Helen Dubiner, Mary Bowser, Tu Vy Nguyen, Steven Williams, Hannah Campos, Tu Tran, Melissa Byrd, Cory Fuller, Christopher Wang
Footnotes
Conflict of Interest
LMZ: Carl Zeiss Meditec, Heidelberg Engineering, Optovue Inc., Topcon Medical System Inc. (financial support)
RA: None
JML: Alcon, Allergan, Bausch & Lomb, Carl Zeiss Meditec, Heidelberg Engineering, Reichert, Valeant Pharmaceuticals (Consultant); Bausch & Lomb, Carl Zeiss Meditec, Heidelberg Engineering, National Eye Institute, Optovue, Reichert, Topcon (financial support)
CAG: National Eye Institute, EyeSight Foundation of Alabama, Research to Prevent Blindness, Carl Zeiss Meditec, Heidelberg Engineering, SOLX (financial support)
RF: None
HD: None
KD: None
MH: None
EWS: None
NH: None
LJS: None
SV: None
KS: None
KR: None
RS: None
DA: None
BCS: None
JP: None
LAA-A: None
SCP: None
CT: None
JC: None
RB: None
PS: None
GAC: None
DB: None
RR: None
NPB: None
LSB: NoneEye
GD: None
FAM: Alcon, Allergan, Bauch + Lomb, Carl Zeiss Meditec, Heidelberg Engineering, Merck, Reichert, Sensimed and Topcon (financial support); Alcon, Allergan, Carl Zeiss Meditec, National Eye Institute, and Reichert (research support); Allergan, Carl Zeiss Meditec, Novartis (consultant)
MCYN: None
SKD: None
NDP: None
JD: None
CDL: None
BIF: None
DWB: None
MAC: None
YIC: None
XG: None
KDT: None
JIR: None
RNW: Aeries Pharmaceutical, Alcon, Allergan, Bausch & Lomb, Eyenovia, Sensimed (Consultant); Heidelberg Engineering, Carl Zeiss Meditec, Genentech, Optovue, Topcon (Financial support)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sommer A, Tielsch JM, Katz J, et al. Racial differences in the cause-specific prevalence of blindness in east Baltimore. N Engl J Med. 1991;325(20):1412–1417. doi: 10.1056/NEJM199111143252004. [DOI] [PubMed] [Google Scholar]
- 2.Wallace J, Lovell HG. Glaucoma and intraocular pressure in Jamaica. Am J Ophthalmol. 1969;67(1):93–100. doi: 10.1016/0002-9394(69)90013-0. [DOI] [PubMed] [Google Scholar]
- 3.Wilson MR. Glaucoma in blacks: where do we go from here? Jama. 1989;261(2):281–282. [PubMed] [Google Scholar]
- 4.Friedman DS, Wolfs RC, O’Colmain BJ, et al. Prevalence of open-angle glaucoma among adults in the United States. Archives of ophthalmology. 2004;122(4):532–538. doi: 10.1001/archopht.122.4.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grant WM, Burke JF., Jr Why do some people go blind from glaucoma? Ophthalmology. 1982;89(9):991–998. doi: 10.1016/s0161-6420(82)34675-8. [DOI] [PubMed] [Google Scholar]
- 6.Hiller R, Kahn HA. Blindness from glaucoma. Am J Ophthalmol. 1975;80(1):62–69. doi: 10.1016/0002-9394(75)90870-3. [DOI] [PubMed] [Google Scholar]
- 7.Wilson R, Richardson TM, Hertzmark E, Grant WM. Race as a risk factor for progressive glaucomatous damage. Ann Ophthalmol. 1985;17(10):653–659. [PubMed] [Google Scholar]
- 8.Munoz B, West SK, Rubin GS, et al. Causes of blindness and visual impairment in a population of older Americans: The Salisbury Eye Evaluation Study. Archives of ophthalmology. 2000;118(6):819–825. doi: 10.1001/archopht.118.6.819. [DOI] [PubMed] [Google Scholar]
- 9.Racette L, Boden C, Kleinhandler SL, et al. Differences in visual function and optic nerve structure between healthy eyes of blacks and whites. Archives of ophthalmology. 2005;123(11):1547–1553. doi: 10.1001/archopht.123.11.1547. [DOI] [PubMed] [Google Scholar]
- 10.Chi T, Ritch R, Stickler D, Pitman B, Tsai C, Hsieh FY. Racial differences in optic nerve head parameters. Archives of ophthalmology. 1989;107(6):836–839. doi: 10.1001/archopht.1989.01070010858029. [DOI] [PubMed] [Google Scholar]
- 11.Mansour AM. Racial variation of optic disc size. Ophthalmic Res. 1991;23(2):67–72. doi: 10.1159/000267091. [DOI] [PubMed] [Google Scholar]
- 12.Tsai CS, Zangwill L, Gonzalez C, et al. Ethnic differences in optic nerve head topography. J Glaucoma. 1995;4(4):248–257. [PubMed] [Google Scholar]
- 13.Varma R, Tielsch JM, Quigley HA, et al. Race-, age-, gender-, and refractive error-related differences in the normal optic disc. Archives of ophthalmology. 1994;112(8):1068–1076. doi: 10.1001/archopht.1994.01090200074026. [DOI] [PubMed] [Google Scholar]
- 14.Zangwill LM, Weinreb RN, Berry CC, et al. Racial differences in optic disc topography: baseline results from the confocal scanning laser ophthalmoscopy ancillary study to the ocular hypertension treatment study. Archives of ophthalmology. 2004;122(1):22–28. doi: 10.1001/archopht.122.1.22. [DOI] [PubMed] [Google Scholar]
- 15.Quigley HA, Brown AE, Morrison JD, Drance SM. The size and shape of the optic disc in normal human eyes. Archives of ophthalmology. 1990;108(1):51–57. doi: 10.1001/archopht.1990.01070030057028. [DOI] [PubMed] [Google Scholar]
- 16.Martin MJ, Sommer A, Gold EB, Diamond EL. Race and primary open-angle glaucoma. Am J Ophthalmol. 1985;99(4):383–387. doi: 10.1016/0002-9394(85)90001-7. [DOI] [PubMed] [Google Scholar]
- 17.Britton RJ, Drance SM, Schulzer M, Douglas GR, Mawson DK. The area of the neuroretinal rim of the optic nerve in normal eyes. Am J Ophthalmol. 1987;103(4):497–504. doi: 10.1016/s0002-9394(14)74271-0. [DOI] [PubMed] [Google Scholar]
- 18.Budde WM, Jonas JB, Martus P, Grundler AE. Influence of optic disc size on neuroretinal rim shape in healthy eyes. J Glaucoma. 2000;9(5):357–362. doi: 10.1097/00061198-200010000-00003. [DOI] [PubMed] [Google Scholar]
- 19.Caprioli J, Miller JM. Optic disc rim area is related to disc size in normal subjects. Archives of ophthalmology. 1987;105(12):1683–1685. doi: 10.1001/archopht.1987.01060120081030. [DOI] [PubMed] [Google Scholar]
- 20.Jonas JB, Gusek GC, Naumann GO. Optic disc, cup and neuroretinal rim size, configuration and correlations in normal eyes. Invest Ophthalmol Vis Sci. 1988;29(7):1151–1158. [PubMed] [Google Scholar]
- 21.Sample PA, Girkin CA, Zangwill LM, et al. The African Descent and Glaucoma Evaluation Study (ADAGES): design and baseline data. Archives of ophthalmology. 2009;127(9):1136–1145. doi: 10.1001/archophthalmol.2009.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Girkin CA, Sample PA, Liebmann JM, et al. African Descent and Glaucoma Evaluation Study (ADAGES): II. Ancestry differences in optic disc, retinal nerve fiber layer, and macular structure in healthy subjects. Archives of ophthalmology. 2010;128(5):541–550. doi: 10.1001/archophthalmol.2010.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Racette L, Liebmann JM, Girkin CA, et al. African Descent and Glaucoma Evaluation Study (ADAGES): III. Ancestry differences in visual function in healthy eyes. Archives of ophthalmology. 2010;128(5):551–559. doi: 10.1001/archophthalmol.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medeiros FA, Leite MT, Zangwill LM, et al. African Descent and Glaucoma Evaluation Study (ADAGES): Rates of Progressive Glaucomatous Visual Field Loss in African Descent and European Descent Patients. Invest Ophthalmol Vis Sci ARVO E: Abstract. 2011 [Google Scholar]
- 25.Khachatryan N, Medeiros FA, Sharpsten L, et al. The African Descent and Glaucoma Evaluation Study (ADAGES): predictors of visual field damage in glaucoma suspects. Am J Ophthalmol. 2015;159(4):777–787. doi: 10.1016/j.ajo.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fazio MA, Grytz R, Bruno L, et al. Regional variations in mechanical strain in the posterior human sclera. Invest Ophthalmol Vis Sci. 2012;53(9):5326–5333. doi: 10.1167/iovs.12-9668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fazio MA, Grytz R, Morris JS, et al. Age-related changes in human peripapillary scleral strain. Biomechanics and modeling in mechanobiology. 2013;13(3):551–563. doi: 10.1007/s10237-013-0517-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fazio MA, Grytz R, Morris JS, Bruno L, Girkin CA, Downs JC. Human Scleral Structural Stiffness Increases More Rapidly with Age in Donors of African Descent Compared to Donors of European Descent. Invest Ophthalmol Vis Sci. 2014;55(11):7189–7198. doi: 10.1167/iovs.14-14894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grytz R, Fazio MA, Girard MJA, et al. Material properties of the posterior human sclera. Journal of the mechanical behavior of biomedical materials. 2013;29:602–617. doi: 10.1016/j.jmbbm.2013.03.027. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhodes LA, Huisingh C, Johnstone J, et al. Variation of laminar depth in normal eyes with age and race. Invest Ophthalmol Vis Sci. 2014;55(12):8123–8133. doi: 10.1167/iovs.14-15251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kankipati L, Johnstone JK, Girkin CA. Changes In Lamina Cribrosa Depth In Response To Increased Intraocular Pressure In Subjects With No Ocular Disease. Invest Ophthalmol Vis Sci ARVO Annual Meeting Abstracts. 2011 Abstact #6371. [Google Scholar]
- 32.Palmer ND, McDonough CW, Hicks PJ, et al. A genome-wide association search for type 2 diabetes genes in African Americans. PloS one. 2012;7(1):e29202. doi: 10.1371/journal.pone.0029202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDonough CW, Palmer ND, Hicks PJ, et al. A genome-wide association study for diabetic nephropathy genes in African Americans. Kidney Int. 2011;79(5):563–572. doi: 10.1038/ki.2010.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iyengar SK, Sedor JR, Freedman BI, et al. Genome-Wide Association and Trans-ethnic Meta-Analysis for Advanced Diabetic Kidney Disease: Family Investigation of Nephropathy and Diabetes (FIND) PLoS Genet. 2015;11(8):e1005352. doi: 10.1371/journal.pgen.1005352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henkin L, Bergman RN, Bowden DW, et al. Genetic epidemiology of insulin resistance and visceral adiposity. The IRAS Family Study design and methods. Ann Epidemiol. 2003;13(4):211–217. doi: 10.1016/s1047-2797(02)00412-x. [DOI] [PubMed] [Google Scholar]
- 36.Wagenknecht LE, Mayer EJ, Rewers M, et al. The insulin resistance atherosclerosis study (IRAS) objectives, design, and recruitment results. Ann Epidemiol. 1995;5(6):464–472. doi: 10.1016/1047-2797(95)00062-3. [DOI] [PubMed] [Google Scholar]
- 37.Das SK, Sharma NK, Elbein SC. Analysis of osteocalcin as a candidate gene for type 2 diabetes (T2D) and intermediate traits in Caucasians and African Americans. Dis Markers. 2010;28(5):281–286. doi: 10.3233/DMA-2010-0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Auer PL, Lettre G. Rare variant association studies: considerations, challenges and opportunities. Genome Med. 2015;7(1):16. doi: 10.1186/s13073-015-0138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83(3):311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89(1):82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee S, Emond MJ, Bamshad MJ, et al. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. 2012;91(2):224–237. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin P, Hartz SM, Zhang Z, et al. A new statistic to evaluate imputation reliability. PloS one. 2010;5(3):e9697. doi: 10.1371/journal.pone.0009697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bild DE, Bluemke DA, Burke GL, et al. Multi-Ethnic Study of Atherosclerosis: objectives and design. Am J Epidemiol. 2002;156(9):871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 44.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 45.Kang HM, Sul JH, Service SK, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42(4):348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conomos MP, Miller MB, Thornton TA. Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet Epidemiol. 2015;39(4):276–293. doi: 10.1002/gepi.21896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conomos MP, Reiner AP, Weir BS, Thornton TA. Model-free Estimation of Recent Genetic Relatedness. American Journal of Human Genetics. 2016;98(1):127–148. doi: 10.1016/j.ajhg.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen H, Wang C, Conomos MP, et al. Control for Population Structure and Relatedness for Binary Traits in Genetic Association Studies via Logistic Mixed Models. American Journal of Human Genetics. 2016;98(4):653–666. doi: 10.1016/j.ajhg.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teikari JM. Genetic factors in open-angle (simple and capsular) glaucoma. A population-based twin study. Acta Ophthalmol (Copenh) 1987;65(6):715–720. doi: 10.1111/j.1755-3768.1987.tb07069.x. [DOI] [PubMed] [Google Scholar]
- 50.Tielsch JM, Katz J, Sommer A, Quigley HA, Javitt JC. Family history and risk of primary open angle glaucoma. The Baltimore Eye Survey. Archives of ophthalmology. 1994;112(1):69–73. doi: 10.1001/archopht.1994.01090130079022. [DOI] [PubMed] [Google Scholar]
- 51.Wolfs RC, Klaver CC, Ramrattan RS, van Duijn CM, Hofman A, de Jong PT. Genetic risk of primary open-angle glaucoma. Population-based familial aggregation study. Archives of ophthalmology. 1998;116(12):1640–1645. doi: 10.1001/archopht.116.12.1640. [DOI] [PubMed] [Google Scholar]
- 52.Drance SM, Schulzer M, Thomas B, Douglas GR. Multivariate analysis in glaucoma. Use of discriminant analysis in predicting glaucomatous visual field damage. Archives of ophthalmology. 1981;99(6):1019–1022. doi: 10.1001/archopht.1981.03930011019007. [DOI] [PubMed] [Google Scholar]
- 53.Wang R, Wiggs JL. Common and rare genetic risk factors for glaucoma. Cold Spring Harbor perspectives in medicine. 2014;4(12):a017244. doi: 10.1101/cshperspect.a017244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burdon KP, Macgregor S, Hewitt AW, et al. Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat Genet. 2011;43(6):574–578. doi: 10.1038/ng.824. [DOI] [PubMed] [Google Scholar]
- 55.Wiggs JL, Lynch S, Ynagi G, et al. A genomewide scan identifies novel early-onset primary open-angle glaucoma loci on 9q22 and 20p12. Am J Hum Genet. 2004;74(6):1314–1320. doi: 10.1086/421533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Consortium CAD, Deloukas P, Kanoni S, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45(1):25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schunkert H, Konig IR, Kathiresan S, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43(4):333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bailey JN, Loomis SJ, Kang JH, et al. Genome-wide association analysis identifies TXNRD2, ATXN2 and FOXC1 as susceptibility loci for primary open-angle glaucoma. Nat Genet. 2016;48(2):189–194. doi: 10.1038/ng.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thorleifsson G, Walters GB, Hewitt AW, et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat Genet. 2010;42(10):906–909. doi: 10.1038/ng.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wiggs JL, Kang JH, Yaspan BL, et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma in Caucasians from the USA. Hum Mol Genet. 2011;20(23):4707–4713. doi: 10.1093/hmg/ddr382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burdon KP, Mitchell P, Lee A, et al. Association of open-angle glaucoma loci with incident glaucoma in the Blue Mountains Eye Study. Am J Ophthalmol. 2015;159(1):31–36. e31. doi: 10.1016/j.ajo.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Writing Committee for the Normal Tension Glaucoma Genetic Study Group of Japan. Glaucoma S, Meguro A, Inoko H, Ota M, Mizuki N, Bahram S. Genome-wide association study of normal tension glaucoma: common variants in SRBD1 and ELOVL5 contribute to disease susceptibility. Ophthalmology. 2010;117(7):1331–1338. e1335. doi: 10.1016/j.ophtha.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 63.Ramdas WD, van Koolwijk LM, Ikram MK, et al. A genome-wide association study of optic disc parameters. PLoS Genet. 2010;6(6):e1000978. doi: 10.1371/journal.pgen.1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Macgregor S, Hewitt AW, Hysi PG, et al. Genome-wide association identifies ATOH7 as a major gene determining human optic disc size. Hum Mol Genet. 2010;19(13):2716–2724. doi: 10.1093/hmg/ddq144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fan BJ, Wang DY, Pasquale LR, Haines JL, Wiggs JL. Genetic variants associated with optic nerve vertical cup-to-disc ratio are risk factors for primary open angle glaucoma in a US Caucasian population. Invest Ophthalmol Vis Sci. 2011;52(3):1788–1792. doi: 10.1167/iovs.10-6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gibson J, Griffiths H, De Salvo G, et al. Genome-wide association study of primary open angle glaucoma risk and quantitative traits. Mol Vis. 2012;18:1083–1092. [PMC free article] [PubMed] [Google Scholar]
- 67.Pasquale LR, Loomis SJ, Kang JH, et al. CDKN2B-AS1 genotype-glaucoma feature correlations in primary open-angle glaucoma patients from the United States. Am J Ophthalmol. 2013;155(2):342–353. e345. doi: 10.1016/j.ajo.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Springelkamp H, Hohn R, Mishra A, et al. Meta-analysis of genome-wide association studies identifies novel loci that influence cupping and the glaucomatous process. Nat Commun. 2014;5:4883. doi: 10.1038/ncomms5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hysi PG, Cheng CY, Springelkamp H, et al. Genome-wide analysis of multi-ancestry cohorts identifies new loci influencing intraocular pressure and susceptibility to glaucoma. Nat Genet. 2014;46(10):1126–1130. doi: 10.1038/ng.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen F, Klein AP, Klein BE, et al. Exome array analysis identifies CAV1/CAV2 as a susceptibility locus for intraocular pressure. Invest Ophthalmol Vis Sci. 2014;56(1):544–551. doi: 10.1167/iovs.14-15204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ozel AB, Moroi SE, Reed DM, et al. Genome-wide association study and meta-analysis of intraocular pressure. Hum Genet. 2014;133(1):41–57. doi: 10.1007/s00439-013-1349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gao X, Gauderman WJ, Liu Y, et al. A genome-wide association study of central corneal thickness in Latinos. Invest Ophthalmol Vis Sci. 2013;54(4):2435–2443. doi: 10.1167/iovs.13-11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lu Y, Dimasi DP, Hysi PG, et al. Common genetic variants near the Brittle Cornea Syndrome locus ZNF469 influence the blinding disease risk factor central corneal thickness. PLoS Genet. 2010;6(5):e1000947. doi: 10.1371/journal.pgen.1000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu Y, Vitart V, Burdon KP, et al. Genome-wide association analyses identify multiple loci associated with central corneal thickness and keratoconus. Nat Genet. 2013;45(2):155–163. doi: 10.1038/ng.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Springelkamp H, Mishra A, Hysi PG, et al. Meta-analysis of Genome-Wide Association Studies Identifies Novel Loci Associated With Optic Disc Morphology. Genet Epidemiol. 2015;39(3):207–216. doi: 10.1002/gepi.21886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nemesure B, Jiao X, He Q, et al. A genome-wide scan for primary open-angle glaucoma (POAG): the Barbados Family Study of Open-Angle Glaucoma. Hum Genet. 2003;112(5–6):600–609. doi: 10.1007/s00439-003-0910-z. [DOI] [PubMed] [Google Scholar]
- 77.Jiao X, Yang Z, Yang X, et al. Common variants on chromosome 2 and risk of primary open-angle glaucoma in the Afro-Caribbean population of Barbados. Proc Natl Acad Sci U S A. 2009;106(40):17105–17110. doi: 10.1073/pnas.0907564106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rotimi CN, Chen G, Adeyemo AA, et al. Genomewide scan and fine mapping of quantitative trait loci for intraocular pressure on 5q and 14q in West Africans. Invest Ophthalmol Vis Sci. 2006;47(8):3262–3267. doi: 10.1167/iovs.05-1537. [DOI] [PubMed] [Google Scholar]
- 79.Liu Y, Hauser MA, Akafo SK, et al. Investigation of known genetic risk factors for primary open angle glaucoma in two populations of African ancestry. Invest Ophthalmol Vis Sci. 2013;54(9):6248–6254. doi: 10.1167/iovs.13-12779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Loomis SJ, Kang JH, Weinreb RN, et al. Association of CAV1/CAV2 genomic variants with primary open-angle glaucoma overall and by gender and pattern of visual field loss. Ophthalmology. 2014;121(2):508–516. doi: 10.1016/j.ophtha.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wiggs JL, Hauser MA, Abdrabou W, et al. The NEIGHBOR consortium primary open-angle glaucoma genome-wide association study: rationale, study design, and clinical variables. J Glaucoma. 2013;22(7):517–525. doi: 10.1097/IJG.0b013e31824d4fd8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Charlson ES, Sankar PS, Miller-Ellis E, et al. The primary open-angle african american glaucoma genetics study: baseline demographics. Ophthalmology. 2015;122(4):711–720. doi: 10.1016/j.ophtha.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Crooks KR, Allingham RR, Qin X, et al. Genome-wide linkage scan for primary open angle glaucoma: influences of ancestry and age at diagnosis. PloS one. 2011;6(7):e21967. doi: 10.1371/journal.pone.0021967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Psaty BM, O’Donnell CJ, Gudnason V, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2(1):73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zelefsky JR, Harizman N, Mora R, et al. Assessment of a race-specific normative HRT-III database to differentiate glaucomatous from normal eyes. J Glaucoma. 2006;15(6):548–551. doi: 10.1097/01.ijg.0000212289.00917.a8. [DOI] [PubMed] [Google Scholar]
- 86.Bryc K, Durand EY, Macpherson JM, Reich D, Mountain JL. The genetic ancestry of African Americans, Latinos, and European Americans across the United States. Am J Hum Genet. 2015;96(1):37–53. doi: 10.1016/j.ajhg.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Collins-Schramm HE, Chima B, Operario DJ, Criswell LA, Seldin MF. Markers informative for ancestry demonstrate consistent megabase-length linkage disequilibrium in the African American population. Hum Genet. 2003;113(3):211–219. doi: 10.1007/s00439-003-0961-1. [DOI] [PubMed] [Google Scholar]
- 88.Seldin MF, Morii T, Collins-Schramm HE, et al. Putative ancestral origins of chromosomal segments in individual african americans: implications for admixture mapping. Genome Res. 2004;14(6):1076–1084. doi: 10.1101/gr.2165904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Frazer KA, Ballinger DG, Cox DR, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Campbell MC, Tishkoff SA. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu Rev Genomics Hum Genet. 2008;9:403–433. doi: 10.1146/annurev.genom.9.081307.164258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Girkin CA, Nievergelt CM, Kuo JZ, et al. Biogeographic Ancestry in the African Descent and Glaucoma Evaluation Study (ADAGES): Association With Corneal and Optic Nerve Structure. Invest Ophthalmol Vis Sci. 2015;56(3):2043–2049. doi: 10.1167/iovs.14-15719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Seldin MF, Pasaniuc B, Price AL. New approaches to disease mapping in admixed populations. Nat Rev Genet. 2011;12(8):523–528. doi: 10.1038/nrg3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sommer A, Tielsch JM, Katz J, et al. Racial differences in the cause-specific prevalence of blindness in east Baltimore. N Engl J Med. 1991;325(20):1412–1417. doi: 10.1056/NEJM199111143252004. [DOI] [PubMed] [Google Scholar]
- 95.Tielsch JM, Sommer A, Katz J, Royall RM, Quigley HA, Javitt J. Racial variations in the prevalence of primary open-angle glaucoma. The Baltimore Eye Survey. Jama. 1991;266(3):369–374. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.