Summary
In recent years, our understanding of the complex number of signals that need to be integrated between a diverse number of receptors present on natural killer (NK) cells and ligands present on target cells has improved. Here, we review the progress made in identifying interactions between dengue viral peptides presented on HLA Class 1 molecules with inhibitory and activating killer‐like immunoglobulin receptors on NK cells, direct interactions of viral proteins with NK cell receptors, the involvement of dengue virus‐specific antibodies in mediating antibody‐dependent cell‐mediated cytotoxicity and the role of soluble factors in modulating NK cell responses. We discuss findings of NK cell activation early after natural dengue infection, and point to the role that NK cells may play in regulating both innate and adaptive immune responses, in the context of our new appreciation of interactions of dengue virus with specific NK cell receptors. With a number of flavivirus vaccine candidates in clinical trials, how NK cells respond to attenuated dengue virus and subunit protein vaccine candidates and shape adaptive immunity will need to be considered.
Keywords: activation, cytokines, innate lymphoid cells, killer cell immunoglobulin‐like receptors, NK cell
Introduction
Dengue viruses (DENV) comprise four distinct serotypes DENV‐1 to DENV‐4, and are a major public health problem in tropical and sub‐tropical countries. DENV belong to the genus flavivirus and include a number of other pathogens, including yellow fever virus, West Nile virus (WNV), Japanese encephalitis virus (JEV) and Zika virus (ZIKV). Clinical symptoms following DENV infection range from fever, myalgia and rash to significant thrombocytopaenia, haemorrhage and plasma leakage into interstitial spaces in the more severe form, dengue haemorrhagic fever (DHF). There are many risk factors associated with developing DHF, which include infection with specific viral strains, host genetics and nutritional status, but prior immunity to DENV is considered a major risk factor.1, 2 Plasma leakage, which is a hallmark of DHF, occurs several days following infection, at the end of viraemia and at a time of significant immune activation. Because infection for a second time with a heterologous serotype of DENV significantly increases your chance of developing DHF, a lot of effort has been spent to understand how components of the adaptive immune response including antibodies, T‐cells and cytokines they secrete contribute to the pathogenesis of secondary infection. Excessive immune activation results in disproportionate cytokine production and increased vascular permeability.
There is ample evidence to indicate that cells of the innate immune system including epidermal and dermal dendritic cells, mast cells, monocytes/macrophages, natural killer (NK) and NK‐like cells are also activated very early during primary and secondary DENV infection.3 Cytokines such as TNF‐α and chemotactic factors including IP‐10, MCP‐1, IL‐8 induced by in vitro and in vivo infection of innate cells with DENV may help eliminate virally infected cells and also recruit other cells.3 Type 1 IFNs elevated in the sera of infected individuals could play an important role in the anti‐viral defense against DENV by curtailing viral dissemination. Cross‐reactive memory B and T lymphocytes are activated earlier during a second DENV infection. As the response amplifies, cells of the innate and adaptive immune system and soluble factors they secrete recruit additional cell types.
One such innate cell type actively involved in the immune response against DENV is NK cells. The ability of NK cells to secrete cytolytic granules has long been recognized, and they are a crucial first line of defense to eliminate DENV‐infected cells. NK cells are regulated by a network of receptors on the cell surface that allows them to distinguish a virally infected cell from a healthy cell. Major NK cell receptors include the activating natural cytotoxicity receptors (NCRs–NKp30, NKp44 and NKp46), inhibitory or activating killer cell immunoglobulin‐like receptors (KIRs), which interact with HLA‐I, inhibitory or activating C‐type lectins (NKG2a/CD94, NKG2d and NKG2c/CD94) and FcγRIIIA (CD16), which binds to the Fc region of an antibody bound to the surface of a virally infected cell and can mediate antibody‐dependent cell‐mediated cytotoxicity (ADCC) (Fig. 1).4 Two major subsets of NK cells are defined based on the expression of CD56 and CD16. CD56brightCD16dim/neg are robust cytokine‐producing cells, and CD56dim/negCD16pos are cytolytic and express KIRs and other inhibitory receptors. Differential expression of inhibitory and activating receptors on subsets of NK cells accounts for some of the specificity of the NK cell response.
Figure 1.
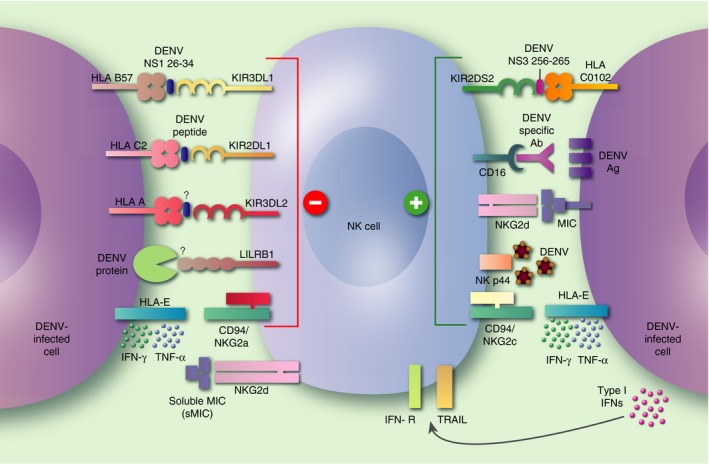
Interactions of surface receptors on natural killer (NK) cells with dengue viruses (DENV). Following DENV infection, viral antigens and viral peptides loaded on HLA molecules are detected on the surface of infected cells. Soluble factors released by DENV‐infected cells can stimulate NK cells. Specific viral peptides on the NS1 and NS3 proteins presented on HLA‐B57 and HLA‐C0102 have been identified and found to interact with KIR3DL1 and KIR2DS2, inhibitory and activating killer cell immunoglobulin‐like receptors (KIRs), respectively. Other yet to be identified DENV peptides will be presented on HLA alleles, and could interact with KIRs and modulate NK cell function. The E protein of DENV can directly interact with activating receptor NKp44. CD16 present on NK cells can bind the Fc portion of antibodies and mediate antibody‐dependent cell‐mediated cytotoxicity (ADCC) by recognizing DENV antigens on the surface of infected cells. DENV‐induced expression of HLA‐E on infected or bystander cells can interact with CD94/NKG2a, and maintain inhibition of NK cells or interact with the CD94/NKG2c receptors and activate NK cells.
Killer cell immunoglobulin‐like receptor interactions with DENV
NK cells are regulated by inhibitory KIRs (denoted by an ‘L’ for its long cytoplasmic tail) that recognize HLA Class I molecules on healthy cells.5 Downregulation of HLA Class I molecules on target cells (as is often seen with viral infections) initiates a signal that the target cell is no longer healthy and leads to the activation of NK cells. Much less is known about the ligands for activating KIRs (usually denoted by an ‘S’ for its short cytoplasmic tail), although KIR2DS1 is known to interact with HLA‐C.6
Genetic population studies in human immunodeficiency virus (HIV)‐infected individuals suggest that KIRs are important in the control of infection.7 More recently, there is genetic evidence that supports the role of KIRs in DENV infections as well. In studies done in Gabon, Brazil and in Western India, a difference in the frequencies of KIR genes such as KIR3DL1, KIR3DS1, KIR2DS1, KIR2DS5 and KIR2DL5 was found in patients with dengue compared with healthy controls.8, 9, 10 Furthermore, the analysis in one study found a possible protective effect against dengue fever in individuals with the AA genotype. The data suggest that the expression of select KIRs on NK cells might preferentially affect its activation and influence the outcome of disease.
Experimental data indicate that KIR molecules interact with DENV peptides presented on HLA molecules. KIR3DL1 interacts with HLA‐B57 and has been implicated in many chronic viral infections, including HIV.11, 12 Townsley et al. demonstrated an interaction between the inhibitory KIR3DL1 and a conserved NS1 peptide on DENV presented by HLA‐B57.13 Tetramers generated against the NS1 peptide bound KIR3DL1 transfectants as well as CD56dim NK cells (known to express KIRs). Using peripheral blood mononuclear cells (PBMCs) from children undergoing acute dengue infection in Thailand, tetramer‐positive NK cells were also found to be most activated during the critical period of illness, several days after ongoing fever and at the end of viraemia. Because DENV upregulates HLA Class I molecules, we speculated that KIR3DL1 was able to interact with HLA‐B57 early during acute DENV infection, when viraemia levels were high, and maintain inhibition of NK cells. As viraemia decreased, KIR3DL1 was able to sense the decrease in HLA Class I expression on target cells, thus activating subsets of NK cells during the critical period of illness.
Petitdemange et al. phenotyped NK cells in patients from Gabon undergoing acute DENV infection.14 They found expanded frequencies of KIR2DL1+ NK cells during acute DENV infection. KIR2DL1 is an inhibitory receptor that interacts with HLA‐C molecules, and the authors speculate that viral peptides presented on HLA‐C2 following DENV‐2 infection can interact with KIR2DL1 and prevent NK cell activation. The DENV peptides presented on HLA‐C2 were not identified in this study.
While HIV and HCV peptides have been shown to modulate NK cell functional responses,15, 16 studies to definitively show that DENV peptides can modulate NK cell function by interacting with inhibitory KIRs are yet to be published. Because self‐peptides can also maintain inhibition in healthy cells, it remains to be proven that the HLA‐restricted DENV peptides can displace self‐peptides and modulate NK cell function in vivo. Cell lines that stabilize the HLA molecule of interest only in the presence of viral peptides are currently being tested in our laboratory. We have experienced technical challenges with some TAP‐deficient cells lines transfected with HLA alleles of interest; HLA‐B57 appears to be incorrectly folded thus preventing significant expression on the cell surface and subsequent functional studies.
Naiyer et al. wanted to determine whether flavivirus peptides bind activating KIRs. They focused their efforts on identifying a conserved target of an activating receptor KIR2DS2 on NK cells. A peptide on the NS3 protein that was conserved in 61/63 flaviviruses, including DENV, stabilized HLA‐C0102 and interacted with KIR2DS2 resulting in the activation of NK cells.17 Presentation of the NS3 peptide in the context of HLA‐C0102 was able to inhibit HCV and DENV replication. HEK cells stably transfected with a DENV replicon and transiently transfected with HLA C0102 but not 0304 were recognized by NK cell lines expressing 2DS2.
Together, the data indicate that DENV peptides interact with both inhibitory and activating KIRs on NK cells, and are likely to play a role in modulating NK cell responses (Fig. 1). For example, if an individual with HLA‐B57 and C0102 alleles is infected with DENV, the NS1 peptide may continue to maintain an inhibitory signal by interacting with KIR3DL1 presented on HLA‐B57 during viraemia, while the conserved NS3 peptide presented on HLA‐C0102 will interact with KIR2DS2 and potentially activate this subset of NK cells. These viral peptides presented on HLA alleles will also activate antigen‐specific T‐cells, many of which express KIRs. How KIRs on DENV‐specific T‐cells modulate responses is unknown. Several other yet to be identified peptides may be presented on HLA Class I molecules and interact with KIRs on NK cells. The outcome of the response will depend on the balance between positive or negative signals following interaction with inhibitory KIRs, activating KIRs and other receptors that interact with ligands that can induce inhibitory and activating signals on NK cells.
Upregulation of HLA‐E following DENV infection
Another major inhibitory receptor on NK cells is CD94a/NKG2a. The ligand for CD94a is HLA‐E. HLA‐E has a dual role in the immune response because it can also interact with activating receptors NKG2c/d on NK cells; however, the interaction with the inhibitory receptor NKG2a is of higher affinity. JEV was recently shown to modestly upregulate HLA‐E on endothelial cells with significant amounts of soluble levels of HLA‐E detected in supernatants of infected cells.18, 19 Our unpublished work indicates that DENV upregulate HLA‐E on the surface of endothelial cells, but the levels of soluble HLA‐E detected in supernatants are modest. DENV may thus be able to modulate NK cell function through the interaction with activating or inhibitory receptors NKG2c/d or NKG2a on NK cells. Low levels of soluble HLA‐E may not be sufficient to block signalling and evade NK cell activation. While HLA‐E has been shown to interact with NKG2a/c on NK cells, HLA‐E can present peptides from pathogens that are recognized by αβ TCRs.20 Peptides derived from Mtb, L. monocytogenes, CMV and EBV presented on HLA‐E can be recognized by CD8+ T‐cells. DENV‐induced HLA‐E has the potential to interact with and impact both the innate and adaptive arms of the immune system.
Interactions that can activate NK cells
The E protein of DENV and WNV interacts directly with NKp44, a natural cytotoxicity receptor expressed predominantly on activated NK cells (Fig. 1).21 NKp44‐expressing NK cells were moderately elevated during the acute phase of dengue infection in a longitudinal study of patients from Gabon, supporting the notion that subsets of NK cells are preferentially activated in response to DENV infection.14 Whether the ligand for NKp44 was elevated in virally infected cells is being pursued. Genetic‐based studies in a large number of Vietnamese children and adults strongly suggest that MIC A and B are a susceptibility locus for severe dengue.22, 23, 24 Because MIC B interacts with NKG2d an activating receptors on NK cells, the genome‐wide association studies support a role for NK cells in shaping the outcome of mild and severe forms of disease; however, functional studies are required to confirm the contribution of MIC B in NK cell activation during acute DENV infections. A clinical study that found elevated levels of soluble MIC B in the sera of infants with dengue infections25 suggests that MIC B in the circulation could potentially block the interaction with NKG2d and prevent activation of NK cells.
CD16 (FcγRIIIA) on NK cells has been shown to mediate ADCC activity of antibodies induced by DENV infection (Fig. 1). DENV‐specific ADCC correlates with cell surface expression of DENV antigens and serotype‐specific neutralizing Ab titres.26, 27, 28 Higher ADCC activity in pre‐secondary DENV‐3 infection plasma samples correlated with lower plasma viraemia levels not seen with pre‐secondary DENV‐2 infection plasma samples.27
Regulation of NK cells by cytokines
In addition to the interaction between receptors on NK cells and ligands on infected cells, NK cells can be activated indirectly through cytokine signalling. Enhanced production of IFN‐α and soluble TRAIL has been found in the sera of patients with mild dengue disease.29, 30 TRAIL has an important role in the anti‐viral response against DENV,31 and can induce NK cell apoptosis of hepatic stellate cells in HCV‐infected patients.32 Gandini et al. determined whether TRAIL was expressed on NK cells during acute dengue, and found a higher frequency of TRAIL+CD16 + NK cells using PBMCs from patients in Brazil with mild disease.33 They developed an in vitro model, where DENV infection of monocytes induced IFN‐α and augmented NK cell cytotoxicity mediated by TRAIL. Blocking Type I IFNs reduced TRAIL expression on NK cells, suggesting partial regulation by IFNs. The authors speculate that TRAIL expression on CD16+ NK cells may be an additional way to mediate cytotoxicity and eliminate virally infected cells.
Activation of NK cells during acute DENV infections
A number of clinical studies indicate that NK cells are activated early after DENV infection and play an important role in the immune response against DENV. These studies found an increased frequency of CD56+ CD69+ cells in patients with severe compared with mild dengue disease in Thailand and Vietnam.34, 35 NK cells in patients with severe disease, DHF, were activated during study entry compared with patients with other febrile illnesses and healthy controls. A more detailed analysis of subsets of NK cells was performed by Petitdemange et al. using PBMCs from patients undergoing DENV‐2 infection in Gabon. The authors found significant activation of NK cells during acute DENV‐2 infection with the production of significant IFN‐γ by NK cells.14 Transient increases in the frequencies of NKG2c+NK cells with lower frequencies of cells expressing NKG2a, NKp30, CD161 were found in DENV patients.
Keawvichit et al. examined frequencies of activated NK cells in PBMCs from patients in Thailand undergoing acute DENV infections.36 As has been previously reported, NK cells were activated during acute dengue. The major difference noted was the decreased frequencies of NK cell subsets that expressed CXCR3 and an increased frequency of cells that expressed CCR10 between healthy subjects and dengue patients, but not between patients with DF compared with patients with DHF.
In a study by Costa et al., IFN‐γ production by human NK cells was found to be important in controlling dengue viral replication in an optimized humanized mouse model.37 Cell–cell contact between monocyte‐derived dendritic cells and NK cells was important for NK cell activation (CD69 and CD25 upregulation) and IFN‐γ secretion. Adhesion molecules including DNAM, 2B4, LFA‐1 and CD2 were more important for mediating activation, while blocking molecules such as NKp30, NKp44 and NKp46 had minimal impact in this model. This study indicates that NK cells can control DENV replication in vivo, and identifies an important antigen‐presenting cell that may be important to activate NK cells.
NK cell responses to vaccination
Efforts to develop an effective dengue vaccine have been ongoing for nearly 50 years, but substantial advances have been made with live attenuated, inactivated and subunit vaccines in the past 10 years.38 The challenges to generate an effective dengue vaccine are significant, as any vaccine would need to produce safe and durable immunity to all four serotypes of DENV. Furthermore, this should be achieved preferably using one or two doses in individuals. Given the fact that over one‐third of the world's population has been exposed to DENV and there is cross‐reactivity among related flaviviruses such as JEV, WNV and ZIKV, the vaccine would need to avoid eliciting pathogenic immune responses in flavi‐immune individuals and also induce robust immunity in flavi‐naïve individuals.39
NK cells have long been recognized to eliminate virally infected cells, but in the past decade our understanding has evolved as NK cells have broad diversity, and even the potential to develop into memory cells and protect against subsequent infections.40, 41 NK cells also have the ability to regulate and stimulate T‐ and B‐cell responses following infection, and therefore strategies to boost stimulatory responses of NK cells and dampen inhibitory functions of NK cells should be considered.42 While eliciting broadly neutralizing antibodies is a goal for an effective DENV vaccine, it is becoming clear that Abs that mediate ADCC can provide protective immunity against HIV and influenza.43 Therefore, evaluating DENV‐specific ADCC Abs should be considered for any emerging DENV vaccine candidate.
Conclusions
Dengue viruses interact with a number of receptors on NK cells. We are now at the stage where we can better evaluate how human NK cells respond to DENV infection and vaccination. There are many questions that can be answered in the coming years with the new tools we have and improved understanding of multiple interactions between inhibitory and activating receptors on NK cells. Are there unique subsets of NK cells that respond to natural primary versus secondary DENV infection? Does the NK cell response to an acute viral infection such as DENV vary significantly from the more well‐characterized chronic viral infections HIV and CMV? Do responses of NK cells in the circulation differ from NK cell responses in tissues? Does vaccination alter the NK cell repertoire in DENV‐immune versus ‐naive individuals? Do multiple prime‐boost strategies being used in some DENV vaccine candidates change the NK cell repertoire? Understanding the dynamics of the NK cell response to DENV and what constitutes a beneficial response is certain to contribute to the development of an efficacious vaccine.
Disclosures
There are no commercial or financial conflicts of interest to declare.
Acknowledgements
This review is supported by grant R21AI113479 from the NIH/NIAID. The author would like to thank Luis Vargas Sanchez for critically reading the review.
References
- 1. Screaton G, Mongkolsapaya J, Yacoub S, Roberts C. New insights into the immunopathology and control of dengue virus infection. Nat Rev Immunol 2015; 15:745–59. [DOI] [PubMed] [Google Scholar]
- 2. Rothman AL. Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nat Rev Immunol 2011; 11:532–43. [DOI] [PubMed] [Google Scholar]
- 3. Srikiatkhachorn A, Mathew A, Rothman AL. Immune‐mediated cytokine storm and its role in severe dengue. Semin Immunopathol 2017; 39:563–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL et al Innate or adaptive immunity? The example of natural killer cells. Science 2011; 331:44–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rajagopalan S, Long EO. Understanding how combinations of HLA and KIR genes influence disease. J Exp Med 2005; 201:1025–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chapel A, Garcia‐Beltran WF, Holzemer A, Ziegler M, Lunemann S, Martrus G et al Peptide‐specific engagement of the activating NK cell receptor KIR2DS1. Sci Rep 2017; 7:2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Holzemer A, Garcia‐Beltran WF, Altfeld M. Natural killer cell interactions with classical and non‐classical human leukocyte antigen Class I in HIV‐1 infection. Front Immunol 2017; 8:1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Petitdemange C, Wauquier N, Jacquet JM, Theodorou I, Leroy E, Vieillard V. Association of HLA Class‐I and inhibitory KIR genotypes in Gabonese patients infected by Chikungunya or Dengue Type‐2 viruses. PLoS ONE 2014; 9:e108798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Beltrame LM, Sell AM, Moliterno RA, Clementino SL, Cardozo DM, Dalalio MM et al Influence of KIR genes and their HLA ligands in susceptibility to dengue in a population from southern Brazil. Tissue Antigens 2013; 82:397–404. [DOI] [PubMed] [Google Scholar]
- 10. Alagarasu K, Bachal RV, Shah PS, Cecilia D. Profile of killer cell immunoglobulin‐like receptor and its human leucocyte antigen ligands in dengue‐infected patients from Western India. Int J Immunogenet 2015; 42:432–8. [DOI] [PubMed] [Google Scholar]
- 11. Fadda L, O'Connor GM, Kumar S, Piechocka‐Trocha A, Gardiner CM, Carrington M et al Common HIV‐1 peptide variants mediate differential binding of KIR3DL1 to HLA‐Bw4 molecules. J Virol 2011; 85:5970–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boulet S, Song R, Kamya P, Bruneau J, Shoukry NH, Tsoukas CM et al HIV protective KIR3DL1 and HLA‐B genotypes influence NK cell function following stimulation with HLA‐devoid cells. J Immunol 2010; 184:2057–64. [DOI] [PubMed] [Google Scholar]
- 13. Townsley E, O'Connor G, Cosgrove C, Woda M, Co M, Thomas SJ et al Interaction of a dengue virus NS1‐derived peptide with the inhibitory receptor KIR3DL1 on natural killer cells. Clin Exp Immunol 2016; 183:419–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Petitdemange C, Wauquier N, Devilliers H, Yssel H, Mombo I, Caron M et al Longitudinal analysis of natural killer cells in dengue virus‐infected patients in comparison to chikungunya and chikungunya/dengue virus‐infected patients. PLoS Negl Trop Dis 2016; 10:e0004499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cassidy SA, Cheent KS, Khakoo SI. Effects of peptide on NK cell‐mediated MHC I recognition. Front Immunol 2014; 5:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cassidy S, Mukherjee S, Myint TM, Mbiribindi B, North H, Traherne J et al Peptide selectivity discriminates NK cells from KIR2DL2‐ and KIR2DL3‐positive individuals. Eur J Immunol 2015; 45:492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Naiyer MM, Cassidy SA, Magri A, Cowton V, Chen K, Mansour S et al KIR2DS2 recognizes conserved peptides derived from viral helicases in the context of HLA‐C. Sci Immunol 2017; 2:pii: eaal5296. [DOI] [PubMed] [Google Scholar]
- 18. Shwetank, Date OS, Kim KS, Manjunath R. Infection of human endothelial cells by Japanese encephalitis virus: increased expression and release of soluble HLA‐E. PLoS ONE 2013; 8:e79197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shwetank, Date OS, Carbone E, Manjunath R. Inhibition of ERK and proliferation in NK cell lines by soluble HLA‐E released from Japanese encephalitis virus infected cells. Immunol Lett 2014; 162:94–100. [DOI] [PubMed] [Google Scholar]
- 20. Joosten SA, Sullivan LC, Ottenhoff TH. Characteristics of HLA‐E restricted T‐cell responses and their role in infectious diseases. J Immunol Res 2016; 2016:2 695 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hershkovitz O, Rosental B, Rosenberg LA, Navarro‐Sanchez ME, Jivov S, Zilka A et al NKp44 receptor mediates interaction of the envelope glycoproteins from the West Nile and dengue viruses with NK cells. J Immunol 2009; 183:2610–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Whitehorn J, Chau TN, Nguyet NM, Kien DT, Quyen NT, Trung DT et al Genetic variants of MICB and PLCE1 and associations with non‐severe dengue. PLoS ONE 2013; 8:e59067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Khor CC, Chau TN, Pang J, Davila S, Long HT, Ong RT et al Genome‐wide association study identifies susceptibility loci for dengue shock syndrome at MICB and PLCE1. Nat Genet 2011; 43:1139–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Garcia G, del Puerto F, Perez AB, Sierra B, Aguirre E, Kikuchi M et al Association of MICA and MICB alleles with symptomatic dengue infection. Hum Immunol 2011; 72:904–7. [DOI] [PubMed] [Google Scholar]
- 25. Libraty DH, Zhang L, Obcena A, Brion JD, Capeding RZ. Circulating levels of soluble MICB in infants with symptomatic primary dengue virus infections. PLoS ONE 2014; 9:e98509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sun P, Morrison BJ, Beckett CG, Liang Z, Nagabhushana N, Li A et al NK cell degranulation as a marker for measuring antibody‐dependent cytotoxicity in neutralizing and non‐neutralizing human sera from dengue patients. J Immunol Methods 2017; 441:24–30. [DOI] [PubMed] [Google Scholar]
- 27. Laoprasopwattana K, Libraty DH, Endy TP, Nisalak A, Chunsuttiwat S, Vaughn DW et al Antibody dependent cellular cytotoxicity in pre‐secondary dengue virus serotype 3 (DV3) but not in DV2 infection plasma samples inversely correlated with viremia levels. J Infect Dis 2007; 195:1108–16. [DOI] [PubMed] [Google Scholar]
- 28. Kurane I, Hebblewaite D, Brandt WE, Ennis FA. Lysis of dengue virus‐infected cells by natural cell‐mediated cytotoxicity and antibody‐dependent cell‐mediated cytotoxicity. J Virol 1984; 52:223–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Becerra A, Warke RV, Martin K, Xhaja K, de Bosch N, Rothman AL et al Gene expression profiling of dengue infected human primary cells identifies secreted mediators in vivo . J Med Virol 2009; 81:1403–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gandini M, Gras C, Azeredo EL, Pinto LM, Smith N, Despres P et al Dengue virus activates membrane TRAIL relocalization and IFN‐alpha production by human plasmacytoid dendritic cells in vitro and in vivo . PLoS Negl Trop Dis 2013; 7:e2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Warke RV, Martin KJ, Giaya K, Shaw SK, Rothman AL, Bosch I. TRAIL is a novel antiviral protein against dengue virus. J Virol 2008; 82:555–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Glassner A, Eisenhardt M, Kramer B, Korner C, Coenen M, Sauerbruch T et al NK cells from HCV‐infected patients effectively induce apoptosis of activated primary human hepatic stellate cells in a TRAIL‐, FasL‐ and NKG2D‐dependent manner. Lab Invest 2012; 92:967–77. [DOI] [PubMed] [Google Scholar]
- 33. Gandini M, Petitinga‐Paiva F, Marinho CF, Correa G, De Oliveira‐Pinto LM, de Souza LJ et al Dengue virus induces NK cell activation through trail expression during infection. Mediators Inflamm 2017; 2017:5 649 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Green S, Pichyangkul S, Vaughn DW, Kalayanarooj S, Nimmannitya S, Nisalak A et al Early CD69 expression on peripheral blood lymphocytes from children with dengue hemorrhagic fever. J Infect Dis 1999; 180:1429–35. [DOI] [PubMed] [Google Scholar]
- 35. Chau TN, Quyen NT, Thuy TT, Tuan NM, Hoang DM, Dung NT et al Dengue in Vietnamese infants–results of infection‐enhancement assays correlate with age‐related disease epidemiology, and cellular immune responses correlate with disease severity. J Infect Dis 2008; 198:516–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Keawvichit R, Khowawisetsut L, Lertjuthaporn S, Tangnararatchakit K, Apiwattanakul N, Yoksan S et al Differences in activation and tissue homing markers of natural killer cell subsets during acute dengue infection. Immunology 2017; 153:455–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Costa VV, Ye W, Chen Q, Teixeira MM, Preiser P, Ooi EE et al Dengue virus‐infected dendritic cells, but not monocytes, activate natural killer cells through a contact‐dependent mechanism involving adhesion molecules. MBio 2017; 8:pii: e00741‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Murphy BR, Whitehead SS. Immune response to dengue virus and prospects for a vaccine. Annu Rev Immunol 2011; 29:587–619. [DOI] [PubMed] [Google Scholar]
- 39. Andrade DV, Harris E. Recent advances in understanding the adaptive immune response to Zika virus and the effect of previous flavivirus exposure. Virus Res 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Freud AG, Mundy‐Bosse BL, Yu J, Caligiuri MA. The broad spectrum of human natural killer cell diversity. Immunity 2017; 47:820–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Geary CD, Sun JC. Memory responses of natural killer cells. Semin Immunol 2017; 31:11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rydyznski CE, Waggoner SN. Boosting vaccine efficacy the natural (killer) way. Trends Immunol 2015; 36:536–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vanderven HA, Jegaskanda S, Wheatley AK, Kent SJ. Antibody‐dependent cellular cytotoxicity and influenza virus. Curr Opin Virol 2017; 22:89–96. [DOI] [PubMed] [Google Scholar]


