Summary
Segmented filamentous bacteria (SFB) are Gram‐positive, spore‐forming, bacteria that primarily colonize the ileum of the small intestine. Upon direct adherence to intestinal epithelial cells, SFB actively stimulate innate and adaptive immune cell activation. The cardinal features of SFB‐induced gut immunity – T helper type 17 (Th17) cell differentiation, IgA production and barrier protection – lead to the containment of SFB and further afford protection against invading pathogens. Th17 cells and interleukin‐17A, however, can also reach peripheral sites and exacerbate autoimmunity. In this review, we highlight salient characteristics of SFB–host interactions and detail the cellular and molecular immune mechanisms involved in coordinating these responses.
Keywords: autoimmunity, immune homeostasis, microbiota, mucosal immunology, segmented filamentous bacteria, Th17 cells
Introduction
Colonization of the mammalian host with microbiota triggers various physiological adaptations including the maturation of the immune system, which is required to maintain a mutually beneficial relationship with the microbiota.1, 2, 3, 4 The host senses and responds to the microbiota through coordinated immune responses that must not be overly aggressive towards commensal microbes, all the while remaining primed to combat invading pathogens. Recent advances in understanding the mechanisms involved in gut microbiota sensing, as well as how distinct species of commensal bacteria can evoke tailored host mucosal immune responses, have provided critical insight into how this balance is maintained. For example, species of Clostridium can induce Foxp3+ regulatory T cells that inhibit intestinal inflammation and IgE production,5, 6 whereas other species of commensal bacteria such as Helicobacter hepaticus can act as pathobionts and exacerbate T helper type 17 (Th17) ‐mediated colitis.7, 8 Segmented filamentous bacteria (SFB) are yet another example of specific commensal bacteria that can potently induce mucosal immune responses,9 as detailed below.
Segmented filamentous bacteria are Gram‐positive commensal bacteria that primarily colonize the ileum of the small intestine at the time of weaning and have been identified in a number of different species including rodents, pigs and other mammals.10, 11 They can be recognized by their characteristic long filamentous appearance and although morphologically similar across species, these different populations of SFB are genetically distinct from one another.12, 13 As such, SFB exhibit restricted host‐specificity and it has been demonstrated that cross‐species colonization between rats and mice does not occur.12 Complete genome sequencing of SFB strains identified these bacteria as a unique member of the Clostridiales that express typical genes for spore formation and flagella, yet lack amino acid biosynthesis enzymes and must rely on the host for provision of essential nutrients.13, 14, 15, 16 Given this need for host nutrients, in mice SFB are found in close association with the absorptive epithelial lining of the terminal ileum where they can make intimate contact with intestinal epithelial cells (IECs) through direct contact and adherence. Adherence is an important feature for imparting the immunomodulatory ability of SFB; however, SFB can still influence certain immune responses without directly adhering to IECs. Of the immunomodulatory effects driven by SFB colonization, the most recognized is the ability to drive postnatal maturation of the intestinal immune system, including IL‐17‐producing CD4+ T cells (Th17 cells), which have antimicrobial effects and promote barrier function to protect the host from infection with extracellular pathogens. SFB also influence a number of other protective immune responses including the development of lymphoid tissue and IgA production,9, 17, 18 and innate lymphoid cell (ILC)‐mediated barrier protection.19, 20, 21 This protection can come at a cost, however, as SFB‐induced Th17 cells can trigger autoimmunity in susceptible hosts.
In this review, we highlight the ability of SFB to dynamically influence different types of immune cells, including Th17 cells, and detail the current understanding of how the mucosal immune system coordinates these responses following SFB colonization. We also highlight the importance of host immunity in directly controlling SFB growth in the intestine and how a failure to do so can generate excessive immune reactions leading to autoimmunity in peripheral tissues.
SFB – potent inducers of Th17 cells
Th17 cells are a subset of CD4+ T helper cells characterized by their expression of the master transcription factor retinoic acid receptor‐related orphan receptor ɣt (RORɣt) and production of the signature cytokines interleukin‐17A (IL‐17A) and IL‐17F.22 Th17 cells are abundant in the lamina propria (LP) of the small intestine, which is recognized as a major effector site for these cells.23, 24, 25 Th17 cells garnered attention as a novel T helper cell lineage distinct from Th1/Th2 cells,26, 27 that were induced by transforming growth factor‐β and IL‐6.22, 28, 29, 30 The first clues that Th17 differentiation may be influenced by the microbiota came from data showing that intestinal Th17 cells are not readily detectable until approximately 3–4 weeks of age in mice.24 This period coincides with the time‐point immediately post‐weaning, a developmental stage largely shaped by the external environment, including diet and the microbiota. In testing the microbiota as a potential inducer of Th17 cells, Ivanov et al. found that genetically identical mice from different animal vendors have different numbers of Th17 in the intestine. Specifically, C57BL/6 mice from The Jackson Laboratory (Jax; Bar Harbor, ME) are largely devoid of these cells while C57BL/6 mice from Taconic Biosciences (Tac; Rensselaer, NY) harbour an abundance of intestinal Th17 cells in the LP.24, 31 Co‐housing Jax and Tac mice was shown to induce Th17 cells in the small intestine of Jax mice and transplanting faeces from Jax and Tac mice into germ‐free (GF) mice revealed that only the Tac microbiota were able to induce intestinal Th17 cells.31 Molecular approaches to dissect the microbiota composition in these mice revealed one of the main differences between mice from these two different vendors was the presence of SFB. Subsequent studies using faeces from SFB‐monoassociated mice confirmed the ability of SFB to induce intestinal Th17 cell responses.31
In coinciding studies, Gaboriau et al.25 took a systematic approach, testing the ability of a range of different bacteria to induce T‐cell responses throughout the intestinal LP of GF mice. They observed that conventional microbiota could induce diverse immune responses in a host‐specific manner, and also that mucosal T‐cell responses are induced by a subset of host‐specific bacterial species. Clostridia appeared to be particularly important in driving T helper cell subset differentiation, and a focus on defined species of Clostridia uncovered a role for SFB in stimulating the maturation of T‐cell subsets, particularly Th17 cells.25
Although it is well‐appreciated that SFB colonization generates Th17 cells in the intestine, it is less clear how Th17 cells function to provide host protection.32, 33 Accumulating evidence supports a role for Th17 cells in controlling the degree of SFB colonization.34 Interleukin‐17A and IL‐22 production by Th17 cells can combine with other sources of IL‐22, including innate lymphoid cells and neutrophils, to promote antimicrobial peptide expression including α‐defensins and reactive oxygen species (ROS),34 and can directly promote mucosal barrier function through the regulation of tight junction proteins.35, 36 These effects of Th17 cells help to bolster host defence against SFB themselves, as well as invading pathogens.37 For example, SFB‐induced immunity can protect the host against the intestinal bacterial pathogens Citrobacter rodentium 31 and Salmonella typhimurium.38 Additionally, Th17 cell responses induced by SFB were found to be sufficient to overcome intestinal permeability defects and maintain protection against acute enteric infection with Toxoplasma gondii or Salmonella typhimurium.39 SFB colonization is also able to confer protection against Salmonella enteritidis in rats,40 enteropathogenic Escherichia coli in rabbits41 as well as the protozoan pathogen Entamoeba histolytica in mice.42 Interestingly, SFB monocolonization in GF mice does not provide protection against C. rodentium, suggesting the importance of an intact microbiota in coordinating a full cache of coordinated immune responses following SFB colonization.31
Adhesion of SFB to IECs as a crucial factor in shaping immunity
The ability of SFB to adhere to the intestinal epithelium is apparent in scanning electron microscopy images of the terminal ileum of mice harbouring SFB.25, 31, 43, 44, 45 SFB intimately attach to IECs of the ileum and cells overlying Peyer's patches (PPs), and this physical interaction is a critical step for the induction of Th17 cell differentiation (Fig. 1). Supporting evidence for this comes from cross‐species colonization experiments demonstrating that mouse‐specific SFB induce Th17 cell responses in mice, while rat‐specific SFB, which do not adhere to the mouse ileum, are unable to mediate this effect.44 SFB mutants lacking the ability to adhere to the epithelium also fail to induce Th17 differentiation in the intestine.44 Examination of the transcriptional programming induced in IECs by SFB adhesion revealed that serum amyloid A (SAA) proteins are important factors in Th17 programming.31, 44 The molecular trigger of SAA following SFB adhesion was reported to include the transcription factor CCAAT/enhancer‐binding protein δ induced by reorganization of the actin skeleton in epithelial cells.18 Epithelial secretion of SAA1 and SAA2 induced by SFB adhesion acts on CD11c+ cells to stimulate cytokine production that shapes the tissue microenvironment to potentiate the induction of Th17 cells. Interleukin‐23 produced by antigen‐presenting cells (APCs) upon recognition of SFB also initiates an IL‐23R/IL‐22 circuit in type 3 ILCs (ILC3) to promote SAA secretion from the epithelium;46 however, this appears to occur independent of SFB adhesion.18
Figure 1.
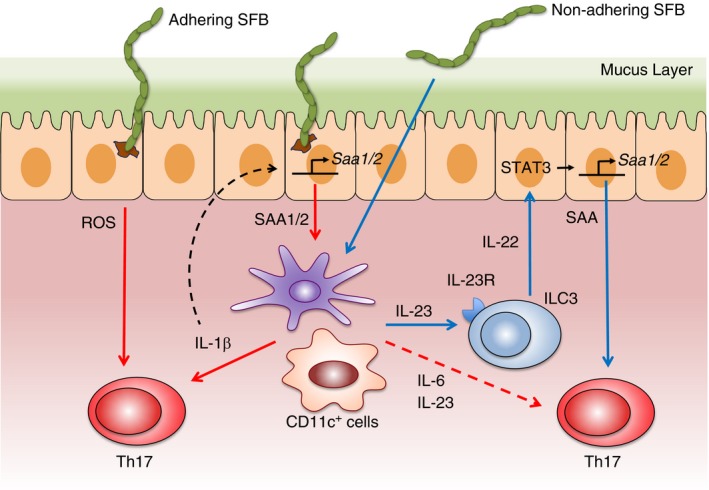
Segmented filamentous bacteria (SFB) adhesion and serum amyloid A (SAA) induction are critical factors in T helper type 17 (Th17) cell differentiation. Adhesion of SFB to the intestinal epithelial cells (IECs) of the ileum induces the production of SAA1 and SAA2. SAA proteins act on CD11c+ cells in the lamina propria (LP) to promote the production of interleukin‐6 (IL‐6) and IL‐23 that foster differentiation of naive CD4+ T cells into Th17 cells. SAA also triggers antigen‐presenting cells (APCs) to secrete IL‐1β that induces Th17 cell differentiation. IL‐1β from APCs can cycle back to act on IECs to elicit further SAA production, creating an amplification loop that favours greater Th17 cell induction. SFB adhesion to IEC also stimulates reactive oxygen species (ROS) that participate in the polarization of Th17 cells. In a separate signalling circuit where adhesion is not vital, SFB can stimulate production of IL‐23 from APCs that subsequently acts on type 3 innate lymphoid cells (ILC3) to secrete IL‐22. IL‐22 from ILC3 signals through IEC in a STAT3‐dependent manner to prompt the secretion of SAA that promotes the production of IL‐17A from primed ROR ɣt+ Th17 cells.
In addition to SAA induction, SFB adhesion also highly up‐regulates levels of the ROS‐generating enzymes dual oxidase 2 (Duox2) and its maturation factor Duoxa2 in IECs. Scavenging of ROS during SFB colonization limits Th17 cell induction, suggesting that ROS also play a role in the ability of SFB adhesion to influence Th17 cell responses.44 Additional factors induced by SFB adhesion may be involved, and it is apparent that Th17 cell induction in response to SFB is controlled by a number of integrated signals. One such factor is host genetic background, as BALB/c mice mount a much weaker Th17 cell induction in response to SFB. This strain‐specific effect may be partially explainable by reduced production of IL‐1β by CD11c+ cells and SAA by IECs in BALB/c mice.18
Sites of induction and Th17 cell specificity in response to SFB
The primary site of SFB adherence is the ileum,16 which is enriched with Th17 cells.46 Differentiation of Th17 cells appears to occur in the LP and associated lymphoid tissue, and a two‐step model has been proposed for this process. First, CD4+ T cells are primed in an antigen‐specific manner in the mesenteric lymph nodes (mLNs). After acquiring RORɣt expression, these T cells migrate back to the ileal LP. Second, RORɣt+ cells that traffic back to the ileum are programmed by the tissue microenvironment to further express IL‐17A.46 Robust expression of IL‐17A in the ileum correlates with SFB adhesion and the induction of SAA proteins.31, 44, 46 SAA production prompted by SFB adhesion may further influence Th17 differentiation through direct and/or indirect mechanisms.44, 46 Interestingly, the priming of Th17 cells in the mLNs is not requisite for SFB‐induced Th17 cell differentiation. Several studies have demonstrated that SFB can induce naive LP CD4+ T cells to become Th17 in the absence of secondary lymphoid tissue, such as mLNs.17, 47, 48 It appears that the priming and induction of Th17 cells may also occur locally in the small intestinal LP where APCs can sample and present antigen to influence CD4+ T cells in situ, probably in isolated lymphoid follicles.
Although not critical to Th17 differentiation, the presentation of SFB peptides in the mLNs is important for the development of antigen‐specific Th17 cells. Examination of the T‐cell receptor (TCR) repertoire of Th17 cells induced during SFB colonization revealed focused specificity towards antigens encoded by SFB and minimal overlap towards other unrelated antigens.49 In addition, TCR hybridomas derived from intestinal Th17 cells recognized antigens encoded by SFB whereas hybridomas from other non‐Th17 intestinal CD4+ T cells failed to recognize SFB‐derived antigens. To further elaborate the mechanisms of SFB‐induced Th17 cell specificity Yang et al. performed elegant experiments where intestinal Th17 cell responses were tracked in mice reconstituted with naive donor T cells expressing SFB‐specific TCRs. When these mice were colonized with SFB almost all donor T cells specific for SFB were found in the small intestinal LP and were positive for RORɣt+ expression; however, these donor cells were completely absent in the small intestinal LP of the mice that were not exposed to SFB. Furthermore, in these mice, simultaneous colonization with SFB and the strong Th1‐inducing bacteria Listeria monocytogenes promoted SFB‐specific T cells to differentiate only into RORɣt+ Th17 cells, suggesting that SFB dominates in its ability to direct Th17 differentiation. It is important to note that when mice harbouring T cells expressing a TCR specific for SFB were orally infected with L. monocytogenes engineered to express an SFB antigen but where void of SFB, SFB‐specific T cells were induced to express T‐bet rather than RORɣt. These striking findings are probably due to a characteristic of L. monocytogenes that favours Th1 responsiveness over a Th17 cell response and although this process is not completely understood, these data suggest that CD4+ T cells are polarized to become either Th1 or Th17 cells depending on which bacterial antigen is involved and the local microenvironment in which they differentiate. Notably, the Th17 responses induced by SFB in the absence of secondary lymphoid tissue are mostly non‐specific, suggesting the ability of SFB to create a Th17‐priming environment irrespective of antigen specificity.17 In support of this concept, the microenvironment conditioned by SFB colonization appeared sufficient to promote Th17 cell differentiation of ovalbumin‐specific CD4+ T cells (OT‐II) on a recombinase activating gene (RAG)‐sufficient background in the presence of cognate antigen,47 although another study found that OT‐II cells on a RAG‐deficient background could not undergo Th17 differentiation in the presence of antigen.50 Overall, it is likely that both antigen specificity and the tissue microenvironment induced by SFB play a role in Th17 cell induction for both SFB‐specific and non‐specific CD4+ T cells.
Antigen‐presenting cells direct SFB‐induced Th17 responses
In the intestine a highly complex network of APCs comprised of dendritic cells (DCs) and macrophages is responsible for acquiring and presenting bacterial antigens to CD4+ T cells. In response to bacterial encounter, DCs and macrophages are actively influenced to secrete cytokines that drive unique CD4+ T‐cell differentiation and expansion in the mLN and LP.51, 52 Important cytokines that promote naive CD4+ T‐cell differentiation into Th17 cells include IL‐1β, IL‐6, IL‐23 and transforming growth factor‐β.32, 53 In vitro approaches using CD11c+ LP DCs have demonstrated that these cytokines may also participate in SFB‐induced Th17 cell induction.31, 44 Additionally, SAA can robustly potentiate Th17 cell differentiation in DC–T‐cell co‐cultures,31, 44 which may be through the ability of SAA to induce IL‐6 and IL‐23 in LP DCs.31 SAA can also prompt the production of IL‐1β by CD11c+ cells, which can act on the epithelium to further stimulate SAA secretion, so creating an amplification loop for the induction of Th17 cells. The tissue microenvironment shaped by SFB colonization appears to programme APCs to induce Th17 cells, as CD11c+ DCs isolated from mice harbouring SFB are more efficient at inducing Th17 cells in vitro than counterparts isolated from mice void of SFB.54 Such in vivo conditioning of DCs by SFB appears to extend outside the intestine as bone‐marrow‐derived DCs from SFB‐colonized mice are also conditioned to produce more IL‐23 and IL‐17A than DCs from SFB‐negative mice.42
In vivo studies have also demonstrated the importance of CD11c+ LP cells in SFB‐induced Th17 cell responses. Specifically, this process requires antigen presentation by CD11c+ cells of the intestinal LP through MHCII to initiate Th17 differentiation in the LP following SFB colonization.47, 50 As CD11c is expressed by both DCs and macrophages in the LP, and these cells participate in the priming and/or maintenance of Th17 cells at mucosal surfaces through their expression of MHCII, it has been a challenge to dissect their unique and overlapping roles.51, 54 Attempting to better define the specific contributions of DCs and macrophages to SFB‐induced Th17 cell responses, Panea et al. took a targeted approach in vivo using a number of different transgenic mice. Interestingly, depletion of the different subsets of DCs in the intestine (CD11b+ CD103+, CD11b− CD103+ and CD11b+ CD103−), had no detectable effect on Th17 cell levels in the LP following SFB colonization. Instead, CX3CR1+ macrophages derived from Ly6C+ CCR2+ monocytes appeared to be most critical for SFB‐induced Th17 cell responses.55 As macrophages are not adept at migrating to lymph nodes, these findings are aligned with evidence showing that secondary lymphoid tissues are dispensable to SFB‐induced Th17 cell differentiation.47, 50 However, the apparent requirement of secondary lymphoid tissue to enforce Th17 specificity towards SFB antigens17, 46 suggests that DCs and macrophages play coordinated roles in this process.
The role of SFB in shaping immune cell function
Following the presentation of SFB antigens by APCs and the subsequent acquisition of RORɣt expression, CD4+ T cells in the LP of the ileum are prompted to produce IL‐17A by SAA secreted from IECs. The IL‐17A derived from Th17 cells is an important effector cytokine that can act directly on the epithelium to induce the production of antimicrobial peptides and other cytokines and chemokines, including those involved in the recruitment of neutrophils. However, not all SFB‐driven responses require Th17 cells and other immune cells can directly respond to SFB through different mechanisms. Some of these responses require SFB adherence to IECs while others occur in a non‐adherent manner and together these immune cell functions, which will be detailed below, mount a coordinated effort to contain SFB growth.
B cells
Mucosal IgA is secreted across the epithelium where it binds to bacteria and limits direct contact with the host.56, 57 Colonization and subsequent adhesion of SFB to IECs, especially those overlying PPs, is capable of inducing and enhancing intestinal IgA responses.9, 17, 18, 44, 58 Monocolonization of GF mice with SFB potently expands PP germinal centres, which are a major site of intestinal IgA production. Coinciding with germinal centre expansion following SFB colonization are increases in B220+ IgA+ B cells and faecal IgA.17, 44 The SFB‐induced IgA response is dependent on SFB adhesion44 and shows gradual accumulation of somatic mutations and large clonal diversity, indicative of a broad IgA response. SFB‐induced IgA is reduced in T‐cell‐deficient mice, suggesting that high‐affinity, antigen‐specific T‐cell‐dependent antibody responses may be important in this response.59 Interestingly, SFB‐induced IgA production does not require PPs as SFB potently stimulates development of isolated lymphoid follicles and tertiary lymphoid tissue that can compensate for the loss PPs. Interactions with other members of the microbiota may also be important for IgA production following SFB colonization as there are significantly more abundant and specific IgA responses in conventional mice colonized with SFB compared with SFB‐monoassociated mice.17 Once produced, IgA is capable of coating SFB59 and may provide protection to the host through immune exclusion and control of the SFB niche. IgA deficiency in the gut leads to robust expansion of SFB, an effect that is reversed by reconstitution of the IgA response.58 In addition to inducing IgA, SFB also augments expression of the polymeric immunoglobulin receptor in IEC,43 which is important for transporting IgA across the epithelium and into the lumen.60, 61
Innate lymphoid cells
An early observation in SFB studies was that along with the induction of IL‐17, SFB could also promote IL‐22 production in the small intestine.31 Although Th17 cells can produce IL‐22, ILC3 are believed to be the major contributor of intestinal IL‐22. The ability of SFB to induce IL‐22 appears to be dependent upon IL‐23 signalling, as IL‐23 secreted following SFB colonization can signal through the IL‐23R on ILC3 to trigger the release of IL‐22. As such, defects in this IL‐23R/IL‐22 axis can lead to expansion of SFB.20, 21 The aryl hydrocarbon receptor also plays a role in this axis by maintaining ILC3 and their production of IL‐22.20 Interestingly, SFB‐induced IL‐22 does not require adherence of SFB to the epithelium, as rat‐specific SFB, which does not adhere in mice, can induce levels of IL‐22 production by ILC3 comparable to that observed with mouse‐specific SFB.44 Interleukin‐22 is well‐appreciated to promote antimicrobial activity, trigger protective epithelial fucosylation,48 and bolster barrier function.19 These effects of SFB‐induced IL‐22 from ILC3 may explain the host protection against enteric pathogens mediated by SFB.
In addition to producing IL‐22 in response to SFB, ILC3 can potentiate Th17 cell differentiation through SAA release triggered by signal transducer and activator of transcription 3‐dependent IL‐22 signalling in the epithelium.46 Conversely, another study found that ILC3 and the IL‐23R/IL‐22 signalling axis can limit Th17 cell responses.21 Although the principle effector mechanism of ILC3 is IL‐22 production, they also express MHCII and can suppress Th17 responses50 that may be associated with their ability to limit commensal bacteria‐specific CD4+ T‐cell responses.62 Future studies are likely to clarify the complex and dynamic interplay between SFB, ILC3 and Th17 cells.
Granulocytes
Consistent with the induction of a neutrophilic response promoted by Th17 cells responding to extracellular pathogens in the lung,32 SFB colonization also leads to a robust influx of neutrophils into the ileum. Interestingly, this neutrophil influx requires adaptive immune cells (probably Th17) and IL‐17A, as RAG‐deficient mice and mice treated wth IL‐17A neutralizing antibody exhibit dramatically impaired neutrophil recruitment in response to SFB colonization.33 The link between Th17 cells, IL‐17A and neutrophil accumulation appears at least partially mediated by the induction of the neutrophil chemokines CXCL1 and CXCL2 by IL‐17A. Once in the ileum, neutrophils recruited in response to SFB are instrumental in the direct and/or indirect production of IL‐22. The IL‐22 can be packaged and delivered from granules in neutrophils and contributes to the production of antimicrobial peptides including RegIIIα, RegIIIβ, RegIIIγ and defensins, which may limit the growth of SFB.33, 63, 64 Failure to mount a neutrophil response following SFB colonization not only leads to overgrowth of SFB, but also leads to accumulation of Th17 cells in the ileum. In this regard, neutrophils constitute a key innate effector arm responsible for limiting SFB expansion and Th17 cells.33 Overall, neutrophils are key early responders that coordinate with other innate and adaptive immune cells and factors to contain SFB and provide critical host protection (Fig. 2).
Figure 2.
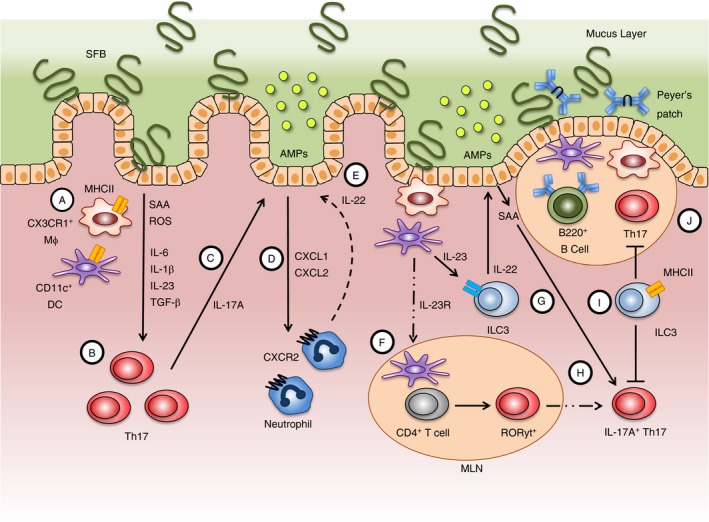
Summarized are the complex immune cell functions in the small intestinal lamina propria (LP) and adjacent lymphoid tissue that are shaped by colonization with SFB. Several mechanisms dictate the T helper type 17 (Th17) cell response towards SFB, including MHCII‐mediated presentation of SFB antigens by CD11c+ cells [CX3CR1+ macrophages (Mϕ) and CD11c+ dendritic cells (DCs)] and the secretion of cytokines into the local environment (A) that promotes the differentiation of naive CD4+ T cells into Th17 cells locally in the LP (B). Interleukin‐17A (IL‐17A) produced from Th17 cells can act on the epithelium to promote barrier function (C) while also stimulating neutrophil recruitment via CXCL1 and CXCL2 (D). IL‐17A from T cells and IL‐22 from neutrophils can act on the epithelium to trigger the release of antimicrobial peptides (AMPs) that can help control SFB growth (E). Naive CD4+ T cells in the mesenteric lymph nodes (MLNs) are also primed to express the transcription factor ROR ɣt by migrating DCs carrying SFB antigen (F). DCs exposed to SFB antigen also produce IL‐23 that binds to the IL‐23R on type 3 innate lymphoid cells (ILC3) to enhance secretion of IL‐22 (G). Serum amyloid A (SAA) released from IECs in response to ILC3‐derived IL‐22 can act on ROR ɣt+ T cells homing back to the gut from the MLN to promote IL‐17A production (H). In addition to providing barrier protection through the production of IL‐22 and subsequent expression of AMPs, ILC3 can also directly inhibit Th17 cell responses through MHCII (I). SFB colonization also robustly potentiates the formation of germinal centres in Peyer's patches and the secretion of IgA from B cells that work as another line of protection in controlling SFB growth (J).
Influence of SFB colonization on extra‐intestinal tissues
The influence of SFB on the immune system extends beyond the intestine and can have effects on peripheral sites. In distal organs where Th17 cells promote host defence, SFB colonization can be beneficial to the host. However, SFB colonization can also favour the progression of diseases where Th17 cell responses play a role in pathogenesis including arthritis and multiple sclerosis. Other immune cells shaped by SFB colonization may play unique roles dependent upon their contribution to the disease pathogenesis (Fig. 3).
Figure 3.
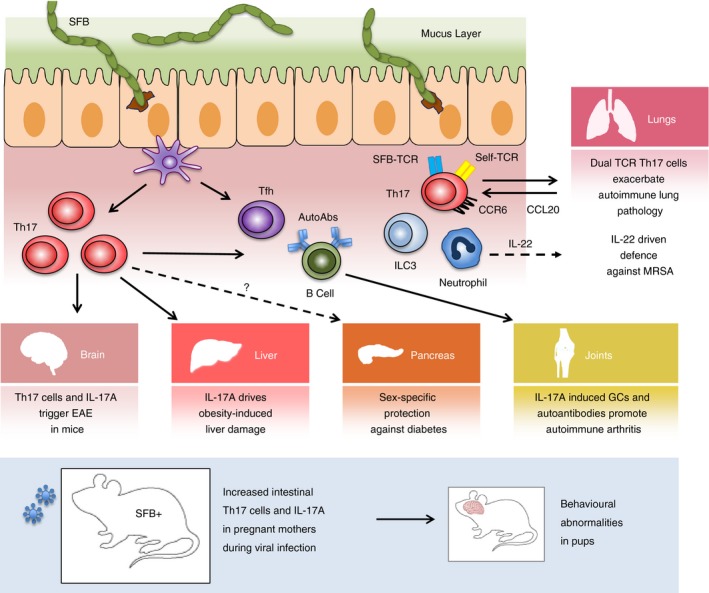
Intestinal segmented filamentous bacteria (SFB) colonization influences immune responses in distant organ sites through various mechanisms. Different and overlapping immune responses triggered by SFB colonization of the intestine can promote autoimmunity and pathology in the brain, liver, joints and lungs. Viral infection in pregnant mothers colonized with SFB can lead to developmental and behavioural abnormalities in offspring through a mechanism involving interleukin‐17A (IL‐17A). Distal responses influenced by SFB can also protect against disease in the pancreas and help fight infections in the lungs.
Joints
The ability of SFB colonization to exacerbate autoimmunity was first demonstrated in a mouse model of autoimmune arthritis. In the K/BxN mouse model, which resembles human inflammatory arthritis, spontaneous disease development is driven by the microbiota and disease is almost completely absent in GF mice.65 Interestingly, mono‐association of GF mice with SFB was capable of rapidly triggering disease that was associated with increased Th17 cells and auto‐antibody production. Increased levels of IL‐17A from intestinal Th17 cells responding to SFB were shown to be important for driving this model, potentially through a direct effect on B cells, which enhances germinal centre formation and the production of autoantibodies.65, 66 SFB colonization is also capable of stimulating T follicular helper (Tfh) cell differentiation in PPs through a mechanism involving suppression of IL‐2 signalling and induction of Bcl6. Once Tfh cells egress from PPs to distal systemic lymphoid tissues they can promote germinal centre responses and increase production of autoantibodies, driving autoimmune arthritis in SFB‐colonized mice.67
Central nervous system
The establishment of the microbiota as an important component in the gut–brain axis has revealed the contributions of microbes, including SFB, in diseases of the central nervous system (CNS). GF mice that are normally resistant to experimental autoimmune encephalomyelitis, a preclinical model of multiple sclerosis in mice, develop robust disease when monocolonized with SFB. In these studies, increased Th17 cell numbers in the intestine paralleled increased CNS disease activity and heightened levels Th17 cells and IL‐17A production in the spinal cord.68 As Th17 cells are the first wave of proinflammatory T cells that infiltrate the CNS in EAE, SFB colonization is sufficient to initiate neurological inflammation in mice.69 Defective barrier defences and failure to contain SFB may also exacerbate EAE through augmented proliferation of Th17 cells responding to SFB overgrowth.34
Recent evidence has also drawn a link between SFB‐induced Th17 cell responses in the intestine and developmental abnormalities in the CNS. For example, offspring born to mothers harbouring SFB following maternal immune activation (MIA), which mimics viral infection, are much more prone to neurodevelopmental abnormalities.70, 71 These effects may be due in part to DCs from pregnant mothers having a greater propensity to produce the Th17‐inducing cytokines IL‐1β, IL‐6 and IL‐23 when compared with non‐pregnant controls. Functional Toll‐like receptor‐3‐signalling in DCs also appears to be required to potentiate intestinal Th17 cell differentiation induced by SFB colonization following MIA.
Lungs
The influence of SFB colonization on immune responses can also traverse the gut–lung axis to impact pulmonary function. Paralleling protection afforded to the gut, colonization with SFB can also protect against certain lung infections. Acquisition of SFB has been shown to protect mice from acute lung infection with methicillin‐resistant Staphylococcus aureus (MRSA).72 This heightened defence against MRSA allotted by SFB colonization does not appear to be related to IL‐17A production per se, and instead seems to correlate with increased infiltration of neutrophils and heightened levels of IL‐22 in the lung. Indeed, neutralization of IL‐22 was capable of blocking the protective effects of SFB, while administration of IL‐22 to non‐SFB‐colonized mice could provide a similar level of protection as SFB‐colonized mice.72 The specific cellular source of IL‐22 in this study was not elucidated but could be produced by T cells, ILCs, or neutrophils. In addition to protecting from MRSA, SFB colonization can also enhance immune defence against the fungi Aspergillus fumigatus through augmenting Th17 cells.73
The effects of SFB on the lungs are not always beneficial; however. SFB colonization can promote lung pathology that often accompanies autoimmune arthritis by enhancing Th17 immune responses. Enhanced Th17 cell responses in SFB colonized mice do not seem to be the result of bystander activation and instead lead to the expansion of Th17 cells co‐expressing TCRs that recognize an SFB epitope and self‐antigen. Systemic inflammation driven by the initiation of arthritis was recently shown to drive expression of the T‐cell chemoattractant CCL20 in the lung, allowing for SFB‐induced Th17 cells expressing dual TCRs to migrate to the lung and provoke pulmonary damage.74 CCL20 production by the lung represents just one mechanism by which SFB‐induced Th17 cells in the intestine can travel to the lung and other mechanisms may regulate migration of Th17 cells to distinct peripheral organs.
Liver and pancreas
Two additional peripheral sites that can be influenced by SFB colonization are the liver and pancreas. In the liver, SFB‐induced IL‐17A signalling can exacerbate obesity‐induced liver damage associated with non‐alcoholic fatty liver disease in mice.75 SFB may also drive immune‐mediated damage in other models of liver disease.76 Interestingly, SFB‐colonization can, to some extent, provide protection against the development of diabetes in mice. Although SFB colonization does not restrain infiltration of T cells into the pancreas (insulitis), SFB can prevent the development of diabetes in predisposed non‐obese diabetic mice. Strikingly, SFB‐induced protection from diabetes appears to be sex‐specific as effects only occurred in females and were correlated with increased Th17 cell numbers in the intestinal LP, but not the pancreas and systemic lymphoid tissue.77 These results provide evidence of SFB‐mediated protection of disease in predisposed mice; however, the exact mechanisms of protection, including those related to sex differences, remain to be determined.
Conclusions
The discovery that SFB colonization robustly induces Th17 cells in the intestine was a notable breakthrough in the understanding of how specific components of the microbiota regulate unique immune responses; however, fundamental questions remain unanswered. First and foremost is whether advances in defining the experimental biology of SFB in mice has translational potential to improve human health. Although SFB can colonize humans, no data currently exist linking SFB to human health or disease. Even if SFB turns out to not be a major contributor to human health, studies defining how SFB influences host immune responses, particularly Th17 responses, may be informative in defining how distinct components of the human gut microbiota may influence intestinal and peripheral immunity. Importantly, it does not appear that Th17 induction is restricted to SFB, as recent evidence has demonstrated that the commensal bacterium Bifidobacterium adolescentis is capable of inducing intestinal Th17 cells in mice.78 Given that SFB are genetically distinct and host‐specific, the search for human equivalents that can foster potent intestinal Th17 cell responses warrants further investigation. Exciting recent findings have indicated that a 20‐member consortium of bacteria isolated from humans can induce Th17 cell differentiation in the intestinal LP of mice,44 providing initial clues that SFB‐like bacteria may indeed exist in humans. Another area where additional studies are needed involves potential interactions of SFB with other members of a complex microbiota, including viruses and fungi. Studies comparing SFB‐induced immune responses in conventionally housed and SFB‐monoassociated mice should help to shed light on how other members of the microbiota influence effects of SFB. Lastly, mechanisms behind the ability of SFB to promote autoimmunity have been uncovered, but how immune responses in the intestine bridge the gap to make their way to distal organs is less well understood and deserving of much more attention. Fortunately, researchers are now equipped with a full cache of tools to dissect these important questions. A better understanding of how immune networks are shaped by SFB, and other members of the microbiota, combined with the knowledge of how these responses are paralleled in humans may provide a fertile landscape for therapeutic discoveries targeted at infectious diseases and autoimmunity.
Disclosures
The authors have no competing interests to disclose.
Acknowledgements
KLF is supported by postdoctoral fellowships from Alberta Innovates and AbbVie/Canadian Association of Gastroenterology/Canadian Institutes for Health Research. TLD is supported by National Institutes of Health grant 1R01DK097256.
References
- 1. Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science 2012; 336:1268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou L, Sonnenberg GF. Essential immunologic orchestrators of intestinal homeostasis. Sci Immunol 2018; 3:pii: eaao1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 2009; 9:313–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature 2016; 535:75–84. [DOI] [PubMed] [Google Scholar]
- 5. Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011; 331:337–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013; 500:232–6. [DOI] [PubMed] [Google Scholar]
- 7. Coccia M, Harrison OJ, Schiering C, Asquith MJ, Becher B, Powrie F, et al IL‐1β mediates chronic intestinal inflammation by promoting the accumulation of IL‐17A secreting innate lymphoid cells and CD4+ Th17 cells. J Exp Med 2012; 209:1595–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Morrison PJ, Bending D, Fouser LA, Wright JF, Stockinger B, Cooke A, et al Th17‐cell plasticity in Helicobacter hepaticus‐induced intestinal inflammation. Mucosal Immunol 2013; 6:1143–56. [DOI] [PubMed] [Google Scholar]
- 9. Klaasen HL, Van der Heijden PJ, Stok W, Poelma FG, Koopman JP, Van den Brink ME, et al Apathogenic, intestinal, segmented, filamentous bacteria stimulate the mucosal immune system of mice. Infect Immun 1993; 61:303–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blumershine RV, Savage DC. Filamentous microbes indigenous to the murine small bowel: a scanning electron microscopic study of their morphology and attachment to the epithelium. Microb Ecol 1977; 4:95–103. [DOI] [PubMed] [Google Scholar]
- 11. Klaasen HL, Koopman JP, Poelma FG, Beynen AC. Intestinal, segmented, filamentous bacteria. FEMS Microbiol Rev 1992; 8:165–80. [DOI] [PubMed] [Google Scholar]
- 12. Tannock GW, Miller JR, Savage DC. Host specificity of filamentous, segmented microorganisms adherent to the small bowel epithelium in mice and rats. Appl Environ Microbiol 1984; 47:441–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Prakash T, Oshima K, Morita H, Fukuda S, Imaoka A, Kumar N, et al Complete genome sequences of rat and mouse segmented filamentous bacteria, a potent inducer of th17 cell differentiation. Cell Host Microbe 2011; 10:273–84. [DOI] [PubMed] [Google Scholar]
- 14. Pamp SJ, Harrington ED, Quake SR, Relman DA, Blainey PC. Single‐cell sequencing provides clues about the host interactions of segmented filamentous bacteria (SFB). Genome Res 2012; 22:1107–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sczesnak A, Segata N, Qin X, Gevers D, Petrosino JF, Huttenhower C, et al The genome of th17 cell‐inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host Microbe 2011; 10:260–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chase DG, Erlandsen SL. Evidence for a complex life cycle and endospore formation in the attached, filamentous, segmented bacterium from murine ileum. J Bacteriol 1976; 127:572–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lecuyer E, Rakotobe S, Lengline‐Garnier H, Lebreton C, Picard M, Juste C, et al Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity 2014; 40:608–20. [DOI] [PubMed] [Google Scholar]
- 18. Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun 1999; 67:1992–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dudakov JA, Hanash AM, van den Brink MR. Interleukin‐22: immunobiology and pathology. Annu Rev Immunol 2015; 33:747–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qiu J, Guo X, Chen ZM, He L, Sonnenberg GF, Artis D, et al Group 3 innate lymphoid cells inhibit T‐cell‐mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity 2013; 39:386–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shih VF, Cox J, Kljavin NM, Dengler HS, Reichelt M, Kumar P, et al Homeostatic IL‐23 receptor signaling limits Th17 response through IL‐22‐mediated containment of commensal microbiota. Proc Natl Acad Sci USA 2014; 111:13942–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, et al The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL‐17+ T helper cells. Cell 2006; 126:1121–33. [DOI] [PubMed] [Google Scholar]
- 23. Esplugues E, Huber S, Gagliani N, Hauser AE, Town T, Wan YY, et al Control of TH17 cells occurs in the small intestine. Nature 2011; 475:514–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, et al Specific microbiota direct the differentiation of IL‐17‐producing T‐helper cells in the mucosa of the small intestine. Cell Host Microbe 2008; 4:337–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gaboriau‐Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, et al The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 2009; 31:677–89. [DOI] [PubMed] [Google Scholar]
- 26. Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, et al Interleukin 17‐producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol 2005; 6:1123–32. [DOI] [PubMed] [Google Scholar]
- 27. Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, et al A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol 2005; 6:1133–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, et al Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 2006; 441:235–8. [DOI] [PubMed] [Google Scholar]
- 29. Mangan PR, Harrington LE, O'Quinn DB, Helms WS, Bullard DC, Elson CO, et al Transforming growth factor‐β induces development of the TH17 lineage. Nature 2006; 441:231–4. [DOI] [PubMed] [Google Scholar]
- 30. Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, et al IL‐6 programs TH‐17 cell differentiation by promoting sequential engagement of the IL‐21 and IL‐23 pathways. Nat Immunol 2007; 8:967–74. [DOI] [PubMed] [Google Scholar]
- 31. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009; 139:485–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weaver CT, Elson CO, Fouser LA, Kolls JK. The Th17 pathway and inflammatory diseases of the intestines, lungs, and skin. Annu Rev Pathol 2013; 8:477–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Flannigan KL, Ngo VL, Geem D, Harusato A, Hirota SA, Parkos CA, et al IL‐17A‐mediated neutrophil recruitment limits expansion of segmented filamentous bacteria. Mucosal Immunol 2017; 10:673–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kumar P, Monin L, Castillo P, Elsegeiny W, Horne W, Eddens T, et al Intestinal interleukin‐17 receptor signaling mediates reciprocal control of the gut microbiota and autoimmune inflammation. Immunity 2016; 44:659–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee JS, Tato CM, Joyce‐Shaikh B, Gulen MF, Cayatte C, Chen Y, et al Interleukin‐23‐independent IL‐17 production regulates intestinal epithelial permeability. Immunity 2015; 43:727–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maxwell JR, Zhang Y, Brown WA, Smith CL, Byrne FR, Fiorino M, et al Differential roles for interleukin‐23 and interleukin‐17 in intestinal immunoregulation. Immunity 2015; 43:739–50. [DOI] [PubMed] [Google Scholar]
- 37. Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi‐Joannopoulos K, Collins M, et al Interleukin (IL)‐22 and IL‐17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med 2006; 203:2271–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, et al Gut immune maturation depends on colonization with a host‐specific microbiota. Cell 2012; 149:1578–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Edelblum KL, Sharon G, Singh G, Odenwald MA, Sailer A, Cao S, et al The microbiome activates CD4 T‐cell‐mediated immunity to compensate for increased intestinal permeability. Cell Mol Gastroenterol Hepatol 2017; 4:285–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Garland CD, Lee A, Dickson MR. Segmented filamentous bacteria in the rodent small intestine: their colonization of growing animals and possible role in host resistance to Salmonella. Microb Ecol 1982; 8:181–90. [DOI] [PubMed] [Google Scholar]
- 41. Heczko U, Abe A, Finlay BB. Segmented filamentous bacteria prevent colonization of enteropathogenic Escherichia coli O103 in rabbits. J Infect Dis 2000; 181:1027–33. [DOI] [PubMed] [Google Scholar]
- 42. Burgess SL, Buonomo E, Carey M, Cowardin C, Naylor C, Noor Z, et al Bone marrow dendritic cells from mice with an altered microbiota provide interleukin 17A‐dependent protection against Entamoeba histolytica colitis. MBio 2014; 5:e01817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schnupf P, Gaboriau‐Routhiau V, Gros M, Friedman R, Moya‐Nilges M, Nigro G, et al Growth and host interaction of mouse segmented filamentous bacteria in vitro . Nature 2015; 520:99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, et al Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 2015; 163:367–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Davis CP, Savage DC. Habitat, succession, attachment, and morphology of segmented, filamentous microbes indigenous to the murine gastrointestinal tract. Infect Immun 1974; 10:948–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ, et al An IL‐23R/IL‐22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell 2015; 163:381–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Geem D, Medina‐Contreras O, McBride M, Newberry RD, Koni PA, Denning TL. Specific microbiota‐induced intestinal Th17 differentiation requires MHC class II but not GALT and mesenteric lymph nodes. J Immunol 2014; 193:431–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Goto Y, Obata T, Kunisawa J, Sato S, Ivanov II, Lamichhane A, et al Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science 2014; 345:1254009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, et al Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature 2014; 510:152–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Goto Y, Panea C, Nakato G, Cebula A, Lee C, Diez MG, et al Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity 2014; 40:594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Denning TL, Wang YC, Patel SR, Williams IR, Pulendran B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17‐producing T cell responses. Nat Immunol 2007; 8:1086–94. [DOI] [PubMed] [Google Scholar]
- 52. Flannigan KL, Geem D, Harusato A, Denning TL. Intestinal antigen‐presenting cells: key regulators of immune homeostasis and inflammation. Am J Pathol 2015; 185:1809–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, et al A validated regulatory network for Th17 cell specification. Cell 2012; 151:289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Denning TL, Norris BA, Medina‐Contreras O, Manicassamy S, Geem D, Madan R, et al Functional specializations of intestinal dendritic cell and macrophage subsets that control Th17 and regulatory T cell responses are dependent on the T cell/APC ratio, source of mouse strain, and regional localization. J Immunol 2011; 187:733–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Panea C, Farkas AM, Goto Y, Abdollahi‐Roodsaz S, Lee C, Koscso B, et al Intestinal monocyte‐derived macrophages control commensal‐specific Th17 responses. Cell Rep 2015; 12:1314–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell‐independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science 2000; 288:2222–6. [DOI] [PubMed] [Google Scholar]
- 57. Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 2004; 303:1662–5. [DOI] [PubMed] [Google Scholar]
- 58. Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, et al Aberrant expansion of segmented filamentous bacteria in IgA‐deficient gut. Proc Natl Acad Sci USA 2004; 101:1981–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, et al Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014; 158:1000–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brandtzaeg P. Mucosal immunity: induction, dissemination, and effector functions. Scand J Immunol 2009; 70:505–15. [DOI] [PubMed] [Google Scholar]
- 61. Brandtzaeg P, Prydz H. Direct evidence for an integrated function of J chain and secretory component in epithelial transport of immunoglobulins. Nature 1984; 311:71–3. [DOI] [PubMed] [Google Scholar]
- 62. Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, et al Innate lymphoid cells regulate CD4+ T‐cell responses to intestinal commensal bacteria. Nature 2013; 498:113–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zindl CL, Lai JF, Lee YK, Maynard CL, Harbour SN, Ouyang W, et al IL‐22‐producing neutrophils contribute to antimicrobial defense and restitution of colonic epithelial integrity during colitis. Proc Natl Acad Sci USA 2013; 110:12768–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chen F, Cao A, Yao S, Evans‐Marin HL, Liu H, Wu W, et al mTOR mediates IL‐23 induction of neutrophil IL‐17 and IL‐22 production. J Immunol 2016; 196:4390–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, et al Gut‐residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 2010; 32:815–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hsu HC, Yang P, Wang J, Wu Q, Myers R, Chen J, et al Interleukin 17‐producing T helper cells and interleukin 17 orchestrate autoreactive germinal center development in autoimmune BXD2 mice. Nat Immunol 2008; 9:166–75. [DOI] [PubMed] [Google Scholar]
- 67. Teng F, Klinger CN, Felix KM, Bradley CP, Wu E, Tran NL, et al Gut microbiota drive autoimmune arthritis by promoting differentiation and migration of Peyer's patch T follicular helper cells. Immunity 2016; 44:875–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T‐cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA 2011; 108(Suppl 1):4615–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kebir H, Kreymborg K, Ifergan I, Dodelet‐Devillers A, Cayrol R, Bernard M, et al Human TH17 lymphocytes promote blood–brain barrier disruption and central nervous system inflammation. Nat Med 2007; 13:1173–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kim S, Kim H, Yim YS, Ha S, Atarashi K, Tan TG, et al Maternal gut bacteria promote neurodevelopmental abnormalities in mouse offspring. Nature 2017; 549:528–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shin Yim Y, Park A, Berrios J, Lafourcade M, Pascual LM, Soares N, et al Reversing behavioural abnormalities in mice exposed to maternal inflammation. Nature 2017; 549:482–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gauguet S, D'Ortona S, Ahnger‐Pier K, Duan B, Surana NK, Lu R, et al Intestinal microbiota of mice influences resistance to Staphylococcus aureus pneumonia. Infect Immun 2015; 83:4003–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. McAleer JP, Nguyen NL, Chen K, Kumar P, Ricks DM, Binnie M, et al Pulmonary Th17 antifungal immunity is regulated by the gut microbiome. J Immunol 2016; 197:97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bradley CP, Teng F, Felix KM, Sano T, Naskar D, Block KE, et al Segmented filamentous bacteria provoke lung autoimmunity by inducing gut–lung axis Th17 cells expressing dual TCRs. Cell Host Microbe 2017; 22(697–704):e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Harley IT, Stankiewicz TE, Giles DA, Softic S, Flick LM, Cappelletti M, et al IL‐17 signaling accelerates the progression of nonalcoholic fatty liver disease in mice. Hepatology 2014; 59:1830–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Celaj S, Gleeson MW, Deng J, O'Toole GA, Hampton TH, Toft MF, et al The microbiota regulates susceptibility to Fas‐mediated acute hepatic injury. Lab Invest 2014; 94:938–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci USA 2011; 108:11548–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Tan TG, Sefik E, Geva‐Zatorsky N, Kua L, Naskar D, Teng F, et al Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc Natl Acad Sci USA 2016; 113:E8141–50. [DOI] [PMC free article] [PubMed] [Google Scholar]


