Abstract
DDX3 is an RNA helicase with oncogenic properties. The small molecule inhibitor RK-33 is designed to fit into the ATP binding cleft of DDX3 and hereby block its activity. RK-33 has shown potent activity in preclinical cancer models. However, the mechanism behind the antineoplastic activity of RK-33 remains largely unknown. In this study we used a dual phosphoproteomic and single cell tracking approach to evaluate the effect of RK-33 on cancer cells. MDA-MB-435 cells were treated for 24 hours with RK-33 or vehicle control. Changes in phosphopeptide abundance were analyzed with quantitative mass spectrometry using isobaric mass tags (Tandem Mass Tags). At the proteome level we mainly observed changes in mitochondrial translation, cell division pathways and proteins related to cell cycle progression. Analysis of the phosphoproteome indicated decreased CDK1 activity after RK-33 treatment. To further evaluate the effect of DDX3 inhibition on cell cycle progression over time, we performed timelapse microscopy of Fluorescent Ubiquitin Cell Cycle Indicators labeled cells after RK-33 or siDDX3 exposure. Single cell tracking indicated that DDX3 inhibition resulted in a global delay in cell cycle progression in interphase and mitosis. In addition, we observed an increase in endoreduplication. Overall, we conclude that DDX3 inhibition affects cells in all phases and causes a global cell cycle progression delay.
Introduction
DEAD box RNA helicases form a large protein family with ATPase dependent helicase activity and are characterized by the presence of a highly conserved D-E-A-D (Asp-Glu-Ala-Asp) motif. Due to their ability to unwind complex RNA structures, they have been linked to virtually all steps of RNA processing: from transcription and translation to the regulation of small non-coding RNA's [1]. DDX3, also known as DDX3X, is a family member which has an oncogenic role in the development of breast [2] and several other types of cancer [3], [4], [5]. DDX3 was demonstrated to have anti-apoptotic properties [6], [7], [8] and to play a role in migration [2], [9] and invasion [10], [11], [12]. In addition, several studies have linked DDX3 to cell cycle progression [13], [14] and DDX3 inhibition has been reported to result in a G1-arrest [3], [4]. Interestingly, the mechanism by which DDX3 regulates these processes is not limited to mRNA translational control. DDX3, like other DEAD box RNA helicases, was found to be multifunctional [15] and for instance directly regulates the kinase activity of CK1ε [16].
A small molecule inhibitor, RK-33, was recently developed to target DDX3 for cancer treatment [17]. RK-33 is designed to inactivate DDX3 by binding to its ATP pocket and was found to block the in vitro helicase activity of the yeast homologue of DDX3, Ded1p. Pull down experiments showed that RK-33 selectively binds DDX3 over other DEAD box RNA helicase family members [4]. Several preclinical models demonstrated RK-33 to have potent anti-cancer activity, both as a monotherapy [18] and as a radiosensitizer [4], [5]. However, better understanding of the mechanism through which RK-33 exerts its effect is needed.
This study aims to further elucidate the working mechanism of DDX3 inhibition with RK-33 by using a dual approach. Given the role of DDX3 in translation and regulation of kinase activity we performed a phosphoproteomics experiment, to monitor the changes after RK-33 treatment on the protein expression level and the protein phosphorylation status. In addition, since DDX3 has been linked to cell cycle progression, and cell cycle status has a strong influence on the phosphoproteomic landscape of the cell, we tracked the cell cycle status of single cells after RK-33 exposure, to shed further light on the influence of DDX3 inhibition on cell cycle progression over time.
Methods
Cell Culture
MCF7 and MDA-MB-435 cells were originally purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cell lines were STR-profiled (Geneprint 10, Promega, Madison, WI, USA, last in November 2015) and mycoplasma tested (Universal Mycoplasma Detection Kit, ATCC, last in January 2016) on a regular basis. For proteomics experiments MDA-MB-435 cells were plated in 100 mm dishes and allowed to attach overnight. Three replicates were exposed to 4.5 μM RK-33 or DMSO for 24 hours and harvested in extraction buffer, containing 1% SDS, 1 mM EDTA, 1 mM sodium orthovanadate, 1 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM sodium fluoride and proteinase inhibitor cocktail. 500 μg of protein was reduced with 5 mM DTT, alkylated with 10 mM Iodoacetaminde, precipitated by TCA/acetone and digested with trypsin in 10 mM TEAB buffer.
Proteomics
Peptides in each sample were labeled with one of 10 unique isobaric tandem mass tags (TMT 10-plex) (ThermoFisher Scientific) and mixed. 500 μg of the combined samples of TMT labeled peptides was fractionated with basic reverse phase chromatography into 24 fractions. 10% of each fraction, or approximately 50 μg of 24 fractions, was used directly for overall protein abundance analysis. The remaining 450 μg of peptides in 24 fractions was enriched with TiO2 for analysis of the phosphorylation at Serine and Threonine residues. Not enriched and phosphor-enriched peptides were separated on a 75 um x 150 mm ProntoSIL-120-5-C18 H column (5 μm, 120 Å (BISCHOFF), www.bischoff-chrom.com) using 2–90% acetonitrile gradient at 300 nl/min over 90 min and analyzed on a Thermo Scientific Q Exactive Plus mass spectrometer interfaced with an Easy-nLC 1000 (Thermo Fisher Scientific). Peptides were identified from isotopically resolved masses in precursor (MS) and fragmentation (MS/MS) spectra at resolutions 70,000 and 35,000, respectively, extracted using 3 nodes (without deconvolution and with deconvolution by Xtract or MS2 Processor) in Proteome Discoverer (PD) software (v1.4, Thermo-Fisher Scientific, San Jose, CA. USA) and searched with Mascot 5.2.1 (Matrix Sciences, Boston, MA, USA) against the RefSeq2015 database. Search criteria included sample species; trypsin as the enzyme, allowing one missed cleavage; cysteine carbamidomethylation and TMT 10-plex labeling of N-termini as fixed modifications; methionine oxidation, asparagine and glutamine deamination, and TMT 10-plex labeling of lysine and phosphorylation of serine and threonine as variable modifications.
Bioinformatics and Statistics; Proteomics
Only peptide matched spectra at <1% false discovery rate (FDR) and < 30% mass isolation interference, with all reporter ions were detected, were included for downstream analyses. Individual protein relative abundances were calculated by: [1] log2-transformation of the reporter ion intensities, [2] subtracting the spectrum medians of the log2 transformed reporter ion intensities (median-polishing), and [3] summarizing all reporter ion intensities that belong to the same protein by their median value. In a final step [4], the channel medians across all proteins were subtracted to correct for potential loading differences, as described previously [19]. Only proteins quantified by reporter ion spectra from more than one peptide were included for statistical downstream analyses. Protein abundances were compared between three RK-33 treated vs DMSO treated samples. Statistical inference between two groups of interest was assessed by moderated t-test statistics [20], [21]. For multiple comparison correction, q-values were calculated from the observed moderated p-values. Proteins with a q < 0.05 and a fold change larger than 1.15 were considered significantly altered.
Gene Set Expression Analysis and Network Analysis
Gene set expression analysis was performed by searching Geneontology Biological Processes database using the Gorilla web tool, with all identified proteins (significantly altered and unaltered) as the background dataset [22]. FDR q-values were calculated using the Benjamini and Hochberg method from unadjusted p-values of hypergeometric enrichment tests. GOterms with a q-value <0.05 were considered significant. Only the most specific GOterm in a family is shown, ignoring less-specific parent terms. Interactions within the group of significantly altered proteins were surveyed by searching the STRING database [23], [24] version 9.1, using a confidence level of 0.9.
Bioinformatics and Statistics; Phosphoproteomics
Phosphopeptide abundances were normalized to the abundance of the corresponding protein in the unenriched analysis and were than compared between three RK-33 treated vs DMSO treated samples. Only unique phosphopeptides with an isolation interference <30% and a PhosphoRS score> 70%, that corresponded to a single protein and for which all reporter ions could be detected, were included in the final analysis.
Motif Analysis
The surrounding sequence (7 amino acids up- and downstream) of all identified phosphorylation sites was downloaded from the RefSeq database. This was not possible for phosphosites in close proximity of the C- or N-terminus, which were therefore excluded from the motif analysis. The motif-x algorithm was used to identify motifs that were enriched among the significantly up- or downregulated phosphopeptides [25]. Only motifs with a P < .01 and a minimum occurrence of 9 times were selected. Enrichment of motifs was calculated against a background dataset with all identified phosphosites. Kinases known to phosphorylate the phosphopeptides of the identified motifs were looked up in the Networkin database [26].
Kinase Enrichment Analysis
Kinase enrichment analysis (KEA) was performed with the KEA2 webtool [27], using both significantly altered individual phosphopeptide sites and phosphorylated proteins as input. Only kinases with a Benjamini-Hochberg corrected p-value smaller than 0.05 were considered statistically significant.
FUCCI Timelapse Microscopy
MDA-MB-435 and MCF7 cells were lentivirally transduced with constructs encoding the Fluorescent Ubiquitin Cell Cycle Indicators (FUCCI) sensors mCherry-hCdt1 (Genbank accession nr: AB512478) and mVenus-hGeminin (Genbank accession nr: AB512479) in the pCSII-EF vector [28]. Double positive cells were selected by fluorescence activated cell sorting. For DDX3 knockdown in MCF7 FUCCI cells, cells were transfected with jetPrime transfection reagent (Polyplus, New York, NY, USA) and 80 nM siControl (non-targeting pool) or siDDX3 sequences (ON-TARGETplus, Dharmacon, Lafayette, CO, USA). For tracking of single cells after DDX3 inhibition, FUCCI transduced cells were grown on collagen-coated, four chamber, glass bottom, 35 mm dishes (Greiner) and allowed to attach overnight. Timelapse imaging was started directly after RK-33 addition or 48 hours after siDDX3 transfection. Seven by three μm z-sections were acquired with five minute intervals during 24 hours in the FITC and TRITC channel and by differential interference contrast, using a Deltavision Elite system (GE Healthcare) controlling a Scientific CMOS camera (pco.edge 5.5.) and an Olympus 20× 0.75 NA air objective. After projection, the movies were assembled and analyzed in FIJI [29]. The duration of each cell cycle of single cells was monitored and compared between treatment conditions. An average of 100 cells per condition for MDA-MB-435 and 60 cells per condition for MCF7 were monitored for signs of extensive vacuolization, apoptosis or necrosis. Cells were declared arrested when no progression to the next cell cycle phase occurred within 24 hours. Mitotic duration was calculated as the time taken from nuclear envelope breakdown until the initiation of anaphase onset. Median time in mitosis was compared by a Mann–Whitney U test. Time to vacuolization or death was compared per cell cycle phase at onset of RK-33 treatment by plotting Kaplan-Meijer curves and performing log-rank tests.
Immunoblotting
All cells were harvested at 50–70% confluency. Cells were lysed in SDS-extraction buffer and sonicated on ice. 30 μg protein was loaded on SDS-PAGE gels for gel-electrophoresis. The blots were probed overnight with primary antibodies against DDX3 (1:1000, mAb AO196) [30], β-actin (1:10000, A5441, Sigma-Aldrich), followed by appropriate secondary antibodies, development with ECL (Bio-Rad, Hercules, CA, USA) and imaging with a G:BOX Chemi XR5 (Syngene, Frederick, MD, USA).
Results
Proteomic Analysis After Treatment with the DDX3 Inhibitor, RK-33
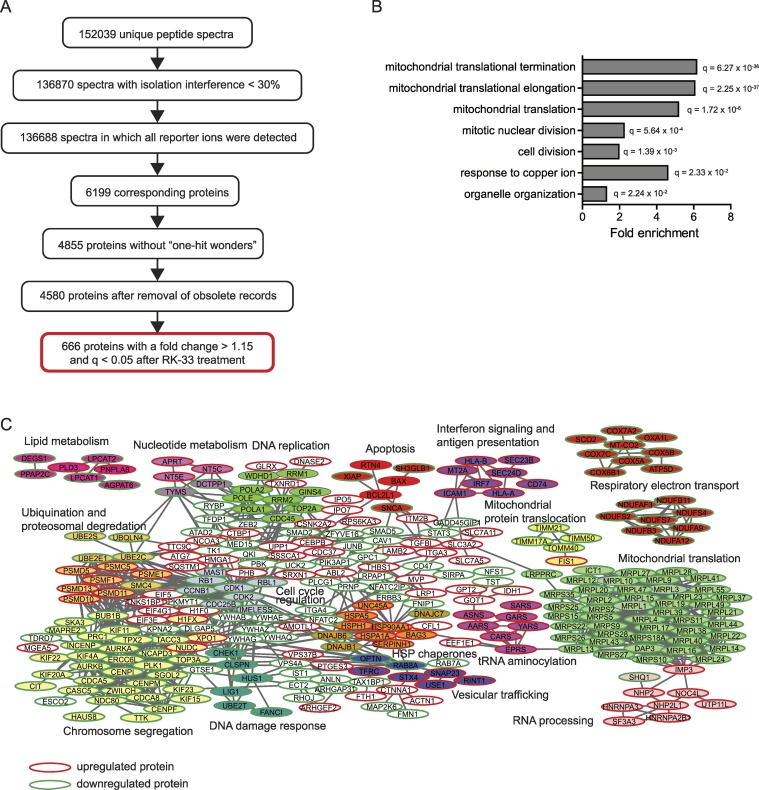
In order to evaluate changes in the protein landscape after RK-33 treatment we performed quantitative proteomics in the metastatic cancer cell line MDA-MB-435 treated with either DMSO as a vehicle control or 4.5 μM RK-33 for 24 hours. 152,039 unique peptide spectra were identified, corresponding to 4580 proteins that could be used for downstream analysis. Of these, 666 were significantly altered with a fold change of larger than 15% and a moderated q-value smaller than 0.05 after exposure to RK-33 (Figure 1A). Gene set enrichment analysis was performed in the Gene Ontology Biological Processes database using the full dataset with 4580 proteins as a background. As shown in Figure 1B, the strongest enrichment was observed for the mitochondrial translation termination (6.2 fold enriched, q = 6.27 x 10−38) and elongation GOterms (6.1 fold enriched, q = 2.25 x 10−37). In addition, proteins involved in mitotic nuclear division (fold enrichment 2.24, q = 5.64 x 10−4) and cell division (fold enrichment 1.96, q = 1.39 x 10−3) were commonly altered.
Figure 1.
The proteomic landscape after DDX3 inhibition with RK-33. A. Flowchart showing bioinformatical processing and analysis of peptide spectra identified by quantitative proteomics after 24 hours DMSO or 4.5 μM RK-33 exposure in MDA-MB-435 cells. B. Bar graph showing significantly enriched GOterms identified by gene-set enrichment analysis among proteins significantly altered after DDX3 inhibition with RK-33 treatment. Q-values are calculated are Benjamini and adjusted p-values generated by hypergeometric enrichment test. C. STRING network analysis showing clusters of interrelated proteins among the significantly altered proteins after RK-33 treatment. Each circle corresponds to a protein and is labeled with the gene symbol. The color fills indicate groups of proteins with a particular function (see labels). Direction of the fold change is indicated by the border color for each protein: green indicates downregulation, red indicates upregulation.
Network Analysis of Affected Proteins
Network analysis using the STRING database [23] revealed several tight networks among the significantly altered proteins (Figure 1C). A clear cluster of downregulated mitochondrial ribosomal proteins was observed. Correspondingly, clusters of respiratory electron transport proteins, which are translated in part by the mitochondrial ribosome, were downregulated as well. We more elaborately investigated the effect of DDX3 inhibition on mitochondrial translation and reported about our findings in a dedicated paper [31].
In addition to a clear effect on mitochondrial translation, several other more closely interrelated protein clusters were identified. With regard to nuclear and cell division a large cluster of proteins involved in chromosome segregation and the mitotic cell cycle was observed (Figure 1C). The expression level of most of these proteins was also decreased after RK-33 exposure. In close relation to this cluster, a cluster of several ubiquitin-conjugating enzymes went down, whereas regulatory subunits of the proteasome mainly increased. With regard to cell cycle progression, proteins involved in DNA damage response and DNA replication both went down. Very central among all these clusters was a cluster with downregulated cell cycle regulation proteins, including CDK1 (fold change 0.81, q = 0.0009) and CDK2 (fold change 0.73, q = 0.0004). Several proteins that play a role in apoptosis clustered together as well, with an increase in pro-apoptotic and a decrease in anti-apoptotic proteins. Given the known role of DDX3 in lipogenesis [32] and RNA processing translation [33], it was also interesting to see clusters that contained proteins involved in lipid metabolism and tRNA aminocylation.
Phosphoproteomic Changes Indicate Decreased CDK1 Activity After RK-33 Treatment
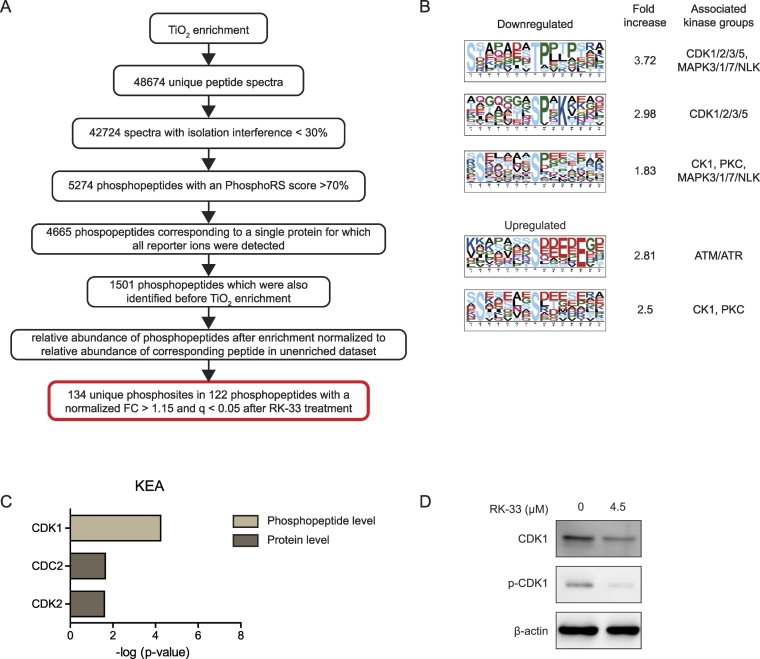
After TiO2 enrichment 5274 phosphopeptides could be reliably identified. Of 1501 phosphopeptides the corresponding protein was also identified in the unenriched analysis, allowing the abundance of the phosphopeptide to be normalized for the abundance of the whole protein (Figure 2A). This normalization ensured that changes in the phosphopeptide abundance reflected changes in phosphorylation status and not changes in relative abundance of the whole protein. 122 phosphopeptides with 134 unique phosphosites were significantly altered after RK-33 treatment.
Figure 2.
Changes in the phosphoproteome after RK-33 treatment indicate altered CDK1 activity. A. Flowchart showing bioinformatical processing and analysis of phospopeptide spectra identified by quantitative phospoproteomics after 24 hours DMSO or 4.5 μM RK-33 exposure in MDA-MB-435 cells. B. Motif-x analysis showing enriched motifs among the significantly up- or downregulated phosphopeptides and their associated kinase groups in the Networkin database. C. Bar graphs showing kinase enrichment analysis (KEA) among kinases known to phosphorylate the phosphopeptides and phosphoproteins significantly altered after RK-33 treatment. Benjamini and Hochberg corrected p-values of hypergeometric enrichment tests. D. Immunoblot showing a decrease in CDK1 and pCDK1 expression after 12 hours 4.5 μM RK-33 exposure in MDA-MB-435 cells.
As shown in Figure 2B, motif analysis of the surrounding amino acids of the downregulated phosphosites found enrichment for the S-X-X-X-X-X-X-t (3.72 fold), s-X-X-K (2.98 Fold) and the S-X-X-X-Xs motif (1.83 Fold), where the capital letters indicate the surrounding amino acid and the non-capitalized letter is the phosphorylated amino acid. Phosphopeptides with these motifs are known to be phosphorylated by kinases from the CDK1/2/3/5 and CK1 kinases, among others (Figure 2B), according to the Networking database [26]. Upregulated phosphosites were enriched for the s-X-X-X-XE motif (2.81 fold), commonly phosphorylated by ATM/ATR kinases [26] and again for the S-X-X-X-X-X-Xs motif (2.5 fold). Kinase enrichment analysis (KEA) at the phosphosite level also indicated enrichment for sites phosphorylated by CDK1 (P = 5.5 x 10−5). Total CDK1 and abundance of phosphorylated CDK1 (pCDK1) was also found to be decreased by immunoblotting in MDA-MB-435 cells 12 hours after RK-33 exposure (Figure 2D). Since not all identified phosphosites were associated with known kinases and the same kinases often phosphorylate proteins at more than one position, we performed KEA at the phosphorylated protein level as well. This analysis indicated that proteins that are known to be phosphorylated by CDC2 (P = .023) or CDK2 (P = .025) were overrepresented among the proteins with an altered phosphorylation status.
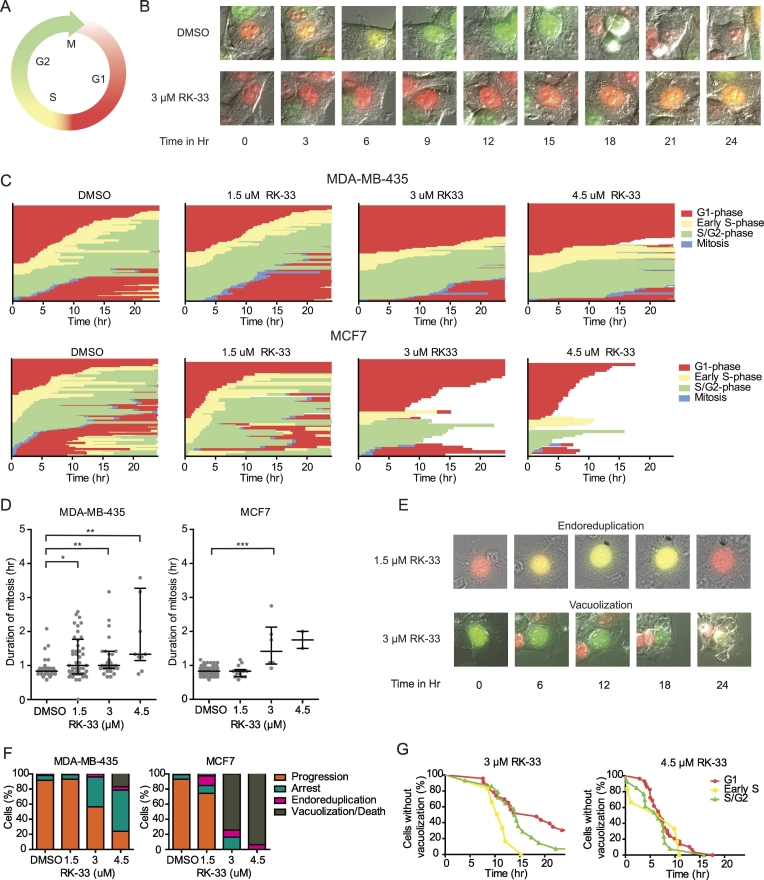
RK-33 Causes a Delay in All Cell Cycle Phases
Both the proteomic and phosphoproteomic changes after 24 hours of RK-33 treatment indicated global changes in proteins directly or indirectly involved in cell cycle progression and cell division, with a central role for CDK1. However, since it is hard to separate cause from effect when analyzing at a static 24 hour time point, we decided to monitor the changes in cell cycle progression and cell division in single cells over time using the FUCCI system and timelapse microscopy (Figure 3A). Interestingly, a delay in all cell cycle phases was seen in both MDA-MB-435 and MCF7 cells (Figure 3B and C). The median time spent in mitosis could be properly estimated, since the relatively short duration of this cell cycle phase allowed for observation of both the beginning and end of this phase. As shown in Figure 3D, mitosis was significantly longer after 1.5 μM (0.96 h, P = .048), 3 μM (2.0 h, P = .003) and 4.5 μM RK-33 (1.17 h, P = .002) in MDA-MB-435 cells, when compared to DMSO treated cells (0.83 h). A similar delay was observed in MCF7 cells, but this was only significantly different after 3 μM RK-33 (1.41 h vs. 0.83 h, P < .001). A delay was also observed for all interphases, but was hard to quantify, since it was uncommon to observe both the beginning and end of these phases.
Figure 3.
Single cell tracking shows RK-33 causes a delay in all cell cycle phases. A. Schematic overview of cell cycle progression with fluorescent changes of the FUCCI constructs mCherry-hCdt1 and mVenus-hGeminin. B. Example merged DIC and fluorescent images showing the color change in a single DMSO and RK-33 treated MDA-MB-435-FUCCI cell over 24 hours. C. Graphs showing delayed cell cycle progression in single FUCCI labeled MDA-MB-435 and MCF7 cells after RK-33 treatment over 24 hours. Each line on the y-axis represents a single cell. D. Dot plot showing the median duration of mitosis after RK-33 treatment in FUCCI labeled MDA-MB-435 and MCF7 cells. Graphs represent median with interquartile range. P-values were calculated by a Mann–Whitney U Test. * P < .05, ** P < .01, *** P < .001. E. Example of endoreduplication and vacuolization occurring after RK-33 treatment in FUCCI-labeled MCF7 cells. F. Bar graphs showing the cell fate of RK-33 treated, FUCCI-labeled MCF7 and MDA-MB-435 cells. G. Kaplan Meijer plots showing the time to vacuolization or cell death for each phase during which the cell (MCF7) was first exposed to RK-33.
Vacuolization and Endoreduplication are Frequent After RK-33 Treatment
Morphological changes were also more commonly observed after RK-33 treatment. Especially in MCF7 extensive vacuolization was frequent (Figure 3E). In MDA-MB-435 vacuolization was less common, but apoptosis (nuclear membrane blebbing) and necrosis (disintegration of the cells) were observed here as well. Since it was hard to determine whether and at what exact point cells died after vacuolization, we lumped vacuolization and cell death when scoring cellular fate. As shown in Figure 3F, both the amount of arrested cells and of cells undergoing vacuolization and/or cell death increased after RK-33 exposure in a dose-dependent manner. The time to vacuolization or cell death in MCF7 was slightly shorter for cells first exposed to 3 μM RK-33 in S-phase, when compared to those first exposed in the G1- or G2 phase (Figure 3G), but this difference was not statistically significant (log rank test P = .229). Another aberrancy that was more frequent in both cell lines after RK-33 treatment was endoreduplication. Endoreduplication is a phenomenon where the cell does not undergo mitosis after replicating its DNA content, but returns to the biochemical state of the G1 phase [34] (Figure 3E & F). In MCF7 6.3–12.8% of cells underwent endoreduplication after 1.5–4.5 μM RK-33, as compared to 1% in of control cells. In MDA-MB-435 the phenotype was less pronounced, but endoreduplication was also more common after 3–4.5 μM RK-33 (4.0–4.5%), when compared to DMSO treated cells (2.1%).
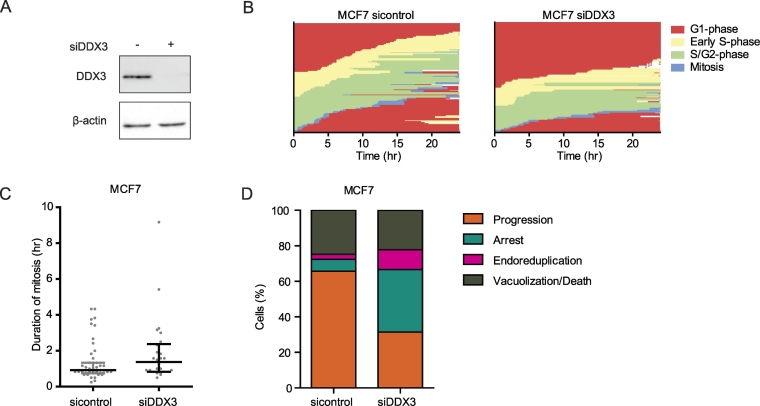
DDX3 Knockdown Slows Down Cell Cycle Progression and Induces Endoreduplication
To evaluate whether a similar effect on the cell cycle could be observed after knockdown of DDX3, we treated MCF7-FUCCI cells with siDDX3 and compared them to siControl treated cells with timelapse microscopy (Figure 4A). As shown in Figure 4B, a similar global delay or arrest in all cell cycle phases was observed after DDX3 knockdown. This was again best quantifiable for the median duration of mitosis, which was longer after siDDX3 (1.38 h), when compared to siControl (0.92 h), but this difference was not statistically significant (P = .080; Figure 4C). A clear increase in arrested cells (35.2% vs 6.7%) and cells undergoing endoreduplication (11.1% vs 2.9%) was observed after siDDX3, as well. The amount of cells undergoing vacuolization and/or cell death in this timeframe was comparable after siControl or siDDX3 treatment (Figure 4D).
Figure 4.
DDX3 knockdown results in a global cell cycle delay and endoreduplication. A. Immunoblot showing DDX3 expression 48 hours after siControl or siDDX3 transfection in MCF7 cells. B. Graphs showing delayed cell cycle progression in single MCF7-FUCCI cells 48 hours to 72 hours after siControl or siDDX3 transfection. Each line on the y-axis represents a single cell. C. Dot plot showing the median duration of mitosis after siDDX3 transfection in FUCCI labeled MCF7 cells. Graphs represent median with interquartile range. P-values were calculated by a Mann–Whitney U Test. D. Bar graphs showing the cell fate of MCF7-FUCCI cells 48 to 72 hours after siDDX3 transfection.
Discussion
In this work, we evaluated the effect of DDX3 inhibition with RK-33. At the protein level RK-33 treatment mainly resulted in altered expression of proteins involved in mitochondrial translation, cell division and cell cycle progression. Reduced CDK1 and CDK2 expression seemed to have a central role in the network of affected cell cycle related proteins. We identified motifs among the phosphopeptides with changed expression levels after RK-33 treatment, of which several were associated with the CDK1 kinase family. KEA also indicated a significant enrichment for phosphopeptides that are phosphorylated by CDK1. Cell cycle evaluation by tracking single FUCCI labeled cells with timelapse microscopy showed a global delay in all cell cycle interphases and mitosis after both RK-33 treatment and DDX3 knockdown. In addition, endoreduplication was more frequently observed after DDX3 inhibition.
Downregulation of CDK1 has been linked to a global delay in cell cycle progression [35] and endoreduplication [36], by other studies as well. There are multiple pathways by which DDX3 inhibition with RK-33 could affect CDK1. Wang, et al. showed that the DEAD box RNA helicase family member DDX6 mediates CDK1 expression through interaction with stem-loop structures in its 3’UTR [37]. Furthermore, a functional internal ribosome entry site (IRES) was identified in the CDK1 5’UTR [38]. DDX3 is known to be involved in translation of both mRNAs with a complex 5’UTR region [39], as well as cap-independent translation of proteins with an IRES [33]. We identified a reduction in CDK1 protein levels after RK-33 treatment with both proteomics and immunoblotting. It is therefore possible that DDX3 is involved in cap- dependent or independent translation of CDK1. Another possibility is that DDX3 directly affects the kinase activity of CDK1, as was observed previously for CK1 family members [16]. Further validation of the decrease in either abundance or activity of CDK1 and the mechanism behind this decrease after DDX3 inhibition is required.
Interestingly, DDX3 inhibition was previously associated with an increased proportion of cells in G1 as measured by flow cytometry, suggestive of a specific G1 arrest [3], [4], [40]. This can be explained cells dying faster in other cell phases than in G1 (Figure 3C and G) and by the possibility that cells are relatively more delayed in G1 than in other phases. It is hard to estimate the relative delay in each cell cycle phase, because the beginning and end of the interphases are frequently not observed in our 24 hour time window, especially not after RK-33 treatment. However, our data clearly indicate a delay in all phases and not one that is limited to the G1 cell cycle phase.
We identified changes after RK-33 treatment in several cellular pathways that were previously linked to functions of DDX3. Proteins involved in nuclear/cell division and chromosome segregation were prominent among the altered proteins. Pek, et al. previously identified DDX3 to be responsible for chromosomal localization of hCAP-H, a condensin 1 component, and found that inhibition of DDX3 resulted in lagging chromosomes [41]. In addition, a recent paper identified that DDX3 prevents multipolar mitosis by epigenetic transcriptional and translation regulation of p53 [42]. Their findings are in line with our observation that the duration of mitosis is increased after RK-33 treatment.
In addition, DDX3 was previously shown to play a regulatory role both in the intrinsic [43] and extrinsic [6], [7] pathways of apoptosis. RK-33 treatment resulted in upregulation of pro-apoptotic proteins (e.g. BAX, BCL2L1) and downregulation of anti-apoptotic proteins (e.g. XIAP), confirming the previously described anti-apoptotic role of DDX3 [7]. However, since we evaluated the proteomic landscape only at a static and late time point (24 hours), it is not possible to say whether the effects on apoptosis, and several other pathways, are a result of direct involvement of DDX3 or occurring as a downstream effect of other cellular perturbations. Vacuolization, ultimately resulting in cell death, was more frequent in MCF7 than MDA-MB-435 cells and is most likely part of an autophagy response we previously observed after DDX3 inhibition with RK-33 in this cell line [31].
Other functions of DDX3 that were identified by our analysis that DDX3 or related DEAD box proteins were previously found to be involved in are; lipid metabolism [32]; the DNA damage response [4], [32] and in tRNA aminocylation [44]. Another interesting group of proteins that was upregulated after RK-33 treatment was the group of tRNA ligases. tRNA ligases function in maturation of tRNAs to facilitate protein translation [44]. DDX1, another DEAD box RNA helicase family member, was recently found to regulate the tRNA ligase complex [44]. Further research on the involvement of DDX3 in this context is warranted.
Overall, the most prominent change in the proteome landscape after RK-33 treatment was mitochondrial translation. Our recent paper explored the effect of DDX3 inhibition on mitochondrial ribosomes, oxidative phosphorylation and the relation to radiosensitizing capacities of RK-33 [31]. It is possible that the effect observed on cell cycle regulation described in the present paper, is partly a result of reduced bioenergetic capacity to fuel cell cycle progression. Further studies will need to confirm whether the effect of RK-33 on cell cycle progression through CDK1 regulation is a separate direct phenomenon, or secondary to the previously described metabolic changes. We conclude that DDX3 inhibition with RK-33 results in a global delay in cell cycle progression, at the proteome level characterized by altered expression of proteins involved in mitochondrial translation, cell division and cell cycle progression and at the phosphoproteomic level by changes indicative of reduced CDK1 activity.
Acknowledgments
Acknowledgements
Funding
This work was supported by the National Institutes of Health grant [5R01CA207208] to VR and by the Dutch Cancer Society (UU2013-5851).
References
- 1.Linder P, Fuller-Pace FV. Looking back on the birth of DEAD-box RNA helicases. Biochim Biophys Acta. 2013;1829(8):750–755. doi: 10.1016/j.bbagrm.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Botlagunta M, Vesuna F, Mironchik Y, Raman A, Lisok A, Winnard P, Jr., Mukadam S, Van Diest P, Chen JH, Farabaugh P. Oncogenic role of DDX3 in breast cancer biogenesis. Oncogene. 2008;27(28):3912–3922. doi: 10.1038/onc.2008.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heerma van Voss MR, Vesuna F, Trumpi K, Brilliant J, Berlinicke C, de Leng W, Kranenburg O, Offerhaus GJ, Burger H, van der Wall E. Identification of the DEAD box RNA helicase DDX3 as a therapeutic target in colorectal cancer. Oncotarget. 2015;6(29):28312–28326. doi: 10.18632/oncotarget.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bol GM, Vesuna F, Xie M, Zeng J, Aziz K, Gandhi N, Levine A, Irving A, Korz D, Tantravedi S. Targeting DDX3 with a small molecule inhibitor for lung cancer therapy. EMBO Mol Med. 2015;7(5):648–669. doi: 10.15252/emmm.201404368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie M, Vesuna F, Tantravedi S, Bol GM, Heerma van Voss MR, Nugent K, Malek R, Gabrielson KL, Van Diest PJ, Tran PT. RK-33 radiosensitizes prostate cancer cells by blocking the RNA helicase DDX3. Cancer Res. 2016 doi: 10.1158/0008-5472.CAN-16-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Wang H, Wang Z, Makhija S, Buchsbaum D, LoBuglio A, Kimberly R, Zhou T. Inducible resistance of tumor cells to tumor necrosis factor-related apoptosis-inducing ligand. receptor 2-mediated apoptosis by generation of a blockade at the death domain function. Cancer Res. 2006;66(17):8520–8528. doi: 10.1158/0008-5472.CAN-05-4364. [DOI] [PubMed] [Google Scholar]
- 7.Sun M, Song L, Li Y, Zhou T, Jope RS. Identification of an antiapoptotic protein complex at death receptors. Cell Death Differ. 2008;15(12):1887–1900. doi: 10.1038/cdd.2008.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shih JW, Wang WT, Tsai TY, Kuo CY, Li HK, Wu Lee YH. Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem J. 2012;441(1):119–129. doi: 10.1042/BJ20110739. [DOI] [PubMed] [Google Scholar]
- 9.Sun M, Song L, Zhou T, Gillespie GY, Jope RS. The role of DDX3 in regulating Snail. Biochim Biophys Acta. 2011;1813(3):438–447. doi: 10.1016/j.bbamcr.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagerstrand D, Tong A, Schumacher SE, Ilic N, Shen RR, Cheung HW, Vazquez F, Shrestha Y, Kim SY, Giacomelli AO. Systematic interrogation of 3q26 identifies TLOC1 and SKIL as cancer drivers. Cancer Discov. 2013;3(9):1044–1057. doi: 10.1158/2159-8290.CD-12-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen HH, Yu HI, Cho WC, Tarn WY. DDX3 modulates cell adhesion and motility and cancer cell metastasis via Rac1-mediated signaling pathway. Oncogene. 2015;34(21):2790–2800. doi: 10.1038/onc.2014.190. [DOI] [PubMed] [Google Scholar]
- 12.Xie M, Vesuna F, Botlagunta M, Bol GM, Irving A, Bergman Y, Hosmane RS, Kato Y, Winnard PT, Jr., Raman V. NZ51, a ring-expanded nucleoside analog, inhibits motility and viability of breast cancer cells by targeting the RNA helicase DDX3. Oncotarget. 2015;6(30):29901–29913. doi: 10.18632/oncotarget.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai MC, Chang WC, Shieh SY, Tarn WY. DDX3 regulates cell growth through translational control of cyclin E1. Mol Cell Biol. 2010;30(22):5444–5453. doi: 10.1128/MCB.00560-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosner A, Rinkevich B. The DDX3 subfamily of the DEAD box helicases: divergent roles as unveiled by studying different organisms and in vitro assays. Curr Med Chem. 2007;14(23):2517–2525. doi: 10.2174/092986707782023677. [DOI] [PubMed] [Google Scholar]
- 15.Fuller-Pace FV. DEAD box RNA helicase functions in cancer. RNA Biol. 2013;10(1):121–132. doi: 10.4161/rna.23312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cruciat CM, Dolde C, de Groot RE, Ohkawara B, Reinhard C, Korswagen HC, Niehrs C. RNA helicase DDX3 is a regulatory subunit of casein kinase 1 in Wnt-beta-catenin signaling. Science. 2013;339(6126):1436–1441. doi: 10.1126/science.1231499. [DOI] [PubMed] [Google Scholar]
- 17.Kondaskar A, Kondaskar S, Kumar R, Fishbein JC, Muvarak N, Lapidus RG, Sadowska M, Edelman MJ, Bol GM, Vesuna F. Novel, Broad Spectrum Anti-Cancer Agents Containing the Tricyclic 5:7:5-Fused Diimidazodiazepine Ring System. ACS Med Chem Lett. 2010;2(3):252–256. doi: 10.1021/ml100281b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilky BA, Kim C, McCarty G, Montgomery EA, Kammers K, DeVine LR, Cole RN, Raman V, Loeb DM. RNA helicase DDX3: a novel therapeutic target in Ewing sarcoma. Oncogene. 2016;35(20):2574–2583. doi: 10.1038/onc.2015.336. [DOI] [PubMed] [Google Scholar]
- 19.Herbrich SM, Cole RN, West KP, Jr., Schulze K, Yager JD, Groopman JD, Christian P, Wu L, O'Meally RN, May DH. Statistical inference from multiple iTRAQ experiments without using common reference standards. J Proteome Res. 2013;12(2):594–604. doi: 10.1021/pr300624g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kammers K, Cole RN, Tiengwe C, Ruczinski I. Detecting Significant Changes in Protein Abundance. EuPA Open Proteom. 2015;7:11–19. doi: 10.1016/j.euprot.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 22.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–15. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lacroix M. Significance, detection and markers of disseminated breast cancer cells. Endocr Relat Cancer. 2006;13(4):1033–1067. doi: 10.1677/ERC-06-0001. [DOI] [PubMed] [Google Scholar]
- 25.Chou MF, Schwartz D. Biological sequence motif discovery using motif-x. Curr Protoc Bioinformatics. 2011:15–24. doi: 10.1002/0471250953.bi1315s35. [Chapter 13:Unit 13] [DOI] [PubMed] [Google Scholar]
- 26.Horn H, Schoof EM, Kim J, Robin X, Miller ML, Diella F, Palma A, Cesareni G, Jensen LJ, Linding R. KinomeXplorer: an integrated platform for kinome biology studies. Nat Methods. 2014;11(6):603–604. doi: 10.1038/nmeth.2968. [DOI] [PubMed] [Google Scholar]
- 27.Lachmann A, Ma'ayan A. KEA: kinase enrichment analysis. Bioinformatics. 2009;25(5):684–686. doi: 10.1093/bioinformatics/btp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132(3):487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 29.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Angus AG, Dalrymple D, Boulant S, McGivern DR, Clayton RF, Scott MJ, Adair R, Graham S, Owsianka AM, Targett-Adams P. Requirement of cellular DDX3 for hepatitis C virus replication is unrelated to its interaction with the viral core protein. J Gen Virol. 2010;91(Pt 1):122–132. doi: 10.1099/vir.0.015909-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heerma van Voss MR, Vesuna F, Bol GM, Afzal J, Tantravedi S, Bergman Y, Kammers K, Lehar M, Malek R, Ballew M. Targeting mitochondrial translation by inhibiting DDX3: a novel radiosensitization strategy for cancer treatment. Oncogene. 2018;37(1):63–74. doi: 10.1038/onc.2017.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Q, Pene V, Krishnamurthy S, Cha H, Liang TJ. Hepatitis C virus infection activates an innate pathway involving IKK-alpha in lipogenesis and viral assembly. Nat Med. 2013;19(6):722–729. doi: 10.1038/nm.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geissler R, Golbik RP, Behrens SE. The DEAD-box helicase DDX3 supports the assembly of functional 80S ribosomes. Nucleic Acids Res. 2012;40(11):4998–5011. doi: 10.1093/nar/gks070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fox DT, Duronio RJ. Endoreplication and polyploidy: insights into development and disease. Development. 2013;140(1):3–12. doi: 10.1242/dev.080531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumper S, Mardakheh FK, McCarthy A, Yeo M, Stamp GW, Paul A, Worboys J, Sadok A, Jorgensen C, Guichard S. Rho-associated kinase (ROCK) function is essential for cell cycle progression, senescence and tumorigenesis. elife. 2016;5 doi: 10.7554/eLife.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ullah Z, Kohn MJ, Yagi R, Vassilev LT, DePamphilis ML. Differentiation of trophoblast stem cells into giant cells is triggered by p 57/Kip2 inhibition of CDK1 activity. Genes Dev. 2008;22(21):3024–3036. doi: 10.1101/gad.1718108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Arribas-Layton M, Chen Y, Lykke-Andersen J, Sen GL. DDX6 Orchestrates Mammalian Progenitor Function through the mRNA Degradation and Translation Pathways. Mol Cell. 2015;60(1):118–130. doi: 10.1016/j.molcel.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marash L, Liberman N, Henis-Korenblit S, Sivan G, Reem E, Elroy-Stein O, Kimchi A. DAP5 promotes cap-independent translation of Bcl-2 and CDK1 to facilitate cell survival during mitosis. Mol Cell. 2008;30(4):447–459. doi: 10.1016/j.molcel.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 39.Lai MC, Lee YH, Tarn WY. The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol Biol Cell. 2008;19(9):3847–3858. doi: 10.1091/mbc.E07-12-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukumura J, Noguchi E, Sekiguchi T, Nishimoto T. A temperature-sensitive mutant of the mammalian RNA helicase, DEAD-BOX X isoform, DBX, defective in the transition from G1 to S phase. J Biochem. 2003;134(1):71–82. doi: 10.1093/jb/mvg126. [DOI] [PubMed] [Google Scholar]
- 41.Pek JW, Kai T. DEAD-box RNA helicase Belle/DDX3 and the RNA interference pathway promote mitotic chromosome segregation. Proc Natl Acad Sci U S A. 2011;108(29):12007–12012. doi: 10.1073/pnas.1106245108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen WJ, Wang WT, Tsai TY, Li HK, Lee YW. DDX3 localizes to the centrosome and prevents multipolar mitosis by epigenetically and translationally modulating p53 expression. Sci Rep. 2017;7(1):9411. doi: 10.1038/s41598-017-09779-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun M, Zhou T, Jonasch E, Jope RS. DDX3 regulates DNA damage-induced apoptosis and p53 stabilization. Biochim Biophys Acta. 2013;1833(6):1489–1497. doi: 10.1016/j.bbamcr.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Popow J, Jurkin J, Schleiffer A, Martinez J. Analysis of orthologous groups reveals archease and DDX1 as tRNA splicing factors. Nature. 2014;511(7507):104–107. doi: 10.1038/nature13284. [DOI] [PMC free article] [PubMed] [Google Scholar]