Abstract
Alzheimer's disease (AD) is characterized by an accumulation of β-amyloid (Aβ42) accompanied by brain atrophy and cognitive decline. Several recent studies have shown that Aβ42 accumulation is associated with gray matter (GM) changes prior to the development of cognitive impairment, in the so-called preclinical stage of the AD (pre-AD). It also has been proved that the GM atrophy profile is not linear, both in normal ageing but, especially, on AD. However, several other factors may influence this association and may have an impact on the generalization of results from different samples. In this work, we estimate differences in rates of GM volume change in cognitively healthy elders in association with baseline core cerebrospinal fluid (CSF) AD biomarkers, and assess to what these differences are sample dependent. We report the dependence of atrophy rates, measured in a two-year interval, on Aβ42, computed both over continuous and categorical values of Aβ42, at voxel-level (p < 0.001; k < 100) and corrected for sex, age and education. Analyses were performed jointly and separately, on two samples. The first sample was formed of 31 individuals (22 Ctrl and 9 pre-AD), aged 60–80 and recruited at the Hospital Clinic of Barcelona. The second sample was a replica of the first one with subjects selected from the ADNI dataset. We also investigated the dependence of the GM atrophy rate on the basal levels of continuous p-tau and on the p-tau/Aβ42 ratio. Correlation analyses on the whole sample showed a dependence of GM atrophy rates on Aβ42 in medial and orbital frontal, precuneus, cingulate, medial temporal regions and cerebellum. Correlations with p-tau were located in the left hippocampus, parahippocampus and striatal nuclei whereas correlation with p-tau/Aβ42 was mainly found in ventral and medial temporal areas. Regarding analyses performed separately, we found a substantial discrepancy of results between samples, illustrating the complexities of comparing two independent datasets even when using the same inclusion criteria. Such discrepancies may lead to significant differences in the sample size needed to detect a particular reduction on cerebral atrophy rates in prevention trials. Higher cognitive reserve and more advanced pathological progression in the ADNI sample could partially account for the observed discrepancies. Taken together, our findings in these two samples highlight the importance of comparing and merging independent datasets to draw more robust and generalizable conclusions on the structural changes in the preclinical stages of AD.
Abbreviations: AD, Alzheimer's disease; Aβ42, amyloid beta; p-tau, phosphorylated tau; t-tau, total tau; Ctrl, control; preAD, preclinical Alzheimer's disease; HCB, Hospital Clinic Barcelona; ADNI, Alzheimer's Disease Neuroimaging Initiative; MMSE, Mini Mental State examination; CDR, Clinical Dementia Rating; ELISA, Enzyme-Linked ImmunoSorbent Assay; VBM, voxel-based morphometry; GM, gray matter; WM, white matter; CSF, Cerebro-Spinal Fluid; PLR, pairwise longitudinal registration; DI, divergences of the longitudinal deformations; ROI, region of interest; TIV, total intracranial volume; FWE, Family Wise Error; L, left; R, right
Keywords: Alzheimer's disease, Preclinical Alzheimer's disease, Longitudinal VBM, CSF biomarkers
Highlights
-
•
GM atrophy rates depends differently on values of CSF Aβ42 than on CSF p-tau in the preclinical stage of AD.
-
•
Discrepant results were obtained. Although nominally equivalent, samples might reflect different time-windows in the AD continuum.
-
•
It is necessary a further effort to standardize CSF-biomarkers measures and thresholds to make different samples to be directly comparable.
1. Introduction
Alzheimer's disease (AD) is a chronic neurodegenerative disorder that slowly progresses over decades and is characterized by progressive neuropathology, brain atrophy and, ultimately cognitive decline. The neuropathological hallmarks of AD are amyloid-β (Aβ42) plaques and neurofibrillary tau tangles. Converging evidence shows that the pathophysiological process of the disease begins decades before the time of clinical diagnosis (Braak and Del Tredici, 2013; Villemagne et al., 2013). Either sampling cerebrospinal fluid (CSF) or with amyloid PET imaging (Landau et al., 2013; Tolboom et al., 2009), Aβ42 alterations can be detected in a substantial percentage of cognitively healthy subjects (Jack et al., 2014; Morris et al., 2010). This has led to the formulation, for research purposes, of the concept of preclinical Alzheimer disease (Dubois et al., 2016; Sperling et al., 2011). While it is not yet known whether all these individuals with asymptomatic cerebral amyloidosis will eventually develop AD, there is consensus that the prognosis for amyloid-positive subjects is worse than for amyloid negative ones (Chételat et al., 2013).
By the time clinical impairment is detectable, substantial neurodegeneration has already taken place (Morris and Price, 2001). However, the relationship between Aβ42 deposition and brain atrophy in preclinical AD is still under debate (Chételat et al., 2013; Fjell et al., 2014, Fjell et al., 2014). In cross-sectional comparisons, amyloid positive non-demented subjects show decreased whole brain (Fagan et al., 2009) or hippocampal volumes (Bourgeat et al., 2010; Dickerson et al., 2009; Mormino et al., 2009; Storandt et al., 2009). Conversely, other studies did not found significant differences in hippocampal volume between amyloid positive and negative healthy individuals (Jack et al., 2010; Vemuri et al., 2009) but some did in parietal and posterior cingulate cortex extending into the precuneus (Becker et al., 2011), or even found increased volume in temporal regions including the hippocampus (Chételat et al., 2010) and parahippocampal areas (Gispert et al., 2015).
Longitudinal studies comparing cerebral atrophy rates have reported increased declines in whole brain, hippocampus, amygdala and posterior cingulate and in temporal parietal and frontal regions in preclinical AD (Doré et al., 2013; Insel et al., 2016; Lorenzi et al., 2015; Mattsson et al., 2014; Schott et al., 2010; Storandt et al., 2009). However, some others were unable to detect such accelerated atrophy rates (Driscoll et al., n.d.; Fotenos et al., 2008). One factor that may partially account for such discrepant results is the dichotomization of cognitively normal samples into two groups based on a threshold of amyloid abnormality. Nevertheless, studying the association between amyloid levels and atrophy rates in pooled samples of cognitively healthy subjects, again some found significant associations in the hippocampus (Andrews et al., 2013; Stricker et al., 2012) and in temporal regions (Doré et al., 2013) whereas some others did not (Driscoll et al., n.d.; Tosun et al., 2010) or the association was only evident in Aβ42 positive subjects, after dichotomizing the sample again (Fjell et al., 2010; Schott et al., 2010). The longitudinal evolution of GM volume has been proved to be non-linear (Fjell et al., 2014, Fjell et al., 2014; Gispert et al., 2015) so the atrophy rate is non constant over-time neither along disease progress. It is still unclear which factors affect, a part from age and gender, the GM atrophy rate progression in cognitively healthy subjects.
Taken together, these reports reflect a complex interaction between Aβ42 levels and cerebral atrophy. Recently, it has been suggested that such an association might be better modeled using nonlinear regression techniques (Becker et al., 2011; Fjell et al., 2014, Fjell et al., 2014; Fortea et al., 2011; Gispert et al., 2015) and that atrophy rates may greatly vary across different brain regions (Insel et al., 2014). The aim of this work is to characterize longitudinal changes, computed in a two-year period, in cognitively healthy (healthy controls and preclinical AD elders and to seek for linear associations between baseline CSF biomarker levels (Aβ42, p-tau and p-tau/Aβ42). The linear association of variables with atrophy rates deals with second order term of the atrophy, acceleration or restrain of atrophy, by looking for variations in the first order term (atrophy rate). We used the publicly available dataset from the Alzheimer's Disease Neuroimaging Initiative (ADNI) to replicate the findings in our sample. Thus, a secondary purpose of the work was to check for the dependence of results on the sample, and hence, to evaluate the equivalence of datasets.
2. Material and methods
2.1. Subjects
In this work, we have studied and compared two independent samples with 2 year longitudinal MRI scans and baseline CSF measurements. The first sample, referred to as HCB, originally consisted of 34 cognitively healthy subjects recruited at Alzheimer's Disease and Other Cognitive Disorders Unit from Hospital Clinic of Barcelona. The sample was a subset of the one described in (Gispert et al., 2015) formed of those subjects who underwent to a second MRI scan in two years' time and who remained cognitively healthy by the time of the second scan. The inclusion criteria were neurologically healthy subjects, aged 60–80 years, that didn't present any evidence of cognitive impairment (Clinical Dementia Rating, CDR = 0). The sample was mostly recruited among relatives of neurological patients who voluntary agreed to take part of the study and few subjects with subjective cognitive decline (SDC) whose cognitive tests had normal scores. Almost all controls began to the first group. Participants underwent complete clinical and neuropsychological examinations and a lumbar puncture to determine CSF Aβ42 and p-tau values. The mean time interval between the lumbar puncture and the first MRI session was 44 ± 35 days, ranged from 1 to 134 days. Aβ42, t-tau and p-tau quantitation was performed using ELISA (Enzyme-Linked ImmunoSorbent Assay kits, Innogenetics, Ghent, Belgium). Specific details about quantitation can be found in (Gispert et al., 2016). The Aβ42 range for abnormality, which defined preclinical AD stage, was set to (Aβ42 < 500 pg/ml) as it was determined for ELISA in a previous work (Antonell et al., 2014). The same MRI protocol was used in both scans: High-resolution structural images by MPRAGE sequence (TR/TE/TI = 2300/2.98/900 ms, respectively, FA = 9°, 240 sagittal slices, 1 × 1 × 1 mm3 voxel) on a 3T TIM TRIO scanner (Siemens, Erlangen, Germany) at the IDIBAPS's Imaging core facilities. Two subjects had to be removed from the sample because a very poor quality of the images (movement artefact) and one preclinical AD was removed from the sample because showing an outlier value of p-tau (213 pg/ml while the mean and standard deviation of the rest of the sample was 56.4 and 47 pg/ml respectively), what biased all the analysis in which p-tau was involved. Then, the final sample consisted of 31 individuals (22 control and 9 preclinical AD). All participants signed a written consent to take part in the study and the Ethical Committee of the hospital approved the research protocol.
The second sample was a replica selected from ADNI dataset (Alzheimer's Disease Neuroimaging Initiative database: adni.loni.usc.edu). The selection criteria were identical to the previous sample: cognitively normal individuals (MMSE > 24 and CDR = 0), aged 60 to 80 years old, who had been submitted to two sessions of MRI delayed two years on a same 3T scanner, and who had Aβ42 value at the time of the first MRI session. The search resulted in 49 subjects, 22 controls and 17 preAD. The selection between them was done by matching age, as main confounding factor, with subjects from HCB. Although picking the youngest ones, there still was a significant difference on age between samples (Table 1). One of the firstly selected preAD subjects was removed of the sample because presenting a massive right temporal atrophy that altered all VBM analyses (outlier) and substituted by the following next in the list. The acquisition parameters depended on site/scanner, being common the matrix (256 × 256 × 240) and voxel size (1 × 1 × 1.2 mm3) for all subjects. Subjects scanned on a Tim TRIO had been acquired with almost the same protocol that the one used in HCB (TR/TE/TI = 2300/2.98/900 ms, respectively, FA = 9°, 240 sagittal slices, 1 × 1 × 1.2 mm3 voxel). In ADNI dataset, the quantitation of Aβ42, p-tau and t-tau levels was performed using xMAP Luminex platform and Innogenetics/Fujirebio AlzBio3 immunoassay kits. Nine of the subjects were preclinical at the time of the first scan because their Aβ42 level was below the threshold of pathology (192 pg/ml for this technique) (Shaw et al., 2009). According (Mattsson et al., 2013), both quantitation techniques are not directly comparable, so the limit for pathology depends on the measure technique.
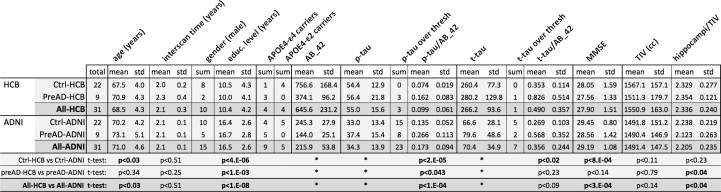
Table 1.
Demographic comparison of samples. Volume of hippocampus is the mean of right and left hippocampus, relative to total intracranial volume and expressed in ‰. Thresholds of p-tau were 75 pg/ml in HCB and 23 pg/ml in ADNI. Thresholds for t-tau were 450 pg/ml in HCB and 93 pg/ml in ADNI. Statistical significance criterion was set to p < 0.05 (in bold significant differences). ⁎Statistical tests were not calculated because different quantitation method was used in each cohort. That made values not to be comparable.
Subjects were separated in 4 groups named Ctrl-HCB, PreAD-HCB, Ctrl-ADNI and PreAD-ADNI, for controls and preclinical subjects in HCB and ADNI cohorts respectively. Similarly, Crtl-All and preAD-All referred to control and preAD groups of the merged sample (HCB + ADNI) and All-HCB and All-ADNI denoted all the subjects of each sample.
2.2. Evaluation of the equivalence of the samples
Visual inspection of images seemed to point toward a greater degree of atrophy in PreAD-ADNI than in PreAD-HCB. To verify this impression and prior to the main analyses, we checked for the equivalence of the samples. We compared equivalence of the p-tau/Aβ42 ratio and relative hippocampal volume distributions among samples. We also performed a few direct voxel-wise two-sample t-tests on GM atrophy rates images between subgroups (Supplementary material). Pre-post differences in total intracranial volume, supposed to be zero since skull size remains invariant along time, were also calculated to check for unbalanced variance related to scanner stability.
To evaluate relative hippocampal volumes, the AAL atlas in the MNI space was warped to every structural image in the native space by using the inverse of the deformation obtained in the segmentation procedure. The adapted atlas was multiplied by the subject's GM mask. The volume of hippocampi was calculated as the number of voxels labeled as hippocampus on the masked AAL atlas multiplied by the size of the voxel. Volumes of left and right hippocampi were averaged and divided by the total intracranial volume.
2.3. Image pre-processing
Longitudinal changes were evaluated by means of pairwise longitudinal registration (PLR) function from SPM12 (Wellcome Trust Center for Neuroimaging; UCL, UK; http://www.fil.ion.ucl.ac.uk/spm/), which provides a high dimensional warping between pre and post images. PLR resulted in an image of divergences (DI) of the longitudinal deformations for every subject, which accounted for local shrinking or expansion of the tissues, and a pre-post average image. The reason of choosing PLR was to avoid the original contrast on the T1 MRI having any influence on images submitted to statistics, so images could be directly compared whatever the scanner and protocol of acquisition was. We assumed that, although GM gray intensity depended on T1 contrasts and consequently on acquisition protocol and scanner, the performance of PLR would not, so potential differences in T1 contrast would not be reflected in DI as long as the very same protocol was used in the pre and post acquisition. Furthermore, statistical analyses were performed directly on DI, instead the more standard DI*GM, in a sort of Deformation Based Morphometry. The aim was to make purely longitudinal analyses unaffected by the cross differences of GM between groups. The inclusion of GM information in the image submitted to statistics could have resulted in a misinterpretation of the results, because there would not have been possible to distinguish longitudinal changes (divergences) and from the cross-sectional (GM maps).
In detail, pre and post images were matched using a two-step procedure: first, by using a standard registration to locate both images in the same position, and later, the PLR to calculate the local deformations occurred during the inter-scan period. PLR resulted in a pre-post average and a DI map for every subject. Since pre-images were taken as reference, the lower the value on DI, the bigger the atrophy rate. Average images were segmented and used as input to create a sample-specific template using Dartel (Ashburner, 2007). Individual segmented pre-post-average images were also employed to remove non-brain voxels from DI images. DI values were preserved just in those voxels whose probability of belonging to GM or WM was bigger than the probability of belonging to any other tissue. The purpose was to avoid that negative values of DI in GM were altered by positive values of DI in CSF in the smoothing procedure; due to a local GM atrophy implies a local CSF expansion. That would result in a cancellation of the expansion/shrinking information in GM voxels close to CSF. Dartel template was normalized to MNI and resulting warps were applied to masked DI, preserving concentrations, and to average images. Normalized DIs were, then, smoothed with a Gaussian kernel of 8 mm. Later, they were divided per the exact interval of time between scans (in years, one decimal) to get atrophy rates and masked with a customized common GM mask to restrict statistical analyses to GM.
The common GM mask was obtained by segmenting the mean of the normalized pre-post average images and looking for those voxels whose probability of belonging to GM was bigger than the probability of belonging to any other tissue. Then it was manually edited to remove most of periventricular voxels wrongly classified as GM, dilated by one voxel to include edge voxels and, finally, completed with a basal ganglia mask, obtained from AAL atlas, to include the thalamus and globus pallidus, which otherwise were partially excluded when using an intensity threshold as single criterion.
2.4. Statistical analyses of images
All voxel-based statistical analyses were performed using age at the first MRI session, gender (Hua et al., 2010) and education (categorical: subjects having or not >15 years of education) as confounding covariates. Ancova was the chosen option to deal with global effects. Because of the small sample size, statistical threshold was set to p < 0.001 uncorrected, with a minimum cluster size of 100 voxels (0.33 cm3) for all analyses. Due to this statistical threshold might be lenient, we remarked those results that also survived a more restrictive p < 0.05 FWE corrected threshold. AAL toolbox (Tzourio-Mazoyer et al., 2002) was used to label statistical maps.
Correlations between atrophy rates and basal CSF biomarkers (positive correlation with Aβ42, and negative with p-tau and p-tau/Aβ42) were also evaluated on HCB, ADNI and on the whole sample (HCB + ADNI). In whole sample analysis, Aβ42 and p-tau values were split in 2 columns and the dataset covariate was included to account for the different CSF quantitation techniques. In correlations with p-tau/Aβ42 ratio, the variable was not split in two columns assuming that, although the different techniques made the values of the CSF-biomarker not directly comparable, the ratio would likely be almost equivalent between samples. The fact the ratio of the respective thresholds of abnormality were close (0.12 and 0.15 for HCB and ADNI respectively) supported this assumption. For the shake of completeness, voxel-based two-sample t-tests were performed to seek for statistical differences between groups (analysis on categorical values of Aβ42) (Supplementary 1). Analysis on categorical values of p-tau could not be performed due to the small incidence of p-tau pathology in HCB cohort.
2.5. Sample sizes
The sample size needed to detect a 25% reduction in the mean annual hippocampal atrophy rate (one-sided t-test; α = 0.05) with 80% or 95% power was also calculated for preclinical subjects in both samples, in a similar approach as in (Risacher et al., 2010). To this end, G*Power v3.1.9.2 was used (Faul et al., 2007) on gray matter atrophy data obtained from a 4 mm-radius sphere at the peak on left hippocampus of the p-tau/Aβ42 correlation after correction by age, sex and educational level.
3. Results
3.1. Demographics
Table 1 shows a demographic comparison of both samples. Age and CSF biomarkers in the table correspond to the time of first scan. Although choosing the youngest subjects of ADNI meeting the selection criteria, HBC cohort was significantly younger (1.5 years on average due to Ctrl-HCB < Ctrl-ADNI, no significant differences between preAD). HCB sample showed significantly lower educational attainment than the ADNI one (all subgroups). Table 1 also shows statistically significant differences in p-tau/Aβ42 ratio, t-tau/Aβ42 ratio (just CtrlHCB > Ctrl-ADNI), MMSE score (All-HCB < All-ADNI because Ctrl-HCB < Ctrl-ADNI, no significant differences between PreAD), and relative hippocampal volumes (All-HCB > All-ADNI, because preAD-HCB > preAD-ADNI, no significant differences between Ctrl).
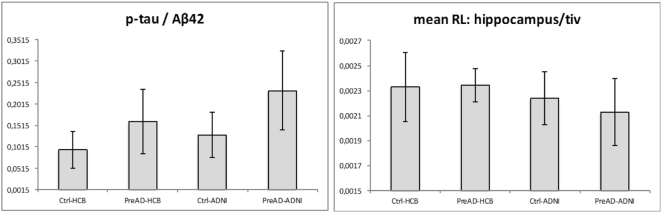
Fig. 1a plots the p-tau/Aβ42 ratio distribution across subgroups corrected for age and gender. A part from those tests mentioned in Table 1, p-tau/Aβ42 distribution showed significant differences, both on original values and values corrected for age and gender, in the following inter-subgroups comparisons: Ctrl-HCB < PreAD-HCB, Ctrl-ADNI < PreAD-ADNI and Ctrl-HCB < PreAD-ADNI. However, the difference Ctrl-ADNI < PreAD-HCB was not significant (p = 0.38). Fig. 1b shows the relative volume of hippocampus (mean right-left divided by total intracranial volume). It can be seen that the degree of hippocampal atrophy of preclinical subjects from ADNI was bigger than the observed atrophy in the rest of the groups. As already mentioned, differences were statistically significant for the tests All-HCB > All-ADNI due to PreAD-HCB > PreAD-ADNI, both cases with and without correction for age and gender. No significant differences were found in the pre-post TIV stability measure between samples, the difference of TIV between samples subgroups.
Fig. 1.
Plots of p-tau/Aβ42 and relative hippocampal volume distributions across groups at pre time point, corrected for age and gender.
3.2. Longitudinal VBM
As expected, since classification as control or preclinical subjects depended only on Aβ42 values, the analysis of the dependence of atrophy rates on continuous (correlation analysis) and categorical (two-sample t-test) Aβ42 values on the whole sample (Fig. 2) gave a similar pattern but correlation analysis showed to be more sensitive.
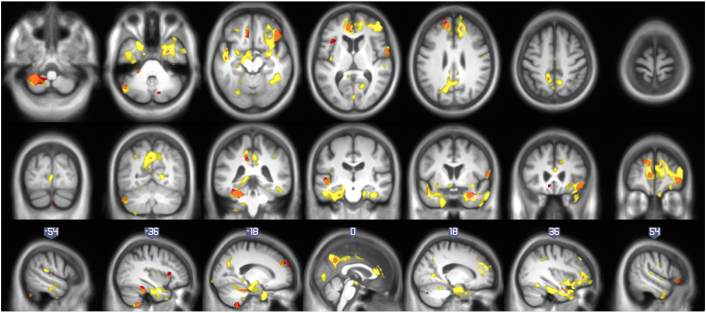
Fig. 2.
Comparison of correlation of longitudinal atrophy (atrophy-rates) with continuous Aβ42 (yellow) with respect to Ctrl vs PreAD two-sample t-test, i.e. categorical Aβ42 (red), on the HCB + ADNI merged sample. Orange areas denote overlap of statistical maps. Correlation with continuous Aβ42 showed to be more sensitive than the two-sample t-test.(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 3 shows statistical maps of correlation of atrophy rates with Aβ42 (first row), p-tau (second row) and p-tau/Aβ42 (third row) on HCB (first column), ADNI (second column) and the whole sample (third column). Table 2, Table 3, Table 4 list the significant areas in the Aβ42 p-tau and p-tau/Aβ42 correlation analyses on the whole sample, respectively. About correlation of atrophy rates with Aβ42, most significant differences were found in right frontal lobe, bilateral temporal lobe and precuneus, and left cerebellum. Other AD relevant areas showed differences with lower statistical significance: bilateral lingual and middle cingulum, right parietal lobe and left occipital lobe. Categorical analysis and other complementary two-sample t-test to check the equivalence of cohorts can be found in Supplementary material.
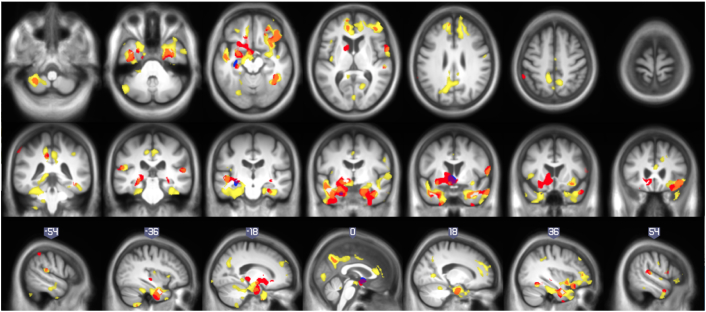
Fig. 3.
Correlation of longitudinal atrophy (atrophy-rates) with CSF-biomarkers: comparison of results in different samples: a) shows correlations with Aβ42. b) displays correlations with p-tau. c) shows correlation with p-tau/Aβ42. Detailed enumeration of regions is provided in Table 2, Table 3, Table 4, Table 5.
Table 2.
Correlation with Aβ42 whole sample. In bold, most reliable results (surviving p < 0.05 FEW correction).
| p cluster FWE | Cluster size | p voxel FWE | T | x (mm) | y (mm) | z (mm) | AAL label of peak |
|---|---|---|---|---|---|---|---|
| <0.001 | 20,205 | 0.003 | 5.95 | 15 | 49.5 | 16.5 | Frontal_Sup_Medial_R |
| 0.008 | 5.59 | 34.5 | 6 | −33 | Temporal_Pole_Mid_R | ||
| 0.010 | 5.52 | 37.5 | 45 | 4.5 | Frontal_Mid_R | ||
| <0.001 | 10,625 | 0.003 | 5.93 | −16.5 | 7.5 | −40.5 | Temporal_Pole_Mid_L |
| 0.007 | 5.62 | −46.5 | −1.5 | −22.5 | Temporal_Mid_L | ||
| 0.011 | 5.47 | −42 | −9 | −30 | Temporal_Inf_L | ||
| <0.001 | 6539 | 0.003 | 5.87 | 3 | −55.5 | 34.5 | Precuneus_R |
| 0.008 | 5.59 | 1.5 | −64.5 | 43.5 | Precuneus_L | ||
| 0.013 | 5.44 | −7.5 | −55.5 | 36 | Precuneus_L | ||
| 0.497 | 318 | 0.032 | 5.14 | 10.5 | 52.5 | −24 | Frontal_Sup_Orb_R |
| 0.148 | 725 | 0.040 | 5.06 | 45 | −48 | −15 | Temporal_Inf_R |
| 0.085 | 4.80 | 36 | −54 | −16.5 | Fusiform_R | ||
| 0.337 | 449 | 0.113 | 4.70 | −49.5 | −31.5 | 16.5 | Temporal_Sup_L |
| 0.976 | 3.38 | −63 | −25.5 | 13.5 | Temporal_Sup_L | ||
| 0.987 | 3.31 | −45 | −19.5 | 16.5 | Rolandic_Oper_L | ||
| 0.145 | 732 | 0.120 | 4.68 | 61.5 | 1.5 | −28.5 | Temporal_Mid_R |
| 0.376 | 4.21 | 51 | 1.5 | −48 | Temporal_Inf_R | ||
| 0.480 | 330 | 0.126 | 4.66 | 18 | −61.5 | 4.5 | Calcarine_R |
| 0.810 | 3.73 | 13.5 | −48 | −1.5 | Lingual_R | ||
| 0.033 | 1278 | 0.142 | 4.61 | −30 | −49.5 | −55.5 | Cerebelum_8_L |
| 0.890 | 3.61 | −13.5 | −55.5 | −57 | Cerebelum_9_L | ||
| 0.974 | 3.39 | −18 | −48 | −64.5 | Cerebelum_8_L | ||
| 0.219 | 593 | 0.211 | 4.46 | −46.5 | −64.5 | −39 | Cerebelum_Crus1_L |
| 0.548 | 284 | 0.294 | 4.32 | 0 | −82.5 | 4.5 | Lingual_L |
| 0.709 | 188 | 0.294 | 4.32 | −28.5 | 39 | −19.5 | Frontal_Inf_Orb_L |
| 0.565 | 273 | 0.449 | 4.13 | 16.5 | −46.5 | −22.5 | Cerebelum_4_5_R |
| 0.940 | 3.51 | 4.5 | −45 | −21 | Vermis_3 | ||
| 0.837 | 116 | 0.511 | 4.06 | −27 | −72 | −13.5 | Fusiform_L |
| 0.612 | 244 | 0.623 | 3.94 | −40.5 | 16.5 | 1.5 | Insula_L |
| 0.982 | 3.35 | −39 | 22.5 | 10.5 | Frontal_Inf_Tri_L | ||
| 0.988 | 3.30 | −49.5 | 15 | −4.5 | Temporal_Pole_Sup_L | ||
| 0.784 | 146 | 0.626 | 3.93 | 57 | −25.5 | 10.5 | Temporal_Sup_R |
| 0.729 | 177 | 0.743 | 3.81 | −24 | −102 | −7.5 | Occipital_Mid_L |
| 0.835 | 117 | 0.781 | 3.76 | −15 | −39 | −45 | Cerebelum_10_L |
| 0.738 | 172 | 0.808 | 3.73 | 37.5 | −43.5 | 39 | Parietal_Inf_R |
| 0.824 | 3.71 | 39 | −36 | 43.5 | SupraMarginal_R | ||
| 0.793 | 141 | 0.815 | 3.72 | 1.5 | −12 | 46.5 | Cingulum_Mid_R |
| 0.978 | 3.37 | 0 | 1.5 | 40.5 | Cingulum_Mid_L |
Table 3.
Correlation with p-tau whole sample. No voxel survived p < 0.05 FEW corrected threshold.
| p cluster FWE | Cluster size | p voxel FWE | T | x (mm) | y (mm) | z (mm) | AAL label of peak |
|---|---|---|---|---|---|---|---|
| 0.251 | 570 | 0.124 | 4.64 | −2 | 3 | −8 | Caudate_L |
| 0.963 | 3.41 | −14 | 10 | −14 | Olfactory_L | ||
| 0.773 | 151 | 0.313 | 4.27 | −30 | −18 | −10 | Hippocampus_L |
| 0.827 | 119 | 0.546 | 3.99 | −22 | −4 | −32 | ParaHippocampal_L |
Table 4.
Correlation with p-tau/Aβ42 whole sample. In bold, most reliable results (surviving p < 0.05 FEW corrected threshold).
| p cluster FWE | Cluster size | p voxel FWE | T | x (mm) | y (mm) | z (mm) | AAL label of peak |
|---|---|---|---|---|---|---|---|
| 0.000 | 9042 | 0.000 | 6.84 | −4.5 | 1.5 | −9 | Pallidum_L |
| 0.005 | 5.71 | −21 | −7.5 | −28.5 | ParaHippocampal_L | ||
| 0.017 | 5.32 | −7.5 | 21 | −9 | Caudate_L | ||
| 0.032 | 1328 | 0.000 | 6.51 | 45 | 22.5 | −13.5 | Frontal_Inf_Orb_R |
| 0.065 | 4.87 | 43.5 | 36 | −18 | Frontal_Inf_Orb_R | ||
| 0.349 | 4.22 | 33 | 12 | −12 | Insula_R | ||
| 0.695 | 197 | 0.024 | 5.21 | 10.5 | 49.5 | −28.5 | Frontal_Sup_Orb_R |
| 0.001 | 2767 | 0.028 | 5.16 | 28.5 | −1.5 | −33 | ParaHippocampal_R |
| 0.029 | 5.15 | 28.5 | −4.5 | −45 | Fusiform_R | ||
| 0.055 | 4.93 | 15 | −9 | −27 | ParaHippocampal_R | ||
| 0.333 | 464 | 0.171 | 4.52 | 3 | −55.5 | 34.5 | Precuneus_R |
| 0.549 | 3.99 | −6 | −64.5 | 39 | Precuneus_L | ||
| 0.477 | 338 | 0.182 | 4.50 | 42 | −43.5 | −13.5 | Fusiform_R |
| 0.560 | 3.98 | 37.5 | −55.5 | −13.5 | Fusiform_R | ||
| 0.739 | 171 | 0.189 | 4.48 | −61.5 | −43.5 | 45 | Parietal_Inf_L |
| 0.771 | 3.75 | −66 | −46.5 | 33 | SupraMarginal_L | ||
| 0.470 | 343 | 0.196 | 4.47 | 25.5 | 40.5 | −12 | Frontal_Mid_Orb_R |
| 0.651 | 223 | 0.264 | 4.35 | −13.5 | 37.5 | −24 | Frontal_Sup_Orb_L |
| 0.564 | 277 | 0.303 | 4.29 | −30 | −51 | −57 | Cerebelum_8_L |
| 0.399 | 401 | 0.308 | 4.28 | 64.5 | 7.5 | 9 | Rolandic_Oper_R |
| 0.956 | 3.44 | 66 | 1.5 | 19.5 | Postcentral_R | ||
| 0.688 | 201 | 0.450 | 4.10 | 54 | −28.5 | 12 | Temporal_Sup_R |
| 0.827 | 120 | 0.569 | 3.97 | 39 | 43.5 | 6 | Frontal_Mid_R |
| 0.802 | 135 | 0.699 | 3.83 | −9 | −39 | 39 | Cingulum_Mid_L |
| 0.832 | 117 | 0.723 | 3.81 | 31.5 | −22.5 | −6 | Hippocampus_R |
| 0.991 | 3.26 | 24 | −27 | 0 | Thalamus_R | ||
| 0.847 | 108 | 0.860 | 3.63 | −54 | −30 | 16.5 | Temporal_Sup_L |
| 0.962 | 3.42 | −61.5 | −27 | 13.5 | Temporal_Sup_L |
Correlations with p-tau (whole sample) showed differences in left hippocampus, parahippocampus, caudate and olfactory areas. However, none of such differences survived a restrictive p < 0.05 FWE threshold. Finally, the analysis of the p-tau/Aβ42 ratio resulted in significant correlation in bilateral parahippocampi, orbito frontal and right fusiform. Other AD related areas with a lower statistical significance were bilateral precuneus, temporal superior, middle cingulate, left parietal lobe and right hippocampus, postcentral, rolandic gyrus and caudate. Table 5 is a summary of the differences between characteristics of HCB and ADNI cohorts and their corresponding correlation maps. It is worthy to remember that no surviving voxels in p-tau correlation in ADNI cohort doesn't mean that there were not atrophy anywhere in the brain, but the atrophy-ratio were independent of p-tau-values. For the shake of completeness, we included in Supplementary material the one sample t-test of both samples to elucidate in which areas of the brain there were a significant atrophy in each subgroup. As expected, those test show a significant atrophy-rate in AD-related (both samples) and other cortical areas (just HCB).
Table 5.
Differences between cohorts. Correlations with continuous Aβ42, p-tau and p-tau/Aβ42 ratio. (*) p < 0.05 FWE corrected at peak or cluster level.
| Cohort | Protective factors | Risk factors | Correlation with Aβ42 | Correlation with p-tau | Correlation with p-tau/Aβ42 |
|---|---|---|---|---|---|
| HCB |
|
|
|
|
|
| ADNI |
|
|
|
|
|
Fig. 4 provides the overlap of correlation maps for the three CSF biomarkers tested. It can be seen than p-tau/Aβ42 shared regions with both p-tau and Aβ42 maps, but at this level of significance, those two last maps were disjoint. The atrophy rate of relevant regions in AD such as hippocampus appeared to be independent to basal Aβ42 level.
Fig. 4.
Correlation of longitudinal atrophy (atrophy-rates) with basal CSF biomarkers in (HCB + ADNI): continuous Aβ42 (yellow), continuous p-tau (blue) and p-tau/Aβ42 (red). Orange and violet areas shows overlap of p-tau/Aβ42 maps with Aβ42 and p-tau maps, respectively. No overlap was found between Aβ42 and p-tau maps at this significance level.(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
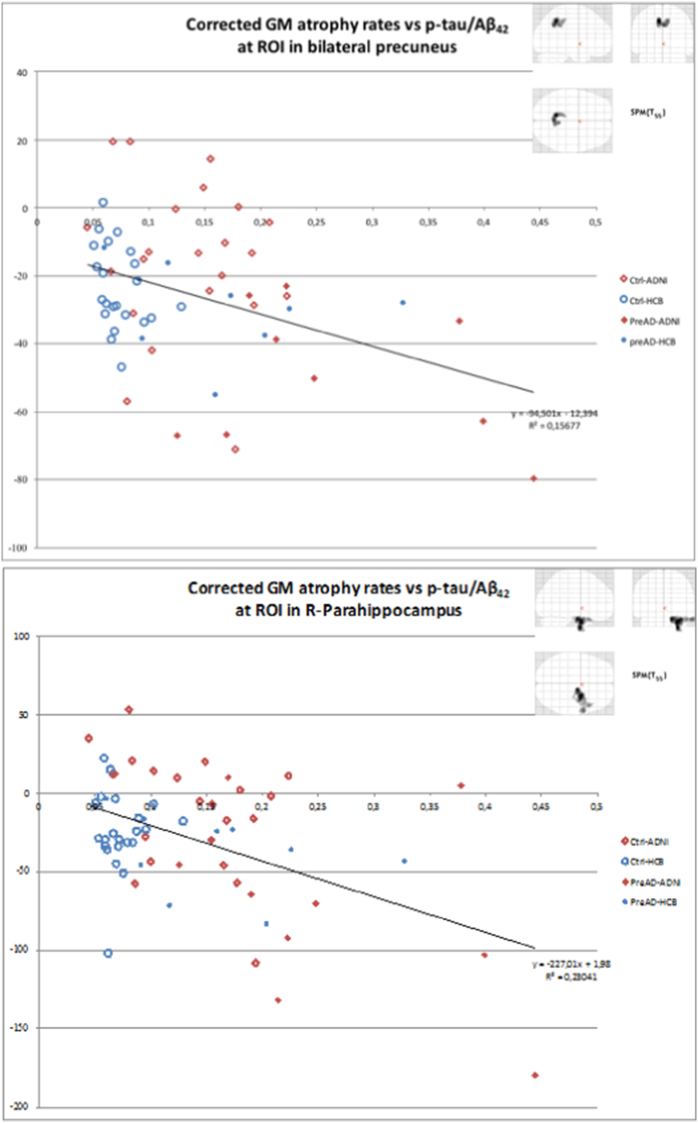
To illustrate the behavior of subgroups in correlation analyses, Fig. 5 displays the scatter plots of the mean GM atrophy rates correlation versus the p-tau/Aβ42 ratio, measured on two ROIs. The first ROI is the significant cluster at the right parahippocampus obtained in the correlation analysis with p-tau/Aβ42 (2767 voxels, 9.34 cm3). The second ROI correspond to the significant cluster at precuneus from the Ctrl-All > PreAD-All test (1331 voxels, 4.49 cm3). The regression lines are overlaid.
Fig. 5.
Plots of mean GM atrophy rate (longitudinal) versus p-tau/Aβ42 for two clusters. Bilateral precuneus ROI corresponds to the significant cluster in that area in the two-sample t-test analysis on the whole sample. R-parahipocampus ROI corresponds to the significant cluster in that region in the correlation of atrophy rates with p-tau/Aβ42 on the whole sample analysis. It contains voxels from adjacent regions.
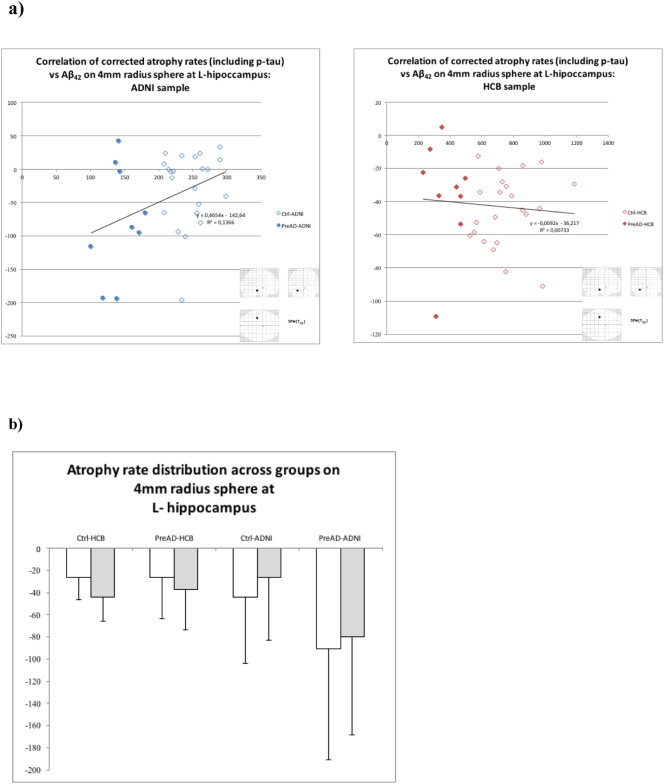
Finally, to elucidate why, although these regions are highly related to AD, no correlation of atrophy rates with Aβ42 were found, we plotted the mean values of atrophy rates, corrected by age, sex, high education, global and p-tau, on a sphere of 4 mm of radius centered at the voxel showing maximum correlation with p-tau/Aβ42 on the left hippocampus (whole sample analysis). Due to the different Aβ42 scaling, this plot was made separately for HCB and ADNI (Fig. 6a). A plot of the distribution across groups of the mean atrophy rates on the same sphere is also included (Fig. 6b).
Fig. 6.
a) Longitudinal atrophy rates versus Aβ42 on a 4 mm-radius sphere at the peak on left hippocampus of p-tau/Aβ42 correlation. Values corrected for age, sex, educational level and p-tau. b) Distribution of mean longitudinal atrophy rates (white without correction, gray with correction for age, gender and education years) on the same sphere across groups.
Finally, the sample size to detect a 25% reduction in the hippocampal atrophy rate in preclinical subjects with 80% (95%) power was found to be 380 (664) for the HCB sample and 240 (419) for the ADNI sample (Table 6). Compared to previous estimates for the same target (Risacher et al., 2010), the sample sizes in the two preclinical populations in our study fall between those reported for stable MCI patients and healthy controls, as expected.
Table 6.
Sample sizes to detect a 25% reduction in the hippocampal atrophy rate in preclinical subjects.
| 80% Power | 95% Power | |
|---|---|---|
| HCB | 380 | 664 |
| ADNI | 240 | 419 |
4. Discussion
In this study we aimed at identifying cerebral structural longitudinal changes across the preclinical stages of AD. To this end, we analyzed two-year volume change rates in association to baseline CSF biomarkers in two independent datasets formed by cognitively healthy subjects who were CSF Aβ42 positive and negative according to established cut-off thresholds. Our main finding is that atrophy rates are positively correlated to basal CSF Aβ42 levels in the precuneus and inferior temporal and frontal areas in concordance with that describe in (Becker et al., 2011; Dickerson et al., 2009; Fjell et al., 2014, Fjell et al., 2014) among others. Interestingly, the hippocampus was spared from this association. CSF Aβ42 is known to change from physiologically values to reach a plateau long before the onset of clinical cognitive decline. On the other hand, in these subjects CSF p-tau was mostly associated to volume reductions in medial temporal areas in line with previous findings (Gispert et al., 2015). In contrast with CSF Aβ42, early changes in CSF tau are known to predict the onset of clinical cognitive decline (Sutphen et al., 2015) and has been shown to be strongly associated to brain atrophy in these areas (Tarawneh et al., 2015).
These results were obtained after merging two independent samples of cognitively healthy subjects. This approach was selected to improve the robustness, reproducibility and generalizability of our findings. However, the comparability of the two samples was not straightforward. First, the HCB sample was significantly younger and reached lower educational attainment. Both variables were included as covariates in the statistical model to correct for their effects. Moreover, PreAD-ADNI presented lower hippocampal volumes than the preAD-HBC. This finding can underpin a lower cognitive reserve in the HCB sample, also assessed by significant differences in MMSE scores (Table 1), that could have resulted in a diagnosis of MCI with milder pathology that in the ADNI sample (Arenaza-Urquijo et al., 2013). Furthermore, Ctrl-HCB showed higher gray matter volume with respect to PreAD-HCB in the precuneus and posterior parietal areas. These areas have been reported to be affected in early-onset AD (Möller et al., 2013), and the precuneus has been described to shown early atrophy with increasingly abnormal CSF levels (Gispert et al., 2015).
A second point of discussion about the equivalence of cohorts is the possible differences in the selection of participants. In one hand, the percentage of APOE4-e4 carriers was abnormally low in Ctrl-HCB (just 1 in 22 - Table 1). This percentage might be low by chance, or because, due to a lower cognitive reserve in HCB, more people with APOE4-ε4 risk factor had become MCI and then excluded from the group submitted to the post scan; as inclusion criteria, they must remained cognitively healthy at the time of the second scan. Another difference is in the percentage of preclinical Alzheimer participants with abnormal p-tau levels, which is higher in the ADNI data set. According to the new research framework (Jack et al., 2017), these are considered to have already AD, while those with only amyloid changes are defined as preclinical participants with Alzheimer pathogenic change and hence are earlier in the disease continuum.
These results, as a whole, suggest that the ADNI sample was more advanced in the AD continuum than the HBC one. This could account for the observed discrepant results of the individual analysis and the lack of an increase in the atrophy-rate of medial temporal areas in the preAD-HCB with respect to Ctrl-HCB. Taken together, such a relative shift in the pathological progression between the two samples also highlights the importance of comparing and merging independent datasets to draw more robust and generable conclusions on the structural changes in the preclinical stages of AD. Importantly, such a shift may have important consequences in the sample size needed to detect a reduction on cerebral atrophy rates in prevention trials. It is worth noting that other unbalanced demographic variables, known or unknown, might have influenced in the disparity of results, as well. For instance, Mediterranean diet (speculative, no information available on alimentary habits of participants) could have protective effects in HCB participants (Titova et al., 2013).
The different degree of progression in the two samples could also explain the increased sensitivity observed in the correlation analysis with continuous CSF Aβ42 than when splitting the sample in preclinical and control groups according to the respective reference cutoff thresholds for amyloid positivity (categorical Aβ42). Even though the regions involved are very similar in the comparison of both groups (Fig. 4) and correlations with CSF Aβ42, the latter showed increased peak and cluster level significance. These findings suggest that correlation with CSF biomarkers, even when having to account for different analytical methods, provide with more robust and comparable results than using reference thresholds to dichotomize the biological continuum of preclinical AD. Please note that these thresholds were mostly derived to show optimal diagnostic capacity between cognitively healthy and impaired subjects and do not have to necessarily correspond to inflexion points with biological significance. We show that these considerations raise important practical implications for using GM changes as efficacy outcome in prevention trials. The different rates of hippocampal atrophy between the two studied samples translated in significantly different estimated sample sizes to detect a 25% reduction. Since the ADNI sample appears to be more advanced in the preclinical course of AD, the needed sample size 37% lower than what would be required for the other cohort.
As a limitation of the methods, we have to state that we had to split Aβ42 and p-tau columns in two in the correlation analysis on the whole sample to deal with the non-equivalence of CSF biomarkers measures between cohorts. As Aβ42 and p-tau scaling depended on the sample, the statistical fitting would outcome a different atrophy - biomarker proportional ratio for each sample (beta values). The contrast employed (1,1) asked for the sum of both betas being significantly greater than zero. The consequence of this was that one sample could have a stronger influence than the other in the whole sample analysis, probably ADNI over HCB since the biomarker values were lower. We had not considered the option of balancing betas in the contrast to deal with this issue, since the leveling coefficients would be difficult to fix because they would also depend on the interaction of CSF biomarkers with the rest of covariates. Instead, we let it as a limitation of the study. This could explain why the results of p-tau correlation on HCB sample were not replicated in the whole sample analysis. Afterward, we tried to standardize biomarkers values in some way. First option was to use of the index described in (Molinuevo et al., 2013). However, the lack of subjects with high levels of Aβ42 in ADNI sample biased its index, making it not to be comparable with HCB's index. Then, the alternative was to use ratios. We decide to use p-tau/Aβ42 because it is widely described in the literature (Harari et al., 2014) and the ratio was potentially independent on the sample. As mentioned, the equivalence of the ratio at the threshold values would suggest the ratio of metabolites could be, at certain degree of accuracy, directly comparable.
There are some additional limitations in our work aside of the previously mentioned one. For instance, the samples sizes were relatively small. For this reason, we had to choose a lenient statistical threshold for all voxel-based analysis. This implied that some results were reliable (p < 0.05 FEW corrected) and some just trending or plausible (p < 0.001 uncorrected). The size of the sample made either impossible to perform a stratified analysis on p-tau that would have increase the value of the work. In this regard, merging independent datasets also eased with this difficulty.
In addition, it should be mentioned that some analyses that could clarify the discrepancy between samples could have not been done because the characteristics of the study. For instance, the cross-sectional two-sample t-test between ADNI and HCB controls at first time point could not be done because the covariate scanner, indispensable in multisite cross-sectional analyses, would be the same that the group factor. In this regard, working with longitudinal data greatly facilitates the pooling data form different sites. In addition, CSF samples were not available at the time of the second scan, so it cannot be discarded that some controls became preclinical AD during the follow-up period. Finally, there is a possible bias due to the different resolution of acquisition between samples. Specifically acquisition voxels in ADNI dataset were not isometric and that could slightly affect the performance of PLR. However, the plausibility of our results in view of existing literature validates the use of this methodology, in our opinion.
5. Conclusions
GM atrophy rates have been calculated from two-year delayed MRI T1 structural images of cognitively normal subjects and correlated them with CSF biomarkers on two samples (one local and the other from ADNI), both separately and jointly. Analyses on the whole sample showed a correlation of GM atrophy rates, adjusted by age, sex and educational level, with basal Aβ42 levels in precuneus, ventral and medial temporal areas and medial frontal areas; with p-tau located in left hippocampus, parahippocampus and striatal nuclei and with p-tau/Aβ42 mainly in ventral and medial temporal areas. Regarding analyses performed separately, we found a substantial discrepancy of results between samples, which may lead to significant differences in the sample size needed to detect a particular reduction on cerebral atrophy rates in prevention trials. We suggest that the most likely explanation of this is that, although sharing the same inclusion criteria, both samples reflected a different stage in the AD continuum. This might suggest that atrophy in the AD continuum first appears in parietal areas and later spreads to medial temporal and frontal areas. Also, it is worthy to mention that an exhaustive characterization of its demographic parameters is needed to replicate a sample. Moreover, further effort has to be done in order to standardize CSF-biomarkers measures and thresholds of pathology to make samples from different center to be directly comparable.
Acknowledgments
The research leading to these results has received support from the Innovative Medicines Initiative Joint Undertaking under grant agreement n° 115568, resources of which are composed of financial contribution from the European Union's Seventh Framework Programme (FP7/2007-2013) and EFPIA companies' in kind contribution. The present communication reflects the authors' view and neither IMI nor the European Union, EFPIA, or any Associated Partners are responsible for any use that may be made of the information contained herein. Lorena Rami was funded by the Miguel Servet grant CP2/00023. Juan D. Gispert holds a ‘Ramón y Cajal’ fellowship (RYC-2013-13054).
Data collection for this paper was partially funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2018.04.016.
Contributor Information
Juan Domingo Gispert, Email: jdgispert@barcelobabeta.org.
José Luis Molinuevo, Email: jlmolinuevo@barcelonabeta.org.
Appendix A. Supplementary data
Complementary image statistical analyses.
References
- Andrews K.A., Modat M., Macdonald K.E., Yeatman T., Cardoso M.J., Leung K.K., Barnes J., Villemagne V.L., Rowe C.C., Fox N.C., Ourselin S., Schott J.M. Atrophy rates in asymptomatic amyloidosis: implications for Alzheimer prevention trials. PLoS One. 2013;8 doi: 10.1371/journal.pone.0058816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonell A., Mansilla A., Rami L., Lladó A., Iranzo A., Olives J., Balasa M., Sánchez-Valle R., Molinuevo J.L. Cerebrospinal fluid level of YKL-40 protein in preclinical and prodromal Alzheimer's disease. J. Alzheimers Dis. 2014;42:901–908. doi: 10.3233/JAD-140624. [DOI] [PubMed] [Google Scholar]
- Arenaza-Urquijo E.M., Molinuevo J.L., Sala-Llonch R., Solé-Padullés C., Balasa M., Bosch B., Olives J., Antonell A., Lladó A., Sánchez-Valle R., Rami L., Bartres-Faz D. Cognitive reserve proxies relate to gray matter loss in cognitively healthy elderly with abnormal cerebrospinal fluid amyloid-β levels. J. Alzheimers Dis. 2013;35:715–726. doi: 10.3233/JAD-121906. [DOI] [PubMed] [Google Scholar]
- Ashburner J. A fast diffeomorphic image registration algorithm. NeuroImage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Becker J.A., Hedden T., Carmasin J., Maye J., Rentz D.M., Putcha D., Fischl B., Greve D.N., Marshall G.a., Salloway S., Marks D., Buckner R.L., Sperling R.a., Johnson K.a. Amyloid-β associated cortical thinning in clinically normal elderly. Ann. Neurol. 2011;69:1032–1042. doi: 10.1002/ana.22333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeat P., Chételat G., Villemagne V.L., Fripp J., Raniga P., Pike K., Acosta O., Szoeke C., Ourselin S., Ames D., Ellis K.A., Martins R.N., Masters C.L., Rowe C.C., Salvado O. Beta-amyloid burden in the temporal neocortex is related to hippocampal atrophy in elderly subjects without dementia. Neurology. 2010;74:121–127. doi: 10.1212/WNL.0b013e3181c918b5. [DOI] [PubMed] [Google Scholar]
- Braak H., Del Tredici K. Amyloid-?? may be released from non-junctional varicosities of axons generated from abnormal tau-containing brainstem nuclei in sporadic Alzheimer's disease: a hypothesis. Acta Neuropathol. 2013;126:303–306. doi: 10.1007/s00401-013-1153-2. [DOI] [PubMed] [Google Scholar]
- Chételat G., La Joie R., Villain N., Perrotin A., De La Sayette V., Eustache F., Vandenberghe R. Amyloid imaging in cognitively normal individuals, at-risk populations and preclinical Alzheimer's disease. NeuroImage Clin. 2013;2:356–365. doi: 10.1016/j.nicl.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chételat G., Villemagne V.L., Pike K.E., Baron J.C., Bourgeat P., Jones G., Faux N.G., Ellis K.a, Salvado O., Szoeke C., Martins R.N., Ames D., Masters C.L., Rowe C.C. Larger temporal volume in elderly with high versus low beta-amyloid deposition. Brain. 2010;133:3349–3358. doi: 10.1093/brain/awq187. [DOI] [PubMed] [Google Scholar]
- Dickerson B.C., Bakkour A., Salat D.H., Feczko E., Pacheco J., Greve D.N., Grodstein F., Wright C.I., Blacker D., Rosas H.D., Sperling R.a, Atri A., Growdon J.H., Hyman B.T., Morris J.C., Fischl B., Buckner R.L. The cortical signature of Alzheimer's disease: regionally specific cortical thinning relates to symptom severity in very mild to mild AD dementia and is detectable in asymptomatic amyloid-positive individuals. Cereb. Cortex. 2009;19:497–510. doi: 10.1093/cercor/bhn113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doré V., Villemagne V.L., Bourgeat P., Fripp J., Acosta O., Chetélat G., Zhou L., Martins R., Ellis K.A., Masters C.L., Ames D., Salvado O., Rowe C.C. Cross-sectional and longitudinal analysis of the relationship between Aβ deposition, cortical thickness, and memory in cognitively unimpaired individuals and in Alzheimer disease. JAMA Neurol. 2013;70:903–911. doi: 10.1001/jamaneurol.2013.1062. [DOI] [PubMed] [Google Scholar]
- Driscoll, I., Zhou, Y., An, Y., Sojkova, J., Davatzikos, C., Michael, A., n.d. Lack of association between 11 C-PiB and longitudinal brain atrophy in non-demented older individuals. Neurobiol. Aging 32, 2123–2130. doi: 10.1016/j.neurobiolaging.2009.12.008.Lack. [DOI] [PMC free article] [PubMed]
- Dubois B., Hampel H., Feldman H.H., Scheltens P., Aisen P., Andrieu S., Bakardjian H., Benali H., Bertram L., Blennow K., Broich K., Cavedo E., Crutch S., Dartigues J.F., Duyckaerts C., Epelbaum S., Frisoni G.B., Gauthier S., Genthon R., Gouw A.A., Habert M.O., Holtzman D.M., Kivipelto M., Lista S., Molinuevo J.L., O'Bryant S.E., Rabinovici G.D., Rowe C., Salloway S., Schneider L.S., Sperling R., Teichmann M., Carrillo M.C., Cummings J., Jack C.R. Preclinical Alzheimer's disease: definition, natural history, and diagnostic criteria. Alzheimers Dement. 2016 doi: 10.1016/j.jalz.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan A.M., Head D., Shah A.R., Marcus D., Mintun M., Morris J.C., Holtzman D.M. Decreased cerebrospinal fluid A?? 42 correlates with brain atrophy in cognitively normal elderly. Ann. Neurol. 2009;65:176–183. doi: 10.1002/ana.21559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faul F., Erdfelder E., Lang A.-G., Buchner A. G*power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods. 2007;39:175–191. doi: 10.3758/bf03193146. [DOI] [PubMed] [Google Scholar]
- Fjell A.M., Amlien I.K., Sneve M.H., Grydeland H., Tamnes C.K., Chaplin T. a, Rosa M.G.P., Walhovd K.B. The roots of Alzheimer's disease: are high-expanding cortical areas preferentially targeted? Cereb. Cortex. 2014 doi: 10.1093/cercor/bhu055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjell A.M., McEvoy L., Holland D., Dale A.M., Walhovd K.B., Alzheimer's Disease Neuroimaging Initiative What is normal in normal aging? Effects of aging, amyloid and Alzheimer's disease on the cerebral cortex and the hippocampus. Prog. Neurobiol. 2014;117:20–40. doi: 10.1016/j.pneurobio.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjell A.M., Walhovd K.B., Fennema-Notestine C., McEvoy L.K., Hagler D.J., Holland D., Brewer J.B., Dale A.M. CSF biomarkers in prediction of cerebral and clinical change in mild cognitive impairment and Alzheimer's disease. J. Neurosci. 2010;30:2088–2101. doi: 10.1523/JNEUROSCI.3785-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortea J., Sala-Llonch R., Bartrés-Faz D., Lladó A., Solé-Padullés C., Bosch B., Antonell A., Olives J., Sanchez-Valle R., Molinuevo J.L., Rami L. Cognitively preserved subjects with transitional cerebrospinal fluid ß-amyloid 1-42 values have thicker cortex in Alzheimer's disease vulnerable areas. Biol. Psychiatry. 2011;70:183–190. doi: 10.1016/j.biopsych.2011.02.017. [DOI] [PubMed] [Google Scholar]
- Fotenos A.F., Mintun M.A., Snyder A.Z., Morris J.C., Buckner R.L. Brain volume decline in aging: evidence for a relation between socioeconomic status, preclinical Alzheimer disease, and reserve. Arch. Neurol. 2008;65:113–120. doi: 10.1001/archneurol.2007.27. [DOI] [PubMed] [Google Scholar]
- Gispert J.D., Rami L., Sánchez-Benavides G., Falcon C., Tucholka A., Rojas S., Molinuevo J.L. Nonlinear cerebral atrophy patterns across the Alzheimer's disease continuum: impact of APOE4 genotype. Neurobiol. Aging. 2015;36 doi: 10.1016/j.neurobiolaging.2015.06.027. [DOI] [PubMed] [Google Scholar]
- Gispert J.D., Suárez-Calvet M., Monté G.C., Tucholka A., Falcon C., Rojas S., Rami L., Sánchez-Valle R., Lladó A., Kleinberger G., Haass C., Molinuevo J.L. Cerebrospinal fluid sTREM2 levels are associated with gray matter volume increases and reduced diffusivity in early Alzheimer's disease. Alzheimers Dement. 2016;12 doi: 10.1016/j.jalz.2016.06.005. [DOI] [PubMed] [Google Scholar]
- Harari O., Cruchaga C., Kauwe J.S.K., Ainscough B.J., Bales K., Pickering E.H., Bertelsen S., Fagan A.M., Holtzman D.M., Morris J.C., Goate A.M. Phosphorylated tau-Aβ42 ratio as a continuous trait for biomarker discovery for early-stage Alzheimer's disease in multiplex immunoassay panels of cerebrospinal fluid. Biol. Psychiatry. 2014;75:723–731. doi: 10.1016/j.biopsych.2013.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X., Hibar D.P., Lee S., Toga A.W., Jack C.R., Weiner M.W., Thompson P.M. Sex and age differences in atrophic rates: an ADNI study with n = 1368 MRI scans. Neurobiol. Aging. 2010;31:1463–1480. doi: 10.1016/j.neurobiolaging.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel P.S., Mattsson N., Donohue M.C., Mackin R.S., Aisen P.S., Jack C.R., Shaw L.M., Trojanowski J.Q., Weiner M.W. The transitional association between β-amyloid pathology and regional brain atrophy. Alzheimers Dement. 2014:1–9. doi: 10.1016/j.jalz.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel P.S., Mattsson N., Mackin R.S., Schöll M., Nosheny R.L., Tosun D., Donohue M.C., Aisen P.S., Jagust W.J., Weiner M.W. Accelerating rates of cognitive decline and imaging markers associated with β-amyloid pathology. Neurology. 2016;86:1887–1896. doi: 10.1212/WNL.0000000000002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Bennett D.A., Blennow K., Dunn B., Elliott C., Budd Haeberlein S., Holtzman D., Jagust W., Jessen F., Karlawish J., Liu E., Masliah E., Luis Molinuevo J., Montine T., Phelps C., Rankin K.P., Rowe C., Scheltens P., Siemers E., Silverberg N., Sperling R. 2017. 2018 NIA-AA Research Framework to Investigate the Alzheimer's Disease Continuum Background: Rationale for Updating 2011 NIA-AA Guidelines for Alzheimer's Disease. [Google Scholar]
- Jack C.R., Wiste H.J., Vemuri P., Weigand S.D., Senjem M.L., Zeng G., Bernstein M.A., Gunter J.L., Pankratz V.S., Aisen P.S., Weiner M.W., Petersen R.C., Shaw L.M., Trojanowski J.Q., Knopman D.S. Brain beta-amyloid measures and magnetic resonance imaging atrophy both predict time-to-progression from mild cognitive impairment to Alzheimer's disease. Brain. 2010;133:3336–3348. doi: 10.1093/brain/awq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Wiste H.J., Weigand S.D., Rocca W.A., Knopman D.S., Mielke M.M., Lowe V.J., Senjem M.L., Gunter J.L., Preboske G.M., Pankratz V.S., Vemuri P., Petersen R.C. Age-specific population frequencies of cerebral β-amyloidosis and neurodegeneration among people with normal cognitive function aged 50–89 years: a cross-sectional study. Lancet Neurol. 2014;13:997–1005. doi: 10.1016/S1474-4422(14)70194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau S.M., Lu M., Joshi A.D., Pontecorvo M., Mintun M.A., Trojanowski J.Q., Shaw L.M., Jagust W.J. Comparing positron emission tomography imaging and cerebrospinal fluid measurements of β-amyloid. Ann. Neurol. 2013;74:826–836. doi: 10.1002/ana.23908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzi M., Pennec X., Frisoni G.B., Ayache N. Disentangling normal aging from Alzheimer's disease in structural magnetic resonance images. Neurobiol. Aging. 2015;36:S42–S52. doi: 10.1016/j.neurobiolaging.2014.07.046. [DOI] [PubMed] [Google Scholar]
- Mattsson N., Andreasson U., Persson S., Carrillo M.C., Collins S., Chalbot S., Cutler N., Dufour-Rainfray D., Fagan A.M., Heegaard N.H.H., Robin Hsiung G.Y., Hyman B., Iqbal K., Lachno D.R., Lleó A., Lewczuk P., Molinuevo J.L., Parchi P., Regeniter A., Rissman R., Rosenmann H., Sancesario G., Schröder J., Shaw L.M., Teunissen C.E., Trojanowski J.Q., Vanderstichele H., Vandijck M., Verbeek M.M., Zetterberg H., Blennow K., Käser S.a. CSF biomarker variability in the Alzheimer's Association quality control program. Alzheimers Dement. 2013;9:251–261. doi: 10.1016/j.jalz.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson N., Tosun D., Insel P.S., Simonson A., Jack C.R., Beckett L.a, Donohue M., Jagust W., Schuff N., Weiner M.W. Association of brain amyloid-β with cerebral perfusion and structure in Alzheimer's disease and mild cognitive impairment. Brain. 2014;137:1550–1561. doi: 10.1093/brain/awu043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinuevo J.L., Gispert J.D., Dubois B., Heneka M.T., Lleo A., Engelborghs S., Pujol J., de Souza L.C., Alcolea D., Jessen F., Sarazin M., Lamari F., Balasa M., Antonell A., Rami L. The AD-CSF-index discriminates Alzheimer's disease patients from healthy controls: a validation study. J. Alzheimers Dis. 2013;36:67–77. doi: 10.3233/JAD-130203. [DOI] [PubMed] [Google Scholar]
- Möller C., Vrenken H., Jiskoot L., Versteeg A., Barkhof F., Scheltens P., van der Flier W.M. Different patterns of gray matter atrophy in early- and late-onset Alzheimer's disease. Neurobiol. Aging. 2013;34:2014–2022. doi: 10.1016/j.neurobiolaging.2013.02.013. [DOI] [PubMed] [Google Scholar]
- Mormino E.C., Kluth J.T., Madison C.M., Rabinovici G.D., Baker S.L., Miller B.L., Koeppe R.a., Mathis C.a, Weiner M.W., Jagust W.J. Episodic memory loss is related to hippocampal-mediated β-amyloid deposition in elderly subjects. Brain. 2009;132:1310–1323. doi: 10.1093/brain/awn320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J.C., Price J.L. Pathologic correlates of nondemented aging, mild cognitive impairment, and early-stage Alzheimer's disease. J. Mol. Neurosci. 2001;17:101–118. doi: 10.1385/jmn:17:2:101. [DOI] [PubMed] [Google Scholar]
- Morris J.C., Roe C.M., Xiong C., Fagan A.M., Goate A.M., Holtzman D.M., Mintun M.a. APOE predicts amyloid-beta but not tau Alzheimer pathology in cognitively normal aging. Ann. Neurol. 2010;67:122–131. doi: 10.1002/ana.21843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risacher S.L., Shen L., West J.D., Kim S., McDonald B.C., Beckett L.A., Harvey D.J., Jack C.R., Weiner M.W., Saykin A.J. Longitudinal MRI atrophy biomarkers: relationship to conversion in the ADNI cohort. Neurobiol. Aging. 2010;31:1401–1418. doi: 10.1016/j.neurobiolaging.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott J.M., Bartlett J.W., Fox N.C., Barnes J. Increased brain atrophy rates in cognitively normal older adults with low cerebrospinal fluid A??1-42. Ann. Neurol. 2010;68:825–834. doi: 10.1002/ana.22315. [DOI] [PubMed] [Google Scholar]
- Shaw L.M., Vanderstichele H., Knapik-Czajka M., Clark C.M., Aisen P.S., Petersen R.C., Blennow K., Soares H., Simon A., Lewczuk P., Dean R., Siemers E., Potter W., Lee V.M.-Y.Y., Trojanowski J.Q. Cerebrospinal fluid biomarker signature in Alzheimer's disease neuroimaging initiative subjects. Ann. Neurol. 2009;65:403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling R.A., Aisen P.S., Beckett L.A., Bennett D.A., Craft S., Fagan A.M., Iwatsubo T., Jack C.R., Kaye J., Montine T.J., Park D.C., Reiman E.M., Rowe C.C., Siemers E., Stern Y., Yaffe K., Carrillo M.C., Thies B., Morrison-Bogorad M., Wagster M.V., Phelps C.H. Toward defining the preclinical stages of Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:280–292. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storandt M., Mintun M.A., Head D., Morris J.C. Cognitive decline and brain volume loss as signatures of cerebral amyloid-beta peptide deposition identified with Pittsburgh compound B: cognitive decline associated with Abeta deposition. Arch. Neurol. 2009;66:1476–1481. doi: 10.1001/archneurol.2009.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker N.H., Dodge H.H., Dowling N.M., Han S.D., Erosheva E.a., Jagust W.J. CSF biomarker associations with change in hippocampal volume and precuneus thickness: implications for the Alzheimer's pathological cascade. Brain Imaging Behav. 2012;6:599–609. doi: 10.1007/s11682-012-9171-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutphen C.L., Jasielec M.S., Shah A.R., Macy E.M., Xiong C., Vlassenko A.G., Benzinger T.L.S., Stoops E.E.J., Vanderstichele H.M.J., Brix B., Darby H.D., Vandijck M.L.J., Ladenson J.H., Morris J.C., Holtzman D.M., Fagan A.M. Longitudinal cerebrospinal fluid biomarker changes in preclinical Alzheimer disease during middle age. JAMA Neurol. 2015:1–14. doi: 10.1001/jamaneurol.2015.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarawneh R., Head D., Allison S., Buckles V., Fagan A.M., Ladenson J.H., Morris J.C., Holtzman D.M. Cerebrospinal fluid markers of neurodegeneration and rates of brain atrophy in early Alzheimer disease. JAMA Neurol. 2015;72:656–665. doi: 10.1001/jamaneurol.2015.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titova O.E., Ax E., Brooks S.J., Sjögren P., Cederholm T., Kilander L., Kullberg J., Larsson E.-M., Johansson L., Ahlström H., Lind L., Schiöth H.B., Benedict C. Mediterranean diet habits in older individuals: associations with cognitive functioning and brain volumes. Exp. Gerontol. 2013;48:1443–1448. doi: 10.1016/j.exger.2013.10.002. [DOI] [PubMed] [Google Scholar]
- Tolboom N., van der Flier W.M., Yaqub M., Boellaard R., Verwey N.a, Blankenstein M.a, Windhorst A.D., Scheltens P., Lammertsma A.a, van Berckel B.N.M. Relationship of cerebrospinal fluid markers to 11C-PiB and 18F-FDDNP binding. J. Nucl. Med. 2009;50:1464–1470. doi: 10.2967/jnumed.109.064360. [DOI] [PubMed] [Google Scholar]
- Tosun D., Schuff N., Truran-Sacrey D., Shaw L.M., Trojanowski J.Q., Aisen P., Peterson R., Weiner M.W. Relations between brain tissue loss, CSF biomarkers, and the ApoE genetic profile: a longitudinal MRI study. Neurobiol. Aging. 2010;31:1340–1354. doi: 10.1016/j.neurobiolaging.2010.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N., Landeau B., Papathanassiou D., Crivello F., Etard O., Delcroix N., Mazoyer B., Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Vemuri P., Wiste H.J., Weigand S.D., Shaw L.M., Trojanowski J.Q., Weiner M.W., Knopman D.S., Petersen R.C., Jack C.R. MRI and CSF biomarkers in normal, MCI, and AD subjects: predicting future clinical change. Neurology. 2009;73:294–301. doi: 10.1212/WNL.0b013e3181af79fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villemagne V.L., Burnham S., Bourgeat P., Brown B., Ellis K.a., Salvado O., Szoeke C., Macaulay S.L., Martins R., Maruff P., Ames D., Rowe C.C., Masters C.L. Amyloid β deposition, neurodegeneration, and cognitive decline in sporadic Alzheimer's disease: a prospective cohort study. Lancet Neurol. 2013;12:357–367. doi: 10.1016/S1474-4422(13)70044-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complementary image statistical analyses.