Abstract
The Gram-negative bacterium Aggregatibacter actinomycetemcomitans, commonly associated with localized aggressive periodontitis (LAP), secretes an RTX (repeats-in-toxin) protein leukotoxin (LtxA) that targets human white blood cells, an interaction that is driven by its recognition of the lymphocyte function-associated antigen-1 (LFA-1) integrin. In this study, we report on the inhibition of LtxA-LFA-1 binding as an antivirulence strategy to inhibit LtxA-mediated cytotoxicity. Specifically, we designed and synthesized peptides corresponding to the reported LtxA binding domain on LFA-1 and characterized their capability to inhibit LtxA binding to LFA-1 and subsequent cytotoxic activity in human immune cells. We found that several of these peptides, corresponding to sequential β-strands in the LtxA-binding domain of LFA-1, inhibit LtxA activity, demonstrating the effectiveness of this approach. Further investigations into the mechanism by which these peptides inhibit LtxA binding to LFA-1 reveal a correlation between toxin-peptide affinity and LtxA-mediated cytotoxicity, leading to a diminished association between LtxA and LFA-1 on the cell membrane. Our results demonstrate the possibility of using target-based peptides to inhibit LtxA activity, and we expect that a similar approach could be used to hinder the activity of other RTX toxins.
Keywords: repeats-in-toxin, RTX, Aggregatibacter actinomycetemcomitans, lymphocyte function-associated antigen-1, leukotoxin, peptide therapeutics, antivirulence
Graphical abstract
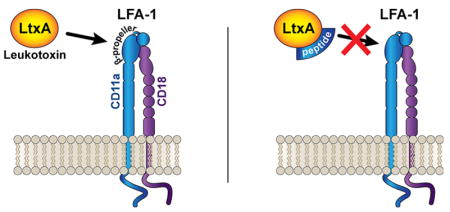
The RTX family of proteins are cytotoxins secreted from Gram-negative bacteria characterized by secretion through the type I secretion system and a highly conserved glycine- and aspartate-rich repeat domain associated with Ca2+ binding.1,2 These cytotoxins are produced by a diverse group of pathogens including Vibrio cholerae, Bordetella pertussis, Pseudomonas aeruginosa, Escherichia coli, Mannheimia hemolytica, and Aggregatibacter actinomycetemcomitans.1–5 A number of RTX toxins have been demonstrated to bind to integrin receptors on the host cell plasma membrane6–8 as an initial step in their toxic mechanism.
The RTX leukotoxin (LtxA) produced by A. actinomycetemcomitans specifically kills leukocytes in the host,9,10 allowing the bacterium to evade the immune response and providing a colonization advantage for the bacterium.11,12 Colonizing the upper aerodigestive tract in humans, A. actinomycetemcomitans is most commonly associated with localized aggressive periodontitis (LAP)13–15 but has also been linked with disseminated infections such as brain and lung abscesses16,17 and infective endocarditis.18 Current treatment methods includes mechanical debridement in combination with systemic antibiotics such as tetracycline;19–21 however, growing antibiotic resistance, including resistance to tetracycline in A. actinomycetemcomitans,22,23 has decreased the effectiveness of this treatment and necessitated the development of alternative treatment strategies.24,25 Toward this end, our lab investigates the use of targeted peptides to inhibit the activity of LtxA to eliminate the colonization advantage provided by the toxin, thus leaving the bacteria susceptible to natural host clearance mechanisms.
Peptides are increasingly finding applications as therapeutic agents in the treatment of a diverse array of diseases including infectious diseases.26–28 Currently, there are over 200 peptides approved by the US Food and Drug Administration for therapeutic use,29 including several peptide antibiotics, such as gramicidin and polymyxin. These peptide-based antibiotics interact with the bacterial cell directly, resulting in cell death. Our approach differs in that we are targeting the virulence mechanisms of the bacterium, promoting bacterial clearance by inhibiting toxin activity through the development of targeted peptides to block specific interactions of the toxin with host cells.
In its interaction with host immune cells, LtxA must bind to both cholesterol30,31 and an integrin receptor, lymphocyte function-associated antigen-1 (LFA-1), which induces cell death through a lysosomal-mediated pathway.8,32–34 Deficiency of either of these components in otherwise targeted cells has been shown to abolish toxin activity.32,35 Recent studies have also suggested that P2X receptor activation plays a role in LtxA cytotoxicity.36,37 Inhibition of the P2X receptor by antagonists was shown to protect both human erythrocytes and monocytes from LtxA-induced lysis. A similar investigation found that antagonist-inhibition of P2X receptors also protected erythrocytes from Staphylococcus aureus hemolysin A (Hla).38 However, a follow-up study revealed that the antagonists bind directly to Hla, not the P2X receptor, to inhibit toxin activity.39 Because the role of P2X receptors and antagonists in LtxA toxicity toward immune cells remains ambiguous, we have focused on inhibiting its required interactions with cholesterol and LFA-1.
LFA-1 is a transmembrane glycoprotein that functions as an adhesion and signaling receptor on the surface of immune cells, which upon activation, binds with its ligand, intercellular adhesion molecule-1 (ICAM-1), preceding cell transmigration into tissue.40,41 The integrin is composed of two noncovalently associated subunits: CD11a (αL) and CD18 (β2). Both subunits are transmembrane proteins consisting of short C-terminal cytoplasmic domains and much larger N-terminal extracellular domains, which undergo a significant conformational shift upon activation.40,42 Within the extracellular region of the CD11a subunit is a domain containing seven four-stranded β-sheet repeats (W1–W7), which folds into the β-propeller domain,43,44 and has been shown to be targeted by LtxA.32
We have previously demonstrated the inhibition of LtxA-mediated cytotoxicity by blocking the LtxA-cholesterol interaction with peptides based on the cholesterol-binding site of LtxA.35,45 In this study, we use an analogous approach to show that small peptides corresponding to sequential β-strands in the β-propeller domain of LFA-1, the reported LtxA binding site, prevent LtxA-mediated cytotoxicity by inhibiting the LtxA-LFA-1 interaction. We demonstrate that the affinity of the peptides for LtxA regulates their inhibitory action. Requisite binding to integrins has been demonstrated for several other RTX toxins;6,7 therefore, we expect that equivalent approaches with receptor-based peptides may be broadly applicable to inhibit RTX toxin activity.
RESULTS AND DISCUSSION
Peptide Design
The domain of human CD11a (hCD11a) that is recognized by LtxA has been shown to consist of a region of the β-propeller comprising the fourth β-strand (S4) of the W1 β-sheet and all four β-strands (S1–S4) of the W2 β-sheet (Figure 1).32 Rather than synthesizing the entire domain of interest as one peptide, we divided the region into its component β-strands to permit reasonable synthesis time scales and yields.46 To identify the locations of the strand boundaries in the sequence, we analyzed previous studies on the folding topology of the β-propeller and predictions of disulfide bonds between the strands,43,44,47 then synthesized five β-strand peptides (Table 1). Each peptide is designated by its β-sheet (W) and strand (S) numbers.
Figure 1.
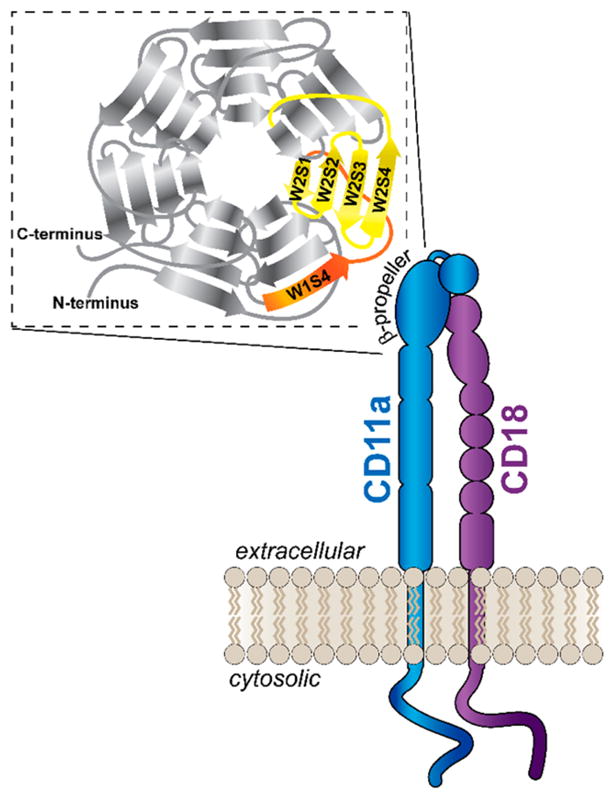
Diagram of the LFA-1 integrin, which is composed of two noncovalently associated subunits, CD11a and CD18. Both proteins are composed of extracellular, transmembrane, and cytosolic domains. The CD11a subunit contains a β-propeller region in its extracellular domain (inset), which is composed of seven β-sheets (W1–W7), each containing four β-strands (S1–S4). The highlighted β-strands are those targeted by LtxA and serve as the basis of the peptides synthesized for LtxA inhibition.
Table 1.
Sequences of Peptides
| Human CD11a-based Peptides | Murine CD11a-based Peptides | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||||||||||||||||||||||||||||||||||
| peptide | Sequence | peptide | Sequence | ||||||||||||||||||||||||||||||||||||||||
| W1S4 | T | G | H | C | L | P | mW1S4 | S | E | F | C | Q | P | ||||||||||||||||||||||||||||||
| W2S1 | V | T | L | R | G | S | N | Y | T | S | K | Y | L | G | M | T | L | A | T | D | P | mW2S1 | V | S | L | H | G | S | N | H | T | S | K | Y | L | G | M | T | L | A | T | D | A |
| W2S2 | T | D | G | S | I | L | A | C | D | P | G | mW2S2 | A | K | G | S | L | L | A | C | D | P | G | ||||||||||||||||||||
| W2S3 | L | S | R | T | C | D | Q | N | T | mW2S3 | Same as human | ||||||||||||||||||||||||||||||||
| W2S4 | Y | L | S | G | L | C | Y | L | F | R | Q | N | L | Q | G | P | M | L | mW2S4 | Not synthesized | |||||||||||||||||||||||
Red underline indicates residues differing from corresponding position in human sequence.
To complement our investigations, we also synthesized a series of peptides corresponding to the analogous region of murine CD11a (mCD11a, Table 1). Earlier work demonstrated that cells expressing chimeric LFA-1 containing the murine β-propeller were not sensitive to LtxA,32 indicating that LtxA is unable to bind to the murine β-propeller. Therefore, we anticipated that at least one of these peptides could be used as a negative control in our experiments. As shown in Table 1, we synthesized three peptides, mW1S4, mW2S1, and mW2S2. The mW2S3 sequence is identical to that of hW2S3 and was therefore not resynthesized.48
Preincubation of LtxA with hCD11a Peptides Inhibits Cytotoxicity
Each of the synthesized peptides was assessed to determine its capacity to inhibit LtxA-mediated cytotoxicity in THP-1 monocytes in suspension. Each peptide was preincubated with LtxA for 1 h at 37 °C before the toxin was added to the cells. We then determined the THP-1 cell viability after 3 h using a Trypan blue exclusion assay.
We found that four of the five hCD11a-based peptides prevented LtxA-mediated cytotoxicity (Figure 2a). When LtxA was pretreated with the W1S4, W2S1, W2S2, or W2S3 peptides, the toxin-mediated cytotoxicity was almost completely inhibited, resulting in cell viabilities significantly greater than that of the LtxA-only positive control. The W2S4 peptide, however, was not effective in blocking LtxA activity, suggesting that the W2S4 peptide does not bind to LtxA. Because the W2S4 peptide was ineffective in blocking LtxA-mediated cytotoxicity, we did not synthesize the negative control, mW2S4 peptide.
Figure 2.
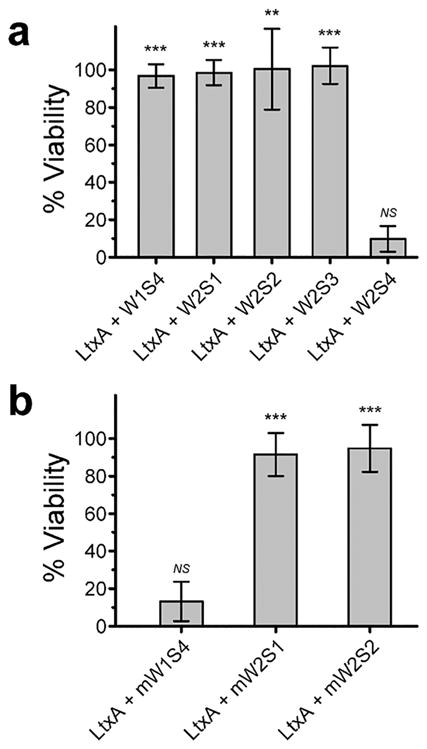
Viability of THP-1 cells after 3 h determined by Trypan blue exclusion. (a) W1S4, W2S1, W2S2, and W2S3 peptides inhibited LtxA-mediated cytotoxicity, but the W2S4 peptide did not. (b) Two of the murine peptides, mW2S1 and mW2S2, inhibited LtxA-mediated cytotoxicity, but the mW1S4 peptide did not. The viabilities were normalized to the positive and negative controls, and the level of significance was determined by a two-sample t-test. NS, P > 0.05; **, P ≤ 0.001; ***, P ≤ 0.0001 versus LtxA control.
Although LtxA has been shown not to recognize the complete murine β-propeller domain,32 we found that two of the three murine peptides (mW2S1 and mW2S2) also significantly inhibited LtxA-mediated cytotoxicity (Figure 2b). This suggests that both peptides retain some affinity for LtxA, and that differences in the sequences compared to the human-based peptide (red underlined in Table 1) does not alter their effectiveness. Conversely, the mW1S4 peptide was not effective in inhibiting LtxA-mediated cytotoxicity, and cell viability after being treated with LtxA and mW1S4 peptide was not significantly different from cells incubated with toxin alone. For this reason, we used the mW1S4 peptide as a negative control in the remainder of our experiments.
As shown in Table 1, the sequences of our hCD11a and mCD11a peptides share a 78% sequence homology. The hW2S3 and mW2S3 sequences are completely identical, and the corresponding human and murine W2S2 and W2S1 peptides vary only slightly. On the other hand, the sequences of the hW1S4 and mW1S4 are almost entirely different. Because the mW1S4 peptide is the only murine peptide that does not inhibit LtxA activity, this small region is likely the domain that confers species specificity to the LtxA-LFA-1 interaction.
In each of our experiments, we found that preincubating the toxin with the peptide was crucial to the inhibition of the toxin activity. Simultaneous addition of LtxA and peptide to THP-1 cells led to diminished inhibitory action for all peptides (Figure S1a,b), suggesting that the peptides interact with the toxin specifically to inhibit LtxA-mediated cytotoxicity. Consequently, we theorized that the inhibition should be dependent on peptide concentration. Using the same concentration of LtxA, we decreased the peptide concentration by an order of magnitude and investigated the cell viability. Our results reveal that the reduced peptide concentration did not significantly inhibit LtxA cytotoxicity (Figure S1c), demonstrating the inhibitory effect is concentration dependent. We therefore hypothesized that the observed inhibition of cytotoxicity originates from binding of the peptides to LtxA to block the receptor-binding domain.
Another mechanism that could explain the decreased cytotoxicity could be that the peptides simply induce toxin aggregation. To explore this possibility, we investigated peptide aggregation by measuring the fluorescence of 8-anilino-1-naphthalenesulfonic acid (ANS) at 350 nm in solutions containing increasing peptide concentrations. A blue shift in the emission maxima indicates a decreased polar environment indicative of peptide aggregation.49,50 Our results show that, of the human peptides that inhibited LtxA activity, aggregation began at peptide concentrations approximately five times that which was used in the cytotoxicity investigations (Figures S2a–d). Additional analysis of each of the peptides incubated with LtxA at the same concentration used in the cytotoxicity investigations also revealed no aggregation of the toxin (Figure S2e). This suggests that aggregation is not a predominant mechanism for the inhibited cytotoxicity and further supports our hypothesis that the peptides act by specifically blocking the receptor-binding domain on LtxA.
Inhibition of LtxA Activity Correlates with Affinity
To measure the binding kinetics between each of the peptides and LtxA, we used surface plasmon resonance (SPR). Sequential injections of LtxA analyte solutions at increasing concentrations were flowed across sensor chips functionalized with peptide to measure the interaction between LtxA and the peptide. Fitting the resulting sensorgrams (Figure S3) to a 1:1 Langmuir binding model, we determined the association and dissociation rate constants (Table S1) and calculated the equilibrium dissociation constants (KD).
We found the KD of LtxA for all five peptides (Figure 3) to be on the order of 10−8 M. However, the KD for LtxA binding to W2S4, the only hCD11a-based peptide that did not inhibit LtxA-mediated cytotoxicity, was approximately twice as large as the values for LtxA binding to the other peptides, indicating that the toxin has a reduced affinity for this peptide. The interaction between LtxA and the mW1S4 peptide, our negative control that does not inhibit LtxA-mediated cytotoxicity, was equivalent to that observed for nonspecific binding, indicating that this peptide has negligible affinity for LtxA.
Figure 3.
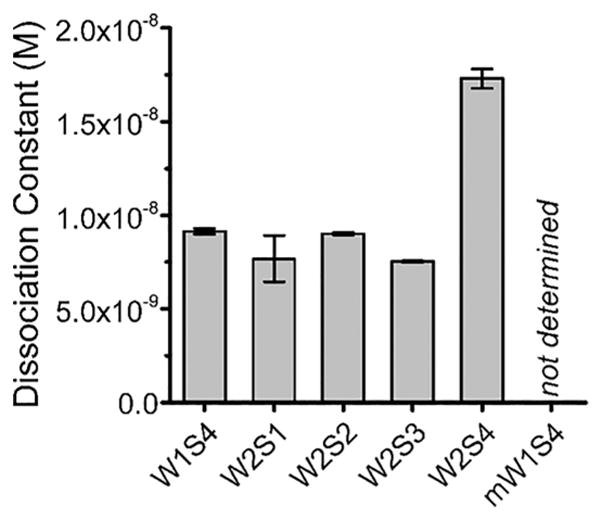
Equilibrium dissociation constants of LtxA binding to each of the peptides, calculated from the association and dissociation rate constants in Table S1. To determine the rate constants for each peptide, four concentrations of LtxA were flowed over a peptide-conjugated SPR chip, and the subsequent sensorgrams were fit to a 1:1 Langmuir binding model. Errors are propagated from the TraceDrawer fitting results.
To further characterize the peptides, we investigated their structure using circular dichroism (CD) to identity any structural differences between the peptides that may be contributing to the LtxA affinity. The domains represented by the peptides are individual β-strands that compose the β-propeller in the native CD11a structure. Taken out of this context, we did not anticipate the peptides to maintain such a secondary structure since the structure of the β-sheet is dependent upon hydrogen bonding between the antiparallel β-strands. As expected, analysis of the CD spectra (Figure S4a) revealed that none of the peptides has a predominantly ordered structure (Figure S4b), suggesting that the affinity of LtxA for the peptides arises from its recognition of the peptide sequence rather than a specific structural motif.
Although the affinity of LtxA for LFA-1 is not known, these results demonstrate that peptides containing only small domains of the reported binding site retain sufficient affinity for the toxin to inhibit LtxA-mediated cytotoxicity. Additionally, these results demonstrate that the affinity of LtxA for the peptides is directly correlated with their inhibitory action.
LFA-1-Based Peptides Inhibit LtxA Association with THP-1 Cells
To demonstrate that the LFA-1-based peptides specifically inhibit the interaction of LtxA with LFA-1 on THP-1 cells, we investigated changes in the colocalization of LtxA and LFA-1 in the presence of the peptides. We focused our investigation on the four hCD11a peptides that inhibited LtxA-mediated cytotoxicity using the nonbinding mW1S4 as a negative control. LtxA was labeled with Alexa Fluor 647 (AF647), then incubated with one of the four peptides, and THP-1 cells were incubated with FITC-tagged TS 2/4 anti-CD11a monoclonal antibody to label the LFA-1. The toxin–peptide solution was added to the labeled cells and imaged using confocal microscopy (Figure S5). We then analyzed the colocalization of the toxin with LFA-1 using Manders’ colocalization coefficients (MCC), which characterized the co-occurrence of the fluorescent probes.51 If the peptides block the CD11a binding domain of LtxA, we would expect a decrease in the colocalization of the labeled LtxA with labeled LFA-1 in the presence of the peptides.
We found that preincubating LtxA with the hCD11a peptides diminished the association of the LtxA with LFA-1 (Figure 4) compared to the LtxA only control. The composite confocal images (Figure 4a) provide a representative qualitative assessment of the diminished association. In the absence of peptide, LtxA is observed within the cells by the 15 min time point; however, a significant fraction remains on the plasma membrane, in the vicinity of LFA-1. In contrast, cells treated with the peptide-LtxA solution exhibit significantly less LtxA on both the cell membrane and within the cell, demonstrated by the decreased mean LtxA-AF647 fluorescence of the cells (Figure S6a), indicating a reduced interaction between the toxin and integrin.
Figure 4.
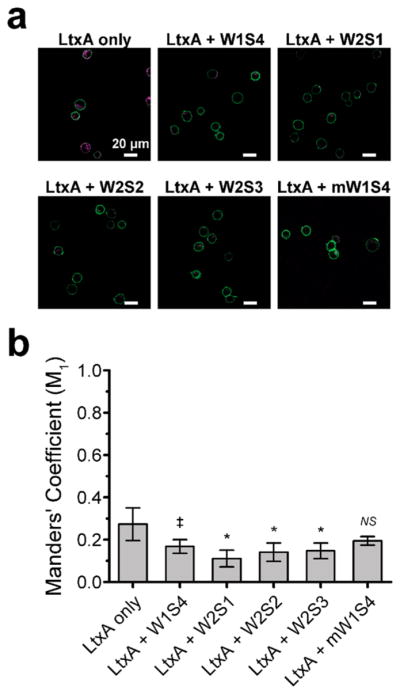
Colocalization analysis of the interaction between LtxA and LFA-1-expressing THP-1 cells. (a) Composite confocal images of the diminished LtxA (magenta) association with LFA-1 (green) after pretreatment of the toxin with the peptides. Scale bar: 20 μm. (b) Mean Manders’ coefficient M1 of labeled LFA-1 (green) and labeled LtxA (magenta) preincubated with peptide. Error bars represent the standard deviation of the mean M1. The level of significance was determined by a two-sample t-test. NS, P > 0.05; ‡, P < 0.05; *, P ≤ 0.01 versus LtxA control.
To quantify these observations, we measured the colocalization between the labeled LtxA and the labeled LFA-1 using MCC. The M1 coefficient values refer to the co-occurrence of LtxA (magenta) in pixels that contain LFA-1 (green). We found a significant decrease in the M1 values for samples that contain LtxA preincubated with hCD11a peptides (Figure 4b) compared to the LtxA only control. We also determined there was no significant difference in colocalization between LtxA and LFA-1 when the toxin was incubated with the mW1S4 peptide, compared to the control, consistent with this peptide’s lack of inhibitory activity. The simultaneous addition of LtxA and peptide resulted in no significant difference in colocalization compared to the LtxA only control (Figure S6b), consistent with our cytotoxicity investigations.
In addition to its binding to LFA-1, LtxA is also known to bind cholesterol with strong affinity, which is shown to be critical to its cytotoxic activity.30,45 Therefore, as a control to demonstrate that the inhibition of toxin association with the membrane is not due to inhibition of cholesterol binding, we investigated the ability of the LFA-1-based peptides to inhibit LtxA binding to LFA-1-free, cholesterol-containing giant unilamellar vesicles (GUVs). The GUV membranes, containing 1% N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (NBD-PE), were treated with AF647-labeled LtxA either alone or pretreated with one of the four peptides (Figure S7). We found that the peptides do not inhibit the colocalization of the LtxA with the GUVs (Figure 5), demonstrating that these peptides do not affect the ability of LtxA to bind cholesterol. As further support, we characterized the ability of LtxA treated with peptides to bind with fluorescent liposomes using a dot blot assay. LtxA preincubated with peptides was dotted on a nitrocellulose membrane and incubated with liposomes composed of POPC, cholesterol, and NBD-PE. Measuring the resulting fluorescence intensity of the dots revealed that none of the peptides significantly inhibited the binding of LtxA to cholesterol (Figure S8). Together, our results suggest that the peptide-mediated inhibition of LtxA binding to the cell membrane (Figure 4) is due to the ability of the peptides to specifically inhibit binding to the toxin’s protein receptor, LFA-1, rather than the plasma membrane lipids. The inability of the peptides to inhibit LtxA binding to these LFA-1-free GUVs (Figure 5) supports our hypothesis that the diminished cytotoxic activity originates from the peptides blocking the LFA-1 binding site on LtxA.
Figure 5.
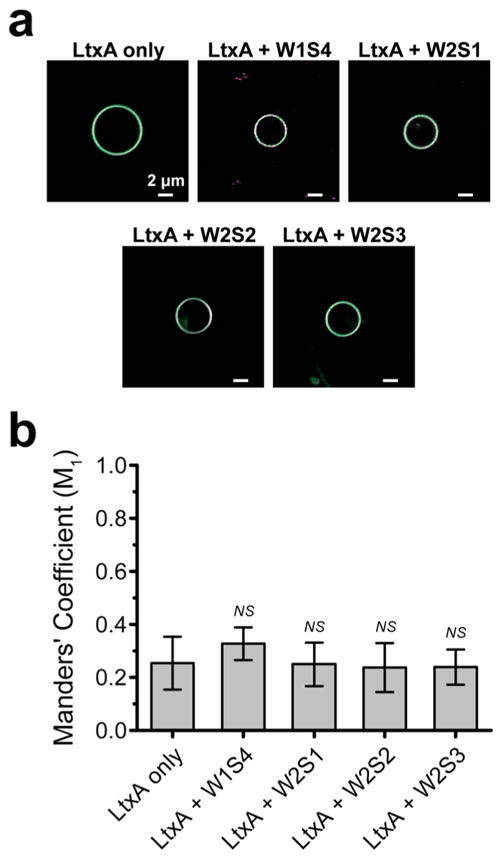
Colocalization analysis of the interaction of LtxA with LFA-1-free GUVs. (a) Composite confocal images of the LtxA (magenta) association with GUV membranes (green). Scale bar: 2 μm. (b) Mean Manders’ colocalization coefficient M1 of labeled phospholipid (NBD-PE, green) and labeled LtxA (magenta) preincubated with peptide. Error bars represent the standard deviation of the mean M1. The level of significance was determined by a two-sample t-test. NS, P > 0.05.
Although receptor-based peptides to inhibit binding have been broadly used in the field of antiviral therapies,52–54 the approach has seen less use in the field of bacterial infections, as most bacteria can be readily treated with antibiotics. However, as antibiotic resistance grows, the development of antivirulence strategies, including design of antitoxin molecules based on the toxin receptor, has increased. For example, cholera toxin (CT) binds with picomolar affinity to the ganglioside GM1,55 and soluble GM1 has been effectively used to inhibit binding of CT to host cells.56 A similar approach was investigated for blocking binding of the heat-labile enterotoxin (LT) from Escherichia coli to host cells using galactose-derived inhibitors targeting the homologous GM1 binding domain.57 In addition, substantial interest has been paid to development of antitoxin molecules against anthrax toxin including a soluble peptide derived from the toxin binding domain of the cell surface receptor.58
CONCLUSION
Our investigation demonstrates the effectiveness of receptor-based peptides to inhibit LtxA cytotoxicity. We designed small, synthetic peptides based on the reported LtxA binding site of LFA-1 to inhibit LtxA-mediated cytotoxicity. Of the five peptides we designed based on the human integrin, four had inhibitory action, demonstrating that only small domains of the CD11a binding domain are required to inhibit the activity of LtxA. We have demonstrated that the peptides bind to LtxA with a strong affinity and specifically disrupt the interaction between LtxA and LFA-1 on the host cell membrane leading to inhibition of toxin-mediated cytotoxicity. Our results demonstrate that target-based peptides can successfully be used to inhibit the cytotoxic activity of LtxA, and we anticipate that an analogous approach could be used to disrupt the binding of other RTX toxins to their integrin receptors as a means to inhibit activity.
METHODS
Chemicals
Unless indicated, all reagents were purchased from Sigma-Aldrich and used without further purification.
Cell Culture
THP-1 monocyte cells from ATCC (Manassas, VA) were cultured in RPMI 1640 (Corning) supplemented with 10% (v/v) fetal bovine serum (FBS, Quality Biological), 1% (v/v) 2-mercaptoethanol, and (v/v) 1% penicillin/streptomycin (Gibco) growth media in 5% CO2 at 37°C.
LtxA Purification
LtxA was purified from the supernatant of the JP2 strain of A. actinomycetemcomitans using size exclusion chromatography as previously described.59 The presence of LtxA was verified by Western blot (Figure S9a) using anti-LtxA monoclonal antibodies,9 and the toxin purity of 91% was determined by Coomassie staining (Figure S9b). The purified protein solution was lyophilized for storage.
Peptide Synthesis
The peptides were prepared using 9-fluorenylmethyloxycarbonyl (Fmoc) solid-phase synthesis as described previously.45 Briefly, the Fmoc group of H-Rink Amide ChemMatrix resin (0.47 mmol/g) (PCAS BioMatrix) was cleaved with a solution of 6% (wt %) piperidine and 1% (wt %) 1-hydroxybenzotriazole monohydrate (HOBt) in dimethylformamide (DMF) for 20 min and subsequently washed with methanol (MeOH) and dichloromethane (DCM). A coupling solution of Fmoc-protected amino acid (4.0 equiv), tetramethyl-O-(1H-benzotriazol-1-yl) uranium hexafluorophosphate (HBTU) (3.9 equiv), and N,N-diisopropylethylamine (DIEA) (8.0 equiv) was prepared in DMF and incubated with the resin for 90 min. After being washed with MeOH and DCM, the successive Fmoc protecting groups were removed using the same deprotection and washing steps used for the resin, and the progress of the synthesis was periodically verified by electrospray ionization (ESI) mass spectrometry (SCIEX 3200 QTRAP, SCIEX, Framingham, MA). At the end of the solid-phase synthesis, the N-terminal amino acid was acetylated with a solution of acetic anhydride and DIEA in DMF. The peptide was then cleaved from the resin with a solution of 2.5% (vol %) triisopropylsilane and 2.5% (vol %) water in trifluoroacetic acid (TFA) for 2 h and precipitated with cold diethyl ether.
Peptide Purification
The peptides were purified by reversed-phase high-performance liquid chromatography (HPLC) on an Agilent 5 Prep-C18 5 μm, 150 mm × 21.20 mm column (Agilent, Santa Clara, CA) (phase A, water and 0.1% TFA; phase B, acetonitrile and 0.1% TFA) using a gradient from 95% A and 5% B to 0% A and 100% B over 12 min. The identity of the peptide was confirmed by matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF) or electrospray ionization (ESI) mass spectrometry, and the purified peptide was then lyophilized and stored at −80 °C.
GUV Preparation
GUVs were prepared by reverse phase evaporation based on a protocol previously described.60 Lipid mixtures containing 79% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC, Avanti Polar Lipids, Alabaster, AL), 20% cholesterol, and 1% N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (NBD-PE, ThermoFisher) were prepared in chloroform with 7% (vol %) methanol. In a round-bottom flask, 100 μL of the lipid mixture was injected below 3 mL of a 1× PBS, 10 mM EDTA solution and placed on a rotary evaporator immersed in a 40 °C water bath. Under vacuum, the evaporation of organic phase spontaneously forms GUVs in the aqueous phase.
Cytotoxicity Assays
LtxA and peptide stocks were prepared by resuspending the lyophilized proteins in PBS buffer. Equal molar concentrations of LtxA and peptide solutions were then coincubated for 1 h at 37 °C before being added to cells. Suspension THP-1 cells at a concentration of 2 × 106 cells/mL in PBS were treated with the LtxA-peptide solution (160 nM final concentration), and the initial cell viability was determined by Trypan blue exclusion assay. After incubation of the cells with the toxin for 3 h at 37 °C, the Trypan blue exclusion assay was repeated to calculate the percent change in cell viability. The changes in cell viability were then normalized to the negative and positive controls, PBS and LtxA without peptide respectively, conducted during each experiment.
ANS Fluorescence
A stock solution of ANS was prepared in PBS at a concentration of 2.1 mM, determined by the absorbance at 350 nm using an extinction coefficient of 5000 M−1 cm−1.61,62 ANS stock was added to peptide and LtxA-peptide samples for a final dye concentration of 50 μM. Samples were incubated at 37 °C for 1 h, then allowed to equilibrate to room temperature prior to measurements. The fluorescence emission spectra were measured with a PTI Quantamaster 400 (Horiba, Edison, NJ) between 460 and 560 nm with an excitation wavelength of 350 nm. The fluorescence data were fit to a bi-Gaussian function with Origin 2016 (OriginLab Corp., Northampton, MA) to determine the wavelength of peak emission intensity.
SPR Investigations
SPR studies were performed with an OpenSPR instrument (Nicoya Lifesciences, Waterloo, Canada) at room temperature with a 100 μL injection loop. Standard gold sensor chips were cleaned with a piranha solution and subsequently functionalized with a 5 mM solution of 12-mercaptododecanoic acid (12-MDA) in ethanol at a flow rate of 20 μL/min. The running buffer was then changed to filtered and degassed 1× PBS, pH 7.4 for the remainder of the experiment. Each peptide was cross-linked to the 12-MDA using N-hydroxysuccinimide (NHS) and N-(3-(dimethylamino)propyl)-N′-ethylcarbodiimide hydrochloride (EDC) chemistry, and any remaining activated conjugation sites were blocked with ethanolamine. The sensor chip was allowed to stabilize for at least 1 h, and the system was primed with four alternating injections of PBS (blank) or 70 nM LtxA at a flow rate of 40 μL/min. Each analyte injection was followed by an injection of regeneration buffer composed of 50 mM each of piperidine, trisodium phosphate, ethanolamine, and glycine, pH 10.3. Finally, a series of injections of LtxA analyte solutions of increasing concentrations were passed over the sensor chip to measure the LtxA-peptide interaction. Each injection was followed by an injection of regeneration buffer to remove the bound LtxA from the sensor. Nonspecific binding (NSB) of LtxA to the chip was assessed using sensor chips functionalized as described, but without peptide immobilization. The NSB was subtracted from the sensorgrams, and the kinetics of the interaction was analyzed with TraceDrawer 1.6.1 (Ridgeview Instruments AB, Vänge, Sweden) by fitting the sensorgrams to a 1:1 Langmuir binding model.
Circular Dichroism
The secondary structure of each peptide was determined based on the circular dichroism (CD) spectra, which were recorded using a Jasco J-815 CD spectrometer (Jasco Inc., Easton, MD). Each peptide was scanned from 260 to 190 nm, with a 1.00 nm bandwidth, 1 nm data pitch, and 50 nm/min scanning speed. All peptides except W2S4 were dissolved in 10 mM phosphate buffer at a peptide concentration of 0.25 mg/mL. W2S4 stock was dissolved in 50% acetonitrile (AcCN) (vol %) in water and further diluted in 10 mM phosphate buffer to 0.25 mg/mL. All the baselines and samples were measured three times, and averaged baselines were subtracted from each sample. The subtracted CD spectra were averaged again to generate the curves shown in Figure S4a. Each subtracted spectrum was further analyzed with DICHROWEB using CONTIN/LL and reference data set seven to determine the secondary structure.63
Fluorescence Labeling
THP-1 cells (2.5 × 106 cells/mL) were washed in PBS and immunolabeled with a 2.5 ng/μL solution of FITC-functionalized TS 2/4 anti-CD11a monoclonal antibody (BioLegend, San Diego, CA) for 15 min. After labeling, the cells were washed twice in PBS and resuspended in phenol red-free media at a concentration of 5 × 105 cells/mL. The cells were plated at a density of 1.25 × 105 cells/mL in poly L-lysine-coated glass bottom dishes for imaging.
Lyophilized LtxA was dissolved at 5 μg/μL in 0.1 M sodium bicarbonate, pH 8.0 and incubated with Alexa Fluor 647 NHS ester (AF647, Thermo Fisher) (9.7 μM final concentration) for 10 min. The labeled LtxA was then centrifuged in a Zeba spin desalting column (Thermo Fisher) to separate unbound dye and exchange the buffer to 1× PBS, pH 7.4 as per the manufacturer’s instructions.
Confocal Microscopy
THP-1 cells shown in Figures 4 and S5 were imaged with a Nikon C1si confocal laser scanning microscope system and NIS Elements software (Nikon Instruments Inc., Melville, NY). Confocal images were acquired utilizing the Apo 60 × /NA 1.40 λs oil objective with 488 and 647 nm diode laser sources.
All GUV samples and the THP-1 cells used in Figure S6b were imaged with a Zeiss LSM 880 confocal laser scanning microscope system combined with the Zeiss Axio Observer 7 inverted microscope and ZEN imaging software (Carl Zeiss, Inc., Thornwood, NY). Confocal images were acquired utilizing the C-Plan-Apochromat 63×/NA 1.4 oil objective and two laser sources: argon (488 nm) and HeNe (633 nm).
Image Processing and Analysis
For colocalization analysis, the background signal in each image was subtracted using a 5625 μm2 background region of interest (ROI) with NIS Elements. For each image, an ROI was defined that encapsulated the membranes of all THP-1 cells, and the MCCs were calculated using the Coloc2 plugin in Fiji.64 An average and standard deviation MCC was calculated from four to six images for each peptide. Using the same confocal images, the mean fluorescence intensities were determined with Fiji by altering the ROIs used in the MCC calculations to include the entire cell interior.
Dot Blot
LtxA was incubated with one of the peptides for 1 h at 37 °C at the same concentrations used for cytotoxicity. Liposomes with the same composition as the GUVs (79% POPC, 20% cholesterol, 1% NBD-PE) were prepared by extrusion. LtxA alone or LtxA–peptide solution was spotted on a nitrocellulose membrane then incubated with the fluorescently labeled cholesterol-containing liposomes. Binding of the toxin to the liposomes was quantified using the resulting fluorescence intensity of each spot.
Statistical Analysis
All statistical analyses were performed in Origin 2016 (OriginLab Corp., Northampton, MA) using the independent two-sample t-test for equal means.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (DE026962 (E.K.), DE022795, and DE025275) and National Science Foundation (1554417). The authors would like to thank Dr. Edward T. Lally for providing the anti-LtxA monoclonal antibodies and Dr. Lesley Chow for the generous assistance with peptide synthesis and purification.
ABBREVIATIONS
- 12-MDA
12-mercaptododecanoic acid
- AcCN
acetonitrile
- AF647
Alexa Fluor 647 NHS ester
- ANS
8-anilino-1-naphthalenesulfonic acid
- CD
circular dichroism
- CT
cholera toxin
- DCM
dichloromethane
- DIEA
N,N-diisopropylethylamine
- DMF
dimethylformamide
- EDC
N-(3-(dimethylamino)propyl)-N′-ethylcarbodiimide hydrochloride
- EDTA
ethylenediaminetetraacetic acid
- ESI
electrospray ionization
- FBS
fetal bovine serum
- FITC
fluorescein isothyocyanate
- Fmoc
9-fluorenylmethyloxycarbonyl
- GUV
giant unilamellar vesicle
- HBTU
tetramethyl-O-(1H-benzotriazol-1-yl) uranium hexafluorophosphate
- hCD11a
human CD11a
- Hla
hemolysin A
- HOBt
1-hydroxybenzotriazole monohydrate
- HPLC
high-performance liquid chromatography
- ICAM-1
intercellular adhesion molecule-1
- KD
equilibrium dissociation constant
- LFA-1
lymphocyte function-associated antigen-1
- MALDI-TOF
matrix-assisted laser desorption/ionization-time-of-flight
- MCC
Manders’ colocalization coefficients
- mCD11a
murine CD11a
- MeOH
methanol
- NBD-PE
N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)-1,2-di-hexadecanoyl-sn-glycero-3 phosphoethanolamine
- NHS
N-hydroxysuccinimide
- NSB
nonspecific binding
- PBS
phosphate buffered saline
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- ROI
region of interest
- RTX
repeats-in-toxin
- SPR
surface plasmon resonance
- TFA
trifluoroacetic acid
Footnotes
Author Contributions
E.K. and A.C.B. designed the research approach. E.K., S.H., E.H.C., and S.Y. performed the experiments, and E.K. and E.H.C. analyzed the data. E.K. and A.C.B. wrote the manuscript. All authors have approved the final version of the manuscript.
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsinfecdis.7b00230.
Viability of THP-1 cells determined by Trypan blue exclusion; fluorescence emission peak of ANS; SPR sensorgrams of LtxA binding to hCD11a and mCD11a peptides; association and dissociation rate constants of LtxA–peptide interactions; CD analysis of peptide structure; confocal microscopy images of FITC labeled THP-1 cells and LtxA labeled with AF647; normalized mean fluorescence of LtxA-AF647 in THP-1 cells and mean MCC from simultaneous addition of LtxA and peptide; confocal microscopy images of fluorescent GUVs and LtxA labeled with AF647; quantification of dot blot to measure LtxA binding to cholesterol-containing liposomes; Western blot and Coomassie staining of purified LtxA (PDF)
References
- 1.Linhartova I, Bumba L, Masin J, Basler M, Osicka R, Kamanova J, Prochazkova K, Adkins I, Hejnova-Holubova J, Sadílkova L, Morova J, Sebo P. RTX proteins: a highly diverse family secreted by a common mechanism. FEMS Microbiol Rev. 2010;34:1076–1112. doi: 10.1111/j.1574-6976.2010.00231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chenal A, Sotomayor-Perez AC, Ladant D. Structure and function of RTX toxins. In: Alouf JE, Popoff MR, editors. The Comprehensive Sourcebook of Bacterial Protein Toxins. 4. Academic Press; Boston: 2015. pp. 677–718. [Google Scholar]
- 3.Aulik NA, Hellenbrand KM, Czuprynski CJ. Mannheimia haemolytica and Its Leukotoxin Cause Macrophage Extracellular Trap Formation by Bovine Macrophages. Infect Immun. 2012;80:1923–1933. doi: 10.1128/IAI.06120-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiles TJ, Mulvey MA. The RTX pore-forming toxin α-hemolysin of uropathogenic Escherichia coli: progress and perspectives. Future Microbiol. 2013;8:73–84. doi: 10.2217/fmb.12.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang L, Conway JF, Thibodeau PH. Calcium-induced Folding and Stabilization of the Pseudomonas aeruginosa Alkaline Protease. J Biol Chem. 2012;287:4311–4322. doi: 10.1074/jbc.M111.310300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeyaseelan S, Kannan MS, Briggs RE, Thumbikat P, Maheswaran SK. Mannheimia haemolytica Leukotoxin Activates a Nonreceptor Tyrosine Kinase Signaling Cascade in Bovine Leukocytes, Which Induces Biological Effects. Infect Immun. 2001;69:6131–6139. doi: 10.1128/IAI.69.10.6131-6139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morova J, Osicka R, Masin J, Sebo P. RTX cytotoxins recognize β2 integrin receptors through N-linked oligosaccharides. Proc Natl Acad Sci U S A. 2008;105:5355–5360. doi: 10.1073/pnas.0711400105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lally ET, Kieba IR, Sato a, Green CL, Rosenbloom J, Korostoff J, Wang JF, Shenker BJ, Ortlepp S, Robinson MK, Billings PC. RTX Toxins Recognize a β2 Integrin on the Surface of Human Target Cells. J Biol Chem. 1997;272:30463–30469. doi: 10.1074/jbc.272.48.30463. [DOI] [PubMed] [Google Scholar]
- 9.DiRienzo JM, Tsai CC, Shenker BJ, Taichman NS, Lally ET. Monoclonal antibodies to leukotoxin of Actinobacillus actinomycetemcomitans. Infect Immun. 1985;47:31–36. doi: 10.1128/iai.47.1.31-36.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lally ET, Golub EE, Kieba IR. Identification and immunological characterization of the domain of Actinobacillus actinomycetemcomitans leukotoxin that determines its specificity for human target cells. J Biol Chem. 1994;269:31289–31295. [PubMed] [Google Scholar]
- 11.Kachlany SC. Aggregatibacter actinomycetemcomitans Leukotoxin: from Threat to Therapy. J Dent Res. 2010;89:561–570. doi: 10.1177/0022034510363682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korostoff J, Yamaguchi N, Miller M, Kieba I, Lally ET. Perturbation of mitochondrial structure and function plays a central role in Actinobacillus actinomycetemcomitans leukotoxin-induced apoptosis. Microb Pathog. 2000;29:267–278. doi: 10.1006/mpat.2000.0390. [DOI] [PubMed] [Google Scholar]
- 13.Faveri M, Figueiredo LC, Duarte PM, Mestnik MJ, Mayer MPA, Feres M. Microbiological profile of untreated subjects with localized aggressive periodontitis. J Clin Periodontol. 2009;36:739–749. doi: 10.1111/j.1600-051X.2009.01449.x. [DOI] [PubMed] [Google Scholar]
- 14.Fine DH, Markowitz K, Furgang D, Fairlie K, Ferrandiz J, Nasri C, McKiernan M, Gunsolley J. Aggregatibacter actinomycetemcomitans and Its Relationship to Initiation of Localized Aggressive Periodontitis: Longitudinal Cohort Study of Initially Healthy Adolescents. J Clin Microbiol. 2007;45:3859–3869. doi: 10.1128/JCM.00653-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johansson A, Kalfas S. Virulence Mechanisms of Leukotoxin from Aggregatibacter actinomycetemcomitans. In: Virdi M, editor. Oral Health Care - Prosthodontics, Periodontology, Biology, Research and Systemic Conditions. InTech; 2012. [Google Scholar]
- 16.Hagiwara SI, Fujimaru T, Ogino A, Takano T, Sekijima T, Kagimoto S, Eto Y. Lung abscess caused by infection of Actinobacillus actinomycetemcomitans. Pediatr Int. 2009;51:748–751. doi: 10.1111/j.1442-200X.2009.02899.x. [DOI] [PubMed] [Google Scholar]
- 17.Rahamat-Langendoen JC, van Vonderen MGA, Engström LJ, Manson WL, van Winkelhoff AJ, Mooi-Kokenberg EANM. Brain abscess associated with Aggregatibacter actinomycetemcomitans: case report and review of literature. J Clin Periodontol. 2011;38:702–706. doi: 10.1111/j.1600-051X.2011.01737.x. [DOI] [PubMed] [Google Scholar]
- 18.Sen Yew H, Chambers ST, Roberts SA, Holland DJ, Julian KA, Raymond NJ, Beardsley J, Read KM, Murdoch DR. Association between HACEK bacteraemia and endocarditis. J Med Microbiol. 2014;63:892–895. doi: 10.1099/jmm.0.070060-0. [DOI] [PubMed] [Google Scholar]
- 19.Akincibay H, Orsal S, Sengun D, Tozum TF. Systemic administration of doxycycline versus metronidazole plus amoxicillin in the treatment of localized aggressive periodontitis: A clinical and microbiologic study. Quintessence Int. 2008;39:e33–39. [PubMed] [Google Scholar]
- 20.Mandell RL, Tripodi LS, Savitt E, Goodson JM, Socransky SS. The Effect of Treatment on Actinobacillus actinomycetemcomitans in Localized Juvenile Periodontitis. J Periodontol. 1986;57:94–99. doi: 10.1902/jop.1986.57.2.94. [DOI] [PubMed] [Google Scholar]
- 21.Oettinger-Barak O, Dashper SG, Catmull DV, Adams GG, Sela MN, Machtei EE, Reynolds EC. Antibiotic susceptibility of Aggregatibacter actinomycetemcomitans JP2 in a biofilm. J Oral Microbiol. 2013;5:20320. doi: 10.3402/jom.v5i0.20320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saxen L, Asikainen S. Metronidazole in the treatment of localized juvenile periodontitis. J Clin Periodontol. 1993;20:166–171. doi: 10.1111/j.1600-051x.1993.tb00339.x. [DOI] [PubMed] [Google Scholar]
- 23.Walker CB. The acquisition of antibiotic resistance in the periodontal microflora. Periodontol 2000. 1996;10:79–88. doi: 10.1111/j.1600-0757.1996.tb00069.x. [DOI] [PubMed] [Google Scholar]
- 24.Deas DE, Mealey BL. Response of chronic and aggressive periodontitis to treatment. Periodontol 2000. 2010;53:154–166. doi: 10.1111/j.1600-0757.2009.00334.x. [DOI] [PubMed] [Google Scholar]
- 25.Mombelli A, Gmur R, Gobbi C, Lang NP. Actinobacillus actinomycetemcomitans in Adult Periodontitis. II Characterization of Isolated Strains and Effect of Mechanical Periodontal Treatment. J Periodontol. 1994;65:827–834. doi: 10.1902/jop.1994.65.9.827. [DOI] [PubMed] [Google Scholar]
- 26.Yu Y, Deng YQ, Zou P, Wang Q, Dai Y, Yu F, Du L, Zhang NN, Tian M, Hao JN, Meng Y, Li Y, Zhou X, Fuk-Woo Chan J, Yuen KY, Qin CF, Jiang S, Lu L. A peptide-based viral inactivator inhibits Zika virus infection in pregnant mice and fetuses. Nat Commun. 2017;8:15672. doi: 10.1038/ncomms15672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Koh JJ, Liu S, Lakshminarayanan R, Verma CS, Beuerman RW. Membrane Active Antimicrobial Peptides: Translating Mechanistic Insights to Design. Front Neurosci. 2017;11:73. doi: 10.3389/fnins.2017.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kratochvil MJ, Yang T, Blackwell HE, Lynn DM. Nonwoven Polymer Nanofiber Coatings That Inhibit Quorum Sensing in Staphylococcus aureus: Toward New Nonbactericidal Approaches to Infection Control. ACS Infect Dis. 2017;3:271–280. doi: 10.1021/acsinfecdis.6b00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Usmani SS, Bedi G, Samuel JS, Singh S, Kalra S, Kumar P, Ahuja AA, Sharma M, Gautam A, Raghava GPS. THPdb: Database of FDA-approved peptide and protein therapeutics. PLoS One. 2017;12:e0181748. doi: 10.1371/journal.pone.0181748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown AC, Balashova NV, Epand RM, Epand RF, Bragin A, Kachlany SC, Walters MJ, Du Y, Boesze-Battaglia K, Lally ET. Aggregatibacter actinomycetemcomitans Leukotoxin Utilizes a Cholesterol Recognition/Amino Acid Consensus Site for Membrane Association. J Biol Chem. 2013;288:23607–23621. doi: 10.1074/jbc.M113.486654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fong KP, Tang HY, Brown AC, Kieba IR, Speicher DW, Boesze-Battaglia K, Lally ET. Aggregatibacter actinomycetemcomitans leukotoxin is posttranslationally modified by addition of either saturated or hydroxylated fatty acyl chains. Mol Oral Microbiol. 2011;26:262–276. doi: 10.1111/j.2041-1014.2011.00617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kieba IR, Fong KP, Tang HY, Hoffman KE, Speicher DW, Klickstein LB, Lally ET. Aggregatibacter actinomycetemcomitans leukotoxin requires β-sheets 1 and 2 of the human CD11a β-propeller for cytotoxicity. Cell Microbiol. 2007;9:2689–2699. doi: 10.1111/j.1462-5822.2007.00989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balashova N, Dhingra A, Boesze-Battaglia K, Lally ET. Aggregatibacter actinomycetemcomitans leukotoxin induces cytosol acidification in LFA-1 expressing immune cells. Mol Oral Microbiol. 2016;31:106–114. doi: 10.1111/omi.12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DiFranco KM, Gupta A, Galusha LE, Perez J, Nguyen TVK, Fineza CD, Kachlany SC. Leukotoxin (Leukothera®) targets active leukocyte function antigen-1 (LFA-1) protein and triggers a lysosomal mediated cell death pathway. J Biol Chem. 2012;287:17618–17627. doi: 10.1074/jbc.M111.314674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown AC, Koufos E, Balashova NV, Boesze-Battaglia K, Lally ET. Inhibition of LtxA toxicity by blocking cholesterol binding with peptides. Mol Oral Microbiol. 2016;31:94–105. doi: 10.1111/omi.12133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fagerberg SK, Jakobsen MR, Skals M, Praetorius HA. Inhibition of P2X Receptors Protects Human Monocytes against Damage by Leukotoxin from Aggregatibacter actinomycetemcomitans and α-Hemolysin from Escherichia coli. Infect Immun. 2016;84:3114–3130. doi: 10.1128/IAI.00674-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munksgaard PS, Vorup-Jensen T, Reinholdt J, Söderström CM, Poulsen K, Leipziger J, Praetorius HA, Skals M. Leukotoxin from Aggregatibacter actinomycetemcomitans causes shrinkage and P2X receptor-dependent lysis of human erythrocytes. Cell Microbiol. 2012;14:1904–1920. doi: 10.1111/cmi.12021. [DOI] [PubMed] [Google Scholar]
- 38.Skals M, Leipziger J, Praetorius HA. Haemolysis induced by α-toxin from Staphylococcus aureus requires P2X receptor activation. Pfluegers Arch. 2011;462:669. doi: 10.1007/s00424-011-1010-x. [DOI] [PubMed] [Google Scholar]
- 39.Schwiering M, Husmann M, Hellmann N. P2X-Receptor Antagonists Inhibit the Interaction of S. aureus Hemolysin A with Membranes. Toxins. 2017;9:332. doi: 10.3390/toxins9100332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hogg N, Patzak I, Willenbrock F. The insider’s guide to leukocyte integrin signalling and function. Nat Rev Immunol. 2011;11:416–426. doi: 10.1038/nri2986. [DOI] [PubMed] [Google Scholar]
- 41.Smith A, Stanley P, Jones K, Svensson L, McDowall A, Hogg N. The role of the integrin LFA-1 in T-lymphocyte migration. Immunol Rev. 2007;218:135–146. doi: 10.1111/j.1600-065X.2007.00537.x. [DOI] [PubMed] [Google Scholar]
- 42.Springer TA, Dustin ML. Integrin inside-out signaling and the immunological synapse. Curr Opin Cell Biol. 2012;24:107–115. doi: 10.1016/j.ceb.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang C, Springer TA. Folding of the β-propeller domain of the integrin alphaL subunit is independent of the I domain and dependent on the β2 subunit. Proc Natl Acad Sci U S A. 1997;94:3162–3167. doi: 10.1073/pnas.94.7.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Springer TA. Folding of the N-terminal, ligand-binding region of integrin α-subunits into a β-propeller domain. Proc Natl Acad Sci U S A. 1997;94:65–72. doi: 10.1073/pnas.94.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koufos E, Chang EH, Rasti ES, Krueger E, Brown AC. Use of a Cholesterol Recognition Amino Acid Consensus Peptide To Inhibit Binding of a Bacterial Toxin to Cholesterol. Biochemistry. 2016;55:4787–4797. doi: 10.1021/acs.biochem.6b00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Straner P, Taricska N, Szabo M, Toth GK, Perczel A. Bacterial expression and/or solid phase peptide synthesis of 20–40 amino acid long polypeptides and miniproteins, the case study of Class B GPCR ligands. Curr Protein Pept Sci. 2016;17:147–155. doi: 10.2174/1389203716666151102105215. [DOI] [PubMed] [Google Scholar]
- 47.Zang Q, Lu C, Huang C, Takagi J, Springer TA. The Top of the Inserted-like Domain of the Integrin Lymphocyte Function-associated Antigen-1 β Subunit Contacts the α Subunit β-Propeller Domain near β-Sheet 3. J Biol Chem. 2000;275:22202–22212. doi: 10.1074/jbc.M002883200. [DOI] [PubMed] [Google Scholar]
- 48.Kaufmann Y, Tseng E, Springer TA. Cloning of the murine lymphocyte function-associated molecule-1 alpha-subunit and its expression in COS cells. J Immunol. 1991;147:369–374. [PubMed] [Google Scholar]
- 49.Gasymov OK, Glasgow BJ. ANS Fluorescence: Potential to Augment the Identification of the External Binding Sites of Proteins. Biochim Biophys Acta, Proteins Proteomics. 2007;1774:403–411. doi: 10.1016/j.bbapap.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider A, Lang A, Naumann W. Fluorescence Spectroscopic Determination of the Critical Aggregation Concentration of the GnRH Antagonists Cetrorelix, Teverelix and Ozarelix. J Fluoresc. 2010;20:1233–1240. doi: 10.1007/s10895-010-0674-5. [DOI] [PubMed] [Google Scholar]
- 51.Dunn KW, Kamocka MM, McDonald JH. A practical guide to evaluating colocalization in biological microscopy. Am J Physiol, Cell Physiol. 2011;300:C723. doi: 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao H, Zhou J, Zhang K, Chu H, Liu D, Poon VKM, Chan CCS, Leung HC, Fai N, Lin YP, Zhang AJX, Jin DY, Yuen KY, Zheng BJ. A novel peptide with potent and broad-spectrum antiviral activities against multiple respiratory viruses. Sci Rep. 2016;6:22008. doi: 10.1038/srep22008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skalickova S, Heger Z, Krejcova L, Pekarik V, Bastl K, Janda J, Kostolansky F, Vareckova E, Zitka O, Adam V, Kizek R. Perspective of Use of Antiviral Peptides against Influenza Virus. Viruses. 2015;7:5428–5442. doi: 10.3390/v7102883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jenssen H. Anti herpes simplex virus activity of lactoferrin/lactoferricin – an example of antiviral activity of antimicrobial protein/peptide. Cell Mol Life Sci. 2005;62:3002–3013. doi: 10.1007/s00018-005-5228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuziemko GM, Stroh M, Stevens RC. Cholera Toxin Binding Affinity and Specificity for Gangliosides Determined by Surface Plasmon Resonance. Biochemistry. 1996;35:6375–6384. doi: 10.1021/bi952314i. [DOI] [PubMed] [Google Scholar]
- 56.Chatterjee D, Chaudhuri K. Association of cholera toxin with Vibrio cholerae outer membrane vesicles which are internalized by human intestinal epithelial cells. FEBS Lett. 2011;585:1357–1362. doi: 10.1016/j.febslet.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 57.Merritt EA, Sarfaty S, Feil IK, Hol WGJ. Structural foundation for the design of receptor antagonists targeting Escherichia coli heat-labile enterotoxin. Structure. 1997;5:1485–1499. doi: 10.1016/s0969-2126(97)00298-0. [DOI] [PubMed] [Google Scholar]
- 58.Bradley KA, Young JAT. Anthrax toxin receptor proteins. Biochem Pharmacol. 2003;65:309–314. doi: 10.1016/s0006-2952(02)01455-7. [DOI] [PubMed] [Google Scholar]
- 59.Diaz R, Ghofaily LA, Patel J, Balashova NV, Freitas AC, Labib I, Kachlany SC. Characterization of leukotoxin from a clinical strain of Actinobacillus actinomycetemcomitans. Microb Pathog. 2006;40:48–55. doi: 10.1016/j.micpath.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 60.Moscho A, Orwar O, Chiu DT, Modi BP, Zare RN. Rapid preparation of giant unilamellar vesicles. Proc Natl Acad Sci U S A. 1996;93:11443–11447. doi: 10.1073/pnas.93.21.11443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cardamone M, Puri NK. Spectrofluorimetric assessment of the surface hydrophobicity of proteins. Biochem J. 1992;282:589–593. doi: 10.1042/bj2820589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stryer L. The interaction of a naphthalene dye with apomyoglobin and apohemoglobin: A fluorescent probe of non-polar binding sites. J Mol Biol. 1965;13:482–495. doi: 10.1016/s0022-2836(65)80111-5. [DOI] [PubMed] [Google Scholar]
- 63.Sreerama N, Woody RW. Estimation of Protein Secondary Structure from Circular Dichroism Spectra: Comparison of CONTIN, SELCON, and CDSSTR Methods with an Expanded Reference Set. Anal Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 64.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


