Abstract
Listeria monocytogenes is an intracellular food-borne pathogen that has evolved to enter mammalian host cells, survive within them, spread from cell to cell, and disseminate throughout the body. A series of secreted virulence proteins from Listeria are responsible for manipulation of host-cell defense mechanisms and adaptation to the intracellular lifestyle. Identifying when and where these virulence proteins are located in live cells over the course of Listeria infection can provide valuable information on the roles these proteins play in defining the host-pathogen interface. These dynamics and protein levels may vary from cell to cell, as bacterial infection is a heterogeneous process both temporally and spatially. No assay to visualize virulence proteins over time in infection with Listeria or other Gram-positive bacteria has been developed. Therefore, we adapted a live, long-term tagging system to visualize a model Listeria protein by fluorescence microscopy on a single-cell level in infection. This system leverages split-fluorescent proteins, in which the last strand of a fluorescent protein (a 16-amino-acid peptide) is genetically fused to the virulence protein of interest. The remainder of the fluorescent protein is produced in the mammalian host cell. Both individual components are nonfluorescent and will bind together and reconstitute fluorescence upon virulence-protein secretion into the host cell. We demonstrate accumulation and distribution within the host cell of the model virulence protein InlC in infection over time. A modular expression platform for InlC visualization was developed. We visualized InlC by tagging it with red and green split-fluorescent proteins and compared usage of a strong constitutive promoter versus the endogenous promoter for InlC production. This split-fluorescent protein approach is versatile and may be used to investigate other Listeria virulence proteins for unique mechanistic insights in infection progression.
Introduction
The facultative intracellular pathogen Listeria monocytogenes is the causative agent of the food-borne disease listeriosis and has emerged as a model system to study host-pathogen interactions on a cellular (1, 2, 3, 4, 5) and organismal level (6, 7). This Gram-positive bacterium can cross the placental, intestinal and blood-brain barrier, making Listeria infections particularly dangerous for immunocompromised patients and pregnant women, for whom it can lead to miscarriage (8). Listeria can enter different types of mammalian cells—including epithelial cells and macrophages—escape the internalization vacuole, replicate within the cytosol while escaping the immune response, and spread from cell to cell (1, 8). Listeria infection progression often displays heterogeneous phenotypes on a single-cell level, including intracellular replication patterns that vary from cell to cell both in space and time (9, 10) and nonsynchronized spread from cell to cell (11). Approaches to investigate single-cell infection dynamics, including fluorescence microscopy assays (12), can provide insights in Listeria infections that cannot be gathered by bulk assays.
To enable and support the intracellular lifecycle, Listeria produces and secretes a series of proteins called virulence proteins during the infection process (13, 14). Over 700 proteins were predicted to have a signal peptide for secretion via Sec (15), the major secretion pathway in Listeria (16). Six other secretion pathways exist in Listeria (13, 17), and protein clients of these pathways continue to be cataloged by proteomic analyses (18, 19). The timing of protein secretion and localization within the host cell often correlates with function. For example, OrfX (20) and LntA (21) localize to the host nucleus, where they interact with host proteins and modulate their function. In the nuclei of macrophages, OrfX directly interacts with and reduces levels of RybP, a regulator of innate immunity, among other functions (20). LntA interacts with BAHD1, a factor involved in chromatin remodeling, which results in immune response modulation (21). The well-characterized virulence protein ActA is secreted to the Listeria surface and promotes actin nucleation to enable Listeria’s spread from cell to cell (11, 22, 23, 24). More recently, it was found that ActA has an additional function in biofilm formation (25, 26). Similarly, new roles of perhaps the most heavily studied Listeria virulence protein, listeriolysin O (27, 28, 29), continue to be discovered (30), including roles in organelle and histone manipulation (31, 32). Together, a picture of complex dynamics with ever-increasing diversity of localization patterns and functionalities for secreted virulence proteins emerges, necessitating approaches to track these proteins on a single-cell level during infection for mechanistic insights.
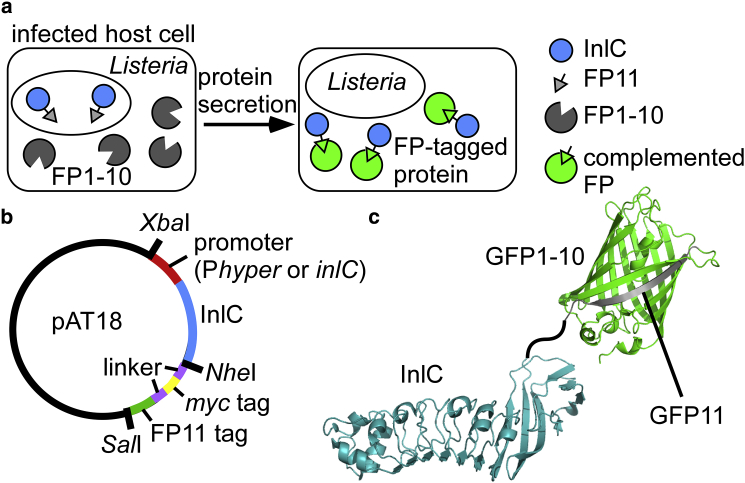
Live-fluorescence-microscopy approaches are ideal to dissect dynamics of bacterial infections on a single-cell level. We and others previously developed live fluorescence tools to visualize and quantify effector proteins secreted from different Gram-negative pathogens in infection, including Salmonella, Shigella, and Escherichia coli (33, 34, 35, 36, 37, 38, 39, 40). The effector proteins labeled by different fluorescent tags in these studies are secreted by the type III secretion system (41), a well-studied and common protein secretion pathway across the inner and outer membrane in Gram-negative bacteria. A particularly powerful tool is the split-green fluorescent protein (GFP) system, in which the last β-strand of GFP (GFP11, 16 amino acids) is genetically fused to the protein of interest via a short and flexible linker (34, 35) (see also Fig. 1 a). The remainder of GFP (called GFP1-10) is produced in the host cell, and the individual components are nonfluorescent. Upon secretion of the GFP11-tagged effector protein, the two components complement and produce fluorescent GFP, allowing visualization and tracking of the effector via GFP. Importantly, by using only GFP11 and not the stable full-length GFP, steric interference with the protein-secretion apparatus is minimized (34, 35). Using this approach, effector dynamics in single infected cells were readily resolved, including visualization of effector dynamics, localization within host cells, and quantification of effector protein levels over the course of infection (33, 34, 35). Live-fluorescence-tagging approaches of secreted proteins from Listeria or other Gram-positive pathogens have not been demonstrated. We sought to test if the split-fluorescent protein system could be adapted to visualize secreted Listeria proteins of interest in the context of infection.
Figure 1.
Overview of strategy for tagging Listeria virulence proteins with split-fluorescent proteins (FPs). (a) An illustration of split-FP application in Listeria infection is shown. Listeria cells produce InlC with a C-terminal FP11 tag (the 11th strand of a fluorescent protein of interest, see Table S3 for sequence information of the tag). The host cell produces the remaining portion of the FP (FP1-10, see Table S4 for sequence information). After secretion of InlC-FP11, the two components of the FP complement, producing a fluorescent protein tag. (b) A plasmid overview for production of InlC-FP11 variants in Listeria is shown. In this study, two different promoters (Phyper and the endogenous inlC promoter) and three different FP11 tags were used (GFP11, mNeonGreen11, and super-folder Cherry11). The FP11 tag is fused to InlC via a flexible linker that includes a myc tag (see Table S3). (c) Structures of InlC (47) and GFP (61) illustrating C-terminal tagging of InlC via a flexible linker are shown. To see this figure in color, go online.
Internalin C (InlC) is a Listeria protein from the internalin family (42) that is secreted via the Sec pathway (15) and controlled by the PrfA transcriptional regulator (43). In whole-animal mice studies, virulence of an inlC deletion strain was significantly attenuated (42), but bacterial uptake and intracellular replication were not affected by deletion of InlC (44). InlC’s role in infection is underscored by the fact that inlC is absent in nonpathogenic Listeria strains and expression is high in infection conditions, namely in the intestine and blood (45). Two functions of InlC have been described in the literature. Gouin et al. demonstrated that InlC interferes with the host innate immune response by directly interacting with IKKα (46), whereas Rajabian et al. found that InlC promotes cell-to-cell spread by relieving membrane tension via interaction with the adaptor protein Tuba (44). We chose InlC as an ideal test case to establish split-fluorescent protein tagging for the following reasons. First, InlC was produced with C-terminal fusions in various contexts, including tagging with a myc tag (46) and usage of an affinity tag for protein purification (47), indicating that a C-terminal tag is unlikely to interfere with protein production or function. Furthermore, the crystal structure of InlC is known (47), and manual inspection suggests that the InlC C-terminus is accessible and that a tag is unlikely to interfere with function (Fig. 1 c). GFP complementation in infection takes ∼2 h (34, 35) and is therefore ideal for robust visualization of virulence proteins at later infection time points. Indeed, several studies indicate high levels of InlC accumulation in infected epithelial cells at late infection stages (>4 h) both by Western blotting analysis (46) and immunofluorescence (46, 48, 49, 50). Lastly, the available immunofluorescence data in the literature (46, 48, 49, 50) allow for assessment of InlC localization patterns by our split-fluorescent protein system. Together, visualizing secretion dynamics of tagged InlC in infection serves as a robust platform to establish the split-fluorescent protein tool in Listeria.
Materials and Methods
Strains and plasmids
An overview of Listeria strains used in this study is presented in Table S1 (see also Fig. 1 b for a schematic). All experiments were done in Listeria monocytogenes EGDe (BUG 1600) (51) or a variant of this strain in which inlC was deleted (EGDe ΔinlC, BUG 2118). Plasmids for production of InlC-fusions in Listeria were generated in E. coli Stellar cells (Takara Bio USA, Mountain View, CA) as follows. A unique XbaI site was introduced upstream of the Phyper promoter in pAT18_cGFP (12) by standard site-directed mutagenesis techniques. A unique SalI site is located immediately after the cGFP stop codon. The region encompassing the Phyper promoter and the cGFP coding sequence was removed via XbaI and SalI restriction digestion and replaced by inserts encoding for various inlC fusions (see Fig. 1 b). Variants of the inserts include two different promoters (Phyper versus the inlC promoter) and the 11th strand of different fluorescent proteins, namely the 11th strand of GFP (GFP11) (34, 35), the 11th strand of mNeonGreen (mNG11) (52), and the 11th strand of super-folder Cherry (sfCh11) (52). These inserts were purchased as g-block (IDT, San Jose, CA) DNA fragments, including XbaI and SalI restriction sites for ligation. Annotated sequences for promoters and the tag sequences are provided in Tables S2 and S3. The DNA and resulting protein coding sequences for the GFP11, mNG11, and sfCh11 tags are shown in Table S3. The promoter and coding sequence for inlC were retrieved from the National Library of Medicine, US, National Center for Biotechnology Information, available from https://www.ncbi.nlm.nih.gov/gene/ (gene ID: 985945, inlC internalin C (Listeria monocytogenes EGD-e)). To ensure optimal expression in Listeria, the codon usage for the tags was optimized manually according to codon usage in Listeria (53) and verified using the %MinMax codon usage quantification algorithm (54) (Fig. S1).
For fluorescent protein complementation, a plasmid encoding GFP1-10 and a nuclear blue fluorescent protein (BFP) marker was used (described earlier (34)). Plasmids to produce strands 1–10 of mNeonGreen (mNG1-10) and super-folder Cherry (sfCh1-10) were a gift from Bo Huang (52). The coding sequences of each gene were ligated into pcDNA3.1(+) using standard cloning methods. Protein sequences of GFP1-10, mNG1-10, and sfCh1-10 are listed in Table S4.
Culture of bacteria
Listeria strains were grown in brain heart infusion (BHI) media (Thermo Fisher Scientific, Waltham, MA) at 37°C. E. coli strains were grown in Luria-Bertani broth (Thermo Fisher Scientific) at 37°C. When required, erythromycin was added at a final concentration of 5 μg/mL for Listeria and at a final concentration of 150 μg/mL for E. coli (see Table S1). Growth curves of Listeria strains were collected as follows. A saturated overnight culture of each strain was diluted to an optical density (O.D.) at 600 nm of 0.001. Wells of a 96-well plate with clear bottom were filled with 300 μL of each culture in triplicate. Growth of bacterial cultures was monitored via O.D. at 600 nm every 30 min at 30°C for 18 h in a Tecan Safire-II plate reader (Tecan, Männedorf, Switzerland) while leaving the plate shaking.
Listeria electroporation
To introduce plasmids into Listeria strains, cells were grown for 2 h at 37°C and shaken at 180 rotations per minute until an O.D. of 0.2 at 600 nm was reached. Penicillin-G was added to a final concentration of 0.12 μg/mL. The Listeria cells were grown for an additional 3 h until the final O.D. was between 0.6 and 0.8 at 600 nm. Cells were harvested by centrifugation for 30 min at 4000 rotations per minute and resuspended in cold (4°C) electroporation buffer consisting of 816 mM sucrose, 1 mM MgCl2 (pH 7). Cells were washed three times using 100, 66, and 33 mL electroporation buffer in subsequent steps. All resuspension steps were performed at 4°C. The washed cells were resuspended at a concentration of ∼1 × 1011 cells/mL in electroporation buffer. Aliquots of 100 μL cells were placed in electroporation cuvettes with a 0.2 cm gap (Bio-Rad, Hercules, CA) along with 5 μL plasmid DNA at a concentration of 1 μg/μL. Cells were electroporated at 2.4 kV, 200 Ohms, and 25 μF. The cells were immediately transferred in 900 μL prewarmed BHI media and incubated at 37°C for 3 h, followed by plating on selective plates and incubation at 37°C overnight.
Mammalian cell culture
HeLa cells (ATCC CCL-2) were used as model epithelial cells in this study. Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Thermo Fisher Scientific) supplemented with 10% (vol/vol) fetal calf serum (FBS) (Thermo Fisher Scientific). Cells were grown at 37°C with 5% CO2. HeLa cells were passaged every 2–4 days and not used past passage 12. To passage cells, they were first rinsed with phosphate-buffered saline (PBS) incubated with trypsin EDTA (Thermo Fisher Scientific) until cells had detached. Trypsin was quenched with DMEM/FBS media, and cells were seeded in new dishes. For high throughput comparison of GFP complementation (Fig. 6), a stable HeLa cell line expressing GFP1-10 and the nuclear marker TagBFP was used as previously described (34).
Figure 6.
High throughout quantification of GFP complementation during Listeria infection. (a) HeLa cells were infected with ΔinlCPhyper-inlC-GFP11 and ΔinlCpInlC-inlC-GFP11 side by side, and infections were visualized over time in parallel in the same experimental setup. A representative infection (ΔinlCpInlC-inlC-GFP11) at 3 h postinfection (left panel) vs. 6.5 h postinfection (right panel) is shown. The inset indicates examples of Listeria cells in the DIC channel. Scale bars, 20 μm. Images in the GFP channel are presented at the same intensity levels. (b) Quantification of GFP fluorescence intensity for the time course shown in (a) is given. The arrows indicate 3 and 6.5 h postinfection. (c) The mean GFP fluorescence signal for all cells in the experiment was determined at 3 h postinfection vs. 6.5 h postinfection (one experiment; n = 128 cells for ΔinlCPhyper-inlC-GFP11 and n = 75 cells for ΔinlCpInlC-inlC-GFP11). Statistical significance was determined using a Tukey honest significant difference ANOVA test. To see this figure in color, go online.
Primary bone-marrow-derived macrophages (BMDMs) were isolated as previously described (55). Briefly, marrow was flushed with PBS from the femurs and tibias of 2–3-month-old SV129S6 mice (Taconic Biosciences, Hudson, NY). The cells were overlaid onto an equal volume of Histopaque-1083 (Sigma-Aldrich, St. Louis, MO) and centrifuged at 500 × g for 25 min. Monocytes at the interface were harvested and incubated for 6–7 days at 37°C in 5% CO2 in DMEM (Sigma-Aldrich) supplemented with FBS (20%), L-glutamine (2 mM), sodium pyruvate (1 mM), beta-mercaptoethanol (50 μM), HEPES (10 mM), and penicillin-streptomycin (50 IU/mL of penicillin and 50 μg/mL of streptomycin) containing recombinant murine macrophage-colony-stimulating factor (10 pg/μL) (PeproTech, Rocky Hill, NJ) to promote monocyte differentiation into macrophages.
Bacterial infections
To visualize fluorescent protein complementation in live infections, 0.15 × 106 HeLa cells were seeded 2 days before the infection in homemade imaging dishes (35 mm) with a 10 mm center hole covered by cover glass (No. 1.5; VWR International, Aurora, CA). DNA encoding for GFP1-10 (or mNG1-10 or sfCh1-10) was transfected on day two, using the TransIT transfection system at a concentration of 2.5 μg per imaging dish according to the manufacturer’s recommendations (Mirus Bio, Madison, WI). One dish was treated equally and used on the day of the infection for counting to determine the multiplicity of infection (MOI). On the day of the infection, a saturated overnight culture of the Listeria strain of interest was diluted 10-fold in BHI media (supplemented with antibiotic as necessary) and grown for about 3 h while periodically measuring the O.D. at 600 nm. When the O.D. at 600 nm reached 0.6–0.8, bacteria were pelleted and rinsed in DMEM media three times. To calculate the MOI, we assumed each O.D. unit at 600 nm corresponded to 109 bacteria/mL. Infections were performed at an MOI of 100. HeLa cells to be infected were rinsed with DMEM media three times. The media was then replaced with DMEM supplemented with Listeria cells. The infected cells were incubated in 37°C and 5% CO2 for 1 h. The media was then exchanged with DMEM media with 10% FBS supplemented with 20 μg/mL of gentamicin (Thermo Fisher Scientific). The cells were then incubated for the remainder of the infection course in 37°C/5% CO2.
For infections without live cell complementation, 0.25 × 106 HeLa cells were seeded, and the infection was performed as described above on the following day.
To monitor infection in macrophages, differentiated primary macrophages at 6 days postisolation were lifted by scraping, and cells were subjected to nucleofection using 2.5 μg of DNA and reaction conditions recommended by the manufacturer for Nucleofector Program Y-001 (Lonza, Allendale, NJ). Nucleofected cells were seeded into 35 mm glass-bottom dishes and incubated at 37°C with 5% CO2 for 6–24 h before infection and imaging. Macrophage cells producing GFP1-10 were infected with the ΔinlCpInlC-inlC-GFP11 strain at an MOI of 5. Infections were allowed to proceed for 45 min at 37°C and 5% CO2 before the media was changed to phenol-red-free DMEM and 20 μg/mL gentamicin. The cells were incubated in 37°C/5% CO2 during the course of the infection and during imaging on the microscope.
For colony-forming unit (CFU) assays, HeLa cells were infected as described above in six-well dishes in triplicates at an MOI of 100. After infection for the specified times, the media was removed, and HeLa cells were lysed by incubation in 0.1% Triton X-100 in water for 20 min at room temperature. To quantify bacteria in each infection, serial dilutions were plated on BHI plates, and CFUs were determined in technical triplicates.
Analysis of cell lysates by protein electrophoresis and Western blotting
To generate Listeria lysates, the equivalent of 1 mL bacterial culture for O.D. at 600 nm of 0.5 was pelleted and resuspended in 40 μL lysozyme buffer (20% sucrose, 10 mg/mL lysozyme, 10 mM Tris (pH 8.0), 50 mM NaCl, 10 mM EDTA). The resuspended pellets were incubated at 55°C for 20 min. After addition of 10 μL of B-PER Bacterial Protein Extraction Reagent (Thermo Fisher Scientific), the samples were incubated at room temperature for 15 min. Each cell lysate was diluted 1:1 with 2× SDS-PAGE loading buffer and spun for 5 min to remove debris, and the supernatant was used for SDS-PAGE.
For SDS-PAGE, O.D. normalized Listeria lysates (10 μL of the lysates prepared as described above) were loaded per well. Protein samples were separated by SDS-PAGE on a 14% gel and stained by Coomassie according to standard protocols.
For Western blotting, the SDS-PAGE gel was transferred onto a polyvinylidene fluoride membrane and probed with primary and secondary antibody as outlined in Table S5. Immunoblots were developed with Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare Life Sciences, Pittsburgh, PA) and imaged on an ImageQuant LAS4000 imaging system (GE Healthcare Life Sciences).
Cell fixation and immunofluorescence
At indicated time points postinfection, cells were fixed in 4% paraformaldehyde (PFA) in water for 10 min, rinsed in PBS, and permeabilized by incubating in 0.2% Triton X-100 in PBS for 5 min. For preserving GFP-complementation fluorescence upon fixation, cells were fixed using 1% PFA and 1% sucrose in PBS, fixed for 20 min, and permeabilized using 0.1% Tween for 15 min. Cells were rinsed three times for 5 min each in PBS, and slides were blocked with 200 μL of 5% FBS in PBS for 20 min. After rinsing in PBS, slides were incubated with the primary antibody in 5% FBS in PBS. Dilutions and sources of all antibodies and stains are listed in Table S5. After rinsing in PBS three times, the slides were incubated with the secondary antibody in 5% FBS in PBS. Alexa 594-phalloidin or Coumarin-phalloidin for actin staining was added together with the secondary antibody. After incubation with the secondary antibody, slides were washed in PBS, rinsed in water, and mounted.
Fluorescence microscopy
Unless otherwise indicated, all fluorescence images were acquired on a Nikon Ti-E widefield fluorescence microscope (Nikon, Tokyo, Japan) equipped with Nikon elements software, Ti-E perfect focus system, an iXon3 EMCCD camera (Andor, Belfast, UK), mercury arc lamp, DAPI (375/5 excitation, 400 dichroic, 470/12 emission), GFP (480/10 excitation, 490 dichroic, 510/10 emission) and mCherry (560/10 excitation, 585 dichroic, 610/25 emission) filter sets. External excitation and emission filter wheels were controlled by a Lambda 10-3 filter changer (Sutter Instruments, Novato, CA), and dichroic mirrors were placed on cubes in the dichroic turret. Images were collected using a 60× oil objective (NA 1.40) and exposures; EM gain settings and neutral density filter settings are listed in Table S6 for each data set in this study. Live cells were maintained at 37°C and 5% CO2 in a LiveCell environment chamber (Pathology Devices, Westminster, MD) during the experiments. For time-course-infection experiments (Fig. 4 b), images were collected every 15 min.
Figure 4.
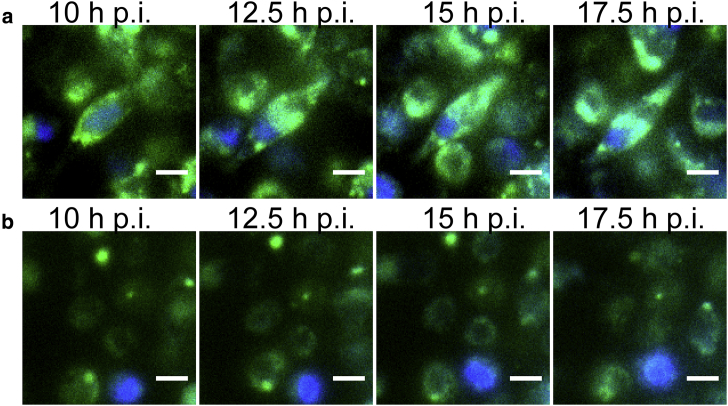
Split-GFP tagging of InlC during infection of epithelial cells. (a) Microscopy images of HeLa cells producing GFP1-10 (see Table S4 for sequence information) and a nuclear TagBFP transfection marker are shown. Shown are cells infected with ΔinlCPhyper-inlC-GFP11 (see Table S3 for tag sequence information) at 18 h postinfection (top panel, two experiments, 15 cells) and an uninfected control (bottom panel). Insets in DIC channel are examples of Listeria. Arrows in the InlC-GFP channel are examples of InlC cell membrane puncta. (b) Microscopy images of live HeLa at different time points postinfection with ΔinlCPhyper-inlC-GFP11 are shown. Listeria entered the cell shown here at 6 h postinfection (see arrows in DIC inset) and accumulate within the host cell throughout the time course. Cells were transfected to produce GFP1-10 and a nuclear transfection marker. Scale bars, 20 μm. To see this figure in color, go online.
For high throughput comparison of GFP complementation (Fig. 6), a Nikon Ti-E HCA widefield fluorescence microscope (Nikon) was used. This system was equipped with Nikon elements software, Ti-E perfect focus system, a digital CMOS camera (Hamamatsu Photonics, Hamamatsu, Japan), a Lumencor Spectra X light engine 500 mW solid-state light source, and DAPI (395 excitation, 475/20 emission), GFP (470 excitation, 540/21 emission), and brightfield-transmitted illumination using a PEKA light engine (Lumencor, Beaverton, OR). Images were collected using a Plan Apo λ 40× Ph2 DM air objective (NA 0.95). Images were acquired at 50% light intensity for 200 ms for each fluorescent or brightfield channel every 15 min over the time course of the experiment. Live cells were maintained at 37°C and 5% CO2 in an Okolab cage incubator (Okolab, Burlingame, CA) during the experiment.
Results and Discussion
A series of inlC fusion constructs were generated to enable live-cell visualization of InlC in the context of infection of mammalian cells. We first fused the 11th strand of GFP with a flexible linker to the C-terminus of InlC (Fig. 1; see Table S3 for the tag sequence), analogous to a previous approach in which secreted effector proteins from the Gram-negative pathogen Salmonella were tagged with GFP11 (33, 34, 35). Production of the resulting nonfluorescent fusion protein was under control of the constitutive Phyper promoter on the multicopy plasmid pAT18 (12) (Fig. 1 b). This initial construct allowed for maximal protein production in Listeria to assess the feasibility of split-GFP complementation in the context of Listeria infections. The remainder of GFP (GFP1-10, see Table S4 for sequence information) is also nonfluorescent and was produced in the mammalian host cell, as was done previously in Salmonella infection systems (33, 34, 35). Upon secretion of InlC-GFP11 in Listeria infection, the GFP11 portion of InlC-GFP11 is predicted to complement with GFP1-10, inducing complemented fluorescent InlC-GFP for visualization of InlC dynamics in the host cell (Fig. 1 a).
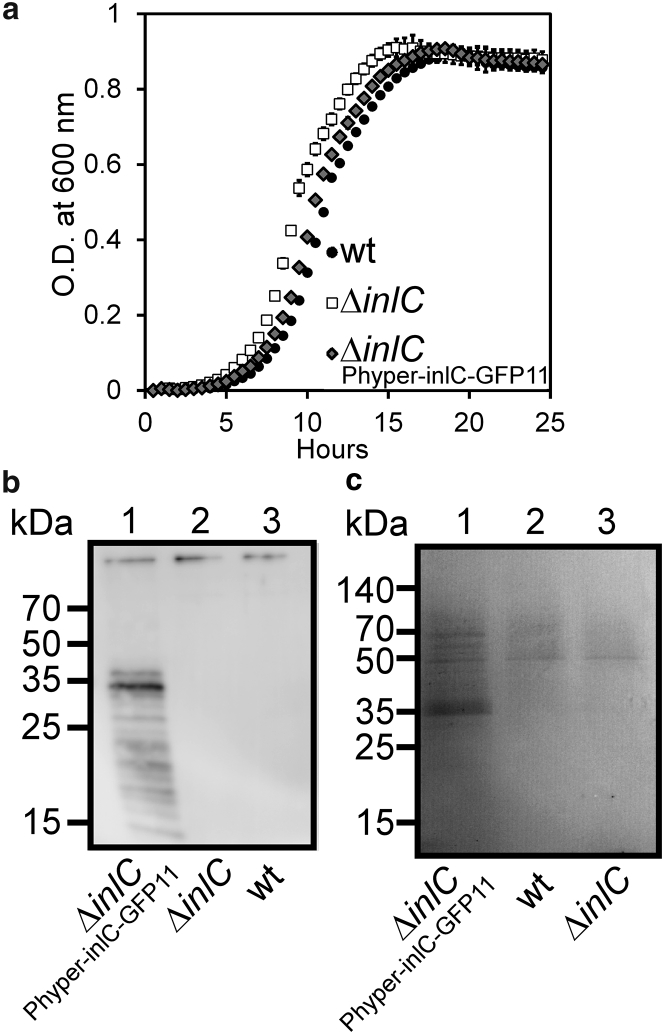
We first assessed whether production of tagged InlC from the pAT18 plasmid significantly alters Listeria growth. We compared the growth of wt (wild-type) Listeria and ΔinlC, as well as a strain in which InlC was produced as a fusion with GFP11 from a plasmid (ΔinlCPhyper-inlC-GFP11, Fig. 2 a). The engineered strain displayed growth comparable to that of the wt strain, leading us to conclude that harboring the plasmid encoding for InlC fusions does not significantly affect cell growth, even in the case in which the InlC-GFP11 fusion is constitutively produced under control of the Phyper promoter.
Figure 2.
Engineered strain ΔinlCPhyper-inlC-GFP11 displays normal in vitro growth, protein secretion, and infectivity. (a) The growth curve of Listeria strains at 30°C is shown. Errors bars indicate STD from n = 3 experiments. (b) An immunoblot of Listeria cell lysates after 25 h growth using a polyclonal anti-InlC antibody is shown. (c) A Coomassie-stained SDS-PAGE gel of secreted proteome of wt, ΔinlC, and ΔinlCPhyper-inlC-GFP11 strains collected by trichloroacetic acid precipitation of supernatants from Listeria cultures after 25 h growth is shown.
Next, we assessed whether production of the fusion protein InlC-GFP11 from the pAT18 plasmid results in InlC secretion and whether the proteome secreted from Listeria is altered in strains harboring a plasmid. The supernatant of overnight cultures of Listeria samples was precipitated with trichloroacetic acid and probed for the presence of the InlC protein using Western blotting via a polyclonal anti-InlC antibody (Fig. 2 b). No band was observed in the ΔinlC strain or wt strain, as would be expected because in the wt strain, InlC is produced under control of its endogenous promoter, which is only active after Listeria has entered host cells (12, 45). However, we observed a band around 35 kDa in the ΔinlCPhyper-inlC-GFP11 strain, consistent with a size of 33.2 kDa for full-length InlC. We concluded that InlC was produced and secreted when under the control of the constitutive Phyper promoter. Whole protein samples of the culture supernatant were then analyzed on a Coomassie-stained SDS-PAGE gel (Fig. 2 c). A band appears around 35 kDa for strain ΔinlCPhyper-inlC-GFP11 (lane 1), confirming production of InlC-GFP11 and in line with the Western blot analysis (Fig. 2 b). As seen for the Western blot, the 35 kDa band corresponding to InlC-GFP11 was absent in the wt strain and the ΔinlC strains (lanes 2, 3). We observed bands of ∼50 and ∼70 kDa across all three strains tested. These bands presumably correspond to other secreted proteins and suggest that there is not a significant change in overall protein secretion in the ΔinlCPhyper-inlC-GFP11 strain.
Bacterial strains harboring plasmids sometimes exhibit decreased infectivity (56). We compared bacterial load of the engineered strain ΔinlCPhyper-inlC-GFP11 with the wt and ΔinlC strains at 7, 13, and 29 h postinfection (Fig. S2). To measure bacterial load, a CFU assay was performed in which HeLa cells were infected with each strain and CFUs were quantified at each time point. At 7 and 13 h, there was a slight decrease in bacterial load for strains harboring plasmids with tagged InlC compared to the wt and ΔinlC strains, but at 29 h, the CFU count between engineered strains and both the wt and ΔinlC control displayed no significant difference. This observation is in line with a previous finding in which no difference in the infectivity of cultured cells was observed for Listeria monocytogenes EGD ΔinlC versus its wt parent (46). We concluded that infectivity in our cell infection assay is not significantly affected for our engineered strains.
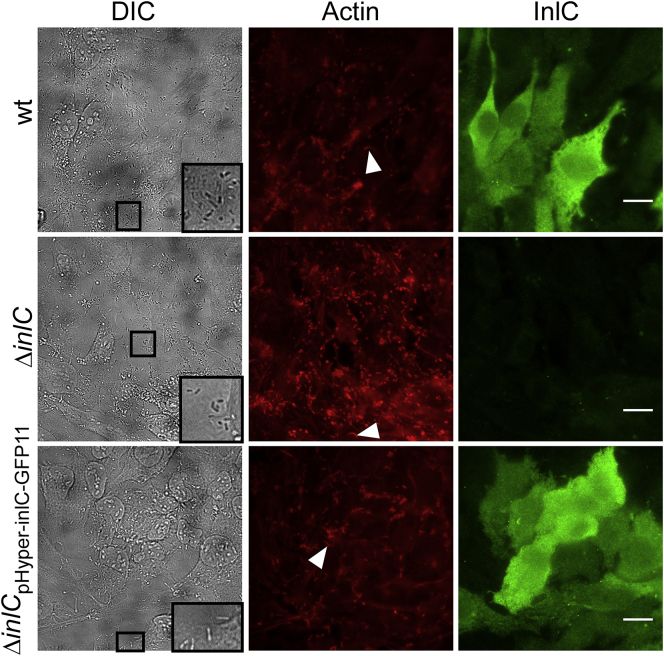
To test whether tagging InlC with a fluorescent protein fragment affects secretion and/or localization phenotypes in infection, we carried out immunofluorescence against InlC in HeLa cells infected with different Listeria strains (Fig. 3). HeLa cells were infected with wt, ΔinlC, and ΔinlCPhyper-inlC-GFP11 strains and fixed 25 h postinfection. Listeria are visible as small rods in the differential interference contrast (DIC) channel (Fig. 3, zoomed-in insets in DIC channel). Representative regions with comparable Listeria infection densities are presented for each strain (Fig. 3). Actin was labeled using red fluorescent phalloidin and localized to the rod-shaped Listeria cells (Fig. 3) because of the ability of Listeria cells to polymerize actin (11, 22). As established in the literature (23, 24), Listeria infection leads to characteristic actin tails (Fig. 3, arrows in red channel). InlC was visualized using an anti-InlC antibody (Fig. 3). InlC levels for wt and ΔinlCPhyper-inlC-GFP11 were comparable, whereas ΔinlC produced only background green fluorescence signal, indicating successful secretion of InlC for wt and ΔinlCPhyper-inlC-GFP11. InlC localized throughout the cytosol as seen previously (46, 48, 49). Together, we concluded that InlC secretion in the ΔinlCPhyper-inlC-GFP11 strain is comparable with secretion of endogenous InlC in the wt strain, and our engineered strain may be used to report on InlC dynamics within the host cell by our live-cell fluorescence complementation approach.
Figure 3.
Tagged InlC is produced and secreted into the HeLa host cell in the context of Listeria infection. HeLa cells were infected with the indicated strains, and cells were fixed 25 h postinfection. Actin was detected via Alexa 594-phalloidin, and InlC was detected by immunofluorescence using a polyclonal anti-InlC antibody (46) and an Alexa-Fluor-488-labeled secondary antibody. Representative Listeria cells are indicated in the insets in the DIC channel, and actin tails are indicated by arrows in the actin channel. Images are presented at the same intensity levels for each channel. Scale bars, 20 μm (wt infection: n = 36 infected cells total; ΔinlC: n = 29 infected cells total; ΔinlCPhyper-inlC-GFP11: n = 63 infected cells total). To see this figure in color, go online.
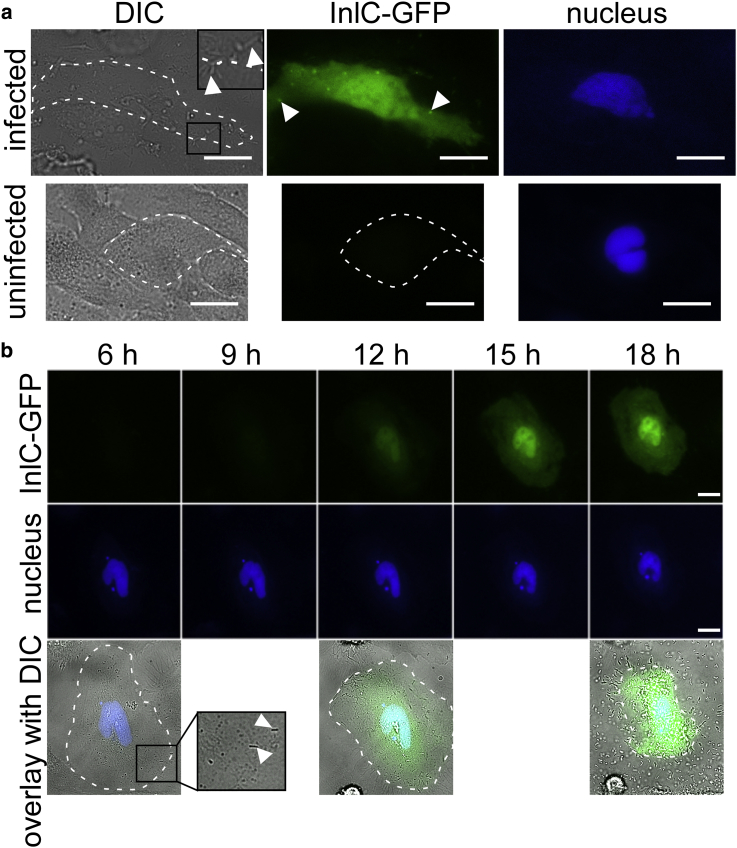
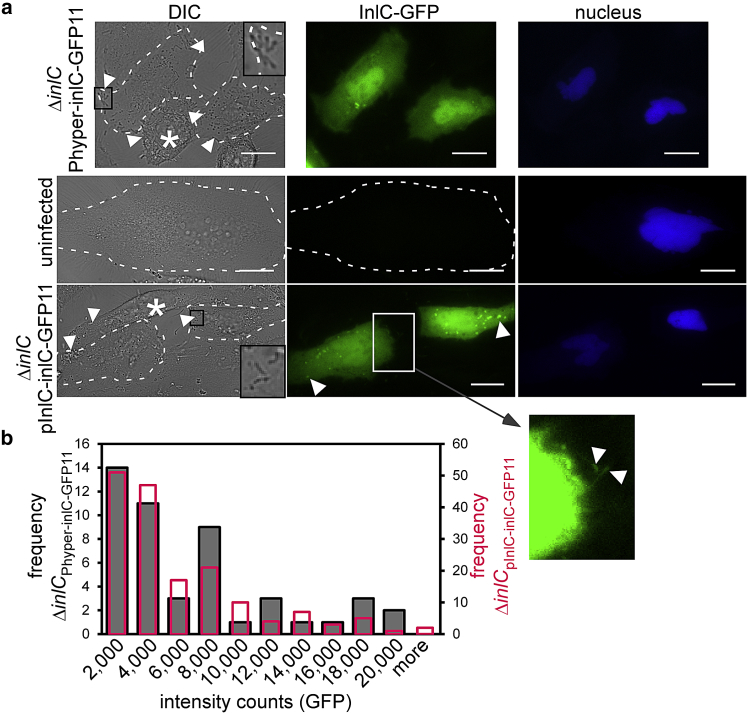
Although secreted Listeria proteins including InlC have been routinely visualized in fixed infections by immunofluorescence (46, 48, 49, 50), no tools have been applied to track virulence proteins in live infections. To determine whether the split-GFP system can be used to track secreted InlC, HeLa cells were transfected with DNA encoding for GFP1-10 before Listeria infection (see Table S4 for the protein sequence of GFP1-10). The plasmid encoding GFP1-10 included a transfection marker producing nuclear TagBFP. Fig. 4 presents the results of cells that were transfected with GFP1-10 and infected with ΔinlCPhyper-inlC-GFP11 (see Table S3 for tag sequence details). Listeria infections were observed in the DIC channel via the distinct rod-like shape of the bacteria (Fig. 4 a, inset). Green fluorescence signal was detected in HeLa cells infected with the ΔinlCPhyper-inlC-GFP11 strain but not for an uninfected control (Fig. 4 a). These results confirm complementation of GFP1-10 and GFP11 upon InlC-GFP11 secretion during Listeria infection. We observed diffuse cytosolic signal of the complemented InlC-GFP (Fig. 4 a), in line with cytosolic InlC signal detected by immunofluorescence by us (Fig. 3) and others (46, 48, 49). Interestingly, studies in the literature also report localization of InlC at cell protrusions that form during Listeria cell-to-cell spread (49). We observed similar protrusions that appear as brighter green puncta in live cells (Fig. 4 a) and occasionally in fixed immunofluorescence images (Fig. S3, arrow). The complemented GFP fluorescence could be preserved upon fixation, and the pattern of localization generally matched the fluorescence signal from anti-InlC immunofluorescence (Fig. S4). When InlC puncta were visible in IF images (via GFP comp or anti-InlC), they were juxtaposed with Listeria (detected via DIC) and actin bundles (detected via phalloidin), consistent with the suggestion that InlC may promote cell-to-cell spreading (Fig. S4). Together, we conclude that our split-GFP tagging system reports on InlC localization and resembles localization patterns of endogenous InlC.
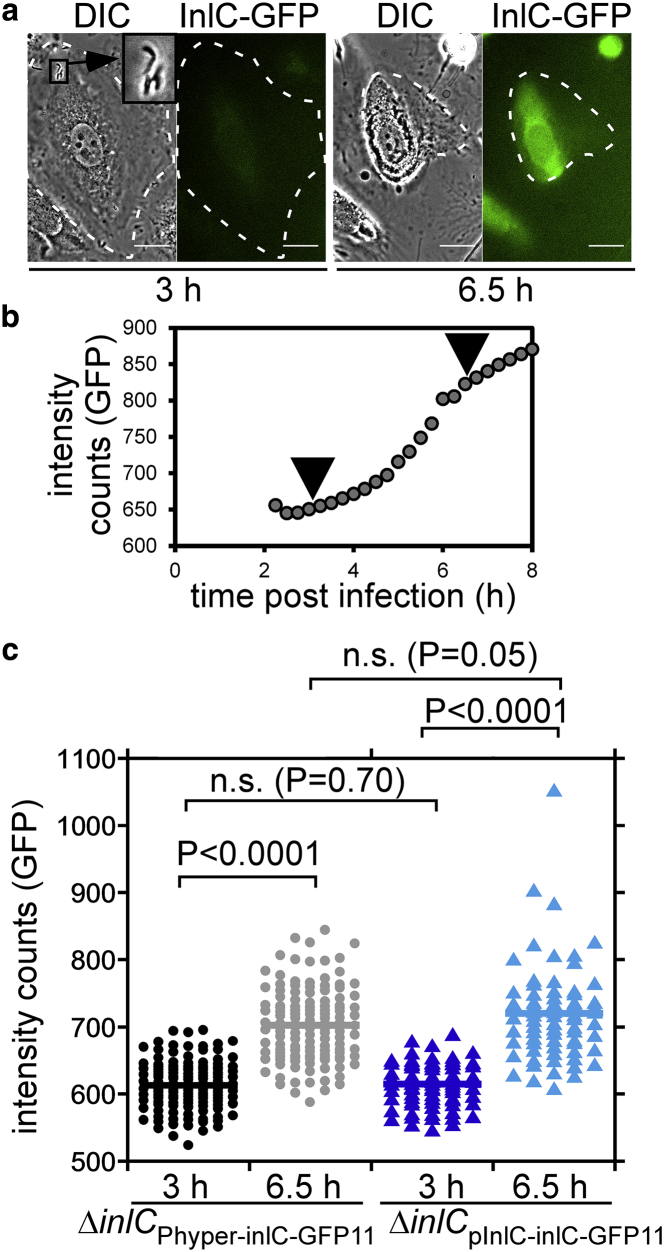
Because the split-GFP system enables time-resolved visualization of tagged protein dynamics, we monitored GFP fluorescence for several hours during infection of epithelial cells (Fig. 4 b). HeLa cells were transfected with GFP1-10 including the nuclear TagBFP transfection marker and infected with the strain ΔinlCPhyper-inlC-GFP11. Because Listeria infections are heterogeneous both in space and time, it is often difficult to track infection progression in a bulk assay. The time course presented in Fig. 4 b shows Listeria that enter a single HeLa cell at 6 h postinfection (see DIC inset in Fig. 4 b), precisely indicating the infection starting point. We monitored the GFP channel for 12 h past this time point and observed an increase of the green fluorescence signal in the cytosol and nucleus, in line with intracellular accumulation of InlC. As seen with Western blot assays in the literature (46), high levels of InlC accumulated late in the infection process, leading us to conclude that our split-GFP approach is suitable for spatiotemporal resolution of secreted proteins in infection.
We next assessed InlC-visualization in live infections when the InlC-GFP11 fusion was produced from the endogenous inlC promoter to closer mimic the wt Listeria strain. For a direct comparison between the inlC and Phyper promoters, we infected HeLa cells transfected with GFP1-10 (Table S4) with strains ΔinlCPhyper-inlC-GFP11 and ΔinlCpInlC-inlC-GFP11 side by side for 24 h (Fig. 5, see Tables S2 and S3 for sequence information of the promoters and tags). As seen before with ΔinlCPhyper-inlC-GFP11 (Fig. 4), we observed diffuse cytosolic signal in the GFP channel for both strains (Fig. 5 a), confirming successful complementation of GFP1-10 with InlC-GFP11. No GFP fluorescence was seen for infected cells that were not transfected with GFP1-10, confirmed by the absence of the nuclear TagBFP marker (Fig. 5 a, stars in DIC images). As before (Fig. 4 a), we observed bright green puncta in the GFP channel in addition to cytosolic GFP fluorescence (Fig. 5 a, arrows in GFP channel). These likely represent protrusions from Listeria during cell-to-cell spread as observed by others (49) and by us in immunofluorescence images (Fig. S3 arrow, Fig. S4 inset). Occasionally, we also observed elongated protrusions in the xy plane in the GFP channel (Fig. 5 a, contrast enhanced inset). We concluded that these structures also represent membrane protrusions as Listeria spreads from cell to cell. To directly confirm that the GFP signal after complementation corresponds to signal from InlC, we repeated the complementation experiment with ΔinlCpInlC-inlC-GFP11, fixed cells, and performed immunofluorescence against InlC (Fig. S4, a and b). Both the overall fluorescence pattern, as well as protrusions at the cell membrane (Fig. S4 a), overlapped for the signal from complemented GFP and InlC. Overall, usage of the endogenous inlC promoter and the Phyper promoter resulted in comparable phenotypic GFP complementation patterns.
Figure 5.
Visualization of complemented InlC-GFP under control of different promoters. (a) HeLa cells were transfected with a plasmid to produce GFP1-10 (Table S4) and a nuclear TagBFP transfection marker and infected with indicated Listeria strains (see sequence information in Tables S2 and S3) for 24 h. Images in the GFP channel are presented at the same intensity levels. Arrows and insets in the DIC channel indicate Listeria cells. Stars in the DIC channel indicate cells that were infected but not transfected (confirmed by absence of the nuclear TagBFP marker). Arrows in the InlC-GFP channel for ΔinlCpInlC-inlC-GFP11 indicate InlC membrane protrusions (as in Fig. 4). Scale bars, 20 μm. The inset for ΔinlCpInlC-inlC-GFP11 shows a contrast-enhanced image indicating elongated membrane protrusions. (b) The mean intensity in the GFP channel was quantified for individual cells 24 h postinfection using identical acquisition settings (gray filled bars: ΔinlCPhyper-inlC-GFP11, three independent experiments, 48 cells; pink open bars: ΔinlCpInlC-inlC-GFP11, two independent experiments, 168 cells). To see this figure in color, go online.
For a quantitative comparison of the inlC and Phyper promoters, we compared the mean cytosolic fluorescence signal in each infected cell for ΔinlCPhyper-inlC-GFP11 and ΔinlCpInlC-inlC-GFP11 at 24 h postinfection (Fig. 5 b). The mean fluorescence intensity for both strains was comparable (average fluorescence intensity in arbitrary units of 5878 counts for ΔinlCPhyper-inlC-GFP11 vs. 5031 counts for ΔinlCpInlC-inlC-GFP11). Median counts were 3411 vs. 3239, respectively. Similarly, although the distribution of intensities between cells was heterogeneous and spanned an order of magnitude for both strains, no difference in intensity distribution was apparent (Fig. 5 b). For an additional robust comparison of the inlC and Phyper promoters, we infected HeLa cells with the ΔinlCPhyper-inlC-GFP11 vs. ΔinlCpInlC-inlC-GFP11 strain and visualized infection and GFP complementation on a high-content analysis microscope (Fig. 6). This experimental setup allowed for parallel imaging of both infections at the same time. The HeLa cell line stably produced GFP1-10 (Table S4) and the nuclear TagBFP marker (34). We quantified GFP fluorescence at two time points during the time course, 3 h postinfection vs. 6.5 h postinfection. As expected, fluorescence counts increased over time for both strains (Fig. 6 c). However, no statistically significant difference in GFP intensity was observed when comparing the two strains at each time point. Although the Phyper promoter is strong and induces constitutive protein expression (12), the inlC promoter is only active in infection for high level InlC production (12, 45). This indicates that both promoter strengths produce comparable translocated InlC levels in our experimental conditions (Figs. 5 and 6). We concluded that the Phyper promoter may serve as a robust promoter for split-GFP tagging of secreted proteins from Listeria and that the system is compatible with using endogenous promoters as well.
In addition to infecting epithelial cells, Listeria also infects macrophages. We recently demonstrated that split-GFP could be used to track Salmonella effector proteins translocated into primary BMDMs (34). To assess whether the system could also be used to monitor Listeria effectors, we transfected BMDMs with GFP1-10 and the nuclear TagBFP and infected them with ΔinlCpInlC-inlC-GFP11. Cells containing GFP1-10 are readily identified via blue nuclear marker expressed from an internal ribosome entry site from the GFP1-10 plasmid. As observed for Salmonella, the fluorescence complementation signal is weaker than in HeLa cells, but is significantly above the background fluorescence (Fig. 7). Because BMDMs in culture are phagocytic, InlC localization is not strictly cytosolic, and the protein seems to accumulate in phagosome and early lysosomes. This system can be used to track Listeria effectors in this physiologically relevant model system.
Figure 7.
BMDMs expressing GFP1-10 and the nuclear TagBFP infected with ΔinlCpInlC-inlC-GFP11. GFP complementation signal increases over the course of infection for infected cells (a) but not for uninfected cells (b). The fluorescence signal was adjusted to have the same thresholds for (a) and (b). GFP complementation may accumulate in nontransfected cells as macrophages consume other transfected and infected cells. Scale bars, 20 μm. To see this figure in color, go online.
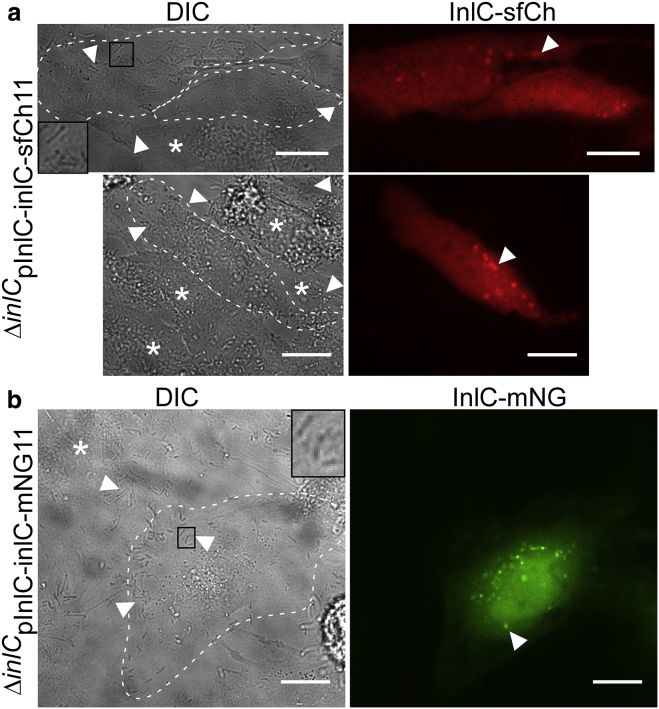
Although split-GFP tagging for secreted bacterial proteins in infection has been used previously in Salmonella infections (33, 34, 35), usage of split-fluorescent proteins of different colors would allow for broad flexibility in multicolor fluorescent imaging assays. Our Listeria expression plasmid is modular, and the split-fluorescent protein tag can be exchanged easily (Fig. 1 b). Guided by the recent optimization of split mNeonGreen and split super-folder Cherry (52, 57), we exchanged the GFP11 tag with the 11th strand of mNeonGreen (called here mNG11, derived from mNG2 in (52)) and the 11th strand of super-folder Cherry (called here sfCh11, derived from sfCherry2 in (52), see Table S3 for sequence information of mNG11 and sfCh11). Both InlC fusions were produced under control of the inlC promoter (Table S2). We transfected HeLa cells with sfCh1-10, encoding for the nonfluorescent strands 1–10 of super-folder Cherry (see Table S4 for sequence information), followed by infection with ΔinlCpInlC-inlC-sfCh11 for 24 h. As expected for a 24 h infection, all HeLa cells in the field of view presented in Fig. 8 a were infected with Listeria (Fig. 8 a, inset and arrows in DIC channel). We observed fluorescence above background in the red fluorescent channel for some cells, consistent with complementation of sfCh11 and sfCh1-10. Some infected cells were nonfluorescent, in line with the assumption that not all cells were transfected with sfCh1-10 (Fig. 8 a, stars in DIC channel indicate infected cells without red fluorescence). Note that sfCh1-10 and mNG1-10 do not include a transfection marker as does GFP1-10. As seen for split-GFP (Figs. 4 and 5), the red fluorescence signal was diffusely distributed throughout the cytosol, and we also observed fluorescent puncta (Fig. 8 a, arrow in InlC-sfCh channel) that resemble cell protrusions found during Listeria cell-to-cell spread. To assess complementation of mNeonGreen, HeLa cells were transfected with DNA encoding for the nonfluorescent mNG1-10 (see Table S4 for sequence information) and infected with ΔinlCpInlC-inlC-mNG11 for 24 h. As for GFP and super-folder Cherry complementation, we readily observed infected HeLa cells displaying bright green fluorescence throughout the cytosol, indicating successful complementation (Fig. 8 b). We also observed bright puncta (Fig. 8 b, arrow in InlC-mNG channel) as observed for GFP and super-folder Cherry complementation, resembling cell protrusions during Listeria spread. Together, we concluded that tagging secreted Listeria proteins with split-fluorescent proteins is highly modular and compatible with proteins of different fluorescent colors, including GFP, mNeonGreen, and super-folder Cherry.
Figure 8.
Visualization of InlC via super-folder Cherry and mNeonGreen complementation in infection. (a) HeLa cells were transfected with sfCh1-10 (see Table S4 for sequence information) and infected with ΔinlCpInlC-inlC-sfCh11(see Tables S2 and S3 for promoter and tag sequence information) for 24 h. Shown are two representative fields of view with sfCh signal above background. Raw fluorescence counts above background are variable (upper panel: ∼2000 cytosolic counts, lower panel: ∼8000 cytosolic counts). Stars mark infected cells that display background sfCh counts and were presumably not transfected with sfCh1-10 (typical background counts: <1000). Arrows and inset in the DIC channel point to Listeria. Arrows in the InlC-sfCh channel indicate InlC cell protrusions as in Figs. 4 and 5. Scale bars, 20 μm (two experiments, 10 cells). (b) HeLa cells were transfected with the nonfluorescent mNG1-10 (see Table S4 for sequence information) and infected with ΔinlCpInlC-inlC-mNG11 (see Tables S2 and S3 for promoter and tag information) for 24 h. Arrows and inset in the DIC channel indicate Listeria cells, and the star in the DIC channel indicates a HeLa cell that is infected but displays background green fluorescence signal. The arrow in the InlC-mNG channel indicates bright puncta presumably corresponding to cell protrusions during Listeria cell-to-cell spread. Scale bars, 20 μm (two experiments, 16 cells). To see this figure in color, go online.
Conclusions
Visualization of secreted virulence proteins in bacterial infections on a single-cell level is a powerful tool to gain insights into the spatiotemporal dynamics of the complex infection process. We have here demonstrated a proof of principle application of tagging the secreted protein InlC from Listeria in epithelial cells and primary macrophages. InlC localization and accumulation throughout the infection can be readily quantified, while also capturing the heterogeneity of the infection between different cells. The split-fluorescent protein tagging strategy features a short and unstructured peptide tag—the 11th strand of different fluorescent proteins—therefore minimizing potential interference with the endogenous protein secretion process. Importantly, we have demonstrated usage of three different split-fluorescent proteins (GFP, mNeonGreen, and super-folder Cherry), allowing tagging with two spectrally diverse fluorescent proteins. This flexibility will allow for multicolor imaging in the future, in which other host and/or pathogen components can be tagged simultaneously by other fluorescent markers.
Usage of the endogenous inlC promoter and a strong general promoter (Phyper) confirm that the system established here may be broadly used for secreted Listeria proteins, either with their endogenous promoters or with the Phyper promoter for robust protein production. We envision usage of this technology for chromosomally integrated fusion proteins or with weaker promoters will be possible for the following reasons. First, the contrast of our images is high (up to 20,000 counts in complementation versus less than 1000 counts in the background, Fig. 5). Additionally, contrast may be improved by using multiple copies of the fluorescent protein tag (52), as was done before for Salmonella effector tagging (34). Together, this platform is robust and represents a versatile tool to gain insights in Listeria infection biology.
We envision that split-fluorescent protein tagging of secreted proteins can be widely applicable in diverse biological contexts. Our previous work on labeling secreted effector proteins from Salmonella (34, 35) was limited to tagging proteins secreted by the type III secretion mechanism, a specialized secretion pathway for virulence proteins in Gram-negative bacteria (41). The application in Listeria secretion demonstrates compatibility with the Sec secretion pathway, a general and ubiquitous secretion pathway in all bacteria with more than one third of the bacterial proteome as clients (58). Besides in-depth investigations of diverse bacterial infections in different mammalian cell systems, dynamic visualization of secreted proteins may be used for various other applications including biofilm formation, labeling of cell wall or membrane proteins, or synthetic biology systems. Lastly, the evolutionary similarity between the bacterial Sec secretion system and mammalian secretion systems in organelles (59) suggests that the split-fluorescent technology may be used to investigate mammalian secretion systems as well.
There are a number of important considerations for using split-fluorescent protein systems. Tagging a protein of interest with the 11th strand of a fluorescent protein (Table S3) is limited to terminal tags, ideally at the C-terminus to avoid interfering with an N-terminal signal sequence. Depending on each protein’s function, C-terminal tagging may interfere with its function, and each protein of interest must be evaluated for this potential problem, a general caveat of fluorescent protein tagging (60). Moreover, complementation of GFP in the context of infection takes at least 2 h (35), limiting the time resolution of detection for early secretion events. Therefore, this technology may be particularly useful for tagging of virulence proteins that accumulate later in the infection, as is the case for InlC. Additionally, we produced tagged InlC from a high-copy plasmid, resulting in overexpression of the fusion. Although our proof of principle live InlC imaging produced results consistent with fixed literature results, subtler biologically relevant phenotypes may be extracted when the tagged virulence protein is integrated in the genome. Finally, the overall brightness of split-fluorescent protein systems is typically less than parent fluorescent proteins, super-folder GFP, mNeonGreen, and super-folder Cherry2. In E. coli, the GFP1-10 system was ∼90% as bright as super-folder GFP, the split mNeonGreen system was ∼60% the fluorescence of mNeonGreen, and split super-folder mCherry2 was equal to the fluorescence of super-folder mCherry (52). In these cases, the 1–10 portion was fused to the 11th strand via a spacer, so whether these numbers accurately reflect the achievable brightness when the two components are expressed in trans is unclear.
Author Contributions
D.B., E.B., and A.E.P. designed research. D.B., E.B., M.M., and D.M.T.N. performed research. D.B., E.B., M.M., and D.M.T.N. analyzed data. P.C. contributed reagents and experimental advice. D.B., E.B., and A.E.P. wrote the manuscript.
Acknowledgments
We thank Fabrizia Stavru for experimental advice.
Plasmids to produce strands 1–10 of mNeonGreen and super-folder Cherry were a generous gift of Professor Bo Huang (University of California San Francisco). We acknowledge financial support from the Human Frontiers Science Project. D.M.T.N. was supported by the NSF IGERT grant #1144807.
Editor: David Piston.
Footnotes
Dilara Batan and Esther Braselmann contributed equally to this work.
Four figures and six tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30382-5.
Supporting Material
References
- 1.Hamon M., Bierne H., Cossart P. Listeria monocytogenes: a multifaceted model. Nat. Rev. Microbiol. 2006;4:423–434. doi: 10.1038/nrmicro1413. [DOI] [PubMed] [Google Scholar]
- 2.Portnoy D.A., Auerbuch V., Glomski I.J. The cell biology of Listeria monocytogenes infection: the intersection of bacterial pathogenesis and cell-mediated immunity. J. Cell Biol. 2002;158:409–414. doi: 10.1083/jcb.200205009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lebreton A., Stavru F., Cossart P. Organelle targeting during bacterial infection: insights from Listeria. Trends Cell Biol. 2015;25:330–338. doi: 10.1016/j.tcb.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Rolhion N., Cossart P. How the study of Listeria monocytogenes has led to new concepts in biology. Future Microbiol. 2017;12:621–638. doi: 10.2217/fmb-2016-0221. [DOI] [PubMed] [Google Scholar]
- 5.Cossart P., Lebreton A. A trip in the “New Microbiology” with the bacterial pathogen Listeria monocytogenes. FEBS Lett. 2014;588:2437–2445. doi: 10.1016/j.febslet.2014.05.051. [DOI] [PubMed] [Google Scholar]
- 6.Disson O., Lecuit M. In vitro and in vivo models to study human listeriosis: mind the gap. Microbes Infect. 2013;15:971–980. doi: 10.1016/j.micinf.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 7.D’Orazio S.E. Animal models for oral transmission of Listeria monocytogenes. Front. Cell. Infect. Microbiol. 2014;4:15. doi: 10.3389/fcimb.2014.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cossart P. Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc. Natl. Acad. Sci. USA. 2011;108:19484–19491. doi: 10.1073/pnas.1112371108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birmingham C.L., Canadien V., Brumell J.H. Listeriolysin O allows Listeria monocytogenes replication in macrophage vacuoles. Nature. 2008;451:350–354. doi: 10.1038/nature06479. [DOI] [PubMed] [Google Scholar]
- 10.Helaine S., Holden D.W. Heterogeneity of intracellular replication of bacterial pathogens. Curr. Opin. Microbiol. 2013;16:184–191. doi: 10.1016/j.mib.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 11.Tilney L.G., Portnoy D.A. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J. Cell Biol. 1989;109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balestrino D., Hamon M.A., Toledo-Arana A. Single-cell techniques using chromosomally tagged fluorescent bacteria to study Listeria monocytogenes infection processes. Appl. Environ. Microbiol. 2010;76:3625–3636. doi: 10.1128/AEM.02612-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desvaux M., Hébraud M. The protein secretion systems in Listeria: inside out bacterial virulence. FEMS Microbiol. Rev. 2006;30:774–805. doi: 10.1111/j.1574-6976.2006.00035.x. [DOI] [PubMed] [Google Scholar]
- 14.Camejo A., Carvalho F., Cabanes D. The arsenal of virulence factors deployed by Listeria monocytogenes to promote its cell infection cycle. Virulence. 2011;2:379–394. doi: 10.4161/viru.2.5.17703. [DOI] [PubMed] [Google Scholar]
- 15.Renier S., Micheau P., Desvaux M. Subcellular localization of extracytoplasmic proteins in monoderm bacteria: rational secretomics-based strategy for genomic and proteomic analyses. PLoS One. 2012;7:e42982. doi: 10.1371/journal.pone.0042982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bierne H., Cossart P. Listeria monocytogenes surface proteins: from genome predictions to function. Microbiol. Mol. Biol. Rev. 2007;71:377–397. doi: 10.1128/MMBR.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lenz L.L., Portnoy D.A. Identification of a second Listeria secA gene associated with protein secretion and the rough phenotype. Mol. Microbiol. 2002;45:1043–1056. doi: 10.1046/j.1365-2958.2002.03072.x. [DOI] [PubMed] [Google Scholar]
- 18.Renier S., Chambon C., Desvaux M. Exoproteomic analysis of the SecA2-dependent secretion in Listeria monocytogenes EGD-e. J. Proteomics. 2013;80:183–195. doi: 10.1016/j.jprot.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 19.Dumas E., Desvaux M., Hébraud M. Insight into the core and variant exoproteomes of Listeria monocytogenes species by comparative subproteomic analysis. Proteomics. 2009;9:3136–3155. doi: 10.1002/pmic.200800765. [DOI] [PubMed] [Google Scholar]
- 20.Prokop A., Gouin E., Dussurget O. OrfX, a nucleomodulin required for Listeria monocytogenes virulence. MBio. 2017;8 doi: 10.1128/mBio.01550-17. e01550-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lebreton A., Lakisic G., Bierne H. A bacterial protein targets the BAHD1 chromatin complex to stimulate type III interferon response. Science. 2011;331:1319–1321. doi: 10.1126/science.1200120. [DOI] [PubMed] [Google Scholar]
- 22.Dabiri G.A., Sanger J.M., Southwick F.S. Listeria monocytogenes moves rapidly through the host-cell cytoplasm by inducing directional actin assembly. Proc. Natl. Acad. Sci. USA. 1990;87:6068–6072. doi: 10.1073/pnas.87.16.6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welch M.D., Iwamatsu A., Mitchison T.J. Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature. 1997;385:265–269. doi: 10.1038/385265a0. [DOI] [PubMed] [Google Scholar]
- 24.Gouin E., Welch M.D., Cossart P. Actin-based motility of intracellular pathogens. Curr. Opin. Microbiol. 2005;8:35–45. doi: 10.1016/j.mib.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 25.Travier L., Guadagnini S., Lecuit M. ActA promotes Listeria monocytogenes aggregation, intestinal colonization and carriage. PLoS Pathog. 2013;9:e1003131. doi: 10.1371/journal.ppat.1003131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Travier L., Lecuit M. Listeria monocytogenes ActA: a new function for a ‘classic’ virulence factor. Curr. Opin. Microbiol. 2014;17:53–60. doi: 10.1016/j.mib.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Kayal S., Charbit A. Listeriolysin O: a key protein of Listeria monocytogenes with multiple functions. FEMS Microbiol. Rev. 2006;30:514–529. doi: 10.1111/j.1574-6976.2006.00021.x. [DOI] [PubMed] [Google Scholar]
- 28.Cossart P., Vicente M.F., Berche P. Listeriolysin O is essential for virulence of Listeria monocytogenes: direct evidence obtained by gene complementation. Infect. Immun. 1989;57:3629–3636. doi: 10.1128/iai.57.11.3629-3636.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schnupf P., Portnoy D.A. Listeriolysin O: a phagosome-specific lysin. Microbes Infect. 2007;9:1176–1187. doi: 10.1016/j.micinf.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 30.Hamon M.A., Ribet D., Cossart P. Listeriolysin O: the Swiss army knife of Listeria. Trends Microbiol. 2012;20:360–368. doi: 10.1016/j.tim.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Hamon M.A., Batsché E., Cossart P. Histone modifications induced by a family of bacterial toxins. Proc. Natl. Acad. Sci. USA. 2007;104:13467–13472. doi: 10.1073/pnas.0702729104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stavru F., Palmer A.E., Cossart P. Atypical mitochondrial fission upon bacterial infection. Proc. Natl. Acad. Sci. USA. 2013;110:16003–16008. doi: 10.1073/pnas.1315784110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McQuate S.E., Young A.M., Palmer A.E. Long-term live-cell imaging reveals new roles for Salmonella effector proteins SseG and SteA. Cell. Microbiol. 2017;19:e12641. doi: 10.1111/cmi.12641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young A.M., Minson M., Palmer A.E. Optimized fluorescence complementation platform for visualizing Salmonella effector proteins reveals distinctly different intracellular niches in different cell types. ACS Infect. Dis. 2017;3:575–584. doi: 10.1021/acsinfecdis.7b00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Engelenburg S.B., Palmer A.E. Imaging type-III secretion reveals dynamics and spatial segregation of Salmonella effectors. Nat. Methods. 2010;7:325–330. doi: 10.1038/nmeth.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Engelenburg S.B., Palmer A.E. Quantification of real-time Salmonella effector type III secretion kinetics reveals differential secretion rates for SopE2 and SptP. Chem. Biol. 2008;15:619–628. doi: 10.1016/j.chembiol.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Engelenburg S.B., Nahreini T., Palmer A.E. FACS-based selection of tandem tetracysteine peptides with improved ReAsH brightness in live cells. ChemBioChem. 2010;11:489–493. doi: 10.1002/cbic.200900689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gawthorne J.A., Audry L., Roe A.J. Visualizing the translocation and localization of bacterial type III effector proteins by using a genetically encoded reporter system. Appl. Environ. Microbiol. 2016;82:2700–2708. doi: 10.1128/AEM.03418-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Enninga J., Mounier J., Tran Van Nhieu G. Secretion of type III effectors into host cells in real time. Nat. Methods. 2005;2:959–965. doi: 10.1038/nmeth804. [DOI] [PubMed] [Google Scholar]
- 40.Young A.M., Palmer A.E. Methods to illuminate the role of Salmonella effector proteins during infection: a review. Front. Cell. Infect. Microbiol. 2017;7:363. doi: 10.3389/fcimb.2017.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moest T.P., Méresse S. Salmonella T3SSs: successful mission of the secret(ion) agents. Curr. Opin. Microbiol. 2013;16:38–44. doi: 10.1016/j.mib.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 42.Engelbrecht F., Chun S.K., Sokolovic Z. A new PrfA-regulated gene of Listeria monocytogenes encoding a small, secreted protein which belongs to the family of internalins. Mol. Microbiol. 1996;21:823–837. doi: 10.1046/j.1365-2958.1996.541414.x. [DOI] [PubMed] [Google Scholar]
- 43.de las Heras A., Cain R.J., Vázquez-Boland J.A. Regulation of Listeria virulence: PrfA master and commander. Curr. Opin. Microbiol. 2011;14:118–127. doi: 10.1016/j.mib.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 44.Rajabian T., Gavicherla B., Ireton K. The bacterial virulence factor InlC perturbs apical cell junctions and promotes cell-to-cell spread of Listeria. Nat. Cell Biol. 2009;11:1212–1218. doi: 10.1038/ncb1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toledo-Arana A., Dussurget O., Cossart P. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 46.Gouin E., Adib-Conquy M., Cossart P. The Listeria monocytogenes InlC protein interferes with innate immune responses by targeting the IkappaB kinase subunit IKKalpha. Proc. Natl. Acad. Sci. USA. 2010;107:17333–17338. doi: 10.1073/pnas.1007765107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Polle L., Rigano L.A., Schubert W.D. Structural details of human tuba recruitment by InlC of Listeria monocytogenes elucidate bacterial cell-cell spreading. Structure. 2014;22:304–314. doi: 10.1016/j.str.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 48.Kühbacher A., Emmenlauer M., Pizarro-Cerdá J. Genome-wide siRNA screen identifies complementary signaling pathways involved in Listeria infection and reveals different actin nucleation mechanisms during Listeria cell invasion and actin comet tail formation. MBio. 2015;6 doi: 10.1128/mBio.00598-15. e00598–e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kühbacher A., Gouin E., Pizarro-Cerdá J. Imaging InlC secretion to investigate cellular infection by the bacterial pathogen Listeria monocytogenes. J. Vis. Exp. 2013;79:e51043. doi: 10.3791/51043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Samba-Louaka A., Pereira J.M., Cossart P. Listeria monocytogenes dampens the DNA damage response. PLoS Pathog. 2014;10:e1004470. doi: 10.1371/journal.ppat.1004470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Glaser P., Frangeul L., Cossart P. Comparative genomics of Listeria species. Science. 2001;294:849–852. doi: 10.1126/science.1063447. [DOI] [PubMed] [Google Scholar]
- 52.Feng S., Sekine S., Huang B. Improved split fluorescent proteins for endogenous protein labeling. Nat. Commun. 2017;8:370. doi: 10.1038/s41467-017-00494-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakamura Y., Gojobori T., Ikemura T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clarke T.F., IV, Clark P.L. Rare codons cluster. PLoS One. 2008;3:e3412. doi: 10.1371/journal.pone.0003412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silva-Herzog E., Detweiler C.S. Salmonella enterica replication in hemophagocytic macrophages requires two type three secretion systems. Infect. Immun. 2010;78:3369–3377. doi: 10.1128/IAI.00292-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Knodler L.A., Bestor A., Steele-Mortimer O. Cloning vectors and fluorescent proteins can significantly inhibit Salmonella enterica virulence in both epithelial cells and macrophages: implications for bacterial pathogenesis studies. Infect. Immun. 2005;73:7027–7031. doi: 10.1128/IAI.73.10.7027-7031.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kamiyama D., Sekine S., Huang B. Versatile protein tagging in cells with split fluorescent protein. Nat. Commun. 2016;7:11046. doi: 10.1038/ncomms11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsirigotaki A., De Geyter J., Karamanou S. Protein export through the bacterial Sec pathway. Nat. Rev. Microbiol. 2017;15:21–36. doi: 10.1038/nrmicro.2016.161. [DOI] [PubMed] [Google Scholar]
- 59.Kunze M., Berger J. The similarity between N-terminal targeting signals for protein import into different organelles and its evolutionary relevance. Front. Physiol. 2015;6:259. doi: 10.3389/fphys.2015.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Specht E.A., Braselmann E., Palmer A.E. A critical and comparative review of fluorescent tools for live-cell imaging. Annu. Rev. Physiol. 2017;79:93–117. doi: 10.1146/annurev-physiol-022516-034055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang F., Moss L.G., Phillips G.N., Jr. The molecular structure of green fluorescent protein. Nat. Biotechnol. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.