Abstract
Nanoparticles used in cellular applications encounter free serum proteins that adsorb onto the surface of the nanoparticle, forming a protein corona. This protein layer controls the interaction of nanoparticles with cells. For nanomedicine applications, it is important to consider how intravenous injection and the subsequent shear flow will affect the protein corona. Our goal was to determine if shear flow changed the composition of the protein corona and if these changes affected cellular binding. Colorimetric assays of protein concentration and gel electrophoresis demonstrate that polystyrene nanoparticles subjected to flow have a greater concentration of serum proteins adsorbed on the surface, especially plasminogen. Plasminogen, in the absence of nanoparticles, undergoes changes in structure in response to flow, characterized by fluorescence and circular dichroism spectroscopy. The protein-nanoparticle complexes formed from fetal bovine serum after flow had decreased cellular binding, as measured with flow cytometry. In addition to the relevance for nanomedicine, these results also highlight the technical challenges of protein corona studies. The composition of the protein corona was highly dependent on the initial mixing step: rocking, vortexing, or flow. Overall, these results reaffirm the importance of the protein corona in nanoparticle-cell interactions and point toward the challenges of predicting corona composition based on nanoparticle properties.
Introduction
Nanomedicine offers the promise of treating human disease with nanoparticles (NPs) that significantly enhance the function of conventional molecular-scale drugs (1). Nanomedicines have the ability to target a specific subpopulation of cells and then release a drug or combination of drugs now localized at these specific cells. Despite intense research in this area, very few NPs have been translated to the clinic. A recent analysis of the literature suggests one reason for the lack of success in the field of cancer nanomedicines, estimating that only 0.7% of injected NPs reach tumors, whereas the vast majority of NPs instead accumulate in the liver and spleen (2). Although 0.7% may be an underestimate, it does highlight the significant challenges facing the advancement of nanomedicine. Successful NP tumor targeting, both passive and active, requires the NPs to stay in circulation long enough to reach and accumulate at the tumors. In comparison, most NPs are rapidly cleared from the bloodstream, accumulating in the liver and spleen after recognition by the reticuloendothelial system. By being cleared to the liver, the NPs lack the circulation time necessary to reach their target. Non-tumor-targeting applications, such as cardiovascular disease, have similar clearance challenges.
Many nanomedicines will be delivered intravenously, subjecting them to the forces associated with blood flow. The flow rates of the circulatory system range from relatively slow speeds in capillaries (0.085 cm/s) to rapid flow in the arteries (∼10 cm/s) with peak velocities of ∼60 cm/s in the aorta (3, 4, 5). The subsequent interaction of the immune system with NPs is mediated by a layer of blood serum proteins that adsorb on the NP surface during circulation. This opsonization, resulting in a “corona” of proteins on the NP surface (6, 7, 8, 9, 10, 11, 12, 13, 14, 15), can be decreased by PEGylation, but complete inhibition remains a challenge (16, 17, 18, 19, 20, 21, 22). The protein corona, described as either a “hard” corona of tightly bound proteins or a “soft” corona of dynamic, weakly bound proteins (23, 24, 25, 26), has been studied extensively as a function of NP diameter, composition, and surface functionalization (6, 7, 8, 9, 10, 11, 12, 13, 14). With few exceptions (27, 28, 29, 30, 31), previous studies have mostly ignored the effect of circulatory shear flow on the composition and structure of the protein corona. This previous work has found that NPs subjected to flow, both gold NPs (13 nm, PEGylated or functionalized with tannic acid) and lipid NPs (140 nm, unmodified and PEGylated), had greater amounts of protein adsorbed on the NP surface (27, 29). Interestingly, a much wider variety of proteins were identified in the coronas of the PEGylated liposomes after flow in comparison to those in static conditions. 207 distinct proteins were found in the corona formed under flow compared to 118 proteins under static conditions, with 108 proteins common to both coronas (31).
Our goal was to use a well-controlled model system of polystyrene NPs under physiologically relevant flow rates to determine how flow affected both the protein corona and the subsequent cellular binding of the protein-NP complexes. NPs, in a solution of blood serum proteins, were subjected to flow with flow rates chosen to mimic capillaries (0.085 cm/s), veins (0.85 cm/s), and arteries (8.5 cm/s) (3, 4, 5). The resulting protein corona was identified with gel electrophoresis and western blotting. We find that the total amount of protein is increased on the surface of NPs that are subjected to flow. Of specific interest is plasminogen, which is enriched on the NP surface in comparison with other serum proteins. Fluorescence and circular dichroism (CD) spectroscopy were used to probe structural changes of plasminogen in the absence of NPs, suggesting a partial denaturation under flow. In addition, protein-NP complexes formed from serum proteins subjected to flow show a slight decrease in cellular binding compared to protein-NP complexes formed under static conditions. These results suggest that flow is a parameter that should be examined in future corona studies and may guide the design of nanomedicines with improved circulation and targeting capabilities.
Materials and Methods
NPs and characterization
Carboxylate-modified polystyrene NPs (“orange,” #F8809, 200 nm, Excite: 540 nm/Emit: 560 nm, ε = 1.80 × 109 Μ−1 cm−1) (Thermo Fisher Scientific, Carlsbad, CA) were used for all experiments in the main text. A comparison with similar carboxylate-modified polystyrene NPs (“yellow-green,” #F8811, 200 nm, Excite: 505 nm/Emit: 515 nm; Thermo Fisher Scientific) is discussed in the Supporting Material. NP molar concentrations were calculated based on the number of NPs per milliliter (N, Eq. 1 provided by the supplier), specifically defined as follows:
| (1) |
with S = percent solids (w/w, provided by supplier, 2% = 0.02 g/mL), PS = density of polymer (1.05 g/mL), and d = average NP diameter (0.20 μm). This number of NPs per mL (N) is converted to a molar concentration using Avogadro’s number.
Dynamic light scattering (DynaPro NanoStar; Wyatt Technology, Santa Barbara, CA) was used to measure the diameter and polydispersity index (PDI) of the NPs in water (Table 1) (32, 33). Zeta potential (ZP) (Table 1) was measured using a Malvern Zetasizer (Nano-Z; Malvern Instruments, Worchestershire, England) (33). Electrophoretic mobility was converted to a ZP using the Smoluchowski approximation. For dynamic light scattering and ZP experiments, an NP concentration of 8 pM was used. Each measurement consisted of 30 runs and was carried out in triplicate with three distinct solutions. For protein corona characterization (flow and static) and western blots, an NP concentration of 20 pM was used. Transmission electron microscopy (TEM) measurements were performed using a JEOL 100 CX II TEM at the Center for Nanostructure Characterization, part of the Materials Characterization Facility at the Institute for Materials at Georgia Tech (Atlanta, GA). Images were obtained at 100 kV with 200 kX magnification. NPs (20 pM) were drop-cast on carbon-coated copper grids and dried at room temperature. Samples were imaged using a charge-coupled device camera (Finger Lakes Instruments, Lima, NY) with Maxim software (Scientific Instruments and Applications, Duluth, GA). Averages and SDs are reported for all measurements.
Table 1.
NP Characterization
| NPs | dh (nm) | PDI | ZP (mV) | TEM (nm) |
|---|---|---|---|---|
| #F8809 (Lot: #1835801) | 275 ± 11 | 0.06 ± 0.002 | −31 ± 5 | 200 ± 10, n = 50 |
Cell culture
HeLa cervical carcinoma cells (CCL-2) (American Type Culture Collection, Manassas, VA) were cultured in minimal essential medium (MEM) (#61100) (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS) (#10437-028) (Life Technologies, Carlsbad, CA) at 37°C and 5% carbon dioxide. Cells were passaged every 3–4 days.
Protein corona formation and characterization
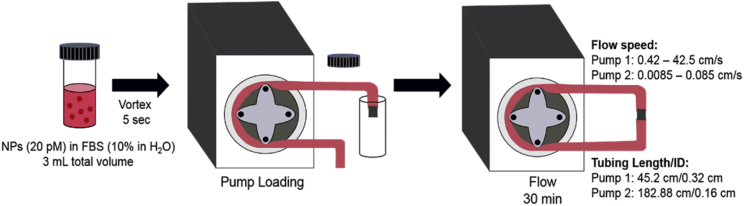
To obtain a protein corona under static conditions, NPs (20 pM) were combined with FBS (#10437-028) (Life Technologies) for a final FBS concentration of 10% v/v in water with 3 mL total volume. The NPs and FBS were briefly vortexed (∼5 s) and then incubated for 30 min at room temperature with no additional mixing. The protein corona formed under flow was investigated at the same NP and FBS concentrations used in the static condition with flow rates controlled by peristaltic pumps with polyvinyl chloride (PVC) tubing (Fig. 1). As with the static experiments, NPs (20 pM) and FBS (10% in water, 3 mL total volume) were briefly vortexed. This mixture was pumped into the tubing of the peristaltic pump, which was then connected to the other end of the tubing with a plastic tube connector (#5463K36 or #5463K38, noted below) (McMaster-Carr, Douglasville, GA). To cover the range of flow rates, two different pumps were used. For faster flow rates (0.42, 0.85, 8.5, and 42.5 cm/s), a Harvard Apparatus peristaltic pump was used (#55-7777, “Pump 1”) (Harvard Apparatus, Holliston, MA) with PVC tubing (1/8” inner diameter (ID), 1/4” outer diameter (OD), #5155T17) (McMaster-Carr) and a plastic tube connector (#5463K38) (McMaster-Carr). Slower flow rates (0.0085, 0.042, and 0.085 cm/s) used a second peristaltic pump (#724048, “Pump 2”) (Harvard Apparatus) and PVC tubing (1/16” ID, 1/18” OD, #5155T12) (McMaster-Carr) with a plastic tube connector (#5463K36) (McMaster-Carr). The following assumptions were made regarding this flow system: 1) the protein-NP mixture is a Newtonian, incompressible fluid, and 2) the flow through the tubing is laminar. Based on these assumptions, we calculate volumetric flow speed, as shown in Eq. 2:
| (2) |
where v is the speed, Q is the volumetric flow set by the pump, and ID is the inner diameter of the tubing. The 3 mL NP-protein mixture, approximately the volume of the tubing, was pumped through the circle of tubing (∼1 ft) for 30 min at room temperature.
Figure 1.
Pump system for protein corona formation under flow conditions. To see this figure in color, go online.
Protein concentration was determined using a bicinchoninic acid (BCA) assay (#23227) (Thermo Fisher Scientific). For BCA assays, unbound and weakly bound soft-corona proteins were removed from the protein-NP mixture by centrifugation (8000 rcf, 15 min, ×3), and the assay was performed using the resuspended protein-NP pellet. The relative abundance of specific corona proteins was analyzed with gel electrophoresis. A detergent, sodium dodecyl sulfate (SDS) (Laemmli loading buffer, #BP-110R) (Boston BioProducts, Ashland, MA) (5 min, vortexing), was used to remove the hard corona from the NPs before loading on the gel. Protein samples (20 μg) were heated for 5 min at 100°C and then loaded onto a gel (tris-glycine SDS gel, #456-1094) (Bio-Rad, Hercules, CA) for sodium dodecyl sulfate polyacrylamide gel electrophoresis (230 V, 35 min). A 10–190 kDa molecular weight marker (ProSieve Color Protein Marker, #50550) (Lonza, Rockland, ME) was included. Gels were stained for 1 h (SimplyBlue Safe Stain, #LC6060) (Thermo Fisher Scientific) and then imaged with an Odyssey Imager (LI-COR Biosciences, Lincoln, NE). Densitometric analysis (Image J; http://rsb.info.nih.gov/ij/) was used to measure any changes in protein concentration. Gel electrophoresis was carried out for both equal total protein concentration, as determined by BCA, and equal NP concentrations. To match NP concentrations, NP absorption at 541 nm was measured with an ultraviolet (UV)-visible spectrophotometer (DU 800) (Beckman Coulter, Brea, CA).
Initial identification of hard corona proteins was based on molecular weight, as determined with gel electrophoresis. Western blotting was used to confirm that the identity of the ∼88 kDa protein was plasminogen. Gel electrophoresis was carried out as described above and then transferred to a polyvinylidene difluoride membrane (100 V, 45 min). The membrane was blocked (#MB-070) (Rockland Immunochemicals, Pottstown, PA) for 1 h at 4°C. The primary antibody, antiplasminogen (1:5000, ab154560) (Abcam, Cambridge, MA), was added and incubated overnight at 4°C in blocking buffer. The membrane was washed with tris-buffered saline (TBS)-Tween (three times, 10 min). Secondary antibody was incubated for 1 h at 4°C in blocking buffer (1:10,000, #926-6802) (LI-COR Biosciences). The membrane was washed twice with TBS-Tween for 10 min and then once with TBS. Blots were imaged with an Odyssey Imager (LI-COR Biosciences).
Fluorescence spectroscopy
The fluorescence emission spectrum of human plasminogen was recorded before and after flow. Measurements were made using a 3-mm path-length quartz cell (105.254-QS) (Hellma, Müllheim, Germany). An aqueous solution of plasminogen (5 μM, ab92924) (Abcam) was prepared, and fluorescence was measured at room temperature (Excite: 280 nm, RF-5301PC Fluorometer) (Shimadzu, Kyoto, Japan). The fluorescence of plasminogen after flow (0.85 and 8.5 cm/s, 30 min) was measured under the same conditions.
CD spectroscopy
CD spectra (J-810) (Jasco, Oklahoma City, OK) were acquired using a 0.1- and 0.5-mm path-length quartz cell (20/O-Q-0.5) (Starna, Atascadero, CA) for plasminogen and bovine serum albumin (BSA), respectively. The bandwidth was set to 2 nm, and the integration time was a function of the photomultiplier tube voltage. Samples were measured in water at 20°C with water used as a blank. Plasminogen (0.099 mg mL−1) and BSA (0.3 mg mL−1) were measured with flow (8.5 cm/s, 30 min) and without flow. All measurements were carried out in triplicate. Spectra were smoothed with a Savitzky-Golay least-squares fit (digital filter = 10). Spectra are an average of three consecutive scans.
Flow cytometry
Cellular binding of NPs was measured using flow cytometry (Accuri C6) (Becton Dickinson, Franklin Lakes, NJ). Cells were cultured in 25 cm2 flasks to 100% confluency. Before the addition of NPs, the cells were washed three times with phosphate-buffered saline (PBS). Cells were incubated with NPs (20 pM) at 4°C for 20 min in MEM supplemented with 10% FBS after flow (8.5 cm/s). As a control experiment, cells were incubated with NPs (20 pM) at 4°C for 20 min in MEM supplemented with 10% FBS that was not subjected to flow. Cells were washed three times with PBS to remove excess NPs. Accutase (A6964) (Sigma-Aldrich, St. Louis, MO) was used to remove the cells from the surface of the culture flasks. After removal, the cell suspension was distributed into 1 mL aliquots and pelleted down (7000 rcf, 10 min, room temperature) to remove Accutase and cell culture medium. The cell pellet was resuspended in PBS and filtered with a cell strainer (SK-06336-63) (Cole-Parmer, Vernon Hills, IL) for flow cytometry. A 488 nm laser was used to excite the NP fluorescence, and fluorescence emission was collected on a 530/30 nm bandpass filter. For each experiment, 15,000 cells in the population of live cells, determined with a propidium iodide assay (2 μg propidium iodide/mL PBS, FL-2 filter), were sampled. Histograms were analyzed using Becton Dickinson C6 software 1.0.264.21. The percent change of the mean intensity of the population was used to measure the change in NP binding. Significance was determined by a two-tailed Student’s t-test. Error bars denote ± SD for n = 3 distinct samples.
Results and Discussion
NP characterization
The carboxylate-modified polystyrene NPs used in these experiments were used as provided by the supplier with no additional modification. To ensure that the NPs met specifications, they were characterized by hydrodynamic diameter (dh), PDI, ZP, and TEM (Fig. S1; Table 1).
Concentration of corona proteins changes in response to flow
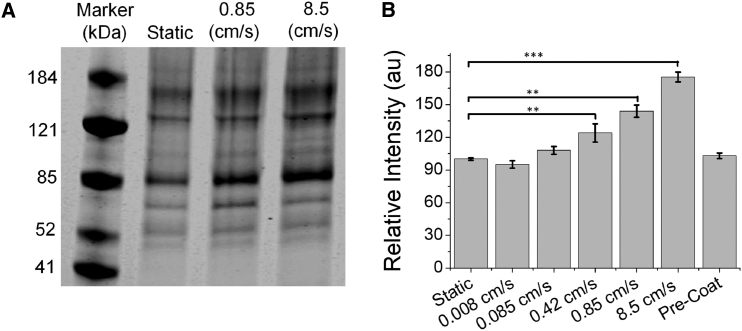
Polystyrene NPs (20 pM, 200 nm, carboxylate-modified) and 10% FBS were subjected to continuous flow (0.008–8.5 cm/s) through a peristaltic pump for 30 min at room temperature. After 30 min, soft corona proteins were removed from the polystyrene NPs using a series of centrifugation and resuspension steps previously optimized in the Payne Laboratory (34). A detergent (SDS) was then used to remove the tightly bound hard corona from the NPs before gel electrophoresis. The hard corona formed under flow, analyzed by BCA assay and gel electrophoresis (Fig. 2), was compared to conventional corona experiments in which the NPs and serum proteins are mixed by briefly vortexing (∼5 s) and then incubated for 30 min. Of specific interest were three flow rates that were representative of capillaries (0.085 cm/s), veins (0.85 cm/s), and arteries (8.5 cm/s) (3, 4, 5).
Figure 2.
Hard corona proteins isolated from the surface of 200 nm polystyrene NPs as a function of flow rate. (A) The relative concentrations of individual proteins were measured with gel electrophoresis. Values were normalized against conventional experiments (static). A representative gel is shown with densitometric analysis in Table 2. (B) Densitometric analysis of the band at 88 kDa (plasminogen, western blot shown in Fig. S2A) as a function of flow rate is shown. n = 7 at each speed. Fig. 2A shows a gel loaded with equal concentrations of protein. Similar results were obtained when gels were loaded with equal NP concentrations (Fig. S2B). Results for precoated NPs were obtained after 30 min flow at 8.5 cm/s. ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and samples with p-values not shown had nonsignificant differences.
A BCA assay showed an increase in the concentration of hard corona proteins on the NP surface for NPs subjected to flow with significant changes detected at flow rates ≥0.42 cm/s (by BCA: 142% ± 7% at 0.85 cm/s and 197% ± 5% at 8.5 cm/s; static = 100%, n = 4, NP concentrations are held constant). No changes in protein concentration were observed at 0.085 cm/s, and subsequent experiments focused on higher speeds. Multiple hard corona proteins were observed by gel electrophoresis (Fig. 2 A, loaded with equal concentrations of protein). Based on molecular weight and previous proteomics analysis of the coronas of identical NPs (33), two bands of the hard corona could be identified: plasminogen (88 kDa) and serum albumin (66 kDa). Based on previous proteomics analysis, antithrombin and assorted complement proteins with their cleavage products are also present in the hard corona but are more difficult to identify based on molecular weight. Gelsolin (86 kDa) was also identified in the hard corona based on proteomics. A western blot was used to confirm the plasminogen assignment (Fig. S2 A). Densitometric analysis (ImageJ) showed that the concentration of plasminogen in the corona increased under flow (Fig. 2 B; Table 2) with relative values of 144 ± 6 and 175 ± 5% at 0.85 and 8.5 cm/s (both n = 7, defined as 100% for static samples, average and SD reported), respectively. In comparison, the concentration of albumin was identical under static and flow conditions (static = 100%, 101 ± 2 and 104 ± 3% at 0.85 and 8.5 cm/s, respectively, both n = 7; Table 2). Unidentified protein bands at ∼125 and 180 kDa showed small increases in response to flow (180 kDa = 111 ± 4 and 116 ± 7% at 0.85 and 8.5 cm/s, both n = 7; 125 kDa = 111 ± 3 and 122 ± 6% at 0.85 and 8.5 cm/s, both n = 7; Table 2). Similar results were obtained when gels were loaded with matched NP concentrations (Fig. S2 B) rather than matched protein concentrations (Fig. 2 A). A series of control experiments were carried out to ensure that these results were independent of the specific pump used (Fig. S3 A; Table S1) or lot of FBS (Fig. S3 B; Table S2).
Table 2.
Percent Change of Observed Corona Proteins as Function of Flow Speed
| Speed (cm/s) | Plasminogen (%) | BSA (%) | 125 kDa (%) | 180 kDa (%) |
|---|---|---|---|---|
| 0.85 | 144 ± 6 | 101 ± 2 | 111 ± 3 | 111 ± 4 |
| 8.5 | 175 ± 5 | 104 ± 3 | 122 ± 6 | 116 ± 7 |
For comparison, experiments were carried out with a corona formed by incubation on a platform rocker (30 min, room temperature, speed = 7 rpm) and on a vortexer (30 min). A corona formed after incubation on the platform rocker was similar to that formed under static conditions (Fig. S2 C), whereas the corona formed with constant vortexing was similar to that formed with 0.85 cm/s flow (Fig. S2 C), with an increased concentration of plasminogen detected by gel electrophoresis (155% ± 2%, n = 3). In addition, we examined the effect of precoating the NPs with a protein corona before flow. Previous work with silica NPs (90 nm, carboxylate-modified) and gold NPs (10 nm, citrate-modified) has shown that precoating NPs alters the ultimate corona composition (28, 35). Preincubation of NPs with FBS (20 pM NPs, 10% FBS, 30 min, static) followed by flow for 30 min (0.85 and 8.5 cm/s) resulted in no change in plasminogen concentration (Figs. 2 B and S2 D). Although precoating NPs with serum proteins could be used in nanomedicine applications to prevent flow-dependent changes in protein corona, possibly taking advantage of the corona (36), it is worthwhile to explore the underlying reasons for flow-dependent changes in plasminogen concentration.
Previous research examined changes in the protein corona for 13 nm gold NPs modified with polyethylene glycol or tannic acid in response to flow at 0.5 cm/s for 24 h (27). Although specific corona proteins were not identified, this previous research also detected an increase in protein concentration, measured with gel electrophoresis and a colorimetric protein assay, for both NP-surface modifications. Similarly, 140 nm lipid NPs, both unmodified and PEGylated, were found to have increased protein coronas after flow at 50 cm/s for 90 min (29). For these lipid NPs, proteins in the corona were identified by mass spectrometry. Plasminogen was not found, but this likely reflects the difference in the NP composition.
Plasminogen structure is altered in response to flow
The observation that plasminogen was enriched on NPs subjected to flow points toward possible changes in protein structure under flow. To test this possibility, we used two complementary spectroscopic methods, fluorescence and CD, to probe protein structure in response to flow. These spectroscopic experiments were carried out in the absence of NPs. Serum albumin, which showed no change in corona concentration in response to flow (Fig. 2), was used for comparison. Fluorescence spectroscopy makes use of the endogenous fluorescence of the two proteins, specifically the 19 tryptophan residues of plasminogen and two tryptophan residues of BSA. Spectra recorded before and after flow (8.5 cm/s) show changes in plasminogen structure and no changes in albumin structure (Fig. 3, A and B). The faster flow rate (8.5 cm/s), similar to that of arteries, was used to maximize possible structural changes. The increased fluorescence emission from 350 to 550 nm observed for plasminogen under flow is similar, but not identical, to the fluorescence emission of plasminogen irradiated with UV light (280 nm, 45 min, 2.3 W/m2), which shows a similar shoulder at 405 nm (37). At a slower flow rate (0.85 cm/s), similar but less pronounced spectral shifts were observed (Fig. S4 A).
Figure 3.
Spectroscopic characterization of serum proteins after flow (8.5 cm/s, 30 min). (A) Fluorescence emission of plasminogen before (blue) and after (red) flow is shown. (B) Fluorescence emission of bovine serum albumin (BSA) before (blue) and after (red) flow is shown. Representative spectra (of three total) are shown. (C) CD spectra of plasminogen before (blue) and after (red) flow is shown. (D) CD spectra of BSA before (blue) and after (red) flow is shown. The SD from three distinct samples is shown by the shaded region of each line. Spectroscopic characterization of serum proteins was carried out in the absence of NPs. To see this figure in color, go online.
CD spectroscopy, which is sensitive to protein secondary structure, shows a similar result (Fig. 3, C and D). Native plasminogen consists of β-strands and random coils (37, 38). After flow (8.5 cm/s), plasminogen shows a decrease absorbance at both 202 and 230 nm, corresponding to a loss of ordered structure. The secondary structure of serum albumin is unchanged by flow. In comparison, treatment with 6-aminohexanoic acid, which is commonly used in studies of plasminogen, does not lead to changes in plasminogen secondary structure based on CD spectroscopy in the 190–220 nm region (38, 39).
To determine if these flow-induced changes in protein structure in the absence of NPs were relevant to the observed changes in protein coronas (Fig. 2; Table 2), a corona was formed using FBS (10%) subjected to flow (8.5 cm/s) incubated with NPs under static conditions (30 min, room temperature, identical to conditions used for Fig. 2; Table 2). The resulting corona, characterized with gel electrophoresis, is nearly identical to that formed under flow (Fig. S5; Table S3).
The observed change in plasminogen structure in response to flow raises the question of physiological relevance. The pH of the solution before (7.41 ± 0.03, n = 3) and after (7.45 ± 0.02, n = 3) flow is nearly identical, suggesting a mechanically, rather than chemically, induced change in structure. As a blood serum protein, plasminogen is constantly subjected to flow, and it is interesting to consider how even small structural changes could affect the biological function, serving as the inactive precursor to plasmin. The plasminogen protein used in these experiments is obtained from human plasma, purified by lysine chromatography, and then resuspended in water for spectroscopic measurement. To determine if plasminogen returns to the native conformation after a period of flow (8.5 cm/s), we incubated the same sample at 37°C in PBS and recorded spectra from 30 min to 6 h. We found a partial but incomplete return to the initial fluorescence spectrum (Fig. S4 B). Previous work examining activation in response to UV-illumination showed that the UV-induced changes in protein structure enhanced the activation of plasminogen by 2.6× (37), suggesting that flow-induced changes in structure could be relevant for plasminogen function.
Cellular binding
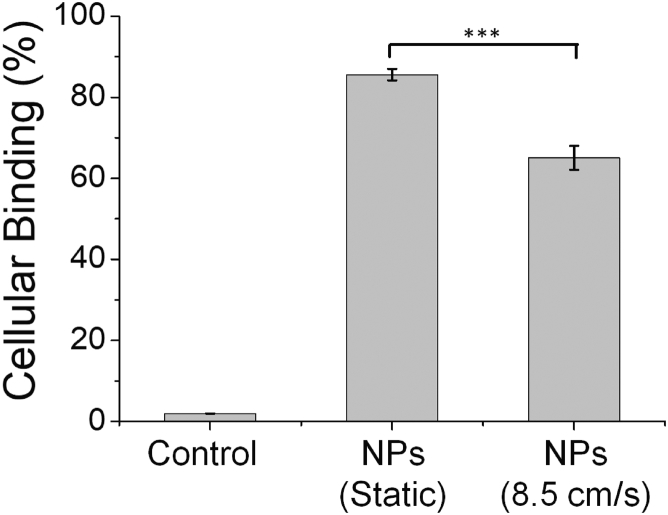
The results described above demonstrate that NPs under flow form a protein corona enriched in plasminogen (Fig. 2) and that plasminogen undergoes structural changes under flow (Fig. 3). To determine if this enrichment and change in structure had any effect on NP-cell interactions, we used flow cytometry to measure the binding of NPs to HeLa cells in the presence of 10% FBS and 10% FBS subjected to flow (8.5 cm/s). NPs (20 pM) and FBS (10%) were added to cells for 20 min of cold-binding at 4°C. At this temperature, protein-NP complexes bind to the cell surface but are not internalized by cells (34, 40, 41, 42, 43, 44, 45), allowing us to separate cellular binding from internalization. Equal concentrations of NPs were used, and protein concentrations were measured with UV-visible immediately before addition to cells to ensure equal concentrations. Flow cytometry was used to quantify cellular fluorescence. These measurements show a small (65.0 ± 3%, compared to 85.6 ± 1%, n = 3 distinct samples) but significant decrease in cellular binding of NPs for cells incubated with serum protein subjected to flow (Fig. 4). Measurements of dh, PDI, and ZP show nearly identical values for hard-corona-NPs with the corona formed under static conditions (30 min, room temperature) or continuous vortexing (30 min, room temperature), similar to flow (Fig. S2 C; Table S4). The small decrease in cellular binding observed for NPs incubated with serum proteins after flow suggests that plasminogen receptors may be responsible for some cellular binding of the protein-NP complex and that a change in the structure of plasminogen disrupts this interaction. In comparison, lipid NPs with a corona formed from circulating FBS showed increased uptake by HeLa cells (29), suggesting increased protein density or structural changes to proteins that affect cellular binding are not limited to polystyrene NPs.
Figure 4.
Flow cytometry was used to measure cellular binding of NPs incubated with 10% FBS under standard conditions and after 30 min of flow (8.5 cm/s). The NPs were added to the proteins after flow. ∗∗∗p ≤ 0.001.
Previous research from our lab showed that the cellular binding of carboxylate-modified polystyrene NPs (“yellow-green,” #F8811) (Thermo Fisher) in the presence of serum proteins was dominated by albumin and that adsorption of albumin on the NP surface led to competition with free BSA for cellular receptors on BS-C-1 and Chinese hamster ovary cells (6, 34, 46). A comparison of the current “orange” NPs and previous “yellow-green” NPs is provided in Fig. S6.
Conclusions
It is becoming increasingly clear that the protein corona controls the interaction of NPs with cells (6, 7, 12, 13, 14). Although the main method of nanomedicine delivery is expected to be intravenous, few studies have addressed the formation of a protein corona under flow (27, 28, 29, 30, 31). The goal of the experiments described above was to determine if shear flow changed the composition of the protein corona and then to determine if these changes affected cellular binding. Colorimetric assays of protein concentration and gel electrophoresis show that NPs subjected to flow have a greater concentration of proteins adsorbed on the NP surface, especially for plasminogen (Fig. 2). In addition, plasminogen, in the absence of NPs, shows changes in structure in response to flow, characterized by fluorescence and CD spectroscopy (Fig. 3). The protein-NP complexes formed from FBS after flow show decreased cellular binding, measured with flow cytometry (Fig. 4). Our studies used polystyrene NPs, but the result appears general. Gold NPs (13 nm) functionalized with polyethylene glycol or tannic acid also showed an increase in protein concentration (27). Similarly, lipid NPs (140 nm), both unmodified and PEGylated, were found to have increased protein coronas in response to flow (29). In addition to the relevance to nanomedicine, these results also highlight the technical challenges of protein corona studies. We observed differences in the protein corona that were highly dependent on the initial mixing step: rocking, vortexing, or flow (Figs. 2 and S2 C).
From a protein biophysics perspective, the structural changes of plasminogen but not BSA in response to flow are an interesting result. Changes to proteins dependent on shear force have been observed previously, measured as changes in enzyme activity or protein structure and determined by measuring changes in exposure of hydrophobic domains or molecular weight (47, 48). The classic example of protein response to flow is the von Willebrand factor, which requires shear-induced proteolysis for its biological activity (49, 50). Because much of the flow-dependent research is driven by bioprocessing and storage of therapeutic proteins (47), the structural changes of plasminogen in response to flow have not been examined previously. However, structural changes of plasminogen have been examined in response to 6-aminohexanoate and hydrostatic pressure (38, 39, 51, 52). This previous work suggests that the resulting structural changes mimic the structural changes that result from plasminogen binding to surfaces, such as fibrin clots, which is the initial step in activation (51, 52). In this case, it is possible that small structural changes due to flow aid in the activation of plasminogen.
Author Contributions
D.T.J. and S.M.P. performed research and analyzed data. R.G.M. and W.A.L. provided technical advice. D.T.J., S.M.P., and C.K.P. designed experiments and wrote the manuscript.
Acknowledgments
The authors thank Georgia Institute of Technology for support of this research.
Editor: Antoine van Oijen.
Footnotes
Christine K. Payne’s present address is Department of Mechanical Engineering and Materials Science, Duke University, Durham, North Carolina.
Six figures and five tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30296-0.
Supporting Material
References
- 1.Wicki A., Witzigmann D., Huwyler J. Nanomedicine in cancer therapy: challenges, opportunities, and clinical applications. J. Control. Release. 2015;200:138–157. doi: 10.1016/j.jconrel.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 2.Wilhelm S., Tavares A.J., Chan W.C. Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 2016;1:16014. [Google Scholar]
- 3.Baran U., Shi L., Wang R.K. Capillary blood flow imaging within human finger cuticle using optical microangiography. J. Biophotonics. 2015;8:46–51. doi: 10.1002/jbio.201300154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dolan J.M., Kolega J., Meng H. High wall shear stress and spatial gradients in vascular pathology: a review. Ann. Biomed. Eng. 2013;41:1411–1427. doi: 10.1007/s10439-012-0695-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gabe I.T., Gault J.H., Braunwald E. Measurement of instantaneous blood flow velocity and pressure in conscious man with a catheter-tip velocity probe. Circulation. 1969;40:603–614. doi: 10.1161/01.cir.40.5.603. [DOI] [PubMed] [Google Scholar]
- 6.Fleischer C.C., Payne C.K. Nanoparticle-cell interactions: molecular structure of the protein corona and cellular outcomes. Acc. Chem. Res. 2014;47:2651–2659. doi: 10.1021/ar500190q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walczyk D., Bombelli F.B., Dawson K.A. What the cell “sees” in bionanoscience. J. Am. Chem. Soc. 2010;132:5761–5768. doi: 10.1021/ja910675v. [DOI] [PubMed] [Google Scholar]
- 8.Lunov O., Syrovets T., Simmet T. Differential uptake of functionalized polystyrene nanoparticles by human macrophages and a monocytic cell line. ACS Nano. 2011;5:1657–1669. doi: 10.1021/nn2000756. [DOI] [PubMed] [Google Scholar]
- 9.Gessner A., Lieske A., Müller R.H. Functional groups on polystyrene model nanoparticles: influence on protein adsorption. J. Biomed. Mater. Res. A. 2003;65:319–326. doi: 10.1002/jbm.a.10371. [DOI] [PubMed] [Google Scholar]
- 10.Brewer S.H., Glomm W.R., Franzen S. Probing BSA binding to citrate-coated gold nanoparticles and surfaces. Langmuir. 2005;21:9303–9307. doi: 10.1021/la050588t. [DOI] [PubMed] [Google Scholar]
- 11.Alkilany A.M., Nagaria P.K., Wyatt M.D. Cellular uptake and cytotoxicity of gold nanorods: molecular origin of cytotoxicity and surface effects. Small. 2009;5:701–708. doi: 10.1002/smll.200801546. [DOI] [PubMed] [Google Scholar]
- 12.Lynch I., Cedervall T., Dawson K.A. The nanoparticle-protein complex as a biological entity; a complex fluids and surface science challenge for the 21st century. Adv. Colloid Interface Sci. 2007;134–135:167–174. doi: 10.1016/j.cis.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 13.Treuel L., Nienhaus G.U. Toward a molecular understanding of nanoparticle-protein interactions. Biophys. Rev. 2012;4:137–147. doi: 10.1007/s12551-012-0072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park S., Hamad-Schifferli K. Nanoscale interfaces to biology. Curr. Opin. Chem. Biol. 2010;14:616–622. doi: 10.1016/j.cbpa.2010.06.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carrillo-Carrion C., Carril M., Parak W.J. Techniques for the experimental investigation of the protein corona. Curr. Opin. Biotechnol. 2017;46:106–113. doi: 10.1016/j.copbio.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Gref R., Domb A., Langer R. The controlled intravenous delivery of drugs using PEG-coated sterically stabilized nanospheres. Adv. Drug Deliv. Rev. 1995;16:215–233. doi: 10.1016/0169-409X(95)00026-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gref R., Lück M., Müller R.H. ‘Stealth’ corona-core nanoparticles surface modified by polyethylene glycol (PEG): influences of the corona (PEG chain length and surface density) and of the core composition on phagocytic uptake and plasma protein adsorption. Colloids Surf. B Biointerfaces. 2000;18:301–313. doi: 10.1016/s0927-7765(99)00156-3. [DOI] [PubMed] [Google Scholar]
- 18.Walkey C.D., Olsen J.B., Chan W.C.W. Nanoparticle size and surface chemistry determine serum protein adsorption and macrophage uptake. J. Am. Chem. Soc. 2012;134:2139–2147. doi: 10.1021/ja2084338. [DOI] [PubMed] [Google Scholar]
- 19.Boulos S.P., Davis T.A., Murphy C.J. Nanoparticle-protein interactions: a thermodynamic and kinetic study of the adsorption of bovine serum albumin to gold nanoparticle surfaces. Langmuir. 2013;29:14984–14996. doi: 10.1021/la402920f. [DOI] [PubMed] [Google Scholar]
- 20.Runa S., Hill A., Payne C.K. Proceedings SPIE, Physical Chemistry of Interfaces and Nanomaterials XIII 9165. SPIE; 2014. PEGylated nanoparticles: protein corona and secondary structure; p. 91651F. [Google Scholar]
- 21.Papi M., Caputo D., Caracciolo G. Clinically approved PEGylated nanoparticles are covered by a protein corona that boosts the uptake by cancer cells. Nanoscale. 2017;9:10327–10334. doi: 10.1039/c7nr03042h. [DOI] [PubMed] [Google Scholar]
- 22.Li Y., Xu Y., Mao H. Impact of anti-biofouling surface coatings on the properties of nanomaterials and their biomedical applications. J. Mater. Chem. B. 2018;6:9–24. doi: 10.1039/C7TB01695F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cedervall T., Lynch I., Linse S. Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc. Natl. Acad. Sci. USA. 2007;104:2050–2055. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milani S., Bombelli F.B., Rädler J. Reversible versus irreversible binding of transferrin to polystyrene nanoparticles: soft and hard corona. ACS Nano. 2012;6:2532–2541. doi: 10.1021/nn204951s. [DOI] [PubMed] [Google Scholar]
- 25.Monopoli M.P., Åberg C., Dawson K.A. Biomolecular coronas provide the biological identity of nanosized materials. Nat. Nanotechnol. 2012;7:779–786. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- 26.Liu W., Rose J., Vidaud C. Protein corona formation for nanomaterials and proteins of a similar size: hard or soft corona? Nanoscale. 2013;5:1658–1668. doi: 10.1039/c2nr33611a. [DOI] [PubMed] [Google Scholar]
- 27.Braun N.J., DeBrosse M.C., Comfort K.K. Modification of the protein corona-nanoparticle complex by physiological factors. Mater. Sci. Eng. C. 2016;64:34–42. doi: 10.1016/j.msec.2016.03.059. [DOI] [PubMed] [Google Scholar]
- 28.Luby A.O., Breitner E.K., Comfort K.K. Preliminary protein corona formation stabilizes gold nanoparticles and improves deposition efficiency. Appl. Nanosi. 2016;6:827–836. [Google Scholar]
- 29.Palchetti S., Pozzi D., Laganà A. Influence of dynamic flow environment on nanoparticle-protein corona: from protein patterns to uptake in cancer cells. Colloids Surf. B Biointerfaces. 2017;153:263–271. doi: 10.1016/j.colsurfb.2017.02.037. [DOI] [PubMed] [Google Scholar]
- 30.Pozzi D., Caracciolo G., Laganà A. The biomolecular corona of nanoparticles in circulating biological media. Nanoscale. 2015;7:13958–13966. doi: 10.1039/c5nr03701h. [DOI] [PubMed] [Google Scholar]
- 31.Palchetti S., Colapicchioni V., Laganà A. The protein corona of circulating PEGylated liposomes. Biochim. Biophys. Acta. 2016;1858:189–196. doi: 10.1016/j.bbamem.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 32.Jayaram D.T., Runa S., Payne C.K. Nanoparticle-induced oxidation of corona proteins initiates an oxidative stress response in cells. Nanoscale. 2017;9:7595–7601. doi: 10.1039/c6nr09500c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Runa S., Khanal D., Payne C.K. TiO2 nanoparticles alter the expression of peroxiredoxin antioxidant genes. J. Phys. Chem. C. 2016;120:20736–20742. [Google Scholar]
- 34.Fleischer C.C., Payne C.K. Nanoparticle surface charge mediates the cellular receptors used by protein-nanoparticle complexes. J. Phys. Chem. B. 2012;116:8901–8907. doi: 10.1021/jp304630q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mirshafiee V., Kim R., Kraft M.L. Impact of protein pre-coating on the protein corona composition and nanoparticle cellular uptake. Biomaterials. 2016;75:295–304. doi: 10.1016/j.biomaterials.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 36.Hamad-Schifferli K. Exploiting the novel properties of protein coronas: emerging applications in nanomedicine. Nanomedicine (Lond.) 2015;10:1663–1674. doi: 10.2217/nnm.15.6. [DOI] [PubMed] [Google Scholar]
- 37.Correia M., Snabe T., Neves-Petersen M.T. Photonic activation of plasminogen induced by low dose UVB. PLoS One. 2015;10:e0116737. doi: 10.1371/journal.pone.0116737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sjöholm I. Studies on the conformational changes of plasminogen induced during activation to plasmin and by 6-aminohexanoic acid. Eur. J. Biochem. 1973;39:471–479. doi: 10.1111/j.1432-1033.1973.tb03146.x. [DOI] [PubMed] [Google Scholar]
- 39.Kornblatt J.A., Rajotte I., Heitz F. Reaction of canine plasminogen with 6-aminohexanoate: a thermodynamic study combining fluorescence, circular dichroism, and isothermal titration calorimetry. Biochemistry. 2001;40:3639–3647. doi: 10.1021/bi001857b. [DOI] [PubMed] [Google Scholar]
- 40.Payne C.K., Jones S.A., Zhuang X. Internalization and trafficking of cell surface proteoglycans and proteoglycan-binding ligands. Traffic. 2007;8:389–401. doi: 10.1111/j.1600-0854.2007.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doorley G.W., Payne C.K. Cellular binding of nanoparticles in the presence of serum proteins. Chem. Commun. (Camb.) 2011;47:466–468. doi: 10.1039/c0cc02618b. [DOI] [PubMed] [Google Scholar]
- 42.Doorley G.W., Payne C.K. Nanoparticles act as protein carriers during cellular internalization. Chem. Commun. (Camb.) 2012;48:2961–2963. doi: 10.1039/c2cc16937a. [DOI] [PubMed] [Google Scholar]
- 43.Hill A., Payne C.K. Impact of serum proteins on MRI contrast agents: cellular binding and T2 relaxation. RSC Advances. 2014;4:31735–31744. doi: 10.1039/C4RA04246H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lesniak A., Salvati A., Åberg C. Nanoparticle adhesion to the cell membrane and its effect on nanoparticle uptake efficiency. J. Am. Chem. Soc. 2013;135:1438–1444. doi: 10.1021/ja309812z. [DOI] [PubMed] [Google Scholar]
- 45.Szymanski C.J., Yi H., Payne C.K. Imaging intracellular quantum dots: fluorescence microscopy and transmission electron microscopy. In: Rosenthal S.J., Wright D.X., editors. Nanobiotechnology Protocols. Humana Press; 2013. pp. 21–23. [DOI] [PubMed] [Google Scholar]
- 46.Fleischer C.C., Payne C.K. Secondary structure of corona proteins determines the cell surface receptors used by nanoparticles. J. Phys. Chem. B. 2014;118:14017–14026. doi: 10.1021/jp502624n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bekard I.B., Asimakis P., Dunstan D.E. The effects of shear flow on protein structure and function. Biopolymers. 2011;95:733–745. doi: 10.1002/bip.21646. [DOI] [PubMed] [Google Scholar]
- 48.Thomas C.R., Geer D. Effects of shear on proteins in solution. Biotechnol. Lett. 2011;33:443–456. doi: 10.1007/s10529-010-0469-4. [DOI] [PubMed] [Google Scholar]
- 49.Sadler J.E. Biochemistry and genetics of von Willebrand factor. Annu. Rev. Biochem. 1998;67:395–424. doi: 10.1146/annurev.biochem.67.1.395. [DOI] [PubMed] [Google Scholar]
- 50.Shim K., Anderson P.J., Sadler J.E. Platelet-VWF complexes are preferred substrates of ADAMTS13 under fluid shear stress. Blood. 2008;111:651–657. doi: 10.1182/blood-2007-05-093021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ponting C.P., Marshall J.M., Cederholm-Williams S.A. Plasminogen: a structural review. Blood Coagul. Fibrinolysis. 1992;3:605–614. [PubMed] [Google Scholar]
- 52.Kornblatt J.A., Kornblatt M.J., Balny C. The effects of hydrostatic pressure on the conformation of plasminogen. Eur. J. Biochem. 1999;265:120–126. doi: 10.1046/j.1432-1327.1999.00695.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.