Abstract
G-protein-coupled receptors (GPCRs) control vital cellular signaling pathways. GPCR oligomerization is proposed to increase signaling diversity. However, many reports have arrived at disparate conclusions regarding the existence, stability, and stoichiometry of GPCR oligomers, partly because of cellular complexity and ensemble averaging of intrareconstitution heterogeneities that complicate the interpretation of oligomerization data. To overcome these limitations, we exploited fluorescence-microscopy-based high-content analysis of single proteoliposomes. This allowed multidimensional quantification of intrinsic monomer-monomer interactions of three class A GPCRs (β2-adrenergic receptor, cannabinoid receptor type 1, and opsin). Using a billion-fold less protein than conventional assays, we quantified oligomer stoichiometries, association constants, and the influence of two ligands and membrane curvature on oligomerization, revealing key similarities and differences for three GPCRs with decidedly different physiological functions. The assays introduced here will assist with the quantitative experimental observation of oligomerization for transmembrane proteins in general.
Introduction
G-protein-coupled receptors (GPCRs) comprise the most abundant family of transmembrane proteins (TMPs) in mammalian cells (1, 2); they control multiple signaling transduction pathways and effect crucial physiological reactions in response to a plethora of endo- and exogenic stimuli (1). Historically, GPCRs were considered monomeric entities, but multiple recent studies indicate that they frequently exist as dimeric or oligomeric assemblies (3). Oligomerization is proposed to increase pharmacological diversity by stabilizing alternative receptor conformations (4) and thereby promoting alternative signaling pathways (5). Thus, a concerted effort has gone into understanding GPCR oligomerization (3). However, many reports have arrived at disparate conclusions regarding the existence, stability, and stoichiometry as well as the influence of endogenous and pharmacological ligands on GPCR oligomers (5, 6, 7, 8).
The majority of GPCR oligomerization studies have been performed in living cells using optical resonance energy transfer (RET) methods to interrogate an ensemble of receptors (see Table 1 from Kasai and Kusumi (9)) or, more lately, single receptor molecules (6). However, cellular complexity, although establishing biological relevance (8), makes it difficult to disentangle the association properties intrinsic to direct receptor monomer-monomer interactions from the influence of multiple effectors that have been shown to impact oligomerization, including confining cytoskeletal elements (10), lipid rafts (11), interacting proteins (12), or extracellular contacts (13). To provide an analysis of GPCR oligomerization unbiased by cellular complexity, we purified and reconstituted GPCRs in proteoliposomes.
Proteoliposomes are model systems that are in principle particularly well adapted to studies of oligomerization because their continuous spherical surface allows TMPs to diffuse perpetually within the plane of the lipid bilayer and oligomerize without any hindrance imposed by physical barriers. However, the reproducible production of homogenous proteoliposomes is a great challenge in proteoliposome reconstitution (14, 15, 16, 17, 18, 19). A recent study demonstrated that individual proteoliposomes within a reconstitution have a highly heterogeneous protein-to-lipid (P/L) ratio that severely skews ensemble measurements of oligomerization (20). In general, heterogeneities are observed for different samples independent of protein type or reconstitution method (14, 15, 16, 17, 18, 19, 20). To overcome this limitation, we exploited fluorescence-microscopy-based high-content analysis (HCA) of single nanoscale proteoliposomes (20). This allowed the accurate multidimensional quantification of pure monomer-monomer interactions unbiased by cellular complexity and intrareconstitution heterogeneities.
Our data provide quantitative comparative insight into the oligomerization of three class A GPCRs: the β2-adrenergic receptor (β2AR), which binds adrenaline and noradrenaline in a variety of tissues to stimulate smooth muscle cell contraction (1); the cannabinoid receptor type 1 (CB1), which binds endocannabinoids in neurons and plays a role in memory, learning, and addiction (21); and opsin, the non-ligand-bound rhodopsin that mediates visual phototransduction in retinal cells (22). To characterize the oligomerization process in-depth, we quantified the number of protein monomers that associate per cluster (stoichiometry), the strength of monomer association (equilibrium constant and related association energy), and the influence of selected ligands and of the geometric structure of the surrounding lipid bilayer. This enabled us to identify key similarities and differences among the three GPCRs with decidedly different physiological functions.
Materials and Methods
Cloning, receptor purification, and labeling
Quantitative fluorescence RET (FRET) microscopy requires that each receptor carry a single fluorescent label; furthermore, to make direct comparisons between GPCRs, the fluorescent labeling site must be placed on the same region as the receptors. We therefore engineered receptor constructs with a specific cysteine labeling site for fluorescent labeling on the surface-exposed part of helix 8 (see Supporting Materials and Methods, Receptor Constructs). For all receptors used in this study, it was confirmed that the single reactive cysteine mutant was functional using ligand binding assays (for details, see β2AR (23), CB1 (24), and opsin (25)), and that fluorescent labeling did not impair functionality. Expression, purification, and fluorescent labeling with either Cy3- or Cy5-maleimide (Amersham Biosciences, Little Chalfont, UK) was carried out as previously described (β2AR (23), CB1 (26), opsin (27)).
Proteoliposome preparation
Fluorescently labeled receptors were reconstituted into proteoliposomes with a total protein to lipid ratio of 1:1000. Oregon Green 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (OG-DHPE) was included at 0.1 mol percentage to fluorescently label all proteoliposomes, and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-poly(ethylene glycol)2000-biotin (DSPE-PEG2000-biotin) was included at 0.1 mol percentage to enable tethering to the passivated microscopy glass surface (see Fluorescence Microscopy). To make direct comparisons between different GPCRs, the receptors were reconstituted into the simplest lipid composition required to maintain receptor functionality (see below). Four different proteoliposome preparations were produced for each receptor: 1) both GPCR-Cy3 and GPCR-Cy5 reconstituted at a 1:1 ratio for quantification of FRET, 2) empty liposomes with no receptors reconstituted, 3) only GPCR-Cy3 reconstituted, and 4) only GPCR-Cy5 reconstituted. Samples 2–4 were prepared as controls to carefully quantify possible intensity signal contaminations in the various microscopy detection channels used in this study (see Quantification of EFRET).
Proteoliposomes were prepared as previously described (20). Briefly, β2AR proteoliposomes were prepared by resuspending a dried lipid film of 1,2-dioleoyl-sn-glycero-3-phosphocholine/cholesteryl hemisuccinate/1,2-dioleoyl-sn-glycero-3-phosphoglycerol/OG-DHPE/DSPE-PEG2000-biotin (79.4:10:10:0.5:0.1) (Avanti Polar Lipids, Alabaster, AL; Steraloids, Newport, RI; Invitrogen, Carlsbad, CA) in buffer (20 mM HEPES, 100 mM NaCl, 1% octylglucoside (pH 7.5)) (20). The lipid detergent mixture was sonicated for 1 h on ice, and subsequently, receptors were added in the desired lipid to protein ratio. After incubation on ice for 2 h, proteoliposomes were formed by the removal of detergent on a Sephadex G-50 (fine) column (25 × 0.8 cm) (Sigma Aldrich, St. Louis, MO).
CB1 and opsin proteoliposomes (28) were prepared by mixing labeled receptor, solubilized in 0.05% n-dodecyl-β-D-maltoside, and lipids 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol/OG-DHPE/DSPE-PEG2000-biotin (59.85:39.85:0.2:0.1) (Avanti Polar Lipids, Steraloids, Invitrogen), solubilized in 0.5 M sodium cholate with approximately two-thirds vol of Bio-Beads SM-2 (Bio-Rad, Hercules, CA) overnight at 4°C. The Bio-Beads were removed by centrifugation (1000 × g, 1 min), yielding proteoliposome preparations in 20 mM HEPES (pH 7.3), 150 mM NaCl, 2 mM MgCl2, and 1 mM EDTA. Both Bio-Beads and size exclusion chromatography are robust methods for removing detergents. Once proteoliposomes are diluted to 0.0025 g/L and added to the glass surface for microscopy imaging (see section below), any remaining detergent will have had ample opportunity to dissociate into the assay buffer before the actual microscopy measurement takes place. We therefore evaluate the likelihood for any trace amounts of leftover detergents to be negligible and not influence the dimerization studies. Also, no lipidomic studies were carried out after reconstitution, as we anticipated the bulk lipids present would comprise the large excess of reconstitution lipids, either 1,2-dioleoyl-sn-glycero-3-phosphocholine/cholesteryl hemisuccinate/1,2-dioleoyl-sn-glycero-3-phosphoglycerol for β2AR or 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol for opsin and CB1.
Fluorescence microscopy
We immobilized the biotinylated proteoliposomes on a poly-L-lysine (PLL)-graft-PEG/PLL-graft-PEG-biotin passivated microscopy glass surface in homebuilt microscope chambers through NeutrAvidin, as described previously (20). Chamber parts were cleaned extensively using ethanol and Milli-Q water. Glass slides (thickness 170 ± 10 μm) were cleaned by consecutive rounds of sonication by 2% (v/v) Helmanex, following 3 × Milli-Q water and 2 × methanol. Glass slides were dried in nitrogen flow, plasma etched for 2 min, mounted in a microscope chamber, and incubated with a mixture of 1000:6 PLL-g-PEG and PLL-g-PEG-biotin (19; SuSoS AG, Dübendorf, Switzerland) (1 g/L) in surface buffer (15 mM HEPES (pH 5.6)) for 30 min. After carefully washing with sample buffer (β2AR: 20 mM HEPES, 100 mM NaCl (pH 7.5); CB1 and opsin: 20 mM HEPES, 100 mM NaCl, 2 mM MgCl2, 1 mM EDTA (pH 7.5)), the surfaces were incubated with 0.1 g/L NeutrAvidin in surface buffer for 10 min, after additional washing 10 × with sample buffer. We controlled proteoliposome surface density by adding samples at a low concentration (0.0025 g/L) to achieve spatial separation between particles and then washed the chamber 10 × with sample buffer. Proteoliposomes were imaged in sample buffer with a Leica TCS-SP5 inverted confocal microscope (Leica, Wetzlar, Germany) and an oil immersion objective HCX PL APO CS × 100 (NA 1.4). 476, 543, and 633 nm laser lines were used to excite the OG-DHPE, Cy3, and Cy5 fluorophores, respectively. The OG intensity signal was filtered out in the range of 486–539 nm and collected by a photomultiplier tube. Cy3 and Cy5 emissions were detected by avalanche photodiodes (APDs) with the emission signals cut off at 625 nm between APD1 and APD2 using a dichroic mirror BS625 (Chroma Technology, Bellows Falls, VT). Cy3 and Cy5 emissions were filtered by bandpass filters BrightLine HC 585/40 (Semrock, Rochester, NY) and HQ675/55M (Chroma Technology), respectively. Images had a resolution of 1024 × 1024 pixels, with a pixel size of 50.5 nm sample length and a bit depth of 16 and were acquired with a scan speed of 400 Hz. All fluorescence microscopy was carried out at room temperature.
Single fluorescent particle characterization
Automated detection and fitting of single fluorescent particles was carried out using software written in Igor Pro v. 6.01 (WaveMetrics, Portland, OR), as previously described (20). Briefly, a two-dimensional Gaussian bell was fitted to each diffraction-limited intensity to assign an xy center and measure the integrated fluorescence intensity. A circularity cutoff of 0.5 (minor axis divided by major axis) was applied to reject spurious intensity signals. Colocalization was defined as particles in separate color channels having centers within a distance of three pixels.
Quantification of EFRET
To accurately determine the efficiency of energy transfer (EFRET), fluorescent signals must be carefully corrected for several signal contaminations. We used three control samples (preparations 2–4 described in Proteoliposome Preparation) and imaged them, employing the exact same microscopy conditions as for the proteoliposomes investigated for FRET (preparation 1). Imaging the single-labeled proteoliposomes allowed us to measure the amount of fluorescence intensity leaking into neighboring emission detection channels (emission bleed-through) and also the amount of unintended fluorescence intensity resulting from excitation by neighboring laser lines (indirect excitation). We identified three such fluorescent signal contaminations and were able to accurately determine EFRET by using the correction factors (ω, α, β) (see Supporting Materials and Methods, Determining Fluorescent Signal Correction Factors (ω, α, β)). In the following section superscript, 0 denotes raw uncorrected intensities. and are the donor and acceptor intensities excited by the donor laser line (543 nm).
The GPCR-Cy3 emission was corrected for OG-DHPE bleed-through (ω) using Eq. 1:
| (1) |
The FRET emission was corrected for two signal contaminations—Cy3 emission bleeding into the acceptor detection channel (β) and the indirect excitation of Cy5 with the donor laser (543 nm) (α)—using Eq. 2:
| (2) |
EFRET was finally calculated according to Eq. 3 (29):
| (3) |
where
| (4) |
was introduced to decouple EFRET from instrumental and photophysical (30) effects. corrected for differences in detection efficiencies and differences in quantum yields of the Cy3 and Cy5 fluorophores. and were determined as carried out previously (20).
Results
Direct imaging of single proteoliposomes enables HCA of compositional heterogeneities
We reconstituted fluorescently labeled GPCRs in proteoliposomes doped with a lipid-coupled dye and produced arrays of single surface-tethered proteoliposomes that could be imaged individually with fluorescence microscopy (Fig. 1 A). We have previously measured nanoscale liposomes at the single particle level and demonstrated that surface immobilization did not compromise either the spherical or the physiochemical integrity of the particles (31, 32). By using single proteoliposomes, we reduced the amount of receptor needed for each assay to ∼6 pg, a 109-fold reduction over traditional assays (see Supporting Materials and Methods, Total Receptor Per Assay). Labeling of both proteins and the membrane phase allowed us to accurately quantify the amount of GPCRs and lipids per proteoliposome and concurrently measure oligomerization by FRET (20).
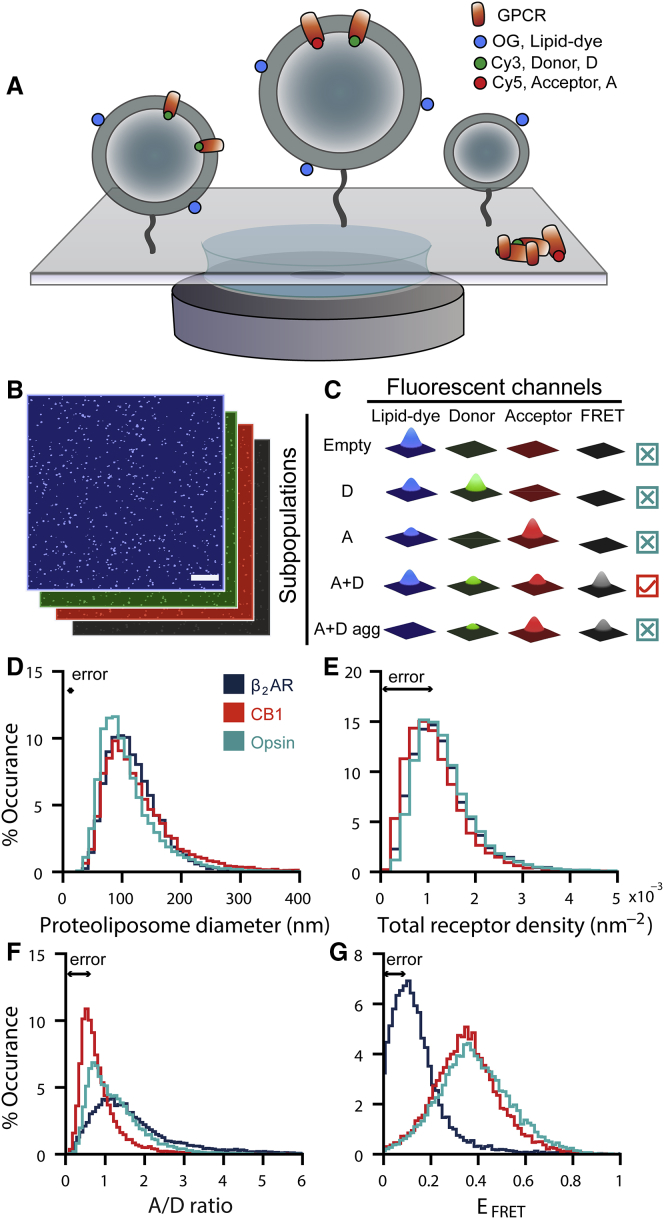
Figure 1.
Single proteoliposome high content analysis allows identification of intrareconstitution compositional heterogeneities. (A) Single proteoliposomes are tethered to a functionalized microscopy glass surface at dilute densities to ensure spatially separated particles. Proteoliposome membranes are labeled with a lipid-coupled dye Oregon Green DHPE (membrane, OG), and reconstituted GPCRs are labeled with either Cy3 (Donor, D) or Cy5 (Acceptor, A) fluorophores. (B) Typical confocal image micrographs of single proteoliposomes are shown. Scale bars, 10 μm. Micrographs are recorded for four fluorescent intensity signals: OG (blue), Cy3 (green), Cy5 (red), and FRET (black). (C) Colocalization of four unique intensity signals per proteoliposome reveals a subset of different particles within a reconstitution sample, namely proteoliposomes without reconstituted GPCRs (Empty), proteoliposomes harboring only donor-labeled or only acceptor-labeled GPCRs (D or A), proteoliposomes harboring both donor-labeled and acceptor-labeled GPCRs (A+D), and protein aggregates (A+D agg). (D–F) Histograms display individual proteoliposome (D) diameters, (E) receptor densities, (F) A/D ratios, and (G) FRET efficiencies (EFRET). Histograms of β2AR and opsin receptor densities overlay one another in (E). For each GPCR, data comprise n > 12,800 single proteoliposomes from >5 technical replicates. Errors shown in (D)–(G) represent the full width of histograms composed from single proteoliposome errors for each parameter. Solely in this figure, data for β2AR are reproduced from (20) to facilitate a direct comparison to CB1 and opsin.
We site-specifically labeled previously well-characterized and functionally active GPCR mutants with either Cy3 or Cy5 fluorophores at single reactive cysteines on the solvent-exposed helix 8 region of each receptor. Receptors were reconstituted in proteoliposomes using standard protocols of gel chromatography and removal of detergent using Bio-Beads (Materials and Methods). Mixing receptors labeled with Cy3 (donor, D) and Cy5 (acceptor, A) allowed us to quantify GPCR homo-oligomerization as the EFRET between D- and A-labeled receptors within single nanoscale proteoliposomes.
Fluorescent confocal microscopy enabled the parallel collection of fluorescent intensities from thousands of individual proteoliposomes (Fig. 1 B). We recorded four intensity signals per proteoliposome—lipid dye, D, A, and FRET (A emission at D excitation) (Fig. 1, B and C)—and extracted the position and integrated intensity for all four signals by fitting a two-dimensional Gaussian function.
Labeling of the proteoliposome lipid phase permitted the discrimination of receptors reconstituted in proteoliposomes from protein aggregates as well as empty liposomes by identifying particles with coexisting lipid and receptor intensities. This approach allowed us to then separate the proteoliposome fraction into the relevant subpopulation for FRET studies containing both D- and A-labeled GPCRs (A+D, Fig. 1 C, red box) from the unwanted populations of singly labeled proteoliposomes. Our colocalization analyses revealed that each receptor reconstitution contained subpopulations with highly different GPCR compositions and that a significant number of particles in each reconstitution were unusable for FRET studies (Fig. S2 A). We selected the relevant proteoliposomes (A+D) from the total population of particles for oligomerization studies: 56 ± 2% for β2AR, 22 ± 2% for CB1, and 18 ± 3% for opsin. Although the mechanisms behind these differences in receptor reconstitution remain unknown, our single-particle approach allows us to isolate this population across different reconstitution samples.
We then quantified intrasample heterogeneities within the selected proteoliposome population. GPCR oligomerization is strongly dependent on receptor concentration; we therefore carefully determined the receptor concentration of each single proteoliposome by quantifying the surface area using a previously published calibration procedure and the absolute number of receptors employing single-molecule photobleaching (see Supporting Materials and Methods, Proteoliposome Size and Receptor Density). We observed proteoliposome diameters from 40 to 400 nm (Fig. 1 D) and absolute receptor surface densities covering a broad range of ∼0.3 − 3.0 × 10−3 receptors/nm2 (Fig. 1 E). Each proteoliposome contained between ∼10 and 200 receptors (Fig. S2 B) with an average of 50 receptors per proteoliposome. Analysis of single proteoliposomes also revealed a broad distribution of A/D ratios (Fig. 1 F) spanning from 0.2 to 5.0 within a single reconstitution sample prepared with a nominal mixing-ratio of 1:1. We propagated the single proteoliposome errors for each parameter in Fig. 1, D–G from the uncertainty associated with quantifying the fluorescence intensity signals from GPCRs and lipids and plotted them as histograms (see Supporting Materials and Methods, Error Propagation). The variation in compositional heterogeneities substantially exceeded the full width of the error histograms (Fig. 1, D–F, black arrows), demonstrating that the variations in diameter (more than 47-fold larger than errors), density (more than fivefold larger than errors), and A/D ratios (more than sixfold larger than errors) can be used for HCA.
Oligomer-specific FRET allows quantification of GPCR oligomerization
FRET association studies require careful controls to verify that the energy transfer signal is specific to physical interactions between receptors. The homogeneous orientation of the receptor in the membrane is crucial to prevent non-natural top-to-tail oligomerization events. We therefore verified that our proteoliposome preparations contained a uniform receptor orientation (β2AR ∼90% outside out (23) and CB1 and opsin >95% inside out) (see Fig. S1 and Supporting Materials and Methods, Orientation of Receptors). Additionally, to avoid measuring artificial receptor crowding, the average distance between receptors must be considerably larger than the Fӧrster radius for the D and A fluorophores. Our calculations reveal that average monomer distances (assuming noninteracting monomers) at the observed high- and low-density extremes (Fig. 1 E) were respectively three- and ninefold larger than the ∼5.6 nm Cy3/Cy5-Fӧrster radius, demonstrating that protein crowding does not contribute substantially to the FRET readout (see Supporting Materials and Methods, Average Monomer Distance).
To demonstrate that we observed specific oligomerization and rule out that EFRET is not contaminated significantly by random collisions of receptors, we have previously used a theoretical scheme showing that EFRET always exceeded the expected EFRET from random encounters (20). Here, we followed the improved experimental guidelines of Lambert and colleagues, who recently used extensive controls to show that RET between noninteracting proteins may only increase as a function of acceptor density, whereas RET resulting from specific association should also be sensitive to donor density (33). To run this test, we plotted EFRET as a function of A density at a low and high D density, respectively (Fig. S2, C–H). We found a relative increase in EFRET at lower D, indicating that EFRET was sensitive to D density. Thus, these results confirmed that EFRET was not influenced by random collisions of receptors in the density ranges investigated here and that all three GPCRs investigated engaged in specific oligomerization.
Having ruled out the contribution of top-to-tail dimerization, receptor crowding, and random collisions, we could now interpret EFRET measurements from more than 12,000 individual proteoliposomes for each receptor. We observed a wide distribution of EFRET (Fig. 1 G) (more than 17-fold larger than errors) as a consequence of the large spread A/D ratios (Fig. 1 F) and protein densities (Fig. 1 E). Interestingly, the CB1 and opsin EFRET peak positions, 0.34 ± 0.001 and 0.37 ± 0.001, respectively, were shifted to higher efficiencies than for the β2AR, 0.13 ± 0.001. These results indicate that the oligomerization of CB1 and opsin differs markedly from that of β2AR because of differences in both association stoichiometry and energy, as we demonstrate further on.
Ensemble-average proteoliposome measurements can severely bias oligomerization assays
Typically, ensemble association studies estimate the average P/L ratio of proteoliposomes from the starting material of reconstitution samples and measure TMP oligomerization with ensemble RET. Having first identified that a significant number of particles were not the relevant A+D proteoliposomes (Fig. 1 C; Fig. S2 A), we next evaluated if an ensemble-average from this sample represented the single proteoliposome distributions of densities, A/D ratios, and EFRET (Fig. 1, E–G). We determined the percentage of single proteoliposomes having values within a 10% error margin around the predicted ensemble-average. We based the 10% error margin on previous reports of RET oligomerization measurements of GPCRs in proteoliposomes (23) and live cells (34).
We calculated a bulk receptor density (ρbulk) under the assumption that all lipid and receptor materials were used to produce proteoliposomes (Table S1). For all three receptors, ρbulk (1.7 ± 0.2 × 10−3 receptors/nm2) represented less than 15% of the single proteoliposomes (Fig. S2 I). Additionally, even though proteoliposomes were prepared at a nominal 1:1 A/D ratio, only 10–12% of single proteoliposomes had an A/D ratio within 10% of the intended A/D = 1 (Fig. S2 I; Table S1). Both observations demonstrate that the common assumption permitting protein density and A/D ratios to be estimated from the amount of starting materials must be used with great caution.
Finally, we estimated an ensemble EFRET average (see Supporting Materials and Methods, Ensemble Proteoliposome EFRET) including signals from A+D proteoliposomes, protein aggregates, and proteoliposomes carrying only D- or A-labeled GPCRs (Fig. S2 I; Table S1). The calculated ensemble EFRET-bulk was similar for all three receptors (β2AR 0.27, CB1 0.27, and opsin 0.22) but deviated greatly from the single proteoliposome EFRET peak positions (see Fig. 1 G) and would incorrectly suggest that the three GPCRs had similar oligomerization behavior. In addition, only 4% (β2AR), 14% (CB1), or 7% (opsin) of single proteoliposomes were within 10% of the predicted EFRET-bulk (Fig. S2 I; Table S1). These calculations demonstrate that protein aggregates and proteoliposomes carrying only D- or A-labeled GPCRs severely alter ensemble-averaged EFRET signals. In contrast to the convoluted and biased ensemble values, single-proteoliposome measurements revealed important differences in the oligomerization propensity of the β2AR, CB1, and opsin.
Identifying unique apparent stoichiometry and association modes for three class A GPCRs
We have previously shown that compositional heterogeneities can be exploited to perform HCA as a function of TMP density and A/D ratio to extract quantitative parameters of oligomerization (20). Here, we went on to quantify both the stoichiometry and association energies for β2AR, CB1, and opsin.
The ratio of D- and A-labeled GPCRs in an oligomer can be used to determine the apparent average oligomer stoichiometry (23, 34, 35, 36, 37, 38). In the simple case of a D-A dimer, the amount of energy transferred by the D- to the A-fluorophore is dependent on the proximity of the two fluorophores (39). In higher-order oligomers, where for example a D-fluorophore may excite multiple acceptors, the A/D ratio will also affect EFRET. To determine the stoichiometry of each GPCR, we used the widely applied method of Veatch and Stryer (38) to relate EFRET to the A/D ratio and extract the apparent average oligomer stoichiometry as a fitting parameter (Fig. 2 A and Supporting Materials and Methods, Receptor Stoichiometry). The Veatch and Stryer theory has been a well-established approach that allows the direct comparison of receptors with stoichiometries greater than dimers; progress is currently being made in the field to improve this classical theory (40).
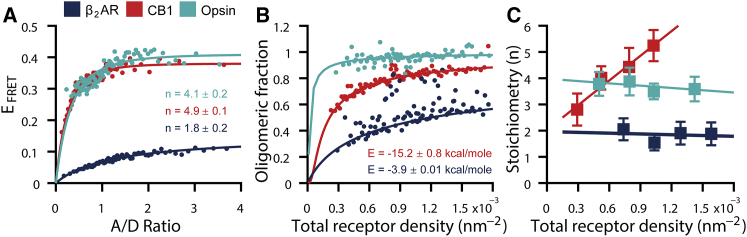
Figure 2.
Quantification of GPCR association strengths and stoichiometry. (A) To quantify the apparent average stoichiometry of each receptor, EFRET is evaluated as a function of A/D ratios and fit with Eq. S3. From the fit, we extract the apparent average stoichiometry (n). The data shown in (A) comprise >12,400 single proteoliposomes. For clear visualization, EFRET values of single proteoliposomes are binned (200 single proteoliposomes per bin), and a weighted average of each bin is displayed. Uncertainties are less than or equal to displayed marker size. (B) To determine the association energies of oligomerization (E), the GPCR oligomeric fraction is evaluated as a function of total receptor density, and data are fitted based on a monomer-dimer (β2AR) or monomer-tetramer (CB1 and opsin) thermodynamic equilibrium using the kinetic theory of FRET (Supporting Materials and Methods). We determine the association energies for β2AR (−3.9 ± 0.007 kcal/mole) and CB1 (−15.2 ± 0.8 kcal/mole) and find that opsin is a high-affinity oligomer. In (B), data were binned (10 proteoliposomes per bin), and a weighted average is shown. For each GPCR, a total of n > 820 single proteoliposomes was included in the analysis. (C) GPCR stoichiometry is shown as a function of total receptor density. Fitting and extracting the apparent average stoichiometry was repeated as in (A) for four proteoliposomes selections with increasing receptor densities while maintaining a constant proteoliposome diameter within ±15 nm of the mean (see Table S1). The apparent average stoichiometry from each density selection is shown. For each GPCR, data comprise n > 2500 single proteoliposomes, where uncertainties represent ±1 SD calculated from the fit of Eq. S3. A linear fit to each data set is included to guide the eye. Our analysis reveals a unique stoichiometry and association mode for each GPCR.
We determined the apparent average stoichiometry for each receptor by fitting this theoretical scheme to all proteoliposomes (n > 12,800) presented in Fig. 1, D–G. The stoichiometry analysis reveals interesting differences between the three receptors. We find that the β2AR forms oligomers of much lower order than CB1 or opsin (Fig. 2 A) with their apparent average stoichiometries being 1.8 ± 0.2, 4.9 ± 0.1, and 4.1 ± 0.2, respectively. Taken together, our results in a simple lipid system reveal that β2AR, CB1, and opsin each have a unique propensity to form oligomers of a certain stoichiometry when oligomerization is solely governed by receptor-receptor interactions.
The direct comparison of our findings with the literature is not straightforward. GPCR oligomerization has been largely measured with ensemble-based RET studies in living cells that have not necessarily arrived at a consensus with respect to individual receptor oligomerization or stoichiometries (7). Given the disparity of information in the literature, it is hard to make a direct comparison between live cell studies and single proteoliposome studies; however, we would like to note that there is existing support in live cells that β2AR can form a dimer (34, 41, 42) and that the smallest repeating unit of rhodopsin in isolated disk membranes is tetrameric (43, 44, 45).
We then proceeded to quantify the association energies of oligomerization. We previously applied the method of Wolber and Hudson (46) to determine the apparent Gibbs free energy of association for β2AR; however, this methodology is restricted to a dimer system and did not satisfyingly fit the CB1 and opsin data. We therefore built upon the advanced kinetic theory of FRET by Raicu (47, 48) and computed the theoretical apparent FRET efficiency as a function of total concentration for two models of a monomer-dimer (β2AR) and monomer-tetramer (CB1 and opsin) reaction (see Supporting Materials and Methods, Receptor Association Energies). To exclude any possible contribution of membrane curvature on protein diffusion (49) and thus oligomerization, we selected proteoliposomes within an extremely narrow diameter range (120–130 nm). We then fitted the ratio of oligomers to free monomers (oligomeric fraction) as a function of total receptor density to the binding curves for each oligomer association model and extracted the association constant of oligomerization Ka.
We determined the apparent Gibbs free energy of association for β2AR, −3.9 ± 0.007 kcal/mole (which is comparable to that found using the classical Wolber and Hudson method, −4.7 ± 0.2 kcal/mole (20)) and for CB1, −15.2 ± 0.8 kcal/mole. The CB1 association was stronger than that of the β2AR, as expected by comparison of the oligomeric fractions of the two receptors in Fig. 2 B (i.e., at 0.6 × 10−3 receptors/nm2, ∼40% of β2AR is oligomeric, whereas ∼75% of CB1 is oligomeric). We could not accurately fit the data for opsin with our association model because the oligomeric fraction of the receptor did not vary substantially within the range of receptor densities that we could measure (Fig. 2 B), indicating that opsin oligomerizes at much lower densities than CB1 or β2AR.
Increasing receptor density has been proposed to shift the equilibrium from monomers to oligomers because of the higher number of protein-protein encounters (42). Thus, one would expect the apparent stoichiometry to increase on average with protein density. The multidimensional heterogeneity of our samples allowed us to test this hypothesis because we were able to quantify EFRET as a function of A/D ratio for multiple receptor densities. In Fig. 2 C, we plot the apparent average GPCR oligomer stoichiometry for four groups of proteoliposomes with increasing surface densities while maintaining a constant proteoliposome diameter. The average apparent stoichiometry of CB1 increased as expected. Interestingly, within the limits of statistical uncertainty, we did not observe a similar change for β2AR (Fig. 2 B). The apparent stoichiometry of opsin was constant over these receptor densities, which was not surprising given the constant oligomeric fraction (Fig. 2 B) of opsin within this range of densities.
Membrane curvature modulates GPCR oligomerization
Upon ligand binding, GPCRs are thought to desensitize by internalization into endocompartments of high membrane curvature (50). However, high membrane curvature has recently attracted considerable attention for being able to regulate multiple biophysical properties of TMPs (49, 51, 52, 53). We therefore set out to explore whether GPCR oligomerization is sensitive to changes in the local membrane shape surrounding the receptor. We did this by evaluating EFRET as a function of liposome diameter (Fig. 3 A).
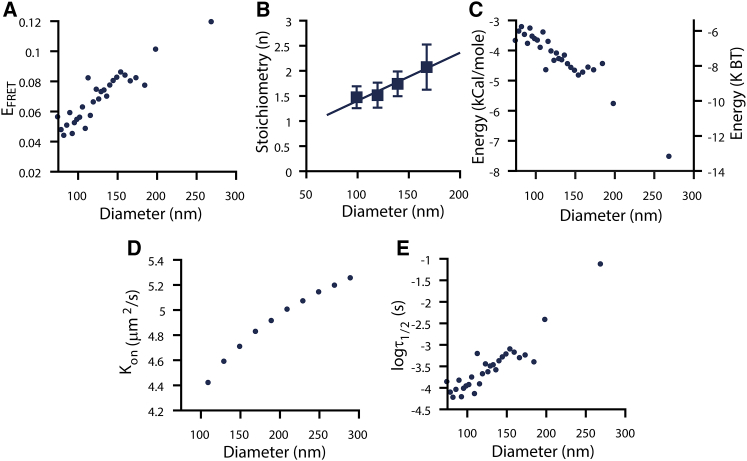
Figure 3.
Membrane curvature modulates GPCR oligomerization. (A) β2AR EFRET is shown as a function of proteoliposome diameter. EFRET increases as proteoliposome diameter increases, and membrane curvature decreases. Data in (A) have a constant receptor density and A/D ratio (Fig. S3). Corresponding data for CB1 and opsin are shown in Fig. S3. In (A), data were binned (75 single proteoliposomes per bin), and a weighted average is shown. Data in (A) comprise a total of n = 2175 single proteoliposomes with uncertainties less than or equal to the displayed marker size. (B) Increasing membrane curvature decreases oligomer stoichiometry. Stoichiometry analysis was repeated as in Fig. 2A for proteoliposome selections with increasing diameters while maintaining a constant receptor density within ±0.2 × 10−3 receptors/nm2 of the mean (see Table S1). Corresponding data for CB1 and opsin are shown in Fig. S4. The apparent average stoichiometry from each diameter selection is shown. Data in (B) comprise n > 2500 single proteoliposomes, where uncertainties represent ±1 SD calculated from the fit of Eq. S3. A linear fit to the data set is included to aid interpretation. (C) Standard Gibbs free β2AR association energy is shown as a function of liposome diameter and membrane curvature. β2AR association energy is decreased by ∼4 kcal/mole from −3.8 to −7.8 kcal/mole in extremely curved versus quasi-planar membrane geometries. (D) Dimerization on-rates (kon) (μm2/s) are shown as a function of liposome diameter. (E) Dimer lifetimes are calculated for the proteoliposome selection of (C). High membrane curvature decreases GPCR oligomerization as well as dimerization on-rates and lifetimes.
Our measurements revealed a pronounced effect on EFRET, changing from ∼0.05 in a highly curved membrane (50 nm in diameter) to ∼0.12 in a more planar membrane (300 nm in diameter) for β2AR (Fig. 3 A). Over the same range of curvatures, EFRET changed from ∼0.30 to 0.40 for both CB1 and opsin (Fig. S3, B and C). These observations suggest that highly bent bilayers reduce the propensity of GPCRs to oligomerize.
To exclude any convolution of the curvature effect with variations in protein density and A/D ratio, we selected proteoliposomes with a constant range of acceptor and donor densities for this analysis (Fig. S3, D–F). Ensuring that both acceptor and donor densities are constant, we can ascribe the observed perturbation in EFRET to solely originate from the influence of membrane curvature. Interestingly, we also found that the apparent stoichiometry of β2AR, CB1, and opsin decreased with high membrane curvature (Fig. 3 B; Fig. S4). Thus, our data reveal that high membrane curvature can destabilize both the propensity of GPCRs to oligomerize and the size of the oligomers.
A major challenge in the membrane curvature field is to design experimental approaches for controlling and measuring the effect of both positive and negative curvature on TMP function (54). Our assay provided an unambiguous assessment of both types of membrane bends on GPCR oligomerization because the receptors favored uniform and different orientations in the proteoliposomes (β2AR, outside out, and CB1 and opsin, inside out). We found that high membrane curvature restrains receptor interactions for both positive (β2AR, outside out) and negative membrane curvatures (CB1 and opsin, inside out). Thus, the sign of the curvature does not influence to a measurable extent the effect of membrane bending on the oligomerization of the three GPCRs that we examined.
Previously, we used the Wolber and Hudson model to determine the standard Gibbs free energy of association and the FRET efficiency within the β2AR dimer (Ebound) to be ∼0.2 for proteoliposomes of 120–130 nm in diameter (20). Assuming that the dimeric complexes on average interact at the same interfaces independently of curvature (i.e., Ebound is constant), we can now estimate the fraction of bound dimers as a function of curvature (see Supporting Materials and Methods, Estimation of β2AR Association Energies as a Function of Membrane Curvature). Our estimate yields a curvature-imposed change in association energy from −7.8 kcal/mole in quasi-planar bilayers to −3.8 kcal/mole in highly curved membranes (Fig. 3 C). A ∼4 kcal/mole change is significantly larger than the cost of structural TMP mutations (55), revealing the pronounced effect of membrane curvature on dimer stability.
To gain insight on the possible underlying mechanisms driving the difference in dimerization at higher membrane curvature, we developed the following theoretical framework. We evaluated the dimerization on-rate constant and its dependence on membrane curvature using protein mobility theory in combination with a diffusion-limited on-rate constant (56) (see Fig. 3 D and Supporting Materials and Methods, Calculation of β2AR On-Rates). We found that the theoretical diffusion constant is much lower in highly curved membranes (1.9 μm2/s) compared to a planar bilayer (3.9 μm2/s). We estimated the dimerization on-rate constant under the assumption that a dimer-binding reaction will take place every time that two receptors collide (diffusion-limited dimerization). Under this assumption, a diffusion constant that increases with the vesicle diameter implies an on-rate constant that decreases in highly curved membranes (Fig. 3 D). Furthermore, by combining our measured association free-energy with the diffusion-limited dimerization model and estimates of the dimerization on-rate, we were able to calculate the approximate dimer lifetimes as a function of membrane curvature (Fig. 3 E). Notably, the combined effect of increased on-rate (faster diffusion) and increased association energy (increased stability of the dimers) predicted a significant increase (by three orders of magnitude) in the dimer lifetime of planar membranes.
Our results demonstrate that the geometry of the membrane environment strongly modulates GPCR oligomerization. Changes in local membrane environments, whether from the dynamic rearrangements of plasma membrane or via cycling to endosomes or nanotubules, are thus predicted to regulate the oligomeric lifetime of GPCRs.
Soluble ligands do not perturb the influence of membrane curvature or GPCR stoichiometry
There has been considerable effort in the GPCR oligomerization field to resolve whether ligands modify class A GPCR receptor oligomerization, with a body of studies with opposing conclusions (compare (23, 57, 58) to (42, 59, 60, 61)). Here, we chose to evaluate the effect of ligands for the β2AR because of its outside-out orientation in proteoliposomes, which allows ligands to bind from solution. We previously tested the influence of the β2AR agonist (isoproterenol (ISO)) and inverse agonist ((2R,3R)-rel-3-isopropylamino-1-(7-methylindan-4-yloxy)-butan-2-ol hydrochloride (ICI) 118,551) by fitting the scaling of EFRET with density (20). We observed an approximately threefold increase of Ka for ICI, suggesting that ligands have the potential to modulate the inherent propensity of β2AR oligomerization.
Here, we first tested the influence of ligand binding on the curvature-dependent oligomerization behavior. We incubated β2AR proteoliposomes with saturating concentrations of ISO (10 μM) and ICI (500 nM) for 30 min before imaging (see Supporting Materials and Methods, Ligands). To evaluate the effect of membrane curvature on oligomerization, we chose a constant protein density (see Fig. S3). The presence of either ligand (Fig. 4 A) did not perturb to a measurable extent the effect of membrane curvature on oligomerization, thus suggesting that the regulation by membrane curvature is unaffected by ligand binding.
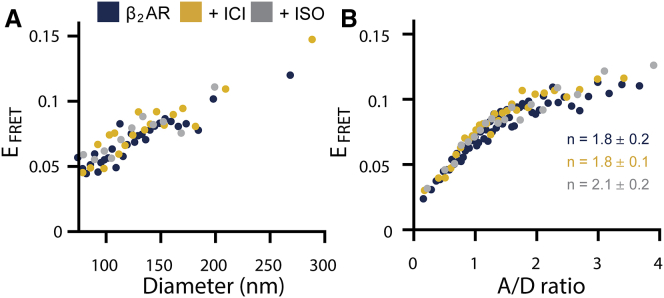
Figure 4.
Effect of ligands on β2AR oligomerization. (A) β2AR oligomerization and (B) apparent average β2AR stoichiometry are shown as a function of membrane curvature under saturating conditions of either agonist isoproterenol (ISO) or inverse agonist ICI 118,551 (ICI). Data in (A) have a constant receptor density and A/D ratio (Fig. S3). In (A), data were binned (75 proteoliposomes per bin), and a weighted average is shown. Data in (A) comprise a total of n > 900 single proteoliposomes for each ligand condition. In (B), data were binned (200 single proteoliposomes per bin), and a weighted average is shown. Data in (B) comprise a total of n > 4000 single proteoliposomes for each ligand condition. Uncertainties in (A) and (B) are less than or equal to the displayed marker size. Taken together, we find that ligands do not perturb the influence of membrane curvature or the stoichiometry of β2AR.
We next tested the effect of ligands on the apparent oligomer stoichiometry, here including all receptor densities and proteoliposome diameters (Fig. 4 B). We found that the apparent average stoichiometry of β2AR 1.8 ± 0.2 did not change in the presence of ICI, 1.8 ± 0.1, or ISO, 2.1 ± 0.2. These results are in line with single-molecule tracking studies in live cells in which ISO was shown not to influence the stoichiometry of β2AR (42).
Although agonist (ISO) and inverse agonist (ICI) binding has been shown to induce different ligand-bound conformational states in β2AR monomers (62, 63), we found here that these ligands did not alter the strong regulatory effect of the membrane or translate into a change in average stoichiometry. Our data suggest that both the influence of membrane curvature and the conserved dimeric β2AR stoichiometry are fundamental to different ligand-bound conformational states of the receptor.
Discussion
The oligomerization of class A GPCRs has been studied extensively, and accumulating evidence support that GPCRs can, in principle, form dimeric or oligomeric assemblies in the plasma membrane (5). However, many reports arrive at conflicting conclusions regarding the existence, stability, and stoichiometry as well as the influence of endogenous and pharmacological ligands on these assemblies (6, 7, 8, 33). The majority of GPCR oligomerization studies have been performed in living cells (9), in which many environmental effectors may influence oligomerization. Here, we exploited a recently developed methodology based on HCA of single proteoliposomes to provide a comprehensive analysis of GPCR oligomerization unbiased by cellular complexity and intrareconstitution proteoliposome heterogeneities. Our results demonstrate that GPCR oligomerization is receptor specific and highly sensitive to environmental effectors, such as local protein density and membrane curvature, and we propose that this could in part explain the inconsistencies present in the published literature.
We directly compared the oligomerization behavior of three GPCRs with distinct biological roles (β2AR, CB1, and opsin) to identify key similarities and differences in their oligomerization propensity. The three receptors associated with vastly different strengths, ranging from the relatively weak association of β2AR to the very strong association of opsin (Fig. 2 B). The apparent Gibbs free energy of association for β2AR (−3.9 ± 0.007 kcal/mole) is slightly higher than what has been reported for the dimerization of a model transmembrane helix in liposomes (−3.0 kcal/mole) (64), which was considered to be reduced to the simplest TMP interaction in lipid membranes. This free energy of association meant that in proteoliposomes, β2AR oligomerization was predominantly modulated for surface densities between 300 and 2000 receptors/μm2 (Fig. 2 B). Interestingly, β2AR in H9C2 cardiomyocyte-like cells (65) is clustered within plasma membrane regions of ∼ 120–160 nm in diameter with very similar densities (∼600–6400 receptors/μm2) (65). It is noteworthy that when receptors are clustered, their density is not uniform over the entire plasma membrane; thus, the average density estimated with ensemble average methods like radio ligand binding can underestimate by a factor of ∼100 or more the real local density (65).
The apparent Gibbs free energy of association for CB1 (−15.2 ± 0.8 kcal/mole) was much lower than that of β2AR, reflecting the propensity of CB1 to form oligomers at much lower receptor densities than β2AR and revealing that CB1 is able to readily form high-energy oligomers in the absence of cellular coagents. We found that opsin had an even lower association energy than CB1, which we could not determine because it remained almost fully oligomerized over the density range covered in our experiment (corresponding to 300–3000 receptors/μm2). The differences that we quantified in oligomer energetics imply that the GPCR monomer-oligomer association reaction can be regulated in a receptor-specific manner. Such specificity would allow cellular effectors to fine-tune the interaction strength and oligomerization state for each receptor in different regions of the cell; thus, providing a means to locally modulate GPCR function.
Dimer dissociation constants in units of receptors/μm2 have been reported for two other class A GPCRs in model cell studies; namely, the N-formyl peptide receptor in Chinese hamster ovary cells (59) (3.6 receptors/μm2) and opsin in COS-7 cells (66) (1010 receptors/μm2). These results suggest that opsin would be the weaker dimer; however, they are obtained in two different cell lines and thus may not be directly comparable. For example, a recent study revealed that β2AR formed clusters in cardiomyocytes but not in HeLa or Chinese hamster ovary cell lines when examined systematically, side by side, by super-resolution photoactivated localization microscopy (67).
It is worth highlighting that the association strengths of interaction for oligomers of different sizes are best compared in units of kilocalories/mole or kilojoules/mole. Considering that the dimer and tetramer reaction schemes [M] + [M] ↔ [D] and [M] + [M] + [M] + [M] ↔ [T] (M denotes monomer, D denotes dimer, and T denotes tetramer), the association constants KA,dimer = [D]/[M]2 and KA,tetramer = [D]/[M]4 have different units of [concentration]−1 and [concentration]−3, respectively. This is also the case when converting association constants into a characteristic length scale such as concentration of receptors per area. Furthermore, converting to a characteristic length scale in the relevant dimension of the membrane solvent is not trivial (68) and would require generalizing assumptions on lipid parameters such as membrane thickness or lipid headgroup area (69), both of which are highly variable between and within cells and influential on association energetics (64). We therefore overall suggest comparing the strengths of interaction on the energy scale in our study.
The apparent average stoichiometry varied greatly among the receptors. β2AR was dimeric, whereas opsin and CB1 were tetrameric and pentameric, respectively (Fig. 2 A), suggesting that CB1 and opsin have multiple interfaces of interaction to accommodate higher-order oligomers. Accumulating evidence from single-molecule studies in live cells suggests that class A GPCR oligomers form reversibly (42, 59, 70, 71) and comprise mixtures of monomers, dimers, and higher-order oligomers (42, 71). We could test the existence of mixed oligomeric species in our samples by monitoring changes in the apparent average stoichiometry as a function of total receptor concentration. The average stoichiometry of CB1 increased with density (Fig. 2 C) as expected, suggesting a shift from predominately dimeric to predominantly pentameric clusters. Surprisingly, we did not observe the same behavior for β2AR, which exhibited a constant (dimeric) average apparent stoichiometry over the entire density range investigated (∼300–3000 receptors/μm2). This is in contrast with a live cell single-particle tracking study of β2AR (42) and suggests that if β2AR forms higher-order oligomers, then additional cellular cofactors may be required. Our findings support that GPCRs can have a preferred average stoichiometry and that the robustness of these stoichiometries to small changes in receptor density is receptor specific.
We further provide, to our knowledge, the first experimental evidence that the geometrical environment of the membrane modulates GPCRs association. Membrane curvature is rapidly regulated in living cells during signaling (e.g., in endocytosis) and is considered crucial in tuning protein recruitment to active signaling sites (72, 73, 74, 75). Interestingly, in addition to their canonical signaling platform in the plasma membrane, GPCRs have recently been observed to function in areas of high membrane curvature such as endosomes (<60–500 nm) (50, 76, 77) and lipid nanotubules (20 − 450 nm) (78, 79) with diameters comparable to proteoliposomes (40 − 400 nm, Fig. 1 D). We found that high membrane curvature decreased the interactions of β2AR, CB1, and opsin (Fig. 3 A; Fig. S3, B and C) and decreased their average stoichiometry (Fig. 3 B). We calculate that membrane curvature perturbs β2AR association energy by ∼4 kcal/mole. A reduction of 4 kcal/mole in association energy is significantly larger (on average >1 kcal/mole higher) than the cost of systematically perturbing TMP helix-helix interactions by structural mutations (55) or the effects of ligands (20), suggesting that modulation by membrane curvature can affect also other TMP assemblies. Furthermore, coupled to the modulation in receptor mobility in curved membranes, the stability change implies order-of-magnitude variations in dimeric lifetimes, which could have profound functional consequences. Our results predict that the geometry of the local membrane environment in living cells, as observed for example in clathrin-coated pits, modulates GPCR oligomerization.
The lipid environment of the membrane likely plays a role in GPCR association in the cell (80). Here, we chose minimal lipid compositions shown to stabilize each GPCR in proteoliposomes to directly compare their oligomerization. The lipid requirements to maintain a functional β2AR were, however, different for CB1 and opsin (Materials and Methods). Nevertheless, we found that the response of receptors to membrane curvature remains intact for all receptors (e.g., oligomerization and stoichiometry decreased for all GPCRs in proteoliposomes with high membrane curvature (see Figs. S3, A–C and S4). Additionally, we found that lipid composition did not overwrite the uniqueness of receptor-receptor interactions for each GPCR, e.g., CB1 and opsin were reconstituted under identical lipid compositions but showed distinct differences in association energies and oligomer stoichiometry as a function of density (Fig. 2, B and C).
Studies of oligomerization in a cellular context undoubtedly enhance the biological relevance of the investigation. However, our findings highlight overall that a quantitative systematic comparison between different receptors is greatly facilitated by experimental conditions that can begin to disentangle cellular complexities and nanoscale heterogeneities to reveal the intrinsic physical propensity of monomers to oligomerize. The assays introduced here are thus expected to assist the quantitative experimental observation of oligomerization for TMPs in general.
Author Contributions
D.S. conceived of the strategy and was responsible for overall project management and supervision. S.M.W., S.M., S.M.C., and D.S. designed all the experiments. S.G.F.R. and J.J.F. expressed, purified, labeled, and reconstituted the β2AR. J.F.F. expressed, purified, labeled, and reconstituted the CB1 and opsin. S.M.W. and S.M. performed the fluorescence measurements, data analysis, and theoretical calculations. C.K. and K.H. determined the association energies. D.P and E.B. performed the theoretical calculations for curvature effects on receptor diffusion and dimer lifetime. S.M.W., S.M., S.M.C., and D.S. wrote the article. All authors discussed the results and commented on the article.
Acknowledements
This work was supported by the Danish Council for Strategic Research (grant number 1311-00002B) and Innovation Fund Denmark (grant number 5184-00048B). Partial funds were provided by National Institutes of Health grants DA038882 and DA026434 (to M.F.). E.B. was supported in part by National Institute on Drug Abuse grant T32 DA007135.
Editor: Charles Deber.
Footnotes
Samuel M. Walsh and Signe Mathiasen contributed equally to this work.
Supporting Materials and Methods, four figures, and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30677-5.
Supporting Citations
References (81, 82, 83, 84, 85, 86, 87, 88, 89) appear in the Supporting Material.
Supporting Material
References
- 1.Rosenbaum D.M., Rasmussen S.G., Kobilka B.K. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Audet M., Bouvier M. Restructuring G-protein- coupled receptor activation. Cell. 2012;151:14–23. doi: 10.1016/j.cell.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Ferré S., Casadó V., Guitart X. G protein-coupled receptor oligomerization revisited: functional and pharmacological perspectives. Pharmacol. Rev. 2014;66:413–434. doi: 10.1124/pr.113.008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levitz J., Habrian C., Isacoff E.Y. Mechanism of assembly and cooperativity of homomeric and heteromeric metabotropic glutamate receptors. Neuron. 2016;92:143–159. doi: 10.1016/j.neuron.2016.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milligan G. The prevalence, maintenance, and relevance of G protein-coupled receptor oligomerization. Mol. Pharmacol. 2013;84:158–169. doi: 10.1124/mol.113.084780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vischer H.F., Castro M., Pin J.P. G protein-coupled receptor multimers: a question still open despite the use of novel approaches. Mol. Pharmacol. 2015;88:561–571. doi: 10.1124/mol.115.099440. [DOI] [PubMed] [Google Scholar]
- 7.Lambert N.A., Javitch J.A. CrossTalk opposing view: weighing the evidence for class A GPCR dimers, the jury is still out. J. Physiol. 2014;592:2443–2445. doi: 10.1113/jphysiol.2014.272997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouvier M., Hébert T.E. CrossTalk proposal: weighing the evidence for class A GPCR dimers, the evidence favours dimers. J. Physiol. 2014;592:2439–2441. doi: 10.1113/jphysiol.2014.272252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kasai R.S., Kusumi A. Single-molecule imaging revealed dynamic GPCR dimerization. Curr. Opin. Cell Biol. 2014;27:78–86. doi: 10.1016/j.ceb.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 10.Chung I., Akita R., Mellman I. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature. 2010;464:783–787. doi: 10.1038/nature08827. [DOI] [PubMed] [Google Scholar]
- 11.Head B.P., Patel H.H., Insel P.A. Interaction of membrane/lipid rafts with the cytoskeleton: impact on signaling and function: membrane/lipid rafts, mediators of cytoskeletal arrangement and cell signaling. Biochim. Biophys. Acta. 2014;1838:532–545. doi: 10.1016/j.bbamem.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berridge M.J. Module 2: cell signalling pathways. Cell Signal. Biol. 2014 Published online October 1, 2014. [Google Scholar]
- 13.Baumgart F., Schütz G.J. Detecting protein association at the T cell plasma membrane. Biochim. Biophys. Acta. 2015;1853:791–801. doi: 10.1016/j.bbamcr.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 14.Mansoor S.E., Palczewski K., Farrens D.L. Rhodopsin self-associates in asolectin liposomes. Proc. Natl. Acad. Sci. USA. 2006;103:3060–3065. doi: 10.1073/pnas.0511010103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rigaud J.L., Lévy D. Reconstitution of membrane proteins into liposomes. Methods Enzymol. 2003;372:65–86. doi: 10.1016/S0076-6879(03)72004-7. [DOI] [PubMed] [Google Scholar]
- 16.Niu S.L., Doctrow B., Mitchell D.C. Rhodopsin activity varies in proteoliposomes prepared by different techniques. Biochemistry. 2009;48:156–163. doi: 10.1021/bi801835s. [DOI] [PubMed] [Google Scholar]
- 17.Larsen J., Hatzakis N.S., Stamou D. Observation of inhomogeneity in the lipid composition of individual nanoscale liposomes. J. Am. Chem. Soc. 2011;133:10685–10687. doi: 10.1021/ja203984j. [DOI] [PubMed] [Google Scholar]
- 18.Chen X., Araç D., Rizo J. SNARE-mediated lipid mixing depends on the physical state of the vesicles. Biophys. J. 2006;90:2062–2074. doi: 10.1529/biophysj.105.071415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura T., Yeliseev A.A., Gawrisch K. Recombinant cannabinoid type 2 receptor in liposome model activates g protein in response to anionic lipid constituents. J. Biol. Chem. 2012;287:4076–4087. doi: 10.1074/jbc.M111.268425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathiasen S., Christensen S.M., Stamou D. Nanoscale high-content analysis using compositional heterogeneities of single proteoliposomes. Nat. Methods. 2014;11:931–934. doi: 10.1038/nmeth.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howlett A.C., Blume L.C., Dalton G.D. CB(1) cannabinoid receptors and their associated proteins. Curr. Med. Chem. 2010;17:1382–1393. doi: 10.2174/092986710790980023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palczewski K. G protein-coupled receptor rhodopsin. Annu. Rev. Biochem. 2006;75:743–767. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fung J.J., Deupi X., Kobilka B.K. Ligand-regulated oligomerization of β(2)-adrenoceptors in a model lipid bilayer. EMBO J. 2009;28:3315–3328. doi: 10.1038/emboj.2009.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fay J.F., Farrens D.L. Purification of functional CB1 and analysis by site-directed fluorescence labeling methods. Methods Enzymol. 2017;593:343–370. doi: 10.1016/bs.mie.2017.06.026. [DOI] [PubMed] [Google Scholar]
- 25.Xie G., Gross A.K., Oprian D.D. An opsin mutant with increased thermal stability. Biochemistry. 2003;42:1995–2001. doi: 10.1021/bi020611z. [DOI] [PubMed] [Google Scholar]
- 26.Fay J.F., Farrens D.L. A key agonist-induced conformational change in the cannabinoid receptor CB1 is blocked by the allosteric ligand Org 27569. J. Biol. Chem. 2012;287:33873–33882. doi: 10.1074/jbc.M112.352328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ridge K.D., Lu Z., Khorana H.G. Structure and function in rhodopsin. Separation and characterization of the correctly folded and misfolded opsins produced on expression of an opsin mutant gene containing only the native intradiscal cysteine codons. Biochemistry. 1995;34:3261–3267. doi: 10.1021/bi00010a016. [DOI] [PubMed] [Google Scholar]
- 28.Tsukamoto H., Sinha A., Farrens D.L. Monomeric rhodopsin is the minimal functional unit required for arrestin binding. J. Mol. Biol. 2010;399:501–511. doi: 10.1016/j.jmb.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Periasamy A., Wallrabe H., Barroso M. Chapter 22: quantitation of protein-protein interactions: confocal FRET microscopy. Methods Cell Biol. 2008;89:569–598. doi: 10.1016/S0091-679X(08)00622-5. [DOI] [PubMed] [Google Scholar]
- 30.McCann J.J., Choi U.B., Bowen M.E. Optimizing methods to recover absolute FRET efficiency from immobilized single molecules. Biophys. J. 2010;99:961–970. doi: 10.1016/j.bpj.2010.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bendix P.M., Pedersen M.S., Stamou D. Quantification of nano-scale intermembrane contact areas by using fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. USA. 2009;106:12341–12346. doi: 10.1073/pnas.0903052106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stamou D., Duschl C., Vogel H. Self-assembled microarrays of attoliter molecular vessels. Angew. Chem. Int. Ed. Engl. 2003;42:5580–5583. doi: 10.1002/anie.200351866. [DOI] [PubMed] [Google Scholar]
- 33.Lan T.H., Liu Q., Lambert N.A. BRET evidence that β2 adrenergic receptors do not oligomerize in cells. Sci. Rep. 2015;5:10166. doi: 10.1038/srep10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mercier J.F., Salahpour A., Bouvier M. Quantitative assessment of β 1- and β 2-adrenergic receptor homo- and heterodimerization by bioluminescence resonance energy transfer. J. Biol. Chem. 2002;277:44925–44931. doi: 10.1074/jbc.M205767200. [DOI] [PubMed] [Google Scholar]
- 35.Meyer B.H., Segura J.M., Vogel H. FRET imaging reveals that functional neurokinin-1 receptors are monomeric and reside in membrane microdomains of live cells. Proc. Natl. Acad. Sci. USA. 2006;103:2138–2143. doi: 10.1073/pnas.0507686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.James J.R., Oliveira M.I., Davis S.J. A rigorous experimental framework for detecting protein oligomerization using bioluminescence resonance energy transfer. Nat. Methods. 2006;3:1001–1006. doi: 10.1038/nmeth978. [DOI] [PubMed] [Google Scholar]
- 37.Adair B.D., Engelman D.M. Glycophorin A helical transmembrane domains dimerize in phospholipid bilayers: a resonance energy transfer study. Biochemistry. 1994;33:5539–5544. doi: 10.1021/bi00184a024. [DOI] [PubMed] [Google Scholar]
- 38.Veatch W., Stryer L. The dimeric nature of the gramicidin A transmembrane channel: conductance and fluorescence energy transfer studies of hybrid channels. J. Mol. Biol. 1977;113:89–102. doi: 10.1016/0022-2836(77)90042-0. [DOI] [PubMed] [Google Scholar]
- 39.Clegg R.M. Fluorescence resonance energy transfer. Curr. Opin. Biotechnol. 1995;6:103–110. doi: 10.1016/0958-1669(95)80016-6. [DOI] [PubMed] [Google Scholar]
- 40.King C., Raicu V., Hristova K. Understanding the FRET signatures of interacting membrane proteins. J. Biol. Chem. 2017;292:5291–5310. doi: 10.1074/jbc.M116.764282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Angers S., Salahpour A., Bouvier M. Detection of β 2-adrenergic receptor dimerization in living cells using bioluminescence resonance energy transfer (BRET) Proc. Natl. Acad. Sci. USA. 2000;97:3684–3689. doi: 10.1073/pnas.060590697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Calebiro D., Rieken F., Lohse M.J. Single-molecule analysis of fluorescently labeled G-protein-coupled receptors reveals complexes with distinct dynamics and organization. Proc. Natl. Acad. Sci. USA. 2013;110:743–748. doi: 10.1073/pnas.1205798110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fotiadis D., Liang Y., Palczewski K. Atomic-force microscopy: rhodopsin dimers in native disc membranes. Nature. 2003;421:127–128. doi: 10.1038/421127a. [DOI] [PubMed] [Google Scholar]
- 44.Jastrzebska B., Fotiadis D., Palczewski K. Functional and structural characterization of rhodopsin oligomers. J. Biol. Chem. 2006;281:11917–11922. doi: 10.1074/jbc.M600422200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gunkel M., Schöneberg J., Al-Amoudi A. Higher-order architecture of rhodopsin in intact photoreceptors and its implication for phototransduction kinetics. Structure. 2015;23:628–638. doi: 10.1016/j.str.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 46.Wolber P.K., Hudson B.S. An analytic solution to the Förster energy transfer problem in two dimensions. Biophys. J. 1979;28:197–210. doi: 10.1016/S0006-3495(79)85171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raicu V. Efficiency of resonance energy transfer in homo-oligomeric complexes of proteins. J. Biol. Phys. 2007;33:109–127. doi: 10.1007/s10867-007-9046-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patowary S., Pisterzi L.F., Raicu V. Experimental verification of the kinetic theory of FRET using optical microspectroscopy and obligate oligomers. Biophys. J. 2015;108:1613–1622. doi: 10.1016/j.bpj.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Domanov Y.A., Aimon S., Bassereau P. Mobility in geometrically confined membranes. Proc. Natl. Acad. Sci. USA. 2011;108:12605–12610. doi: 10.1073/pnas.1102646108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Irannejad R., Tomshine J.C., von Zastrow M. Conformational biosensors reveal GPCR signalling from endosomes. Nature. 2013;495:534–538. doi: 10.1038/nature12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tonnesen A., Christensen S.M., Stamou D. Geometrical membrane curvature as an allosteric regulator of membrane protein structure and function. Biophys. J. 2014;106:201–209. doi: 10.1016/j.bpj.2013.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Callan-Jones A., Sorre B., Bassereau P. Curvature-driven lipid sorting in biomembranes. Cold Spring Harb. Perspect. Biol. 2011;3:a004648. doi: 10.1101/cshperspect.a004648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosholm K.R., Leijnse N., Stamou D. Membrane curvature regulates ligand-specific membrane sorting of GPCRs in living cells. Nat. Chem. Biol. 2017;13:724–729. doi: 10.1038/nchembio.2372. [DOI] [PubMed] [Google Scholar]
- 54.Prévost C., Zhao H., Bassereau P. IRSp53 senses negative membrane curvature and phase separates along membrane tubules. Nat. Commun. 2015;6:8529. doi: 10.1038/ncomms9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doura A.K., Fleming K.G. Complex interactions at the helix-helix interface stabilize the glycophorin A transmembrane dimer. J. Mol. Biol. 2004;343:1487–1497. doi: 10.1016/j.jmb.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 56.Torney D.C., McConnell H.M. Diffusion-limited reaction rate theory for two-dimensional systems. Proc. Math. Phys. Eng. Sci. 1983;387:147–170. [Google Scholar]
- 57.Alvarez-Curto E., Ward R.J., Milligan G. Ligand regulation of the quaternary organization of cell surface M3 muscarinic acetylcholine receptors analyzed by fluorescence resonance energy transfer (FRET) imaging and homogeneous time-resolved FRET. J. Biol. Chem. 2010;285:23318–23330. doi: 10.1074/jbc.M110.122184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ganguly S., Clayton A.H., Chattopadhyay A. Organization of higher-order oligomers of the serotonin1(A) receptor explored utilizing homo-FRET in live cells. Biophys. J. 2011;100:361–368. doi: 10.1016/j.bpj.2010.12.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kasai R.S., Suzuki K.G., Kusumi A. Full characterization of GPCR monomer-dimer dynamic equilibrium by single molecule imaging. J. Cell Biol. 2011;192:463–480. doi: 10.1083/jcb.201009128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Herrick-Davis K., Grinde E., Mazurkiewicz J.E. Oligomer size of the serotonin 5-hydroxytryptamine 2C (5-HT2C) receptor revealed by fluorescence correlation spectroscopy with photon counting histogram analysis: evidence for homodimers without monomers or tetramers. J. Biol. Chem. 2012;287:23604–23614. doi: 10.1074/jbc.M112.350249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goin J.C., Nathanson N.M. Quantitative analysis of muscarinic acetylcholine receptor homo- and heterodimerization in live cells: regulation of receptor down-regulation by heterodimerization. J. Biol. Chem. 2006;281:5416–5425. doi: 10.1074/jbc.M507476200. [DOI] [PubMed] [Google Scholar]
- 62.Rasmussen S.G., Choi H.J., Kobilka B.K. Structure of a nanobody-stabilized active state of the β(2) adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Manglik A., Kim T.H., Kobilka B.K. Structural insights into the dynamic process of β2-adrenergic receptor signaling. Cell. 2015;161:1101–1111. doi: 10.1016/j.cell.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yano Y., Matsuzaki K. Measurement of thermodynamic parameters for hydrophobic mismatch 1: self-association of a transmembrane helix. Biochemistry. 2006;45:3370–3378. doi: 10.1021/bi0522854. [DOI] [PubMed] [Google Scholar]
- 65.Ianoul A., Grant D.D., Pezacki J.P. Imaging nanometer domains of β-adrenergic receptor complexes on the surface of cardiac myocytes. Nat. Chem. Biol. 2005;1:196–202. doi: 10.1038/nchembio726. [DOI] [PubMed] [Google Scholar]
- 66.Comar W.D., Schubert S.M., Smith A.W. Time-resolved fluorescence spectroscopy measures clustering and mobility of a G protein-coupled receptor opsin in live cell membranes. J. Am. Chem. Soc. 2014;136:8342–8349. doi: 10.1021/ja501948w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scarselli M., Annibale P., Radenovic A. Cell type-specific β2-adrenergic receptor clusters identified using photoactivated localization microscopy are not lipid raft related, but depend on actin cytoskeleton integrity. J. Biol. Chem. 2012;287:16768–16780. doi: 10.1074/jbc.M111.329912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fleming K.G. Standardizing the free energy change of transmembrane helix-helix interactions. J. Mol. Biol. 2002;323:563–571. doi: 10.1016/s0022-2836(02)00920-8. [DOI] [PubMed] [Google Scholar]
- 69.Provasi D., Johnston J.M., Filizola M. Lessons from free energy simulations of δ-opioid receptor homodimers involving the fourth transmembrane helix. Biochemistry. 2010;49:6771–6776. doi: 10.1021/bi100686t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hern J.A., Baig A.H., Birdsall N.J. Formation and dissociation of M1 muscarinic receptor dimers seen by total internal reflection fluorescence imaging of single molecules. Proc. Natl. Acad. Sci. USA. 2010;107:2693–2698. doi: 10.1073/pnas.0907915107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cai X., Bai B., Chen J. Apelin receptor homodimer-oligomers revealed by single-molecule imaging and novel G protein-dependent signaling. Sci. Rep. 2017;7:40335. doi: 10.1038/srep40335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Henne W.M., Boucrot E., McMahon H.T. FCHo proteins are nucleators of clathrin-mediated endocytosis. Science. 2010;328:1281–1284. doi: 10.1126/science.1188462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Antonny B. Mechanisms of membrane curvature sensing. Annu. Rev. Biochem. 2011;80:101–123. doi: 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
- 74.Hatzakis N.S., Bhatia V.K., Stamou D. How curved membranes recruit amphipathic helices and protein anchoring motifs. Nat. Chem. Biol. 2009;5:835–841. doi: 10.1038/nchembio.213. [DOI] [PubMed] [Google Scholar]
- 75.Aimon S., Callan-Jones A., Bassereau P. Membrane shape modulates transmembrane protein distribution. Dev. Cell. 2014;28:212–218. doi: 10.1016/j.devcel.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McMahon H.T., Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2011;12:517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- 77.Krauss M., Haucke V. Shaping membranes for endocytosis. Rev. Physiol. Biochem. Pharmacol. 2009;161:45–66. doi: 10.1007/112_2008_2. [DOI] [PubMed] [Google Scholar]
- 78.Nikolaev V.O., Moshkov A., Gorelik J. β2-adrenergic receptor redistribution in heart failure changes cAMP compartmentation. Science. 2010;327:1653–1657. doi: 10.1126/science.1185988. [DOI] [PubMed] [Google Scholar]
- 79.Ibrahim M., Gorelik J., Terracciano C.M. The structure and function of cardiac t-tubules in health and disease. Proc. Biol. Sci. 2011;278:2714–2723. doi: 10.1098/rspb.2011.0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Soubias O., Gawrisch K. The role of the lipid matrix for structure and function of the GPCR rhodopsin. Biochim. Biophys. Acta. 2012;1818:234–240. doi: 10.1016/j.bbamem.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fay J.F., Dunham T.D., Farrens D.L. Cysteine residues in the human cannabinoid receptor: only C257 and C264 are required for a functional receptor, and steric bulk at C386 impairs antagonist SR141716A binding. Biochemistry. 2005;44:8757–8769. doi: 10.1021/bi0472651. [DOI] [PubMed] [Google Scholar]
- 82.Kunding A.H., Mortensen M.W., Stamou D. A fluorescence-based technique to construct size distributions from single-object measurements: application to the extrusion of lipid vesicles. Biophys. J. 2008;95:1176–1188. doi: 10.1529/biophysj.108.128819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ulbrich M.H., Isacoff E.Y. Subunit counting in membrane-bound proteins. Nat. Methods. 2007;4:319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Press W.H., Flannery B.P., Vetterling W.T. Cambridge University Press; Cambridge, UK: 1989. Numerical Recipes in C: The Art of Scientific Computing. [Google Scholar]
- 85.King C., Sarabipour S., Hristova K. The FRET signatures of noninteracting proteins in membranes: simulations and experiments. Biophys. J. 2014;106:1309–1317. doi: 10.1016/j.bpj.2014.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kenworthy A.K., Edidin M. Distribution of a glycosylphosphatidylinositol-anchored protein at the apical surface of MDCK cells examined at a resolution of <100 A using imaging fluorescence resonance energy transfer. J. Cell Biol. 1998;142:69–84. doi: 10.1083/jcb.142.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marrink S.J., De Vries A.H., Mark A.E. Coarse grained model for semiquantitative lipid simulations. J. Phys. Chem. B. 2004;108:750–760. [Google Scholar]
- 88.Saffman P.G., Delbrück M. Brownian motion in biological membranes. Proc. Natl. Acad. Sci. USA. 1975;72:3111–3113. doi: 10.1073/pnas.72.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Henle M.L., Levine A.J. Hydrodynamics in curved membranes: the effect of geometry on particulate mobility. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2010;81:011905. doi: 10.1103/PhysRevE.81.011905. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.