Abstract
Purpose
To determine the significant genomic alterations in patients with metastatic breast cancer (MBC) and survival outcomes in common genotypes.
Patients and Methods
High-depth next-generation sequencing was performed for 202 genes in tumor and normal DNA from 257 patients with MBC, including 165 with estrogen receptor/progesterone receptor–positive and human epidermal growth factor receptor 2 (HER2 [hormone receptor positive (HR+)])–positive, 32 with HER2-positive, and 60 with triple-negative (estrogen receptor/progesterone receptor–negative and HER2-negative) disease. Kaplan-Meier survival analysis was performed in the discovery set, in patients with breast cancer analyzed in The Cancer Genome Atlas, and in a separate cohort of 98 patients with MBC who underwent clinical genomic testing.
Results
Significantly mutated genes (SMGs) varied by histology and tumor subtype, but TP53 was an SMG in all three subtypes. The most SMGs in patients with HR+ cancer were PIK3CA (32%), TP53 (29%), GATA3 (15%), CDH1 (8%), MAP3K1 (8%), PTEN (5%), TGFBR2 (4%), AKT1 (4%), and MAP2K4 (4%). TP53 mutations were associated with shorter recurrence-free survival (P = .004), progression-free survival (P < .001), and overall survival (P = .003). Furthermore, TP53 status was prognostic among patients with HR+ cancer with PIK3CA mutations. TP53 mutations were associated with poorer overall survival in the 442 patients with HR+ breast cancer analyzed in The Cancer Genome Atlas (P = .042) and in an independent set of 96 patients with HR+ MBC who underwent clinical sequencing (P < .001).
Conclusion
SMGs differ by tumor subtype, but TP53 is significantly mutated in all three breast cancer subtypes. TP53 mutations are associated with poor prognosis in HR+ breast cancer and should be considered in the design and interpretation of precision oncology trials.
INTRODUCTION
There is growing interest in genomic profiling for cancer therapy. Emerging data have shown that the targeting of some of these alterations, such as AKT and HER2 mutations, may have antitumor efficacy.1,2 Most proof-of-principle genomically selected trials are conducted in the metastatic setting, whereas many molecular characterization efforts, such as The Cancer Genome Atlas (TCGA), are performed in operable breast cancer.3 To design and interpret genotype-selected trials effectively, the determination of the genomic profile of patients with metastatic breast cancer (MBC), the frequency of genomic alterations and co-alterations, and the effect of common alterations on prognosis is critical.
We determined the genomic profile of patients with MBC in a prospective study and report the significantly altered genes in various breast cancer subtypes. Furthermore, we report the effect of common genotypes on prognosis in hormone receptor–positive (HR+) breast cancer. We validated the prognostic role of TP53 mutations in two additional HR+ cohorts.
PATIENTS AND METHODS
Patient Selection and Enrollment
Two hundred fifty-seven patients with MBC and an adequate amount of archival tumor tissue underwent next-generation sequencing (NGS) in an institutional review board–approved prospective protocol for genomic profiling (ClinicalTrials.gov identifier: NCT01772771). An additional cohort of 98 patients with HR+ MBC who underwent clinical genomic testing were identified as a validation cohort; patients had undergone testing with FoundationOne (Foundation Medicine, Cambridge, MA), Ion AmpliSeq Comprehensive Cancer Panel (Thermo Fisher Scientific, Waltham, MA), or Oncomine Comprehensive Assay (Thermo Fisher Scientific). These clinical records were reviewed with an institutional review board–approved study with waiver of consent.
Genomic Analysis
Samples were evaluated by hematoxylin and eosin staining and macrodissected. DNA was extracted using QIAamp DNA FFPE Tissue Kit (QIAGEN, Santa Clarita, CA) and quantified by Qubit (Invitrogen, Carlsbad, CA). NGS of 202 genes (T200 platform; Data Supplement) was performed on tumor and normal DNA samples as previously described.4 Assays were performed while blinded to the clinical outcomes. Reporting was done consistent with Reporting Recommendations for Tumor Marker Prognostic Studies guidelines.5 Molecular inversion probe (MIP) arrays were performed as previously described.6,7
ESR1 mutation status was tested using the QX200 Droplet Digital PCR System (Bio-Rad Laboratories, Hercules, CA) with primers to assess the following four ESR1 mutations: Y537C (1980A>G), Y537N (1979T>A), Y537S (1980A>C), and D538G (1983A>G; Data Supplement). Positive and negative controls were included in each run. Samples were run in triplicate, with wild-type and mutant ESR1 controls. Quantitative analysis was performed using QuantaSoft software (Bio-Rad Laboratories).
Bioinformatics Analysis
Comprehensive methods for bioinformatics analysis has been previously published.4 For copy number calls, high amplification and high deletion was defined as an estimated copy number of five and 0.6 on NGS analysis and five and one on MIP analysis. Alterations potentially targetable with approved or investigational therapeutics directly or indirectly (eg, inhibiting downstream signaling) were considered actionable. The actionable genes are designated by asterisks in the Data Supplement. The therapeutic implications of these actionable genes are listed in the Data Supplement.
Statistical Analysis
Categorical variables were summarized in frequency tables. Mutation rates were compared with that observed in TCGA. Discrete Independence Statistic Controlling for Observations with Varying Event Rates (DISCOVER), a statistical test for detecting co-occurrence and mutual exclusivity in cancer genomics data, was used.8 Unlike traditional approaches such as Fisher’s exact test, DISCOVER is based on a null model that takes into account the overall tumor-specific alteration rates when deciding whether alterations co-occur more or less often than expected by chance. Multiple testing was adjusted using false discovery rate (FDR).
Recurrence-free survival (RFS) was calculated from the date of initial breast cancer diagnosis to the date of first local or distant relapse, death, or last follow-up. Progression-free survival (PFS) was calculated from the date of treatment start in the metastatic setting to date of treatment end as a result of progression. Overall survival (OS) was calculated from the date of MBC diagnosis.
RESULTS
Somatic Alterations
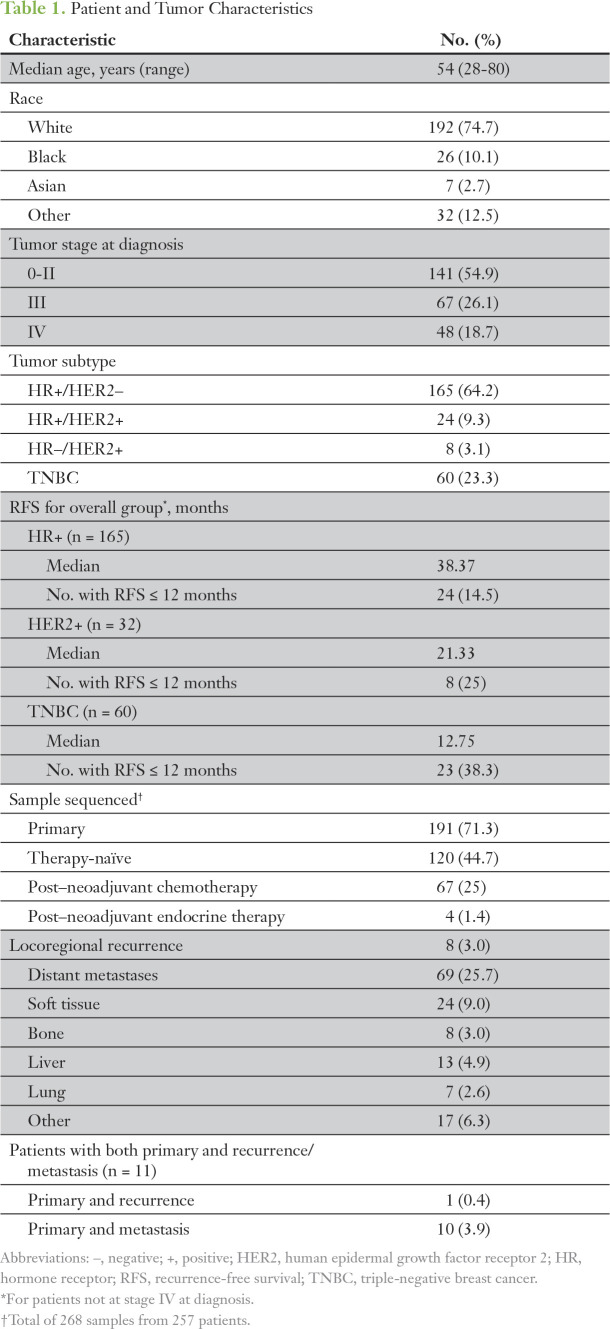
Two hundred sixty-eight samples from 257 patients were sequenced (Table 1). Distribution by tumor subtype was as follows: 165 patients (64.2%) with estrogen receptor/progesterone receptor–positive (ER/PR+) and human epidermal growth factor 2–negative (HER2−) HR+ breast cancer; 60 with triple-negative breast cancer (TNBC); and 32 with HER2+ breast cancer (24 with ER/PR+ and HER2+ and eight with ER/PR– and HER2+). Forty-eight patients (18.7%) had stage IV disease at presentation.
Table 1.
Patient and Tumor Characteristics
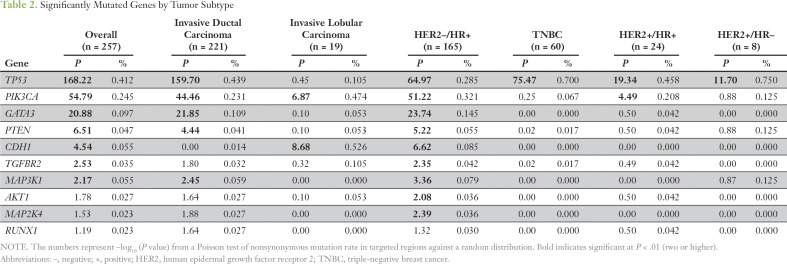
A heat map of the top 50 mutated genes and 50 copy number–altered (CNA) genes are shown in the Data Supplement. Significantly mutated genes (SMGs) varied with histology and tumor subtype (Table 2). TP53 was an SMG in all subtypes but was more frequently mutated in HR–tumors. SMGs in patients with HR+ tumors were PIK3CA (32%), TP53 (29%), GATA3 (15%), CDH1 (8%), MAP3K1 (8%), PTEN (5%), TGFBR2 (4%), AKT1 (4%), and MAP2K4 (4%).
Table 2.
Significantly Mutated Genes by Tumor Subtype
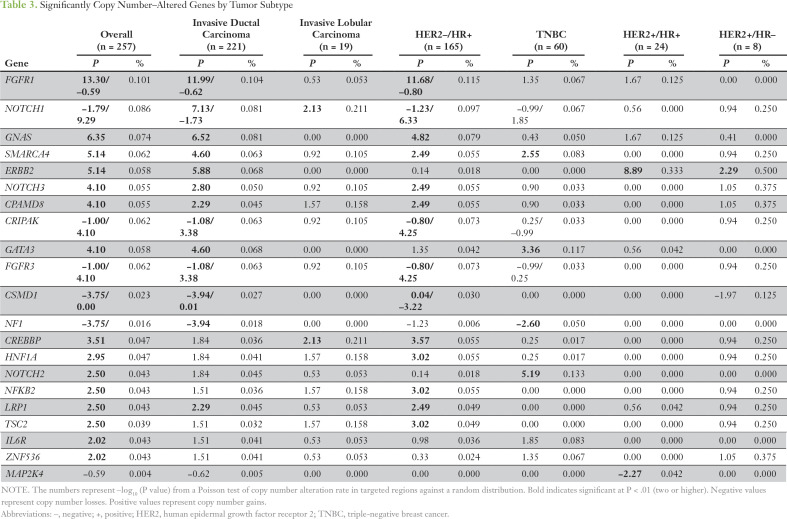
The most significant CNA genes on targeted exome sequencing are listed in Table 3. In TNBC, a gain of NOTCH2, SMARCA4, and GATA3 and a loss of NF1 was observed. In HER2– HR+ breast cancer, a significant gain was seen of FGFR1, GNAS, SMARCA4, CPAMD8, CREBBP, FGFR3, HNF1A, LRP1, and NFKB2 and a loss of CSMD1. We also assessed CNA genes with MIPs in 32 samples from 29 patients by selecting patients with at least one CNA gene on NGS. Of the 36 amplifications detected by NGS, 22 were confirmed by MIP arrays (Data Supplement), including all four patients with FGFR1 amplification, all five with GNAS amplification, three of four with SMARCA4 amplification, and all four with NOTCH2 amplifications. Of 11 deletions detected by NGS, nine were confirmed by MIP arrays, including all three patients with NF1 and all three with PTEN deletions.
Table 3.
Significantly Copy Number–Altered Genes by Tumor Subtype
Alterations in Actionable Genes
Overall, 244 patients (94.9%) had an alteration in at least one potentially actionable gene.9 Of note, mutations differ in their functional consequences; thus, not all mutations may be actionable.9,10 Furthermore, a genomic profile may not be considered actionable because of co-alterations or other patient variables. Actionable alterations included well-recognized alterations, such as PIK3CA mutations (24%) and FGFR amplifications (10%), as well as less-frequent but clinically compelling alterations, such as AKT1 mutations (3%) and HER2 mutations (3%). In addition, there were potentially actionable rarer alterations, such as an inactivating mutation in PTCH1, an activating mutation in IDH1, and high-level amplification of EGFR. Of note, 117 patients (72%) with alterations in an actionable gene had a co-alteration in another potentially actionable gene.
We previously reported that most BRCA1/2 alterations in breast cancer are germline.11,12 However, we observed potentially deleterious somatic alterations in DNA damage repair genes BRCA1/2, PALB2, ATM, and RAD51. Furthermore, we observed alterations in several genes associated with the switch/sucrose nonfermentable complex or other epigenetic processes, including BAP1, ARID1A, DNMT3A, and EP300.
Genomic Alterations in Primary and Recurrent/Metastatic Tumors
Genomic alterations in 191 primary versus 77 recurrent/metastatic tumors were compared (Data Supplement); no significant differences were found by Fisher’s exact test. In addition, we found no differences on the basis of sites of metastases. We had matched primary versus recurrent/metastatic samples from only 11 patients (10 with metastasis and one with locoregional recurrence). All 10 patients who had somatic mutations had additional alterations in their recurrent/metastatic sample not detected in the primary tumor (Data Supplement). Of the HR+ primary tumors, 78 were chemotherapy-naïve, 39 were post–neoadjuvant chemotherapy, and 43 were metastatic samples. No differences were found among these cohorts.
We compared the alterations seen in this study with that in the TCGA breast cohort (Data Supplement). The most common alterations in this series are shown in Fig 1. The MBC cohort was enriched for some alterations, such as TP53 mutations, compared with the TCGA series.
Fig 1.
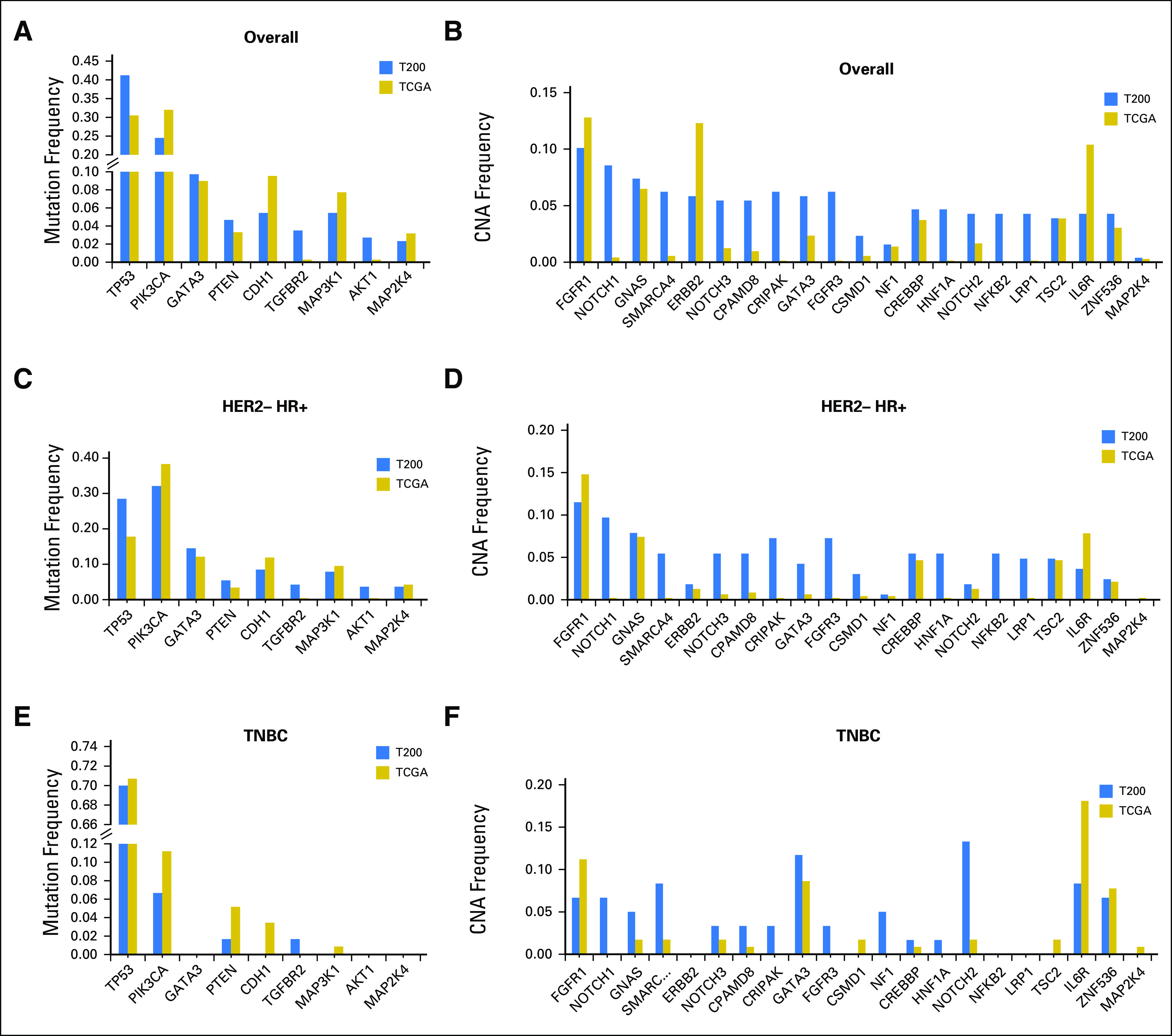
Frequency of the most common alterations in the cohort (T200) versus The Cancer Genome Atlas (TCGA). (A) Overall mutation frequency. (B) Overall copy number alteration (CNA) frequency. (C) Human epidermal growth factor receptor 2–negative (HER2–) hormone receptor–positive (HR+) breast cancer mutation frequency. (D) HER2– HR+ breast cancer CNA frequency. (E) Triple-negative breast cancer (TNBC) mutation frequency. (F) TNBC CNA frequency.
By NGS, 251 of 257 patients had ESR1 sequencing; however, only 151 patients had adequate coverage of ESR1. Of these, 114 were HR+, and only 26 were distant metastasis samples. Only one tumor (4%) had an ESR1 mutation, which was from a patient with HR+ breast cancer who had received letrozole in the metastatic setting. Two other patients with HR+ breast cancer whose primary tumors did not show an ESR1 mutation subsequently had NGS of a new distant metastatic lesion not included in this analysis, and this testing uncovered ESR1 mutations.
Because a 4% ESR1 mutation rate in MBC is lower than what we and others have reported,13,14 we also used droplet digital polymerase chain reaction for four hot spot mutations (ESR1 Y537S, Y537C, Y537N, and D538G) in 49 patients with DNA from metastatic tumor samples.13,14 Thirty-eight patients had endocrine therapy before the biopsy of the metastasis (31 in the adjuvant setting, seven for recurrent disease). Three patients (6.1%) were found to have an ESR1 mutation. Two of these patients also had T200 sequencing, and one was found to have the same ESR1 mutation (D538G), whereas the other, although the same DNA was used, did not have the mutation detected, which suggests that droplet digital polymerase chain reaction may be more sensitive for detection. One patient had four lines of endocrine therapy in the metastatic setting, whereas the other had adjuvant tamoxifen.
Genomic Profile and Prognosis in HR+ Breast Cancer
A heat map and bar plot of the 50 most commonly altered genes in HR+ breast cancer are shown in Fig 2. We tested for the co-occurrence between the top 50 altered genes in HR+ breast cancer using the DISCOVER algorithm. The P and Q values for all the gene pairs are listed in the Data Supplement. With an FDR of 0.1, we found co-alterations in FGFR3 and CRIPAK, which are colocalized on chromosome 4; co-alterations in CPAMD8, SMARCA4, and NOTCH3, which are colocalized on chromosome 19; and co-alterations in CREBBP and TSC2, which are colocalized on chromosome 16. We tested for mutual exclusivity among the top 50 altered genes in HR+ breast cancer (Data Supplement). With an FDR of 0.1, GATA3 alterations were mutually exclusive with TP53 alterations.
Fig 2.
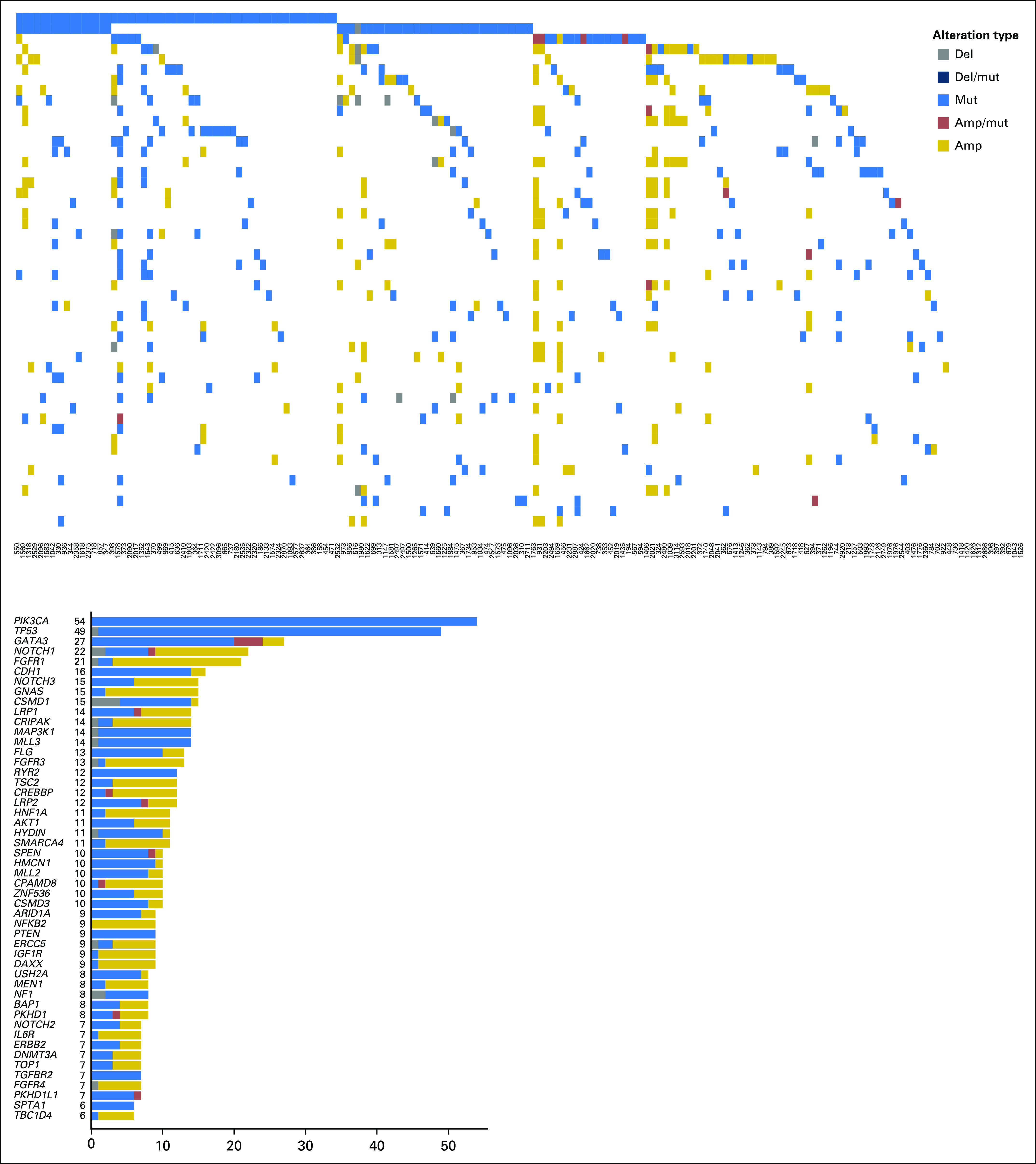
Heat map and bar plot of the alterations in the 50 most commonly altered genes from patients with hormone receptor–positive breast cancer. The samples are presented in order, with the most common alterations on the left. amp, amplification; del, deletion; mut, mutation.
In precision oncology trials, treatment often is given to patients with selected alterations; thus, we assessed the effect of common genomic alterations on PFS in patients with HR+ breast cancer. Of common alterations, the most prominent prognostic effect was attributable to TP53. TP53 mutations were significantly more common in patients with an RFS of ≤ 24 months by Fisher’s exact test (P = .0025). TP53 mutations were associated with a shorter RFS by Kaplan-Meier method (P = .003; Fig 3A). The types and locations of TP53 mutations seen in patients with HR+ breast cancer are depicted in the Data Supplement. Patients with missense TP53 mutations had a longer RFS than those with other types of TP53 mutations, but this difference was not significant (P = .055; Fig 3B).
Fig 3.
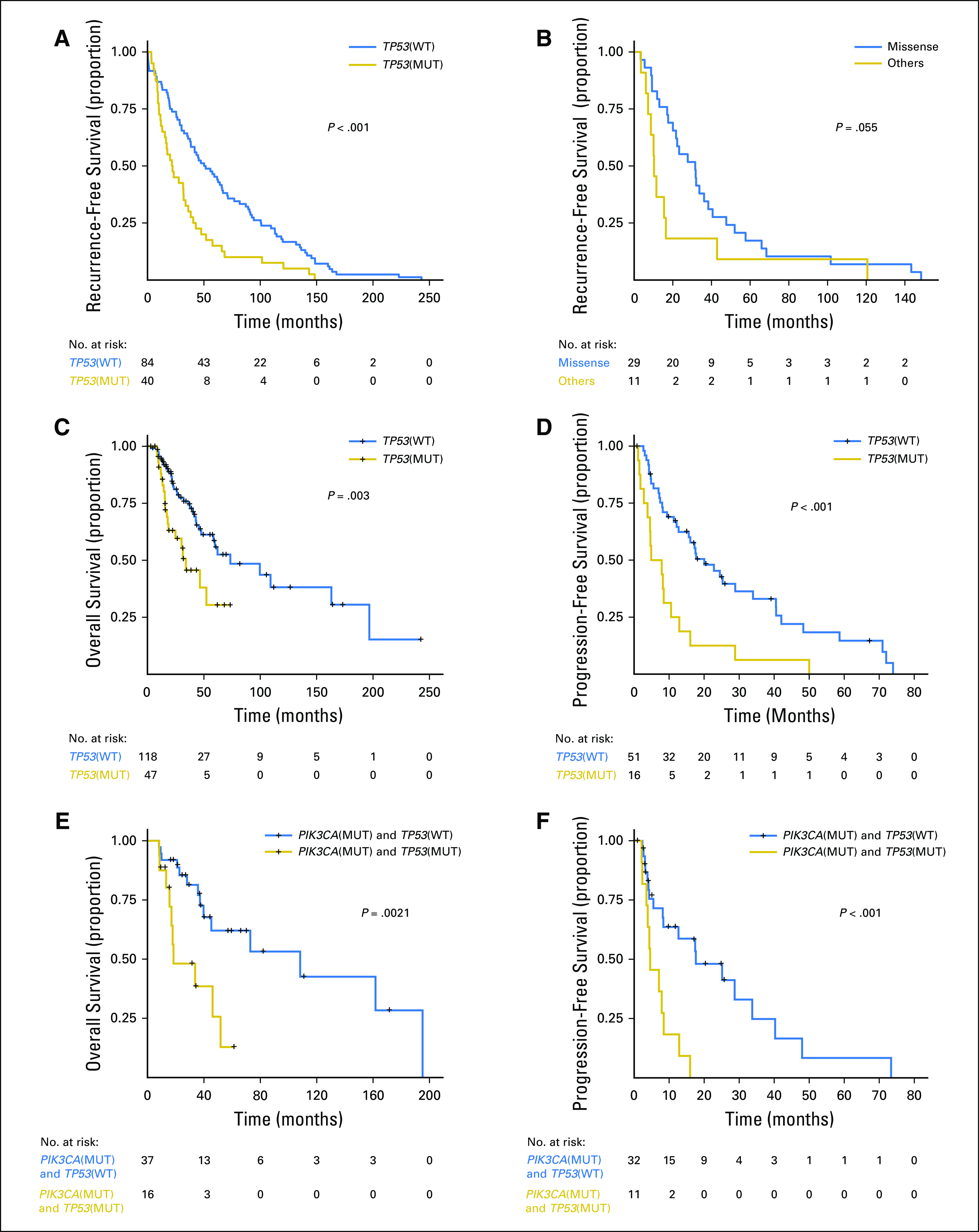
Kaplan-Meier survival analysis for patients with hormone receptor–positive (HR+) metastatic breast cancer (MBC) by TP53 genotype. (A) Recurrence-free survival for patients with HR+ MBC. (B) Recurrence-free survival for patients with HR+ MBC with TP53 mutations by TP53 mutation type (missense v other). (C) Overall survival for patients with HR+ MBC by TP53 mutation status. (D) Progression-free survival on first-line endocrine therapy for patients with HR+ MBC. (E) Overall survival for patients with HR+ PIK3CA-mutant MBC by TP53 mutation status. (F) Progression-free survival for patients with HR+ PIK3CA-mutant MBC by TP53 mutation status. MUT, mutant; WT, wild type.
TP53 mutations were associated with a shorter OS in patients with HR+ breast cancer (P = .003; Fig 3C). TP53 mutations were not associated with survival in patients with TNBC or HER2+ cancer; however, these cohorts were smaller. TP53 mutations were associated with a significantly shorter PFS in patients with HR+ breast cancer who received any first-line metastatic therapy (median, 4.57 v 16.07 months; P < .001; data not shown) as well as in patients who received endocrine therapy only (median, 6.4 v 20.1 months; P < .001; Fig 3D). TP53 mutation type (missense v other) was not associated with OS or PFS on first-line endocrine therapy.
When patients with PIK3CA mutation/amplification, AKT1 mutation/amplification, PTEN mutation/deletion, FGFR1/3 amplifications, GATA3 mutations, or MAP3K1/MAP2K4 mutation/deletion were compared with patients who lack these alterations, no significant difference was found in PFS in the first-line metastatic setting on any therapy (Data Supplement) as well as in patients treated with endocrine therapy (Data Supplement). TP53 mutations were also associated with decreased PFS and OS among patients with HR+ breast cancer with PIK3CA mutations (P < .001 and P = .002, respectively; Figs 3E and 3F).
To validate the prognostic role of TP53, we first, evaluated the 442 patients with HR+ breast cancer in the TCGA. TP53 mutations were associated with a decreased RFS (P = .042; Fig 4A) with a hazard ratio of 2.02 (TP53 mutant v not, 95% CI, 0.997 to 4.095). We next evaluated OS in an independent sample of 96 patients with HR+ MBC who underwent clinical genomic testing and had endocrine therapy as first-line therapy. Patients with TP53 mutations had a significantly shorter survival (median, 56 v 145 months; P < .001; Fig 4B).
Fig 4.
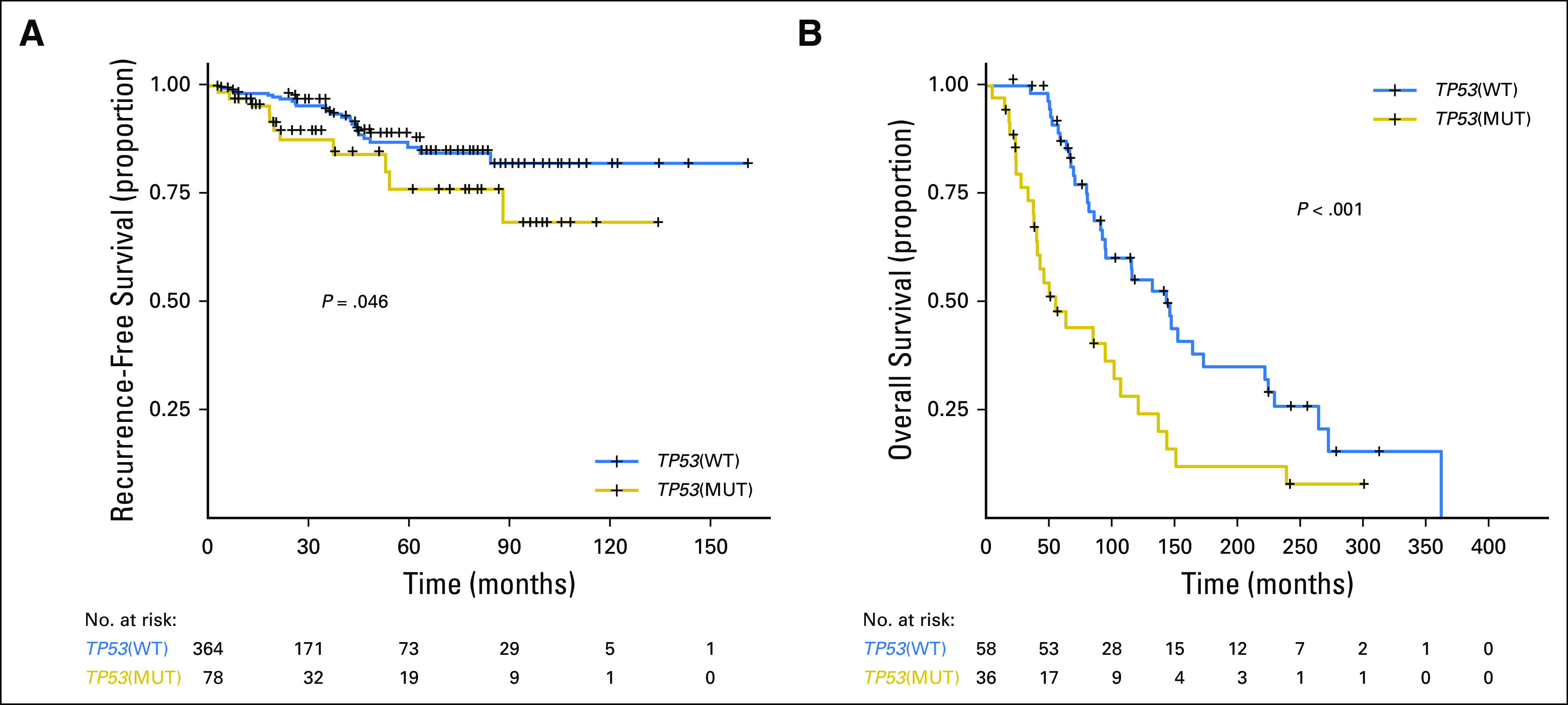
Kaplan-Meier survival analysis for patients with hormone receptor–positive metastatic breast cancer by TP53 genotype in validation cohorts. (A) Recurrence-free survival in The Cancer Genome Atlas by TP53 mutation status. (B) Overall survival in an independent cohort of 98 patients with hormone receptor–positive metastatic breast cancer by TP53 mutation status. MUT, mutant; WT, wild type.
DISCUSSION
TP53 was an SMG in all breast cancer subtypes, but mutations were more frequent in HR–tumors. TP53 mutations were seen in 41% of patients with MBC in our sample compared with 30% in the TCGA, which represents patients with earlier-stage disease (Fig 1) and higher rates of TP53 mutations in HR+ breast cancer (29% v 18%). TP53 mutations are already known to be a harbinger of poor prognosis in breast cancer.15-18 Type and position of mutations may affect cancer outcomes19; this requires additional study. Given the effect of TP53 on prognosis, genotype-selected trials that stratify for TP53 may be considered. In the current study, patients with TP53 mutations had a shorter OS. Furthermore, patients with HR+ tumors with TP53 mutations treated with endocrine therapy in the first-line metastatic setting had a significantly shorter PFS. Of note, when Ellis et al20 compared aromatase inhibitor–sensitive versus aromatase inhibitor–resistant tumors in the neoadjuvant setting (ClinicalTrials.gov identifier: NCT00265759), the TP53 signaling was enriched in resistant tumors (38% of the aromatase inhibitor–resistant and 17% in the sensitive group). The authors concluded that HR+ tumors with TP53 mutations are mostly aromatase inhibitor resistant and would be more appropriately treated with other modalities. However, we do not know whether other regimens would be more effective for these tumors or whether TP53 mutations would equally confer resistance to other agents. However, emerging therapeutics have been targeting mutant p53.21,22 An urgent need exists for novel therapies for TP53 mutant tumors.
In the current study, patients had a variety of genomic alterations. Alterations in the PI3K pathway, including PIK3CA, PTEN, and AKT1 mutations, are already well recognized. The frequency of alterations in this pathway may differ on the basis of patient population (tumor subtype and histology and other variables) as well as the assay and bioinformatics pipeline. In most breast cancer series, this potentially actionable pathway is the most frequently altered; thus, these alterations are being actively pursued in trials with PI3K/AKT/mTOR inhibitors.2,23,24 CDH1 mutations, as expected, were almost exclusively found in invasive lobular carcinoma. CDH1 loss is pathognomonic for lobular carcinomas; that we found CDH1 mutations in only 56% of patients suggests that CDH1 loss also may be mediated through nongenomic mechanisms. Mutations in MAP3K1 and MAP2K4 already have been reported in HR+ breast cancer.20 Ellis et al20 reported a frequency of 15.5% for MAP3K1 and MAP2K4 in ER+ breast cancer. Inactivating mutations in MAP3K1 and MAP2K4 are predicted to abrogate signaling pathways that activate JUN kinases. Therapeutic implications of these alterations have not been well elucidated.
GATA3 mutations are commonly noted in HR+ breast cancer. Ellis et al20 found that GATA3 mutations were enriched in HR+ tumors that exhibited greater neoadjuvant aromatase inhibitor sensitivity in at least one studied cohort. This finding, although preliminary, suggests that GATA3 mutations may be a positive predictive marker for aromatase inhibitor response. In the current study patients with HR+ breast cancer, those with GATA3 mutations trended to have an improved OS, but this difference was not significant (P = .07). The prognostic and predictive value of GATA3 needs to be evaluated further.
Although our NGS platform primarily analyzes commonly mutated genes in cancer, it can provide copy number information.4 Indeed, we identified common CNAs, such as gain in FGFR1 and HER2. We recently reported that when NGS demonstrates high-level amplification, we are able to validate CNAs on an orthogonal platform, such as fluorescent in situ hybridization.25 NGS-based detection of CNAs is limited to high-level losses or gains; thus, we may have underestimated the frequency of copy number changes. However, several of these CNAs, such as NOTCH alterations and NF1 loss, have therapeutic implications and need additional study.
We had too few matched primary and recurrence samples to systematically study genomic evolution in this series. Many patients had primary tumors but not metastatic samples available for profiling. In another study, we reported that in 33 matched primary and recurrent tumors, 97 (87%) of 112 somatic mutations were concordant.26 More recently, Lefebvre et al27 reported the genomic profiling results of patients who underwent a biopsy of MBC in the context of the SAFIR-01, SAFIR-02, SHIVA (Tumor Molecular Profiling Versus Conventional Therapy in Patients With Refractory Cancer), or MOSCATO (Molecular Screening for Cancer Treatment Optimization) prospective trials. A significant overlap exists between the SMG observed in their study and ours. However, in Lefebvre et al, eight genes (ESR1, FSIP2, FRAS1, OSBPL3, EDC4, PALB2, IGFN1, and AGRN) were more frequently mutated in MBC than in early breast cancer profiles in TCGA, which suggests that systematic assessment of metastatic tissue in MBC may lead to identification of additional genomic alterations.
The current study had some additional limitations. The patients were under active treatment for MBC, which represents differing subtypes and receipt of a variety of treatments. Selection bias may have existed in patients chosen for testing. We may not have captured molecular profiles of patients who rapidly progressed while on therapy and succumbed to their disease, or alternately, those who responded well were not perceived as needing molecular characterization. Furthermore, we performed an NGS of a predefined panel of genes. This had the advantage of depth to detect subclonal as well as clonal events but limited our ability to discover novel genomic alterations. Our panel also did not include MDM2 and MDM4, two genes that could be amplified to negatively regulate the TP53 axis in patients with wild-type TP53.
In conclusion, genomic profiling has identified multiple potentially actionable alterations. PIK3CA/TP53 and GATA3 mutations are the most common alterations in HR+ MBC, and TP53 was prognostic in three different HR+ cohorts. The prognostic effect of genotypes should be considered in the design of precision oncology trials. As NGS becomes more commonly used clinically, TP53 may be considered a stratification factor in future randomized trials given the significant effect on outcome. Additional study is needed to determine the role of genomic classification on sensitivity to endocrine therapy given in conjunction with CDK4/6 inhibitors and emerging agents (eg, PI3K pathway inhibitors) in adjuvant endocrine therapy and new endocrine combinations as well as to determine optimal novel therapies that can therapeutically leverage TP53 mutations.
ACKNOWLEDGMENT
We thank Kristin Hargraves and the Khalifa Institute for Personalized Cancer Therapy clinical research team for assistance with data curation and Kurt Evans for technical assistance.
Footnotes
Supported by the Sheikh Khalifa Al Nahyan Ben Zayed Institute for Personalized Cancer Therapy, an AstraZeneca Foundation grant, National Cancer Institute Grant No. U01 CA180964, National Center for Advancing Translational Sciences Grant No. UL1 TR000371 (Center for Clinical and Translational Sciences), the Nellie B. Connally Breast Cancer Research Endowment, Cancer Prevention Research Institute of Texas Precision Oncology Decision Support Core RP150535, the Bosarge Foundation, and MD Anderson Cancer Center support Grant No. P30 CA016672.
AUTHOR CONTRIBUTIONS
Conception and design: Funda Meric-Bernstam, Chetna Wathoo, Lauren Brusco, Jordi Rodon, John Mendelsohn, Kenna R. Mills Shaw, Gordon B. Mills
Financial support: Funda Meric-Bernstam, Lauren Brusco, Tae-Beom Kim, Kenna R. Mills Shaw
Administrative support: Funda Meric-Bernstam, Lauren Brusco, John Mendelsohn, Kenna R. Mills Shaw
Provision of study materials or patients: Funda Meric-Bernstam, Maryam Shariati, Chetna Wathoo, Lauren Brusco, Mehmet Esat Demirhan, Filip Janku, Kenna R. Mills Shaw, Gordon B. Mills
Collection and assembly of data: Funda Meric-Bernstam, Chetna Wathoo, Lauren Brusco, Mehmet Esat Demirhan, Agda Karina Eterovic, Reva K. Basho, Filip Janku, Aysegul Sahin, Jordi Rodon, Russell Broaddus, Ken Chen
Data analysis and interpretation: Funda Meric-Bernstam, Xiaofeng Zheng, Maryam Shariati, Senthil Damodaran, Lauren Brusco, Coya Tapia, Naoto T. Ueno, Russell Broaddus, Tae-Beom Kim, Debu Tripathy, Gordon B. Mills, Ken Chen
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center .
Funda Meric-Bernstam
Honoraria: Dialecta, Sumitomo Group
Consulting or Advisory Role: Genentech, Inflection Biosciences, Pieris Pharmaceuticals, ClearLight Diagnostics, Darwin Health, GRAIL
Research Funding: Novartis, AstraZeneca, Taiho Pharmaceutical, Genentech, Calithera Biosciences, Debiopharm Group, Bayer AG, Aileron Therapeutics, Puma Biotechnology, CytomX Therapeutics, Jounce Therapeutics, Zymeworks, Curis, Pfizer, eFFECTOR Therapeutics
Xiaofeng Zheng
No relationship to disclose
Maryam Shariati
No relationship to disclose
Senthil Damodaran
No relationship to disclose
Chetna Wathoo
No relationship to disclose
Lauren Brusco
Employment: Celgene
Mehmet Esat Demirhan
No relationship to disclose
Coya Tapia
Research Funding: ARMO BioSciences
Travel, Accommodations, Expenses: Incyte
Agda Karina Eterovic
No relationship to disclose
Reva K. Basho
Employment: DaVita (I), Castlewood Treatment Center (I)
Honoraria: Insight Strategy Advisors, Navigant Consulting
Consulting or Advisory Role: Heron Therapeutics, Puma Biotechnology
Naoto T. Ueno
No relationship to disclose
Filip Janku
Stock and Other Ownership Interests: Trovagene
Consulting or Advisory Role: Deciphera, Trovagene, Novartis, Sequenom, Foundation Medicine, Guardant Health, Immunome
Research Funding: Novartis, BioMed Valley Discoveries, Roche, Agios Pharmaceutical, Astellas Pharma, Deciphera, Symphogen, Plexxikon, PIQUR Therapeutics, Fujifilm
Other Relationship: Bio-Rad Laboratories
Aysegul Sahin
No relationship to disclose
Jordi Rodon
Consulting or Advisory Role: Novartis, Eli Lilly, ImClone Systems, SERVIER, Orion, Peptomyc
Research Funding: Novartis, Bayer AG
Russell Broaddus
No relationship to disclose
Tae-Beom Kim
No relationship to disclose
John Mendelsohn
Leadership: Merrimack
Stock and Other Ownership Interests: Merrimack
Patents, Royalties, Other Intellectual Property: Royalty payments from University of California, San Diego
Travel, Accommodations, Expenses: Merck
Kenna R. Mills Shaw
No relationship to disclose
Debu Tripathy
Consulting or Advisory Role: Novartis, Nektar, Pfizer, Puma Biotechnology
Research Funding: Genentech (Inst), Roche (Inst), Novartis (Inst)
Travel, Accommodations, Expenses: Novartis
Gordon B. Mills
Stock and Other Ownership Interests: Catena Pharmaceuticals, ImmunoMet, SignalChem, Spindle Top Ventures, Tarveda
Honoraria: Symphogen, Nuevolution, AstraZeneca, Ionis Pharmaceuticals, Eli Lilly, Novartis, Immunome
Consulting or Advisory Role: AstraZeneca, Catena Pharmaceuticals, Critical Outcome Technologies, ImmunoMET, Ionis, Medimmune, Nuevolution, Pfizer, Precision Medicine, Signalchem Lifesciences, Symphogen, Takeda/Millennium Pharmaceuticals, Tarveda
Research Funding: AbbVie, Adelson Medical Research Foundation, AstraZeneca, Breast Cancer Research Foundation, Critical Outcomes Technology, Horizon Diagnostics, Illumina, Immunomet, Ionis, Karus Therapeutics, Komen Research Foundation, Nanostring, Pfizer, Takeda/Millennium Pharmaceuticals, Tesaro
Patents, Royalties, Other Intellectual Property: Licensed technology HRD assay to Myriad Genetics
Travel, Accommodations, Expenses: AstraZeneca, Symphogen, Ionis Pharmaceuticals, MedImmune, Eli Lilly, Novartis, Immunome, Pfizer
Ken Chen
No relationship to disclose
REFERENCES
- 1.Hyman D, Piha-Paul S, Rodón J, et al. : Neratinib for ERBB2 mutant, HER2 non-amplified, metastatic breast cancer: Preliminary analysis from a multicenter, open-label, multi-histology phase II basket trial. Cancer Research 76:PD5, 2016 [Google Scholar]
- 2.Hyman DM, Smyth LM, Donoghue MTA, et al. : AKT inhibition in solid tumors with AKT1 mutations. J Clin Oncol 35:2251-2259, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Network : Comprehensive molecular portraits of human breast tumours. Nature 490:61-70, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen K, Meric-Bernstam F, Zhao H, et al. : Clinical actionability enhanced through deep targeted sequencing of solid tumors. Clin Chem 61:544-553, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McShane LM, Altman DG, Sauerbrei W, et al. : REporting recommendations for tumour MARKer prognostic studies (REMARK). Br J Cancer 93:387-391, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh RR, Mehrotra M, Chen H, et al. : Comprehensive screening of gene copy number aberrations in formalin-fixed, paraffin-embedded solid tumors using molecular inversion probe-based single-nucleotide polymorphism array. J Mol Diagn 18:676-687, 2016 [DOI] [PubMed] [Google Scholar]
- 7.Chen H, Singh RR, Lu X, et al. : Genome-wide copy number aberrations and HER2 and FGFR1 alterations in primary breast cancer by molecular inversion probe microarray. Oncotarget 8:10845-10857, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canisius S, Martens JW, Wessels LF: A novel independence test for somatic alterations in cancer shows that biology drives mutual exclusivity but chance explains most co-occurrence. Genome Biol 17:261, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meric-Bernstam F, Johnson A, Holla V, et al. : A decision support framework for genomically informed investigational cancer therapy. J Natl Cancer Inst 107:107, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson A, Khotskaya YB, Brusco L, et al. : Clinical use of precision oncology decision support. JCO Precis Oncol doi:10.1200/PO.17.00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez-Angulo AM, Timms KM, Liu S, et al. : Incidence and outcome of BRCA mutations in unselected patients with triple receptor-negative breast cancer. Clin Cancer Res 17:1082-1089, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meric-Bernstam F, Brusco L, Daniels M, et al. : Incidental germline variants in 1000 advanced cancers on a prospective somatic genomic profiling protocol. Ann Oncol 27:795-800, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeselsohn R, Yelensky R, Buchwalter G, et al. : Emergence of constitutively active estrogen receptor-α mutations in pretreated advanced estrogen receptor-positive breast cancer. Clin Cancer Res 20:1757-1767, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schiavon G, Hrebien S, Garcia-Murillas I, et al. : Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med 7:313ra182, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Basho RK, de Melo Gagliato D, Ueno NT, et al. : Clinical outcomes based on multigene profiling in metastatic breast cancer patients. Oncotarget 7:76362-76373, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobes P, Podhorec J, Coufal O, et al. : Influence of mutation type on prognostic and predictive values of TP53 status in primary breast cancer patients. Oncol Rep 32:1695-1702, 2014 [DOI] [PubMed] [Google Scholar]
- 17.Eikesdal HP, Knappskog S, Aas T, et al. : TP53 status predicts long-term survival in locally advanced breast cancer after primary chemotherapy. Acta Oncol 53:1347-1355, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silwal-Pandit L, Vollan HK, Chin SF, et al. : TP53 mutation spectrum in breast cancer is subtype specific and has distinct prognostic relevance. Clin Cancer Res 20:3569-3580, 2014 [DOI] [PubMed] [Google Scholar]
- 19.Petitjean A, Achatz MI, Borresen-Dale AL, et al. : TP53 mutations in human cancers: Functional selection and impact on cancer prognosis and outcomes. Oncogene 26:2157-2165, 2007 [DOI] [PubMed] [Google Scholar]
- 20.Ellis MJ, Ding L, Shen D, et al. : Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 486:353-360, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bauer MR, Joerger AC, Fersht AR: 2-Sulfonylpyrimidines: Mild alkylating agents with anticancer activity toward p53-compromised cells. Proc Natl Acad Sci U S A 113:E5271-E5280, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Synnott NC, Murray A, McGowan PM, et al. : Mutant p53: A novel target for the treatment of patients with triple-negative breast cancer? Int J Cancer 140:234-246, 2017 [DOI] [PubMed] [Google Scholar]
- 23.Kim SB, Dent R, Im SA, et al. : Ipatasertib plus paclitaxel versus placebo plus paclitaxel as first-line therapy for metastatic triple-negative breast cancer (LOTUS): A multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol 18:1360-1372, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baselga J, Im SA, Iwata H, et al. : Buparlisib plus fulvestrant versus placebo plus fulvestrant in postmenopausal, hormone receptor-positive, HER2-negative, advanced breast cancer (BELLE-2): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 18:904-916, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arango NP, Brusco L, Mills Shaw KR, et al. : A feasibility study of returning clinically actionable somatic genomic alterations identified in a research laboratory. Oncotarget 8:41806-41814, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meric-Bernstam F, Frampton GM, Ferrer-Lozano J, et al. : Concordance of genomic alterations between primary and recurrent breast cancer. Mol Cancer Ther 13:1382-1389, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lefebvre C, Bachelot T, Filleron T, et al. : Mutational profile of metastatic breast cancers: A retrospective analysis. PLoS Med 13:e1002201, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]