Abstract
Single-cell transcriptome analysis through next-generation sequencing (single-cell RNA-seq) has been used broadly to address important biological questions. It has proved to be very powerful, and many exciting new biological discoveries have been achieved in the last decade. Its application has greatly improved our understanding of diverse biological processes and the underlying molecular mechanisms, an understanding that would not have been achievable by conventional analysis based on bulk populations. However, so far, single-cell RNA-seq analysis has been used mostly for higher organisms. For microorganisms, single-cell RNA-seq has not been widely used, mainly because the stiff cell wall prevents effective lysis, much less starting RNA material is obtained, and the RNA lacks polyadenylated tails for universal priming of mRNA molecules. In general, the detection efficiency of current single-cell RNA-seq technologies is very low, and further development or improvement of these technologies is required for exploring the microbial world at single-cell resolution. Here, we briefly review recent developments in single-cell RNA-seq of microorganisms and discuss current challenges and future directions.
Main Text
The pioneer work on single-cell transcriptomics was reported as early as 1992 (1), but single-cell next-generation sequencing (RNA-seq) technology was reported for the first time in 2009 (2). Since then, single-cell RNA-seq technology has brought tremendous applications for globally and precisely elucidating the molecular mechanisms and cellular properties of many biological processes, including embryo development, stem cell differentiation, and tumor invasion, at the finest resolution—single cells (3, 4, 5, 6). However, so far, single-cell RNA-seq has been applied mainly to higher organisms and has not been used widely for microorganisms. This is partially because of the small amount of starting RNA material and lack of polyadenylated tails on mRNAs from single microbial cells compared to single cells from higher organisms. The low sensitivity of current single-cell RNA-seq protocols also hinders their applications in microorganisms. Nevertheless, it is of vital importance to investigate the microbial world at single-cell resolution if the technology can be made feasible.
First, many millions of cells are required for conventional bulk population analysis of microorganisms to decipher the molecular mechanisms and underlying biological pathways. However, the average effect caused by such ensemble measurements will be biased, and features of subpopulations or specific patterns will be buried among the whole population of cells. Many studies have demonstrated the existence of cell-to-cell heterogeneity within a whole population of isogenic cells (7, 8, 9). A cell population has to heterogeneously express certain genes to respond and adapt to dramatic changes in environmental conditions. To precisely and accurately dissect the diversity and heterogeneity of such complex microbial systems and provide reasonable explanations of associated biological phenomena, single-cell transcriptome analysis by RNA-seq is necessary.
Second, many traditional bulk-population-analysis approaches require the microbial cells to be cultivated in the laboratory, but so far, only a limited number of microbial species have been cultured successfully. This excludes the possibility of fully profiling hidden species, which have been referred to as “microbial dark matter” (10). Before the development of single-cell RNA-seq, metatranscriptomics was widely used to investigate microbial communities at the population level. But identifying strain-specific heterogeneity in a mixed population is not yet achievable (11, 12), and such ensemble transcriptome analysis is not reflective of individual cells from a whole population. Single-cell RNA-seq, together with metatranscriptomics, could both globally and specifically uncover the complexity of microbial systems.
Third, some microbial cells are very rare and hard to obtain, thus making bulk-input analysis difficult if not impossible. To avoid the need to harvest and purify very rare types of microbial cells from diverse samples, single-cell RNA-seq could be an ideal tool. For example, in replicative aging research of budding yeast Saccharomyces cerevisiae, the replicative lifespan of a single yeast cell was defined as the number of daughter cells it produced before it ceased budding (13). Among a population of yeast cells growing exponentially either on a solid agar plate or in liquid media, the cell mixture will consist of a large proportion of virgin cells and cells budded only once, and the proportion of older cells will decline dramatically. Therefore, it is extremely challenging to efficiently and precisely purify enough aged yeast cells at certain time points from a population of cells with mixed ages. A number of different methods have been developed to address this problem (14, 15, 16, 17, 18), but the quantity and purity of isolated old yeast cells are yet to be satisfactory simultaneously. Single-cell sequencing could resolve this previously unresolved problem. Instead of purifying millions of impure aged cells, a few dozen yeast cells at very specific ages would probably be enough to probe deeply into the molecular mechanisms of aging by single-cell RNA-seq.
Single-cell RNA-seq technology has created a brand-new avenue for modern biological research. Sooner or later, it will be applied to shed light on microbiology research in an unprecedented way. In this short perspective, we briefly summarize the single-cell RNA-seq technologies already being used for microorganisms and discuss current challenges and future directions.
Recent developments in single-cell RNA-seq of microorganisms
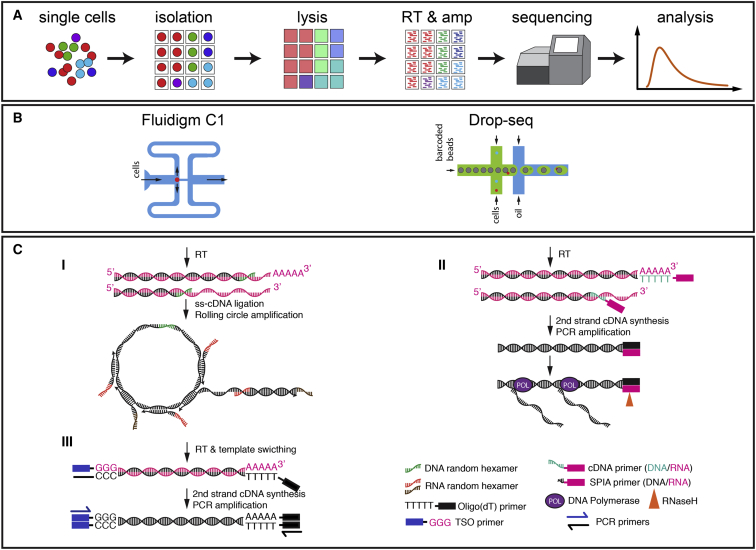
A complete single-cell RNA-seq analysis includes sequential procedures for the isolation of single cells, single-cell lysis, cDNA synthesis and amplification, library preparation and sequencing, and RNA-seq data interpretation. A few recently published review articles cover most of these details (19, 20, 21, 22). Here, we focus mainly on single-cell lysis, cDNA synthesis and amplification, and RNA-seq data interpretation, which are critical to single-cell RNA-seq of microorganisms. Five single-cell transcriptomic or RNA-seq methods that have been used recently in microorganisms are elaborated below: Rolling circle amplification (RCA), RNA-based, single-primer, isothermal amplification (Ribo-SPIA), Smart-seq2, Drop-seq, and Integrated Fluidic Circuit-C1 System (IFC-C1) (Fig. 1; Table 1).
Figure 1.
Workflow, single-cell sequencing platforms, and representative cDNA synthesis and amplification approaches for microorganisms. (A) A summarized pipeline showing the essential steps required to generate a cDNA library for next-generation sequencing is shown. (B) Two popular microfluidic-based sample processing platforms for single-cell sequencing are shown: Fluidigm IFC-C1 and Drop-seq. (C) The procedures used for global amplification of transcripts for RNA-seq of single eukaryote and prokaryote cells are shown (for further details, see text). They are as follows: (I) Rolling circle amplification (RCA); (II) RNA-based, single-primer, isothermal amplification (Ribo-SPIA); (III) Template-switching. To see this figure in color, go online.
Table 1.
Principal Characteristics of Different cDNA Synthesis and Amplification Approaches for Microorganisms
RCA
RCA was the first method to be reported for single-cell transcriptome analysis in prokaryotic cells (23, 24). In the first report in 2011, single bacterium Burkholderia thailandensis was lysed, then 5′-phosphate-dependent exonuclease was applied to remove rRNA and tRNA molecules for the enrichment of mRNA. The cDNA synthesis was performed with the Moloney murine leukemia virus reverse transcriptase and random DNA hexamers. After digestion of chromosomal DNA by McrBC and DpnI enzymes, single-strand cDNA molecules were self-ligated using ssDNA ligase, followed by RCA using φ29 polymerase and random primers. By this method, 20–35 μg cDNA products were obtained from picograms of starting RNA. Subsequent microarray analysis showed low bias and less than 6% dropouts with no contamination, which makes this approach suitable for prokaryotes. It would be interesting to see the RCA results from sequencing instead of microarrays.
Ribo-SPIA
Ribo-SPIA was used for both the first single-cell transcriptome analysis of eukaryotic microbial cells and the first single-cell RNA-seq analysis of prokaryotic cells (25, 26). Ribo-SPIA is a linear and isothermal amplification method that uses DNA/RNA chimeric primers (27). Initially, first-strand cDNA is synthesized by reverse transcription using a unique first-strand DNA/RNA chimeric primer mix. The primers have a 3′ DNA portion that hybridizes to either the polyadenylated tails or to random sequences in the transcripts. The 5′ portion of the primers is a unique RNA sequence that is not complementary to the RNA molecules of the target cell. After first-strand cDNA synthesis, an RNA-dependent DNA polymerase is added, and second-strand cDNA synthesis is carried out till the 5′ RNA portion of the chimeric primer and the double-strand cDNA product with a unique DNA/RNA heteroduplex at one end is formed. Subsequently, the SPIA-based DNA amplification is applied by adding an SPIA DNA/RNA chimeric primer, DNA polymerase with strand-displacement activity, and RNase H. The 5′ RNA portion in the DNA/RNA heteroduplex is cleaved by RNase H, allowing the SPIA DNA/RNA chimeric primer to anneal for further DNA synthesis. The amplification reaction cycles through RNA cleavage, SPIA DNA/RNA chimeric primer binding, and DNA replication and strand displacement, which generates multiple copies of cDNA products with sequence complementary to the original RNA.
In the first study of single-cell transcriptome analysis of eukaryotic microbial cells in 2011, Ribo-SPIA was applied for cDNA synthesis and amplification of single Aspergillus niger cells starting with only ∼1 picogram of RNA per cell (25). The commercial WT-Ovation One-Direct RNA Amplification System (NuGEN, San Carlos, CA) with both oligo(deoxythymidine (dT)) and random primers was used for cDNA synthesis. However, the use of microarray analysis instead of RNA-seq resulted in only ∼4–7% gene coverage of the A. niger genome.
In the first study of single-cell RNA-seq analysis of prokaryotic cells in 2015, Ribo-SPIA was reported to be very powerful, with the expression of up to 98% of all putative genes being detected from single cells of cyanobacterium Synechocystis starting with only 5–7 femtograms of total RNA (26). Such high capture efficiency is surprising, because empirically this would exceed even the typical coverage from bulk RNA-seq experiments using millions of cells. For example, an earlier report of RNA-seq in a bulk population of Salmonella captured only 63–74% of all genes (28). A single-cell RNA-seq study of malaria reported a capture efficiency of only 33% of all genes using Smart-seq2, and a single-cell RNA-seq study of S. cerevisiae also reported a similar but reasonable capture efficiency using an IFC-C1 system (29, 30). However, all of these results were reported by different laboratories using different microbial species and different approaches. A simple comparison without careful dissection may not be enough to draw a final conclusion for the moment.
Smart-seq2
Smart-seq2 uses a template switching mechanism called SMART (switching mechanism at the 5′ end of the RNA transcript) and is a modified version of Smart-seq (31, 32). This method relies on the nontemplate terminal elongation activity of Moloney murine leukemia virus reverse transcriptase. Initially mRNA is primed with a modified oligo-dT primer carrying a specific anchor sequence. Next, reverse transcription starts and proceeds along the RNA template. When the reverse transcriptase reaches the 5′ end of the RNA template, two to five cytosines are added to the 3′ end of the newly synthesized cDNA strand. Then, a universal template molecule (template-switching oligonucleotide (TSO)) with two riboguanosines and one locked nucleic acid (LNA)-modified guanosine at its 3′ end are bound to the newly synthesized cytosines on the cDNA so that the reverse transcriptase can switch to the TSO template and add a universal sequence to the end of the cDNA (33). Subsequently, all cDNA molecules are amplified by polymerase chain reaction (PCR) using high-fidelity DNA polymerase.
Recently, Smart-seq2 with minor modifications was applied for the first single-cell RNA-seq analysis of unicellular eukaryotic microbial cells to investigate transcriptional variation across individual malaria parasites during their asexual growth (29). An average of 1981 genes per cell was detected, representing 33% of the total genes in the malaria genome. A discontinuous program of transcription during asexual growth, which was previously masked by bulk analysis, and novel variations in the expression of gene families among sexual stage parasites, which were important in host-parasite interactions, were uncovered by the single-cell RNA-seq analysis of malaria.
Drop-seq
Drop-seq, along with a few other similar approaches, uses emulsion droplets with nanoliter volumes generated by a sophisticated microfluidic system in which single-cell lysis and cDNA synthesis are carried out. Drop-seq requires a cell suspension and co-encapsulates each single cell with a DNA-barcoded bead in nanoliter-scale droplets. On the beads, there are tremendous oligonucleotides that contain a common sequence for subsequent PCR amplification, followed by a cell barcode, a unique molecular index, and an oligo-dT sequence to capture mRNA molecules. The cell barcode is shared by all the oligonucleotides on the same bead, whereas the unique molecular indices are different among all the oligonucleotides on each bead. After being isolated in droplets, the single cells are first lysed, and then the polyadenosine (poly(A)) tails of the mRNAs are hybridized to the oligo-dT tails on the beads, forming single-cell transcriptomes attached to microparticles. Next, the droplets are broken, and reverse transcription is performed in a single tube. The single-strand cDNA is then converted to double-strand cDNA, which is amplified for library preparation. However, only the 3′ terminal fragments can be used for sequencing. Drop-seq allows the preparation of thousands or more single-cell libraries per day at a very low cost for library construction per cell, which is ∼100-fold cheaper than the former methods. However, the capture efficiency was reported to be around 10–20%, which means only transcripts with more than 20 copies per cell were reliably detected (34).
Despite this, Drop-seq has been applied successfully to study malaria parasites at single-cell resolution (35). In contrast to the previous work on single-cell RNA-seq of malaria using Smart-seq2 (29), only 200–800 genes were detected per cell due to the low capture efficiency of Drop-seq. However, by analyzing more than 18,000 single parasite transcriptomes, the signature of sexual commitment during the proceeding cell cycle of malaria parasites was revealed. This indicates that increasing the number of cells that are analyzed can counteract the negative effect of low capture efficiency of single-cell RNA-seq analysis, at least to some extent.
Integrated microfluidics
Integrated microfluidic devices, such as the IFC-C1 manufactured by Fluidigm (San Francisco, CA), allow the automated capture of single cells. Using IFC-C1, single cells are immobilized in specifically designed microchambers on IFCs and sequentially imaged and lysed. Then, the RNA molecules are converted to cDNA, which is amplified by PCRs based on TSO-mediated priming. The advantage of using IFC-C1 is all reactions are carried out in a completely automated way and in nanoliter-scale volumes. This brings beneficial effects regarding reaction efficiency and saving of reagents. Recently, a single-cell RNA-seq study of S. cerevisiae was performed using IFC-C1 (30). Among the 163 single cells that were analyzed, a median of 2351 genes per cell was detected, with 5667 transcripts covered by at least five reads in more than 5% of the single cells. Both intrinsic and extrinsic regulatory heterogeneities were revealed in the response of the yeast to salt stress. This also implies that, because of the low detection efficiency of current single-cell RNA-seq, typically several hundreds of single cells need to be analyzed to elucidate the biological phenomena or molecular mechanisms of individual cells within isogenic populations. A recent study that compared single-cell RNA-seq with single-molecule RNA fluorescence in situ hybridization (FISH) provided more insights into characterizing the effect of number of cells to be analyzed and transcriptome coverage on the accuracy of gene expression estimations (36).
Current challenges and future directions
Although a few studies on single-cell RNA-seq of microorganisms have been reported using the methods described above, several technical challenges remain. Single-cell lysis and cDNA synthesis and amplification are essential steps for single-cell RNA-seq, and they are particularly challenging when dealing with microorganisms. Unlike higher organisms, microorganisms generally have a rigid cell wall that can impede lysis (37, 38). Whereas a single cell from higher organisms usually contains picograms of mRNA, a single microbial cell usually contains only subpicograms or even femtograms of mRNA (23, 24, 25, 26, 31). Further, the mRNA molecules in microorganisms often lack polyadenylated tails, so suitable approaches for cDNA synthesis and amplification need to be selected (39). Moreover, because of the low detection efficiency of current single-cell RNA-seq methods, the interpretation of microbial single-cell RNA-seq data needs more efforts.
How to effectively lyse a single microbial cell?
In general, there are three types of lysis strategies—chemical, enzymatic, and physical—and each method has its strengths and weaknesses (40, 41, 42). A few studies have reported optimizing lysis protocols for microorganisms, including using various concentrations of sodium dodecyl sulfate and Triton X-100 coupled with different heating regimes and using different commercial lysis kits (37, 38). These studies were targeted at microbial cell lysis for experiments using bulk samples, and the effects of different lysis protocols on single microbial cells remain unclear. In the first report of single-cell microbial transcriptomic analysis (by microarray, not RNA-seq), Triton X-100 and lysozyme were used to lyse the single bacterium B. thailandensis (23, 24). In the first bacterial single-cell RNA-seq analysis, a commercial RNA isolation kit (RNA MicroPrep; ZYMO, Irvine, CA) was used to lyse single Synechocystis cells (26). In the first single eukaryotic microbial cell transcriptomic analysis (by microarray, not RNA-seq), chemical reagents combined with snap freezing and mechanical force were used to lyse single A. niger cells (25). In a recent unicellular eukaryotic microbial RNA-seq analysis, snap freezing and enzymatic treatment were used to lyse single S. cerevisiae cells (30). However, although harsh lysis protocols can effectively lyse a single microbial cell, such treatment may lead to severe degradation of the RNA. Therefore, there has to be a tradeoff between effective single microbial cell lysis and minimal RNA degradation. In future studies, different lysis protocols need to be carefully evaluated and optimized at the single-cell level for each targeted microbial species.
Buffer compatibility also should be considered for the combination of protocols for single microbial cell lysis and downstream cDNA synthesis, especially when there is a template purification step between the procedures. Because most commercial reverse transcriptases are sensitive to detergent, high salt, or EDTA, we suggest that careful control of the chemical lysis conditions is essential. For example, detergents are commonly used in cell lysis protocols, but SuperScript II reverse transcriptase cannot tolerate more than 0.0025% (w/v) sodium dodecyl sulfate. Enzymatic or physical lysis may be better tolerated by downstream cDNA synthesis and amplification procedures. When the reagents for single-cell lysis and cDNA synthesis and amplification are incompatible, gel embedment may be an alternative option for the isolation of single cells (43). Actually, it is advantageous that a hydrogel-based virtual system can compartmentalize templates and reaction products by hydrogel-limited diffusion. Moreover, the physical characteristics of the hydrogel environment may even enhance the coverage of microbial single-cell RNA-seq.
How to efficiently synthesize and amplify cDNA from ultralow amounts of starting mRNA molecules from a single microbial cell?
Besides effective lysis of a single microbial cell, efficient cDNA synthesis and amplification is also important because only subpicogram or even femtogram amounts of mRNA exist in a single microbial cell. The cDNA synthesis and amplification with ultralow mRNA input is very sensitive to contamination, and the low detection efficiency is currently a bottleneck in the application of single-cell RNA-seq of microorganisms. In the past decade, tremendous progress on single-cell transcriptome analysis has been made, especially for increasing the throughput and lowering the cost, but the detection efficiency has been improved only moderately (2, 3, 20, 31, 32, 33, 34, 44, 45, 46, 47, 48, 49, 50). Among the few studies on single-cell transcriptome analysis in microorganisms that have been reported recently, the detection efficiency was mostly less than 10% (23, 24, 25, 26, 29, 30, 35), implying that only transcripts with more than 10 copies per cell can be reliably detected. This excludes the possibility of using single-cell RNA-seq in the majority of microorganisms, including Escherichia coli, because most genes have fewer than 10 transcripts per cell (51).
The current methods of cDNA synthesis and amplification need to be improved dramatically to reliably capture transcripts with very low copy numbers in a single microbial cell (52, 53). A recent study compared six different single-cell RNA-seq methods and found that Smart-seq2 had the highest sensitivity and accuracy (54). This suggests that single-cell RNA-seq of microorganisms may be feasible with better detection efficiency based on Smart-seq2 or other similar methods, if they are modified properly. Before this is achieved, previously established approaches (51, 55, 56) such as single-molecule imaging may provide a more efficient way of describing the stochastic expression dynamics of certain genes in most single microbial cells compared to RNA-seq or other multiplexed approaches. Another approach is the development of in situ methods. Indeed, single-molecule FISH, together with imaging-based methods, enabled single-cell analysis of a small number of genes (57), and was far more efficient than current single-cell RNA-seq methods (up to 95%). With the recent progress in FISH-seq and in situ sequencing technologies, it would be sufficiently multiplexed to cover a whole microbial transcriptome (58, 59). Such spatially resolved approaches also provide additional information, orthogonal to conventional sequencing, and make single-cell expression analysis more informative than spatially nonresolved methods.
How to deal with the nonpolyadenylated mRNA in microbial cells?
Most methods that have been developed for single-cell RNA-seq are based on the 3′ polyadenylated sequences that are ubiquitously present in mRNA molecules of higher organisms (2, 3, 31, 32, 34, 44, 45, 46, 47, 48, 49, 50). However, the mRNA molecules from most prokaryotic cells do not have polyadenylated tails. Therefore, these methods need to be modified before they can be applied for single-cell RNA-seq of microorganisms. Random primers for synthesizing cDNA can be used, but solely using random primers can lead to the inclusion of rRNA and tRNA in the resulting transcriptome library, which will subsequently compromise the overall coverage of mRNA. In addition, in cases in which a mammalian host cell harbors a pathogenic bacterium, elaborately designed primers are required for both polyadenylated and nonpolyadenylated RNA reverse transcription and further cDNA synthesis so that the mutual interaction of host and intracellular pathogen can be studied at single-cell resolution (60). A method called single-cell universal poly(A)-independent RNA sequencing (SUPeR-seq), which was initially designed to analyze linear and circular RNAs in mouse preimplantation embryos, was applied to sequence both polyadenylated and nonpolyadenylated RNAs (61). In this method, a primer containing an adaptor sequence (AdaptorX), a 15-mer dT sequence, and a 6-mer random sequence was used to reverse transcribe both polyadenylated and nonpolyadenylated RNA molecules into first-strand cDNA. After adding a poly(A) tail to first-strand cDNA, another primer containing a different adaptor sequence (AdaptorY) and a 24-mer dT sequence was used for second-strand cDNA synthesis. Then, the double-strand cDNA was amplified by PCR using the AdaptorX and AdaptorY primers. SUPeR-seq has the potential to be applied for studies of single prokaryotic microorganisms and host-pathogen interactions when coupled with the rRNA and tRNA depletion method mentioned above (see RCA) (23, 24, 61). Interestingly, the SUPeR-seq protocol showed low efficiency in amplifying rRNA, offering the possibility of omitting the depletion step.
How to interpret microbial single-cell RNA-seq data?
The interpretation of microbial single-cell RNA-seq data is challenging because of the ultralow amount of starting mRNA from a single microbial cell and the low detection efficiency of the currently available methods. Recent reports have demonstrated that single-cell RNA-seq experiments using different methods, operated by different persons, carried out on different instruments, and even conducted on different days can cause batch effects that confound the interpretation of single-cell RNA-seq data (62, 63). Therefore, a careful and elaborate experimental design—including the choice of single-cell RNA-seq methods, the number of single cells to be sequenced, the sequencing depth, and even some specific workflow such as distributing certain different type of cells into different batches of experiments—is crucial to eliminate or reduce the batch effect (20). Due to the low detection efficiency of single-cell RNA-seq methods and ultralow amount of starting mRNA of single microbial cell, it may require more cycles of PCR for cDNA amplification, but this will result in more amplification bias that distorts the accuracy of RNA-seq data. Technical noise is also inevitably generated during the whole process of single-cell RNA-seq, which would further worsen the precision of RNA-seq data. So, aside from a beautiful experimental design, the data obtained by single-cell RNA-seq of microorganism need to be processed and quality filtered to reduce technical noise before downstream analysis. Single-cell RNA-seq data of microorganisms have zero read counts for many more genes than those of higher organisms or bulk RNA-seq data mainly due to the drop out of many low-expressed genes. Thus, appropriate normalization of the RNA-seq data is also very important, especially for microorganisms, to avoid introducing artifacts that bias downstream data analysis (19, 20). Further, the definition of differentially expressed genes within or between individual microbial cells should be carefully considered in the context of unavoidable technical noise, because in single-cell RNA-seq data of microbial cells, technical noise may prevail over and even bury real biological signals. To eliminate the negative effect of technical noise, it is recommended that spike-in RNAs, such as the External RNA Controls Consortium (64), are added into each sample as a control. And with the help of bioinformatics, the technical noise would probably be deconvolved based on mathematical and statistical model analysis (65, 66), and subsequently the inherent biological variation would be inferred. Notably, increasing the number of single microbial cells to be sequenced can also help to identify differentially expressed genes as well as to explore the heterogeneity of whole populations, at least to some extent (53). However, the current detection efficiency of single-cell RNA-seq needs to be greatly improved before more reliable interpretations of microbial single-cell RNA-seq data become possible (36).
Conclusions
During the past decade, single-cell RNA-seq of many higher organisms has demonstrated the power of studying the transcriptome landscape at single-cell resolution. Many innovative studies using single-cell RNA-seq in higher organisms have been boosted by the fast development of new technologies. However, because of current technical challenges, these single-cell RNA-seq technologies are not yet widely applied to microorganisms. Note that microorganisms are the most abundant and diverse living systems and play vital roles in the entire ecosystem of Earth; we hope that this short perspective will attract more attention and effort to the field of single-cell RNA-seq of microorganisms. We believe that in the near future, the development of new methods will generate an unprecedentedly huge amount of data from single-cell RNA-seq analysis of microorganisms. This will have a profound impact on understanding the fundamental unit of microbial life and lay the foundation for both basic research and clinical science that ultimately will benefit human health.
Acknowledgments
The authors thank all members of the J.W. and Y.H. groups for helpful comments.
The work was supported by the Ministry of Science and Technology of China (2016YFC0900100 to J.W.), the National Natural Science Foundation of China (21675098 to J.W.; 21327808 and 21525521 to Y.H.), the Beijing Advanced Innovation Center for Genomics, and the State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences.
Editor: Antoine van Oijen.
Footnotes
Yi Zhang and Jiaxin Gao contributed equally to this work.
Contributor Information
Yanyi Huang, Email: yanyi@pku.edu.cn.
Jianbin Wang, Email: jianbinwang@mail.tsinghua.edu.cn.
References
- 1.Eberwine J., Yeh H., Coleman P. Analysis of gene expression in single live neurons. Proc. Natl. Acad. Sci. USA. 1992;89:3010–3014. doi: 10.1073/pnas.89.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang F., Barbacioru C., Surani M.A. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 3.Tang F., Barbacioru C., Surani M.A. RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat. Protoc. 2010;5:516–535. doi: 10.1038/nprot.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan L., Yang M., Tang F. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- 5.Tirosh I., Venteicher A.S., Suvà M.L. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539:309–313. doi: 10.1038/nature20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng C., Zheng L., Zhang Z. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169:1342–1356.e16. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 7.Lidstrom M.E., Meldrum D.R. Life-on-a-chip. Nat. Rev. Microbiol. 2003;1:158–164. doi: 10.1038/nrmicro755. [DOI] [PubMed] [Google Scholar]
- 8.Davis K.M., Isberg R.R. Defining heterogeneity within bacterial populations via single cell approaches. BioEssays. 2016;38:782–790. doi: 10.1002/bies.201500121. [DOI] [PubMed] [Google Scholar]
- 9.Qi Z., Pei G., Zhang W. Single-cell analysis reveals gene-expression heterogeneity in syntrophic dual-culture of Desulfovibrio vulgaris with Methanosarcina barkeri. Sci. Rep. 2014;4:7478. doi: 10.1038/srep07478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rinke C., Schwientek P., Woyke T. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 11.Yoon H.S., Price D.C., Bhattacharya D. Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science. 2011;332:714–717. doi: 10.1126/science.1203163. [DOI] [PubMed] [Google Scholar]
- 12.Aylward F.O., Eppley J.M., DeLong E.F. Microbial community transcriptional networks are conserved in three domains at ocean basin scales. Proc. Natl. Acad. Sci. USA. 2015;112:5443–5448. doi: 10.1073/pnas.1502883112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mortimer R.K., Johnston J.R. Life span of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- 14.Lindstrom D.L., Gottschling D.E. The mother enrichment program: a genetic system for facile replicative life span analysis in Saccharomyces cerevisiae. Genetics. 2009;183:413–422. doi: 10.1534/genetics.109.106229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie Z., Zhang Y., Li H. Molecular phenotyping of aging in single yeast cells using a novel microfluidic device. Aging Cell. 2012;11:599–606. doi: 10.1111/j.1474-9726.2012.00821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee S.S., Avalos Vizcarra I., Heinemann M. Whole lifespan microscopic observation of budding yeast aging through a microfluidic dissection platform. Proc. Natl. Acad. Sci. USA. 2012;109:4916–4920. doi: 10.1073/pnas.1113505109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y., Luo C., Li H. Single cell analysis of yeast replicative aging using a new generation of microfluidic device. PLoS ONE. 2012;7:e48275. doi: 10.1371/journal.pone.0048275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jo M.C., Liu W., Qin L. High-throughput analysis of yeast replicative aging using a microfluidic system. Proc. Natl. Acad. Sci. USA. 2015;112:9364–9369. doi: 10.1073/pnas.1510328112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu A.R., Wang J., Huang Y. Single-Cell transcriptional analysis. Annu. Rev. Anal. Chem. (Palo Alto, Calif.) 2017;10:439–462. doi: 10.1146/annurev-anchem-061516-045228. [DOI] [PubMed] [Google Scholar]
- 20.Grün D., van Oudenaarden A. Design and analysis of single-cell sequencing experiments. Cell. 2015;163:799–810. doi: 10.1016/j.cell.2015.10.039. [DOI] [PubMed] [Google Scholar]
- 21.Saliba A.E., Westermann A.J., Vogel J. Single-cell RNA-seq: advances and future challenges. Nucleic Acids Res. 2014;42:8845–8860. doi: 10.1093/nar/gku555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Z., Chen L., Zhang W. Tools for genomic and transcriptomic analysis of microbes at single-cell level. Front. Microbiol. 2017;8:1831. doi: 10.3389/fmicb.2017.01831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang Y., Norris M.H., Hoang T.T. Transcript amplification from single bacterium for transcriptome analysis. Genome Res. 2011;21:925–935. doi: 10.1101/gr.116103.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang Y., McMillan I., Hoang T.T. Single prokaryotic cell isolation and total transcript amplification protocol for transcriptomic analysis. Nat. Protoc. 2015;10:974–984. doi: 10.1038/nprot.2015.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Bekker C., Bruning O., Wösten H.A. Single cell transcriptomics of neighboring hyphae of Aspergillus niger. Genome Biol. 2011;12:R71. doi: 10.1186/gb-2011-12-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J., Chen L., Zhang W. RNA-seq based transcriptomic analysis of single bacterial cells. Integr. Biol (Camb.) 2015;7:1466–1476. doi: 10.1039/c5ib00191a. [DOI] [PubMed] [Google Scholar]
- 27.Kurn N., Chen P., Wang S. Novel isothermal, linear nucleic acid amplification systems for highly multiplexed applications. Clin. Chem. 2005;51:1973–1981. doi: 10.1373/clinchem.2005.053694. [DOI] [PubMed] [Google Scholar]
- 28.Kröger C., Colgan A., Hinton J.C. An infection-relevant transcriptomic compendium for Salmonella enterica Serovar Typhimurium. Cell Host Microbe. 2013;14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Adam J.R., Arthur M., Lawniczak M.K.N. Single-cell transcriptomics of malaria parasites. bioRxiv. 2017 [Google Scholar]
- 30.Gasch A.P., Yu F.B., McClean M.N. Single-cell RNA sequencing reveals intrinsic and extrinsic regulatory heterogeneity in yeast responding to stress. PLoS Biol. 2017;15:e2004050. doi: 10.1371/journal.pbio.2004050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Picelli S., Björklund A.K., Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods. 2013;10(11):1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 32.Ramsköld D., Luo S., Sandberg R. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Y.Y., Machleder E.M., Siebert P.D. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. Biotechniques. 2001;30:892–897. doi: 10.2144/01304pf02. [DOI] [PubMed] [Google Scholar]
- 34.Macosko E.Z., Basu A., McCarroll S.A. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poran A., Nötzel C., Kafsack B.F.C. Single-cell RNA sequencing reveals a signature of sexual commitment in malaria parasites. Nature. 2017;551:95–99. doi: 10.1038/nature24280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torre E., Dueck H., Raj A. Rare cell detection by single-cell RNA sequencing as guided by single-molecule RNA FISH. Cell Syst. 2018;6:171–179.e5. doi: 10.1016/j.cels.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khan I.U., Yadav J.S. Development of a single-tube, cell lysis-based, genus-specific PCR method for rapid identification of mycobacteria: optimization of cell lysis, PCR primers and conditions, and restriction pattern analysis. J. Clin. Microbiol. 2004;42:453–457. doi: 10.1128/JCM.42.1.453-457.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heera R., Sivachandran P., Yin L.S. Efficient extraction of small and large RNAs in bacteria for excellent total RNA sequencing and comprehensive transcriptome analysis. BMC Res. Notes. 2015;8:754. doi: 10.1186/s13104-015-1726-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blainey P.C. The future is now: single-cell genomics of bacteria and archaea. FEMS Microbiol. Rev. 2013;37:407–427. doi: 10.1111/1574-6976.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kvist T., Ahring B.K., Westermann P. Specific single-cell isolation and genomic amplification of uncultured microorganisms. Appl. Microbiol. Biotechnol. 2007;74:926–935. doi: 10.1007/s00253-006-0725-7. [DOI] [PubMed] [Google Scholar]
- 41.Lai H.H., Quinto-Su P.A., Allbritton N.L. Characterization and use of laser-based lysis for cell analysis on-chip. J. R. Soc. Interface. 2008;5(Suppl 2):S113–S121. doi: 10.1098/rsif.2008.0177.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thornhill A.R., McGrath J.A., Handyside A.H. A comparison of different lysis buffers to assess allele dropout from single cells for preimplantation genetic diagnosis. Prenat. Diagn. 2001;21:490–497. doi: 10.1002/pd.109. [DOI] [PubMed] [Google Scholar]
- 43.Xu L., Brito I.L., Blainey P.C. Virtual microfluidics for digital quantification and single-cell sequencing. Nat. Methods. 2016;13:759–762. doi: 10.1038/nmeth.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sasagawa Y., Nikaido I., Ueda H.R. Quartz-Seq: a highly reproducible and sensitive single-cell RNA sequencing method, reveals non-genetic gene-expression heterogeneity. Genome Biol. 2013;14:R31. doi: 10.1186/gb-2013-14-4-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Islam S., Kjällquist U., Linnarsson S. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21:1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hashimshony T., Wagner F., Yanai I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Reports. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 47.Hashimshony T., Senderovich N., Yanai I. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 2016;17:77. doi: 10.1186/s13059-016-0938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaitin D.A., Kenigsberg E., Amit I. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soumillon M., Cacchiarelli D., Mikkelsen T.S. Characterization of directed differentiation by high-throughput single-cell RNA-Seq. bioRxiv. 2014 [Google Scholar]
- 50.Klein A.M., Mazutis L., Kirschner M.W. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taniguchi Y., Choi P.J., Xie X.S. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee J.H., Daugharthy E.R., Church G.M. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat. Protoc. 2015;10:442–458. doi: 10.1038/nprot.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu A.R., Neff N.F., Quake S.R. Quantitative assessment of single-cell RNA-sequencing methods. Nat. Methods. 2014;11:41–46. doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ziegenhain C., Vieth B., Enard W. Comparative analysis of single-cell RNA sequencing methods. Mol. Cell. 2017;65:631–643.e4. doi: 10.1016/j.molcel.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 55.Elowitz M.B., Levine A.J., Swain P.S. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 56.Yu J., Xiao J., Xie X.S. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- 57.Raj A., van den Bogaard P., Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ke R., Mignardi M., Nilsson M. In situ sequencing for RNA analysis in preserved tissue and cells. Nat. Methods. 2013;10:857–860. doi: 10.1038/nmeth.2563. [DOI] [PubMed] [Google Scholar]
- 59.Lee J.H., Daugharthy E.R., Church G.M. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014;343:1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saliba A.E., Li L., Vogel J. Single-cell RNA-seq ties macrophage polarization to growth rate of intracellular Salmonella. Nat. Microbiol. 2016;2:16206. doi: 10.1038/nmicrobiol.2016.206. [DOI] [PubMed] [Google Scholar]
- 61.Fan X., Zhang X., Huang Y. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 2015;16:148. doi: 10.1186/s13059-015-0706-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schurch N.J., Schofield P., Barton G.J. How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA. 2016;22:839–851. doi: 10.1261/rna.053959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peixoto L., Risso D., Abel T. How data analysis affects power, reproducibility and biological insight of RNA-seq studies in complex datasets. Nucleic Acids Res. 2015;43:7664–7674. doi: 10.1093/nar/gkv736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baker S.C., Bauer S.R., Zadro R., External RNA Controls Consortium The external RNA controls consortium: a progress report. Nat. Methods. 2005;2:731–734. doi: 10.1038/nmeth1005-731. [DOI] [PubMed] [Google Scholar]
- 65.Grün D., Kester L., van Oudenaarden A. Validation of noise models for single-cell transcriptomics. Nat. Methods. 2014;11:637–640. doi: 10.1038/nmeth.2930. [DOI] [PubMed] [Google Scholar]
- 66.Brennecke P., Anders S., Heisler M.G. Accounting for technical noise in single-cell RNA-seq experiments. Nat. Methods. 2013;10:1093–1095. doi: 10.1038/nmeth.2645. [DOI] [PubMed] [Google Scholar]