Abstract
Atrial fibrillation (AF) is a serious cardiovascular disease with the phenomenon of beating irregularly. It is the major cause of variety of heart diseases, such as myocardial infarction. Automatic AF beat detection is still a challenging task which needs further exploration. A new framework, which combines modified frequency slice wavelet transform (MFSWT) and convolutional neural networks (CNNs), was proposed for automatic AF beat identification. MFSWT was used to transform 1 s electrocardiogram (ECG) segments to time-frequency images, and then, the images were fed into a 12-layer CNN for feature extraction and AF/non-AF beat classification. The results on the MIT-BIH Atrial Fibrillation Database showed that a mean accuracy (Acc) of 81.07% from 5-fold cross validation is achieved for the test data. The corresponding sensitivity (Se), specificity (Sp), and the area under the ROC curve (AUC) results are 74.96%, 86.41%, and 0.88, respectively. When excluding an extremely poor signal quality ECG recording in the test data, a mean Acc of 84.85% is achieved, with the corresponding Se, Sp, and AUC values of 79.05%, 89.99%, and 0.92. This study indicates that it is possible to accurately identify AF or non-AF ECGs from a short-term signal episode.
1. Introduction
Atrial fibrillation (AF) is the most common type of arrhythmia in clinical disease and gradually becomes the world's rising healthcare burden [1]. According to Framingham heart study, lifetime risk of AF is about 25% [2]. The disease shows that the atrial activity is irregular, and the resulting complications such as stroke and myocardial infarction (MI) [3], endanger the health and lives of humans seriously [4]. Therefore, developing automatic AF detection algorithm is of great clinical and social significance [5, 6].
Generally, AF is significantly different from normal heart rhythm on electrocardiogram (ECG) signals [7]. During AF, RR interval is absolutely irregular and the P-wave is replaced by the continuous irregular F-wave, which is an important feature of AF [8]. Many scholars proposed diverse methods based on RR interval feature, but the accuracy of AF is not sufficient due to the complication of the ECG signals [9], and the pattern recognition ability of the existing statistical and general method is not satisfactory owing to a variety of noise interference [10].
In recent years, AF detection algorithms based on the time domain characteristics have been developed rapidly. Chen et al. [11] developed a multiscale wavelet entropy-based method for paroxysmal atrial fibrillation (PAF) recognition. In their work, recognition and prediction used support vector machine- (SVM-) based method. Fifty recordings from the MIT-BIH PAF Prediction Database were chosen to test the proposed algorithm, with an average sensitivity of 86.16% and average specificity of 89.68%. Maji et al. [12] used empirical mode decomposition (EMD) to extract P-wave mode components and corresponding parameters to determine the occurrence of AF. This proposed algorithm was tested with a total of 110 cycles of normal rhythm and 68 cycles of AF rhythm from the MIT-BIH AF Database. An average sensitivity of 92.89% and an average specificity of 90.48% were achieved. Ladavich and Ghoraani [13] constructed the Gaussian mixture model of the P-wave feature space. The model was then used to detect AF, with an average sensitivity of 88.87% and average specificity of 90.67%, while the positive predictive value was only 64.99%. Although relatively fine detection performances were achieved by the aforementioned methods, problems and questions exist. First, different methods used different signal length for AF identification. How will be the accuracy if performed on a very short-term (such as 1 s) ECG segment? This can show the ability for transient AF detection. In addition, ECG waveforms have various morphology and the abnormal waveforms are different when AF occurs, leading to poor generalization capability of the developed machine learning-based model. Thus, how to improve the model generalization capability is a key issue.
Convolutional neural networks (CNNs) [14] can extract features automatically without manual intervention and expert priori knowledge. Meanwhile, time-frequency (T-F) technology as a preprocess operation is to convert 1D ECG signals to 2D T-F features which can be used to transfer to a classifier. There are many common T-F methods at present, such as the short-time Fourier transform (STFT), the Wigner–Ville distribution (WVD), and the continuous wavelet transform (CWT) [15]. Luo et al. [16] presented a modified frequency slice wavelet transform (MFSWT) in 2017. MFSWT follows the rules of producing T-F representation and contains the information of ECG signals in both time and frequency domains, such as P-wave, QRS-wave, and T-wave. Additionally, MFSWT can locate the above characters accurately and avoid the complexity of setting parameters. The spectrogram of MFSWT can be expressed as images, while the combination of CNN and images is one of the most excellent choices. For example, Liu et al. [17] proposed the method to learn conditional random fields (CRFs) using structured SVM (SSVM) based on features learned from a pretrained deep CNN for image segmentation. Ravanbakhsh et al. [18] introduced a feature representation for videos that outperforms state-of-the-art methods on several datasets for action recognition. Lee and Kwon [19] built a fully connected CNN, which trained on relatively sparse training samples, and a newly introduced learning approach called residual learning for hyperspectral image (HSI) classification.
In this study, MFSWT was adopted to acquire T-F images for short-term AF and non-AF ECG segments from the MIT-BIH Atrial Fibrillation Database (MIT-BIH AFDB). A deep CNN with a total of 12 layers was developed to train an AF/non-AF classification model. Indices including accuracy, sensitivity, specificity, and the area under the curve were used for model evaluation based on a 5-fold cross validation method [20], to evaluate the stability and generalization ability of the proposed method in comparison with the existing methods. The existing research has achieved very good performance, but there is no validation for large data. In this paper, we used all the data in the database to increase the generalization ability of the model.
2. Methods
2.1. Modified Frequency Slice Wavelet Transform
In our previous work, modified frequency slice wavelet transform (MFSWT) [16] was proposed for heartbeat time-frequency spectrum generation, with following the major principle of frequency slice wavelet transform (FSWT) [21]. The modified transform generates T-F representation from the frequency domain, and a bound signal-adaptive frequency slice function (FSF) was introduced to serve as a dynamic frequency filter. The window size of FSF smoothly changes with energy frequency distribution of signal in low-frequency area. The MFSWT has good performance for low-frequency ECG signals, and its advantages include signal-adaptive, accurate time-frequency component locating. The reconstruction is also independent of FSF, and it is readily accepted by clinicians.
Assume is the Fourier transform of f(t). The MFSWT can be defined as follows:
| (1) |
where t and ω are observed time and frequency, respectively, “∗” represents conjugation operator, and is the frequency slice function (FSF):
| (2) |
q is defined as a scale function of ,
| (3) |
It makes the transform to incorporate signal-adaptive property. In (3), δ corresponds to maximum . ∇(·) is a differential operator, and sign(·) means signum function, which returns 1 if the input is greater than zero, 0 if it is zero, or −1 if it is less than zero. In (2), , according to the claim in [21], then the original signal can be reconstructed as follows:
| (4) |
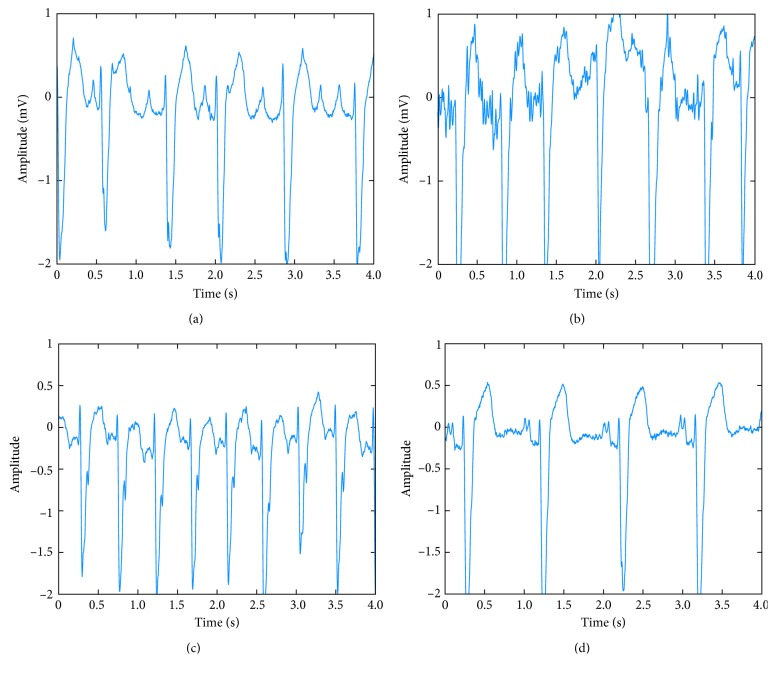
Figure 1 shows 4 s normal ECG, atrial fibrillation signals, and their corresponding MFSWT spectra, respectively, from 06426 recording. By the MFSWT, the time domain characteristics in ECG signal wave, such as P-wave, QRS-wave, and T-wave, have accurately been located in the signal spectrum. At the same time, each component of the spectrum of the T-F space distribution is corresponded well with the ECG signal frequency before.
Figure 1.

Examples from a 4 s normal ECG segment and a 4 s AF ECG segment, as well as their corresponding MFSWT spectra: (a) normal ECG signal; (b) MFSWT spectrum of the normal ECG signal; (c) AF ECG signal; (d) MFSWT spectrum of the AF ECG signal.
In this study, the MFSWT is used as a tool to generate spectrograms of an ECG signal for CNN-based classification. The 1 s window, centered at the detected R-peaks (0.4 s before and 0.6 s after), was used to segment each heartbeat. Subsequently, the T-F spectrograms with the size of 250 × 90 (corresponded 1 s time interval and 0–90 Hz frequency range) were produced by the proposed MFSWT. This is then followed by data reducing. An average 5 × 2 template operator reduces the size of spectrograms to 50 × 45.
2.2. Convolutional Neural Networks (CNNs)
Deep CNN was improved by Lecun et al. [22]. CNN had breakthrough performance over the last few decades for solving pattern recognition problems [23], especially in image classification [24]. It has become a popular method for feature extraction and classification without requiring preprocessing and pretraining algorithm [25].
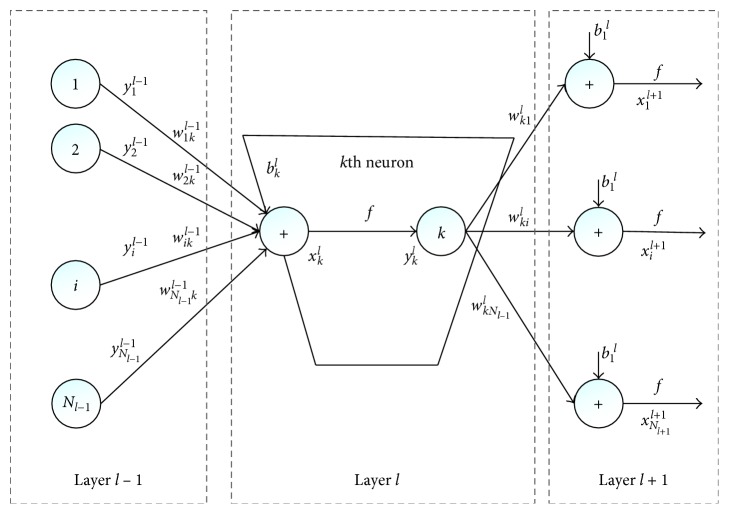
CNN is a composition of sequences of functions or layers that maps an input vector to an output vector. The input xkl is expressed as follows:
| (5) |
Similarly, bkl and wikl−1 are the bias and kernel of the kth neuron at layer l, respectively. sil−1 is the output of the ith neuron at layer l−1, and conv2D(. , .) means a regular 2D convolution without zero padding on the boundaries. So, the output ykl can be described as follows:
| (6) |
Besides, CNN also involves back-propagation (BP), in order to adjust the delta error of the kth neuron at layer l. Assuming that the corresponding output vector of the input is [y1L, y2L,…, yNLL], and its ground truth class vector is [t1, t2,…, tNL], we can write the mean absolute error (MSE) as follows:
| (7) |
Thus, the delta error can be concluded as
| (8) |
The implementation of CNN [14] is as shown in Figure 2.
Figure 2.
The implementation of CNNs.
By the MFSWT, we have converted the signals to characteristic waves in a 2D space. Then, we use CNN to learn relevant information from the characteristic waves in a 2D space and achieve classification. The input to the CNN is characteristic waves in a 2D space computed from the exacted signals. The CNN was implemented using the Neural Network Toolbox in Matlab R2017a.
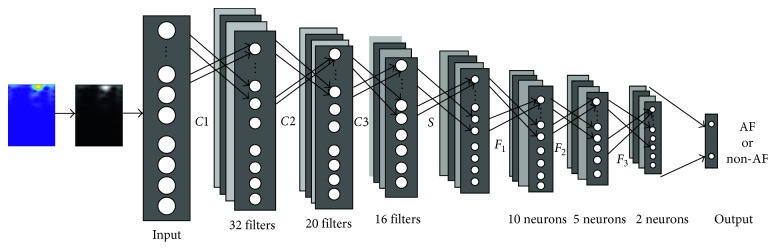
In this paper, we use CNN to automatically extract the features of the labeled image and calculate the scores to classify the predicted image. A 12-layer network structure is developed, which contains 3 convolution layers, 3 ReLU layers, a max pooling layer, and 3 full-connection layers besides the input and output layers. We tested the effect of number of filters in each layer and obtained these values by running a grid search approach. Figure 3 illustrates the architecture of the implemented network and its detailed components for each layer.
Figure 3.
The architecture of the network.
3. Experiment Design
3.1. Database
The database was from the MIT-BIH AFDB [26]. The MIT-BIH AFDB contains a large number of ECG data that have been annotated by a professional cardiologist, which is the authoritative ECG database in the classification of arrhythmia. This database may be useful for development and evaluation of atrial fibrillation/flutter detectors that rely on timing information only. It consists of 25 recordings (obtained from ambulatory ECG recordings of 25 subjects). The individual recordings are each 10 hours 15 min in duration and contain two ECG signals; each sampled at 250 samples per second with 12-bit resolution over a range of ±10 millivolts. The reference manual annotation files contain rhythm change annotations (with the suffix.atr) [27] and the rhythm annotations of types: AF, AFL (atrial flutter), J (AV junctional rhythm), and N (used to indicate all other rhythms) [28]. In our experiment, AFL, J, and N are attributed to non-AF category. Figure 4 shows the ECG examples (each 4 s) of the four rhythm types (normal, AF, atrial flutter, and AV junction) from the “06426” recording.
Figure 4.
ECG examples of the four rhythm types: (a) normal, (b) atrial fibrillation, (c) atrial flutter, and (d) AV junction rhythm.
3.2. Signal Preprocessing
There are a total of 25 recordings in AFDB, and two of them (numbers 00735 and 03665) have no relevant ECG data. Thus, 23 recordings are included in the experiment. In order to split the dataset equally, we divide the 23 recordings into five groups, and the basis for grouping is to reduce the differences of number in the two classes. The recording numbers for 5 groups are 5, 4, 5, 5, 4, subsequently. This experiment uses 5-fold cross validation for evaluation. For example, when using the first group to test, it means that the whole data in 04015, 04126, 04936, 07879, and 08405 recordings are used to verify, and the remaining 17 records are all used to train the model. Detailed recordings of grouping conditions are shown in Table 1.
Table 1.
Recordings of grouping conditions.
| Fold | Recordings | ||||
|---|---|---|---|---|---|
| 1 | 04015 | 04126 | 04936 | 07879 | 08405 |
| 2 | 04043 | 04048 | 07859 | 07910 | |
| 3 | 04746 | 05261 | 08215 | 08378 | 08455 |
| 4 | 04908 | 06426 | 07162 | 08219 | 08434 |
| 5 | 05091 | 05121 | 06453 | 06995 | |
We employ a balanced image dataset to train the model. That is, we choose the same number of AF samples from the non-AF category for training, while all the samples in the remaining fold for test. For example, for testing the fold 1, there are 415,109 normal images and 294,136 AF images as training data from the folds 2–5. We use all the 294,136 AF images and then randomly select 294,136 normal images, resulting in 588,272 images as training CNN model. Then, we test the performance of the developed CNN model using all data in fold 1, that is, 123,083 normal images and 90,403 AF images. Table 2 presents the detailed numbers for each fold testing.
Table 2.
Numbers of the images for testing each fold.
| Testing fold | Training | Balanced training | Test | |||
|---|---|---|---|---|---|---|
| Non-AF | AF | Non-AF | AF | Non-AF | AF | |
| 1 | 415,109 | 294,136 | 294,136 | 294,136 | 123,083 | 90,403 |
| 2 | 422,935 | 302,194 | 302,194 | 302,194 | 115,257 | 82,345 |
| 3 | 424,919 | 300,509 | 300,509 | 300,509 | 113,273 | 84,030 |
| 4 | 441,739 | 314,916 | 314,916 | 314,916 | 96,453 | 69,623 |
| 5 | 448,066 | 326,401 | 326,401 | 326,401 | 90,126 | 58,138 |
4. Results
4.1. Epoch Number of the CNN
The grid search method [29] is applied to select the optimal epoch number of the CNN. Figure 5 shows the AUC of the test set (AUC is defined as the area under the ROC curve, often used to evaluate the classifier with imbalance data) at varying epoch number, and Figure 6 shows training and test accuracies at varying epoch number. We can see from Figure 5 that the AUC of test set is at a high level while epoch number is 15, and the wave becomes stable in Figure 6. Therefore, we choose the number of epochs as 15.
Figure 5.
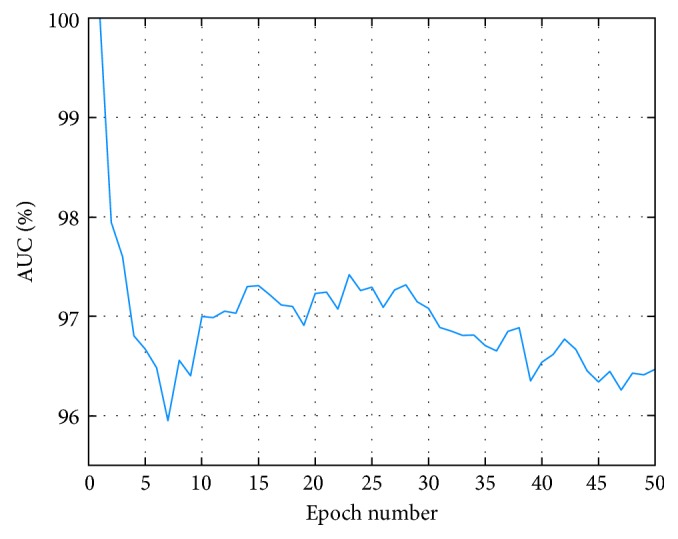
The AUC of test set at varying epoch number.
Figure 6.
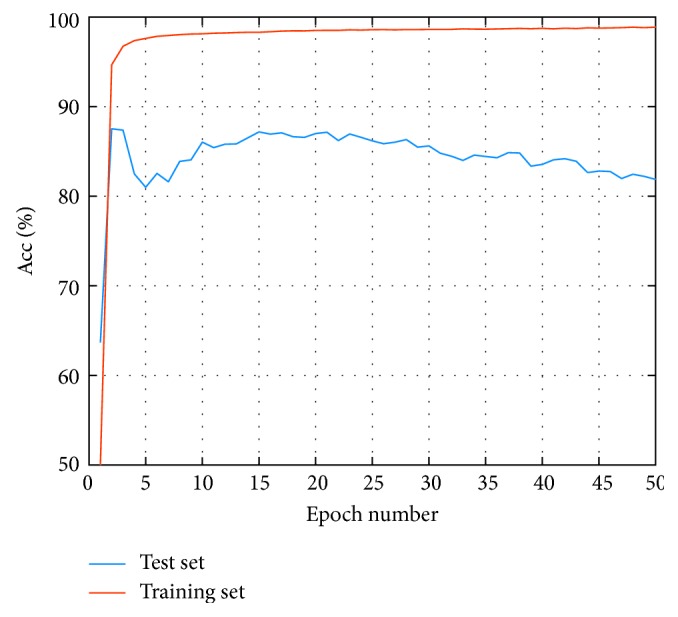
Test Acc and training Acc at varying epoch number.
According to the introduction of Section 2.2 on CNN architecture, the input layer (layer 0) is for images with the size of 50 × 45 × 1 and is convolved with a kernel size of 10 × 9 to produce the layer 1. Layer 2 is the ReLU layer. The output of layer 2 is convolved with a kernel size of 8 × 7 to develop the layer 3 and layer 4. Similarly, a subsequent feature maps are convolved with a kernel size of 9 to acquire the layer 5 and layer 6. A max pool layer with the stride of 2 (layer 7) is applied to the generated characteristics. Then, the feature has been extracted. Finally, features are transported to layer 8 with 10 neurons and connected to 5 and 2 neurons in layer 9 and layer 10, respectively, to classify. In addition, we select the epoch number as 15, and the learning rate initial value is set to 0.001 while the number of minimal batches is 256. In addition, specific parameter is shown in Table 3.
Table 3.
The optimal CNN specifications designed for the ECG classification problem.
| Parameters | Values |
|---|---|
| Learning rate | 0.001 |
| First convolutional layer kernel size | 10 ∗ 9 |
| Number of feature maps in the first convolutional and subsampling layer | 32 |
| Second convolutional layer kernel size | 8 ∗ 7 |
| Number of feature maps in the second convolutional and subsampling layer | 20 |
| Third convolutional layer kernel size | 9 |
| Number of feature maps in the third convolutional and subsampling layer | 16 |
| Subsampling layer kernel size | 2 |
| Number of neurons in the first fully connected layer | 10 |
| Number of neurons in the second fully connected layer | 5 |
| Number of neurons in the third fully connected layer | 2 |
| Number of epochs | 15 |
| Number of minimal batches | 256 |
4.2. Performance Metrics
This research uses the MIT-BIH AFDB to verify the proposed method to detect AF performance. Four widely used metrics, that is, sensitivity (Se), specificity (Sp), accuracy (Acc), and the area under the curve (AUC), were used (and defined below) for assessment of classification performance. In addition, AUC and Acc refer to the overall system performance, while the remaining indexes are specific to each class, and they measure the generalization ability of the classification algorithm to differentiate events.
Moreover, the Acc includes both test set accuracy and training set accuracy. According to the attribute of the label (positive or negative), the result can generate four basic indexes: true positive (TP), false positive (FP), true negative (TN), and false negative (FN), and in this case, Acc is the radio of the number of correct predicted labels and total number of the labels, thus Acc=(TP+TN)/(TP+TN+FP+FN). Se is the true positive rate and is the probability of incorrectly diagnosing into positive among all positive patients, so Se=TP/(TP+FN). Sp is proportion of incorrectly diagnosing into negative among all negative patients, so Sp=TN/(TN+FP). ROC curve is based on a series of different ways of binary classification (boundary value or decision threshold), with true positive rate (Se) as the ordinate, the false positive rate (1−Sp) as the abscissa, and AUC is defined as the area under the ROC curve, often used to evaluate the classifier with imbalance data. Each fold is tested by a specific classifier with the same parameters as shown in Section 4.1; besides, we also selected the average and standard deviation (SD) of the experimental results to be evaluated, and the results are summarized in Table 4.
Table 4.
The experimental results.
| Fold | Test data | Training data | ||||||
|---|---|---|---|---|---|---|---|---|
| Acc (%) | Se (%) | Sp (%) | AUC | Acc (%) | Se (%) | Sp (%) | AUC | |
| 1 | 86.63 | 77.95 | 95.97 | 0.95 | 97.59 | 96.68 | 98.54 | 0.99 |
| 2 | 86.82 | 84.28 | 88.62 | 0.93 | 98.21 | 97.81 | 98.62 | 0.99 |
| 3 | 83.55 | 78.90 | 87.38 | 0.91 | 97.80 | 96.77 | 98.88 | 0.99 |
| 4 | 65.93 | 58.59 | 72.08 | 0.70 | 98.26 | 98.51 | 98.00 | 0.98 |
| 5 | 82.41 | 75.06 | 87.99 | 0.90 | 98.40 | 98.56 | 98.24 | 0.99 |
| Mean | 81.07 | 74.96 | 86.41 | 0.88 | 98.05 | 97.67 | 98.46 | 0.99 |
| SD | 8.68 | 9.74 | 8.73 | 0.10 | 0.34 | 0.91 | 0.34 | 0 |
| Mean# | 84.85 | 79.05 | 89.99 | 0.92 | 98.00 | 97.46 | 98.57 | 0.99 |
| SD# | 1.92 | 3.34 | 3.48 | 0.02 | 0.32 | 0.78 | 0.23 | 0 |
#The results only from the average of the folds 1, 2, 3, and 5.
From Table 4, a mean Acc of 81.07% from 5-fold cross validation is achieved for the test data. The corresponding Se, Sp, and AUC results are 74.96%, 86.41%, and 0.88. It is worth to note that the results from the fourth fold are low. This is because there is an extremely poor signal quality ECG recording in the fourth fold divided as shown in Table 1, which has significantly different time-frequency features compared to the clean ECG signals. So the results from the folds 1, 2, 3, and 5 are recalculated as shown in Table 4 to exclude the low-quality signal effect. Herein, a mean Acc of 84.85% is achieved for the test data, with the corresponding Se, Sp, and AUC values of 79.05%, 89.99%, and 0.92.
5. Discussion and Conclusion
An ECG is widely used in medicine to monitor small electrical changes on the skin of a patient's body arising from the activities of the human heart. Due to the variability and difficulty of AF, traditional detection algorithm cannot be extracted to distinguish obvious characteristics accurately.
In our work, we present a unique architecture of CNN to distinguish AF beats from all other types of ECG beats. MFSWT is adopted to acquire the T-F images of AF and non-AF, respectively, and then, we divide all the data in the MIT-BIH AFDB into training set and test set different from the existing models and build a deep CNN with a total of 12 layers to extract the characters of training set. Finally, the test set is evaluated by the trained model to obtain the performance indexes (including Acc, Se, Sp, and AUC).
Compared with other studies, the difference is that we use all ECG recordings in the MIT-BIH AFDB. However, other studies only selected a part of the recordings for training and testing, and Table 5 shows the comparison between our study and other studies. Obviously, the proposed method does not improve the Acc, Se, and Sp significantly, but this paper uses lots of data to train the model in order to improve the generalization ability of the model. Moreover, we use an important indicator called AUC to evaluate the unbalanced data model and obtained a good evaluation standard.
Table 5.
Comparison with reference studies.
| Algorithm | Data | Acc (%) | Se (%) | Sp (%) | AUC |
|---|---|---|---|---|---|
| Chen et al. [11] | 50 signals | — | 89.68 | 86.16 | — |
| Maji et al. [12] | 178 cycles | — | 90.48 | 92.89 | — |
| Ladavich and Ghoraani [13] | 14,600 beats | — | 90.67 | 88.87 | — |
| Proposed method | All recordings | 81.07 | 74.96 | 86.41 | 0.88 |
| Proposed method | All recordings but excluding one fold with extreme noisy recording | 84.85 | 79.05 | 89.99 | 0.92 |
In short, we proposed a protocol for AF beat detection as follows. (1) Use all the recordings of MIT-BIH Atrial Fibrillation Database for algorithm development and validation. (2) Use 5-fold cross validation to examine the algorithm performance. The results have been registered for Acc, Se, Sp, and AUC. Group the folds by recordings rather than heartbeats to prevent heartbeats of the same patient from appearing in both training and test sets. (3) Use a separate database, for instance AF Database, as an independent test to evaluate the generalization ability of the algorithm. We believe that accurate AF beat recognition can facilitate the detection of AF rhythm. As the first step, AF beat is particularly important. Only by improving the accuracy and generalization of AF beat detection can we more effectively implement AF surveillance.
In addition, more data could be used to evaluate the proposed method; for example, we only focus on one ECG lead, and the study can be extended to two ECG leads. We also can try to use more databases for verification. This algorithm can be used for monitoring and prevention of AF, which has great practical meaning.
Acknowledgments
The project was partly supported by the National Natural Science Foundation of China (61571113 and 61671275), the Key Research and Development Programs of Jiangsu Province (BE2017735), the Natural Science Foundation of Shandong Province in China (2014ZRE2733), and the Fundamental Research Funds for the Central Universities in Southeast University (2242018k1G010). The authors thank the support from the Southeast-Lenovo Wearable Intelligent Lab.
Contributor Information
Shoushui Wei, Email: sswei@sdu.edu.cn.
Chengyu Liu, Email: bestlcy@sdu.edu.cn.
Data Availability
The data used to support the findings of this study are available from the open-access MIT-BIH Atrial Fibrillation Database.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Rieta J. J., Ravelli F., Sörnmo L. Advances in modeling and characterization of atrial arrhythmias. Biomedical Signal Processing and Control. 2013;8(6):956–957. doi: 10.1016/j.bspc.2013.10.008. [DOI] [Google Scholar]
- 2.Lloydjones D. M. Lifetime risk for development of atrial fibrillation: the Framingham Heart Study. Circulation. 2004;110(9):1042–1046. doi: 10.1161/01.CIR.0000140263.20897.42. [DOI] [PubMed] [Google Scholar]
- 3.Soliman E. Z., Lopez F., O’Neal W. T., et al. Atrial fibrillation and risk of ST-segment elevation versus non-ST segment elevation myocardial infarction: the atherosclerosis risk in communities (ARIC) study. Circulation. 2015;131(21):1843–1850. doi: 10.1161/circulationaha.114.014145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Langley P., Dewhurst M., Di Marco L. Y., et al. Accuracy of algorithms for detection of atrial fibrillation from short duration beat interval recordings. Medical Engineering and Physics. 2012;34(10):1441–1447. doi: 10.1016/j.medengphy.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Bruser C., Diesel J., Zink M. D. H., Winter S., Schauerte P., Leonhardt S. Automatic detection of atrial fibrillation in cardiac vibration signals. IEEE Journal of Biomedical and Health Informatics. 2013;17(1):162–171. doi: 10.1109/titb.2012.2225067. [DOI] [PubMed] [Google Scholar]
- 6.Lip G. Y. H., Brechin C. M., Lane D. A. The global burden of atrial fibrillation and stroke: a systematic review of the epidemiology of atrial fibrillation in regions outside North America and Europe. Chest. 2012;142(6):1489–1498. doi: 10.1378/chest.11-2888. [DOI] [PubMed] [Google Scholar]
- 7.Linker D. T. Accurate, automated detection of atrial fibrillation in ambulatory recordings. Cardiovascular Engineering and Technology. 2016;7(2):182–189. doi: 10.1007/s13239-016-0256-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y., Tang X., Wang A., Tang H. Probability density distribution of delta RR intervals: a novel method for the detection of atrial fibrillation. Australasian Physical and Engineering Sciences in Medicine. 2017;40(3):707–716. doi: 10.1007/s13246-017-0554-2. [DOI] [PubMed] [Google Scholar]
- 9.Petrenas A., Sornmo L., Marozas V., Lukosevicius A. A noise-adaptive method for detection of brief episodes of paroxysmal atrial fibrillation. Proceedings of Computing in Cardiology Conference; September 2013; Zaragoza, Spain. pp. 739–742. [Google Scholar]
- 10.Larburu N., Lopetegi T., Romero I. Comparative study of algorithms for atrial fibrillation detection. Proceedings of Computing in Cardiology Conference; September 2011; Hangzhou, China. pp. 265–268. [Google Scholar]
- 11.Chen Y., Wang Z., Li Q. Multi-scale wavelet entropy based method for paroxysmal atrial fibrillation recognition. Space Medicine and Medical Engineering. 2013;26(5):352–355. [Google Scholar]
- 12.Maji U., Mitra M., Pal S. Automatic detection of atrial fibrillation using empirical mode decomposition and statistical approach. Procedia Technology. 2013;10(1):45–52. doi: 10.1016/j.protcy.2013.12.335. [DOI] [Google Scholar]
- 13.Ladavich S., Ghoraani B. Rate-independent detection of atrial fibrillation by statistical modeling of atrial activity. Biomedical Signal Processing and Control. 2015;18:274–281. doi: 10.1016/j.bspc.2015.01.007. [DOI] [Google Scholar]
- 14.Kiranyaz S., Ince T., Gabbouj M. Real-time patient-specific ECG classification by 1-D convolutional neural networks. IEEE Transactions on Biomedical Engineering. 2016;63(3):664–675. doi: 10.1109/tbme.2015.2468589. [DOI] [PubMed] [Google Scholar]
- 15.Yan Z., Miyamoto A., Jiang Z., Liu X. An overall theoretical description of frequency slice wavelet transform. Mechanical Systems and Signal Processing. 2010;24(2):491–507. doi: 10.1016/j.ymssp.2009.07.002. [DOI] [Google Scholar]
- 16.Luo K., Li J., Wang Z., Cuschieri A. Patient-specific deep architectural model for ECG classification. Journal of Healthcare Engineering. 2017;2017:13. doi: 10.1155/2017/4108720.4108720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu F., Lin G., Shen C. CRF Learning with CNN Features for Image Segmentation. Amsterdam, Netherlands: Elsevier Science Inc.; 2015. [Google Scholar]
- 18.Ravanbakhsh M., Mousavi H., Rastegari M., Murino V., Davis L. S. Action recognition with image based CNN features. http://arxiv.org/abs/1512.03980.
- 19.Lee H., Kwon H. Going deeper with contextual CNN for hyperspectral image classification. IEEE Transactions on Image Processing. 2017;26(10):4843–4855. doi: 10.1109/tip.2017.2725580. [DOI] [PubMed] [Google Scholar]
- 20.Kohavi R. A study of cross-validation and bootstrap for accuracy estimation and model selection. Proceedings of International Joint Conference on Artificial Intelligence; August 1995; Montreal, QC, Canada. pp. 1137–1143. [Google Scholar]
- 21.Yan Z., Miyamoto A., Jiang Z. Frequency slice algorithm for modal signal separation and damping identification. Computers and Structures. 2011;89(1-2):14–26. doi: 10.1016/j.compstruc.2010.07.011. [DOI] [Google Scholar]
- 22.Lecun Y., Bottou L., Bengio Y., Haffner P. Gradient-based learning applied to document recognition. Proceedings of the IEEE. 1998;86(11):2278–2324. doi: 10.1109/5.726791. [DOI] [Google Scholar]
- 23.Jiang X., Pang Y., Li X., Pan J. Speed up deep neural network based pedestrian detection by sharing features across multi-scale models. Neurocomputing. 2016;185:163–170. doi: 10.1016/j.neucom.2015.12.042. [DOI] [Google Scholar]
- 24.Krizhevsky A., Sutskever I., Hinton G. E. ImageNet classification with deep convolutional neural networks. Proceedings of International Conference on Neural Information Processing Systems; December 2012; Lake Tahoe, NV, USA. pp. 1097–1105. [Google Scholar]
- 25.Chaib S., Yao H., Gu Y., Amrani M. Deep feature extraction and combination for remote sensing image classification based on pre-trained CNN models. International Conference on Digital Image Processing; July 2017; Prague, Czech. p. p. 104203D. [Google Scholar]
- 26.Jiang K., Huang C., Ye S., Chen H. High accuracy in automatic detection of atrial fibrillation for Holter monitoring. Journal of Zhejiang University. 2012;13(9):751–756. doi: 10.1631/jzus.b1200107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murgatroyd F. D., Xie B., Copie X., Blankoff I., Camm A. J., Malik M. Identification of atrial fibrillation episodes in ambulatory electrocardiographic recordings: validation of a method for obtaining labeled R-R interval files. Pacing and Clinical Electrophysiology Pace. 1995;18(6):p. 1315. doi: 10.1111/j.1540-8159.1995.tb06972.x. [DOI] [PubMed] [Google Scholar]
- 28.Petrėnas A., Sörnmo L., Lukoševičius A., Marozas V. Detection of occult paroxysmal atrial fibrillation. Medical and Biological Engineering and Computing. 2015;53(4):287–297. doi: 10.1007/s11517-014-1234-y. [DOI] [PubMed] [Google Scholar]
- 29.Brito J. A. A., Mcneill F. E., Webber C. E., Chettle D. R. Grid search: an innovative method for the estimation of the rates of lead exchange between body compartments. Journal of Environmental Monitoring. 2005;7(3):p. 241. doi: 10.1039/b416054a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the open-access MIT-BIH Atrial Fibrillation Database.