Abstract
Folate‐mediated one‐carbon metabolism (FOCM) is a key pathway essential for nucleotide synthesis, DNA methylation, and repair. This pathway is a critical target for 5‐fluorouracil (5‐FU), which is predominantly used for colorectal cancer (CRC) treatment. A comprehensive assessment of polymorphisms in FOCM‐related genes and their association with prognosis has not yet been performed. Within 1,739 CRC cases aged ≥30 years diagnosed from 2003 to 2007 (DACHS study), we investigated 397 single nucleotide polymorphisms (SNPs) and 50 candidates in 48 FOCM‐related genes for associations with overall‐ (OS) and disease‐free survival (DFS) using multiple Cox regression (adjusted for age, sex, stage, grade, BMI, and alcohol). We investigated effect modification by 5‐FU‐based chemotherapy and assessed pathway‐specific effects. Correction for multiple testing was performed using false discovery rates (FDR). After a median follow‐up time of 5.0 years, 585 patients were deceased. For one candidate SNP in MTHFR and two in TYMS, we observed significant inverse associations with OS (MTHFR: rs1801133, C677T: HR het = 0.81, 95% CI: 0.67–0.97; TYMS: rs1001761: HR het = 0.82, 95% CI: 0.68–0.99 and rs2847149: HR het = 0.82, 95% CI: 0.68–0.99). After FDR correction, one polymorphism in paraoxonase 1 (PON1; rs3917538) was significantly associated with OS (HR het = 1.28, 95% CI: 1.07–1.53; HR hzv = 2.02, 95% CI:1.46–2.80; HR logAdd = 1.31, pFDR = 0.01). Adjusted pathway analyses showed significant associations for pyrimidine biosynthesis (P = 0.04) and fluorouracil drug metabolism (P < 0.01) with significant gene–chemotherapy interactions, including PON1 rs3917538. This study supports the concept that FOCM‐related genes could be associated with CRC survival and may modify effects of 5‐FU‐based chemotherapy in genes in pyrimidine and fluorouracil metabolism, which are relevant targets for therapeutic response and prognosis in CRC. These results require confirmation in additional clinical studies.
Keywords: Colorectal cancer, one‐carbon metabolism, polymorphisms, survival
Introduction
Colorectal cancer (CRC) incidence and mortality rates have been decreasing during the past decade; yet it remains the second leading cause of cancer deaths in the United States 1.
The antimetabolite 5‐fluorouracil (5‐FU) is the most frequently used chemotherapy in CRC treatment, targeting thymidylate synthase (TYMS) in folate‐mediated one‐carbon metabolism (FOCM) 2. 5‐FU directly inhibits purine synthesis by inhibiting TYMS, resulting in decreased DNA replication and repair 3. There are large interindividual differences, however, in the effectiveness and tolerability of 5‐FU, which may be due to genetic variation in FOCM‐related genes 4, 5, 6. An overview of folate‐mediated one‐carbon metabolism and the genes and metabolites involved in that pathway is presented in Fig. 1 6.
Figure 1.
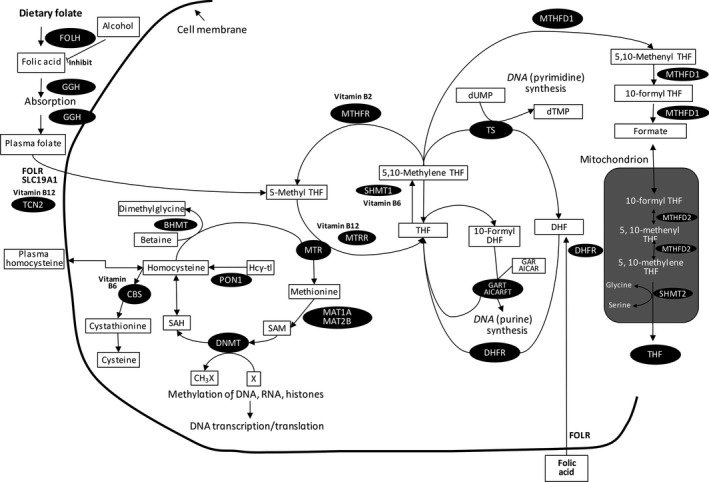
Overview of folate‐mediated one‐carbon metabolism, links to methylation reactions and nucleotide synthesis (by Cheng TY et al. 2015). THF = tetrahydrofolate; DHF = dihydrofolate; DNMT = DNA methyltransferases; GGH = gamma‐glutamyl‐hydrolase; RFC = reduced folate carrier; Hcy‐tl = homocysteine thiolactone; hFR = human folate receptor; MTHFR = 5,10‐methylenetetrahydrofolate reductase; DHFR = dihydrofolate reductase; GART = glycinamide ribonucleotide transformylase; AICARFT = 5‐amino‐imidazole‐4‐carboxamide ribonucleotide transformylase; AICAR = 5‐aminoimidazole‐4‐carboxamide ribonucleotide; GAR = glycinamide ribonucleotide; SAM (AdoMet) = S‐adenosylmethionine; SAH (AdoHcy) = S‐adenosylhomocysteine; dUMP = deoxyuridine monophosphate; dTMP = deoxythymidine monophosphate; MS = methionine synthase; TS = thymidylate synthase; DNMT = DNA methyltransferases; MTRR = methionine synthase reductase; X = a variety of substrates for methylation
Prior studies investigating FOCM‐related genes and response to 5‐FU‐based chemotherapy in CRC patients were largely limited to TYMS and MTHFR variants yielding inconsistent results 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19. While earlier data linked low expression of TYMS to worse response to 5‐FU‐based chemotherapy 17, subsequent studies have related low TYMS expression to improved response rates in patients with CRC 13, 18. In vitro data have linked 5‐FU sensitivity to MTHFR A1298C but not MTHFR C677T. Yet, clinical studies 9, 10, 11, 16, 19 associated 5‐FU sensitivity with MTHFR C677T but not with MTHFR A1298C. Understanding the interaction of TYMS and MTHFR genotypes with 5‐FU‐based chemotherapy could help identify patients who are more likely to respond to 5‐FU‐based chemotherapy, using personalized information to tailor chemotherapy.
To date, no comprehensive assessment of genetic variability in FOCM and association with CRC prognosis has been performed. Therefore, we aimed to assess whether single genetic variants as well as a priori defined pathways in FOCM (e.g., folate, pyrimidine synthesis, and fluorouracil pathway) were associated with overall‐ and disease‐free survival in patients from a large cohort of prospectively followed CRC patients. Finally, we evaluated interactions between genetic variants and 5‐FU‐based chemotherapy on overall‐ and disease‐free survival.
Materials and Methods
Study population
Our study population comprised 1,739 CRC patients who participated in an ongoing population‐based study “Darmkrebs: Chancen der Verhütung durch Screening” (DACHS) from Germany with long‐term follow‐up of patients 20. CRC patients with a primary, confirmed diagnosis of CRC, recruited from hospitals in the Rhein‐Neckar‐Odenwald region between 1 January 2003 and December 2007 were included. Patients were eligible if they were ≥30 years of age, resident in the study region, and able to complete an in‐person interview. Extensive information on sociodemographic characteristics, medical history and lifestyle factors was collected by trained interviewers using standardized questionnaires to collect information on established and suggested CRC risk and prognostic factors. A blood sample (>99% of the analyzed patients) or mouthwash for DNA extraction was taken. Clinical and histological data were extracted from medical and pathological records.
Follow‐up information on overall survival (OS) and disease‐free survival (DFS; defined as cancer recurrence) was collected 3 and 5 years after diagnosis. For all patients, vital status, date, and cause of death through the end of 2012 were ascertained via local population registries. Causes of death were verified by death certificates and coded based on ICD‐10 classifications. Information on therapy (at 3‐year follow‐up) and recurrences (at 3‐ and 5‐year follow‐up) was collected from clinical providers.
The study was approved by the ethics committee of the University of Heidelberg and conducted in agreement with the Helsinki Declaration. Written informed consent was provided from all participants for future use of research purposes.
SNPs and functional non‐SNP polymorphisms
Altogether, 1,754 cases were genotyped. Based on functional data and literature, we selected 48 genes in the FOCM pathway: AARS, ABCC4, ADH1B, ADH1C, BHMT, BHMT2, CBS, DHFR, DNMT1, DNMT3A, DNMT3B, DPYD, DPYS, DUT, EHMT1, EHMT2, FDXR, FOLH1, FOLR1, FPGS, GGH, GNMT, MAT1A, MAT2B, MTHFD1, MTHFD2, MTHFR, MTR, MTRR, NFKB1, NME1, NME2, PON1, PRDM2, RRM1, RRM2, SHMT1, SHMT2, SLC19A1, SLC29A1, TK1, TCN2, TYMP, TYMS, UMPH2, UMPK, UMPS, and UNG (Table S1).
Polymorphisms that may affect protein levels and/or function are referred to as candidate polymorphisms. We selected 50 candidates (Table S2), including, among others, five polymorphisms in TYMS (rs1001761, rs10502289, rs503296 including two intronic variants (rs2847149, rs2853533)), five TCN candidates (rs1131603, rs1801198, rs4820889, rs9606756, and rs9621049), and two MTHFR candidates (rs1801131(C677T), rs1801133(A1298C)). Additionally, two non‐SNP variants in the TYMS gene were selected: an insertion/deletion (indel) of 6 bp at position 1494 (3′ UTR indel) and a variable number of tandem repeats of a 28‐bp sequence (TSER) 21.
We used a comprehensive approach to investigate 397 tagSNPs, which represent genetic variation across the selected genes (Table S1). The tagging approach exploits the linkage disequilibrium (LD; nonrandom correlation between SNPs) across the human genome by selecting tagSNPs, which serve as proxies for correlated SNPs in specific regions. Hence, a subset of SNPs may be sufficient to cover most of the genetic variation within a specific region. Data from the HapMap Project were used with a pairwise tagging approach applying r 2 = 0.80 as cutoff 22.
Genotyping
Genomic DNA was extracted from EDTA blood or mouthwash samples using the FlexiGene DNA kit (Qiagen GmbH, Hilden, Germany) and quantified using Quant‐iT Pico Green dsDNA reagent kit (Invitrogen/Life Technologies, Darmstadt, Germany). Of 492 selected SNPs, 447 passed quality control after genotyping (for 45, success rate was below 95%, seven were not in Hardy–Weinberg equilibrium (HWE) and were selected to be genotyped on the customized GoldenGate assay (Illumina, San Diego, CA) 23. The iPLEX assay (Sequenom, Hamburg, Germany) for the MassArray system was used to genotype five SNPs that failed genotyping on the Illumina GoldenGate platform 24. Quality of genotyping was high with concordance of duplicates from Centre d'Etude du Polymorphisme Humain (Paris, France) and control samples above 98%. The two selected non‐SNP polymorphisms in the TYMS gene were genotyped using fragment analysis and single‐strand conformation polymorphism in the laboratory of Dr. Ulrich at the National Center for Tumor Diseases in Heidelberg, Germany.
Statistical analysis
Differences in baseline characteristics between deceased and nondeceased patients and deviation from HWE were evaluated using chi‐square statistic. Imputation of environmental factors was made by single imputation due to only few missing values (<1%) except for grade (11%). Imputation of grade was compared with best case (all missings set to grade 1) and worst case (all missings set to grade 4), resulting in similar effect estimates (<3% difference).
Genotype imputation was performed using IMPUTE2 and the 1000 Genomes reference panel 25. Follow‐up time was calculated as time from diagnosis to the event of interest or censoring (date of last information).
Cox proportional hazards models were used to estimate hazard ratios (HR) for OS and DFS, and their 95% confidence intervals (CIs) were associated with the genetic variants. We considered three types of inheritance models: codominant (if each of the three genotypes as derived from the biallelic SNPs had frequencies ≥5), dominant (if at least one genotype as derived from the biallelic SNPs had frequencies <5), and log‐additive model.
Multivariable models were determined using a backward elimination procedure on the interaction terms based on Akaike's information criterion, forcing clinical variables and all main effects into the model. Analyses were adjusted for age (<60, 60–70, 70–80, 80+), sex, stage (I, II, III, IV), grade (1/2 vs. 3/4), BMI (<18.5, 18.5–25, 25–30, 30+ kg/m2) and alcohol intake (0, 0–6.1, 6.1–15.6, 15.6–32.6, >32.6 g/day).
We investigated effect modification by 5‐FU‐based chemotherapy in the associations of all investigated SNPs with OS and DFS. The interaction terms between SNPs and 5‐FU‐based chemotherapy were derived from a comparison of the model with and without interaction terms using the likelihood ratio test. For pathway analysis, polymorphisms in high LD (r 2 > 0.5) within each gene were summarized to discard redundant information. Remaining polymorphisms were standardized 26, and genewise principal component analysis was applied explaining 95% of the variance in the data. The SNPs and the two non‐SNP polymorphisms were entered into a multivariable global test using Cox regression modeling 27. The Molecular Signatures Database v3.1 of the Broad Institute was used to identify subpathways (i.e., gene sets) searching for one‐carbon, folate, and 5‐FU‐based chemotherapy. YY KEGG and YY GO pathways were extracted 28 (Table S3). Candidate variants were not adjusted for multiple testing as they were selected based on functional data and independent hypotheses. TagSNPs or pathway analyses were corrected using the false discovery rates (FDR) for main effects and interaction tests.
All statistical analyses were two‐sided (significance level: P < 0.05) and performed using SAS (v9.4, SAS Institute, Cary, NC) and R (v3.1, R Foundation for Statistical Computing, Vienna, Austria).
Results
During a median follow‐up time of 5.0 years (range: 0.01–6.4 years), 585 of the 1,739 patients died, 420 due to CRC. Patients were on average 68.2 ± 10.4 years old at diagnosis (Table 1). Deceased patients were more likely to be older, have a higher tumor stage and grade compared to nondeceased patients, and also more likely to have received adjuvant chemotherapy (especially 5‐FU and folic acid therapy). Almost two thirds of the patients had a colon carcinoma compared to one third of patients diagnosed with rectal cancer.
Table 1.
Selected characteristics of deceased and nondeceased patients.a
| Deceased (n = 585) | Nondeceased (n = 1,154) | P‐Value | |
|---|---|---|---|
| Age (%) | |||
| <60 | 84 (14.4) | 249 (21.6) | <0.01 |
| 60–70 | 173 (29.6) | 428 (37.1) | |
| 70–80 | 189 (32.3) | 363 (31.5) | |
| 80+ | 139 (23.8) | 114 (9.9) | |
| Sex (%) | |||
| Female | 259 (44.3) | 465 (40.3) | 0.11 |
| Male | 326 (55.7) | 689 (59.7) | |
| Site (%) | |||
| Colon | 359 (61.4) | 700 (60.7) | 0.77 |
| Rectum | 226 (38.6) | 454 (39.3) | |
| CRC first‐degree family history (%) | |||
| No | 502 (85.8) | 979 (84.8) | 0.59 |
| Yes | 83 (14.2) | 175 (15.2) | |
| Stage (%) | |||
| I | 61 (10.4) | 366 (31.7) | <0.01 |
| II | 115 (19.7) | 413 (35.8) | |
| III | 201 (34.4) | 344 (29.8) | |
| IV | 208 (35.6) | 31 (2.7) | |
| Grade (%) | |||
| 1,2 | 369 (63.1) | 877 (76.0) | <0.01 |
| 3,4 | 216 (36.9) | 277 (24.0) | |
| Smoking (%) | |||
| Never | 304 (52.0) | 533 (46.2) | 0.08 |
| Former | 200 (34.2) | 442 (38.3) | |
| Current | 81 (13.8) | 179 (15.5) | |
| BMI [kg/m2] (%) | |||
| <18 | 22 (3.8) | 18 (1.6) | <0.01 |
| 18–25 | 235 (40.2) | 384 (33.3) | |
| 25–30 | 231 (39.5) | 513 (44.5) | |
| >30 | 97 (16.6) | 239 (20.7) | |
| Alcohol intake, [g/day] (%) | |||
| 0 | 213 (36.4) | 310 (26.9) | <0.01 |
| >0–6.1 | 94 (16.1) | 217 (18.8) | |
| >6.1–15.6 | 94 (16.1) | 204 (17.7) | |
| >15.6–32.6 | 91 (15.6) | 214 (18.5) | |
| >32.6 | 93 (15.9) | 209 (18.1) | |
| Radiotherapy (%) | |||
| No | 468 (80) | 942 (81.6) | 0.03 |
| Adjuvant | 54 (9.2) | 90 (7.8) | |
| Neo‐adjuvant | 55 (9.4) | 119 (10.3) | |
| Chemotherapy (%) | |||
| No | 230 (39.3) | 697 (60.4) | <0.01 |
| Adjuvant | 312 (53.3) | 379 (32.8) | |
| Neo‐adjuvant | 33 (5.6) | 75 (6.5) | |
| 5‐FU‐based chemotherapy (%) | |||
| No | 40 (6.8) | 52 (4.5) | 0.91 |
| Yes | 283 (48.4) | 359 (31.1) | |
| Not available | 253 (43.2) | 742 (64.3) | |
Percentages may not add up to 100.
P‐values in bold are statistically significant.
Selected results are presented in Tables 2 and 3. All results for the two non‐SNP variants in the TYMS gene are presented in Table S4a,b,c. The pathway analyses are presented in Table S5. Sensitivity analyses restricting the dataset to patients who received 5‐FU / 5‐FU + FA are presented in Table S6. The results of all survival analyses are presented in Table S7 and Table S8 (overall survival and disease‐free survival) and Table S9 and Table S10 (overall survival and disease‐free survival stratified by 5‐FU based chemotherapy). We have clearly defined hypotheses for each of the selected n = 50 candidate SNPs and consider the unadjusted P‐values as the relevant ones for this study. However, we have decided to present the FDR‐adjusted P‐values for the candidate SNPs as well. We observed significant inverse associations with OS for three candidate SNPs: one SNP in MTHFR (rs1801133, C677T: HRhet = 0.81, 95% CI: 0.67–0.97) and two candidates in TYMS (rs1001761: HRhet = 0.82, 95% CI: 0.68–0.99 and rs2847149: HRhet = 0.82, 95% CI: 0.68–0.99). A polymorphism in the paraoxonase 1 (PON1) gene (tag SNP rs3917538) was significantly associated with OS after FDR adjustment: HRhzv = 2.02, 95% CI: 1.46–2.80; HRhet = 1.28, 95% CI: 1.07–1.53; HRlogAdd = 1.31, pFDR < 0.01). Nominally significant associations were observed for two SNPs in PON1 (rs3917527, rs757158) and one in TYMS (rs2244500). Significant inverse associations were observed for one candidate SNP in EHMT2 with DFS: rs2736428 (HRhet = 0.80, 95% CI: 0.66–0.98). However, in the more recent HapMap database 22, this SNP is located on SLC44A4. Thus, we decided not to consider it further. Nominally significant associations were observed for 19 tagSNPs, but diminished after FDR adjustment.
Table 2.
Associations between selected polymorphisms in FOCM‐related genes and overall‐ and disease‐free survival
| Gene | SNP | Genotype | HR(95%‐CI)h | P a | pFDR b | pTrend c | pTrend‐FDR d | pGenewide‐FDR e | |
|---|---|---|---|---|---|---|---|---|---|
| Overall Survival | PON1 | rs3917538 | C/C | ref | <0.01 | <0.01 | <0.01 | ||
| C/T | 1.18 (0.97–1.43) | 0.09 | |||||||
| T/T | 2.02 (1.46–2.80) | <0.01 | |||||||
| C/T or T/Tf | 1.28 (1.07–1.53) | <0.01 | 0.59 | 0.04 | |||||
| TYMS | rs1001761g | C/C | ref | 0.04 | 0.73 | 0.11 | |||
| C/T | 0.84 (0.68–1.02) | 0.08 | |||||||
| T/T | 0.77 (0.59–1.00) | 0.05 | |||||||
| C/T or T/Tf | 0.82 (0.68–0.99) | 0.04 | 0.59 | 0.09 | |||||
| TYMS | rs2847149g | G/G | ref | 0.04 | 0.73 | 0.11 | |||
| G/A | 0.84 (0.68–1.02) | 0.08 | |||||||
| A/A | 0.77 (0.59–1.00) | 0.02 | |||||||
| G/A or A/Af | 0.82 (0.68–0.99) | 0.04 | 0.59 | 0.09 | |||||
| TYMS | rs495139 | C/C | ref | 0.07 | 0.79 | 0.11 | |||
| C/G | 1.48 (1.20–1.82) | <0.01 | |||||||
| G/G | 1.17 (0.89–1.53) | 0.27 | |||||||
| C/G or G/Gf | 1.39 (1.14–1.69) | <0.01 | 0.45 | <0.01 | |||||
| Disease‐free survival | MAT2B | rs6882306 | T/T | ref | <0.01 | 0.45 | 0.01 | ||
| T/C | 1.31 (1.05–1.62) | 0.01 | |||||||
| C/C | 1.91 (1.15–3.16) | 0.01 | |||||||
| T/C or C/Cf | 1.35 (1.10–1.66) | <0.01 | 0.84 | 0.02 | |||||
| UMPS | rs1162 | A/A | ref | <0.01 | 0.60 | 0.02 | |||
| A/G | 1.22 (0.99–1.50) | 0.06 | |||||||
| G/G | 1.57 (1.15–2.13) | <0.01 | |||||||
| A/G or G/Gf | 1.29 (1.06–1.57) | 0.01 | 0.84 | 0.07 |
p:P‐value for log‐additive and dominant model.
pFDR:FDR‐adjusted.
pTrend:P‐value trend.
pTrend‐FDR:FDR‐adjusted trend.
pGenewide‐FDR:FDR‐adjusted genewide effect.
Dominant model, (HRhet).
Candidate, FDR‐adjusted cutoff for significance of P‐value = 0.01.
Adjusted for age, sex, stage, grade, BMI, alcohol intake.
P‐values in bold are statistically significant.
Table 3.
Associations between selected polymorphisms in FOCM‐related genes and overall‐ and disease‐free survival stratified by 5‐FU‐based chemotherapy.a
| Gene | SNP | Genotype | No 5‐FU‐based chemotherapy | Received 5‐FU‐based chemotherapy | P‐values Interaction | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alive | Deceased | HR(95% CI) | Alive | Deceased | HR(95%‐CI) | |||||||
| n | n | n | n | ptrend b | ptrend‐FDR c | pFDR‐Genewide d | ||||||
| Overall survival | PON1 | rs3917538 | C/C | 37 | 20 | ref | 231 | 132 | 1.21 (0.71–2.06) | 0.36 | 0.59 | 0.04 |
| C/T | 18 | 12 | 1.55 (0.69–3.47) | 137 | 112 | 1.54 (0.89–2.64) | ||||||
| T/T | 3 | 2 | 0.55 (0.07–4.30) | 9 | 26 | 2.97 (1.51–5.85) | ||||||
| C/T or T/Te | 21 | 14 | 1.30 (0.59–2.83) | 146 | 138 | 1.65 (0.97–2.83) | ||||||
| Disease‐free survival | MAT2B | rs12655857 | G/G | 24 | 21 | ref | 189 | 170 | 0.80 (0.47–1.35) | 0.01 | 0.99 | 0.04 |
| G/T | 31 | 9 | 0.36 (0.15–0.88) | 128 | 119 | 0.91 (0.53–1.55) | ||||||
| T/T | 5 | 2 | 0.22 (0.03–1.67) | 24 | 17 | 0.78 (0.38–1.60) | ||||||
| G/T or T/Te | 36 | 11 | 0.33 (0.14–0.79) | 152 | 136 | 0.89 (0.52–1.52) | 0.01 | 0.85 | 0.03 | |||
| TCN2 | rs9621049f | C/C | 54 | 25 | ref | 264 | 256 | 1.65 (1.03–2.67) | 0.02 | 0.99 | 0.15 | |
| C/T | 6 | 6 | 3.33 (1.22–9.10) | 73 | 47 | 1.41 (0.80–2.45) | ||||||
| T/T | 0 | 1 | 0.63 (0.09–4.60) | 4 | 3 | 1.05 (0.14–7.98) | ||||||
| C/T or T/Te | 6 | 7 | 3.34 (1.22–9.11) | 77 | 50 | 1.39 (0.80–2.43) | 0.02 | 0.85 | 0.08 | |||
Adjusted for age, sex, stage, grade, BMI, alcohol intake.
ptrend: P‐value for trend.
ptrend‐FDR:FDR‐adjusted trend.
pFDR‐Genewide:FDR‐adjusted genewide effect.
Dominant model (HRhet).
Candidate, FDR‐adjusted cutoff for significance of P‐value = 0.02.
P‐values in bold are statistically significant.
Selected results of effect modification analyses are presented in Table 3. We observed nominally significant interactions between 5‐FU‐based chemotherapy and 12 tagSNPs with OS (pInter < 0.05; data not shown). Significant interactions in relation to DFS were observed for three candidate SNPs, two on TCN2 (rs1801198: pInter = 0.02 and rs9621049: pInter = 0.02), and one on SHMT1 (rs9909104:pInter = 0.04). For DFS, we identified 17 nominally significant interactions between tagSNPs and 5‐FU‐based chemotherapy (P < 0.05). There was no significant association with OS or DFS (Table S4a) or effect modification for TYMS 3′ UTR 1494 del with OS or DFS (Table S4b,c). The TSER 2R/2R genotype was associated with a marginal nearly threefold increase in risk of death in patients receiving 5‐FU‐based chemotherapy (HRhzv = 2.97, 95% CI: 0.96–9.26) compared to chemonaïve patients (HRhzv = 2.17, 95% CI: 0.99–4.73pInter = 0.06; Table S4c). FDR‐adjusted analyses showed genewide effects on OS for PON1 and TYMS (both pFDRGene < 0.01) and significant interaction with 5‐FU‐based chemotherapy (e.g., PON1 pFDRGeneInter = 0.04 and TYMS pFDRGeneInter = 0.01).
Genewide effects on DFS and significant interaction with 5‐FU‐based chemotherapy were observed for MAT2B (pFDRGene = 0.01, pFDRGeneInter = 0.04) and UMPS (pFDRGene = 0.02, pFDRGeneInter = 0.02; data not shown). 5‐FU‐based chemotherapy is often combined with other drugs that do not target the folate pathway. Yet, it is possible that drugs such as oxaliplatin or irinotecan may affect 5‐FU‐based–SNP interactions. To address this question, we performed sensitivity analyses restricting the dataset to patients who received 5‐FU / 5‐FU + FA (Table S6). Due to the limited statistical power, results need to be interpreted with caution. For the dominant genotype of rs3917538 (PON1), we have observed similar associations with overall survival between patients receiving 5‐FU‐based chemotherapy compared to patients who have received 5‐FU / 5‐FU + FA: (5‐FU‐based chemotherapy: HRhzv = 2.97, 95% CI: 0.96–9.26; 5‐FU / 5‐FU + FA: HRhzv = 2.84, 95% CI: 1.31–6.16). The same was observed for the associations for rs12655857 (MAT2B) and DFS. For rs9621049 (TCN2) restricting the dataset to patients who have received 5‐FU / 5‐FU + FA revealed a statistically significant reduced risk of death among patients with the CT/TT genotype: (5‐FU / 5‐FU + FA: HRhet/hzv = 0.55, 95% CI: 0.32–0.96)pinteraction = 0.03.
In global pathway analyses, we observed global significance for OS in the “fluorouracil” (P = 0.01) and pyrimidine pathway (P = 0.04), but not in “folate,” “methionine,” or “purine” pathways (Table S5).
Discussion
Our study provides, for the first time, a comprehensive pathway analysis of genetic variants in FOCM and their role in overall‐ and disease‐free survival in patients with CRC. Data from our interaction analyses support the importance of genetic variants as modifiers of response to 5‐FU‐based chemotherapy and the prognostic impact in patients with CRC. Pathway effects were observed for genes in pyrimidine biosynthesis and fluorouracil drug metabolism, which are relevant targets for therapeutic response and CRC prognosis.
Prior studies primarily investigated TYMS and MTHFR candidate gene variants; however, with inconsistent and limited results in that, only a few FOCM‐related genes were evaluated 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19. In agreement with prior research, we have shown an inverse association of rs1801133 (MTHFR, C677T) 29, 30, 31, rs1001761, and rs284714912, 17, 18 with OS in CRC patients. Numerous tagSNPs in FOCM‐related genes were nominally associated with OS (e.g., DPYD, DPYS). After FDR correction, only PON1 (rs3917538; intronic, C/T) remained significant. Notably, prior studies have shown increased serum PON1 activity in patients with CRC compared to healthy controls 32, 33. Genetic variation in PON1 has also been linked to prostate 34 and ovarian cancer 35. There are no prior studies on rs3917538. This SNP, however, is highly correlated with rs662 (LD r 2 = 0.70), a missense mutation within 450‐kb distance of rs3917538. Rs662 has been linked to prognosis in metastatic gastric cancer 36.
For PON1 and TYMS, we observed genewide significance (PON1: pFDRGene = 0.04; TYMS: pFDRGene < 0.01).
We did not observe significant associations of SNPs with DFS after FDR adjustment. Genewide significance after FDR adjustment was observed for MAT2B (pFDRGene = 0.02) and UMPS (pFDRGene < 0.01). MAT2B belongs to the methionine adenosyltransferase family and catalyzes the biosynthesis of S‐adenosylmethionine (SAM). SAM is essential in FOCM and has been linked to induced growth of human colon cancer cells in vitro 37. The gene UMPS encodes uridine 5′‐monophosphate synthase, an enzyme that catalyzes the final steps of de novo pyrimidine biosynthetic pathway. While the activity of this pathway is low in resting cells, it is indispensable in proliferating cells and is invariably upregulated in neoplastic cells and tumors 38.
In stratified analyses by 5‐FU‐based chemotherapy, we did not observe significant interactions with a priori selected candidate SNPs and OS. Significant genewide interactions with 5‐FU‐based chemotherapy were observed for PON1 and TYMS (pFDR‐INT = 0.04, pFDR‐INT < 0.01, respectively). Prior research has linked the response rate and toxicity of 5‐FU‐based chemotherapy to thymidylate synthase 18. In fact, higher expression of TYMS in tumors has been associated with poor prognosis and worse response to 5‐FU‐based chemotherapy regimens 39. While there is strong evidence for the role of TYMS in response to 5‐FU‐based chemotherapy 39, this is the first study linking PON1 to chemotherapy response in CRC. Prior data in metastatic gastric cancer show poor OS in patients with PON1 rs662 AA/AG genotype that have received a combined regimen of 5‐FU‐based chemotherapy, epirubicin, and oxaliplatin 36. This is consistent with our findings.
Significant interaction with 5‐FU‐based chemotherapy and two TCN2 candidates—rs9621049 and rs1801198—was observed for DFS. Prior research has linked rs1801198 to CpG island methylator phenotype high status 40, which is increasingly being recognized as an independent predictor of response to 5‐FU‐based chemotherapy 41, 42.
After FDR adjustment, we observed genewide significant interaction between 5‐FU‐based chemotherapy and DFS for MAT2B that catalyzes SAM biosynthesis. SAM modulates the anticancer effect of 5‐FU, but not other cytotoxic agents such as cisplatin 43. We did observe genewide significance for the association of UMPS with DFS, without effect modification by 5‐FU‐based chemotherapy. This is surprising as mutations of UMPS have been linked to 5‐FU resistance in CRC 44.
This is the most comprehensive study to date investigating the role of FOCM in relation to CRC survival. In addition, we evaluated interactions between FOCM genes and 5‐FU‐based chemotherapy and their impact on CRC prognosis. The pathway analysis approach covered all genetic variants simultaneously; thus, it accounts for interactions between genes assessing the association between a pathway and disease prognosis. All events of interest were ascertained actively and verified using death certificates, medical records, and information from attending physicians. Therefore, misclassification in the outcome variable is highly unlikely. The majority of patients were residents of Central Europe, which is indicative for a homogeneous study population.
Several limitations should be noted. False‐positive results might have occurred when we investigated the gene‐5‐FU interactions although we used FDR to minimize this possibility. The generalizability of our discoveries from a population free of folic acid fortification to populations where fortification is mandatory may be limited as folic acids can impact several aspects of FOCM 45. Further investigations in clinical populations are warranted to replicate findings and validate the clinical importance of the present results.
In conclusion, genetic variation in FOCM appears to be, to some extent, associated with CRC prognosis. Notably, effects were observed for genes in pyrimidine biosynthesis and fluorouracil drug metabolism, which are relevant therapeutic targets. Further investigations in clinical populations are warranted to replicate findings and validate the clinical importance of the present results.
Conflict of Interest
There are no conflict of interest disclosures from the authors.
Supporting information
Table S1. Polymorphisms in Folate‐mediated One‐Carbon Metabolism by Gene.
Table S2. Candidate SNPs.
Table S3. Selected subpathways and genes included.
Table S4. (a) Associations between non‐SNP TYMS polymorphisms with overall and disease‐free survival. (b) Associations between non‐SNP TYMS polymorphisms with overall survival stratified by 5‐FU chemotherapy. (c) Associations between non‐SNP TYMS polymorphisms with disease‐free survival stratified by 5‐FU chemotherapy.
Table S5. Global test on different pathways.
Table S6. Associations between selected polymorphisms in FOCM‐related genes and overall‐ and disease‐free survival stratified by 5‐FU‐based chemotherapy.
Table S7. Associations between polymorphisms in FOCM‐related genes and overall survival.
Table S8. Associations between selected polymorphisms in FOCM‐related genes and disease‐free survival.
Table S9. Associations between polymorphisms in FOCM‐related genes and overall survival stratified by 5‐FU‐based chemotherapy
Table S10. Associations between polymorphisms in FOCM‐related genes and disease‐free survival stratified by 5‐FU‐based chemotherapy*.
Acknowledgments
We thank all participants of the DACHS study, the interviewers, the physicians and staff of the following hospitals and cooperating institutions: Chirurgische Universitätsklinik Heidelberg, Klinik am Gesundbrunnen Heilbronn, St. Vincentiuskrankenhaus Speyer, St. Josefskrankenhaus Heidelberg, Chirurgische Universitätsklinik Mannheim, Diakonissenkrankenhaus Speyer, Krankenhaus Salem Heidelberg, Kreiskrankenhaus Schwetzingen, St. Marien‐ und St. Annastiftkrankenhaus Ludwigshafen, Klinikum Ludwigshafen, Stadtklinik Frankenthal, Diakoniekrankenhaus Mannheim, Kreiskrankenhaus Sinsheim, Klinikum am Plattenwald Bad Friedrichshall, Kreiskrankenhaus Weinheim, Kreiskrankenhaus Eberbach, Kreiskrankenhaus Buchen, Kreiskrankenhaus Mosbach, Enddarmzentrum Mannheim, Kreiskrankenhaus Brackenheim, and the Cancer Registry of Rhineland‐Palatinate. We thank the microarray unit of the Genomics and Proteomics Core Facility of the German Cancer Research Center for genotyping, particularly Matthias Schick, and thank Muhabbet Celik, Ursula Eilber, Sabine Behrens, Petra Bächer, and Ute Handte‐Daub for their excellent technical assistance.
Cancer Medicine 2018; 7(7):2797–2807
References
- 1. Siegel, R. L. , Miller K. D., and Jemal A.. 2016. Cancer statistics, 2016. CA Cancer J. Clin. 66:7–30. [DOI] [PubMed] [Google Scholar]
- 2. Scartozzi, M. , Maccaroni E., Giampieri R., Pistelli M., Bittoni A., Del Prete M., et al. 2011. 5‐Fluorouracil pharmacogenomics: still rocking after all these years? Pharmacogenomics 12:251–265. [DOI] [PubMed] [Google Scholar]
- 3. Ulrich, C. M. , Robien K., and McLeod H. L.. 2003. Cancer pharmacogenetics: polymorphisms, pathways and beyond. Nat. Rev. Cancer 3:912–920. [DOI] [PubMed] [Google Scholar]
- 4. Ulrich, C. M. , Rankin C., Toriola A. T., Makar K. W., Altug‐Teber Ö., Benedetti J. K., et al. 2014. J Polymorphisms in folate‐metabolizing enzymes and response to 5‐fluorouracil among patients with stage II or III rectal cancer (INT‐0144; SWOG 9304). Cancer 120:3329–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jennings, B. A. , and Willis G.. 2015. How folate metabolism affects colorectal cancer development and treatment; a story of heterogeneity and pleiotropy. Cancer Lett. 356:224–230. [DOI] [PubMed] [Google Scholar]
- 6. Cheng, T. Y. D. , Makar K. W., Neuhouser M. L., Miller J. W., Song X., Brown E. C., et al. 2015. Folate‐mediated one‐carbon metabolism genes and interactions with nutritional factors on colorectal cancer risk: Women's Health Initiative Observational Study. Cancer 121:3684–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wisotzkey, J. K. , Toman J., Bell T., Monk J. S., and Jones D.. 1999. MTHFR (C677T) polymorphisms and stage III colon cancer: response to therapy. Mol. Diag. 4:95–99. [DOI] [PubMed] [Google Scholar]
- 8. Etienne, M. C. , Ilc K., Formento J. L., Laurent‐Puig P., Formento P., Cheradame S., et al. 2004. Thymidylate synthase and methylenetetrahydrofolate reductase gene polymorphisms: relationships with 5‐fluorouracil sensitivity. Br. J. Cancer 90:526–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jakobsen, A. , Nielsen J. N., Gyldenkerne N., and Lindeberg J.. 2005. Thymidylate synthase and methylenetetrahydrofolate reductase gene polymorphism in normal tissue as predictors of fluorouracil sensitivity. J. Clin. Oncol. 23:1365–1369. [DOI] [PubMed] [Google Scholar]
- 10. Suh, K. W. , Kim J. H., Kim Y. B., Lee C., and Choi S.. 2006. Which gene is a dominant predictor of response during FOLFOX chemotherapy for the treatment of metastatic colorectal cancer, the MTHFR or XRCC1 gene? Ann. Surg. Oncol. 13:1379–1385. [DOI] [PubMed] [Google Scholar]
- 11. Cohen, V. , Panet‐Raymond V., Sabbaghian N., Morin I., Batist G., and Rozen R.. 2003. Methylenetetrahydrofolate reductase polymorphism in advanced colorectal cancer: a novel genomic predictor of clinical response to fluoropyrimidine‐based chemotherapy. Clin. Cancer Res. 9:1611–1615. [PubMed] [Google Scholar]
- 12. Gusella, M. , and Padrini R.. 2007. G>C SNP of thymidylate synthase with respect to colorectal cancer. Pharmacogenomics 8:985–996. [DOI] [PubMed] [Google Scholar]
- 13. Marcuello, E. , Altes A., del Rio E., Cesar A., Menoyo A., and Baiget M.. 2004. Single nucleotide polymorphism in the 5' tandem repeat sequences of thymidylate synthase gene predicts for response to fluorouracil‐based chemotherapy in advanced colorectal cancer patients. Int. J. Cancer 112:733–737. [DOI] [PubMed] [Google Scholar]
- 14. Lurje, G. , Zhang W., Yang D., Groshen S., Hendifar A. E., Husain H., et al. 2007. Thymidylate synthase haplotype is associated with tumor recurrence in stage II and stage III colon cancer patients. in preparation. [DOI] [PubMed]
- 15. Afzal, S. , Jensen S. A., Vainer B., Vogel U., Matsen J. P., Sørensen J. B., et al. 2009. MTHFR polymorphisms and 5‐FU‐based adjuvant chemotherapy in colorectal cancer. Ann. Oncol. 20:1660–1666. [DOI] [PubMed] [Google Scholar]
- 16. Etienne‐Grimaldi, M. C. , Milano G., Maindrault‐Gœbel F., Chibaudel B., Formento J. L., Francoual M., et al. 2010. Methylenetetrahydrofolate reductase (MTHFR) gene polymorphisms and FOLFOX response in colorectal cancer patients. Br. J. Clin. Pharmacol. 69:58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Edler, D. , Glimelius B., Hallström M., Jakobsen A., Johnston P. G., Magnusson I., et al. 2002. Thymidylate synthase expression in colorectal cancer: a prognostic and predictive marker of benefit from adjuvant fluorouracil‐based chemotherapy. J. Clin. Oncol. 20:1721–1728. [DOI] [PubMed] [Google Scholar]
- 18. Lurje, G. , Manegold P. C., Ning Y., Pohl A., Zhang W., and Lenz H. J.. 2009. Thymidylate synthase gene variations: predictive and prognostic markers. Mol. Cancer Ther. 8:1000–1007. [DOI] [PubMed] [Google Scholar]
- 19. Marcuello, E. , Altes A., Menoyo A., Rio E. D., and Baiget M.. 2006. Methylenetetrahydrofolate reductase gene polymorphisms: genomic predictors of clinical response to fluoropyrimidine‐based chemotherapy? Cancer Chemother. Pharmacol. 57:835–840. [DOI] [PubMed] [Google Scholar]
- 20. Brenner, H. , Chang‐Claude J., Jansen L., Knebel P., Stock C., and Hoffmeister M.. 2014. Reduced risk of colorectal cancer up to 10 years after screening, surveillance, or diagnostic colonoscopy. Gastroenterology 146:709–717. [DOI] [PubMed] [Google Scholar]
- 21. Ulrich, C. M. , Curtin K., Potter J. D., Bigler J., Caan B., and Slattery M. L.. 2005. Polymorphisms in the reduced folate carrier, thymidylate synthase, or methionine synthase and risk of colon cancer. Cancer Epidemiol. Biomarkers Prev. 14(Pt 1):2509–2516. [DOI] [PubMed] [Google Scholar]
- 22. Barrett, J. C. , Fry B., Maller J., and Daly M. J.. 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265. [DOI] [PubMed] [Google Scholar]
- 23. Fan, J. B. , Oliphant A., Shen R., Kermani B. G., Garcia F., Gunderson K. L., et al. 2003. Highly parallel SNP genotyping. Cold Spring Harb. Symp. Quant. Biol. 68:69–78. [DOI] [PubMed] [Google Scholar]
- 24. Gabriel, S. , Ziaugra L., and Tabbaa D.. 2009. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr. Protoc. Hum. Genet. Chapter 2: unit2 12. [DOI] [PubMed] [Google Scholar]
- 25. Howie, B. N. , Donnelly P., and Marchini J.. 2009. A flexible and accurate genotype imputation method for the next generation of genome‐wide association studies. PLoS Genet. 5:e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Price, A. L. , Patterson N. J., Plenge R. M., Weinblatt M. E., Shadick N. A., and Reich D.. 2006. Principal components analysis corrects for stratification in genome‐wide association studies. Nat. Genet. 38:904–909. [DOI] [PubMed] [Google Scholar]
- 27. Chen, Y. A. , Almeida J. S., Richards A. J., Muller P., Carroll R. J., and Rohrer B.. 2010. A nonparametric approach to detect nonlinear correlation in gene expression. J. Comput. Graph Stat. 19:552–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Johnson, A. D. , Handsaker R. E., Pulit S. L., Nizzari M. M., O'Donnell C. J., and de Bakker P. I.. 2008. SNAP: a web‐based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 24:2938–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Figueiredo, J. C. , Levine A. J., Crott J. W., Baurley J., and Haile R. W.. 2013. Folate‐genetics and colorectal neoplasia: what we know and need to know next. Mol. Nutr. Food Res. 57:607–627. [DOI] [PubMed] [Google Scholar]
- 30. Yeh, C. C. , Lai C. Y., Chang S. N., Hsieh L. L., Tang R., Sung F. C., et al. 2017. Polymorphisms of MTHFR C677T and A1298C associated with survival in patients with colorectal cancer treated with 5‐fluorouracil‐based chemotherapy. Int. J. Clin. Oncol. 22:484–493. [DOI] [PubMed] [Google Scholar]
- 31. Yehuda‐Shnaidman, E. , and Schwartz B.. 2012. Mechanisms linking obesity, inflammation and altered metabolism to colon carcinogenesis. Obes. Rev. 13:1083–1095. [DOI] [PubMed] [Google Scholar]
- 32. Afsar, C. U. , Gunaldı M., Okuturlar Y., Gedikbası A., Tiken E. E., Kahraman S., et al. 2015. Paraoxonase‐1 and arylesterase activities in patients with colorectal cancer. Int. J. Clin. Exp. Med. 8:21599–21604. [PMC free article] [PubMed] [Google Scholar]
- 33. Bulbuller, N. , Eren E., Ellidag H. Y., Oner O. Z., Sezer C., Aydin O., et al. 2013. Diagnostic value of thiols, paraoxonase 1, arylesterase and oxidative balance in colorectal cancer in human. Neoplasma 60:419–424. [DOI] [PubMed] [Google Scholar]
- 34. Antognelli, C. , Del Buono C., Ludovini V., Gori S., Talesa V. N., Crinò L., et al. 2009. CYP17, GSTP1, PON1 and GLO1 gene polymorphisms as risk factors for breast cancer: an Italian case‐control study. BMC Cancer 9:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Camuzcuoglu, H. , Arioz D. T., Toy H., Kurt S., Celik H., and Erel O.. 2009. Serum paraoxonase and arylesterase activities in patients with epithelial ovarian cancer. Gynecol. Oncol. 112:481–485. [DOI] [PubMed] [Google Scholar]
- 36. Geng, R. , Chen Z., Zhao X., Qiu L., Liu X., Liu R., et al. 2014. Oxidative stress‐related genetic polymorphisms are associated with the prognosis of metastatic gastric cancer patients treated with epirubicin, oxaliplatin and 5‐fluorouracil combination chemotherapy. PLoS ONE 9:e116027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen, H. , Xia M., Lin M., Yang H., Kuhlenkamp J., Li T., et al. 2007. Role of methionine adenosyltransferase 2A and S‐adenosylmethionine in mitogen‐induced growth of human colon cancer cells. Gastroenterology 133:207–218. [DOI] [PubMed] [Google Scholar]
- 38. Garavito, M. F. , Narvaez‐Ortiz H. Y., and Zimmermann B. H.. 2015. Pyrimidine metabolism: dynamic and versatile pathways in pathogens and cellular development. J. Gene. Genomics 42:195–205. [DOI] [PubMed] [Google Scholar]
- 39. Popat, S. , Matakidou A., and Houlston R. S.. 2004. Thymidylate synthase expression and prognosis in colorectal cancer: a systematic review and meta‐analysis. J. Clin. Oncol. 22:529–536. [DOI] [PubMed] [Google Scholar]
- 40. Hazra, A. , Fuchs C. S., Kawasaki T., Kirkner G. J., Hunter D. J., and Ogino S.. 2010. Germline polymorphisms in the one‐carbon metabolism pathway and DNA methylation in colorectal cancer. Cancer Causes Control 21:331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Donada, M. , Bonin S., Barbazza R., Pettirosso D., and Stanta G.. 2013. Management of stage II colon cancer ‐ the use of molecular biomarkers for adjuvant therapy decision. BMC Gastroenterol. 13:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Han, S. W. , Lee H. J., Bae J. M., Cho N. Y., Lee K. H., Kim T. Y., et al. 2013. J Methylation and microsatellite status and recurrence following adjuvant FOLFOX in colorectal cancer. Int. J. Cancer 132:2209–2216. [DOI] [PubMed] [Google Scholar]
- 43. Ham, M. S. , Lee J. K., and Kim K. C.. 2013. S‐adenosyl methionine specifically protects the anticancer effect of 5‐FU via DNMTs expression in human A549 lung cancer cells. Mol. Clin. Oncol. 1:373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Griffith, M. , Mwenifumbo J. C., Cheung P. Y., Paul J. E., Pugh T. J., Tang M. J., et al. 2013. Novel mRNA isoforms and mutations of uridine monophosphate synthetase and 5‐fluorouracil resistance in colorectal cancer. Pharmacogenomics J 13:148–158. [DOI] [PubMed] [Google Scholar]
- 45. Bae, S. , Ulrich C. M., Bailey L. B., Malysheva O., Brown E. C., Maneval D. R., et al. 2014. Impact of folic acid fortification on global DNA methylation and one‐carbon biomarkers in the Women's Health Initiative Observational Study cohort. Epigenetics 9:396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Polymorphisms in Folate‐mediated One‐Carbon Metabolism by Gene.
Table S2. Candidate SNPs.
Table S3. Selected subpathways and genes included.
Table S4. (a) Associations between non‐SNP TYMS polymorphisms with overall and disease‐free survival. (b) Associations between non‐SNP TYMS polymorphisms with overall survival stratified by 5‐FU chemotherapy. (c) Associations between non‐SNP TYMS polymorphisms with disease‐free survival stratified by 5‐FU chemotherapy.
Table S5. Global test on different pathways.
Table S6. Associations between selected polymorphisms in FOCM‐related genes and overall‐ and disease‐free survival stratified by 5‐FU‐based chemotherapy.
Table S7. Associations between polymorphisms in FOCM‐related genes and overall survival.
Table S8. Associations between selected polymorphisms in FOCM‐related genes and disease‐free survival.
Table S9. Associations between polymorphisms in FOCM‐related genes and overall survival stratified by 5‐FU‐based chemotherapy
Table S10. Associations between polymorphisms in FOCM‐related genes and disease‐free survival stratified by 5‐FU‐based chemotherapy*.


