Abstract
The association between adult height and risk of lung cancer has been investigated by epidemiology studies, but the results are inconsistent. Mendelian randomization (MR) analyses with individual‐level data from two genome‐wide association studies, including a total of 7127 lung cancer cases and 6818 controls, were carried out to explore whether adult height is causally associated with risk of lung cancer. A weighted genetic risk score (wGRS) was created based on genotypes of 101 known height‐associated genetic variants. Association between the wGRS and risk of lung cancer was analyzed by logistic regression for each study separately. The combined effect was calculated using fixed effect meta‐analysis. MR analyses showed that increased risk of lung cancer (OR = 1.19, 95%CI: 1.05‐1.35, P = 0.006) associated with taller genetically determined height. Compared with individuals in the lowest tertile of the height‐associated wGRS, those in the highest tertile had 1.10‐fold (95% CI: 1.01‐1.20) increased risk of developing lung cancer. Sensitivity analyses excluding BMI‐associated genetic variants demonstrated consistent association. Our study suggested that genetically taller height was associated with increased risk of lung cancer in East Asian population, indicating that increasing height may have a causal role in lung cancer carcinogenesis.
Keywords: Adult height, East Asian population, Lung cancer, Mendelian randomization
1. INTRODUCTION
Lung cancer is one of the leading causes of cancer morbidity and mortality worldwide.1 It is estimated that there were 1.8 million incident cases of lung cancer and 1.6 million cause‐specific deaths, accounting for nearly one‐fifth of total cancer deaths in 2013.2 As known, lung cancer is a multifactorial disease involving both environmental and genetic factors. Tobacco smoking is the main risk factor for lung cancer, relating to approximately 90% of lung cancer cases.3 Other known risk factors for lung cancer include exposure to occupational and environmental carcinogens such as asbestos and outdoor pollution.4, 5
Human anthropometric indicators are associated with multiple diseases, including cancers. Over the past decade, plenty of epidemiological studies have investigated the associations between body‐mass index (BMI) and cancer risk6, 7; however, the relationship between adult height and cancer risk has received much less attention. Adult height is a complex and highly heritable trait that is determined by both genetic and environmental factors. The heritability of height has been estimated to be up to 80%‐90%.8, 9, 10 Nutrition, diseases, as well as socioeconomic status, are important environmental factors that might affect body height in adulthood.11 Moreover, height measurement is noninvasive, cost‐efficient, and accurate in population‐based studies, which makes adult height become a potential tool for monitoring health conditions.12
Several previous studies have investigated the association of adult height with risk of lung cancer, but the results are inconsistent. A Korea cohort study reported each 5‐cm increment in height was associated with increased risk of lung cancer.13 Similar associations were also recorded in two recent meta‐analyses14, 15; however, the results from Million Women Study in UK did not find significant associations between height and risk of lung cancer.16 It remains unclear whether the observed association reflects a causal effect of adult height on lung cancer, or is due to confounding or biases inherent in conventional epidemiological studies.
Mendelian randomization (MR) is a technique of using genetic variants to estimate the causal effect of a modifiable risk factor from observational data.17 As genotypes generate through alleles randomly assort at gamete formation and segregate randomly at conception, associations between genotypes and outcome are not generally confounded by environmental factors and therefore can avoid reverse causation.
In this study, we used MR approach to assess the association between height and risk of lung cancer using individual‐level data from 13 945 subjects of East Asian population. We derived a weighted genetic risk score (wGRS) comprising 101 height‐related single nucleotide polymorphisms (SNP) identified by previous genome‐wide association studies (GWAS) in East Asian‐ancestry populations and analyzed whether there is a causal relationship between height and risk of lung cancer.
2. MATERIALS AND METHODS
2.1. Study subjects
We used two existing data from previously published lung cancer GWAS studies, that is, the Nanjing Medical University (NJMU) and the Female Lung Cancer Consortium in Asia (FLCCA). The details of these two studies were described elsewhere.18, 19, 20 In brief, the NJMU study included 2331 cases and 3077 controls from Nanjing, Shanghai, Beijing, and Wuhan in China. The FLCCA study was obtained via the database of Genotypes and Phenotypes (dbGAP), and included 4922 cases and 3959 controls from mainland China, South Korea, Japan, Singapore, Taiwan and Hong Kong. For the FLCCA study, we also excluded the overlapped subjects between FLCCA GWAS and NJMU GWAS, thus, 4796 cases and 3741 controls were included in following analyses. Adult height data of the controls were only available in the NJMU Nanjing Study. Height measurement followed standard procedure and was measured to the nearest 0.1 cm. All study participants provided their written informed consent, and the study protocols were approved by the relevant Institutional Review Boards. The demographic characteristics of study population are summarized in Table S1. In total, our analysis consisted of 7127 lung cancer cases and 6818 controls from samples of East Asian descent. The lung cancer cases included 4773 lung adenocarcinoma, 1482 lung squamous cell carcinoma and 872 other lung cancer types.
2.2. Genotype imputation
Full details of the genotyping, quality control and imputation have been reported previously.18, 19, 20 Briefly, the NJMU GWAS was conducted using Affymetrix Genome‐Wide Human SNP Array 6.0 with standard GWAS quality‐control procedures. The FLCCA GWAS was conducted using Illumina Human610_Quadv1_B and Human660W‐Quad_v1_A whole genome genotyping array and downloaded from the database of dbGaP database (dbGaP Study Accession: phs000716.v1.p1). After initial quality control, we excluded individuals with low call rates (<95%), familial relationships and extreme heterozygosity rates, and removed SNPs with low call rates (<95%), minor allele frequency (MAF) <5% and P < 1 × 10−6 for the Hardy‐Weinberg equilibrium test in controls. Genotype imputation was performed with IMPUTE2 (V.2.2.2) software using the 1000 Genomes Project Phase 3 as a reference. Population structure was evaluated by principal components analysis using the software package EIGENSTRAT 3.0 for the NJMU study and the FLCCA study separately.
2.3. Identification of SNPs associated with height
We selected SNPs associated with adult height from three previously published GWAS studies among individuals of East Asian ancestry.21, 22, 23 We identified 110 SNPs showing genome‐wide significance level (P < 5 × 10−8) in East Asian population. To infer adult height more effectively, we further included 26 additional SNPs which were previously reported to be associated with height in populations of European ancestry and replicated with significance level (P < 1 × 10−5) in East Asian studies. Two variants, rs234886 and rs12680655, were not available in our lung cancer GWAS data and were replaced by high linkage disequilibrium (LD) SNPs, rs2237886 and rs733254 (r 2 > 0.9 in 1000 Genomes Project Phase 3 East Asian population). Thirty‐five SNPs were in moderate LD (r 2 > 0.1) with other SNPs based on 1000 Genomes Project Phase 3 data and then excluded in subsequent analyses. Finally, a total of 101 independent height‐related SNPs were used as instrumental variables in our study. In sensitivity analyses, we further excluded SNPs which were associated with BMI or had moderate LD (r 2 > 0.1) with BMI‐related SNPs in East Asian population. Thirty‐seven BMI‐related SNPs were selected from a recently published BMI MR study.24
2.4. MR assumptions
The genetic variants used as instrumental variables in MR need to meet three assumptions25: (1) the genetic variants are associated with the exposure; (2) the genetic variants affect the outcome only via the exposure; (3) the genetic variants are not associated with any confounders that may affect the exposure‐outcome association. Based on the second assumption, a SNP’s association with height should be proportional to its association with cancer risk. Therefore, we assessed the potential violation (pleiotropic effect) of the latter two assumptions using pleiotropy test in “gtx” package (v0.0.8) in R software. Six SNPs in NJMU study and three SNPs in FLCCA study were excluded for further analyses (Tables S2 and S3). Finally, 95 SNPs in NJMU study and 98 SNPs in FLCCA study were used in our study.
2.5. Statistical analysis
We calculated the wGRS for height‐related associated variants as an instrumental variable. To create wGRS for the i‐th subject from the height‐associated variants the following formula was used:
Here, x ij is the number of risk alleles for the j‐th SNP in the i‐th subject (x ij = 0, 1 or 2 for wild‐type homozygous, heterozygous, or homozygous for the effect allele associated with taller height) and w j is the weight or coefficient for the j‐th SNP. The weights or coefficients were the height‐associated β‐estimates scaled to the z‐score‐transformed height per tall allele, obtaining from the previously published GWAS studies. The wGRS could be used as an instrumental variable to represent z‐score‐transformed height in individual level. A higher wGRS indicates taller height, whereas a lower wGRS for shorter height, using association estimate for the tall allele. The effect of wGRS was estimated separately for two studies using logistic regression adjusted for age, sex, pack‐years, and first principal component in NJMU study and adjusted for age, and three principal components in FLCCA study. The combined effect and heterogeneity of height‐associated wGRS on lung cancer was calculated by meta‐analysis.
Besides the wGRS approach, we estimated the effect of height on risk of lung cancer using inverse‐variance weighting (IVW) method, which is a summary data based on MR method. Summary association estimates were used in IVW to calculate causal effects, which was described in greater detail by Burgess et al26 The potential causal association between height and risk of lung cancer was modeled using height‐related SNPs’ β‐estimates obtaining from published GWAS studies. The Cochran’s Q test was used to assess the pleiotropic effect for the genetic variants. The IVW method was conducted in R software using “gtx” package (v0.0.8).
To further examine the relationship between height and lung cancer, we categorized height‐related wGRS into three groups based on its tertile distribution in all participants. We accessed potential non‐linear trends between height and risk of lung cancer using a restricted cubic spline analysis.27 Same statistic methods with wGRS were used to estimate the effect and heterogeneity of SNPs. To identify common biologic processes that might explain the association between height and lung cancer, we performed KEGG pathway analysis using R package “clusterProfiler” (v3.0.5). The genes that 101 height‐associated SNPs locate in or nearest were involved in pathway analysis (Table S2).
All statistical analyses were performed by R version 3.3.1. All statistical tests were two‐sided with P < 0.05 considered statistically significant.
3. RESULTS
3.1. Height‐associated variants and lung cancer
The associations between 101 height‐related SNPs and lung cancer are shown in Table S2. Most of the height‐related SNPs were not significant in both two studies. However, there were 15 SNPs in NJMU study and 13 SNPs in FLCCA study had nominally significant associations (P < 0.05), and only one SNP, rs174547, in NJMU study survived Bonferroni correction (P < 4.95 × 10−4). However, rs174547 was removed after pleiotropy test.
3.2. wGRS and observed height in controls
We first evaluate the relationship between observed height and the wGRS in 1146 controls. As shown in Figure 1, the increasing wGRS was significantly associated with higher observed height in both male (r 2 = 0.014, P = 7.19 × 10−4) and female (r 2 = 0.021, P = 9.41 × 10−3) participants, the estimated effect was in the positive direction (male β estimate = 2.37, female β estimate = 2.92). Therefore, this provides evidence that the wGRS of height‐associated variants has utility in predicting adult height in East Asian populations.
Figure 1.
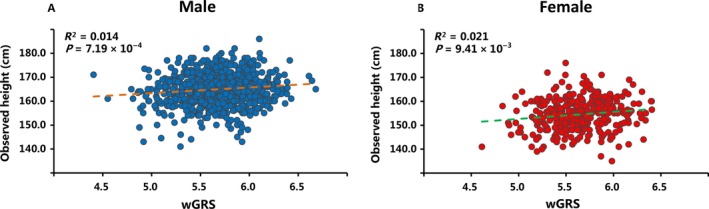
Relation of height‐associated variants wGRS with observed height from 1146 controls in the NJMU study Nanjing samples. Best‐fit lines (dash line) are drawn for the relationship of measured height with height‐associated variants wGRS for (A) male (N = 820) and (B) female (N = 326)
3.3. Genetically determined height associates with risk of lung cancer
As shown in Table 1, the meta‐analysis indicated that higher genetically predicted height was associated with increased risk of all lung cancer (OR = 1.19, 95% CI = 1.05‐1.35, P = 0.006). The significant associations were also found in lung adenocarcinoma (OR = 1.18, 95% CI = 1.03‐1.36, P = 0.020) and lung squamous cell carcinoma (OR = 1.29, 95% CI = 1.04‐1.61, P = 0.021). The results of two studies are presented in Table S4. No significant heterogeneity was observed among two studies. When modeled as a tertile of height‐associated wGRS, wGRS was significantly associated with an increased risk of all lung cancer (per tertile OR = 1.05, 95% CI = 1.01‐1.09, trend P = 0.028) and lung squamous cell carcinoma (per tertile OR = 1.08, 95% CI = 1.01‐1.17, trend P = 0.035); however, not significant in lung adenocarcinoma (per tertile OR = 1.04, 95% CI = 0.99‐1.09, trend P = 0.094). Compared with individuals in the lowest tertile of wGRS, those in the highest tertile had a 1.10‐fold (95% CI = 1.01‐1.20, P = 0.029), 1.09‐fold (95% CI = 0.99‐1.19, P = 0.094), and 1.18‐fold (95% CI = 1.01‐1.37, P = 0.035) increased risk of developing all lung cancer, lung adenocarcinoma and lung squamous cell carcinoma, respectively (Table 2). Additionally, we did not observe a significant non‐linear association (P non‐linear > 0.05) between the height‐related wGRS and risk of lung cancer among two studies (Figure S1), suggesting a potential linear effect of genetic determined height on risk of lung cancer. Furthermore, we estimated the potential causal effect of height on lung cancer using the MR IVW method with summary statistics of each height‐related SNPs. Compared with that using the wGRS method, similar associations between height and risk of lung cancer were observed among all lung cancer, lung adenocarcinoma and lung squamous cell carcinoma (Table 1).
Table 1.
Effect estimates for associations of genetic instruments with height and lung cancer risk
| Genetic instruments | Lung adenocarcinoma | Lung squamous cell carcinoma | All lung cancer | |||
|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | |
| wGRSa | 1.18 (1.03, 1.36) | 0.020 | 1.29 (1.04, 1.61) | 0.021 | 1.19 (1.05, 1.35) | 0.006 |
| IVWa | 1.18 (1.03, 1.36) | 0.018 | 1.30 (1.04, 1.62) | 0.021 | 1.19 (1.05, 1.35) | 0.006 |
| Unweighted GRS | 1.01 (1.00, 1.01) | 0.007 | 1.01 (1.00, 1.02) | 0.029 | 1.01 (1.00, 1.01) | 0.003 |
| wGRS unrelated to BMIb | 1.15 (1.00, 1.33) | 0.046 | 1.26 (1.01, 1.57) | 0.045 | 1.16 (1.02, 1.32) | 0.020 |
| IVW unrelated to BMIb | 1.16 (1.00, 1.33) | 0.045 | 1.26 (1.00, 1.57) | 0.046 | 1.16 (1.03, 1.32) | 0.019 |
| Heterogeneityc | I 2 = 19.3% | 0.266 | I 2 = 0 | 0.821 | I 2 = 15.8% | 0.276 |
The OR and P value were calculated from meta‐analysis of two studies.
The OR and P value were calculated with SNPs after excluding BMI‐related SNPs.
Heterogeneity test was conducted for wGRS of two studies.
Table 2.
Associations of categories of genetic risk score (wGRS) predicting between height and lung cancer risk
| wGRS categories | Lung adenocarcinomaa | Lung squamous cell carcinomaa | All lung cancera | |||
|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | |
| Q1 | Ref. | Ref. | Ref. | |||
| Q2 | 1.06 (0.97, 1.17) | 0.209 | 1.08 (0.92, 1.25) | 0.345 | 1.08 (1.00, 1.18) | 0.062 |
| Q3 | 1.09 (0.99, 1.19) | 0.094 | 1.18 (1.01, 1.37) | 0.035 | 1.10 (1.01, 1.20) | 0.029 |
| P for Trend | 0.094 | 0.035 | 0.028 | |||
The OR and P value were calculated from meta‐analysis of two studies.
3.4. Sensitivity analyses
To determine whether the analyses were robust to the choice of weights used in the genetic risk score calculation, we analyzed the causal association of height and risk of lung cancer using an unweighted GRS. As expected, the association of unweighted GRS remained statistically significant (Table 1), though, the effect size was attenuated. Nor did the results of wGRS change substantially when we additionally excluded the BMI‐related SNPs which might interfere the genetic risk of adult height (Table 1).
3.5. Pathway analysis
To further clarify the potential mechanism of genetically determined height in the development of cancer, we performed pathway analysis. A total of eight significant enriched KEGG pathways (Q value < 0.05) were identified (Table 3). The most significant enriched pathway was transforming growth factor (TGF)‐β signaling pathway, an essential biological pathway of growth and development processes. We also found another two pathways, transcriptional misregulation in cancer and proteoglycans in cancer, directly related to cancer. Most of the significant enriched KEGG pathways, such as TGF‐β, MAPK, Hippo and Hedgehog signaling pathway were important in both development and carcinogenesis.
Table 3.
Biologic pathways identified by KEGG of height‐associated genes
| KEGG ID | Description | GeneRatioa | P value | Q valueb | Genes |
|---|---|---|---|---|---|
| hsa04350 | TGF‐beta signaling pathway | 0.14 | 8.30 × 10−6 | 0.001 | LTBP1/RBL1/MYC/TGFB2/BMP2/BMP6 |
| hsa04913 | Ovarian steroidogenesis | 0.09 | 1.96 × 10−4 | 0.011 | IGF1R/BMP6/IGF1/CYP19A1 |
| hsa05202 | Transcriptional misregulation in cancer | 0.14 | 6.94 × 10−4 | 0.025 | MEF2C/HMGA2/MYC/IGF1R/IGF1/SUPT3H |
| hsa05205 | Proteoglycans in cancer | 0.14 | 1.10 × 10−3 | 0.030 | CCND1/MYC/TGFB2/IGF1R/IGF1/PTCH1 |
| hsa04010 | MAPK signaling pathway | 0.16 | 1.46 × 10−3 | 0.032 | MEF2C/MYC/TGFB2/MAP3K3/IGF1R/IGF1/GNA12 |
| hsa04390 | Hippo signaling pathway | 0.12 | 1.96 × 10−3 | 0.035 | CCND1/MYC/TGFB2/BMP2/BMP6 |
| hsa04630 | Jak‐STAT signaling pathway | 0.12 | 2.44 × 10−3 | 0.035 | CCND1/GHR/SOCS2/MYC/STAT2 |
| hsa04340 | Hedgehog signaling pathway | 0.07 | 2.56 × 10−3 | 0.035 | CCND1/HHIP/PTCH1 |
The gene ratio is the proportion of the genes in the KEGG pathway that were part of the input list for the height‐related genes.
The Q value was calculated for determining the false discovery rate.
4. DISCUSSION
In this study, we applied the MR approach to investigate the relationship between genetically determined height and risk of lung cancer. We found that the increase of height wGRS was roughly linear associated with increased risk of lung cancer, as well as lung adenocarcinoma and lung squamous cell carcinoma subtypes. The findings were robust in analyses used MR IVW method, an unweighted GRS method, or additionally excluded BMI‐associated genetic variants.
Several large prospective cohort studies have assessed the associations between adult height and risk of cancers. Increased adult height has been reported positively associated with the risk of all cancer combined and several site‐specific cancers.13, 16, 28, 29 Green et al16 observed significant positive associations of adult height with up to 10 cancer types based on UK Million Women Study, but not including lung cancer. By contrast, Sung et al13 observed a significant positive association of adult height with lung cancer in a male Korean population. The inconsistent result may be caused by different genetic background between populations or the underlying confounding in observational studies. A recent meta‐analysis concluded that per 10‐cm increase in height was positively associated with 6% increased risk of lung cancer. Modest association between genetically determined height and risk of lung cancer, based on European population, was reported using MR analysis. Moreover, the increased risk for lung adenocarcinoma appears to be stronger than lung squamous cell carcinoma.14 Our results are in line with previous observational study in Korean population and the height MR study. For the analysis presented here, we provide novel evidence of potential causal effect of adult height on risk of lung cancer in East Asian population.
Although numerous previous studies have proved that height may increase cancer risk, the underlying biological mechanisms are still unclear. Adult height is not only determined by a large proportion of genetic factors, but also affected by environmental factors such as nutrition during early life and adolescence, infections and diseases. One possible common mechanism is that hormone levels, especially growth factors like insulin‐like growth factors (IGFs), may play an important role in height‐related increasing cancer risk.30, 31 IGF‐I levels in childhood and adolescence are strongly related to skeletal growth,32 what is more, circulating levels of IGF‐1 has been identified as a risk factor of breast, prostate and colorectum cancer.33, 34, 35, 36 Another possible explanation is that taller people have more cells, thus a greater opportunity for mutations leading to malignant transformation.37, 38 Shared genetic factors may also be a possible explanation. Studies have found that genes related to increased height are also linked with oncogenic pathways, such as p53, c‐Myc, and SMAD3.39 Several height‐related SNPs have also been reported to be associated with risk of testicular and prostate cancer.40 In our analysis, we found height‐related genes were enriched in eight KEGG pathways which play important roles in both development and carcinogenesis. TGF‐β, MARK and Hedgehog signal pathway have been identified by GWAS to be associated with adult height.21, 40, 41 Meanwhile, the pathways mentioned above are also involved in the pathogenesis of cancer.42, 43, 44 For instance, activation of the TGF‐β signaling pathway induces potent cell‐cycle arrest in healthy noncancerous cells and in early‐stage cancerous cells, suggesting that this pathway plays a prominent role in tumor suppression. Therefore, height and cancer development may share the same genetic susceptibility.
Mendelian randomization is an alternative way to estimate the causal effect of exposure of interest on diseases. Compared with observational studies, MR can avoid both reverse causation bias and potential confounding bias. MR analysis can only be used to infer causal effect correctly when three key assumptions are satisfied (Methods). For the first assumption, we selected genetic variants which showed genome‐wide significant association with height in East Asian population or those identified in European population, but can be validated in East Asian population. In accordance with the strict selection criteria, the associations between SNPs and adult height are reliable. For the second assumption, it is possible that height‐related genetic variants are associated with other risk factors that influence the cancer development. That is, those SNPs have pleiotropic effect and the effect of those SNPs on the risk of height that are independent of any effect through height. After excluding those SNPs failed in pleiotropy test using “gtx” package, the pleiotropy test of remained SNPs was not significant, indicating that there was no pleiotropic effect to disturb the causal inference in MR analysis. For the third assumption, we did not observe significant associations of wGRS with potential confounding, like age, gender, or pack‐year. Therefore, we believe the MR analysis performed in current study basically satisfied the three key assumptions and the estimate of causal effect was highly reliable.
To our knowledge, this is the first study to investigate association between adult height and risk of lung cancer in East Asian population. The main strength of our study is that we used MR method to analyze genetic determined height and lung cancer. Using MR method can avoid the reverse causation bias and confounding bias due to traditional epidemiological studies. In addition, we test the linear trend of genetic determined height based on individual wGRS. We also examined the association in two main lung cancer subtypes, lung adenocarcinoma and lung squamous cell carcinoma. However, our study has several limitations. Firstly, all the GWAS identified SNPs only explained a small amount of variance in adult height. Secondly, our study included two previously published GWAS data, one from Chinese population and another from nonsmoking women in Asian. There might be geographic difference between the two populations, although the heterogeneity test was not significant. The subgroup analysis of gender and smoke status was not conducted because one of the studies involved only women and nonsmokers. Thus, female take a large proportion of all samples, the results might not be representative.
In summary, our findings from the MR analysis provide strong evidence that increasing height is associated with increased risk of lung cancer in East Asian population. It is shown that height may be a causal risk factor in lung cancer development. Our study suggests the biologic pathways that genetically determined adult height may also involve in the etiology of lung cancer. However, further investigations are needed to clarify the underlying mechanism.
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
Supporting information
ACKNOWLEDGMENTS
This work was supported by National Natural Science of China (81521004, 81530088), the Priority Academic Program for the Development of Jiangsu Higher Education Institutions (Public Health and Preventive Medicine) and Top‐notch Academic Programs Project of Jiangsu Higher Education Institutions (PPZY2015A067).
Wang L, Huang M, Ding H, et al. Genetically determined height was associated with lung cancer risk in East Asian population. Cancer Med. 2018;7:3445–3452. 10.1002/cam4.1557
Lu Wang and Mingtao Huang are contributed equally to this work.
Contributor Information
Feng Chen, Email: fengchen@njmu.edu.cn.
Hongbing Shen, Email: hbshen@njmu.edu.cn.
REFERENCES
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87‐108. [DOI] [PubMed] [Google Scholar]
- 2. Fitzmaurice C, Dicker D, Pain A, et al. The Global Burden of Cancer 2013. JAMA Oncol. 2015;1:505‐527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hecht SS. Tobacco carcinogens, their biomarkers and tobacco‐induced cancer. Nat Rev Cancer. 2003;3:733‐744. [DOI] [PubMed] [Google Scholar]
- 4. Markowitz SB, Levin SM, Miller A, Morabia A. Asbestos, asbestosis, smoking, and lung cancer. New findings from the North American insulator cohort. Am J Respir Crit Care Med. 2013;188:90‐96. [DOI] [PubMed] [Google Scholar]
- 5. Hamra GB, Guha N, Cohen A, et al. Outdoor particulate matter exposure and lung cancer: a systematic review and meta‐analysis. Environ Health Perspect. 2014;122:906‐911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Renehan AG, Tyson M, Egger M, Heller RF, Zwahlen M. Body‐mass index and incidence of cancer: a systematic review and meta‐analysis of prospective observational studies. Lancet. 2008;371:569‐578. [DOI] [PubMed] [Google Scholar]
- 7. Bhaskaran K, Douglas I, Forbes H, dos‐Santos‐Silva I, Leon DA, Smeeth L. Body‐mass index and risk of 22 specific cancers: a population‐based cohort study of 5.24 million UK adults. Lancet. 2014;384:755‐765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Macgregor S, Cornes BK, Martin NG, Visscher PM. Bias, precision and heritability of self‐reported and clinically measured height in Australian twins. Hum Genet. 2006;120:571‐580. [DOI] [PubMed] [Google Scholar]
- 9. Silventoinen K, Sammalisto S, Perola M, et al. Heritability of adult body height: a comparative study of twin cohorts in eight countries. Twin Res. 2003;6:399‐408. [DOI] [PubMed] [Google Scholar]
- 10. Silventoinen K, Magnusson PK, Tynelius P, Kaprio J, Rasmussen F. Heritability of body size and muscle strength in young adulthood: a study of one million Swedish men. Genet Epidemiol. 2008;32:341‐349. [DOI] [PubMed] [Google Scholar]
- 11. Silventoinen K. Determinants of variation in adult body height. J Biosoc Sci. 2003;35:263‐285. [DOI] [PubMed] [Google Scholar]
- 12. Perkins JM, Subramanian SV, Davey Smith G, Ozaltin E. Adult height, nutrition, and population health. Nutr Rev. 2016;74:149‐165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sung J, Song YM, Lawlor DA, Smith GD, Ebrahim S. Height and site‐specific cancer risk: a cohort study of a korean adult population. Am J Epidemiol. 2009;170:53‐64. [DOI] [PubMed] [Google Scholar]
- 14. Khankari NK, Shu XO, Wen W, et al. Association between adult height and risk of colorectal, lung, and prostate cancer: results from meta‐analyses of prospective studies and mendelian randomization analyses. PLoS Med. 2016;13:e1002118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang F, Xu X, Yang J, Min L, Liang S, Chen Y. Height and lung cancer risk: a meta‐analysis of observational studies. PLoS ONE. 2017;12:e0185316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Green J, Cairns BJ, Casabonne D, Wright FL, Reeves G, Beral V. Height and cancer incidence in the Million Women Study: prospective cohort, and meta‐analysis of prospective studies of height and total cancer risk. Lancet Oncol. 2011;12:785‐794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smith GD, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1‐22. [DOI] [PubMed] [Google Scholar]
- 18. Hu Z, Wu C, Shi Y, et al. A genome‐wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nat Genet. 2011;43:792‐796. [DOI] [PubMed] [Google Scholar]
- 19. Dong J, Hu Z, Wu C, et al. Association analyses identify multiple new lung cancer susceptibility loci and their interactions with smoking in the Chinese population. Nat Genet. 2012;44:895‐899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lan Q, Hsiung CA, Matsuo K, et al. Genome‐wide association analysis identifies new lung cancer susceptibility loci in never‐smoking women in Asia. Nat Genet. 2012;44:1330‐1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. He M, Xu M, Zhang B, et al. Meta‐analysis of genome‐wide association studies of adult height in East Asians identifies 17 novel loci. Hum Mol Genet. 2015;24:1791‐1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hao Y, Liu X, Lu X, et al. Genome‐wide association study in Han Chinese identifies three novel loci for human height. Hum Genet. 2013;132:681‐689. [DOI] [PubMed] [Google Scholar]
- 23. Okada Y, Kamatani Y, Takahashi A, et al. A genome‐wide association study in 19 633 Japanese subjects identified LHX3‐QSOX2 and IGF1 as adult height loci. Hum Mol Genet. 2010;19:2303‐2312. [DOI] [PubMed] [Google Scholar]
- 24. Mao Y, Yan C, Lu Q, et al. Genetically predicted high body mass index is associated with increased gastric cancer risk. Eur J Hum Genet. 2017;25:1061‐1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Didelez V, Sheehan N. Mendelian randomization as an instrumental variable approach to causal inference. Stat Methods Med Res. 2007;16:309‐330. [DOI] [PubMed] [Google Scholar]
- 26. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658‐665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Durrleman S, Simon R. Flexible regression models with cubic splines. Stat Med. 1989;8:551‐561. [DOI] [PubMed] [Google Scholar]
- 28. Engeland A, Tretli S, Hansen S, Bjorge T. Height and body mass index and risk of lymphohematopoietic malignancies in two million Norwegian men and women. Am J Epidemiol. 2007;165:44‐52. [DOI] [PubMed] [Google Scholar]
- 29. Kabat GC, Anderson ML, Heo M, et al. Adult stature and risk of cancer at different anatomic sites in a cohort of postmenopausal women. Cancer Epidemiol Biomarkers Prev. 2013;22:1353‐1363. [DOI] [PubMed] [Google Scholar]
- 30. Gunnell D, Okasha M, Smith GD, Oliver SE, Sandhu J, Holly JM. Height, leg length, and cancer risk: a systematic review. Epidemiol Rev. 2001;23:313‐342. [DOI] [PubMed] [Google Scholar]
- 31. Batty GD, Shipley MJ, Gunnell D, et al. Height, wealth, and health: an overview with new data from three longitudinal studies. Econ Hum Biol. 2009;7:137‐152. [DOI] [PubMed] [Google Scholar]
- 32. Clayton PE, Banerjee I, Murray PG, Renehan AG. Growth hormone, the insulin‐like growth factor axis, insulin and cancer risk. Nat Rev Endocrinol. 2011;7:11‐24. [DOI] [PubMed] [Google Scholar]
- 33. Renehan AG, Zwahlen M, Minder C, O’Dwyer ST, Shalet SM, Egger M. Insulin‐like growth factor (IGF)‐I, IGF binding protein‐3, and cancer risk: systematic review and meta‐regression analysis. Lancet. 2004;363:1346‐1353. [DOI] [PubMed] [Google Scholar]
- 34. Rowlands MA, Gunnell D, Harris R, Vatten LJ, Holly JM, Martin RM. Circulating insulin‐like growth factor peptides and prostate cancer risk: a systematic review and meta‐analysis. Int J Cancer. 2009;124:2416‐2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Key TJ, Appleby PN, Reeves GK, Roddam AW. Insulin‐like growth factor 1 (IGF1), IGF binding protein 3 (IGFBP3), and breast cancer risk: pooled individual data analysis of 17 prospective studies. Lancet Oncol. 2010;11:530‐542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rinaldi S, Cleveland R, Norat T, et al. Serum levels of IGF‐I, IGFBP‐3 and colorectal cancer risk: results from the EPIC cohort, plus a meta‐analysis of prospective studies. Int J Cancer. 2010;126:1702‐1715. [DOI] [PubMed] [Google Scholar]
- 37. Albanes D, Winick M. Are cell number and cell proliferation risk factors for cancer? J Natl Cancer Inst. 1988;80:772‐774. [DOI] [PubMed] [Google Scholar]
- 38. Trichopoulos D, Lipworth L. Is cancer causation simpler than we thought, but more intractable? Epidemiology. 1995;6:347‐349. [PubMed] [Google Scholar]
- 39. Tripaldi R, Stuppia L, Alberti S. Human height genes and cancer. Biochim Biophys Acta. 2013;1836:27‐41. [DOI] [PubMed] [Google Scholar]
- 40. Wood AR, Esko T, Yang J, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46:1173‐1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lango Allen H, Estrada K, Lettre G, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832‐838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Colak S, Ten Dijke P. Targeting TGF‐beta Signaling in Cancer. Trends Cancer. 2017;3:56‐71. [DOI] [PubMed] [Google Scholar]
- 43. Burotto M, Chiou VL, Lee JM, Kohn EC. The MAPK pathway across different malignancies: a new perspective. Cancer. 2014;120:3446‐3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Skoda AM, Simovic D, Karin V, Kardum V, Vranic S, Serman L. The role of the Hedgehog signaling pathway in cancer: a comprehensive review. Bosn J Basic Med Sci. 2018;18:8‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


