Abstract
Introduction
White light cystoscopy (WLC), often supported by urine cytology, is considered the ‘goldstandard’ in the diagnosis and follow-up of bladder cancer (BCa). In recent years, urine microRNA (miRNA) tests have been performed for the detection of bladder cancer.
Material and methods
A systematic review of the PubMed platform was performed by searching for articles in which miRNA in the urine was used for the detection of BCa.
Results
The greatest sensitivity (86.6%) in BCa detection was achieved for multi-miRNA in urine sediment. The greatest specificity (85.3%) was achieved for multi-miRNA from voided urine. There were significant differences (p <0.01) between single-miRNA (OR 8.96; CI 6.37–12.59) and the multi-miRNA group (OR 19.95; CI 13.35–29.81). There were no differences among the specimens (voided urine, supernatant, sediment) used for the test.
Conclusions
Urine miRNAs have the potential to be a valid marker for bladder cancer detection. They can successfully compete with other non-invasive diagnostic tests.
Keywords: bladder cancer ‹›, miRNA ‹›, microRNA ‹›, biomarkers ‹› urine
INTRODUCTION
Bladder cancer (BCa) is the ninth most commonly diagnosed cancer worldwide and the 13th most frequent cause of death [1]. At the time of presentation, most patients have non-muscle invasive bladder cancer (NMIBC), which is characterized by high recurrence rates of up to 70% within five years of initial endoscopic treatment [2]. Gross, painless hematuria is the main symptom in 82.4% of patients with first-time BCa. Although 18.9% of patients with macroscopic hematuria will eventually be diagnosed with urinary tract malignancy, another 8.8% will have a benign disease (e.g. stone, inflammation) [3]. According to urological guidelines, all patients presenting with painless hematuria should undergo a full diagnostic evaluation to identify any potential malignancy of the urinary tract [4]. Currently, white light cystoscopy (WLC), often supported by urine cytology, is considered the ‘gold-standard’ in the diagnosis and follow-up of BCa [5]. However, cystoscopy is a painful and expensive invasive procedure which is not free of complications, and one that requires a doctor, usually a specialist in urology. Despite its non-invasiveness and high specificity, urine cytology cannot be used as a sole test because it has low sensitivity, especially in detecting low grade tumors [6].
Cystoscopy and cytology have yet to be replaced in clinical practice, as the biomarkers investigated thus far offer insufficient sensitivity and specificity in detecting bladder malignancies [7]. BCa is characterized by a long survival period, and it has the highest cost per patient of all cancers from diagnosis to death due to the need for lifelong routine monitoring and treatment; much of these costs are spent on diagnosis and usually lifelong follow-up treatment [8]. The discovery of a comprehensive marker for BCa could not only lead to a reduction in the costs associated with the management of the disease, but also to an improvement in the care of patients.
MicroRNAs (miRNAs) are short (20–24 nucleotides), non-coding, single chain RNAs that occur in eukaryotes [9]. By binding to complementary sequences of mRNAs they play a role in the post-transcription regulation of gene expression [10]. MicroRNAs take part in many physiological and pathological processes like angiogenic signaling, cell proliferation and differentiation, apoptosis and tumorigenesis [11–14]. Some of them function as oncogenes, and others as tumor suppressor genes in cancers [15]. Both the pro- and anti-tumorigenic functions of miRNAs have also been observed in bladder cancer cells [16, 17, 18]. MicroRNA can be found in the tumor tissue itself and in body fluids like urine, blood, saliva and peritoneal fluid [19]. The acquisition of urine offers two key advantages for the patient: urine is easier and less invasive to obtain than blood, and its miRNA content may better represent the local stage of the disease due to it having direct contact with the tumor tissue in the urinary tract [20]. Due to their short length, miRNAs are less vulnerable to degradation than mRNA chains and can be stored for up to 48 hours at room temperature [21, 22].
The aim of the present article is to compare current studies in which the levels of miRNA in urine were assessed for their value in the detection of BCa and to determine their utility as a diagnostic tool.
MATERIAL AND METHODS
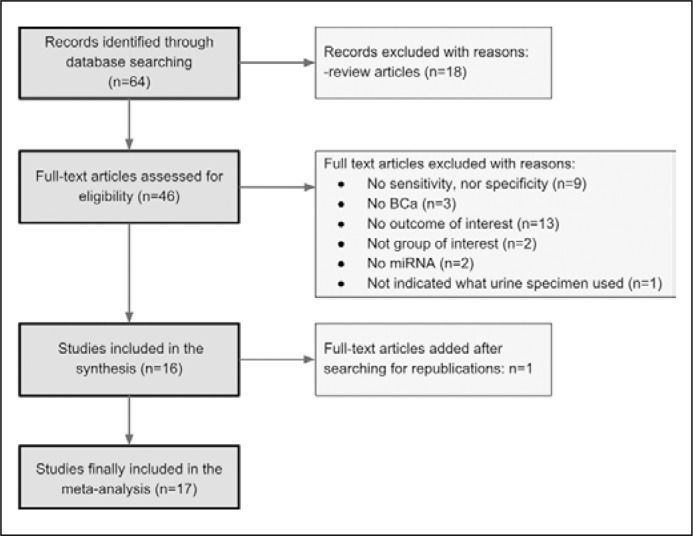
A systematic review of the PubMed platform was performed with the use of following terms: ‘microRNA + bladder cancer + urine’. We included to the analysis original papers which assessed the utility of miRNA extracted from urine for BCa detection. The search was restricted to data published between 01.03.2010 and 28.02.2017. A total of 64 articles met the search criteria. Of those, the following articles were excluded: review articles, those whose methodology did not indicate which part of the urine was used for RNA extraction, those whose results did not detail the sensitivity or specificity for test assessment, and those in which miRNAs were not used for diagnostic purposes. This filtering stage rejected 48 articles. The references of the remaining 16 articles were searched for further relevant publications, resulting in the addition of one more study for the meta-analysis (Figure 1).
Figure 1.
Flow diagram outlining search results, final included and excluded studies.
The meta-analysis and groups comparison was carried out using the statistical program PQStat version 1.6.4.121. The test probability was assumed to be significant at p <0.05 and test probability at p <0.01 was considered to be highly significant.
RESULTS
Table 1. summarizes the main characteristics (including the name of the first author, year of publication, number of patients and control group, specimen, evaluated miRNA, test sensitivity and specificity) of studies included in the meta-analysis. Of the 17 publications, seven used urine supernatant for DNA extraction, five used urine sediment and five used voided urine. In 17 articles, 29 studies evaluated the usefulness of single-miRNA profiling while another 9 used tests based on combinations of various miRNA (multi-miRNA).
Table 1.
Main characteristics of the studies included in the meta-analysis
| Ref | Author | Year | Specimen | No. patients | No. controls | miRNA | SN (%) | SP (%) |
|---|---|---|---|---|---|---|---|---|
| [23] | Zhou | 2014 | Supernatant | 112 | 78 | 106-b | 76.8 | 72.4 |
| [24] | Pospisilova 1 | 2016 | Supernatant | 46 | 13 | 125 | 59.3 | 95.7 |
| 2 | 46 | 13 | 99-a | 74.1 | 82.6 | |||
| [25] | Yun 1 | 2012 | Supernatant | 138 | 144 | 145 | 77.8 | 61.1 |
| 2 | 69 | 144 | 145 | 84.1 | 61.1 | |||
| [26] | Zhang X | 2015 | Supernatant | 162 | 152 | 155 | 80.2 | 84.6 |
| [27] | Wang | 2015 | Supernatant | 192 | 169 | 214 | 90.5 | 65.6 |
| [28] | Zhang D 1 | 2014 | Supernatant | 50 | 21 | 125-b | 84.8 | 76.2 |
| 2 | 50 | 21 | 99-a | 78 | 85.7 | |||
| 3 | 50 | 21 | 99-a/125-b | 86.7 | 81.1 | |||
| [29] | Long | 2015 | Supernatant | 85 | 45 | 26-a/93/191/940 | 70 | 84 |
| [30] | Yamada 1 | 2011 | Sediment | 100 | 74 | 96 | 71 | 89.2 |
| 2 | 100 | 74 | 183 | 74 | 77.3 | |||
| [31] | Eissa | 2014 | Sediment | 94 | 90 | 96 | 72.3 | 88.9 |
| [32] | Miah 1 | 2012 | Sediment | 68 | 53 | 135-b/15-b/1224-3p | 94.1 | 51 |
| 2 | 68 | 53 | 15a | 51.7 | 72 | |||
| 68 | 53 | 15b | 67.8 | 81.3 | ||||
| 68 | 53 | 4-1 | 60 | 58.5 | ||||
| 68 | 53 | 27b | 60.3 | 81.1 | ||||
| 68 | 53 | 100 | 60.4 | 78.7 | ||||
| 68 | 53 | 135b | 71.2 | 74.4 | ||||
| 68 | 53 | 203 | 66.1 | 66 | ||||
| 68 | 53 | 212 | 54.2 | 64 | ||||
| 68 | 53 | 328 | 55.4 | 86.8 | ||||
| 68 | 53 | 1224-3p | 75.9 | 82.4 | ||||
| [33] | Urquidi | 2016 | Sediment | 61 | 60 | combination of 25miRNA | 87 | 100 |
| [34] | Shimizu | 2012 | Sediment | 86 | 20 | 137/124-2/124-3/9-3 | 81 | 89 |
| [35] | Mengual | 2013 | Sediment | 151 | 126 | 187/18a/25/142-3p/ 140-5p/204 | 84.4 | 86.5 |
| [36] | Snowdon | 2012 | Voided urine | 8 | 5 | 125-b/126 | 80 | 100 |
| [37] | Tolle 1 | 2013 | Voided urine | 36 | 19 | 520e | 70 | 63.2 |
| 2 | 36 | 19 | 618 | 70 | 68.4 | |||
| 3 | 36 | 19 | 1255b-5p | 85 | 68.4 | |||
| [38] | Dudziec 1 | 2010 | Voided urine | 68 | 53 | 152/212/328/1224-3p | 81 | 75 |
| 2 | 68 | 53 | 152 | 88 | 58 | |||
| 3 | 68 | 53 | 212 | 40 | 66 | |||
| 4 | 68 | 53 | 328 | 59 | 91 | |||
| 5 | 68 | 53 | 1224-3p | 84 | 58 | |||
| [39] | Hanke | 2010 | Voided urine | 29 | 11 | 126/152 | 72 | 82 |
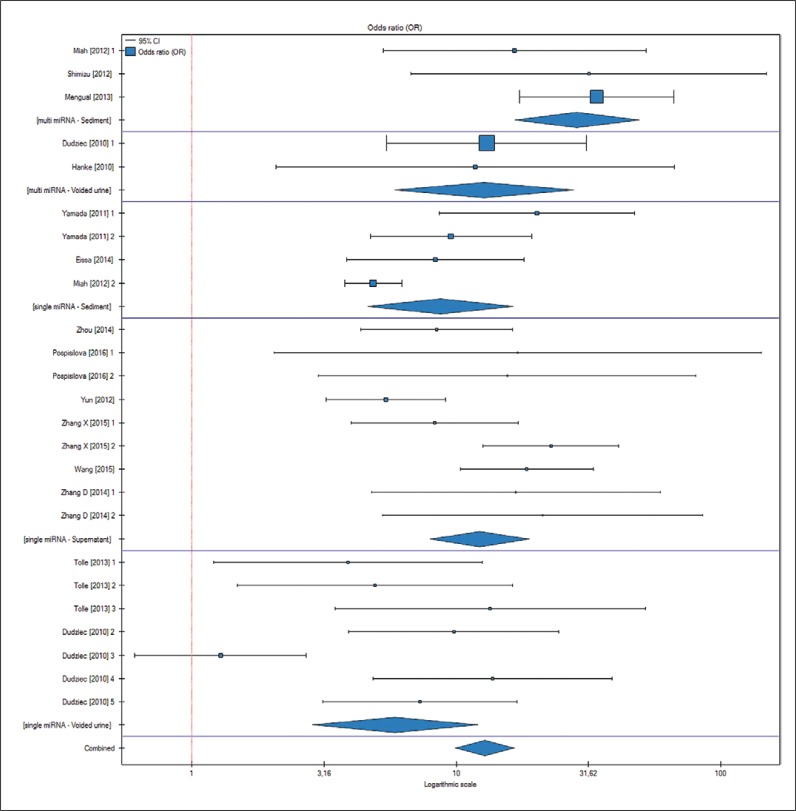
The results of published studies were divided depending on the type of specimen used for miRNA extraction (voided urine, urine sediment or supernatant) and amount of profiled miRNAs (single vs. multi). For each subgroup, sensitivity and specificity was evaluated. The results are presented in Table 2 and Table 3. Sensitivity for BCa detection of single-miRNA testing was lower than the multi-miRNA test, 71.3% vs. 80.9% (p <0.05). Specificity also was in favor of multi-miRNA tests; however, the results were not statistically significant. miRNA tests performed from urine supernatant have greater (78.4%) sensitivity than voided urine (74.3%) and urine sediment (75.6%). At the same time urine supernatant has the highest specificity among all tests: 79.4% compared to 76.5% and 79.3%. An analysis of all subgroups revealed that greatest sensitivity (86.6%) was reached when only multi-miRNA in urine sediment was tested. The greatest specificity (85.3%) was achieved for multi-miRNA from voided urine. A further meta-analysis comparing all 6 groups revealed that the odds ratio for BCa detection was highly significant (p <0.01; OR 5.87–28.66) (Figure 2) for all the groups; however, there were no statistically significant differences (p >0.05) among them.
Table 2.
Median sensitivity (SN) and specificity (SP) in single and multiple miRNA tests – overall and in 3 different specimens used for the test (voided urine, urine supernatant, and sediment)
| Single miRNA | Multi miRNA | |||||
|---|---|---|---|---|---|---|
| SN (%) | Voided urine | Supernatant | Sediment | Voided urine | Supernatant | Sediment |
| 70.9 | 78.4 | 64.6 | 77.6 | 78.4 | 86.6 | |
| Overall: 71.3 | Overall: 80.9 | |||||
| SP (%) | Voided urine | Supernatant | Sediment | Voided Urine | Supernatant | Sediment |
| 67.6 | 76.1 | 77 | 85.3 | 82.6 | 81.6 | |
| Overall: 73.6 | Overall: 83.1 |
Table 3.
Median sensitivity (SN) and specificity (SP) in different urine specimens overall and when only single or multiple miRNAs were used for the study
| Voided urine | Supernatant | Sediment | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Single miRNA | Multi miRNA | Overall | Single miRNA | Multi miRNA | Overall | Single miRNA | Multi miRNA | Overall | |
| SN (%) | 70.9 | 77.6 | 74.3 | 78.4 | 78.4 | 78.4 | 64.6 | 86.6 | 75.6 |
| SP (%) | 67.6 | 85.3 | 76.5 | 76.1 | 82.6 | 79.4 | 77 | 81.6 | 79.3 |
Figure 2.
Forrest plot comparing 6 groups of markers (each consisting of paired: number of miRNA used for the test and kind of urine specimen used) for risk of bladder cancer detection. See Table 1 for more information about the studies qualified for the analysis.
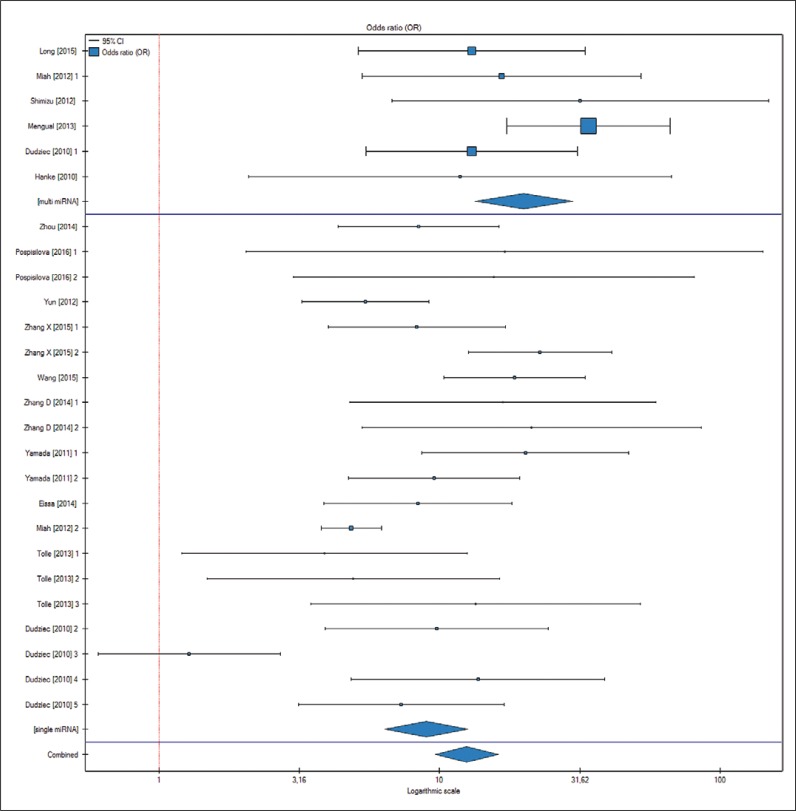
When the influence of number of miRNAs used for the tests and kind of specimen were analyzed separately it revealed that there were significant differences (p <0.01) between single-miRNA (OR 8.96; CI 6.37–12.59) and multi-miRNA group (OR 19.95; CI 13.35–29.81) (Figure 3) for confirming the diagnosis of BCa. There were no differences in terms of the specimen (voided urine, supernatant, sediment) used for the test.
Figure 3.
Forrest plot comparing 2 groups of markers (single vs. multiple microRNA tests) for risk of bladder cancer detection. See Table 1 for more information about the studies qualified for the analysis.
DISCUSSION
Although many biomarkers have been assessed as potential diagnostic tools of BCa, none of them have reached sufficient accuracy to replace cystoscopy and cytology [40]. As there is still a need to find a new marker of tumorigenesis for BCa, further studies focusing on miRNA profiling have been conducted. The first report about altered miRNA expression in bladder cancer was published in 2007 [41]. The lack of invasiveness and stability of miRNA in body fluids (eg. urine) are the main advantages of using it as a potential biomarker [42].
The miRNA profiling in urine specimens has evolved over time. Since 2014, no original study has been performed using voided urine for miRNA detection, and recent years have seen growing interest in tests based on profiling miRNA from urine supernatant. This trend has arguably arisen in response to studies by Wang et al., which found that many cell types are present in urine sediment, including renal tubular cells, urothelial cells, lymphocytes, erythrocytes and tumor cells, which may influence the received test results. As these cells are not present in urine supernatant, their miRNA is therefore free of such contamination [43, 44]. Despite the attempts to increase sensitivity and specificity by trying different specimens used for the test, it seems that this action is of less impact than number and kind of miRNA used for the analysis.
Our meta-analysis confirms that miRNA profiling performed from the three specimens does not differ between the groups (p = 0.23). The highest specificity (86.6%) was established for multi-miRNA from voided urine. The specificity levels may be even higher as miRNA alterations occur before the histological and morphological onset of malignancy, and these results in turn may be interpreted as false positive when only cystoscopy is used to check the obtained results [45].
The simultaneous increase of both sensitivity and specificity when single-miRNA tests are compared to multi-miRNA (Tables 2 and 3) may not be intuitive. A large proportion of the examined studies profile only an individual miRNA or a small combination thereof. In addition, most studies use different miRNAs for the profiling. Indeed, only three articles [28, 32, 38] evaluated the sensitivity and specificity of a few single miRNAs and then calculated how the results changed when a combination of previously-assessed single miRNAs was used for bladder cancer detection (a creation of multi-miRNA test from several single-miRNA tests). The direct comparison of these three studies revealed that sensitivity significantly increased from 71.4% for the single test to 87.1% for the multi-miRNA test, while specificity declined from 87.1% for the single-miRNA test to 66.1% for multi-miRNA test.
In the period from 2010 to 2013, the median sensitivity of elaborated miRNA tests for BCa detection was 71.6%. In more recent studies, dated 2014 to 2016, sensitivity was found to increase to 77.6%, and specificity also improved from 72.3% to 80.2%. This growing increase in the accuracy of miRNA tests is a good sign, as most of newly introduced biomarkers often lose their initially promising results in following studies [46].
It seems reasonable to compare the current sensitivity and specificity of miRNA tests with those demonstrated by some previously assessed markers. To this end, miRNA has been added to a ranking of bladder cancer biomarkers devised by Rhijn [46] (Tables 4 and 5). Cytology, with a median specificity of 94%, offers better specificity than the performed miRNA tests, as do microsatellite and UBC. The highest specificity obtained in multi-miRNA from voided urine ranks it in fourth place out of the 18 ranked tests. Tests based on a combination of different miRNAs extracted from urine sediment offer greater sensitivity for BCa detection than many other tests (86.6%). As noted in Table 5, CYFRA21-1 and Cytokeratin20 have only slightly worse results.
Table 4.
The median sensitivity (SN) of bladder cancer (BCa) diagnostic tests. The numbers in white fields borrowed from Rhijn's systematic review [46]
| Marker | Median SN (%) | Range (min–max) |
|---|---|---|
| miRNA multi-sediment | 86.6 | 81.4-94.1 |
| CYFRA21-1 | 85 | 75–88 |
| Cytokeratin20 | 85 | 79–87 |
| Microsatellite | 82 | 75–92 |
| miRNA multi | 80.9 | 70-94.1 |
| FISH | 79 | 70–86 |
| miRNA supernatant | 78.4 | 70–90.5 |
| miRNA sediment | 75.6 | 51.7–94.1 |
| LewisX | 75 | 68–79 |
| miRNA voided urine | 74.3 | 40-88 |
| miRNA single | 71.3 | 40–90.5 |
| NMP22 | 71 | 47–100 |
| BTA trak | 71 | 60–83 |
| ImmunoCyt | 67 | 52–100 |
| UBC | 60 | 21–80 |
| Cytometry | 60 | 45–85 |
| BTA stat | 58 | 29–74 |
| Cytology | 35 | 13–75 |
BTA – bladder tumor antigen; FISH – fluorescence in situ hybridization; NMP22 – nuclear matrix protein 22; UBC – urinary bladder cancer antigen; CYFRA21-1 – cytokeratin 19 fragments
Table 5.
The median specificity (SP) of bladder cancer (BCa) diagnostic tests. The numbers in white fields borrowed from Rhijn's systematic review [46]
| Marker | Median SP (%) | Range (min–max) |
|---|---|---|
| Cytology | 94 | 85–100 |
| Microsatellite | 89 | 79–100 |
| UBC | 87 | 72–95 |
| miRNA multi-voided urine | 85.3 | 75–100 |
| LewisX | 85 | 67–86 |
| miRNA multi | 83.1 | 51–100 |
| CYFRA21-1 | 82 | 73–95 |
| Cytometry | 82 | 50–92 |
| miRNA supernatant | 79.4 | 61.1–95.7 |
| miRNA sediment | 79.3 | 51–100 |
| miRNA voided urine | 76.5 | 58–100 |
| Cytokeratin20 | 76 | 76 |
| ImmunoCyt | 75 | 62–82 |
| miRNA single | 73.6 | 58–95.7 |
| NMP22 | 73 | 55–98 |
| BTA stat | 73 | 56–86 |
| FISH | 70 | 66–93 |
| BTA trak | 66 | 60–79 |
BTA – bladder tumor antigen; FISH – fluorescence in situ hybridization; NMP22 – nuclear matrix protein 22; UBC – urinary bladder cancer antigen; CYFRA21-1 – cytokeratin 19 fragments
The high sensitivity and moderate specificity of miRNA tests indicate that they may be especially useful in the primary diagnosis of patients with a suspicion of urinary tract malignancy. When attempting to rule out the disease, credible negative results are necessary because they provide few false negatives. However, if the aim is cost reduction, a high positive predictive value (PPV) is desirable. To increase the PPV of a diagnostic tool, it is necessary to choose tests which provide greater specificity [47]. This in turn may lead to emerging two different kind of tests, one for evaluation of gross painless hematuria, and the second used in follow-up of BCa.
While some of the markers (e.g. BTA stat, UBC) provide instant results, those which use miRNA require additional laboratory tests. With currently-available technology, such measurements can prove to be expensive, time-consuming and demanding; however, these limitations will slacken in the near future. For example, by analogy with human genome sequencing, the costs may be reduced by as much as 99.8% over the next 13 years [48]. However, even if these disadvantages are resolved, miRNA tests still need to undergo many validation steps (including the standardization of urine collection or RNA sequencing) before they can be used in everyday clinical practice.
There are several limitations related to the methodology. Firstly, it should be taken into consideration that mainly positive results are being published. Studies that do not show the positive impact of the studied marker on disease diagnosis at initial phases of research are of less interest, which may be a cause of a shift in results. Secondly, the study only includes articles that state both the sensitivity and specificity of miRNAs on BCa detection, which may result in studies with both positive and negative effects on the results being omitted. Finally, each original study was performed by the use of a different miRNA or panel of miRNAs. Thus, a meta-analysis of all the articles gives only a general view of the topic, even though it may show future directions for research. What is considered to be one of the limitations of meta-analyses of miRNA studies is at the same time a great advantage of miRNA tests over the other markers (e.g. cytology), as researchers may add, remove or replace the miRNAs used in the diagnostic panel to optimize the test.
CONCLUSIONS
The results of our analysis show that urine miRNAs have the potential to act as a proper marker for bladder cancer detection. They can successfully compete with other non-invasive diagnostic tests. The chance of choosing many different miRNAs used in the panel, adding the next ones to the existing tests to improve their sensitivity and specificity (which is not possible in 'traditional' tests) should be used to create a biomarker that can finally be accepted by the Urological society. Although many up-to-date studies have been conducted, there is still a need to perform clinical trials to validate the aforementioned data before miRNA use can be adopted in everyday clinical practice.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
We thank Mr. Bas van Rhijn for permission to use his tables, which greatly improved the manuscript.
This study presents independent research partly-funded by the Medical University of Łódź with grant No. 502-03/5-138-02/502-54-146.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Quek ML, Stein JP, Clark PE, et al. Natural history of surgically treated bladder carcinoma with extravesical tumor extension. Cancer. 2003;98:955–961. doi: 10.1002/cncr.11569. [DOI] [PubMed] [Google Scholar]
- 3.Edwards TJ, Dickinson AJ, Natale S, Gosling J, Mcgrath JS. A prospective analysis of the diagnostic yield resulting from the attendance of 4020 patients at a protocol - driven haematuria clinic. BJU Int. 2006;97:301–305. doi: 10.1111/j.1464-410X.2006.05976.x. [DOI] [PubMed] [Google Scholar]
- 4.Wood DP., Jr . Urothelial Tumors of the Bladder. In: Wein AJ, Kavoussi LR, editors. Campbell-Walsh Urology. 10th edn. Philadelphia: 2012. p. 2327. Vol. Chapt 80. [Google Scholar]
- 5.Witjes JA. Bladder carcinoma in situ in 2003: state of the art. Eur Urol. 2004;45:142–146. doi: 10.1016/j.eururo.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 6.Planz B, Jochims E, Deix T, Caspers HP, Jakse G, Boecking A. The role of urinary cytology for detection of bladder cancer. Eur J Surg Oncol. 2005;31:304–308. doi: 10.1016/j.ejso.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 7.Babjuk M, Burger M, Comperat E, et al. EAU Guidelines. 2017. Non-muscle invasive bladder cancer (TaT1 and CIS) p. 11. Chapter 5.6, [Google Scholar]
- 8.Botteman MF, Pashos CL, Redaelli A, Laskin B, Hauser R. The health economics of bladder cancer. Pharmacoeconomics. 2003;21:1315–1330. doi: 10.1007/BF03262330. [DOI] [PubMed] [Google Scholar]
- 9.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid dead. Send to Proc Natl Acad Sci USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 11.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. P Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 13.Ambros V . MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 14.Carlsson J, Davidsson S, Helenius G, et al. A miRNA expression signature that separates between normal and malignant prostate tissues. Cancer Cell Int. 2011;11:14. doi: 10.1186/1475-2867-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen CZ. MicroRNAs as oncogenes and tumor suppressors. New Engl J Med. 2005;353:1768–1771. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 16.Uchida Y, Chiyomaru T, Enokida H, et al. MiR-133a induces apoptosis through direct regulation of GSTP1 in bladder cancer cell lines. Urol Oncol. 2013;31:115–123. doi: 10.1016/j.urolonc.2010.09.017. [DOI] [PubMed] [Google Scholar]
- 17.Yoshino H, Chiyomaru T, Enokida H, et al. The tumour-suppressive function of miR-1 and miR-133a targeting TAGLN2 in bladder cancer. Br J Cancer. 2011;104:808–818. doi: 10.1038/bjc.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de la Peña FA, Kanasaki K, Kanasaki M, Tangirala N, Maeda G, Kalluri R. Loss of p53 and acquisition of angiogenic microRNA profile are insufficient to facilitate progression of bladder urothelial carcinoma in situ to invasive carcinoma. J Biol Chem. 2011;286:20778–20787. doi: 10.1074/jbc.M110.198069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang X, Liang M, Dittmar R, Wang L. Extracellular microRNAs in urologic malignancies: chances and challenges. Int J Mol Sci. 2013;14:14785–14799. doi: 10.3390/ijms140714785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valadi H, Ekstrom K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 22.Miah S, Dudziec E, Rosario DJ, Hamdy FC, Catto JW. Urinary microrna in bladder cancera potential diagnostic tool. Eur Urol Suppl. 2011;10:81. [Google Scholar]
- 23.Zhou X, Zhang X, Yang Y, et al. Urinary cell-free microRNA-106b as a novel biomarker for detection of bladder cancer. Med Oncol. 2014;31:197. doi: 10.1007/s12032-014-0197-z. [DOI] [PubMed] [Google Scholar]
- 24.Pospisilova S, Pazourkova E, Horinek A, et al. MicroRNAs in urine supernatant as potential noninvasive arkers for bladder cancer detection. Neoplasma. 2016;63:799–808. doi: 10.4149/neo_2016_518. [DOI] [PubMed] [Google Scholar]
- 25.Yun SJ, Jeong P, Kim WT, et al. Cell-free microRNAs in urine as diagnostic and prognostic biomarkers of bladder cancer. Int J Oncol. 2012;41:1871–1878. doi: 10.3892/ijo.2012.1622. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Zhang Y, Liu X, et al. Direct quantitative detection for cell-free miR-155 in urine: a potential role in diagnosis and prognosis for non-muscle invasive bladder cancer. Oncotarget. 2016;7:3255–3266. doi: 10.18632/oncotarget.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Zhang X, Wang L, et al. Downregulation of urinary cel - free microRN - 214 as a diagnostic and prognostic biomarker in bladder cancer. J Surg Oncol. 2015;111:992–999. doi: 10.1002/jso.23937. [DOI] [PubMed] [Google Scholar]
- 28.Zhang DZ, Lau KM, Chan ES, et al. Cell-free urinary microRNA-99a and microRNA-125b are diagnostic markers for the non-invasive screening of bladder cancer. PloS one. 2014;9:e100793. doi: 10.1371/journal.pone.0100793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Long J, Sullivan TB, Humphrey J, et al. A non-invasive miRNA based assay to detect bladder cancer in cell-free urine. Am J Transl Res. 2015;7:2500. [PMC free article] [PubMed] [Google Scholar]
- 30.Yamada Y, Enokida H, Kojima S, et al. Mi-96 and miR-183 detection in urine serve as potential tumor markers of urothelial carcinoma: correlation with stage and grade, and comparison with urinary cytology. Cancer Sci. 2011;102:522–529. doi: 10.1111/j.1349-7006.2010.01816.x. [DOI] [PubMed] [Google Scholar]
- 31.Eissa S, Habib H, Ali E, Kotb Y. Evaluation of urinary miRNA-96 as a potential biomarker for bladder cancer diagnosis. Med Oncol. 2015;32:413. doi: 10.1007/s12032-014-0413-x. [DOI] [PubMed] [Google Scholar]
- 32.Miah S, Dudziec E, Drayton RM, et al. An evaluation of urinary microRNA reveals a high sensitivity for bladder cancer. Brit J Cancer. 2012;107:123–128. doi: 10.1038/bjc.2012.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urquidi V, Netherton M, Gomes-Giacoia E, et al. A microRNA biomarker panel for the noninvasive detection of bladder cancer. Oncotarget. 2016;7:86290. doi: 10.18632/oncotarget.13382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimizu T, Suzuki H, Nojima M, Kitamura H, et al. Methylation of a panel of microRNA genes is a novel biomarker for detection of bladder cancer. Eur Urol. 2013;63:1091–1100. doi: 10.1016/j.eururo.2012.11.030. [DOI] [PubMed] [Google Scholar]
- 35.Mengual L, Lozano JJ, Ingelmo-Torres M, Gazquez C, Ribal MJ, Alcaraz A. Using microRNA profiling in urine samples to develop a non-invasive test for bladder cancer. Int J Cancer. 2013;133:2631–2641. doi: 10.1002/ijc.28274. [DOI] [PubMed] [Google Scholar]
- 36.Snowdon J, Boag S, Feilotter H, Izard J, Siemens DR. A pilot study of urinary microRNA as a biomarker for urothelial cancer. Can Urol Assoc. 2013;7:28. doi: 10.5489/cuaj.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tölle A, Jung M, Rabenhorst S, Kilic E, Jung K, Weikert S. Identification of microRNAs in blood and urine as tumour markers for the detection of urinary bladder cancer. Oncol Rep. 2013;30:1949–1956. doi: 10.3892/or.2013.2621. [DOI] [PubMed] [Google Scholar]
- 38.Dudziec E, Miah S, Choudhry H, et al. Hypermethylation of CpG islands and shores around specific microRNAs and mirtrons is associated with the phenotype and presence of bladder cancer. Clin Cancer Res. 2011;17:1287–1296. doi: 10.1158/1078-0432.CCR-10-2017. [DOI] [PubMed] [Google Scholar]
- 39.Hanke M, Hoefig K, Merz H, et al. A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol Oncol. 2010;28:655–661. doi: 10.1016/j.urolonc.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 40.D'Costa JJ, Goldsmith JC, Wilson JS, Bryan RT, Ward DG. A systematic review of the diagnostic and prognostic value of urinary protein biomarkers in urothelial bladder cancer. Bladder Cancer. 2016;2:301–317. doi: 10.3233/BLC-160054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gottardo F, Liu CG, Ferracin M, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;25:387–392. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 42.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 43.Wang G, Tam LS, Kwan BCH, et al. Expression of miR-146a and miR-155 in the urinary sediment of systemic lupus erythematosus. Clin Rheumatol. 2012;31:435–440. doi: 10.1007/s10067-011-1857-4. [DOI] [PubMed] [Google Scholar]
- 44.Yun SJ, Jeong P, Kim WT, et al. Cell-free microRNAs in urine as diagnostic and prognostic biomarkers of bladder cancer. Int J Oncol. 2012;41:1871–1878. doi: 10.3892/ijo.2012.1622. [DOI] [PubMed] [Google Scholar]
- 45.Catto JW, Miah S, Owen HC, et al. Distinct microRNA alterations characterize high-and lowgrade bladder cancer. Cancer Res. 2009;69:8472–8481. doi: 10.1158/0008-5472.CAN-09-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Rhijn BW, Van der Poel HG, van Der Kwast TH. Urine markers for bladder cancer surveillance: a systematic review. Eur Urol. 2005;47:736–748. doi: 10.1016/j.eururo.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 47.Walker P, Subasinghe RP. DNA-based molecular diagnostic techniques: research needs for standardization and validation of the detection of aquatic animal pathogens and diseases. FAO; 2000. [Google Scholar]
- 48.Wetterstrand KA. DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP) Available at: www.genome.gov/sequencingcostsdata Accessed 31.01.2018.