Plants can establish de novo DNA methylation via the RNA-directed DNA methylation (RdDM) pathway, which involves a complex array of proteins as well as small interfering RNAs (siRNAs) and scaffold RNAs. In the canonical RdDM pathway that is best understood in Arabidopsis, complementary pairing between 24-nt siRNAs and nascent scaffold RNAs, together with protein–protein interactions, recruits the protein machinery for DNA methylation. Production of nearly all 24-nt siRNAs depends on the plant-specific RNA polymerase Pol IV, which is thought to generate single-stranded non-coding RNAs that serve as templates for RDR2 (RNA-dependent RNA polymerase 2) transcription. RDR2 transcription results in double-stranded RNAs (dsRNAs) that are subsequently cleaved by DCL3 (Dicer-like 3) to produce 24-nt siRNAs. The siRNAs are then incorporated into Argonaute proteins, mainly AGO4 and AGO6, for recognition of complementary scaffold RNA and recruitment of downstream factors including DNA methyltransferases [1]. In addition to the Pol IV-RDR2-DCL3 pathway that generates 24-nt siRNAs, de novo DNA methylation also involves alternative modules for siRNA production, including (1) Pol II, which not only generates siRNAs and scaffold RNAs at some RdDM loci, but also exhibits spatially distinctive associations with different AGO proteins when compared with Pol IV and Pol V [2]; (2) RDR1 and RDR6 that are involved in 21–22-nt siRNA production while being clearly involved in DNA methylation [3]; and (3) DCL2 and DCL4 that mainly produce 22-nt and 21-nt siRNAs, respectively, while being functionally redundant with DCL3 in mediating RdDM [3].
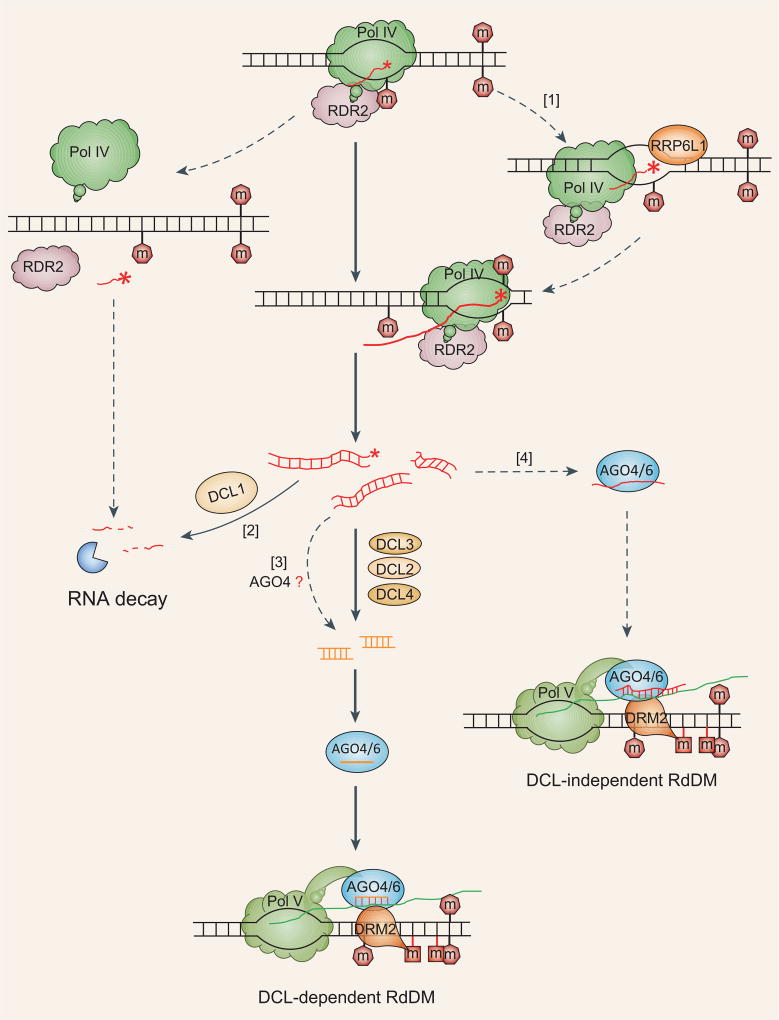
Pol IV-transcribed non-coding RNAs (P4 RNAs) have long been proposed as key initiators of RdDM but were not identified until recently, possibly because of their low abundance and/or rapid processing. In recent P4 RNA studies, researchers performed deep sequencing of small RNAs in the Arabidopsis dcl2/3/4 triple mutant, in which the cleavage of Pol IV-dependent long-precursor RNAs into 24-nt siRNAs theoretically would be blocked. In the dcl2/3/4 mutant, the abundance of 24-nt siRNAs was greatly reduced as expected, and this reduction was accompanied by a significant accumulation of mostly 26- to 50-nt RNAs [4–8]. These latter RNAs are considered to be P4 RNAs because they are Pol IV-dependent and can be processed by exogenous DCL3 into 24-nt siRNAs. The P4 RNAs are similar to the 24-nt siRNAs in terms of genome-wide distribution, abundance, strand bias and 5′-adenine preference. The P4 RNAs are also characterized by 5′-monophosphate and sometimes by 3′-misincorporation that preferentially occurs at positions of methylated cytosines on the template DNA strand. These reports also highlighted a model in which each 24-nt siRNA is produced from one slicing of a precursor P4 RNA. The identification of P4 RNAs, observation of DCL-independent RdDM and a series of other exciting discoveries have increased our understanding of RdDM and raised several interesting questions regarding the RdDM pathway (Fig. 1).
Figure 1.
Pol IV-transcribed non-coding RNAs (P4 RNAs) in the RNA-directed DNA-methylation (RdDM) pathway. In addition to the canonical Pol IV-RDR2-DCL3 module, the Pol IV-dependent RdDM pathway may include the following branches as marked: (1) AtRRP6L1 may positively regulate P4 RNA accumulation by removing P4 RNA 3′ misincorporation; (2) DCL1 negatively regulates P4 RNA accumulation; (3) production of DCL-independent 24-nt siRNAs may involve AGO4 and/or RTL family proteins; and (4) P4 RNAs may directly mediate DCL-independent DNA methylation at some RdDM loci. Speculations are indicated by dashed lines. Red asterisks (*) indicate 3′ misincorporation. ‘m’ indicates a methylated cytosine. P4 RNAs are indicated by red wavy lines; 24 nt siRNAs are shown as orange straight lines; scaffold RNAs are shown as green lines. For simplicity, only those proteins that are discussed in the main text are shown.
HOW ARE DCL-INDEPENDENT 24-nt siRNAs GENERATED?
In Arabidopsis, 24-nt siRNAs account for most of the genome-wide siRNAs and play a prominent role in mediating RdDM. In the Pol IV-RDR2-DCL3 module, P4 RNAs are finally processed by DCL3 to yield 24-nt siRNAs. P4 RNAs are mostly 26–50 nt in length [4,6,7], while DCL3 preferentially cleaves 30- to 50-nt short dsRNAs in vitro [9]. In addition, P4 RNAs isolated from the dcl2/3/4 mutant can be digested by purified DCL3 protein, resulting in 24-nt siRNA products [4]. These observations are all consistent with the notion that DCL3 is the factor responsible for cleaving Pol IV-dependent precursor dsRNA into 24-nt siRNAs. However, 24-nt siRNAs are still detectable in the dcl2/3/4 mutant plants, although their levels are significantly decreased compared to those in the wild-type (WT) [4,6–8]. In dcl2/3/4, the proportions of 24-nt siRNAs, at least at some loci such as AtREP2, are comparable to the proportions of small RNAs of other sizes [8]. A small proportion of 24-nt siRNAs can apparently be produced independently of DCLs 2/3/4 by some unidentified factors. DCL1 may also cleave P4 RNAs, as indicated by the higher levels of P4 RNAs in the dcl1/2/3/4 quadruple mutant than in the dcl2/3/4 mutant [7,8]. While production of 21-nt miRNAs is a signature of small RNA processing by DCL1, it is possible that 24-nt siRNAs are generated when P4 RNAs are processed by DCL1. However, the contribution of DCL1 to 24-nt siRNA production would probably be negligible, because levels of 24-nt siRNAs are similar in dcl1/2/3/4 and dcl2/3/4 [7]. In any case, the presence of 24-nt siRNAs in dcl1/2/3/4 demonstrates that at least some 24-nt siRNAs can be totally DCL-independent.
A possible producer of DCL-independent 24-nt siRNAs is AGO4, which is known to possess slicer activity [10]. AGO2 in animals can cleave miRNA precursors to yield mature miRNAs in a DCL-independent manner [11]. Similarly, Arabidopsis AGO4 may cleave P4 RNAs and directly produce 24-nt siRNAs such that one P4 precursor RNA yields one 24-nt siRNA. AGO4 in association with an siRNA can cleave target RNAs, thereby generating secondary siRNAs [10]. Such a scenario may apply to AGO4-mediated complementary pairing between siRNA and Pol V-generated scaffold RNAs, i.e. AGO4 may cleave scaffold RNAs and thereby generate 24-nt siRNAs to reinforce RdDM. In support of this inference, Pol V contributes to 24-nt siRNA accumulation at many loci. As an alternative to using siRNAs as the initial trigger to cleave the target RNAs, AGO4 may also use P4 RNAs to locate corresponding target RNAs, which can be subsequently sliced to produce shorter RNAs including 21- to 24-nt siRNAs. Loss-of-function mutation of AGO4 may not be suitable for examining the potential role of AGO4 in generating DCL-independent 24-nt siRNAs because pre-existing DNA methylation may be required for Pol V recruitment to RdDM loci [12]. However, a catalytically inactive form of AGO4 in the dcl2/3/4 or dcl1/2/3/4 background will help elucidate how AGO4 may produce DCL-independent 24-nt siRNAs, because the slicer function is unnecessary for AGO4-mediated DNA methylation at some loci [10].
In addition to the DCL family, Arabidopsis has five RNase III-like (RTL) proteins, among which RTL1, RTL2 and RTL3 harbor both dsRNA-binding and RNase III domains. Although the function of RTL proteins is largely unknown, there is evidence for the differential involvement of RTL1 and RTL2 in siRNA production. RTL1 was shown to cleave dsRNA substrates, and its over-expression resulted in almost complete loss of siRNAs that are dependent on DCL2, DCL3 or DCL4 [13]. Thus, RTL1 has been proposed to function as a general suppressor of siRNA production, being upstream of DCL proteins by destroying siRNA precursors [13]. RTL2 modulates 24-nt siRNA production and the associated DNA methylation, positively at some loci while negatively at some other loci, likely by cutting double-stranded siRNA precursors into shorter fragments for further processing by DCL proteins into mature 24-nt siRNAs [14]. It would be useful to determine whether RTL1 competes with DCL3 for P4 RNAs as substrates, and whether RTL2 can contribute to the production of DCL-independent 24-nt siRNAs.
WHAT IS THE ROLE OF AtRRP6L1 IN P4 RNA PRODUCTION?
Theoretically, 24-nt siRNAs may also be generated directly from 5′-3′ or 3′–5′ exoribonulease activities that would prune short P4 RNAs into siRNAs. Arabidopsis RRP6L1 is a putative 3′–5′ exoribonuclease that binds to and positively regulates some long non-coding RNAs, including Pol V-dependent scaffold RNAs, in the RdDM pathway [15]. AtRRP6L1-dependent DNA methylation can also be attributed to its positive regulation of 24-nt siRNAs levels [8,15]. Researchers recently showed that AtRRP6L1 is required for P4 RNA accumulation [8]. The contribution of AtRRP6L1 to 24-nt siRNA generation therefore appears to be due to enhanced P4 RNA production and/or stability rather than to the direct pruning of P4 RNAs into siRNAs.
One possible way by which AtRRP6L1 facilitates P4 RNA production is via RNA retention. Like its homologues in yeast and mammals, AtRRP6L1 mediates chromatin retention of its target RNAs, such as Pol V-dependent scaffold RNAs, for siRNA recruitment [15]. P4 RNA levels are equally dependent on Pol IV and RDR2 [4–8]. Because RDR2 interacts with Pol IV in vivo and because RDR2 is non-functional without Pol IV [16], it appears that RDR2 favors Pol IV transcripts that are retained in the transcription complex; one can therefore speculate that, if AtRRP6L1 also helps retain Pol IV transcripts in the transcription complex, it may facilitate the function of RDR2 in P4 RNA production. In addition, AtRRP6L1 is not required for generation of siRNAs that do not need RNA retention for their production [15]. Recent findings revealed that a portion of P4 RNAs tend to have misincorporation at their 3′ ends, which is correlated with Pol IV transcription termination at positions of methylated cytosines on the template DNA strand [4,6]. This indicates that Pol IV may frequently face transcriptional arrest at sites where DNA methylation causes RNA misincorporation. It is unknown whether Pol IV possesses exoribonuclease activity that can remove 3′ misincorporation. Alternatively, transcriptional arrest can cause the RNA polymerase to backtrack, leaving the 3′ end of the RNA extruded [17]. Although the mechanisms of Rrp6-mediated RNA retention remain unclear, AtRRP6L1 dysfunction decreased the chromatin occupancy of Pol V, and the conserved exoribonuclease domain of AtRRP6L1 is required for its positive regulation of RdDM [15]. These finding suggest the possibility that AtRRP6L1 may function in proofreading during Pol IV transcription by enzymatic removal of 3′ misincorporated nucleotides, thereby allowing for more successful Pol IV transcription. In such a scenario, loss of AtRRP6L1 function would result in fewer P4 RNAs (as has been observed [8]), while the remaining P4 RNA pool would be characterized by smaller RNA sizes in general and by an increased 3′ misincorporation rate.
DO P4 RNAs MEDIATE DNA METHYLATION?
The small sizes of P4 RNAs suggest the following question: like siRNAs, can P4 RNAs mediate DNA methylation? DCL-independent DNA methylation was reported at about one-quarter of RdDM loci where DNA methylation levels are similar between the dcl1/2/3/4 mutant and WT plants [7]. DNA methylation at these loci, however, is normally controlled by DCLs 2/3/4, because methylation levels at these loci are significantly lower in the dcl2/3/4 mutant than in the WT [7]. This conditional independence of DCLs indicates that DNA methylation at these special RdDM loci is mainly regulated by DCL2/3/4 when DCL1 is present; when DCL1 is absent, however, an unknown factor is released from DCL1 suppression and overcomes the consequence of DCLs 2/3/4 mutations, assuring normal methylation levels at these loci. P4 RNA levels are higher in dcl1/2/3/4 than in dcl2/3/4, indicating that P4 RNAs may be the DCL1-repressed factor that is responsible for the restored DNA methylation. RdDM efficiency is affected by the quality of the trigger RNA, i.e. sequence identity between the trigger and the target [18]. Interestingly, in contrast to the dramatic reduction in DNA methylation in dcl2/3/4 compared to the WT, P4 RNA levels in dcl2/3/4 are comparable to 24-nt siRNA levels in the WT [4,6–8]. It therefore seems that P4 RNAs are less efficient than siRNAs in mediating DNA methylation, so that P4RNAabundance must be higher to compensate for the reduction in siRNAs, as was observed in dcl1/2/3/4 relative to dcl2/3/4 [7]. Consistently with this hypothesis, P4 RNA levels are significantly lower at RdDM loci where DNA hypomethylation in dcl2/3/4 cannot be restored in dcl1/2/3/4 than at loci where DNA methylation can be restored by DCL1 mutation [7].
AGO4-bound RNAs are mainly 24-nt siRNAs in WT plants; in the dcl2/3/4 mutant, however, AGO4 still selectively binds to the remaining 22- to 24-nt siRNAs but not to P4 RNAs, despite the concomitant increase in P4 RNA levels and decrease in 24-nt siRNA levels [6]. Consistently with this finding, another report indicated that the overwhelming majority of AGO4-bound RNAs in the dcl2/3/4 mutant are no longer than 26 nt [8]. These observations seem to argue against a role of P4 RNAs in mediating RdDM. It is unclear, however, whether the percentage of P4 RNAs in AGO4-bound RNAs can be higher in dcl1/2/3/4 than dcl2/3/4. It is also unclear whether other AGO proteins, such as AGO6, which is also known to mediate RdDM, may have a different affinity than AGO4 for P4 RNAs. In addition, histone modifications at RdDM loci may alter the efficacy of the AGO-P4 RNA complex in mediating DNA methylation, because repressive histone marks are particularly enriched at loci where DNA hypomethylation in dcl2/3/4 can be restored in dcl1/2/3/4, while euchromatic marks are characteristic of loci where DNA methylation is fully DCL-dependent [7].
Restoration of DNA methylation at the conditionally DCL-independent loci is probably unrelated to the remnant 24-nt siRNAs in dcl1/2/3/4, because the levels of 22- to 24-nt siRNAs appear similar in dcl1/2/3/4 and dcl2/3/4 [8]. P4 RNA production depends on Pol IV and RDR2 [4–8]. Therefore, an additional null mutation of Pol IV or RDR2 in dcl1/2/3/4 should abolish P4 RNA production, and the comparison between these quintuple mutants and dcl1/2/3/4 should reveal whether P4 RNAs are necessary for DCL-independent DNA methylation. Moreover, exogenous application of synthetic P4 RNAs to nrpd1 dcl1/2/3/4 and rdr2 dcl1/2/3/4 quintuple mutants may directly establish a causative role of P4 RNAs in DCL-independent DNA methylation, given that P4 RNAs are not much longer than siRNAs that have been shown to be mobile in plants.
DO P4 RNAs AND siRNAs ACT IN AN INTERCHROMOSOMAL MANNER?
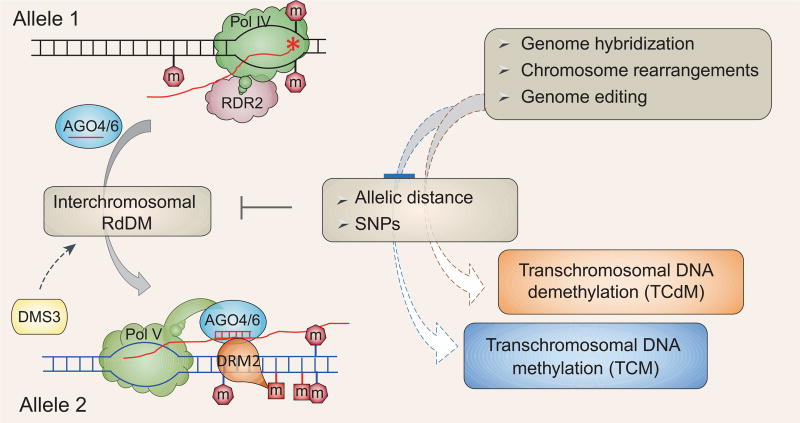
At a given RdDM locus, it would seem that Pol V transcription of scaffold RNAs would have to occur after Pol IV transcription and not vice versa, such that chromatin retention of Pol V-dependent scaffold RNAs would not impede Pol IV-dependent siRNA generation. This assumption, however, may not be necessary because the Arabidopsis genome is diploid. Because the genome is diploid, two alleles of the same genetic locus may each be separately and simultaneously transcribed by Pol IV and Pol V, such that siRNAs or P4 RNAs generated from one allele can efficiently lead to DNA methylation on the other allele. Cooperative RdDM activities between the two alleles may be observable when two parental alleles interact during genetic hybridization. In support of this view, plant hybrids are known to exhibit epigenome interactions, as shown by non-additive increases or decreases in DNA methylation in the F1 progeny compared to the parents [19,20]. Arabidopsis DNA methylome interactions were abolished in the absence of Pol IV and Pol V, suggesting that RdDM is responsible for this interesting phenomenon [20].
In Arabidopsis, cooperative RdDM activities between two chromosomes might explain DNA methylome interactions, which are characterized by transchromosomal DNA methylation (TCM) and transchromosomal DNA demethylation (TCdM). DNA methylation levels at TCM loci are higher and those at TCdM loci are lower in F1 plants than the mid-parent-values. The physical distance between two alleles would affect the efficacy of AGO-siRNA or AGO-P4 RNA complexes in mediating transallelic DNA methylation, thereby resulting in either TCM or TCdM (Fig. 2). Production of Pol V-dependent scaffold RNAs is facilitated by DMS3, which is a putative structural maintenance of chromosomes (SMC) protein [1]. SMC proteins are dynamic linkers of the genome, controlling higher-order chromosome structure and dynamics, such as chromatid pairing and chromosome condensation. Therefore, DMS3 may help mediate chromosome interactions in the RdDM pathway. Genome hybridization may introduce SNPs (single-nucleotide polymorphisms) into the two RdDM alleles; as a result, siRNAs or P4 RNAs from one allele could be less successful in pairing with scaffold RNAs on the other allele, thereby leading to TCdM in the F1 compared to the parents (Fig. 2). This possibility is consistent with the finding that genetic variations between the parents at TCdM loci appear to be greater than those at loci without DNA methylation interactions [20].
Figure 2.
Interchromosomal RdDM as a potential mechanism for DNA methylome interactions. In diploid genomes, siRNAs or P4 RNAs from one allele may act in trans to guide DNA methylation on the other allele. This interchromosomal RdDM process is presumably limited by the physical distance between the two alleles and by the frequency of SNPs. Alterations in allele distance and/or SNP frequency may cause TCM and TCdM events, which are characteristic of epigenome interactions. For simplicity, only those RdDM components that are discussed in the main text are shown.
Although the discovery of P4 RNAs and DCL-independent RdDM has greatly increased our understanding of the RdDM pathway, it has also increased our awareness of many unknowns concerning RNA-mediated epigenetic regulation (Fig. 1). In addition to raising the questions discussed earlier in this report, some findings of recent RdDM studies have deviated from the current RdDM model. For instance, although RDR2 has been assumed to function downstream of Pol IV, recent findings show that RDR2 and Pol IV are equally important for P4 RNA accumulation [4–8], indicating that our understanding of the mode of action of RDR2 remains incomplete. Recent work has also revealed specific but interdependent functions of AGO4 and AGO6 in RdDM [2], suggesting that RdDM is mediated by distinct, spatially regulated combinations of AGO proteins and RNA polymerases; a detailed knowledge of the subnuclear spatial distribution of RdDM components, however, is lacking. Research on these questions and unknowns will lead to improved models of RdDM.
Acknowledgments
We apologize to those colleagues whose work is not cited due to space constraints. H.Z. is supported by the Chinese Academy of Sciences (CAS Hundred Talents Program). J.-K.Z. is supported by the CAS and by the US National Institutes of Health (R01GM070795).
References
- 1.Matzke MA, Mosher RA. Nat Rev Genet. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 2.Duan CG, Zhang H, Tang K, et al. EMBOJ. 2015;34:581–92. doi: 10.15252/embj.201489453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stroud H, Greenberg MV, Feng S, et al. Cell. 2013;152:352–64. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blevins T, Podicheti R, Mishra V, et al. Elife. 2015;4:e09591. doi: 10.7554/eLife.09591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li S, Vandivier LE, Tu B, et al. Genome Res. 2015;25:235–45. doi: 10.1101/gr.182238.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhai J, Bischof S, Wang H, et al. Cell. 2015;163:445–55. doi: 10.1016/j.cell.2015.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang DL, Zhang G, Tang K, et al. Cell Res. 2016;26:66–82. doi: 10.1038/cr.2015.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye R, Chen Z, Lian B, et al. Mol Cell. 2016;61:222–35. doi: 10.1016/j.molcel.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagano H, Fukudome A, Hiraguri A, et al. Nucleic Acids Res. 2014;42:1845–56. doi: 10.1093/nar/gkt1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qi Y, He X, Wang XJ, et al. Nature. 2006;443:1008–12. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- 11.Cheloufi S, Dos Santos CO, Chong MM, et al. Nature. 2010;465:584–9. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson LM, Du J, Hale CJ, et al. Nature. 2014;507:124–8. doi: 10.1038/nature12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shamandi N, Zytnicki M, Charbonnel C, et al. PLoS Biol. 2015;13:e1002326. doi: 10.1371/journal.pbio.1002326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elvira-Matelot E, Hachet M, Shamandi N, et al. Plant Cell. 2016;28:406–25. doi: 10.1105/tpc.15.00540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, Tang K, Qian W, et al. Mol Cell. 2014;54:418–30. doi: 10.1016/j.molcel.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haag JR, Ream TS, Marasco M, et al. Mol Cell. 2012;48:811–18. doi: 10.1016/j.molcel.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komissarova N, Kashlev M. Proc Natl Acad Sci USA. 1997;94:1755–60. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalakouras A, Dadami E, Wassenegger M, et al. Plant J. 2016;87:202–14. doi: 10.1111/tpj.13193. [DOI] [PubMed] [Google Scholar]
- 19.Greaves IK, Groszmann M, Ying H, et al. Proc Natl Acad Sci USA. 2012;109:3570–5. doi: 10.1073/pnas.1201043109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Q, Wang D, Lang Z, et al. Proc Natl Acad Sci USA. 2016;113:E4248–56. doi: 10.1073/pnas.1607851113. [DOI] [PMC free article] [PubMed] [Google Scholar]