Abstract
Ancestral environmental exposures such as toxicants, abnormal nutrition or stress can promote the epigenetic transgenerational inheritance of disease and phenotypic variation. These environmental factors induce the epigenetic reprogramming of the germline (sperm and egg). The germline epimutations can in turn increase disease susceptibility of subsequent generations of the exposed ancestors. A variety of environmental factors, species and exposure specificity of this induced epigenetic transgenerational inheritance of disease is discussed with a consideration of generational toxicology. The molecular mechanisms and processes involved in the ability of these inherited epimutations to increase disease susceptibility are discussed. In addition to altered disease susceptibility, the potential impact of the epigenetic inheritance on phenotypic variation and evolution is considered. Observations suggest environmentally induced epigenetic transgenerational inheritance of disease is a critical aspect of disease etiology, toxicology and evolution that needs to be considered.
Keywords: epigenetics, transgenerational, non-genetic inheritance, disease etiology, evolution, review
Introduction
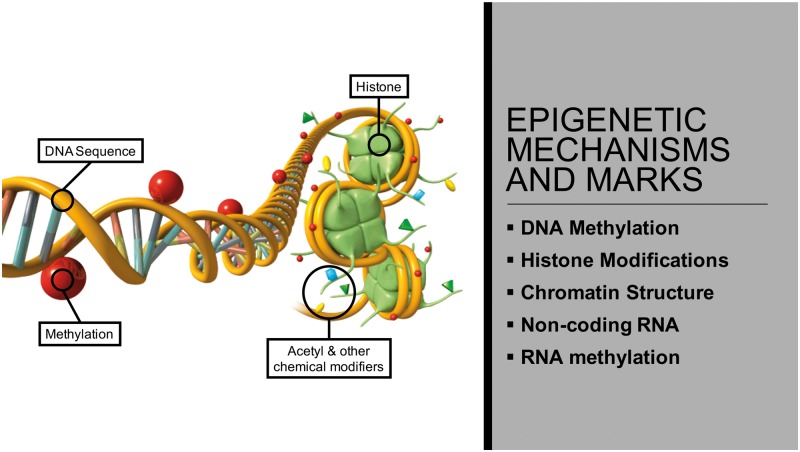
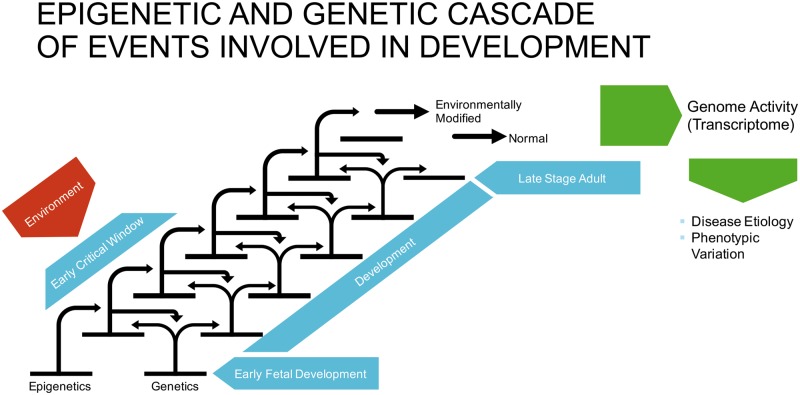
The term epigenetics was originally coined by C.H. Waddington in the 1940s in relation to his studies of gene–environment interactions involving non-Mendelian inherited phenomena [1, 2]. More recent molecular oriented definitions are that epigenetics refers to ‘the molecular factors and processes around the DNA that regulate genome activity independent of DNA sequence, and that are mitotically stable’ [3] (Table 1). These molecular factors include DNA methylation [4], histone modifications [5], non-coding RNAs [6, 7], chromatin structure [8], and RNA methylation [9] (Fig. 1). The complex integration of epigenetic modifications is referred to as the ‘epigenome’. The first whole epigenome analysis was accomplished in 2005, mapping histone acetylation and methylation in yeast [10]. Epigenetic processes are critical for allowing an organism to respond to its environment with changes in gene expression. In addition, epigenetic mechanisms allow a stem cell type to develop into a differentiated cell type [3, 11, 12] (Fig. 2). Therefore, epigenetic processes are an integral part of normal biology.
Table 1:
glossary terms and definitions
| Glossary term | Definition |
|---|---|
| Epigenetics | Molecular factors and processes around DNA that regulate genome activity independent of DNA sequence, and are mitotically stable |
| Epigenetic transgenerational inheritance | Germline mediated inheritance of epigenetic information between generations in the absence of continued direct environmental influences |
| Multigenerational | Direct exposure of multiple generations |
| Epimutation | Environmentally induced differential presence of epigenetic alterations that can lead to altered genome activity when compared to organisms not having the exposure |
Figure 1:
epigenetic mechanisms and processes (marks). Modified from [122]
Figure 2:
epigenetic and genetic cascade of events involved in development. Cascade of genetic and epigenetic stages interacting to promote differentiated cells. The critical window of exposure allows environmental factors to alter the epigenetic cascade to obtain a modified differentiated site and to cause altered transcriptomes to increase disease susceptibility and phenotypic variation. Modified from [3]
Molecular Epigenetic Mechanisms
There are a variety of epigenetic factors that act around the DNA in a cell to regulate gene expression and genome activity. DNA methylation is the most extensively studied epigenetic factor. DNA methylation involves a small (methyl) chemical group being attached to DNA, primarily at the cytosine base when it is adjacent to a guanine residue [4, 13] to produce 5-methylcytosine (5mC). Other chemical modifications of cytosine bases in DNA have also been described. The TET (ten-eleven translocation) family of enzymes can oxidize 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) [14]. In broad terms, the presence of 5mC often represses DNA transcription, while 5hmC is permissive to transcription [15, 16]. However, one of the main functions appears in the DNA methylation erasure during early development [17]. N(6)-methyladenine is an epigenetic modification to the adenine base of DNA that was once thought to only be present in prokaryotic organisms, but has now been described in mammalian embryonic stem cells [18].
The histone proteins that DNA is wrapped around create the nucleosome and can be chemically modified to alter gene expression. There are many different histone post-translational modifications including lysine acetylation, lysine and arginine methylation, arginine citrullination, lysine ubiquitination, lysine sumoylation, ADP-ribosylation, proline isomerization, and serine/threonine/tyrosine phosphorylation [19]. These modifications can change chromatin structure or recruit transcriptional cofactors to DNA in order to regulate gene expression. Alternatively, they can act as repressive marks to reduce gene expression in major regions of the genome such as heterochromatin. In broad terms, histone acetylation can increase transcription, while methylation can be repressive to transcription.
Non-coding RNA molecules can act as epigenetic factors [20]. These are small and long RNA molecules that do not code for a protein, but rather function as RNA to regulate gene expression. The non-coding RNA molecules that act as epigenetic factors are not DNA sequence dependent, so the majority do not rely on having a nucleotide sequence that is complimentary to a specific DNA or RNA region in order to function. Long non-coding RNAs (lncRNAs) [21] and transfer RNA-derived small RNAs (tsRNAs) [22] are examples of RNA classes that are present in sperm and can act as epigenetic factors that affect subsequent generations [23].
RNA molecules can themselves be epigenetically modified and so affect translation and gene expression [24]. The most prevalent reversible modification to the internal sequence of mRNA is methylation of adenosine to form N(6)-methyladenosine (m(6)A). m(6)A mRNA methylation is associated with post-transcriptional regulation [25, 26]. Cytosine methylation (m3C) in both mRNA and tRNA also occurs [27, 28]. Methylation of tRNA inhibits processing of tRNA into tsRNA halves, which themselves affect transcription [22, 29, 30]. Therefore, RNA methylation is the most recent epigenetic molecular factor identified.
The coiling, looping, and general structure of DNA is termed chromatin structure and is also an epigenetic factor [8]. The three-dimensional structure of DNA can make certain regions of the genome accessible to transcription machinery, such as bring enhancer regions near gene promoters to affect gene expression. Therefore, epigenetic molecular processes include DNA methylation, histone modifications, non-coding RNAs, RNA methylation, and chromatin structure.
Epigenetic Transgenerational Inheritance
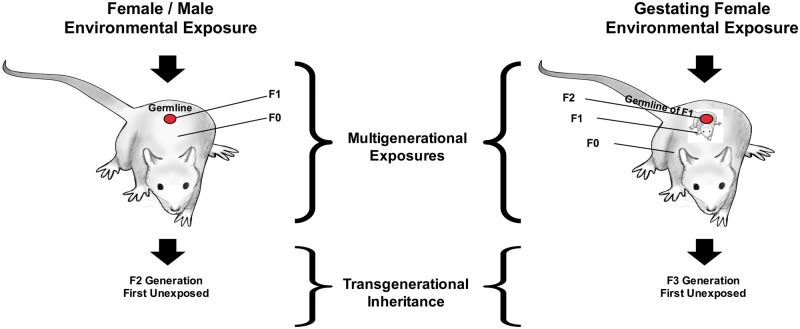
The definition of epigenetic transgenerational inheritance is ‘germline-mediated inheritance of epigenetic information between generations in the absence of continued direct environmental influences that leads to phenotypic variation’ [3, 31] (Table 1). Multigenerational exposures, in contrast, refer to observed effects in subsequent generations that are the result of direct exposure [31] (Table 1; Fig. 3). Direct environmental exposure of the parents, considered to be the F0 generation, can also affect the germline (sperm or eggs) of either parent. Therefore, the next generation (F1) derived from this germline is still considered exposed, and so is not truly transgenerational. For preconception parental exposures the F2 generation offspring is considered the first transgenerational unexposed generation (Fig. 3). The situation is different when a gestating female is exposed, because then the fetus and the fetus’ germline are directly exposed as well. In that case, the F3 generation is the first unexposed transgenerational offspring [31] (Fig. 3).
Figure 3:
environmentally induced transgenerational epigenetic inheritance. Schematic of multigenerational versus transgenerational environmental exposures. Modified from [31]
The Agouti mouse model is an example of multigenerational inheritance [32–34]. When pregnant Agouti mice are exposed to a methyl donor in their diet, they experience increased methylation on an allele of their Agouti gene, which leads to a coat color change in their offspring. Generally, this change is not passed on to future generations. Instead the normal process of epigenetic reprogramming in the germline and early embryo returns the DNA methylation state to its original setting.
An increasing number of examples of transgenerational inheritance of disease are present in the literature (Table 2). Some of the first experiments to establish the potential for epigenetic transgenerational inheritance were performed by Conrad Waddington, who coined the term ‘epigenetic’ [1, 35]. In these studies, it was found that a heat shock induced wing structure change in Drosophila melanogaster persisted for more than seven generations [35]. An even earlier study in Guinea pigs demonstrated transgenerational inheritance of decreased fertility and increased mortality for four generations after ancestral exposure to ethanol vapor, although this was not attributed to epigenetic inheritance at the time [36]. One of the first studies to associate molecular epigenetic changes with transgenerational inheritance of disease in mammals was an investigation of the effects of treating pregnant rats with the agricultural fungicide vinclozolin [37]. The F3 generation (great-grand offspring) demonstrated reproductive abnormalities such as increased testicular germ cell apoptosis and decreased sperm motility. These transgenerational phenotypes were correlated with changes in DNA methylation in the F3 generation sperm [37].
Table 2:
examples of transgenerational inheritance from specific exposures and specific effects
| Exposure | Effects | Reference |
|---|---|---|
| Environmental toxicants | ||
| Vinclozolin | Impaired male fertility; prostate, kidney disease, tumors, immune and reproductive pathologies | [37, 78, 94] |
| Vinclozolin | Gender-specific changes in anxiety-like behavior | [85] |
| Methoxychlor | Impaired male fertility; kidney disease, ovary disease, and obesity | [37, 86] |
| Permethrin/DEET | Prostate, kidney disease | [81] |
| Dioxin | Prostate, kidney disease, reduced fertility, negative effects on pregnancy outcome | [80, 123] |
| BPA/phthalates | Prostate, kidney disease; obesity | [43] |
| Hydrocarbon mixture (jet fuel) | Prostate, kidney disease; obesity; immune and reproductive pathologies | [46] |
| Vinclozolin, permethrin/DEET, plastics, dioxin, jet fuel | Polycystic ovaries, reduced primordial follicle pool | [82] |
| DDT | Obesity | [45] |
| Phthalate | Disruption of testicular germ cell organization and spermatogonial stem cell function, changes in hormones and behavior | [40, 124] |
| Phthalate | Disrupted ovarian function | [41] |
| Tributyltin | Increase in fat depot size | [38] |
| BPA | Cardiac disease; reduced fertility | [48, 72] |
| BPA | Changes in social behavior and neural gene expression | [42] |
| Atrazine | Testicular disease, early puberty, lean phenotype | [125] |
| Benzo[a]pyrene | Behavioral and physiological deficits | [50] |
| Mercury | Behavior change | [49] |
| Other exposures | ||
| Caloric restriction | Cardiovascular mortality | [56, 77] |
| High-fat diet | Increased body size; reduced insulin sensitivity, increased mammary cancer | [57–59] |
| Folate | Congenital malformations | [126] |
| Stress | Reduced social interaction; increased stress resilience; disrupted neural connectivity; physiology changes; increased anxiety | [51–55] |
| Drought | DNA methylation changes | [127] |
| Heat/salt stress | Accelerated flowering, increased salt tolerance | [128] |
| Prediabetes/diabetes | Impaired glucose tolerance; reduced insulin sensitivity, male subfertility | [61, 62] |
| Smoking | Abnormal pulmonary function | [129] |
| Ethanol | Neurological defects; decreased fertility | [36, 47, 130] |
| Heat stress | Increased Hsp70 production and tolerance to heat stress; wing structure changes | [131, 132] |
Several environmental toxicants including vinclozolin, DDT (dichlorodiphenyltrichloroethane), methoxychlor, plastic derived compounds, hydrocarbons, atrazine, tributyltin have been shown to promote the transgenerational inheritance of increased disease susceptibility in rodent models [38, 39] (Table 2). The diseases that were increased transgenerationally included testis, prostate and kidney disease, obesity, polycystic ovaries, reduced oocyte number in the ovaries, and cancer [39]. For the purposes of this review, more recently published investigations of epigenetic transgenerational inheritance of disease will be highlighted, (Table 2). Exposure of mice to the phthalate plastic derived compound DEHP (di(2-ethylhexyl) phthalate) has been shown to result in transgenerational changes to stress hormones, behavior [40], and ovarian function [41]. Earlier studies in mice [42] showed that ancestral exposure to the plastic derived compound bisphenol A (BPA) caused changes in social behavior in juvenile mice and changes in expression of neural genes such as oxytocin and vasopressin. Earlier studies in rats have shown that exposure to a mixture of BPA and phthalates induces transgenerational increases in pubertal abnormalities, testis disease, and ovarian disease [43]. Ancestral exposure of mice to the toxicant tributyltin results in a transgenerational increase in obesity [38, 44]. Earlier investigations in pregnant rat exposures to DDT, jet fuel hydrocarbons, or a BPA/phthalates mix will also increase obesity transgenerationally [43, 45, 46]. Other recently published investigations indicate that ethanol exposure of pregnant mice can cause transgenerational neurological changes in descendants that resemble those of Fetal Alcohol Spectrum Disorders [47]. In zebrafish, BPA exposure of males can result in the transgenerational inheritance of heart disorders in the F2 generation [48]. Zebrafish exposure to mercury [49] or to the industrial pollutant benzopyrene [50] induces the transgenerational inheritance of abnormal neurobehaviors that are correlated with epigenetic changes (i.e. epimutations) in sperm (Table 2) [49, 50].
Exposure to environmental factors other than toxicants can also induce transgenerational inheritance (Table 2). The stress of maternal separation in mice transgenerationally disrupts functional connectivity throughout the brain [51], as well as both impairing social interactions and cognition and making the descendant mice more stress resilient [52]. Mice subjected to restraint stress also transmitted reduced anxiety levels to their transgenerational descendants [53]. Conversely, social hierarchy stress in mice was shown to increase anxiety behaviors transgenerationally [54]. This raises the possibility that several psychological stressors can induce different transgenerational effects. In pregnant rats, the stressors of forced swim and restraint induce transgenerational inheritance of physiological changes such as alterations in catecholamine biosynthesis and immune response [55].
Other examples of transgenerational inheritance have been observed with caloric restriction or high fat diets. The Överkalix study by Bygren et al. [56] shows how cardiovascular mortality in humans can be influenced by reduced childhood and adolescent food supply. Effects were shown to reach into the second generation. Maternal high fat diet in mice can increase body size and reduce insulin sensitivity in F3 generation female offspring [57], although Masuyama et al. [58] demonstrated that a normal diet in utero for three subsequent generations can return glucose and lipid metabolism to normal. In addition, a maternal high fat diet in mice can transgenerationally increase mammary cancer risk [59]. Previous studies with rats demonstrated that exposure of pregnant animals to the environmental toxicant vinclozolin also promoted a transgenerational increase in tumors [60]. Interestingly, diabetes in mice can induce transgenerational inheritance of male subfertility [61]. A paternal prediabetic condition in mice can be inherited transgenerationally as shown by impaired glucose tolerance and decreased insulin sensitivity [62]. Similarly, male rats fed a high fat diet promoted transgenerational inheritance of impaired glucose tolerance in F2 generation offspring [63].
Species Diversity of Epigenetic Transgenerational Inheritance
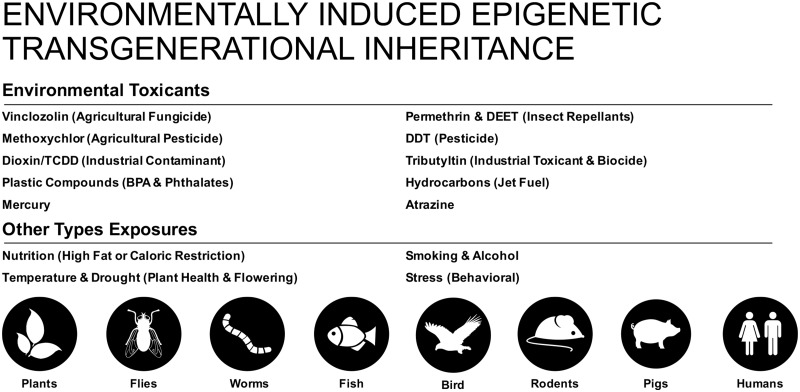
Epigenetic transgenerational inheritance has been identified to occur in a wide variety of organisms (Fig. 4). This review focuses on examples of epigenetic transgenerational inheritance of disease or abnormalities in different animal species. A number of studies have demonstrated the environment (e.g. heat and drought) can induce the epigenetic transgenerational inheritance of phenotypic variation in plants [64]. In the nematode worm Caenorhabditis elegans increased longevity that is associated with the histone modification H3K4me3 methylation can be transgenerationally inherited for up to three generations [65]. As mentioned previously, Waddington performed early experiments using the model insect species D. melanogaster and demonstrated that a heat shock induced wing structure changes that persisted for more than seven generations [1, 35] and now for hundreds of generations in today’s stocks. In more recent examples, it has been found that a high-sugar maternal fly diet can alter the larval body composition for the next two generations [66]. Similarly, a high fat larval diet in fruit flies can cause transgenerational alterations to F2 generation pupal and egg size [67]. Manipulations of the protein levels in the diet of fruit flies can affect longevity and reproduction for three subsequent generations, and this effect is associated with histone modifications [68, 69]. In another arthropod species, the crustacean Daphnia magna, exposure to the toxicant 5-azacytidine results in decreased body length and reduced levels of DNA methylation in non-exposed subsequent generations [70].
Figure 4:
environmentally induced epigenetic transgenerational inheritance. Various exposures and species investigated
Several species of fish have shown epigenetic transgenerational inheritance of disease. Zebrafish exposed to the environmental toxicants benzo(a)pyrene [50], methylmercury [49] or dioxin [71] transmit to their grand-offspring behavioral changes, visual defects, increased body mass, skeletal abnormalities and/or decreased fertility, sometimes associated with changes in DNA methylation. Medaka exposed to the endocrine disruptors BPA or ethinylestradiol produce grand-offspring and great-grand-offspring with reduced fertility [72].
Some bird species have shown evidence of epigenetic transgenerational inheritance. In a study with quail eggs exposed to the environmental estrogen genistein [73] the great-grand offspring age at which the first egg was laid was significantly greater. In ducks, feeding a methionine-deficient diet produces grand-offspring with altered weight gain and changes in metabolic parameters [74].
In mammals most studies of epigenetic transgenerational inheritance have occurred in rodents [75]. Another experimental mammal involves pigs and abnormal nutritional induced epigenetic transgenerational inheritance [76]. Examples of transgenerational inheritance of increased susceptibility to diseases have been outlined above for rats, mice and Guinea pigs [36, 37, 41, 44, 45]. Evidence of epigenetic transgenerational inheritance of disease in humans comes from retrospective studies such as those including the Dutch and Swedish famines [56, 77]. As previously mentioned, the descendants of people exposed to famine conditions as children 9–12 years of age in Sweden were investigated and it was found that men whose grandfathers had been exposed to famine had an increased risk of mortality due to diabetes, and similarly women whose grandmothers were exposed had increased risk [31]. Due to the conservation of environmentally induced epigenetic transgenerational inheritance from plants to humans all organisms will utilize epigenetic inheritance to facilitate environmental adaptation and response.
Phenotypic Diversity of Transgenerationally Inherited Diseases
Studies of the effects of ancestral exposure to an array of toxicants (Table 2) demonstrate epigenetic transgenerational inheritance of a variety of diseases and abnormalities, including testis disease [37], prostate and kidney disease [43, 46, 78–82], mammary tumors [78], immune and reproductive pathologies [46, 78, 83, 84], obesity [45, 46], behavioral effects [85] and many others listed in Table 2. The disease phenotypes observed in these experiments often depend on the specific exposure of the F0 generation. For example, increased obesity risk in rats is inherited transgenerationally after ancestral exposure to DDT, plastic compounds, hydrocarbons and methoxychlor [43, 45, 86], but not dioxins. Jet fuel hydrocarbons induce an elevated rate of luteal ovarian cyst formation in F3 females [46, 82], a phenotype not observed with other exposures. On the other hand, some ovarian disorders such as polycystic ovaries and reduction of the primordial follicle pool size have been shown to be inherited transgenerationally after exposure of the F0 generation to many of the toxicants studied [84, 87]. The explanation for this phenomenon may be that some developmental processes, in this case ovarian follicle development, are more sensitive to epigenetic and gene expression changes in their developmental regulatory networks, and so will be more easily affected than those of other cells and tissues (Fig. 2).
Epigenetic processes are major mechanisms by which organisms respond and adapt to their environment. Therefore, how can environmental epigenetic insults result in transgenerational inheritance of increased disease susceptibility? Since this is a maladaptive response one possible answer may be seen in the predictive adaptive response hypothesis [88]. In this hypothesis an environmental stressor like famine may epigenetically promote an adaptive (thrifty) phenotype in subsequent generations. If the current environment of those descendants has more than adequate nutrients, diseases such as diabetes and obesity are promoted. Another possibility is that an environmental insult, such as exposure to a toxicant, may interfere with the normal molecular epigenetic machinery and result in stochastic and/or directed epigenetic changes that could be considered epimutations. The term epimutation is defined as ‘the environmentally induced differential presence of epigenetic alterations that can lead to altered genome activity, when compared to organisms not having exposure’ (Table 1). If these epimutations occur in germ cells they can lead to transgenerational inheritance of a wider range of phenotypes in the progeny. Some of those phenotypes may be poorly adapted and develop disease. This would explain an increase in disease susceptibility in organisms whose ancestors were exposed to environmental insults. However, phenotypic variation is the ‘raw material’ upon which natural selection acts. Therefore, the increased phenotypic variation may also result in some individuals who are better adapted to an altered environment which can facilitate natural selection and evolution [89].
Developmental Etiology of Epigenetic Transgenerational Inheritance
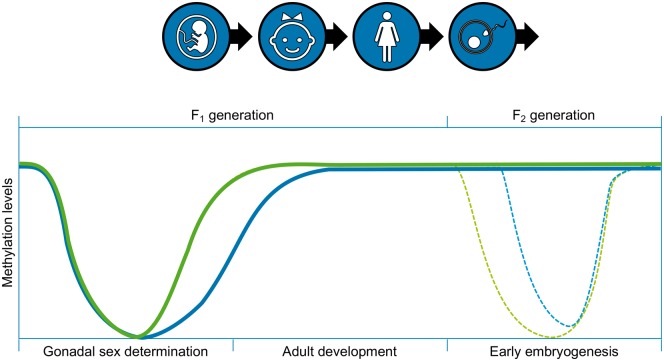
A number of reproductive processes involve DNA methylation changes that normally will be reset by genome-wide DNA methylation reprogramming events. The two main developmental periods are in the early embryo after fertilization and during germ cell specification at gonadal sex determination [90, 91] (Fig. 5). This phenomenon allows embryonic stem cells to develop by removing epigenetic constraints to promote pluripotency. Some parental epigenetic changes, such as imprinted genes, are protected from being reprogrammed during these developmental periods. In contrast, some parent specific imprints are established during this epigenetic reprogramming [92]. Environmentally induced DNA methylation alterations called differential DNA methylation regions (DMRs) [93] present in germ cells behave as imprinted-like genes in the way their methylation patterns persist. By definition, true imprinted genes display ‘parent-of-origin allelic transmission with monoallelic gene expression’. DMRs often demonstrate parent-of-origin allelic transmission, but monoallelic gene expression has not been demonstrated. Differentially methylated sites connected with transgenerational inheritance are called ‘imprinted-like’ [94]. The transmission of epigenetic information to future generations via germ cells can alter the epigenome of the developing embryonic stem cells which would be expected to promote changes to the epigenetic and transcriptomic programming of all derived somatic cells [95]. Those tissues that are sensitive to alterations in their epigenomes and transcriptomes may show increased susceptibility and prevalence of disease development [93, 96] (Fig. 2). Normal biology requires alterations of epigenetics for the development of stem cell populations and subsequent somatic cell differentiation. The epigenetic transgenerational inheritance molecular process is directly linked to these epigenetic reprogramming processes in the germline and the developing embryo.
Figure 5:
epigenetic reprogramming during primordial germ cell development at gonadal sex determination and following fertilization in the early embryo. Modified from [94]
Germline Epimutations
It is a prerequisite for environmentally induced epigenetic transgenerational inheritance that there be epigenetic changes (i.e. epimutations) in the germline, because the germ cells (sperm and egg) are the only cells that can transmit molecular information between generations from the parents to their offspring. Early studies investigating transgenerational epimutations in germ cells used pregnant rats exposed to vinclozolin during the period of gonadal sex determination when epigenetic reprogramming of the fetal germ cells occurs. A genome-wide promoter analysis was applied to look for epigenetic changes in the sperm DNA and approximately 50 differential DMRs were identified in gene promoters in vinclozolin lineage F3 generation sperm DNA versus control lineage [97]. Similar experiments have been performed in rats using a number of additional toxicants including dioxin [80], a mixture of permethrin and DEET (N,N-diethyl-meta-toluamide) [81], BPA and phthalates [43] and jet fuel (hydrocarbons) [46]. All these toxicants were found to promote transgenerational inheritance of both disease and sperm DMRs. Interestingly, it was observed that each toxicant produced an exposure-specific set of DNA methylation changes in the sperm, and comparisons between the different toxicant exposures demonstrated negligible overlap between them [82]. This raises the possibility that these ‘epigenetic signatures’ may be used in the future as a diagnostic tool to determine if an individual has had a particular environmental toxicant exposure in their ancestry. The examination of the genomic features of all these DMRs identified a low CpG density termed CpG deserts [98] and a DNA sequence motif called Environmental Induced Differential Methylation Consensus Sequence 1 (EDM1). Nearly all the DMRs identified with numerous exposures had these genomic features [39]. A machine learning analysis used this data to identify approximately 40 000 potential genome-wide DMR sites susceptible to environmental alterations [99]. Further studies are needed to determine the utility of these potential epimutation sites as biomarkers for exposure and disease.
Comparisons have been made of the DMRs induced in the direct exposure F1 generation and transgenerational F3 generation vinclozolin lineage male sperm [100]. As described above, when the gestating female is directly exposed to a toxicant the F1 generation fetus is also directly exposed, as are the developing germ cells within the F1 generation fetus that will generate the F2 generation. The F3 generation animals are the first non-exposed transgenerational descendants (Fig. 3). Therefore, the molecular mechanisms of inducing epigenetic changes is different in the direct exposure F1 generation, and in the F1 generation germ cells (sperm) that will produce the F2 generation, when compared with mechanisms by which epimutations are induced in the transgenerational F3 generation. In a study involving vinclozolin exposure of gestating female rats there was a distinct set of DNA methylation changes in the F1 generation sperm that was different from the set of methylation changes in the transgenerational F3 generation sperm [100]. This suggests that the direct exposure induced F1 generation sperm epimutations promote epigenetic alterations during germ cell development in subsequent generations that lead to the different DMRs in the F3 generation. This mechanism appears to be associated with altered early embryonic development of the stem cells.
In addition to DNA methylation, other epigenetic factors such as non-coding RNA (ncRNA) can also contribute to epigenetic transgenerational inheritance. Small ncRNAs of the microRNA class are altered in the sperm of stressed vs. un-stressed mice and have been shown experimentally to promote a change in the hypothalamic–pituitary–adrenal stress axis reactivity of offspring [101]. Another class of small non-coding RNAs associated with transgenerational sperm are 5′ halves of tRNAs [102]. These stRNA 5′ halves and microRNAs are transgenerationally altered in the F3 generation sperm of rats ancestrally exposed to vinclozolin during pregnancy [102]. A number of studies have demonstrated the potential role of ncRNA in epigenetic transgenerational inheritance [103].
Another epigenetic factor present in sperm and associated with transgenerational inheritance is the retention of histone proteins [104]. During spermatogenesis in vertebrates the histone cores that DNA is wrapped around in most somatic cell types are replaced by protamines, allowing for more tightly compacted DNA in sperm heads [105]. However, 1–10% of histones are retained in mammals, depending on species [106]. These retained sperm histones have been implicated in regulating gene expression in the resulting offspring [107]. In a recent transgenerational study using rats, Ben Mammar et al. [108] demonstrated that a specific set of histones are retained in F3 generation control lineage sperm. This same set of histones is retained in F3 generation rats ancestrally exposed to vinclozolin or DDT, but additional sites of histone retention are induced in the vinclozolin and DDT lineage sperm [108]. Therefore, histone retention also appears to be associated with sperm mediated transgenerational inheritance of disease following ancestral DDT or vinclozolin exposure [104, 108].
Since post-translational modifications of histones are known to be an epigenetic factor that regulates gene expression studies have investigated histone modifications present in sperm. Histone H3 methylation changes in retained sperm histones have been correlated with fertility in humans [109] and with survival of offspring in mice [110]. Histone modifications have been correlated with epigenetic transgenerational inheritance of altered phenotypes in C. elegans [111], Drosophila [112], and recently in mammals [104].
Previous transgenerational studies have focused on epigenetic factors and epimutations in sperm due to the relative ease of obtaining large numbers of sperm cells. Several studies have shown that epigenetic transgenerational inheritance is mediated through the female germline [45, 86]. Eggs cannot be obtained in large enough quantity to allow traditional molecular analysis. Future studies with single cell analyses may be needed to document the role of epimutations in eggs. Epigenetic factors in eggs appear to play an equally important role in epigenetic inheritance, but remain to be investigated. The epigenetic transgenerational inheritance of disease following environmental exposures will likely be mediated by the integrated actions and combination of different epigenetic factors present in gametes. A recent study in rats demonstrated that after treatment of gestating females with DDT or vinclozolin there were concurrent transgenerational alterations in F3 generation sperm in DNA methylation, histone retention, and non-coding RNAs [108, 113]. Therefore, transgenerational alterations in all the different epigenetic processes appear to be involved in the epigenetic transgenerational inheritance phenomenon.
Transgenerational Gene Expression Changes
Transgenerational inheritance of environmentally induced epigenetic changes requires transmission through the germ line from parents to future generations. However, epigenetic changes themselves would not cause disease, rather they must manifest as changes in gene expression. Ensuing disease such as cancer, prostate or kidney abnormalities, and obesity are brought on by disturbances in gene expression in the pertinent somatic cells. The hypothesis is that the epimutations in the germline alter the epigenome of the embryonic stem cells that then affect all subsequent somatic cell epigenomes and transcriptomes [87, 95] (Fig. 2). These cell and tissue specific epimutations promote tissue specific alterations in transcriptomes [96]. These aberrant transcriptomes could then lead to a susceptibility for physiological abnormalities and disease (Fig. 2).
Exposure of F0 generation animals to environmental toxicants will affect and change the transcriptomes of potentially all tissues in future generations [96]. In a study of rats ancestrally exposed to vinclozolin the transcriptomes of 11 different tissue types from adult male and female animals were examined [96]. It was found that there were gene expression differences between control and vinclozolin lineage animals in the different tissues with minimal overlap in the differentially expressed genes between tissues. However, there was significant overlap in the physiological pathways and cellular processes that were affected by gene expression changes in different tissues. For example, both prostate and liver tissues were enriched for genes in transcription and focal adhesion processes, but the specific genes altered were not the same in each tissue [96]. These observations warranted a closer look at the genomic locations of epimutations and differentially expressed genes. Looking across the different tissue types it was found that there were regions of the genome that had statistically over-represented clusters of gene expression changes [96]. These regions in the genome were called epigenetic control regions (ECR). These ECR are 2–5 megabase in size and have clusters of genes. Within these ECRs are DNA methylation epimutations and long non-coding RNA (ncRNA) expression sites [114]. The long ncRNAs play a role in regulation of distal gene transcription and epigenetic regulation [115, 116]. Observations suggest that within an ECR many of the genes are epigenetically regulated up or down as a block. Therefore, in one cell type those genes within the ECR normally expressed would be regulated while in another cell type a different set of genes within the ECR normally expressed would be affected. Epigenetic alterations within the ECR can influence gene expression in a variety of cell types differently [96]. Interestingly, the location of ECRs has been shown to co-localize with clusters of transgenerational epimutations (e.g. DMRs) found after ancestral toxicant exposures [117].
Several studies have suggested how the molecular mechanisms of environmentally induced transgenerational inheritance may lead to tissue specific disease occurrence. As mentioned earlier, two ovarian disorders, polycystic ovarian syndrome, and primary ovarian insufficiency (premature reduction of the primary follicle pool) were both induced transgenerationally by a number of environmental toxicants [83]. Analysis of this phenomenon involved the isolation of a specific cell type from the tissue that is associated with the disease in the vinclozolin lineage animals. The granulosa cells were isolated from the ovarian follicles of young female rats prior to disease onset. The epigenomes and transcriptomes of these granulosa cells from control and vinclozolin lineages were analyzed and compared [87]. Granulosa cells from F3 generation vinclozolin lineage rats had differences in both the epigenome and the transcriptome compared with the control lineage. Interestingly, some of the affected genes had been previously shown to be associated with polycystic ovarian syndrome and primary ovarian insufficiency [87]. Similar results were obtained when the molecular basis of transgenerational male infertility in rats was examined. As above, changes in the epigenome and transcriptome were found in testicular Sertoli cells of F3 generation rats after ancestral vinclozolin exposure [95]. Several of the differentially regulated genes identified were known to be associated with male infertility, such as HDAC1 and HSP90AA1 [118, 119]. In addition, a number of Sertoli cell genes associated with pyruvate production were down-regulated and this is known to impact spermatogenic cell survival and promote germ cell apoptosis, which is one of the testis pathology phenotypes observed [95]. Therefore, the environmentally induced transgenerational changes in the somatic cell epigenomes are associated with transgenerational changes in gene expression, which are related to the increases in disease development observed.
Experimental and Technical Approach Limitations
One of the main experimental design issues and limitations is a consideration of what constitutes a multigenerational or intergenerational direct exposure versus a true non-exposed transgenerational generation. A number of past studies have referred to F1 generation fetal exposure or F1 generation germline that will generate the F2 generation as transgenerational experiments (Fig. 3). Many previous reports have not carefully considered this issue and misinterpreted the results as transgenerational. A multigenerational or intergenerational exposure experiment is important and helps elucidate risk of exposure on multiple generations physiology and pathology. However, the mechanisms involved are distinct and impacts different than transgenerational generations [31]. This non-genetic form of inheritance needs to be distinguished from multiple generation exposure that is due to direct exposures and toxicities.
Another experimental design issue is the use of mixed cell populations for an epigenetic analysis [120]. Every cell type in the body has the same DNA sequence, so for genetic analysis a mixed cell population does not affect the data or observations. In contrast, each cell type in the organism has a very distinct epigenome to allow the cell type to have its unique cell biology and physiology. The reason a neuron is distinct from a hepatocyte is not the genetic sequence, but the epigenetic differences between the cell types that regulate the unique gene expression. Therefore, an epigenetic analysis of mixed cell populations are influenced by small changes in specific cell population numbers which will alter the epigenetic data experimentally observed without an actual change in molecular epigenetics [120]. A number of epigenetic studies have used whole blood which contains over 20 different cell populations to do epigenetics. Twin studies using this approach have not been revealing due to the variation in cell populations in the blood and inability to dissect out specific epigenetic changes. Purifying a specific cell type such as monocytes from the blood will be far more useful for epigenetic analyses than use of the mixed cell population. Therefore, epigenetic analysis optimally requires purified cell populations [120].
Epigenetic molecular procedures have dramatically developed over the past decade to provide greater accuracy and precision. The technology of next generation sequencing is superior to array technology and previous biochemical procedures. The current procedures for DNA methylation, ncRNA and histone modifications use next generation sequencing which should be considered the optimal approach for any genome-wide analysis. If a few selected sites are examined then the array technology or biochemical approach can be used and are less costly. For the genome-wide approaches, the different DNA methylation approaches are methylated DNA immunoprecipitation (MeDIP) sequencing (MeDIP-Seq) and bisulfite sequencing (BS-Seq). The MeDIP-Seq is biased to low density CpG <20% while the BS-Seq is biased to high density CpG. All these procedures are efficient, but the limitation in CpG bias needs to be considered in the interpretation of the data obtained. The RNA-Seq and chromatin immunoprecipitation ChIP-Seq approaches are the optimal procedures currently available with few alterations. Third generation sequencing that may be able to assess epigenetic modifications during the sequencing will be a future technology to elucidate the DNA methylation CpG density bias, but remains to be optimized. The rate at which epigenetic technology is developing suggests within the next five years we will likely be using new technologies. The research in this area needs to consider the limitations of some of the technology currently used.
Conclusions
Research in the area of environmentally induced epigenetic transgenerational inheritance of disease and phenotypic variation has provided evidence of transgenerational inheritance of epimutations in plants, worms, flies, fish, birds, pigs, mice, rats, and humans [121] (Fig. 4). Ancestral exposure to environmental influences such as toxicants, abnormal nutrition, or stress can induce changes in the germline epigenome that are transmitted to descendants. These epimutations caused by individual exposures must occur in the germline in order to be transmitted. When these germline epigenetic changes become imprinted-like and escape the normal processes of epigenetic reprogramming, then epigenetic transgenerational inheritance can occur. Since the embryonic stem cells develop an altered epigenome, these epimutations subsequently induce somatic cell alterations in the epigenome and transcriptome, which will increase disease susceptibility in the offspring. Therefore, these ancestral exposures to environmental toxicants can lead to transgenerational changes in the epigenome and transcriptome of future generations and lead to an increased incidence of disease. Although DNA methylation is the most thoroughly studied epigenetic mechanism, other epigenetic processes are equally important. Future research will need to investigate the multiple epigenetic mechanisms and how they integrate. The developmental aspects of how the epigenetic transgenerational inheritance of disease develops are still unclear. How epimutations in sperm result in epigenetic changes in the resultant embryo needs to be investigated. How the derived embryonic stem cell changes can lead to epigenetic and transcriptome changes in the function of an adult organ associated with disease also remain to be elucidated on a molecular level. The potential role these ancestral exposures and epigenetic transgenerational inheritance have on disease etiology needs to be seriously considered. In addition, it may be clinically useful to determine what epimutation patterns or signatures are associated with specific disease and/or ancestral exposures in humans. Epigenetic biomarker signatures may be used in the future as a diagnostic tool to assess if an individual has a specific disease susceptibility or environmental toxicant exposures. This will facilitate preventative medicine and therapeutic approaches to mitigate associated disease risks.
Acknowledgements
We apologize to authors whose studies on the topic were not presented or referenced, but the increase in research in the area has grown significantly. We acknowledge the assistance of Dr Millissia Ben Maamar for critical review of the manuscript and Ms Heather Johnson in preparing the manuscript. This was supported by a John Templeton Foundation grant (50183) and NIH (ES012974) grant to M.K.S.
Conflict of interest statement. None declared.
References
- 1. Waddington CH. Organisers and Genes. Cambridge: Cambridge University Press, 1940. [Google Scholar]
- 2. Van Speybroeck L. From epigenesis to epigenetics: the case of C. H. Waddington. Ann N Y Acad Sci 2002;981:61–81. [PubMed] [Google Scholar]
- 3. Skinner MK. Environmental epigenetic transgenerational inheritance and somatic epigenetic mitotic stability. Epigenetics 2011;6:838–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Holliday R, Pugh JE.. DNA modification mechanisms and gene activity during development. Science 1975;187:226–32. [PubMed] [Google Scholar]
- 5. Turner BM. Histone acetylation as an epigenetic determinant of long-term transcriptional competence. Cell Mol Life Sci 1998;54:21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jodar M, Selvaraju S, Sendler E, Diamond MP, Krawetz SA.. The presence, role and clinical use of spermatozoal RNAs. Hum Reprod Update 2013;19:604–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet 2009;5:e1000459.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yaniv M. Chromatin remodeling: from transcription to cancer. Cancer Genet 2014;207:352–7. [DOI] [PubMed] [Google Scholar]
- 9. Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F.. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev 2010;24:1590–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA.. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 2005;122:517–27. [DOI] [PubMed] [Google Scholar]
- 11. Boland MJ, Nazor Kl, Loring JF.. Epigenetic regulation of pluripotency and differentiation. Circ Res 2014;115:311–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Avgustinova A, Benitah SA.. Epigenetic control of adult stem cell function. Nat Rev Mol Cell Biol 2016;17:643–58. [DOI] [PubMed] [Google Scholar]
- 13. Singer J, Roberts-Ems J, Riggs AD.. Methylation of mouse liver DNA studied by means of the restriction enzymes msp I and hpa II. Science 1979;203:1019–21. [DOI] [PubMed] [Google Scholar]
- 14. Kriaucionis S, Tahiliani M.. Expanding the epigenetic landscape: novel modifications of cytosine in genomic DNA. Cold Spring Harb Perspect Biol 2014;6:a018630.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. An J, Rao A, Ko M.. TET family dioxygenases and DNA demethylation in stem cells and cancers. Exp Mol Med 2017;49:e323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mellen M, Ayata P, Heintz N.. 5-hydroxymethylcytosine accumulation in postmitotic neurons results in functional demethylation of expressed genes. Proc Natl Acad Sci USA 2017;114:E7812–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang WW, Dietmann S, Irie N, Leitch HG, Floros VI, Bradshaw CR, Hackett JA, Chinnery PF, Surani MA.. A unique gene regulatory network resets the human germline epigenome for development. Cell 2015;161:1453–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K, Liu Y, Byrum SD, Mackintosh SG, Zhong M, Tackett A, Wang G, Hon LS, Fang G, Swenberg JA, Xiao AZ.. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature 2016;532:329–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rothbart SB, Strahl BD.. Interpreting the language of histone and DNA modifications. Biochim Biophys Acta 2014;1839:627–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kornfeld JW, Bruning JC.. Regulation of metabolism by long, non-coding RNAs. Front Genet 2014;5:57.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wei JW, Huang K, Yang C, Kang CS.. Non-coding RNAs as regulators in epigenetics (Review). Oncol Rep 2017;37:3–9. [DOI] [PubMed] [Google Scholar]
- 22. Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng GH, Peng H, Zhang X, Zhang Y, Qian J, Duan E, Zhai Q, Zhou Q.. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 2016;351:397–400. [DOI] [PubMed] [Google Scholar]
- 23. Chen Q, Yan W, Duan E.. Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nat Rev Genet 2016;17:733–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sibbritt T, Patel HR, Preiss T.. Mapping and significance of the mRNA methylome. Wiley Interdiscip Rev RNA 2013;4:397–422. [DOI] [PubMed] [Google Scholar]
- 25. Yue Y, Liu J, He C.. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev 2015;29:1343–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fu Y, Dominissini D, Rechavi G, He C.. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet 2014;15:293–306. [DOI] [PubMed] [Google Scholar]
- 27. Xu L, Liu X, Sheng N, Oo KS, Liang J, Chionh YH, Xu J, Ye F, Gao YG, Dedon PC, Fu XY.. Three distinct 3-methylcytidine (m(3)C) methyltransferases modify tRNA and mRNA in mice and humans. J Biol Chem 2017;292:14695–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lyons SM, Fay MM, Akiyama Y, Anderson PJ, Ivanov P.. RNA biology of angiogenin: current state and perspectives. RNA Biol 2017;14:171–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, Paramor M, Gleeson JG, Odom DT, Ule J, Frye M.. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep 2013;4:255–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saikia M, Hatzoglou M.. The many virtues of tRNA-derived stress-induced RNAs (tiRNAs): discovering novel mechanisms of stress response and effect on human health. J Biol Chem 2015;290:29761–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Skinner MK. What is an epigenetic transgenerational phenotype? F3 or F2. Reprod Toxicol 2008;25:2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Blewitt ME, Vickaryous NK, Paldi A, Koseki H, Whitelaw E.. Dynamic reprogramming of DNA methylation at an epigenetically sensitive allele in mice. PLoS Genet 2006;2:e49.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Waterland RA, Travisano M, Tahiliani KG.. Diet-induced hypermethylation at agouti viable yellow is not inherited transgenerationally through the female. FASEB J 2007;21:3380–5. [DOI] [PubMed] [Google Scholar]
- 34. Blewitt M, Whitelaw E.. The use of mouse models to study epigenetics. Cold Spring Harb Perspect Biol 2013;5:a017939.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Waddington CH. Canalisation of development and the inheritance of acquired characters. Nature 1942;150:563–5. [Google Scholar]
- 36. Stockard CR, Papanicolaou GN.. Further studies on the modification of the germ-cells in mammals: the effect of alcohol on treated Guinea pigs and their descendants. J Exp Zool 1918;26:119–226. [Google Scholar]
- 37. Anway MD, Cupp AS, Uzumcu M, Skinner MK.. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 2005;308:1466–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chamorro-Garcia R, Sahu M, Abbey RJ, Laude J, Pham N, Blumberg B.. Transgenerational inheritance of increased fat depot size, stem cell reprogramming, and hepatic steatosis elicited by prenatal exposure to the obesogen tributyltin in mice. Environ Health Perspect 2013;121:359–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Skinner MK. Endocrine disruptor induction of epigenetic transgenerational inheritance of disease. Mol Cell Endocrinol 2014;398:4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Quinnies KM, Doyle TJ, Kim KH, Rissman EF.. Transgenerational effects of di-(2-ethylhexyl) phthalate (DEHP) on stress hormones and behavior. Endocrinology 2015;156:3077–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rattan S, Brehm E, Gao L, Niermann S, Flaws JA.. Prenatal exposure to di(2-ethylhexyl) phthalate disrupts ovarian function in a transgenerational manner in female mice. Biol Reprod 2018;98:130–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wolstenholme JT, Edwards M, Shetty SR, Gatewood JD, Taylor JA, Rissman EF, Connelly JJ.. Gestational exposure to bisphenol A produces transgenerational changes in behaviors and gene expression. Endocrinology 2012;153:3828–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner M.. Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS One 2013;8:1–18, e55387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chamorro-Garcia R, Diaz-Castillo C, Shoucri B, Kaech M, Leavitt H, Shioda RT, Blumberg B.. Ancestral perinatal obesogen exposure results in a transgenerational thrifty phenotype in mice. Nat Commun 2017;8:2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Skinner MK, Manikkam M, Tracey R, Guerrero-Bosagna C, Haque MM, Nilsson E.. Ancestral dichlorodiphenyltrichloroethane (DDT) exposure promotes epigenetic transgenerational inheritance of obesity. BMC Med 2013;11:228, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tracey R, Manikkam M, Guerrero-Bosagna C, Skinner M.. Hydrocarbons (jet fuel JP-8) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. Reprod Toxicol 2013;36:104–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Abbott CW, Rohac DJ, Bottom RT, Patadia S, Huffman KJ.. Prenatal ethanol exposure and neocortical development: a transgenerational model of FASD. Cereb Cortex 2017;1–14, bhx168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lombo M, Fernandez-Diez C, Gonzalez-Rojo S, Navarro C, Robles V, Herraez MP.. Transgenerational inheritance of heart disorders caused by paternal bisphenol A exposure. Environ Pollut 2015;206:667–78. [DOI] [PubMed] [Google Scholar]
- 49. Carvan MJI, Kalluvila TA, Klingler RH, Larson JK, Pickens M, Mora-Zamorano FX, Connaughton VP, Sadler-Riggleman I, Beck D, Skinner MK.. Mercury-induced epigenetic transgenerational inheritance of abnormal neurobehavior is correlated with sperm epimutations in zebrafish. PLoS One 2017;12:e0176155–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Knecht AL, Truong L, Marvel SW, Reif DM, Garcia A, Lu C, Simonich MT, Teeguarden JG, Tanguay RL.. Transgenerational inheritance of neurobehavioral and physiological deficits from developmental exposure to benzo[a]pyrene in zebrafish. Toxicol Appl Pharmacol 2017;329:148–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Razoux F, Russig H, Mueggler T, Baltes C, Dikaiou K, Rudin M, Mansuy IM.. Transgenerational disruption of functional 5-HT1AR-induced connectivity in the adult mouse brain by traumatic stress in early life. Mol Psychiatry 2017;22:519–26. [DOI] [PubMed] [Google Scholar]
- 52. Franklin TB, Linder N, Russig H, Thony B, Mansuy IM.. Influence of early stress on social abilities and serotonergic functions across generations in mice. PLoS One 2011;6:e21842.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. He N, Kong QQ, Wang JZ, Ning SF, Miao YL, Yuan HJ, Gong S, Cui XZ, Li CY, Tan JH.. Parental life events cause behavioral difference among offspring: adult pre-gestational restraint stress reduces anxiety across generations. Sci Rep 2016;6:39497.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Saavedra-Rodriguez L, Feig LA.. Chronic social instability induces anxiety and defective social interactions across generations. Biol Psychiatry 2013;73:44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kiss D, Ambeskovic M, Montina T, Metz GA.. Stress transgenerationally programs metabolic pathways linked to altered mental health. Cell Mol Life Sci 2016;73:4547–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bygren LO, Tinghog P, Carstensen J, Edvinsson S, Kaati G, Pembrey ME, Sjostrom M.. Change in paternal grandmothers' early food supply influenced cardiovascular mortality of the female grandchildren. BMC Genet 2014;15:12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dunn GA, Bale TL.. Maternal high-fat diet effects on third-generation female body size via the paternal lineage. Endocrinology 2011;152:2228–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Masuyama H, Mitsui T, Nobumoto E, Hiramatsu Y.. The effects of high-fat diet exposure in utero on the obesogenic and diabetogenic traits through epigenetic changes in adiponectin and leptin gene expression for multiple generations in female mice. Endocrinology 2015;156:2482–91. [DOI] [PubMed] [Google Scholar]
- 59. Nguyen NM, de Oliveira Andrade F, Jin L, Zhang X, Macon M, Cruz MI, Benitez C, Wehrenberg B, Yin C, Wang X, Xuan J, de Assis S, Hilakivi-Clarke L.. Maternal intake of high n-6 polyunsaturated fatty acid diet during pregnancy causes transgenerational increase in mammary cancer risk in mice. Breast Cancer Res 2017;19:77.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Skinner MK, Anway MD.. Epigenetic transgenerational actions of vinclozolin on the development of disease and cancer. Crit Rev Oncog 2007;13:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pavlinkova G, Margaryan H, Zatecka E, Valaskova E, Elzeinova F, Kubatova A, Bohuslavova R, Peknicova J.. Transgenerational inheritance of susceptibility to diabetes-induced male subfertility. Sci Rep 2017;7:4940.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wei Y, Yang CR, Wei YP, Zhao ZA, Hou Y, Schatten H, Sun QY.. Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proc Natl Acad Sci USA 2014;111:1873–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. de Castro Barbosa T, Ingerslev LR, Alm PS, Versteyhe S, Massart J, Rasmussen M, Donkin I, Sjogren R, Mudry JM, Vetterli L, Gupta S, Krook A, Zierath JR, Barres R.. High-fat diet reprograms the epigenome of rat spermatozoa and transgenerationally affects metabolism of the offspring. Mol Metab 2016;5:184–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Quadrana L, Colot V.. Plant transgenerational epigenetics. Annu Rev Genet 2016;50:467–91. [DOI] [PubMed] [Google Scholar]
- 65. Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, Lim JP, Benayoun BA, Shi Y, Brunet A.. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature 2011;479:365–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Buescher JL, Musselman LP, Wilson CA, Lang T, Keleher M, Baranski TJ, Duncan JG.. Evidence for transgenerational metabolic programming in Drosophila. Dis Model Mech 2013;6:1123–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dew-Budd K, Jarnigan J, Reed LK.. Genetic and sex-specific transgenerational effects of a high fat diet in Drosophila melanogaster. PLoS One 2016;11:e0160857.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Xia B, de Belle JS.. Transgenerational programming of longevity and reproduction by post-eclosion dietary manipulation in Drosophila. Aging Aging 2016;8:1115–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Xia B, Gerstin E, Schones DE, Huang W, Steven de Belle J.. Transgenerational programming of longevity through E(z)-mediated histone H3K27 trimethylation in Drosophila. Aging (Aging 2016;8:2988–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Vandegehuchte MB, Lemiere F, Vanhaecke L, Vanden Berghe W, Janssen CR.. Direct and transgenerational impact on Daphnia magna of chemicals with a known effect on DNA methylation. Comp Biochem Physiol C Toxicol Pharmacol 2010;151:278–85. [DOI] [PubMed] [Google Scholar]
- 71. Baker TR, Peterson RE, Heideman W.. Using zebrafish as a model system for studying the transgenerational effects of dioxin. Toxicol Sci 2014;138:403–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bhandari RK, vom Saal FS, Tillitt DE.. Transgenerational effects from early developmental exposures to bisphenol A or 17alpha-ethinylestradiol in medaka, Oryzias latipes. Sci Rep 2015;5:9303.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Leroux S, Gourichon D, Leterrier C, Labrune Y, Coustham V, Riviere S, Zerjal T, Coville JL, Morisson M, Minvielle F, Pitel F.. Embryonic environment and transgenerational effects in quail. Genet Sel Evol 2017;49:14.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Brun JM, Bernadet MD, Cornuez A, Leroux S, Bodin L, Basso B, Davail S, Jaglin M, Lessire M, Martin X, Sellier N, Morisson M, Pitel F.. Influence of grand-mother diet on offspring performances through the male line in Muscovy duck. BMC Genet 2015;16:145.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Skinner MK, Manikkam M, Guerrero-Bosagna C.. Epigenetic transgenerational actions of endocrine disruptors. Reprod Toxicol 2011;31:337–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Braunschweig M, Jagannathan V, Gutzwiller A, Bee G.. Investigations on transgenerational epigenetic response down the male line in F2 pigs. PLoS One 2012;7:e30583.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Veenendaal MV, Painter RC, de Rooij SR, Bossuyt PM, van der Post JA, Gluckman PD, Hanson MA, Roseboom TJ.. Transgenerational effects of prenatal exposure to the 1944-45 Dutch famine. BJOG 2013;120:548–53. [DOI] [PubMed] [Google Scholar]
- 78. Anway MD, Leathers C, Skinner MK.. Endocrine disruptor vinclozolin induced epigenetic transgenerational adult-onset disease. Endocrinology 2006;147:5515–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Anway MD, Skinner MK.. Transgenerational effects of the endocrine disruptor vinclozolin on the prostate transcriptome and adult onset disease. Prostate 2008;68:517–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner MK.. Dioxin (TCDD) induces epigenetic transgenerational inheritance of adult onset disease and sperm epimutations. PLoS One 2012;7:e46249–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner M.. Pesticide and insect repellent mixture (Permethrin and DEET) induces epigenetic transgenerational inheritance of disease and sperm epimutations. Reprod Toxicol 2012;34:708–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Manikkam M, Guerrero-Bosagna C, Tracey R, Haque MM, Skinner MK.. Transgenerational actions of environmental compounds on reproductive disease and identification of epigenetic biomarkers of ancestral exposures. PLoS One 2012;7:1–12, e31901.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Nilsson EE, Anway MD, Stanfield J, Skinner MK.. Transgenerational epigenetic effects of the endocrine disruptor vinclozolin on pregnancies and female adult onset disease. Reproduction 2008;135:713–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Nilsson EE, Skinner MK.. Environmentally induced epigenetic transgenerational inheritance of reproductive disease. Biol Reprod 2015;93:145.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Skinner MK, Anway MD, Savenkova MI, Gore AC, Crews D.. Transgenerational epigenetic programming of the brain transcriptome and anxiety behavior. PLoS One 2008;3:1–11, e3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Manikkam M, Haque MM, Guerrero-Bosagna C, Nilsson E, Skinner MK.. Pesticide methoxychlor promotes the epigenetic transgenerational inheritance of adult onset disease through the female germline. PLoS One 2014;9:1–19, e102091.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Nilsson E, Larsen G, Manikkam M, Guerrero-Bosagna C, Savenkova M, Skinner M.. Environmentally induced epigenetic transgenerational inheritance of ovarian disease. PLoS One 2012;7:1–18, e36129.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bateson P, Gluckman P, Hanson M.. The biology of developmental plasticity and the predictive adaptive response hypothesis. J Physiol 2014;592:2357–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Skinner MK. Environmental epigenetics and a unified theory of the molecular aspects of evolution: a neo-lamarckian concept that facilitates neo-Darwinian evolution. Genome Biol Evol 2015;7:1296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hackett JA, Surani MA.. Beyond DNA: programming and inheritance of parental methylomes. Cell 2013;153:737–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A.. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 2012;484:339–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Constancia M, Pickard B, Kelsey G, Reik W.. Imprinting mechanisms. Genome Res 1998;8:881–900. [DOI] [PubMed] [Google Scholar]
- 93. Skinner MK, Manikkam M, Guerrero-Bosagna C.. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol Metab 2010;21:214–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Jirtle RL, Skinner MK.. Environmental epigenomics and disease susceptibility. Nat Rev Genet 2007;8:253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Guerrero-Bosagna C, Savenkova M, Haque MM, Nilsson E, Skinner MK.. Environmentally induced epigenetic transgenerational inheritance of altered sertoli cell transcriptome and epigenome: molecular etiology of male infertility. PLoS One 2013;8:1–12, e59922.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Skinner MK, Manikkam M, Haque MM, Zhang B, Savenkova M.. Epigenetic transgenerational inheritance of somatic transcriptomes and epigenetic control regions. Genome Biol 2012;13:R91.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Guerrero-Bosagna C, Settles M, Lucker B, Skinner M.. Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS One 2010;5:1–17, e13100.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Skinner MK, Guerrero-Bosagna C.. Role of CpG deserts in the epigenetic transgenerational inheritance of differential DNA methylation regions. BMC Genomics 2014;15:692.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Haque MM, Holder LB, Skinner MK.. Genome-wide locations of potential epimutations associated with environmentally induced epigenetic transgenerational inheritance of disease using a sequential machine learning prediction approach. PLoS One 2015;10:1–25, e0142274.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Beck D, Sadler-Riggleman I, Skinner MK.. Generational comparisons (F1 versus F3) of vinclozolin induced epigenetic transgenerational inheritance of sperm differential DNA methylation regions (epimutations) using MeDIP-Seq. Environ Epigenetics 2017;3:1–12, dvx016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Rodgers AB, Morgan CP, Leu NA, Bale TL.. Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proc Natl Acad Sci USA 2015;112:13699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Schuster A, Skinner MK, Yan W.. Ancestral vinclozolin exposure alters the epigenetic transgenerational inheritance of sperm small noncoding RNAs. Environ Epigenet 2016;2:1–10, dvw001.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Gapp K, Jawaid A, Sarkies P, Bohacek J, Pelczar P, Prados J, Farinelli L, Miska E, Mansuy IM.. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci 2014;17:667–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Skinner MK, Ben Maamar M, Sadler-Riggleman I, Beck D, Nilsson E, McBirney M, Klukovich R, Xie Y, Tang C, Yan W.. Alterations in sperm DNA methylation, non-coding RNA and histone retention associate with DDT-induced epigenetic transgenerational inheritance of disease. Epigenet Chromatin 2018;4:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Bao J, Bedford MT.. Epigenetic regulation of the histone-to-protamine transition during spermiogenesis. Reproduction 2016;151:R55–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Rathke C, Baarends WM, Awe S, Renkawitz-Pohl R.. Chromatin dynamics during spermiogenesis. Biochim Biophys Acta 2014;1839:155–68. [DOI] [PubMed] [Google Scholar]
- 107. Ihara M, Meyer-Ficca ML, Leu NA, Rao S, Li F, Gregory BD, Zalenskaya IA, Schultz RM, Meyer RG.. Paternal poly (ADP-ribose) metabolism modulates retention of inheritable sperm histones and early embryonic gene expression. PLoS Genet 2014;10:e1004317.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Ben Maamar M, Sadler-Riggleman I, Beck D, Skinner MK.. Epigenetic transgenerational inheritance of altered sperm histone retention sites. Sci Rep 2018;8:5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Hammoud SS, Nix DA, Hammoud AO, Gibson M, Cairns BR, Carrell DT.. Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men. Hum Reprod 2011;26:2558–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Siklenka K, Erkek S, Godmann M, Lambrot R, McGraw S, Lafleur C, Cohen T, Xia J, Suderman M, Hallett M, Trasler J, Peters AH, Kimmins S.. Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science 2015;350: 1–14, aab2006. [DOI] [PubMed] [Google Scholar]
- 111. Kelly WG. Transgenerational epigenetics in the germline cycle of Caenorhabditis elegans. Epigenetics Chromatin 2014;7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Ruden DM, Lu X.. Hsp90 affecting chromatin remodeling might explain transgenerational epigenetic inheritance in Drosophila. Cg 2008;9:500–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Ben Maamar M, Sadler-Riggleman I, Beck D, McBirney M, Nilsson E, Klukovich R, Xie Y, Tang C, Yan W, Skinner MK.. Alterations in sperm DNA Methylation, Non-Coding RNA expression, and histone retention mediate Vinclozolin induced epigenetic transgenerational inheritance of disease. Environmental Epigenetics 2018;4:1–19, dvy010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Kung JT, Colognori D, Lee JT.. Long noncoding RNAs: past, present, and future. Genetics 2013;193:651–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Saha P, Verma S, Pathak RU, Mishra RK.. Long noncoding RNAs in Mammalian development and diseases. Adv Exp Med Biol 2017;1008:155–98. [DOI] [PubMed] [Google Scholar]
- 116. Liu KS, Li TP, Ton H, Mao XD, Chen YJ.. Advances of long noncoding RNAs-mediated regulation in reproduction. Chin Med J 2018;131:226–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Haque MM, Nilsson EE, Holder LB, Skinner MK.. Genomic clustering of differential DNA methylated regions (epimutations) associated with the epigenetic transgenerational inheritance of disease and phenotypic variation. BMC Genomics 2016;17:418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Pastuszak AW, Lamb DJ.. The genetics of male fertility—from basic science to clinical evaluation. J Androl 2012;33:1075–84. [DOI] [PubMed] [Google Scholar]
- 119. Matzuk MM, Lamb DJ.. The biology of infertility: research advances and clinical challenges. Nat Med 2008;14:1197–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Skinner MK. Differential DNA methylation analysis optimally requires purified cell populations. Fertil Steril 2016;106:551.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Hanson MA, Skinner MK.. Developmental origins of epigenetic transgenerational inheritance. Environ Epigenet 2016;2:1–9, dvw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Skinner MK. A new kind of inheritance. Sci Am 2014;311:44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Bruner-Tran KL, Osteen KG.. Developmental exposure to TCDD reduces fertility and negatively affects pregnancy outcomes across multiple generations. Reprod Toxicol 2011;31:344–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Doyle TJ, Bowman JL, Windell VL, McLean DJ, Kim KH.. Transgenerational effects of di-(2-ethylhexyl) phthalate on testicular germ cell associations and spermatogonial stem cells in mice. Biol Reprod 2013;88:112.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. McBirney M, King SE, Michelle Pappalardo M, Houser E, Unkefer M, Nilsson E, Sadler-Riggleman I, Beck D, Winchester P, Skinner MK.. Atrazine induced epigenetic transgenerational inheritance of disease, lean phenotype and sperm epimutation pathology biomarkers. PLoS One 2017;12:e0184306–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Padmanabhan N, Jia D, Geary-Joo C, Wu X, Ferguson-Smith AC, Fung E, Bieda MC, Snyder FF, Gravel RA, Cross JC, Watson ED.. Mutation in folate metabolism causes epigenetic instability and transgenerational effects on development. Cell 2013;155:81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Zheng X, Chen L, Li M, Lou Q, Xia H, Wang P, Li T, Liu H, Luo L.. Transgenerational variations in DNA methylation induced by drought stress in two rice varieties with distinguished difference to drought resistance. PLoS One 2013;8:e80253.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Suter L, Widmer A.. Environmental heat and salt stress induce transgenerational phenotypic changes in Arabidopsis thaliana. PLoS One 2013;8:e60364.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Rehan VK, Liu J, Sakurai R, Torday JS.. Perinatal nicotine-induced transgenerational asthma. Am J Physiol Lung Cell Mol Physiol 2013;305:L501–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Govorko D, Bekdash RA, Zhang C, Sarkar DK.. Male germline transmits fetal alcohol adverse effect on hypothalamic proopiomelanocortin gene across generations. Biol Psychiatry 2012;72:378–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Norouzitallab P, Baruah K, Vandegehuchte M, Van Stappen G, Catania F, Vanden Bussche J, Vanhaecke L, Sorgeloos P, Bossier P.. Environmental heat stress induces epigenetic transgenerational inheritance of robustness in parthenogenetic Artemia model. FASEB J 2014;28:3552–63. [DOI] [PubMed] [Google Scholar]
- 132. Waddington CH. Gene assimilation of an acquired character. Evolution 1953;7:118–26. [Google Scholar]