A marked increased in linezolid resistance among invasive Staphylococcus epidermidis was due to dissemination of a cfr-containing clone of S. epidermidis that dominated the gastrointestinal microbiota of patients following receipt of linezolid.
Keywords: Staphylococcus epidermidis, whole-genome sequencing, linezolid resistance, cfr, microbiome
Abstract
Background
Pathobionts, bacteria that are typically human commensals but can cause disease, contribute significantly to antimicrobial resistance. Staphylococcus epidermidis is a prototypical pathobiont as it is a ubiquitous human commensal but also a leading cause of healthcare-associated bacteremia. We sought to determine the etiology of a recent increase in invasive S. epidermidis isolates resistant to linezolid.
Methods
Whole-genome sequencing (WGS) was performed on 176 S. epidermidis bloodstream isolates collected at the MD Anderson Cancer Center in Houston, Texas, between 2013 and 2016. Molecular relationships were assessed via complementary phylogenomic approaches. Abundance of the linezolid resistance determinant cfr was determined in stool samples via reverse-transcription quantitative polymerase chain reaction.
Results
Thirty-nine of the 176 strains were linezolid resistant (22%). Thirty-one of the 39 linezolid-resistant S. epidermidis infections were caused by a particular clone resistant to multiple antimicrobials that spread among leukemia patients and carried cfr on a 49-kb plasmid (herein called pMB151a). The 6 kb of pMB151a surrounding the cfr gene was nearly 100% identical to a cfr-containing plasmid isolated from livestock-associated staphylococci in China. Analysis of serial stool samples from leukemia patients revealed progressive staphylococcal domination of the intestinal microflora and an increase in cfr abundance following linezolid use.
Conclusions
The combination of linezolid use plus transmission of a multidrug-resistant clone drove expansion of invasive, linezolid-resistant S. epidermidis. Our results lend support to the notion that a combination of antibiotic stewardship plus infection control measures may help to control the spread of a multidrug-resistant pathobiont.
Mitigating the impact of antimicrobial-resistant microbes is a global public health priority [1]. It is increasingly appreciated that commensal microbiota play a key role in antimicrobial resistance (AMR), both by providing protection against colonization by antibiotic-resistant pathogens and by serving as a reservoir for AMR genes [2]. Moreover, organisms that are typically commensal can both acquire AMR and be pathogenic, particularly in the healthcare setting [3]. Although such organisms, termed pathogenic symbionts or pathobionts, are significant contributors to AMR, there is a dearth of understanding of the mechanisms underlying antimicrobial-resistant pathobiont emergence and dissemination [4].
Staphylococcus epidermidis is a prototypical pathobiont as it is a ubiquitous commensal of humans and a major cause of healthcare-associated infections [5, 6]. Moreover, S. epidermidis can acquire or develop resistance to numerous classes of antimicrobials and transfer such elements to its more pathogenic relative Staphylococcus aureus [7]. A major concern of AMR in S. epidermidis is resistance to linezolid, an oxazolidinone antibiotic that targets the ribosomal peptidyl transferase center and is widely used to treat staphylococcal infections [8–11]. Linezolid resistance is typically mediated by ribosomal mutations or acquisition of the 23S ribosomal RNA (rRNA) methyltransferase protein Cfr [12]. Mutations in ribosomal proteins are thought to arise in strains adapting to linezolid whereas the cfr gene can be transferred via mobilizable elements and plasmids [13]. Linezolid-resistant (LR) S. epidermidis is increasingly reported worldwide and often associated with the presence of cfr on transferable elements in isolates recovered from humans and animals [8–11, 13, 14].
Beginning in the late 2000s, rare cases of LR S. epidermidis were identified in our institution (a major cancer hospital in the United States). Via a targeted genetic approach, it was found that these strains were multilocus sequence type 2 (ST2), harbored the 23S rRNA G2576T mutation, and lacked cfr [14]. To better design measures to combat LR invasive S. epidermidis infections, we launched a whole-genome-based initiative in combination with patient specific antimicrobial use data to test the hypothesis that linezolid resistance emerged in a broad number of S. epidermidis clones adapting to linezolid exposure.
MATERIALS AND METHODS
Complete methodologies are presented in the Supplementary Materials.
Specimen Collection
A waiver of informed consent to collect clinical data and analyze serial invasive S. epidermidis strains isolated between 2013 and 2015 was provided by the MD Anderson Cancer Center Institutional Review Board (number PA16-0066). Linezolid susceptibility was determined initially by Vitek and then confirmed by Etest if the Vitek result showed a minimum inhibitory concentration (MIC) ≥4 mg/L.
Genome Assembly
Paired-end (PE) whole-genome sequencing (WGS) was performed on the Illumina MiSeq instrument. Multilocus sequence typing (MLST) was performed in silico, and clonal complexes were assigned via Eburst. The complete genome of strain MB151 was assembled using a combination of long-read PacBio and PE short-read data.
Phylogenomic Computations
Three distinct core single-nucleotide polymorphism (SNP)–based methods were used to reconstruct WGS-based phylogenies: (1) kSNP version 3.0, (2) Harvest Suite version 1.0, and (3) an in-house-developed GATK inspired pipeline. In brief, trimmed PE reads were mapped to the MB151 reference genome with Bowtie2 version 2.2.3. Maximum likelihood–based phylogenetic reconstruction was performed with RAxML version 8.2.10 using the generalised time reversible (GTR) + Γ nucleotide substitution model [15]. One hundred bootstrap replicates were evaluated to determined branch support.
Characterization of cfr Location in Strain MB151, Plasmid Transfer, and Plasmid Presence Characterization
S1 nuclease assays were used to detect and estimate the size of cfr-carrying large bacterial plasmids in S. epidermidis isolates as described [16]. Conjugative transfer of cfr was performed by filter mating [17] using S. epidermidis MB151 as donor and S. aureus RN4220RF as recipient.
Linezolid Use Data
Our in-house pharmacy informatics database was queried to identify all administrations of linezolid and daptomycin. Cumulative exposure (defined as the number of unique days on which a drug was administered) and any use (defined as at least 1 dose) was considered for each drug at 3 time points: 30, 60, and 90 days preceding the first isolation of S. epidermidis.
Linezolid Resistance Mechanisms
The presence or absence of cfr was determined by a local Blastn search. Additionally, mutations in the 23S rRNA, L3, L4, and L22 proteins were also catalogued.
Microbiome Data Analysis
The origin of stool samples for 16S rRNA–based microbiome analyses from leukemia patients has been previously described, with all patients providing written informed consent [18]. Staphylococcal emergence in the gastrointestinal microbiome was defined as patients having at least 2 consecutive stool samples in which ≥30% of the 16S rRNA reads mapped to the staphylococcal genus [19] and in which the baseline stool sample had ≤10% of reads mapping to Staphylococcus. Cfr abundance in stool samples was measured by SYBR Green reverse-transcription quantitative polymerase chain reaction (PCR).
Statistical Analyses
A change in the rate of linezolid resistance over time was assessed using the Cochran-Armitage test for trend. Individual use of daptomycin and linezolid was analyzed using Fisher exact test and cumulative exposure was assessed using the Wilcoxon rank-sum test. A P value ≤.05 was considered statistically significant. All analyses were performed using Stata version 13.1 software.
RESULTS
Emergence of Linezolid Resistance Among S. epidermidis and Use of Linezolid
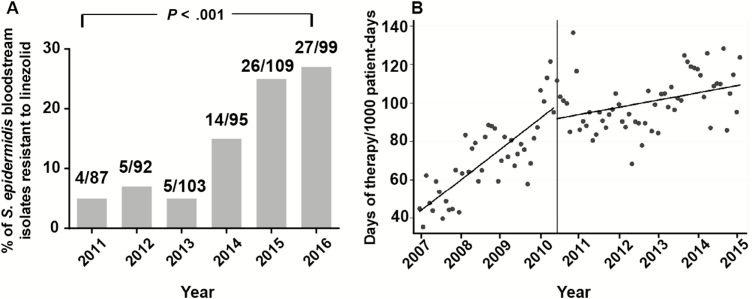
Our clinical microbiology laboratory began routine species designation of coagulase-negative staphylococci in 2011. Thus, we analyzed the rate of linezolid resistance among S. epidermidis bloodstream isolates starting in 2011 and found a statistically significant increase beginning in 2014 and continuing through 2016 (Figure 1A). Given that linezolid use has previously been shown to correlate with linezolid resistance in S. epidermidis [14], we determined the total amount of linezolid use at our hospital starting in 2007. There was a significant increase in linezolid use over time, particularly between 2007 and 2010 (Figure 1B).
Figure 1.
Linezolid resistance and linezolid use over time. A, Percentage of Staphylococcus epidermidis bloodstream isolates resistant to linezolid, stratified by year of isolation. 2011 was the first year at our institution where coagulase-negative staphylococci were routinely identified to the species level. Inset numbers refer to number of resistant isolates/total isolates. P value refers to χ2 test for trend. B, Monthly consumption of linezolid, 2007–2015. Curves correspond to predicted values based on the segmented regression analysis before and after June 2010. The difference in slope, as determined by a linear combination of regression parameter estimates, is –1.0 (95% confidence interval, –1.4 to –.6; P < .0001).
Linezolid Resistance Was Primarily Found in ST5 Isolates
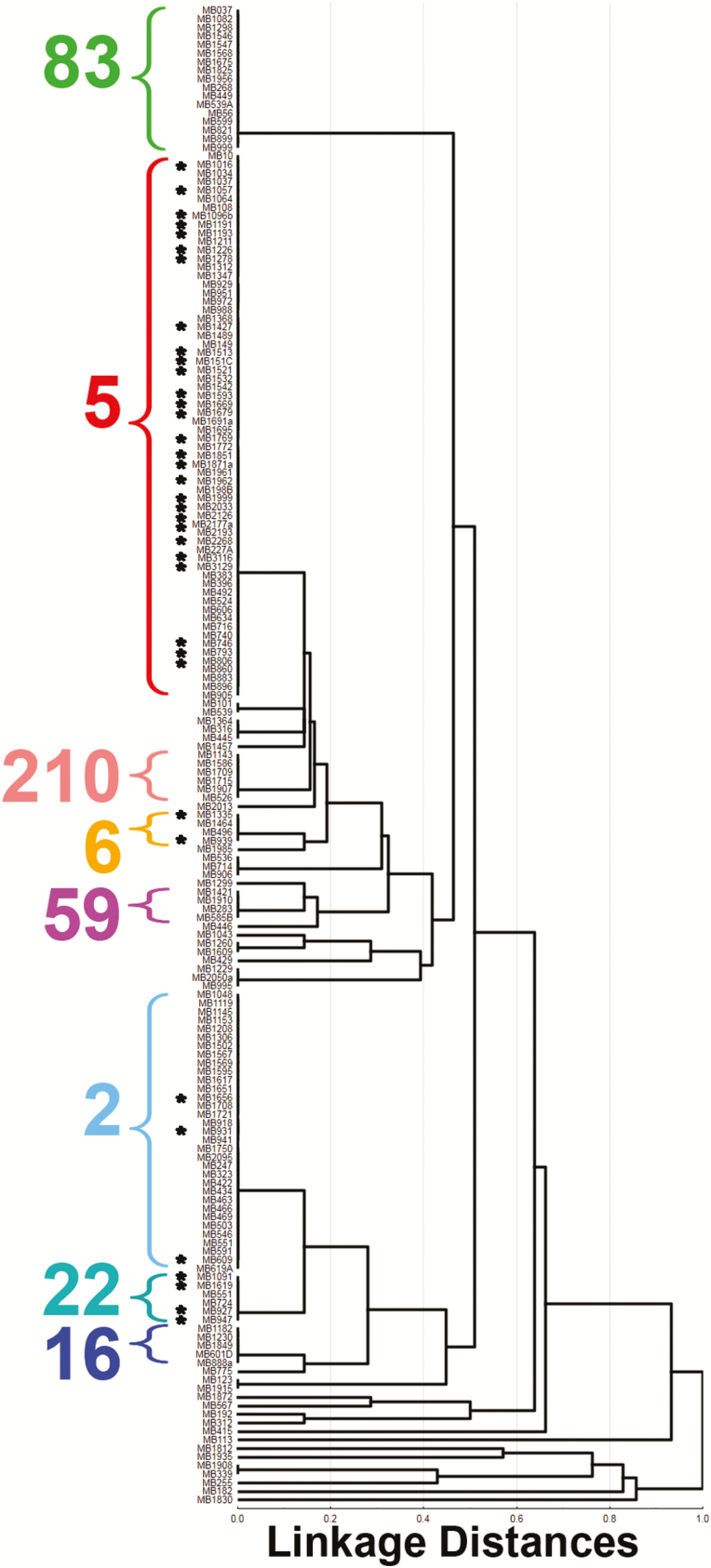
To determine whether linezolid resistance was restricted to a specific S. epidermidis subtype, we determined the MLST of our 176 invasive isolates. Consistent with published data [20], 3 major clonal complexes (CCs) accounted for the majority of infections (CC2, CC5, and CC83) (Supplementary Figure 1 and Supplementary Table 1). However, CC5 strains made up nearly 50% of our isolates, an unusually high percentage compared with other reports [20, 21]. Linezolid resistance was identified in 39 strains (22%) with the vast majority of resistance occurring in ST5 isolates, with the remainder present in ST2, ST6, and ST22 strains (Figure 2). The 90% MIC for the LR isolates was ≥256 mg/L, consistent with high-level linezolid resistance.
Figure 2.
Multilocus sequence typing dendrogram of Staphylococcus epidermidis isolates included in the study. Cluster analysis was performed using the unweighted pair-group method with arithmetic averages and the percentage disagreement distance measure (Statistica version 13; StatSoft, Tulsa, Oklahoma). Linezolid-resistant isolates are shown with black asterisks. Major sequence types are as indicated.
Whole-Genome Characterization of a cfr-Containing LR ST5 S. epidermidis
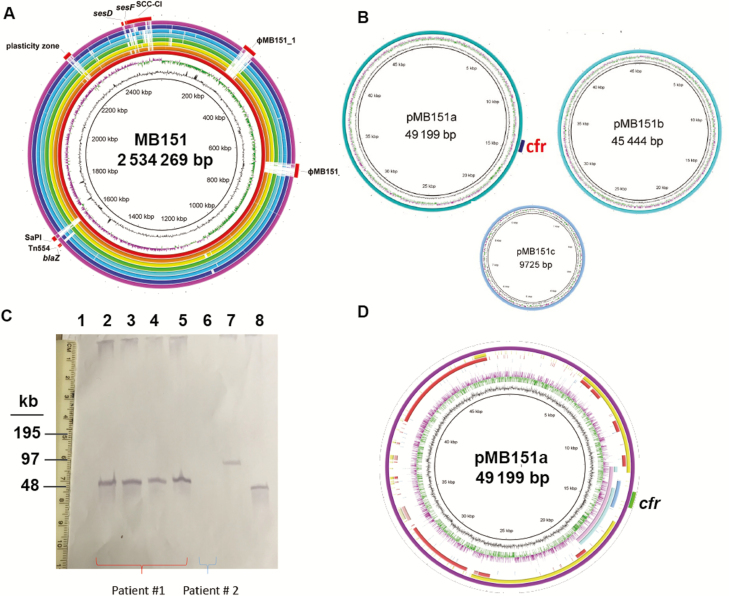
Because the MLST data showed both linezolid-susceptible and -resistant ST5 strains, we could not distinguish between adaptation of previously susceptible strains to linezolid vs spread of a LR clone. Thus, to determine the evolutionary and genomic mechanisms that contributed to the proliferation of LR ST5 S. epidermidis, we determined the complete genome of strain MB151, which was the first ST5 LR strain from our collection. The MB151 genome is comprised of an approximately 2.5-mb circular chromosome along with 3 plasmids (Figure 3A and 3B). MB151 differs from other complete S. epidermidis genomes primarily in its phage content and in the region of the staphylococcal chromosomal cassette (Figure 3A). We identified cfr on a 49-kb plasmid, pMB151a, which was confirmed by S1 nuclease digestion followed by hybridization (Figure 3C; Supplementary Figure 2). pMB151a showed significant sequence similarity to various plasmids previously reported from staphylococci including near 100% identity over 6000 bp in the cfr-containing region with plasmid pSS-01 from Staphylococcus cohnii isolated from swine in China (Figure 3D) [13]. Additional antibiotic resistance genes present in pMB151a included the bifunctional aaC(6ʹ)-Ie-aph(2ʹʹ)-Ia gene mediating aminoglycoside resistance and dfrC, which encodes for resistance to trimethoprim-sulfamethoxazole (Supplementary Figure 3).
Figure 3.
Characterization of MB151 and localization of cfr to pMB151a. A, Genome atlas for the reference sequence type (ST) 5 linezolid resistance strain MB151. Genome scale in megabases (Mbp) is given in the innermost circle (circle 1). Guanine-cytosine (GC) content is displayed in circle 2 with values above (outward directed) or below (inward directed) average indicated. Circle 3 shows GC skew, calculated as (G – C) / (G + C) and averaged over a moving window of 10000 bp showing excess G (green) and C (purple). Rings depict BLASTN comparisons of MB151a and publicly available completed Staphylococcus epidermidis genomes. The innermost ring shows the genome scale (Mbp), and subsequent rings (innermost to outermost) show BLASTN comparisons in order of decreasing homology for ATCC12228 (orange), PR62A (yellow), PM221 (green), SEI (light blue), 14.1.R1 (turquoise), 1457 (dark blue), and BPH0662 (pink). Reference genome landmarks for MB151 are labeled including the staphylococcal chromosomal cassette composite island (SCC-CI). B, Three plasmids identified by sequencing including one with cfr. C, S1 nuclease assay followed by cfr hybridization with cfr band identified at ~49 kb as predicted by sequencing data. Lanes 2–5 are serial isolates of MB151 collected during prolonged bacteremia. Lane 6 is an ST5 S. epidermidis strain that lacked cfr by sequencing data. Lanes 7 and 8 are strains harboring cfr in plasmids. Lane 7 is Enterococcus faecalis 603-50427X [29] and lane 8 is Staphylococcus aureus 004-737X [30]. D, Atlas for cfr-containing plasmid pMB151a. Innermost circles are as detailed for panel (A). Areas of significant homology between pMB151a are identified for the following plasmids: Staphylococcus cohnii pSS-01 (pink), S. epidermidis 426-3147L (light blue), Staphylococcus scuri pSCFS1 (dark blue), S. epidermidis SAP108A (red), and S. aureus pMI (yellow). Green indicates location of cfr.
Evolution of Clonal LR S. epidermidis Revealed Through WGS
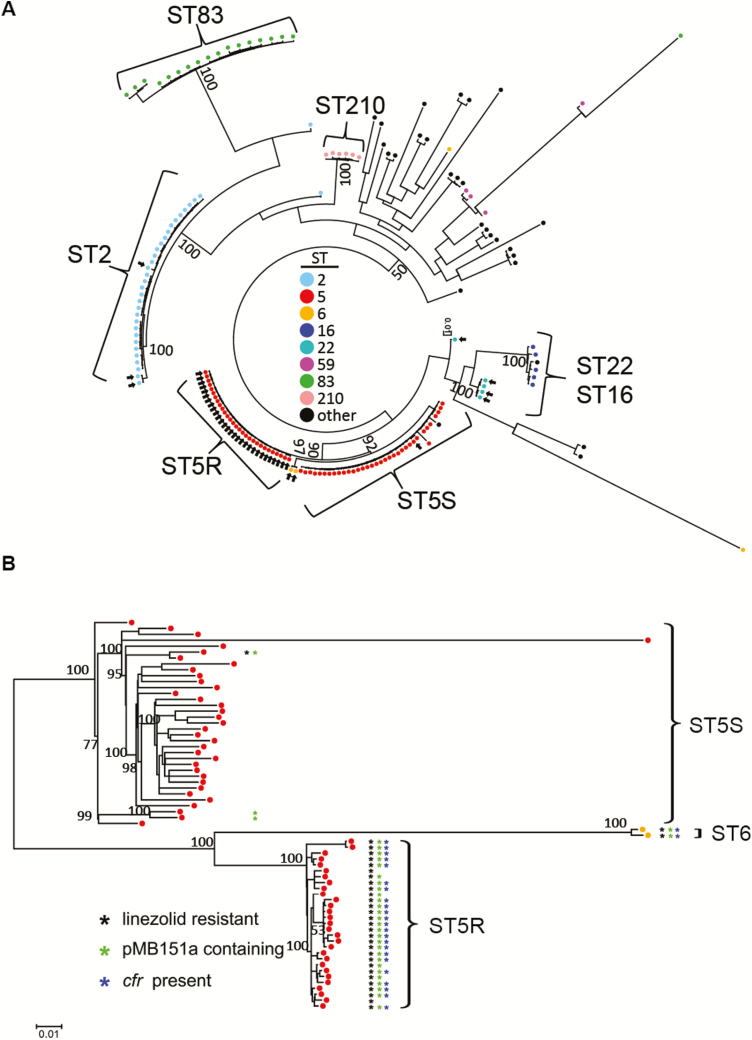
Using strain MB151 as a reference, we constructed a whole-genome phylogeny of all 176 isolates, which clearly revealed a dominant, LR subclade of 29 ST5 strains, which we have called ST5R for resistant (Figure 4A). Strains within the ST5R lineage were separated from each other by an average of <2 SNPs. Two LR ST6 strains also clustered with the ST5R isolates (Figure 4B), as ST6 varies from ST5 by only a single-nucleotide variant in the gtr gene. Linezolid-susceptible ST5 strains, which we have termed ST5S for susceptible, also clustered tightly and were clearly separated from the ST5R strains using multiple methodologies (Figure 4B; Supplementary Figures 4 and 5). Finally, we detected ST2, ST5, and ST22 strains, which were genetically most closely related to a linezolid-susceptible strain and distinct from the ST5R strains, suggesting that these strains had evolved from genetically diverse parental strains (see isolated arrows in Figure 4A). The ST5R isolates exhibited multidrug resistance including to methicillin, levofloxacin, trimethoprim-sulfamethoxazole, and gentamicin (Supplementary Figure 6). Thus, the vast majority of LR S. epidermidis was due to a clone that had likely been transferred to patients on multiple occasions rather than independently arising from linezolid-susceptible isolates.
Figure 4.
Linezolid-resistant (LR) Staphylococcus epidermidis strains form a distinct sequence type (ST) 5 lineage, here named ST5R. A, Whole-genome phylogeny of 166 strains. Maximum-likelihood (ML) tree was reconstructed on an alignment of 56124 core single-nucleotide polymorphisms (SNPs). Colors show major STs as indicated. Arrows show LR strains. B, Whole-genome phylogeny of ST5 strains. ML tree was reconstructed on an alignment of 4565 core SNPs. ST5 strains separated into LR (ST5R) and linezolid-susceptible clusters (ST5S). Two ST6 isolates that clustered with the ST5R isolates are also shown. The presence of linezolid resistance plasmid pMB151a (green asterisk) and cfr (blue asterisk) is indicated among LR (ST5R) strains (black asterisk). ML trees were inferred with the generalised time reversible model with gamma shape parameter (ɑ = 1000). Bootstrap support values of major clades are shown above the branches. Scale indicates the number of nucleotide substitutions per site.
Nontransferrable cfr-Containing Plasmid Is Limited to ST5R Strains
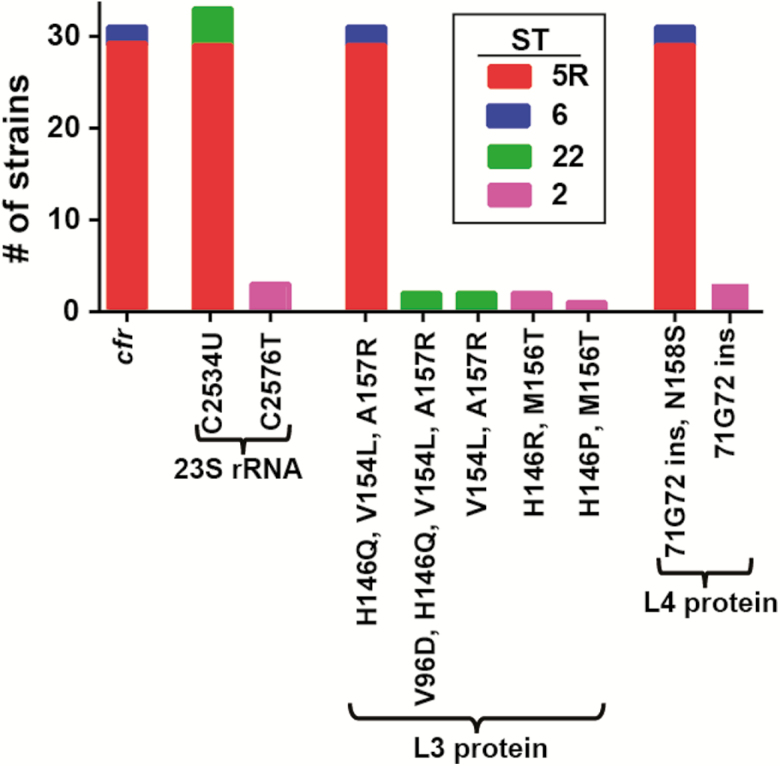
All LR isolates contained mutations in the 23S rRNA and the L3 and L4 proteins, with distinct mutations identified among the various STs (Figure 5). The cfr gene was only present in the ST5R strains and the closely related ST6 isolates. Additionally, cfr was only identified in strains that also contained plasmid pMB151a, but ST5R strains with pMB151a did not always contain cfr (Figure 4B). Analysis of pMB151a showed that the area surrounding the cfr gene, including IS256 elements, could be absent, indicating that the cfr gene can excise from plasmid pMB151a (Supplementary Figure 3). Of interest, in vitro transfer of the cfr-containing plasmid to S. aureus RN4220RF was unsuccessful (data not shown). Consistent with this result, the full conjugative type IV secretion system (T4SS) machine that includes proteins ArtA and TraABCDEFGHIJKLM [22] was absent in pMB151a or in the other 2 plasmids present in MB151. Only 3 proteins were detected, ArtA (Tra regulator), TraA (relaxase), and TraB (surface factor adhesion family protein). Insertion sequences flank these open reading frames, suggesting a significant amount of gene rearrangement that could have altered the transfer ability of pMB151a (Supplementary Figure 3). These results suggest that cfr dissemination by horizontal transfer was unlikely and is consistent with the limited presence of cfr in the highly clonal ST5R and ST6 isolates.
Figure 5.
Linezolid resistance mechanisms identified in our cohort. Shown are genetic changes previously associated with linezolid resistance that were identified in the linezolid-resistant isolates. Colors refer to sequence type (ST) as indicated in the inset legend. Note that all ST5R strains have the same linezolid resistance mechanisms consistent with their clonal nature. Abbreviations: rRNA, ribosomal RNA; ST, sequence type.
Clinical Correlation Between Linezolid Use and Infection by ST5R Strains
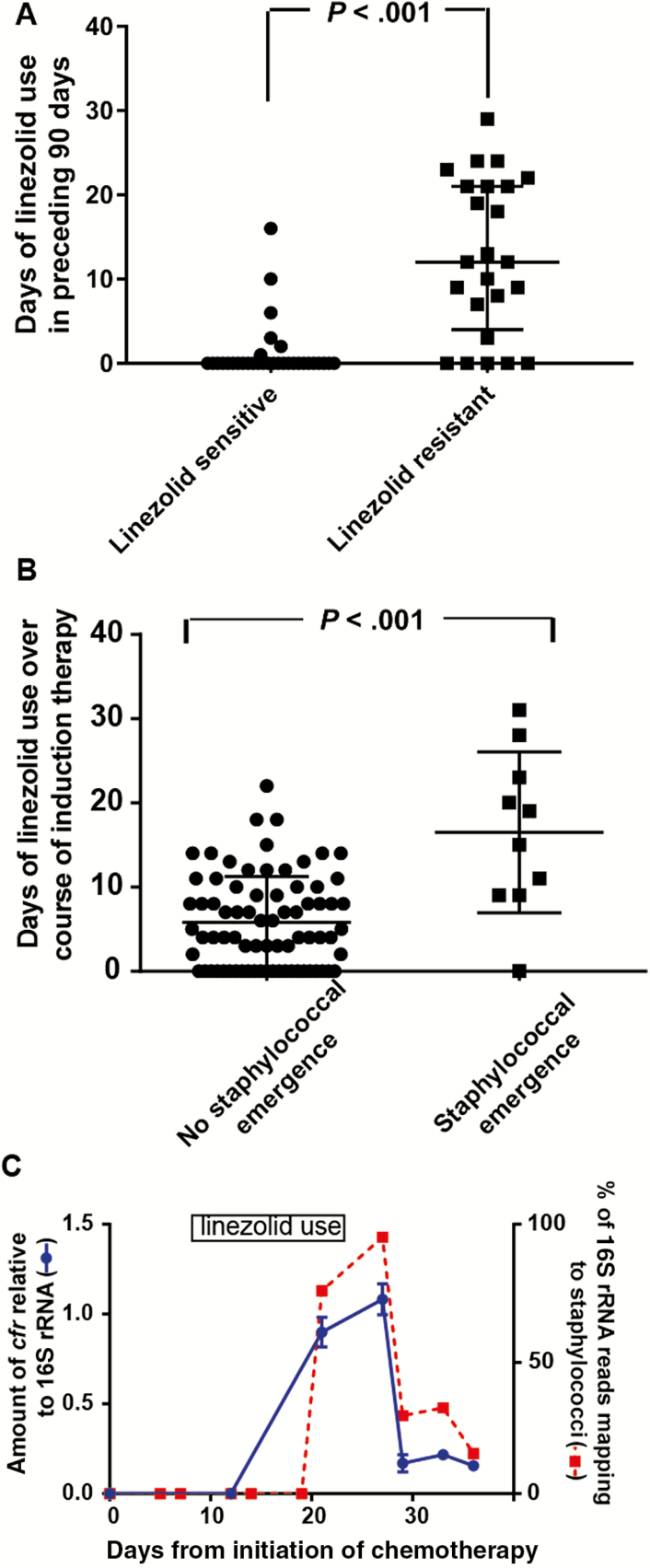
The near-genetic identity of the ST5R strains suggested that coming into contact with the strain, rather than linezolid exposure, might be the main contributing factor to infection with LR S. epidermidis. Indeed, all but 2 of the patients infected by an ST5R isolate had leukemia, which is treated in a distinct area of our hospital. To examine the potential role of linezolid use, we compared the amount of linezolid received at 30, 60, and 90 days preceding infection. We limited our analysis to ST5 strains as nearly all of the patients infected by these strains had a hematologic malignancy and thus would be expected to have similar clinical characteristics. Among the 56 ST5 isolates, 79% of patients with a LR isolate received linezolid within the preceding 90 days, in comparison to 19% of those with linezolid-susceptible isolates (P < .001). Similarly, the cumulative exposure to linezolid in the preceding 90 days was significantly higher among patients with LR isolates (median, 12 days) compared with those with sensitive isolates (median, 0 days; P < .001; Figure 6A). No significant difference in categorical or cumulative daptomycin exposure was observed (data not shown). Similar findings were observed at 30 and 60 days for both antimicrobials (data not shown). Together with our whole-genome analyses, these data suggest that both exposure to an ST5R strain and prior linezolid use drive infection by ST5R isolates rather than adaption of a previously colonizing S. epidermidis strain to linezolid.
Figure 6.
Linezolid use and emergence of staphylococci in the gastrointestinal (GI) microbiome. A, Comparison of linezolid use in the 90 days prior to infection onset for patients with sequence type 5 S. epidermidis stratified by linezolid resistance. P value refers to Mann-Whitney U test. B, Comparison of duration of linezolid use during induction remission chemotherapy for 98 patients with acute myelogenous leukemia stratified by emergence of staphylococci in serial stool samples. P value refers to Mann-Whitney U test. C, Example of patient who had emergence of staphylococci in their GI microbiome. The x-axis shows day of sampling relative to start of chemotherapy. Right y-axis and red squares with dotted red line show percentage of 16S ribosomal RNA (rRNA) reads mapping to staphylococcal genus from the stool samples. Left y-axis and squares with solid blue line depict amount of cfr present in stool samples relative to total 16S rRNA as determined by reverse-transcription quantitative polymerase chain reaction, with data graphed being mean ± standard deviation of samples analyzed in triplicate. The time of linezolid use is shown in the rectangle.
Identification of cfr Expansion in the Microbiome of Leukemia Patients Treated With Linezolid
Our data to this point led us to hypothesize that linezolid exposure in patients with leukemia could result in a proliferation of staphylococci in the commensal microbiota that might ultimately lead to an infection with LR S. epidermidis [19]. Thus, we analyzed 16S rRNA data of serial stool samples from our previously published cohort of patients with acute myelogenous leukemia [18]. We sought to identify patients whose gastrointestinal microbiome became dominated by staphylococci over the course of the study (see Materials and Methods for definition of staphylococcal emergence). Of 98 patients, 10 met our criteria for staphylococcal emergence. The median duration of linezolid use in the patients with staphylococcal emergence was significantly higher compared with patients who did not experience staphylococcal emergence (P < .001 by Mann-Whitney test; Figure 6B). Importantly, all of the patients with staphylococcal emergence associated with linezolid therapy received linezolid prior to having staphylococcal proliferation (examples are shown in Figure 6C and Supplementary Figure 7). We observed no significant relationship between daptomycin use and staphylococcal emergence (data not shown). As our 16S rRNA data could not determine which staphylococcal species was emerging, we tested the stool samples using quantitative PCR with primers specific for S. epidermidis [23] and identified 6 patients in whom staphylococcal dominance was due to S. epidermidis (data not shown). Next, we measured the amount of cfr in the stool samples and found a marked increase in cfr during or following linezolid therapy in 5 of the 6 cases (example shown in Figure 6C, with all patients shown in Supplementary Figure 7). The single patient who had staphylococcal emergence but did not have cfr identified in their stool had not received linezolid. Taken together, we conclude that domination of the gastrointestinal microbiome by cfr-containing S. epidermidis can occur following linezolid therapy.
DISCUSSION
Here, we sought to merge the strengths of molecular and clinical epidemiology by performing WGS on a large number of invasive S. epidermidis isolates combined with patient-specific data to gain insights into the mechanisms underlying proliferation of LR S. epidermidis. Our data indicate that a combination of a significant increase in linezolid use along with transmission of a LR S. epidermidis clone resulted in high rates of linezolid resistance among invasive S. epidermidis. Our approach of combining microbiome and genomic analyses to investigate the emergence of a multidrug-resistant pathobiont provides a pathway for investigating the spread of a broad array of antimicrobial-resistant organisms in the healthcare setting.
Given the high prevalence of S. epidermidis as a human commensal and the diverse genetic background of S. epidermidis isolates [24], the highly clonal nature of our invasive isolates was surprising. For example, we identified numerous instances of invasive S. epidermidis strains differing by 0 or 1 SNPs over the 2.5 million base pair genome. However, less sensitive genetic epidemiology methods such as pulsed-field gel electrophoresis and MLST have also indicated that invasive S. epidermidis strains can be highly clonal, with ST2, ST5, and ST83 being most common [11, 20, 25]. A recent report from France that used WGS also indicated clonal transmission of LR S. epidermidis, although it failed to distinguish among the ST5R and ST5S genotypes, possibly due to the small number of isolates characterized [8]. Conversely, analysis of the skin data from the Human Microbiome Project, which sampled healthy human volunteers, showed highly diverse S. epidermidis strains and only identified a single instance of a colonizing ST5 strain and no ST2 or ST83 strains [26]. Recent information helps reconcile this discrepancy as serial sampling of patients admitted from the community found a lack of hospital-associated S. epidermidis clones upon admission but rapid colonization of patients’ skin with such organisms after 3 days in the hospital [27]. Thus, patients are likely exposed to, colonized by, and infected with isolates belonging to clonal, hospital-adapted lineages, supporting the assumption of a healthcare-transmitted origin that may be susceptible to strict infection control practices.
In addition to being exposed to a LR S. epidermidis clone, linezolid use seems to play an important role in dissemination of resistant strains. Indeed, our microbiome and antibiotic use data show that prolonged linezolid exposure is strongly associated with proliferation of S. epidermidis in the microbiome (Figure 6C) and with infection by an LR strain of S. epidermidis (Figure 6A). Together with our identification of highly related clones of LR S. epidermidis, these data lead us to postulate that linezolid removes competing microflora, resulting in a permissive environment for proliferation of a LR S. epidermidis clone. Moreover, our microbiome data do not indicate that linezolid use removes linezolid-susceptible staphylococci, as we observed essentially no staphylococci in the commensal microbiota prior to linezolid initiation (Figure 6C). Identification of the precise microflora critical to controlling staphylococcal emergence is an ongoing effort in our laboratory that could help explain susceptibility to or even be used to prevent invasive S. epidermidis disease.
The ST5 clone causing the majority of our infections carries a heretofore undescribed plasmid that has significant homology to numerous staphylococcal plasmids, including pSS-01 from S. cohnii containing cfr [13]. pSS-01 was originally identified in a strain isolated in 2010 from swine in China and has since been described in various coagulase-negative staphylococci causing human infections in China [28]. Like pMB151a, in pSS-01 cfr is flanked by IS256-like elements that appear capable of mediating excision and integration of cfr. Recently, LR S. aureus strains causing human infection and containing a similar Cfr-encoding DNA element were identified in China [28], indicating that coagulase-negative staphylococci could serve as a source of linezolid resistance in S. aureus. Fortunately, it appears that pMB151a is not capable of such dissemination. However, the ability of the ST5R clone to cause numerous invasive infections over the past several years suggests that it could either be present in or spread to other healthcare settings, leading to a marked increase in linezolid resistance.
In summary, we present here the largest whole-genome analysis of invasive S. epidermidis isolates to date. Our results provide a framework for understanding how antimicrobials can promote the emergence of AMR in typically commensal organisms that can become major healthcare-associated pathogens.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank the clinical microbiology laboratory staff at MD Anderson Cancer Center (MDACC) for their assistance in collecting isolates. The sequences reported in this article have been deposited in the National Center for Biotechnology BioProject database (accession number PRJNA434275).
Financial support. This work was supported by the Jeanne F. Shelby Scholarship Fund (Clark Fellow Grant at MDACC to S. A. S.). DNA sequencing was performed at the MDACC DNA sequencing facility, which is supported by the National Cancer Institute (grant number P30-CA016672 via the Bioinformatics Shared Resource).
Potential conflicts of interest. All authors: No potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: American Society for Microbiology Microbe Meeting, New Orleans, Louisiana, June 2017.
References
- 1. Chioro A, Coll-Seck AM, Høie B, et al. Antimicrobial resistance: a priority for global health action. Bull World Health Organ 2015; 93:439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pamer EG. Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science 2016; 352:535–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Willems RJ, van Schaik W. Transition of Enterococcus faecium from commensal organism to nosocomial pathogen. Future Microbiol 2009; 4:1125–35. [DOI] [PubMed] [Google Scholar]
- 4. Hornef M. Pathogens, commensal symbionts, and pathobionts: discovery and functional effects on the host. ILAR J 2015; 56:159–62. [DOI] [PubMed] [Google Scholar]
- 5. Otto M. Staphylococcus epidermidis—the ‘accidental’ pathogen. Nat Rev Microbiol 2009; 7:555–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. See I, Freifeld AG, Magill SS. Causative organisms and associated antimicrobial resistance in healthcare-associated, central line-associated bloodstream infections from oncology settings, 2009–2012. Clin Infect Dis 2016; 62:1203–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Méric G, Miragaia M, de Been M, et al. Ecological overlap and horizontal gene transfer in Staphylococcus aureus and Staphylococcus epidermidis. Genome Biol Evol 2015; 7:1313–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dortet L, Glaser P, Kassis-Chikhani N, et al. Long-lasting successful dissemination of resistance to oxazolidinones in MDR Staphylococcus epidermidis clinical isolates in a tertiary care hospital in France. J Antimicrob Chemother 2018; 73:41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tewhey R, Gu B, Kelesidis T, et al. Mechanisms of linezolid resistance among coagulase-negative staphylococci determined by whole-genome sequencing. MBio 2014; 5:e00894–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bonilla H, Huband MD, Seidel J, et al. Multicity outbreak of linezolid-resistant Staphylococcus epidermidis associated with clonal spread of a cfr-containing strain. Clin Infect Dis 2010; 51:796–800. [DOI] [PubMed] [Google Scholar]
- 11. Bender J, Strommenger B, Steglich M, et al. Linezolid resistance in clinical isolates of Staphylococcus epidermidis from German hospitals and characterization of two cfr-carrying plasmids. J Antimicrob Chemother 2015; 70:1630–8. [DOI] [PubMed] [Google Scholar]
- 12. Mendes RE, Deshpande LM, Jones RN. Linezolid update: stable in vitro activity following more than a decade of clinical use and summary of associated resistance mechanisms. Drug Resist Updat 2014; 17:1–12. [DOI] [PubMed] [Google Scholar]
- 13. Wang Y, Zhang W, Wang J, et al. Distribution of the multidrug resistance gene cfr in Staphylococcus species isolates from swine farms in China. Antimicrob Agents Chemother 2012; 56:1485–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mulanovich VE, Huband MD, McCurdy SP, et al. Emergence of linezolid-resistant coagulase-negative Staphylococcus in a cancer centre linked to increased linezolid utilization. J Antimicrob Chemother 2010; 65:2001–4. [DOI] [PubMed] [Google Scholar]
- 15. Stamatakis A, Ludwig T, Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 2005; 21:456–63. [DOI] [PubMed] [Google Scholar]
- 16. Barton BM, Harding GP, Zuccarelli AJ. A general method for detecting and sizing large plasmids. Anal Biochem 1995; 226:235–40. [DOI] [PubMed] [Google Scholar]
- 17. Tomita H, Pierson C, Lim SK, Clewell DB, Ike Y. Possible connection between a widely disseminated conjugative gentamicin resistance (pMG1-like) plasmid and the emergence of vancomycin resistance in Enterococcus faecium. J Clin Microbiol 2002; 40:3326–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Galloway-Peña JR, Smith DP, Sahasrabhojane P, et al. The role of the gastrointestinal microbiome in infectious complications during induction chemotherapy for acute myeloid leukemia. Cancer 2016;122:2186–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Taur Y, Xavier JB, Lipuma L, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis 2012; 55:905–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mendes RE, Deshpande LM, Costello AJ, Farrell DJ. Molecular epidemiology of Staphylococcus epidermidis clinical isolates from U.S. hospitals. Antimicrob Agents Chemother 2012; 56:4656–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Du X, Zhu Y, Song Y, et al. Molecular analysis of Staphylococcus epidermidis strains isolated from community and hospital environments in China. PLoS One 2013; 8:e62742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goessweiner-Mohr N, Arends K, Keller W, Grohmann E. Conjugation in gram-positive bacteria. Microbiol Spectr 2014; 2:PLAS-0004-2013. [DOI] [PubMed] [Google Scholar]
- 23. Martineau F, Picard FJ, Roy PH, Ouellette M, Bergeron MG. Species-specific and ubiquitous DNA-based assays for rapid identification of Staphylococcus epidermidis. J Clin Microbiol 1996; 34:2888–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Conlan S, Mijares LA, Becker J, et al. ; NISC Comparative Sequencing Program Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates. Genome Biol 2012; 13:R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cherifi S, Byl B, Deplano A, Nonhoff C, Denis O, Hallin M. Comparative epidemiology of Staphylococcus epidermidis isolates from patients with catheter-related bacteremia and from healthy volunteers. J Clin Microbiol 2013; 51:1541–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zolfo M, Tett A, Jousson O, Donati C, Segata N. MetaMLST: multi-locus strain-level bacterial typing from metagenomic samples. Nucleic Acids Res 2017; 45:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Widerström M, Wiström J, Edebro H, et al. Colonization of patients, healthcare workers, and the environment with healthcare-associated Staphylococcus epidermidis genotypes in an intensive care unit: a prospective observational cohort study. BMC Infect Dis 2016; 16:743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cai JC, Hu YY, Zhou HW, Chen GX, Zhang R. Dissemination of the same cfr-carrying plasmid among methicillin-resistant Staphylococcus aureus and coagulase-negative staphylococcal isolates in China. Antimicrob Agents Chemother 2015; 59:3669–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Diaz L, Kiratisin P, Mendes RE, Panesso D, Singh KV, Arias CA. Transferable plasmid-mediated resistance to linezolid due to cfr in a human clinical isolate of Enterococcus faecalis.Antimicrob Agents Chemother 2012; 56:3917–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mendes RE, Deshpande LM, Castanheira M, DiPersio J, Saubolle MA, Jones RN. First report of cfr-mediated resistance to linezolid in human staphylococcal clinical isolates recovered in the United States. Antimicrob Agents Chemother 2008; 52:2244–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.