Abstract
The brain can be considered a network, existing of multiple interconnected areas with various functions. MRI provides opportunities to map the large-scale network organization of the brain. We tap into the neurobiochemical dimension of these networks, as neuronal functioning and signal trafficking across distributed brain regions relies on the release and presence of neurotransmitters. Using high-field MR spectroscopic imaging at 7.0 T, we obtained a non-invasive snapshot of the spatial distribution of the neurotransmitters GABA and glutamate, and investigated interregional associations of these neurotransmitters. We demonstrate that interregional correlations of glutamate and GABA concentrations can be conceptualized as networks. Furthermore, patients with epilepsy display an increased number of glutamate and GABA connections and increased average strength of the GABA network. The increased glutamate and GABA connectivity in epilepsy might indicate a disrupted neurotransmitter balance. In addition to epilepsy, the ‘neurotransmitter networks’ concept might also provide new insights for other neurological diseases.
Keywords: GABA, Glutamate, MR spectroscopic imaging, Networks, 7T
Highlights
-
•
We introduce the concept of “neurotransmitter networks”.
-
•
Ultrahigh field spectroscopic images of the brain were obtained.
-
•
Interregional correlations of Glutamate and GABA are conceptualized as networks.
-
•
Increased glutamate and GABA connectivity are shown in patients with epilepsy.
-
•
Neurotransmitter networks might provide new insights for neurological diseases.
1. Introduction
The brain can be considered a network, existing of multiple interconnected brain areas with various functions (van Straaten, 2012). Several studies have shown that measures of connectivity between these areas can be associated with cognitive functions (Song et al., 2008; van den Heuvel et al., 2009), and that the connections are affected in several neurological diseases (Fox and Greicius, 2010; Spencer, 2002). Characterization of brain networks gained large interest in both neuroscience and neurological studies and different methods are currently being employed. First of all, methods such as functional MRI and EEG can be applied to assess the so-called ‘functional networks’: areas are linked together and characterized based on simultaneous brain activity. Other methods characterize brain networks based on structural connectivity, by employing diffusion MRI, thereby visualizing fiber bundles in the brain (Jones, 2008), or over individuals assessing shared distributions of cortical thickness (He et al., 2007).
One particular disease that is often studied using network theory is localization-related epilepsy (i.e. epilepsy with a presumed focal structural cause that cannot be identified historically or be seen with current imaging techniques). Although traditionally considered a focal disease, studies applying functional MRI and diffusion MRI have provided convincing evidence that localization-related epilepsy exhibits profound alterations in both local and distributed functional and structural networks (Bernhardt et al., 2013). Proper neuronal functioning and signal trafficking across distributed brain regions also rely on the release and presence of chemical components, in particular neurotransmitters. Functional and diffusion MRI cannot provide direct information on defective neurons or the associated neurotransmitter disbalance, which underlies abnormal neuronal activity, an essential feature of seizures. Insights into the neurotransmitter network dysfunction in localization-related epilepsy might be of great value to eventually better understand the neuronal network characteristics of epilepsy and also other brain diseases.
Proton MR Spectroscopy (1H-MRS) enables the non-invasive detection and quantification of metabolite concentrations in the brain, thereby offering a window on brain cell metabolism (Rae, 2014). Traditionally, neurometabolites such as N-acetyl-aspartate (NAA; surrogate marker of neuronal density, primarily localized in the central and peripheral nervous system) and creatine (Cr; involved in energy metabolism, often regarded as suitable in vivo concentration reference) are being measured. Aberrant levels of these neurometabolites have been found in epilepsy, but also other neurological diseases or in aging (Öz et al., 2014; Petroff, 2005). More advanced studies also focus on measurements of the inhibitory and excitatory neurotransmitters GABA and glutamate, which can be associated with neural activity (Duncan et al., 2014). However, these studies traditionally consider only local metabolite concentrations, while healthy brain functioning does not only rely on individual brain areas, but also requires proper signal trafficking, and thus relations between distant brain areas (Park and Friston, 2013). Using high-field 1H-MRS imaging at 7.0 T, it is possible to obtain a snapshot of the spatial distribution of GABA and glutamate with a high (mL) spatial resolution.
Therefore, the concept of ‘neurotransmitter networks’ is introduced in this study. This new method relates to the assessment of coordinated spatial variations in neurotransmitter concentrations in the brain across individuals, and might be able to provide additional information on the underlying metabolic changes which affect neuronal functions. We primarily focussed on glutamate and GABA, due to their roles as important excitatory and inhibitory neurotransmitters in the brain. Additionally, NAA networks were considered, as NAA is the neurometabolite that is easiest to measure, although not directly involved in signaling. We assessed the construction and first applicability of ‘neurotransmitter networks’. The concept is first applied healthy participants, and subsequently compared between patients with epilepsy and healthy participants.
2. Materials and methods
2.1. Study procedures
Two groups of participants, healthy volunteers and patients with epilepsy, were included. The exclusion criteria for both groups were all contraindications for MR scanning (such as metal implants or pregnancy), and a medical history with (other) neurological diseases. Additional exclusion criteria for the patients with epilepsy were MRI visible lesions (seen on clinical 3 T scans), changes in antiepileptic drugs (medication or dose) in the last twelve months, or a seizure frequency higher than once a month.
All participants provided written informed consent before participation. Ethical approval for this study was obtained from the medical ethical committee academic hospital Maastricht/Maastricht University, and the study was registered at the Dutch Trial Register with registration number NTR4878.
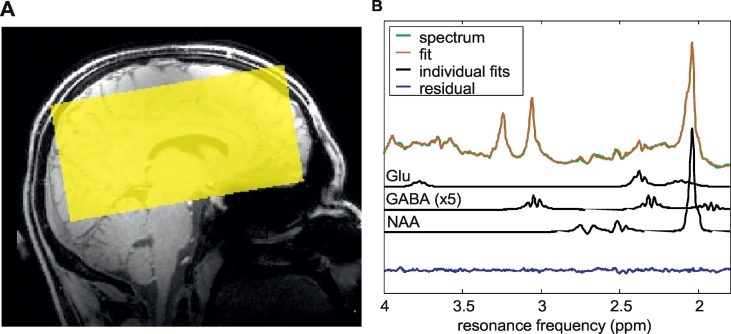
Each participant underwent a 7 Tesla MR examination (Magnetom, Siemens Healthineers, Erlangen, Germany) with a 32-channel head coil. The scanning procedure included whole brain T1-weighted imaging (MP2RAGE (Marques et al., 2010), TR/TE 4500/2.39 ms, TI1/TI2 900/2750 ms, FOV 173 × 230 × 230 mm3, cubic voxel size 0.9 × 0.9 × 0.9 mm3), a whole brain fluid attenuation inversion recovery (FLAIR) sequence (TR/TE 8000/303 ms, TI 2330 ms, FOV 166.4 × 224 × 256 mm3, cubic voxel size 0.8 × 0.8 × 0.8 mm3), and an MRSI acquisition. For the latter, a semi-LASER sequence was applied, which combines conventional RF pulses for slice excitation with orthogonal adiabatic refocusing pulses for volume selection (Scheenen et al., 2008). Frequency offset corrected inversion (cFOCI) pulses were included in this sequence to limit chemical shift artifacts (Ordidge et al., 1996). Other parameters were TR/TE 5520/38 ms, VAPOR water suppression, FOV 150 × 150 × 100 mm3 (Fig. 1), and voxel size 9.4 × 9.4 × 12.5 mm3 (1.1 mL).
Fig. 1.
A. Field-of-view of the MRSI image. B. Typical example of a measured spectrum with the quantitative (LCModel) fit, and individual glutamate (glu), GABA and N-acetylaspartate (NAA) fits.
Five of the healthy controls were scanned twice, with a seven-day interval, to assess the inter-scan reproducibility of the MRSI measures. Both T1-weighted and FLAIR images were checked by a neuroradiologist (P.A.M.H.) for abnormalities.
2.2. Data analysis
2.2.1. Metabolite concentrations
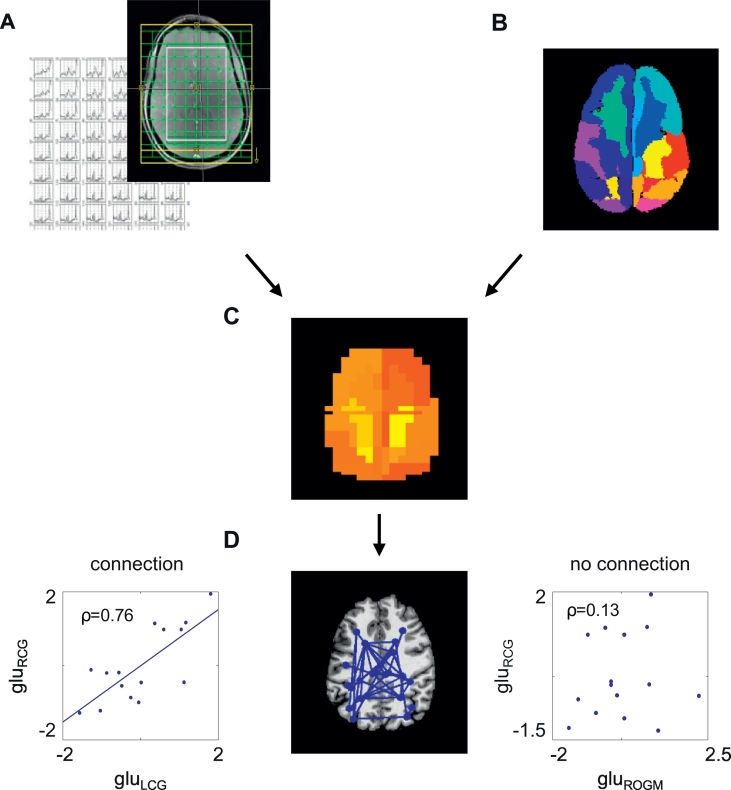
Before constructing the metabolite networks, the metabolite concentrations per brain area were computed. For this purpose, information from the anatomical scan and the spectra were preprocessed and analyzed (Fig. 2). With the MP2RAGE sequence, images were obtained for two inversion times: TI1 (GRETI1) and TI2 (GRETI2), which were combined to create a quantitative longitudinal relaxation time T1-weighted image (Marques et al., 2010). The GRETI2 scan was skull stripped using the brain extraction tool (BET) of FMRIB Software Library (FSL, version 5.0.1) (Jenkinson et al., 2012; Smith, 2002). The T1 map was segmented in grey matter, white matter, and cerebrospinal fluid with FAST, part of FSL (Zhang et al., 2001).
Fig. 2.
Analysis of the metabolite networks. The spectra were first analyzed in LCModel (A) and aligned with the structural image (B), and mean metabolite concentrations were calculated per area (C). Connections between areas were defined as correlated neurometabolite concentrations (D). Glu: standardized glutamate concentration; RCG: right cingulate gyrus; LCG: left cingulate gyrus; ROGM: right occipital grey matter.
To parcellate the brain into a number of standard areas, the atlas was transformed to the skull stripped T1-weighted image (Fig. 2). The atlas was defined by thirty non-overlapping brain areas (Table 1) and was created in MNI space, by combining information from the Harvard-Oxford cortical and subcortical atlases and the ICBM 2009c nonlinear symmetric template (Desikan et al., 2006; Fonov et al., 2009; Frazier et al., 2005). In addition to GM, also WM regions are included, as recent developments in the field indicate that WM has other functions than pure neuron-to-neuron communication (Butt et al., 2014). Furthermore, the presence of neurotransmitters is not only related to neuronal signaling or local synaptic activity. It has for example been shown that neurotransmitters released from axons during action potential propagation acting on glial receptors regulate the homeostatic functions of astrocytes and myelination by oligodendrocytes. Astrocytes also release neurotransmitters, maintaining signaling along potentially long axon tracts. The co-existence of multiple neurotransmitters in the WM are indicative of diverse functions important for information processing. The skull stripped T1 maps were non-linearly transformed to the MNI brain using FNIRT (FSL, version 5.0.1) (Andersson et al., 2007; Jenkinson et al., 2002; Jenkinson and Smith, 2001). The inverse non-linear transformation was then applied to transform an atlas to each individual brain.
Table 1.
Atlas areas, the number of included spectroscopic voxels per area, and percentage of subjects with corresponding data (based on the combined patient and control group).
| Area |
No. of voxels (mean ± standard deviation) |
Percentage subjects with good-quality data |
||
|---|---|---|---|---|
| Hemisphere | Left | Right | Left | Right |
| Thalamus | 4.3 ± 2.1 | 4.5 ± 1.8 | 100% | 100% |
| Basal ganglia | 4.3 ± 2.7 | 3.8 ± 2.3 | 92%a | 92% |
| Hippocampus/amygdala | 1.2 ± 1.2 | 0.7 ± 0.8 | 64%a | 44%a |
| Prefrontal GM | 13.9 ± 7.3 | 11.9 ± 6.3 | 100% | 100% |
| Prefrontal WM | 19.8 ± 6.7 | 20.3 ± 8.0 | 100% | 100% |
| Insula | 7.5 ± 3.8 | 7.6 ± 4.5 | 100% | 88%a |
| Premotor GM | 6.4 ± 4.8 | 4.9 ± 4.4 | 96% | 84%a |
| Premotor WM | 11.4 ± 4.7 | 12.6 ± 4.6 | 100% | 100% |
| Temporal GM | 3.1 ± 3.6 | 2.1 ± 2.9 | 68%a | 56%a |
| Temporal WM | 11.0 ± 4.9 | 10.5 ± 6.0 | 100% | 96%b |
| Parietal GM | 12.0 ± 4.7 | 13.4 ± 5.2 | 100% | 100% |
| Parietal WM | 16.8 ± 4.6 | 15.4 ± 3.8 | 100% | 100% |
| Occipital GM | 9.4 ± 6.1 | 10.5 ± 7.7 | 96%b | 92%b |
| Occipital WM | 0.9 ± 1.2 | 1.0 ± 1.1 | 48%a | 64%a |
| Cingulate gyrus | 5.4 ± 3.1 | 9.2 ± 2.9 | 100% | 100% |
These areas were excluded in the final analyses, because not all participants had good-quality data in these areas.
These areas were additionally excluded in the comparison between patients and healthy participants, because of missing patient data.
Metabolite spectra were analyzed using LCModel (version 6.3, Fig. 1) with a simulated basis set of 20 metabolites. The 20 model spectra (Appendix A) of acetate (Ace), ascorbate/vitamin C (Asc), aspartate (Asp), Cr, GABA, glycine (Gly), glutamate (Glu), glutamine (Gln), glutathione (GSH), glycerophosphocholine (GPC), myo-inositol (Ins), NAA, Nacetylaspartylglutamate (NAAG), phosphocholine (PCh), phosphocreatine (PCr), phosphoethanolamine (PE), taurine (Tau), scyllo-inositol (Sci), macromolecule 2.0 ppm (MM20), and the correction term for Cr (—CrCH2) were generated using VESPA (Versatile Simulation, Pulses and Analysis package, (Soher et al., 2011)). Which applies previously reported chemical shifts and coupling constants initially published by (Govindaraju et al., 2000), which were later updated (Govind et al., 2015), and refined by others including (Kreis and Bolliger, 2012; Tkac, 2008). Spectra were generated using Lorentzian lines, 1 Hz broadened, 4 kHz bandwidth, and 2048 points. Verification of the basis set included phantom experiments with varying concentrations of GABA and glutamate and visual inspection of spectra. A macromolecule spectrum acquired with a sLASER sequence (TE = 36 ms) at 7 T was initially added to the basis set, but not included in the final analysis after visual inspection. Lipids and other macromolecules were estimated using the spline function in LCModel.
Voxelwise spectra were excluded when the signal-to-noise ratio (SNR) was below 20, the Cramér-Rao lower bounds (CRLBs) of the N-acetylaspartate plus N-acetylaspartylglutamate concentrations (tNAA) was higher than 3, the CRLB of creatine plus phosphocreatine (tCr) was higher than 10, or the CRLB of GABA or glutamate was higher than 20. SNR was defined as the ratio of the maximum in the spectrum-minus-Baseline over the Analysis Window to twice the root mean square Residuals (Provencher, 2014). These quality criteria were derived using an extensive iterative process, in which we evaluated many options to obtain the best criteria for selection and rejection of spectra. Line width was not included in these criteria, due to its close interrelationship with SNR and CRLB (Tkac et al., 2009), which are already taken into account. The spectra were also excluded if the total grey and white matter fraction of the corresponding voxel was lower than 50%. As we did not acquire a separate water spectrum due to time restrictions, we did not have the option to perform the preferred absolute quantification (Jansen et al., 2006). Hence, all concentrations were expressed relative to the tCr concentration of the corresponding voxel.
Each MRSI voxel first aligned with the T1-weighted image, and then assigned to the predefined atlas area that showed largest overlap with the voxel. The average concentration per area was corrected for grey and white matter content, age and sex with linear regression analysis, in which the metabolite concentrations were used as the dependent variable (across all participants), and the grey/white matter content, age and sex as independent variables. Grey/white matter content was defined as the fraction of grey matter divided by the grey plus white matter fraction. The corrected metabolite concentrations were equal to the residuals of the linear regression analysis (Eq. (1)):
| (1) |
with Conc the measured (i.e. uncorrected) metabolite concentration per area, Conccorr the corrected metabolite concentration, GM the percentage grey matter and WM the percentage white matter. The Conccorr can be interpreted as the metabolite concentration corrected for variations in grey/white matter content, age, and sex over the participants (similar to (He et al., 2007)). The regression coefficients are displayed as β, which were not used for subsequent analysis.
Due to the relatively strong intensity inhomogeneities that can be particularly severe in MRI at ultra-high field strengths (e.g. 7 T), it is common for connectivity analyses to apply a certain bias field correction (Li et al., 2009). Therefore as final preprocessing step, standardized concentrations were calculated over all healthy participants (Eq. (2)):
| (2) |
with Conccorr the corrected and Concscaled the standardized and corrected metabolite concentrations, and sd the standard deviation. To avoid confusion due to the substantial correction and processing steps, corrected metabolite concentrations are from now on referred to as “metabolite indices”.
2.2.2. Brain metabolite networks and measures
A connection between two brain areas was considered present, when the neurometabolite indices of these two areas correlated significantly over the subjects. We assume that areas support concerted brain activity if an increased neurometabolite concentration in one area is accompanied with an altered concentration of that neurometabolite in another area. Areas with correlated neurometabolite indices among participants are therefore considered ‘metabolically connected’. To obtain the neurometabolite connectivity matrix, the Pearson's correlation coefficient between the metabolite indices of each pair of areas was calculated. The weighted connectivity between pairs of areas was equal to the correlation coefficient ρ if the correlation was below a certain statistically significant threshold (i.e., p < 0.05) and 0 otherwise.
A prerequisite for the analyses is that none of the participants had areas without good-quality data. Therefore, some areas and participants were excluded from the final analysis (Table 1).
2.2.3. Group comparison
To compare the neurometabolite networks in the patients with epilepsy with the control-group, two basic network characteristics were evaluated: the density (i.e. the fraction of actual connections divided by total number of possible connections), and the average network strength. The density was calculated in a range of varying thresholds (varying from ρ = 0.1–0.9) for both subject groups. In this comparison, the Pearson correlation coefficient ρ was applied as threshold, as it is independent of group size, in contrast with the p-value of a correlation.
For the average network strength, a fixed set of nodes was defined, which was based on the combined results of the two groups per neurometabolite. The number of connections and the set of possible connections was fixed across the group of individuals (i.e. a fixed network) to rule out the influence of network density on the computation and comparison of graph metrics across groups (van den Heuvel et al., 2017). The included regions for the fixed analysis can be found in appendix B. The density of the network was set at 0.2, which corresponded to a threshold of approximately p = 0.05 in the healthy subject-group (Fig. 4A). Subsequently, metabolite networks were created for patients and controls, using the fixed network, and the connection strengths of these networks were statistically compared using a binomial test. A consequence of using the covariance method (correlating over subjects within a group) to construe networks is that only one network can be obtained per group, preventing the applicability of traditional Student t-tests. To overcome this problem, we evaluated whether the entire set of connection strengths of these networks differed, by evaluating the distribution of connections stronger in patients and connections stronger in controls, in relationship with the expected probability based on the total number of connections. For this approach a binomial test is appropriate (Ansari-Lari, 2004). For this, the number of connections that were stronger in the patients with epilepsy, and the number of connections stronger in the healthy participants were tallied. The probability of this distribution is approximately binomially distributed, thus the probability of the found distribution can be computed by:
| (3) |
with k the number of connections that is larger in one group, n the total number of connections, and p the probability that a single connection is higher in one group than the other (p = 0.5).
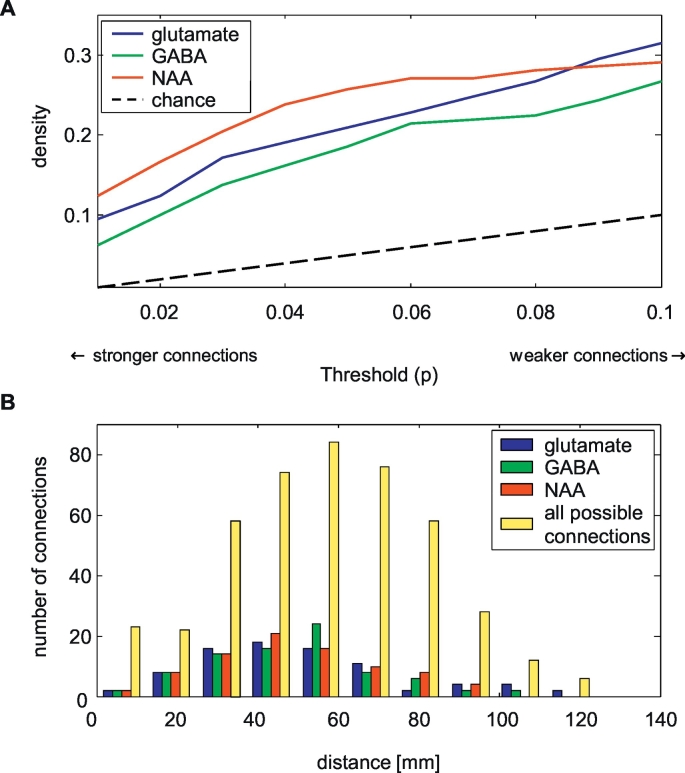
Fig. 4.
Characteristics of the neurometabolite networks. In A, the density of the networks is displayed at different thresholds (i.e. the p-value of the correlation). It can be observed that the density of the networks is much higher than would be expected by chance only. B displays the distribution of distances between the nodes, including the distances between all possible connections. The neurometabolite networks include both long- and short distance connections. The density of networks was set to 0.2 for all three metabolites.
The p-value is then given by the cumulative distribution:
| (4) |
In these analyses, the correction for grey/white matter, age and sex, and the standardization (as described in Section 2.2.1), was applied over the pooled group containing both controls and patients.
To test the robustness for within-subject variations, this analysis was repeated with replacing the data from the 5 healthy participants who were scanned twice with the ‘second measurements’.
3. Results
3.1. Study population
Thirty participants were included in this study (20 participants without and 10 with epilepsy). Data of two healthy participants were excluded because of an overall low spectral quality. Therefore, data of 18 healthy participants (10/8 male/female, age 39.6 ± 16.8 years, age range: 22–65) were considered for further analyses. All ten patients with localization-related epilepsy (7/3 male/female, age 40.1 ± 16.5 years, age range: 20–69 years) were included in these analyses. In none of the participants, epileptogenic abnormalities were identified in the 7 T images by our expert neuroradiologist.
3.2. Metabolite concentrations
Concentrations of GABA, glutamate, and NAA were measured in thirty different brain areas (Table 1). No significant differences were observed for the GABA, glutamate, or NAA concentrations between the patients and controls in any of these areas (Student's t-test, p > 0.05). The average SNR per area across all participants was 47 ± 8 (mean ± SD), while the average CRLBs of glutamate, GABA and NAA were 4.1 ± 0.9, 13.5 ± 0.8, and 2.1 ± 0.2, respectively. The coefficients of variation of the concentration estimates (for the 5 healthy volunteers who were scanned twice) were 8.4 ± 4.2% for glutamate, 11.4 ± 3.8% for GABA, and 7.8 ± 3.1% for NAA (Appendix C). Furthermore, measured (i.e. uncorrected) concentrations are in the same range as reported in previous studies, except for GABA, which was twice as high, possibly due to macromolecule contamination (Appendix D) (O'Gorman et al., 2011; van Veenendaal et al., 2016). In Appendix E, quantitative data and measures of quality for the Occipital GM are provided, indicating similar quality for both groups. The glutamate and NAA concentrations were negatively associated with age in respectively 77% and 73% of the brain areas (linear regression analysis, p < 0.05), while the GABA concentrations were only negatively associated with age in 20% of the brain areas (linear regression analysis, p < 0.05). As no missing data were allowed in the network analyses, data from fifteen healthy participants and 21 areas were included in these analyses (Table 1). In case of the analyses in patients, ten patients and eighteen areas were included.
3.3. Organization of metabolic brain networks
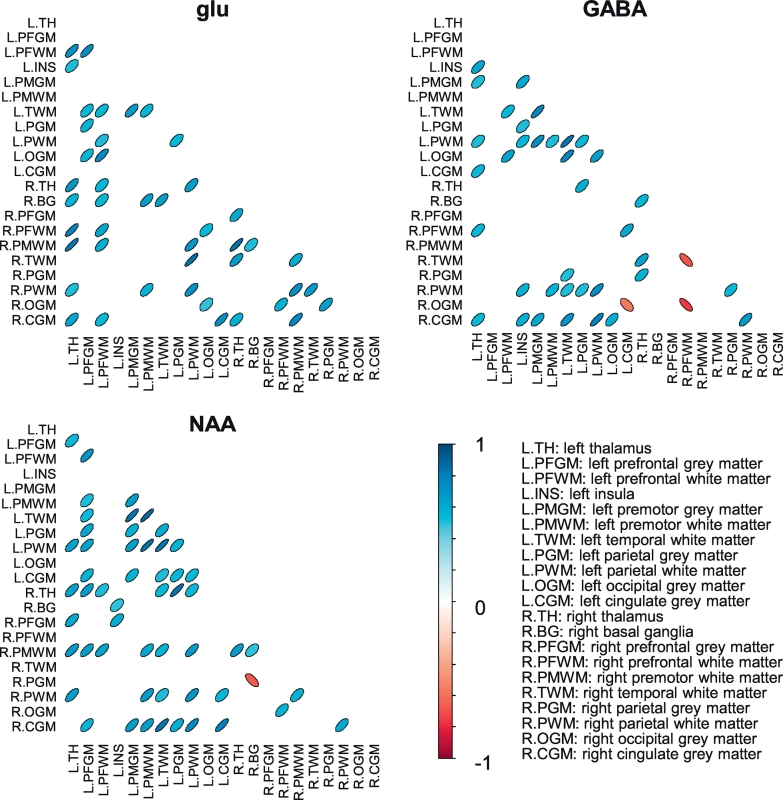
Of the possible connections, 21%, 19%, and 26% showed a significant glutamate, GABA, and NAA correlation, respectively, (p < 0.05) across all fifteen healthy participants (Fig. 3). The density of the networks is higher than expected purely based on chance (i.e. the p-value) for all three metabolites (Fig. 4A). The maximum node degree (number of connections per node) was nine; this was observed in the left prefrontal white matter (glutamate) or left parietal white matter (GABA, NAA). All nodes were connected in the glutamate network, while the GABA network had three unconnected nodes (left and right prefrontal grey matter, and the right premotor white matter). The NAA network had two unconnected nodes (left occipital grey matter and right temporal white matter), and another component (with only two nodes: right prefrontal white matter and right occipital grey matter) which was unconnected to the other nodes. The vast majority of connections was positive: no significant, negative connections were present in the glutamate network, while 11% and 4% of the respectively GABA and NAA connections was negative.
Fig. 3.
Correlation matrices of the glutamate (glu), GABA and N-acetylaspartate (NAA) networks in healthy participants. The shape and color of the ovals indicate the correlations coefficient (ρ) between each pair of areas, i.e. the strength of the connections between those areas and the sign of the correlations (red: negative, blue: positive). Only connections with a p < 0.05 are displayed. Figures were created in R version 3.2.2 (Team, 2015; Wright, 2016). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The relation between the number of connections and distance between the areas is illustrated in Fig. 4B. It can be observed that the distances between all areas are roughly normally distributed, which is in close agreement with the distribution of anatomical distances observed for cortical thickness networks (He et al., 2007). The neurometabolite networks include both long- and short distance connections.
3.4. Metabolite networks in epilepsy
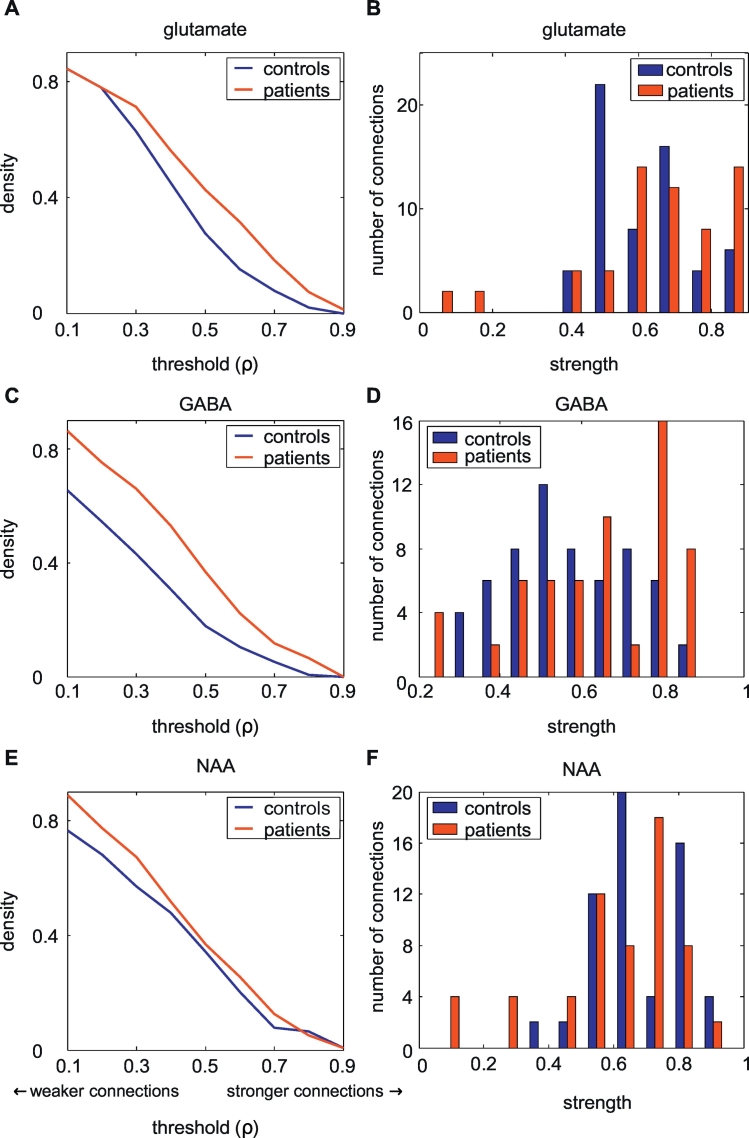
The density of the glutamate and GABA networks were higher in the patients with epilepsy compared with the healthy controls, over a considerable range of rho thresholds (Fig. 5, Table 2). Also the connection strength of the GABA networks was higher in the patients than the controls. The average connection strength of the glutamate networks was comparable between the patients and controls. The NAA networks showed a comparable density and node strength in patients and controls. When comparing the individual connections strengths, the patients with epilepsy displayed a larger Pearson correlation coefficient ρ (i.e. strength) compared with the healthy participants in 97 of the 153 glutamate connections (p < 0.001), 103 of the 152 GABA connections (p < 0.001), and 83 of the 153 NAA connections (p = 0.17).
Fig. 5.
Network density in the healthy-subject group and patient group (A, C, E). In both glutamate and GABA networks, the density in patients is higher than that of the healthy participants. B, D, and F show how the distributions of connection strengths of both groups in the glutamate, GABA, and NAA networks. While the glutamate and NAA networks show similar strength distributions, the GABA network shows increased connection strengths in patients with epilepsy.
Table 2.
Comparison of network characteristics between patients and controls. The density is given at a threshold of ρ = 0.6, whereas the average connection strength is given at a density of 0.2. Because of missing data in the patient group, fewer areas (n = 18) were included in these analyses than those described in Fig. 4.
| Density |
Average connection strength |
|||
|---|---|---|---|---|
| Controlsa | Patients | Controlsa | Patients | |
| Glutamate | 0.15 (0.20) | 0.31 | 0.62 (0.66) | 0.61 |
| GABA | 0.10 (0.07) | 0.22 | 0.54 (0.47) | 0.64 |
| NAA | 0.20 (0.19) | 0.25 | 0.65 (0.68) | 0.64 |
Numbers between brackets display the results of the analyses with the five second measurements and give an indication of the robustness for within-subject variations.
4. Discussion
In this study, the concept of neurotransmitter networks was introduced. A spatial relation between glutamate, GABA, and NAA indices of different areas was shown, suggesting the presence of neurometabolite networks. These networks included both short-range and long-range connections. Furthermore, some areas were characterized as so-called “hub areas”: areas with many connections to other areas, while other areas showed fewer to no connections. Finally, these networks displayed significantly different properties in patients with epilepsy in comparison with healthy participants.
4.1. Interpretation of neurotransmitter networks
An important question is how to interpret these metabolic networks, or, on a smaller scale, metabolic connections. The conceptual idea is that areas that are connected, tend to have similar characteristics, an idea known in social sciences as the “homophily principle” (Simsek and Jensen, 2008). This principle has previously been applied to neurosciences, for instance in correlated cortical thickness (He et al., 2007), gene expression in the brain (Richiardi et al., 2015) or serotonin-receptor binding measured with positron emission tomography (PET) (Erritzoe et al., 2010).
Although neurotransmitter network correlations should not be considered as a direct measure of connectivity, it has been shown previously in cultured hippocampal neurons that synchronous neuronal firing promotes network-based synaptogenesis (Bi and Poo, 1999). Bi et al. showed that localized stimulation can modify synapses that are remote from the stimulated neuron. Changes in polysynaptic pathways could be accounted for by correlating pre- and postsynaptic excitation at distant synaptic sites. Hence, anatomical regions that are “connected” might share similar ways of signal trafficking that rely on synaptic modifications. It is currently unknown whether these regions share a similar receptor architecture or gene expression profile. A straightforward answer to this question may not be possible, as the precise relationships among different regions of the brain are yet to be fully characterized. Future studies, combining connectivity measures from different modalities (e.g. functional MRI, diffusion MRI, MR spectroscopy) might provide meaningful insights these relationships. Additionally, comparisons with quantitative receptor autoradiography, which can provide regional distribution of neurotransmitter receptors in the cerebral cortex (Amunts et al., 2010), might provide additional insights.
Several previous studies already assessed interregional correlations in glutamate, GABA, and NAA concentrations (Grachev and Apkarian, 2001; Greenhouse et al., 2016; Kraguljac et al., 2012; Waddell et al., 2011). It is difficult to compare their results with ours, as they used different methods (e.g. field strength, brain areas). Our study showed the existence of interregional correlations for all three neurometabolites, but the specific coherence depended on the areas being compared; not all areas showed a correlation, which might explain the contrasting results seen in different studies.
An explanation for the biological mechanisms behind these connections remains speculative. Beside its function as neurotransmitter, glutamate is available as metabolic pools in the neuron and linked to glucose metabolism (Rae, 2014). GABA, on the other hand, is most likely related to the GABAergic tone (i.e. the level of continuous GABAergic activity, inducing tonic inhibition, which is not necessarily inhibitory activity) (Rae, 2014). Several studies suggested that neuronal connectivity is related to GABA- and glutamatergic function. At neuronal level, it has been shown that the probability of a connection between two types of neurons, correlates with the average synaptic strength of those two neuron types (Jiang et al., 2015). At brain level, positive correlations have been shown between glutamate concentrations and functional connectivity, and negative between GABA and functional connectivity (Duncan et al., 2014).
NAA is considered a surrogate marker for neuronal integrity and associated with cognitive function (Rae, 2014). It can be hypothesized that when areas are connected, damage in one brain area is accompanied with a decreased neuronal integrity of the other brain area.
Although individual connections are important, these connections are not isolated, as there is a constant interaction between different brain areas and their connections. Therefore, it might be beneficial to assess these brain networks as a whole, which was investigated in this study. Previous structural and functional studies found that brain networks appear so-called “small-world networks”, meaning that they combine a high integration as well as segregation in the network (Bullmore and Sporns, 2009). At this moment, such network characteristics could not be assessed, because of the relatively low number of nodes and the presence of both positive and negative weights, but this should be explored in further studies.
4.2. Neurotransmitter networks in epilepsy
A higher GABA and glutamate connectivity was shown in patients with epilepsy compared with a healthy control group, as indicated by the number of connections and the strengths of the connections. This increased neurotransmitter connectivity seems to be associated with the pathophysiology in epilepsy, as epileptic seizures likely disrupt neurotransmitter networks. Previous studies showed an increased functional and structural connectivity in patients around the epileptic zone, but a decreased connectivity in other brain areas, and it has been suggested that brain networks are reorganized as a result of recurrent epileptic seizures (Englot et al., 2016). Also, antiepileptic drugs might alter glutamate and GABA concentrations (van Veenendaal et al., 2015), and may result in aberrant metabolic brain networks, but these effects were beyond the scope of the current study.
Previous studies showed significant correlations between NAA concentrations in the hippocampus, thalamus, and putamen in patients with temporal lobe epilepsy, while these correlations were absent in healthy subjects (Hetherington et al., 2007; Pan et al., 2012). No changes in NAA connectivity were observed in the current study, but this connectivity might be location-specific. While these previous studies studied connectivity between specific brain areas, we choose to study overall brain networks.
4.3. Study considerations
This explorative study aimed to introduce the concept of neurotransmitter networks and should be considered as proof-of-concept. Employing a correlation over the group as an outcome measure leads to some methodological considerations, as it restricts the possibilities for statistical testing. This was solved by comparing individual connection strengths between groups. Future, larger studies might adopt statistical methods applied in cortical thickness studies, that employ similar outcome measures, such as permutation tests (Bassett et al., 2008; He et al., 2008) or methods to calculate individual measures (Saggar et al., 2015). Unfortunately, the group size in this pilot study is not sufficient to employ these methods. Finally, the applied MRSI sequence has some disadvantages, such as a limited spatial resolution and coverage, and vulnerability to artifacts. The spatial resolution of our MRSI experiment is relatively coarse (1.1 mL) in comparison to structural and functional network experiments, which prompted the use of a brain atlas with relatively large regions, to ensure a sufficient sampling (i.e. sufficient number of overlapping voxels) per region for all subjects. Therefore, the nodes in the network are relatively large in volume, which ensured that it was more robust to select the same region over different subjects. As the obtained neurotransmitter networks do show realistic properties (e.g. both intra and inter-hemispheric connections), we are confident regarding the validity of our approach. Some of the disadvantages could be solved with other sequences, such as GluCEST, which enables whole-brain glutamate measurements with a high resolution (Cai et al., 2012; Cai et al., 2013). Future studies with larger group sizes and dedicated spectroscopic imaging protocols are thus necessary to evaluate the presented concept in more detail. These future studies might also provide a better understanding of the concept of a neurotransmitter network. Additionally, in future studies also comparisons with other MRI-derived networks (e.g. fMRI or DTI based connectivity) could be performed. Lastly, also alternative strategies regarding the MRS analysis, in terms of quality criteria (Kreis, 2016) and use of simulated basis sets (Bhogal et al., 2017) should be explored in future studies.
4.4. Future implications
The results are expected to provide a better understanding of the role of neurotransmitter dysfunction in epilepsy and other neurodegenerative diseases. Non-invasive imaging of neurotransmitter networks might provide an early biomarker of patients at risk (i.e. before the onset of overt symptoms). Lastly, it could aid the development of novel effective treatment options, and be used to assess the efficacy of pharmacological interventions.
4.5. Conclusion
This study presents the novel concept of metabolic brain networks using MR spectroscopic imaging. We showed interregional correlations of glutamate, GABA, and NAA measurements, which can be conceptualized as networks. We showed the applicability of this concept in patients with epilepsy; however, it might also provide new insights for other neurological diseases.
Acknowledgements
The authors thank Esther Steijvers, Lotty Huijboom, and Chris Wiggins for help with data acquisition, and Vincent Boer for help with metabolite spectra simulations. We also would like to acknowledge Alfons Kessels and Dave Langers for statistical support. This study was funded by the University Fund Limburg/SWOL (S.2014.14).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2018.04.006.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
References
- Amunts K., Lenzen M., Friederici A.D., Schleicher A., Morosan P., Palomero-Gallagher N., Zilles K. Broca's region: novel organizational principles and multiple receptor mapping. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson J.L., Jenkinson M., Smith S. FMRIB Analysis Group of the University of Oxford 2. 2007. Non-linear registration, aka spatial normalisation FMRIB technical report TR07JA2. [Google Scholar]
- Ansari-Lari M. Binomial distribution and estimation of the true prevalence of infected animals when pooled samples are used. Can. Vet. J. 2004;45:719. (author reply 720) [PMC free article] [PubMed] [Google Scholar]
- Bassett D.S., Bullmore E., Verchinski B.A., Mattay V.S., Weinberger D.R., Meyer-Lindenberg A. Hierarchical organization of human cortical networks in health and schizophrenia. J. Neurosci. 2008;28:9239–9248. doi: 10.1523/JNEUROSCI.1929-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt B.C., Hong S., Bernasconi A., Bernasconi N. Imaging structural and functional brain networks in temporal lobe epilepsy. Front. Hum. Neurosci. 2013;7:624. doi: 10.3389/fnhum.2013.00624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogal A.A., Schur R.R., Houtepen L.C., van de Bank B., Boer V.O., Marsman A., Barker P.B., Scheenen T.W.J., Wijnen J.P., Vinkers C.H., Klomp D.W.J. (1) H-MRS processing parameters affect metabolite quantification: the urgent need for uniform and transparent standardization. NMR Biomed. 2017;30 doi: 10.1002/nbm.3804. [DOI] [PubMed] [Google Scholar]
- Bi G., Poo M. Distributed synaptic modification in neural networks induced by patterned stimulation. Nature. 1999;401:792–796. doi: 10.1038/44573. [DOI] [PubMed] [Google Scholar]
- Bullmore E., Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- Butt A.M., Fern R.F., Matute C. Neurotransmitter signaling in white matter. Glia. 2014;62:1762–1779. doi: 10.1002/glia.22674. [DOI] [PubMed] [Google Scholar]
- Cai K., Haris M., Singh A., Kogan F., Greenberg J.H., Hariharan H., Detre J.A., Reddy R. Magnetic resonance imaging of glutamate. Nat. Med. 2012;18:302–306. doi: 10.1038/nm.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai K., Singh A., Roalf D.R., Nanga R.P., Haris M., Hariharan H., Gur R., Reddy R. Mapping glutamate in subcortical brain structures using high-resolution GluCEST MRI. NMR Biomed. 2013;26:1278–1284. doi: 10.1002/nbm.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R.S., Ségonne F., Fischl B., Quinn B.T., Dickerson B.C., Blacker D., Buckner R.L., Dale A.M., Maguire R.P., Hyman B.T. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Duncan N.W., Wiebking C., Northoff G. Associations of regional GABA and glutamate with intrinsic and extrinsic neural activity in humans-a review of multimodal imaging studies. Neurosci. Biobehav. Rev. 2014;47C:36–52. doi: 10.1016/j.neubiorev.2014.07.016. [DOI] [PubMed] [Google Scholar]
- Englot D.J., Konrad P.E., Morgan V.L. Regional and global connectivity disturbances in focal epilepsy, related neurocognitive sequelae, and potential mechanistic underpinnings. Epilepsia. 2016;57:1546–1557. doi: 10.1111/epi.13510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erritzoe D., Holst K., Frokjaer V.G., Licht C.L., Kalbitzer J., Nielsen F.A., Svarer C., Madsen J., Knudsen G. A nonlinear relationship between cerebral serotonin transporter and 5-HT(2A) receptor binding: an in vivo molecular imaging study in humans. J. Neurosci. 2010;30:3391–3397. doi: 10.1523/JNEUROSCI.2852-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonov V.S., Evans A.C., McKinstry R.C., Almli C., Collins D. Unbiased nonlinear average age-appropriate brain templates from birth to adulthood. NeuroImage. 2009;47:S102. [Google Scholar]
- Fox M.D., Greicius M. Clinical applications of resting state functional connectivity. Front. Syst. Neurosci. 2010;4:19. doi: 10.3389/fnsys.2010.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier J.A., Chiu S., Breeze J.L., Makris N., Lange N., Kennedy D.N., Herbert M.R., Bent E.K., Koneru V.K., Dieterich M.E. Structural brain magnetic resonance imaging of limbic and thalamic volumes in pediatric bipolar disorder. Am. J. Psychiatr. 2005;162:1256–1265. doi: 10.1176/appi.ajp.162.7.1256. [DOI] [PubMed] [Google Scholar]
- Govind V., Young K., Maudsley A.A. Corrigendum: proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. 2015;28:923–924. doi: 10.1002/nbm.3336. [DOI] [PubMed] [Google Scholar]
- Govindaraju V., Young K., Maudsley A.A. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. 2000;13:129–153. doi: 10.1002/1099-1492(200005)13:3<129::aid-nbm619>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Grachev I.D., Apkarian A.V. Chemical network of the living human brain: evidence of reorganization with aging. Cogn. Brain Res. 2001;11:185–197. doi: 10.1016/s0926-6410(00)00068-9. [DOI] [PubMed] [Google Scholar]
- Greenhouse I., Noah S., Maddock R.J., Ivry R.B. Individual differences in GABA content are reliable but are not uniform across the human cortex. NeuroImage. 2016;139:1–7. doi: 10.1016/j.neuroimage.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Chen Z.J., Evans A.C. Small-world anatomical networks in the human brain revealed by cortical thickness from MRI. Cereb. Cortex. 2007;17:2407–2419. doi: 10.1093/cercor/bhl149. [DOI] [PubMed] [Google Scholar]
- He Y., Chen Z., Evans A. Structural insights into aberrant topological patterns of large-scale cortical networks in Alzheimer's disease. J. Neurosci. 2008;28:4756–4766. doi: 10.1523/JNEUROSCI.0141-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington H., Kuzniecky R., Vives K., Devinsky O., Pacia S., Luciano D., Vasquez B., Haut S., Spencer D., Pan J. A subcortical network of dysfunction in TLE measured by magnetic resonance spectroscopy. Neurology. 2007;69:2256–2265. doi: 10.1212/01.wnl.0000286945.21270.6d. [DOI] [PubMed] [Google Scholar]
- van den Heuvel M.P., Stam C.J., Kahn R.S., Hulshoff Pol H.E. Efficiency of functional brain networks and intellectual performance. J. Neurosci. 2009;29:7619–7624. doi: 10.1523/JNEUROSCI.1443-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel M.P., de Lange S.C., Zalesky A., Seguin C., Yeo B.T.T., Schmidt R. Proportional thresholding in resting-state fMRI functional connectivity networks and consequences for patient-control connectome studies: issues and recommendations. NeuroImage. 2017;152:437–449. doi: 10.1016/j.neuroimage.2017.02.005. [DOI] [PubMed] [Google Scholar]
- Jansen J.F., Backes W.H., Nicolay K., Kooi M.E. 1H MR spectroscopy of the brain: absolute quantification of metabolites. Radiology. 2006;240:318–332. doi: 10.1148/radiol.2402050314. [DOI] [PubMed] [Google Scholar]
- Jenkinson M., Smith S. A global optimisation method for robust affine registration of brain images. Med. Image Anal. 2001;5:143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- Jenkinson M., Bannister P., Brady M., Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Jenkinson M., Beckmann C.F., Behrens T.E., Woolrich M.W., Smith S.M. Fsl. NeuroImage. 2012;62:782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Jiang X., Shen S., Cadwell C.R., Berens P., Sinz F., Ecker A.S., Patel S., Tolias A.S. Principles of connectivity among morphologically defined cell types in adult neocortex. Science. 2015;350 doi: 10.1126/science.aac9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.K. Studying connections in the living human brain with diffusion MRI. Cortex. 2008;44:936–952. doi: 10.1016/j.cortex.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Kraguljac N.V., Reid M.A., White D.M., den Hollander J., Lahti A.C. Regional decoupling of N-acetyl-aspartate and glutamate in schizophrenia. Neuropsychopharmacology. 2012;37:2635–2642. doi: 10.1038/npp.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreis R. The trouble with quality filtering based on relative Cramer-Rao lower bounds. Magn. Reson. Med. 2016;75:15–18. doi: 10.1002/mrm.25568. [DOI] [PubMed] [Google Scholar]
- Kreis R., Bolliger C.S. The need for updates of spin system parameters, illustrated for the case of gamma-aminobutyric acid. NMR Biomed. 2012;25:1401–1403. doi: 10.1002/nbm.2810. [DOI] [PubMed] [Google Scholar]
- Li C., Xu C., Anderson A.W., Gore J.C. MRI tissue classification and bias field estimation based on coherent local intensity clustering: a unified energy minimization framework. Inf. Process. Med. Imaging. 2009;21:288–299. doi: 10.1007/978-3-642-02498-6_24. [DOI] [PubMed] [Google Scholar]
- Marques J.P., Kober T., Krueger G., van der Zwaag W., Van de Moortele P.F., Gruetter R. MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field. NeuroImage. 2010;49:1271–1281. doi: 10.1016/j.neuroimage.2009.10.002. [DOI] [PubMed] [Google Scholar]
- O'Gorman R.L., Michels L., Edden R.A., Murdoch J.B., Martin E. In vivo detection of GABA and glutamate with MEGA-PRESS: reproducibility and gender effects. J. Magn. Reson. Imaging. 2011;33:1262–1267. doi: 10.1002/jmri.22520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordidge R.J., Wylezinska M., Hugg J.W., Butterworth E., Franconi F. Frequency offset corrected inversion (FOCI) pulses for use in localized spectroscopy. Magn. Reson. Med. 1996;36:562–566. doi: 10.1002/mrm.1910360410. [DOI] [PubMed] [Google Scholar]
- Öz G., Alger J.R., Barker P.B., Bartha R., Bizzi A., Boesch C., Bolan P.J., Brindle K.M., Cudalbu C., Dinçer A. Clinical proton MR spectroscopy in central nervous system disorders. Radiology. 2014;270:658–679. doi: 10.1148/radiol.13130531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J.W., Spencer D.D., Kuzniecky R., Duckrow R.B., Hetherington H., Spencer S.S. Metabolic networks in epilepsy by MR spectroscopic imaging. Acta Neurol. Scand. 2012;126:411–420. doi: 10.1111/j.1600-0404.2012.01665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.J., Friston K. Structural and functional brain networks: from connections to cognition. Science. 2013;342 doi: 10.1126/science.1238411. [DOI] [PubMed] [Google Scholar]
- Petroff O. Biochemistry for magnetic resonance spectroscopy. In: Kuzniecky R.I., Jackson G.D., editors. Magnetic Resonance in Epilepsy: Neuroimaging Techniques. Academic Press; 2005. [Google Scholar]
- Provencher S. 2014. LCModel & LCMgui User's Manual. (LCModel Version, 6.3-1H). [Google Scholar]
- Rae C.D. A guide to the metabolic pathways and function of metabolites observed in human brain 1H magnetic resonance spectra. Neurochem. Res. 2014;39:1–36. doi: 10.1007/s11064-013-1199-5. [DOI] [PubMed] [Google Scholar]
- Richiardi J., Altmann A., Milazzo A.-C., Chang C., Chakravarty M.M., Banaschewski T., Barker G.J., Bokde A.L., Bromberg U., Büchel C. Correlated gene expression supports synchronous activity in brain networks. Science. 2015;348:1241–1244. doi: 10.1126/science.1255905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saggar M., Hosseini S.M., Bruno J.L., Quintin E.M., Raman M.M., Kesler S.R., Reiss A.L. Estimating individual contribution from group-based structural correlation networks. NeuroImage. 2015;120:274–284. doi: 10.1016/j.neuroimage.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheenen T.W., Heerschap A., Klomp D.W. Towards 1H-MRSI of the human brain at 7 T with slice-selective adiabatic refocusing pulses. MAGMA. 2008;21:95–101. doi: 10.1007/s10334-007-0094-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek O., Jensen D. Navigating networks by using homophily and degree. Proc. Natl. Acad. Sci. U. S. A. 2008;105:12758–12762. doi: 10.1073/pnas.0800497105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.M. Fast robust automated brain extraction. Hum. Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soher B., Semanchuk P., Todd D., Steinberg J., Young K. Proceedings of 19th Annual Meeting ISMRM. 2011. VeSPA: integrated applications for RF pulse design, spectral simulation and MRS data analysis; p. 1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M., Zhou Y., Li J., Liu Y., Tian L., Yu C., Jiang T. Brain spontaneous functional connectivity and intelligence. NeuroImage. 2008;41:1168–1176. doi: 10.1016/j.neuroimage.2008.02.036. [DOI] [PubMed] [Google Scholar]
- Spencer S.S. Neural networks in human epilepsy: evidence of and implications for treatment. Epilepsia. 2002;43:219–227. doi: 10.1046/j.1528-1157.2002.26901.x. [DOI] [PubMed] [Google Scholar]
- Stam C.J., van Straaten E.C. The organization of physiological brain networks. Clin. Neurophysiol. 2012;123:1067–1087. doi: 10.1016/j.clinph.2012.01.011. [DOI] [PubMed] [Google Scholar]
- Team R.C. Austria; Vienna: 2015. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Tkac I. Ontario, Canada; Toronto: 2008. Refinement of Simulated Basis Set for LCModel Analysis. Proceedings of the 16th Annual Meeting of ISMRM; p. 1624. [Google Scholar]
- Tkac I., Oz G., Adriany G., Ugurbil K., Gruetter R. In vivo 1H NMR spectroscopy of the human brain at high magnetic fields: metabolite quantification at 4T vs. 7T. Magn. Reson. Med. 2009;62:868–879. doi: 10.1002/mrm.22086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veenendaal T.M., IJff D.M., Aldenkamp A.P., Hofman P.A., Vlooswijk M.C., Rouhl R.P., de Louw A.J., Backes W.H., Jansen J.F. Metabolic and functional MR biomarkers of antiepileptic drug effectiveness: a review. Neurosci. Biobehav. Rev. 2015;59:92–99. doi: 10.1016/j.neubiorev.2015.10.004. [DOI] [PubMed] [Google Scholar]
- van Veenendaal T.M., Ijff D.M., Aldenkamp A.P., Lazeron R.H.C., Puts N.A.J., Edden R.A.E., Hofman P.A.M., de Louw A.J.A., Backes W.H., Jansen J.F.A. Glutamate concentrations vary with antiepileptic drug use and mental slowing. Epilepsy Behav. 2016;64:200–205. doi: 10.1016/j.yebeh.2016.08.027. [DOI] [PubMed] [Google Scholar]
- Waddell K.W., Zanjanipour P., Pradhan S., Xu L., Welch E.B., Joers J.M., Martin P.R., Avison M.J., Gore J.C. Anterior cingulate and cerebellar GABA and Glu correlations measured by (1)H J-difference spectroscopy. Magn. Reson. Imaging. 2011;29:19–24. doi: 10.1016/j.mri.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright K. 2016. corrgram: Plot a Correlogram. R Package Version 1.10. [Google Scholar]
- Zhang Y., Brady M., Smith S. Medical Imaging, IEEE Transactions on. Vol. 20. 2001. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm; pp. 45–57. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2