Summary
Mycobacterium tuberculosis (Mtb) adaptation to hypoxia is considered crucial to its prolonged latent persistence in humans. Mtb lesions are known to contain physiologically heterogeneous microenvironments that bring about differential responses from bacteria. Here we exploit metabolic variability within biofilm cells to identify alternate respiratory polyketide quinones (PkQs) from both Mycobacterium smegmatis (Msmeg) and Mtb. PkQs are specifically expressed in biofilms and other oxygen-deficient niches to maintain cellular bioenergetics. Under such conditions, these metabolites function as mobile electron carriers in the respiratory electron transport chain. In the absence of PkQs, mycobacteria escape from the hypoxic core of biofilms and prefer oxygen-rich conditions. Unlike the ubiquitous isoprenoid pathway for the biosynthesis of respiratory quinones, PkQs are produced by type III polyketide synthases using fatty acyl-CoA precursors. The biosynthetic pathway is conserved in several other bacterial genomes, and our study reveals a redox-balancing chemicocellular process in microbial physiology.
Introduction
Tuberculosis (TB) has been recognized since antiquity and remains one of the leading causes of infectious disease mortality. According to a World Health Organization (WHO) estimate, about two billion people are infected asymptomatically with Mycobacterium tuberculosis (Mtb), the causative agent of TB (Zumla et al., 2013). About 5%–10% of these individuals develop active TB, and many are infected with multi-drug resistant (MDR) strains. The underlying challenge is to find ways to eliminate the asymptomatic population, which will prevent reactivation of latent TB cases into active cases. Recent studies reveal the significance of physiological heterogeneity in the microbial population, which enables a subpopulation of bacteria to withstand immune and antibiotic pressures and may, therefore, contribute toward persistence and reactivation (Lidstrom and Konopka, 2010; Manina et al., 2015).
Biofilm formation, an adaptive feature common to several microbes, has been proposed to form gradients of differential nutrient availability and also of varying concentrations of signaling molecules, oxygen, and protons (Vaupel et al., 1989). In such aggregated architectures, diversity within microbial populations has been proposed to drive bacterial community robustness and construct a multicellular lifestyle (Hall-Stoodley et al., 2004; O’Toole et al., 2000). Much of the present knowledge of biofilm formation has emerged from fast-growing, genetically tractable bacterial species like Bacillus, Vibrio, and Pseudomonas (Harmsen et al., 2010; Teschler et al., 2015; Vlamakis et al., 2013). An important requirement for bacterial biofilm formation is a gradual controlled decrease in oxygen levels, providing time for cells to adapt (de Beer et al., 1994). Such microaerophilic conditions impose it on bacterial cells to remodel their respiratory metabolism because oxygen is the terminal electron acceptor during aerobic respiration (Poole and Cook, 2000; Rao et al., 2008). Interestingly, some of the features observed during biofilm formation resemble the physiological adaptation of non-replicating persisters of Mtb during hypoxic conditions (Zhang et al., 2012). In fact, Mtb biofilms have been utilized recently as a cellular screen to identify lead compounds against MDR strains (Wang et al., 2013).
Mycobacteria form a variety of aggregated assemblages such as clumped colonies, granulomas, and both floating (pellicles) as well as surface-adhered biofilms (Guirado and Schlesinger, 2013; Islam et al., 2012; Ramakrishnan, 2012; Zambrano and Kolter, 2005). These biofilms have been reported to accumulate extracellular free mycolates and are also known to harbor drug-tolerant bacilli (Ojha et al., 2008). Tubercle bacilli are known to maintain a respiratory-competent energized membrane in oxygen-depleted niches. However, the intracellular ATP concentration is reduced severalfold (Eoh and Rhee, 2013; Rao et al., 2008). In low oxygen concentrations, Mtb utilizes other respiratory components, and there is a shift to alternate NADH dehydrogenases and cytochrome oxidases (Kana et al., 2001; Shi et al., 2005). Surprisingly, the only known respiratory electron carrier quinone from mycobacteria, menaquinone (MK), is downregulated markedly (Honaker et al., 2010). Although several Gram-negative bacteria are known to switch between ubiquinone (CoQ) and MK with decreasing oxygen concentrations, this mode of exchange between isoprenoid quinones is absent in Gram-positive species (Meganathan, 2001; Schoepp-Cothenet et al., 2009).
In this study, we identify alternate electron shuttling molecules in mycobacteria that support bacterial survival in oxygen-depleted habitats. Unlike canonical respiratory quinones, which are derived from the isoprenoid pathway, these molecules are produced by a polyketide synthase-based pathway. This biosynthetic system is conserved in all mycobacteria as well as in several Actinobacteria and a few Proteobacteria, suggesting a key role of these quinones in bacterial adaptability.
Results
The Transition from a Free-Living to a Microbial Community Induces Hypoxic Cellular Responses in Mycobacteria
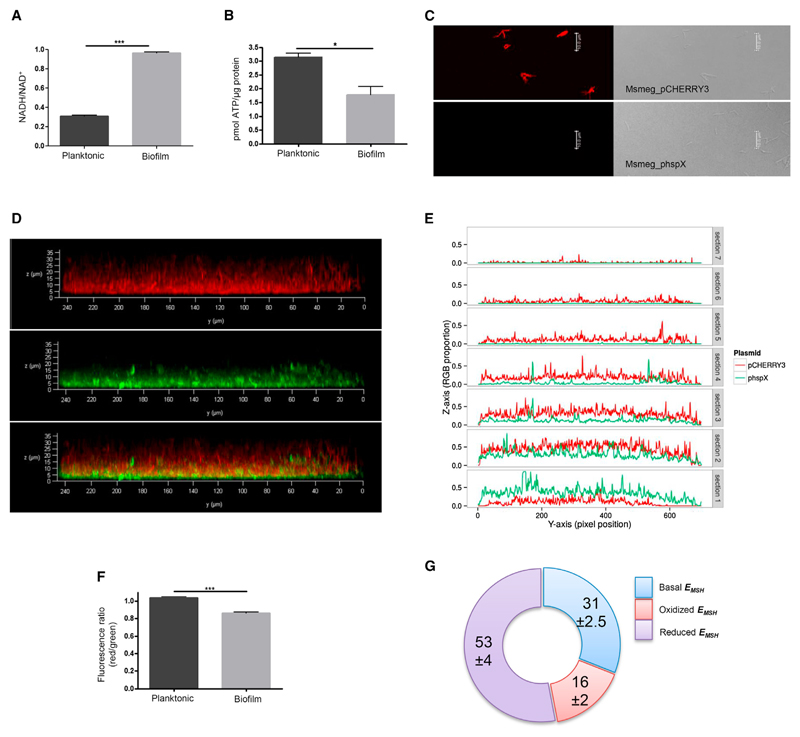
Physiological heterogeneity within biofilms has been largely attributed to decreased oxygen levels in Staphylococcus and Pseudomonas (Borriello et al., 2004; Rani et al., 2007; Williamson et al., 2012). Sustained hypoxic conditions can slow down respiratory electron transport, augmenting the reduced state of electron carriers (Mailloux et al., 2007). To understand the overall physiological status of mycobacterial cells in biofilms, we measured the concentrations of two key cofactors: ATP and NADH. Analysis of the NADH/NAD+ ratio between planktonic and biofilm conditions showed an approximately 3-fold increase in the biofilms (Figure 1A). At the same time, total cellular ATP levels in biofilms were also reduced to half in comparison with planktonic conditions (Figure 1B). The accumulation of NADH and decreased ATP levels in biofilms suggest impaired respiration and, taken together, are indicative of augmented reductive stress. To assess oxygen levels within biofilm layers, we followed the promoter activity of hspX, which has been used previously to estimate hypoxia in mycobacteria (Desjardin et al., 2001; Sherman et al., 2001). Although hspX promoter-driven expression of GFP was completely absent in planktonic cells, significant expression could be observed in the biofilm (Figures 1C and 1D). To mark different layers of mycobacterial biofilms, we separately expressed red fluorescent protein (RFP) under a constitutive promoter in Mycobacterium smegmatis (Msmeg) cells. A clear expression of RFP could be seen even under planktonic conditions (Figure 1C). We then combined these two mycobacterial strains (green and red) in equal proportions and developed biofilms (Figure 1D). Analysis of the biofilm using confocal microscopy in the red and green channel along with co-localization is shown in Figure 1E. A z stack analysis showed red fluorescence throughout the thickness of the biofilm, whereas green fluorescence was limited to the inner layers. Color intensity measurements across seven stacks (each 5 μm) of biofilm showed RFP throughout the biofilm. In contrast, GFP expression was primarily observed in the bottom three stacks that were far from the air boundary of the biofilm and almost no GFP expression in the top three layers of the biofilm (Figure 1E). These data provide evidence for a gradient of oxygen depletion across the biofilm layers.
Figure 1. Hypoxic Cellular Responses in Mycobacterial Biofilm.
(A) Estimation of intracellular NADH/NAD+ levels in planktonic and biofilm cultures.
(B) Level of intracellular ATP in planktonic and biofilm cultures.
(C) Fluorescence and bright-field images of Msmeg WT-pCHERRY3 and Msmeg WT-hspX under planktonic conditions.
(D) A z stack view of biofilm formed by a mixture of Msmeg WT-pCHERRY3 and Msmeg WT-phspX. The top shows the red channel, the center shows the green channel, and the bottom displays co-localization of the two.
(E) Quantitative assessment of the fluorescence of pCHERRY3 and hspX in the mixed-culture biofilm.
(F) Assessment of the membrane potential of cells in planktonic and biofilm modes of growth based on potential dependent internalization of the carbocyanine dye 3,3′-diethyloxacarbocyanine iodide.
(G) Estimation of EMSH of the cells in biofilm compared with planktonic cells.
Statistical significance was analyzed by two-tailed Student’s t test. *p < 0.05, ***p < 0.001. All error bars represent ± SEM. See also Figure S1.
Low oxygen concentrations, along with an impaired respiratory pathway, have been shown to alter the redox status in mycobacterial cells (Davidson and Schiestl, 2001; Leistikow et al., 2010). We therefore probed the membrane potential in Msmeg biofilms using internalization of the carbocyanine dye DiOC2. Ratiometric analysis did indeed reveal a relative decrease in the membrane potential of the biofilm cells compared with planktonic cultures (Figure 1F). The redox homeostasis in mycobacteria is primarily maintained by mycothiol (MSH) (Newton et al., 2008). Because perturbation in the levels of MSH will affect redox homeostasis, we examined the ability of Msmeg mutants of the MSH pathway to form biofilms. Msmeg mutants in the MSH pathway—ΔmshA (MSH glycosyltransferase), ΔmshD (MSH acetyltransferase), Δmca (MSH S-conjugate amidase), and Δmtr (MSH disulfide reductase)—all showed a poor ability to form biofilm pellicles (Figure S1A). Each of these mutants, however, showed no significant differences in their planktonic growth (Figure S1B). This phenotypic analysis was further substantiated by quantitatively measuring the mycothiol redox potential (EMSH) using the MSH-specific redox sensor Mrx1-roGFP2 (Bhaskar et al., 2014). The ratio of emission at 510 nm after excitation at 405 and 488 nm was calculated to estimate differences in planktonic and biofilm cells. Interestingly, approximately 70% of mycobacterial cells showed perturbed MSH homeostasis, with more than 50% showing reduced EMSH (Figure 1G). 31% of biofilm cells showed EMSH similar to planktonic cells. The results, taken together, show an altered redox environment and cellular bioenergetics because of differential oxygen partial pressure across the mycobacterial biofilm.
The Quest for a Novel Quinone Metabolite in Msmeg
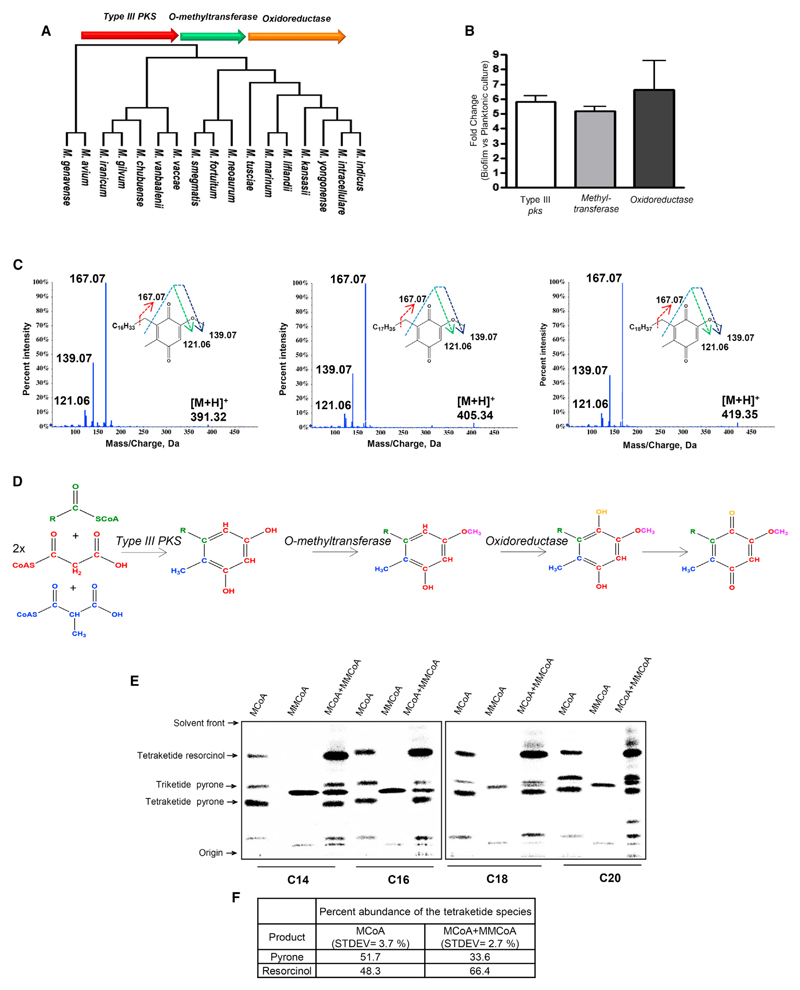
Microbes dissipate terminal electrons through a variety of extracellular electron acceptors. However, the transfer of electrons between various respiratory complexes requires intermediate isoprenoid quinone electron carriers. The downregulation of the central electron carrier MK under hypoxic condition raised questions regarding the maintenance of the respiratory electron chain. Deactivation of the aerophilic branch of respiration in mycobacteria results in constitutive activation of the microaerophilic branch, but the gene involved in the nonmevalonate pathway of isoprenoid biosynthesis is downregulated drastically (Matsoso et al., 2005). This further confounds the MK dependence of mycobacterial respiration. In a recent study in Streptomyces griseus, a new pathway for producing phenolic lipids has been reported (Funabashi et al., 2008). Interestingly, these metabolites contain a benzoquinone or a pyrone scaffold with a long alkyl chain. The benzoquinone ring is derived from an alkyl resorcinol formed by type III polyketide synthase (PKS) that is modified by two downstream enzymes, all of which are present in an operon. However, the functional role of these metabolites could not be ascribed because these could only be isolated from recombinant bacteria by heterologous expression of the three-gene operon (Funabashi et al., 2008). Computational analysis identified a homologous operon, MSMEG_0808 (type III pks), MSMEG_0809 (methyltransferase), and MSMEG_0811 (oxidoreductase), in Msmeg. We therefore hypothesized that Msmeg could also produce a similar metabolite. Although this biosynthetic operon was conserved across several mycobacterial species, this cluster was surprisingly missing from the three major pathogenic strains Mtb, Mycobacterium leprae, and Mycobacterium bovis (Figure 2A).
Figure 2. Type III pks Cluster-Derived Alkyl benzoquinones from Msmeg Biofilm.
(A) Mycobacterial species containing a type III pks operon in their genomes.
(B) Real-time expression analysis of the pks10 cluster in Msmeg biofilm relative to planktonic culture.
(C) Tandem-MS characterization of the alkyl benzoquinones isolated from Msmeg biofilm.
(D) Biosynthetic pathway for the alkyl benzoquinones.
(E) Autoradiogram of radio-TLC showing product formed by Msmeg PKS10 using different fatty acyl-CoAs as the starter unit and radiolabeled MCoA and/or MMCoA as the extender unit(s).
(F) Quantitation of tetraketide pyrone and tetraketide resorcinol products formed by PKS10 using only MCoA or a mixture of MCoA and MMCoA.
See also Figures S2–S5.
Our initial attempts to isolate phenolic lipids in shake-flask planktonic cultures were unsuccessful. Overexpression of all three proteins in Msmeg also did not yield the expected metabolites. In a previous study, we reported a novel type III PKS from mycobacteria that utilize long-chain acyl coenzyme A (CoA) substrates (Saxena et al., 2003). Because exogenous supplementation of precursors is known to enhance the production of metabolites, we added palmitate to these cultures. Analysis of ethyl acetate extracts of the recombinant strain showed several new peaks in the high-performance liquid chromatography (HPLC) chromatograms (Figure S2B). High-resolution electrospray ionization mass spectrometry (HR-ESI-MS) characterization revealed the presence of triketide and tetraketide pyrones and tetraketide resorcinol (Figure S2C). Surprisingly, none of these metabolites were modified by the two downstream enzymes to produce quinones. Feeding with oleate or arachidonate produced metabolites that incorporated the respective fatty acyl chain (Figures S3A–S3D). The requirement of overexpression as well as feeding of precursors strongly suggested that this pathway might not be operational during shake-flask culture conditions.
Characterization of Alkyl benzoquinones from Msmeg Biofilms
The altered respiratory metabolism of mycobacteria in biofilms prompted us to explore expression of these PKS-derived quinones in these aggregated assemblages. Comparative transcriptional profiling of the type III pks three-gene cluster MSMEG_0808 (pks10), MSMEG_0809 (O-methyltransferase), and MSMEG_0811 (oxidoreductase) under planktonic and biofilm conditions showed significant upregulation of all three genes in biofilms (Figure 2B). Ethyl acetate extracts of cultures identified three peaks specifically in the biofilm extracts (Figure S4A). LC-HR-ESI-MS analysis identified molecular ion peaks of m/z 391.3199, 405.3355, and 419.3512 Da, all of which show common fragmentation pattern in tandem MS analysis producing [M+H+] ions of 167.07, 139.07 and 121.06 Da (Figure 2C). These signature peaks are identical to a previously reported characterization of alkyl benzoquinones, and the difference of 14 Da corresponds to “-CH2-” unit differences in the alkyl chain. The two-dimensional [13C, 1H] heteronuclear single quantum coherence (HSQC) nuclear magnetic resonance (NMR) spectrum showed a complete fingerprint of this benzoquinone metabolite, further confirming the structure of the metabolites (Figure S4B). Chemical syntheses of two analogs corresponding to an alkyl chain length of 16 and 18 carbons further provided conclusive chemical identification (Figures S4C and S4D).
Biochemical Characterization of Msmeg Type III PKS
The logic for alkyl benzoquinone biosynthesis suggested that alkyl resorcinol produced by type III PKS may involve two different types of extender units. Although utilization of two different extender units is rather unusual for iterative PKS systems, a similar mode of catalysis has also been reported for S. griseus PKS10 (Funabashi et al., 2008). We cloned and expressed MSMEG_0808 (pks10) in Escherichia coli. Cell-free enzymatic assays with purified PKS10 protein using medium- and long-chain aliphatic-CoA as the starter unit and radioactive malonyl-CoA (MCoA) or methyl malonyl-CoA (MMCoA) as the extender unit provided the expected products, as analyzed by radioactive thin-layer chromatography (radio-TLC) (Figure 2E) and mass spectrometry (Figure S5). Although MCoA-derived metabolites produced both acyl pyrone and alkyl resorcinol, reactions with MMCoA primarily produced methylated pyrones. Interestingly, when both MCoA and MMCoA were used together in the assay, there was a substantial increase in the amount of resorcinolic product (Figures 2E and 2F). Our study therefore provides evidence for the formation of alkyl benzoquinones by mycobacterial type III PKS and shows that the methyl-branching on the ring arises from the specific incorporation of MMCoA during iterative catalysis.
Alkyl benzoquinone Biosynthesis in Mtb
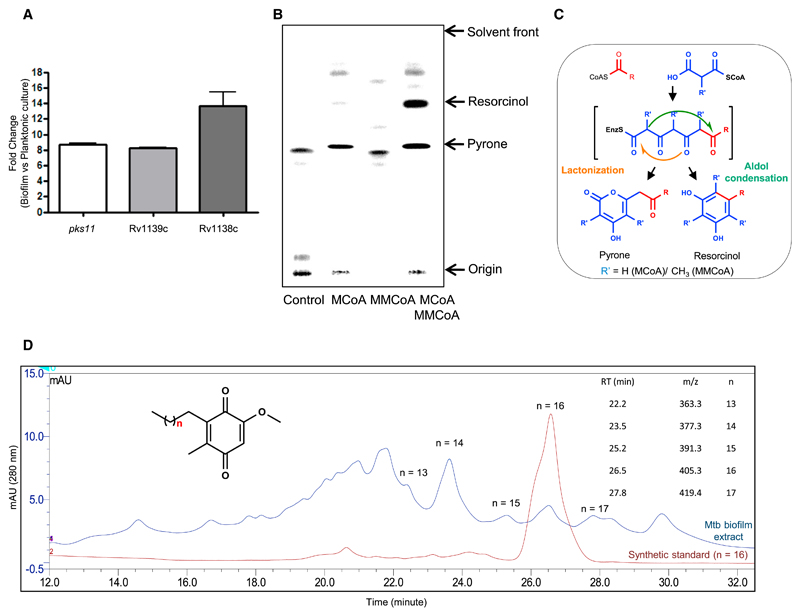
The absence of an alkyl benzoquinone biosynthetic cluster in pathogenic mycobacteria was intriguing because biofilm formation is a common feature to all mycobacterial species. Protein sequence analysis revealed close homologs of Msmeg enzymes in the Mtb genome. The putative O-methyltransferase (Rv1139c) and oxidoreductase (Rv1138c) are present together in the Mtb genome and show ~64% and 48% sequence identity with cognate Msmeg enzymes (MSMEG_0809 and MSMEG_0811, respectively). The Msmeg PKS10 homolog identified three type III PKS proteins in the Mtb genome, with a probability of the alignment to occur by chance for pks10 (Rv1660), pks11 (Rv1665), and pks18 (Rv1372) of 8e−169, 6e−172, and 5e−44, respectively. Transcriptional analysis showed upregulation of all three Mtb type III pks genes along with Rv1139c and Rv1138c in biofilms relative to planktonic cultures (Figure 3A; Figure S6). Interestingly, pks11, Rv1139c, and Rv1138c showed a similar (more than 8-fold) increase at the transcript level (Figure 3A). We had reported earlier that PKS18 and PKS11 produce alkyl and acyl pyrones by using long-chain acyl-CoA as starter and MCoA as extender units (Saxena et al., 2003). Careful analysis of the active-site residues of Msmeg PKS10, Mtb PKS10, Mtb PKS11, and Mtb PKS18 indicated the presence of a Trp residue at position 230 for Msmeg PKS10 and Mtb PKS11, whereas Mtb PKS10 and Mtb PKS18 contain Tyr and Leu at this position, respectively. Trp at this position in ArsB from Azotobacter has been shown to be crucial to biosynthesizing resorcinols (Satou et al., 2013). We therefore re-investigated the in vitro products formed by PKS11 and PKS18 in the presence of both MCoA and MMCoA. The product profile of Mtb PKS11 was completely identical to that of Msmeg PKS10, and substantial alkyl resorcinol synthesis could be observed when both MCoA and MMCoA were used simultaneously as extender units (Figure 3B). There was no difference in the activity of PKS18 even after addition of MMCoA to the reaction mixture. Incidentally, resorcinol and pyrone ring systems are both produced by the same tetraketide intermediate following different cyclizations (Figure 3C). It is interesting to note that the extra methyl group from MMCoA condensation in the last step favors cyclization via an aldol condensation. All of these bioinformatic studies as well as cell-free assays suggest that alkyl benzoquinones could also be formed by Mtb.
Figure 3. Type III pks Cluster-Derived Alkyl benzoquinones from Mtb Biofilm.
(A) Real-time expression analysis of the pks11, Rv1139c, and Rv1138c genes of Mtb.
(B) Autoradiogram of radio-TLC showing products formed by Mtb PKS11 using stearoyl-CoA as a starter unit and radiolabeled MCoA and/or MMCoA as an extender unit(s).
(C) Reaction mechanism for type III PKS-catalyzed formation of alkyl pyrone and alkyl resorcinol.
(D) Ultra-performance liquid chromatography (UPLC) chromatogram for alkyl benzoquinones isolated from Mtb biofilm. mAU, milliabsorbance unit.
See also Figure S6.
We therefore analyzed Mtb biofilm cultures for the presence of alkyl benzoquinones using LC-HR-ESI-tandem mass spectrometry (MS/MS). Gratifyingly, we could identify a series of alkyl benzoquinones with the alkyl chain length varying from C15 to C19 having m/z values of 363.2891, 377.3052, 391.3208, 405.3372, and 419.3524 (Figure 3D). All of these metabolites showed similar retention times as the Msmeg molecules and also had identical fragmentation spectra. The difference of 14 units corresponded to the additional -CH2- unit difference in the alkyl chain. Moreover, the retention time for the C18 compound was identical to the synthetic standard.
Physiological Significance of Alkyl benzoquinones
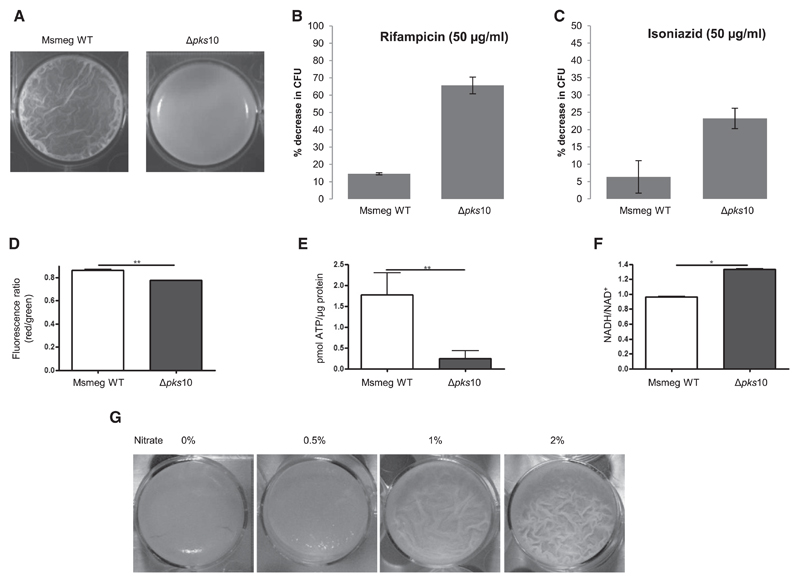
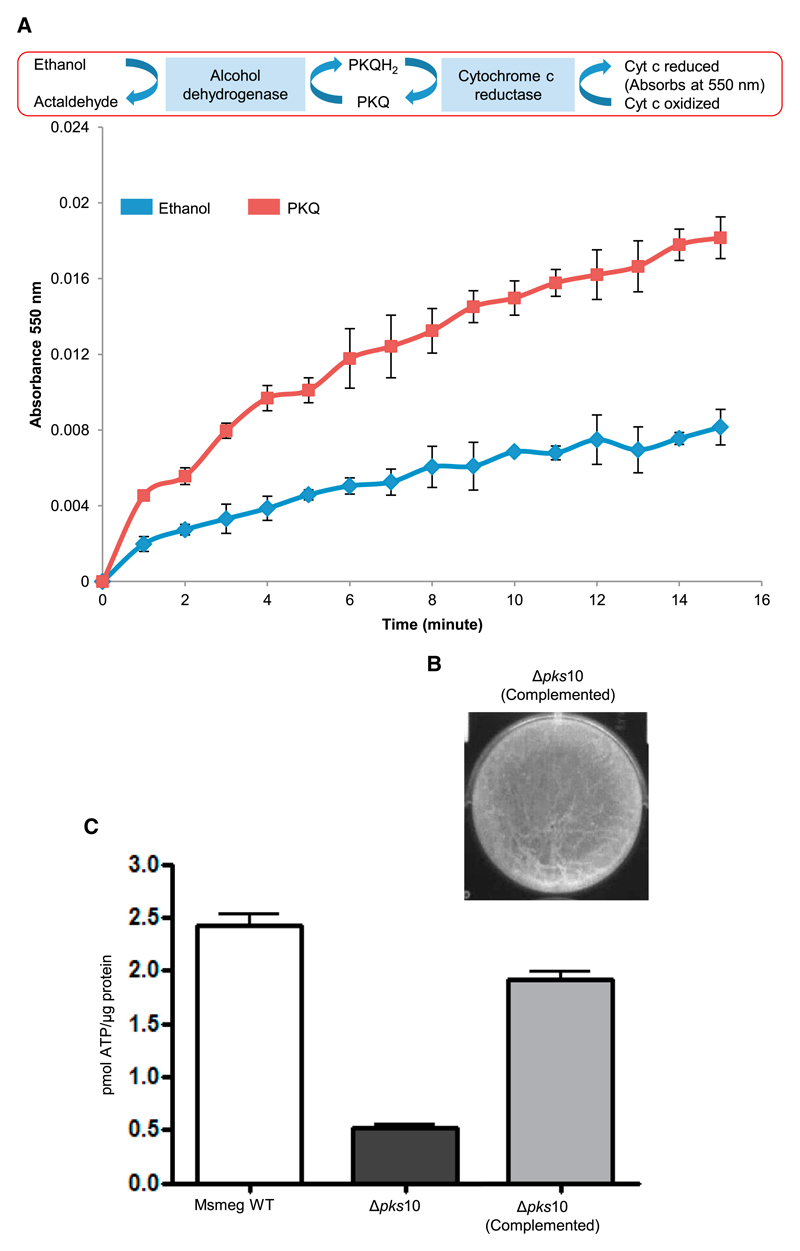
To establish the physiological role of alkyl benzoquinones, we generated a pks10 knockout (Δpks10) strain in Msmeg (Figures S7A and S7B). As anticipated, the mutant Δpks10 strain showed no growth profile defect under planktonic conditions (Figure S7C). However, the mutant strain showed stark differences under biofilm conditions and formed a fragile film lacking the characteristic reticulation of wild-type (WT) biofilm (Figure 4A). Scanning electron microscopy studies also confirmed a non-porous clumpy surface for Δpks10, indicating collapse of the community structure (Figure S7D). The role of alkyl benzoquinones in biofilms was also examined by performing antibiotic sensitivity comparisons between WT and Δpks10 strains. Antibiotic sensitivity assays with rifampicin and isoniazid showed no significant differences for the WT and mutant Msmeg under planktonic conditions (Figure S7E). In contrast, the Δpks10 strain showed an approximately 4-fold higher sensitivity toward both antibiotics (Figures 4B and 4C).
Figure 4. Effect of pks10 Gene Knockout on the Mycobacterial Biofilm.
(A) Biofilm formed by the WT and Δpks10 strains.
(B) Percent decrease in colony-forming units (CFUs) upon treatment of Msmeg biofilm with rifampicin.
(C) Percent decrease in CFUs upon treatment of Msmeg biofilm with isoniazid.
(D) Assessment of the membrane potential of cells in biofilm cultures of the WT and Δpks10 strain based on the potential dependent internalization of the carbocyanine dye 3,3′-diethyloxacarbocyanine iodide.
(E) Level of intracellular ATP in biofilm cultures of the WT and Δpks10 strain.
(F) Estimation of intracellular NADH/NAD+ levels in biofilm cultures of the WT and Δpks10 strains.
(G) Effect of exogenous addition of nitrate on the biofilm of the Msmeg Δpks10 strain.
Statistical significance was analyzed by two-tailed Student’s t test. *p < 0.05, **p < 0.01. All error bars represent ± SEM. See also Figure S7.
We then examined the levels of essential cofactors between mutant and wild-type strains during planktonic and biofilm culture conditions (Figures 4D–4F; Figures S7F–S7H). Mutant biofilms showed decreased membrane potential and a several fold decrease in ATP levels compared with the WT (Figures 4D and 4E). The ratios of NADH/NAD+ were also elevated significantly in the Δpks10 strain, indicating accumulation of reducing equivalents in the absence of these quinones (Figure 4F). These comparative analyses between WT and Δpks10 suggested the requirement of alkyl benzoquinones for maintaining cellular respiration in biofilms. Furthermore, we examined whether exogenous addition of alternate electron acceptors such as nitrates could improve biofilm formation. Addition of nitrates recovered biofilm robustness, stability, and reticulation pattern (Figure 4G), probably through dissipation of excess accumulated electrons.
The Role of Alkyl benzoquinones in Respiration
The role of alkyl benzoquinones in electron transport was established biochemically by performing in vitro reconstitution of a component of the electron transport chain. In this cell-free assay, cytochrome c reduction kinetics were monitored by measuring absorbance at 550 nm (Figure 5A). To maintain a continuous supply of electrons to alkyl benzoquinone, alcohol dehydrogenase-mediated oxidation of ethanol was coupled to the reduction of cytochrome c reductase. Electrons transferred from alkyl benzoquinone to cytochrome c reductase, in turn, resulted in the reduction of cytochrome c. This coupled assay provided strong evidence for the biochemical potential to function as an electron carrier in the respiratory electron transport chain. Furthermore, chemical complementation of mutant biofilms with a synthetic alkyl benzoquinone standard restored biofilm architecture and also resulted in more than a 75% recovery in ATP level (Figures 5B and 5C). In contrast to ubiquitous respiratory isoprenoid quinones, these metabolites are derived from a PKS biosynthetic pathway, and we name them here polyketide quinones (PkQs).
Figure 5. Physiological Significance of Alkyl benzoquinones.
(A) Cytochrome c (Cyt c) reduction by alkyl benzoquinone mediated via cytochrome c reductase.
(B) Restoration of the reticulated surface phenotype upon complementation with an alkyl benzoquinone.
(C) Recovery of the intracellular ATP level in biofilm cultures of the Δpks10 strain upon complementation with an alkyl benzoquinone.
The Importance of Polyketide Quinones in General Hypoxia
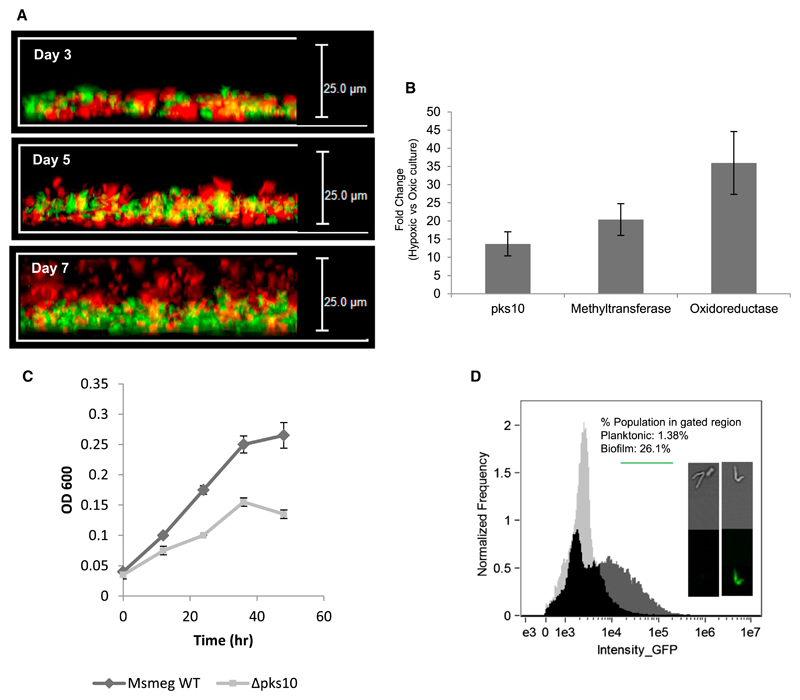
Our results, in conjunction with several recent studies with biofilms from other bacteria (Borriello et al., 2004; Rani et al., 2007; Williamson et al., 2012), provide compelling evidence for the presence of hypoxic conditions in biofilms. Because PkQ restores microaerophilic respiration, we reasoned that the Δpks10 mutant may not be able to inhabit the core oxygen-deficient zones. To study this property, we mixed WT and mutant strains of Msmeg and examined their preferential localization in biofilms. To demarcate WT and Δpks10 cells, we episomally expressed GFP and RFP proteins, respectively, in these strains. The kinetics of the biofilm formation was followed by examining the distribution of mutant and WT cells in different layers by using fluorescence microscopy. Remarkably, with progressive growth of the biofilm, the mutant strain began to segregate from the WT strain and eventually occupied the upper layers on day 7 (Figure 6A). Although this study demonstrates distinct fitness differences between the WT and mutant Msmeg strains in the hypoxic biofilm, the mechanism of this segregation remains unknown. The absence of WT Msmeg from the top layers could also be due to the formation of a loose layer of mutant cells that fail to form a sufficiently dense structural matrix barrier to gaseous diffusion.
Figure 6. Hypoxia and the type III pks Gene Cluster.
(A) Localization of Msmeg WT (green) and Msmeg Δpks10 (red) strains along the vertical layers of biofilm.
(B) qRT-PCR expression analysis of the type III pks gene cluster under hypoxic conditions.
(C) Planktonic growth curve of the Msmeg WT and Δpks10 strains under hypoxic conditions.
(D) pks10 promoter activity under planktonic and biofilm conditions. The frequency versus intensity plots for the two cultures were overlapped to show the exclusive GFP-positive population of the biofilm (dark gray).
The expression of PkQ under oxygen-depleted conditions was also examined by growing planktonic cultures of Msmeg under hypoxic conditions. We observed a remarkable increase in transcripts of the PkQ biosynthetic cluster, suggesting a role of PkQ under these microaerophilic conditions (Figure 6B). The Δpks10 strain, which earlier showed no growth difference under aerobic conditions, showed a clear growth defect in this hypoxic environment compared with Msmeg WT (Figure 6C). To further provide support for the expression of PkQ under oxygen-limiting conditions, we probed pks10 promotor-driven expression of GFP. We replaced the hsp60 promotor in the pMV261-HA vector with a 500-base pair (bp) upstream sequence of Msmeg pks10. The expression of GFP was monitored using an Amnis imaging flow cytometer. The analysis of the gated population in planktonic and biofilm cultures showed a clear enhancement (18-fold) of GFP-positive cells (Figure 6D), again providing support for the enhanced expression of the PkQ biosynthetic cluster in biofilms. We then examined pks10 promoter-driven GFP expression under hypoxic planktonic growth conditions. Imaging flow cytometry analysis again showed a more than 4-fold increase in the expression of green cells. Together, our studies demonstrate that PkQs are also expressed under oxygen-depleted conditions to support the growth of mycobacteria.
Phylogenetic Conservation of Polyketide Quinone Biosynthetic Machinery
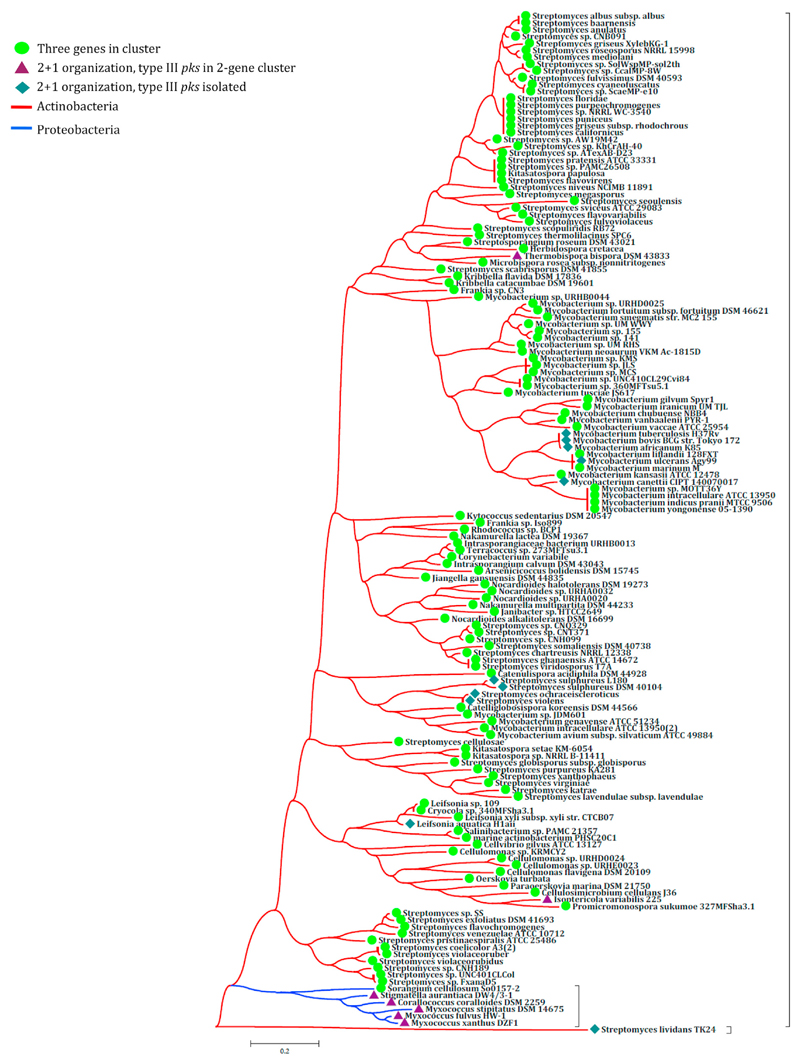
Formation of multicellular communities by microbes is a common feature of several bacteria. We therefore examined whether this type III pks biosynthetic cluster is conserved in other bacteria. Analysis of other mycobacterial genomes revealed the presence of a similar biosynthetic cluster in more than 14 mycobacterial species, including Mycobacterium marinum, and Mycobacterium avium. Genome organization similar to Mtb is also observed in M. bovis, whereas these are present as pseudogenes in M. leprae. The genome of M. leprae, in fact, has been suggested to have undergone massive gene decay, particularly in the anaerobic and microaerophilic respiratory modules (Cole et al., 2001). We then queried genome databases for these two types of genomic organization and examined phylogenetic conservation based on the criterion of 60% protein sequence coverage alignment with an e value of less than 10−5. Our analysis revealed conservation of this pathway in several Actinobacteria and a few Proteobacteria (Figure 7). Three different types of genomic organization for this cluster were observed. In some species, all three genes were clustered together, whereas several species had only two genes next to each other in the genomic locus. The two-gene organization differed in the juxtaposed genes; either a type III PKS and methyltransferase were together, or methyltransferase and oxidoreductase were present next to one another.
Figure 7. Phylogenetic Conservation of the type III pks Gene Cluster.
The phylogenetic tree depicts the selective enrichment of the three-membered type III pks gene cluster in different bacterial classes. Circles, triangles, or diamonds at each node denote different genome organizations of the three genes of the type III PKS cluster.
Discussion
Mtb is anticipated to encounter a variety of intracellular stresses. Among the many challenging conditions, differential oxygen levels can elicit comprehensive changes in mycobacterial physiology (Barry et al., 2009; Boshoff and Barry, 2005). During these adaptations, Mtb has to sustain respiration and generate energy from various sources (Eoh and Rhee, 2013; Shi et al., 2005). Not surprisingly, the mycobacterial genome harbors a variety of dehydrogenases and oxidases to regenerate reducing equivalents and also genes that can support anaerobic metabolism (Cole et al., 1998; Cook et al., 2014). In addition to this endogenous redundancy, the heterogeneity within a bacterial population is emerging to be another crucial facet of bacterial adaptation (Lidstrom and Konopka, 2010; Manina et al., 2015). Biofilm development, in which a conglomeration of cells forms a multi-layered structure, is considered to be another mode of multicellular adaptation (Stewart and Franklin, 2008; Webb et al., 2003). This three-dimensional architecture generates islands of heterogeneity with differential oxygen availability that are recalcitrant to host immune-clearance mechanisms (Gilbert et al., 2002). Interestingly, Mtb granulomas are also similar complex aggregates with hypoxic microenvironments and could provide multiple advantages to community survival (Ramakrishnan, 2012; Via et al., 2008). To understand the energetics of mycobacterial respiration under such oxygen-depleted hypoxic conditions, we examined mycobacterial survival in biofilms.
Previous studies have shown that the Mtb membrane remains energized in hypoxic environments, albeit with reduced ATP generation (Rao et al., 2008). As a ubiquitous oxidant in nature, oxygen levels can significantly affect the pH, redox state, and reduction potential in many diverse environments. We examined aggregates of mycobacterial pellicles at air-liquid interfaces by using the hspX promoter-driven expression of GFP. Although previous studies in other bacterial species have demonstrated variations in oxygen concentration across biofilm layers using microelectrodes, GFP expression operated like an intracellular sensor of the extent of hypoxic stress (Dietrich et al., 2013). A z stack confocal analysis showed remarkable heterogeneity in GFP expression within biofilm layers, and maximum expression was observed in the deeper layers of biofilms. This is in agreement with the extracellular measurements of oxygen concentration in other bacterial biofilms. Sustained hypoxia is marked by increased reactive oxygen species (ROS) production and can perturb the cellular redox balance (Korge et al., 2015; Murphy, 2009). Our analysis of the measurement of the redox potential of MSH substantiates the redox heterogeneity in biofilms. In comparison with planktonic cultures, approximately 50% of cells showed reduced EMSH, which largely suggests a more reducing microenvironment in the biofilm. It is interesting to note that a small subpopulation also shows oxidizing EMSH and that the status of other subpopulations are identical to planktonic cells. This amalgamation of populations probably provides a strategic advantage in terms of cooperation and may indeed be a crucial mechanism to mitigate a variety of external threats.
The downregulation of the isoprenoid biosynthetic pathway upon activation of the microaerophilic branch of the respiratory chain and the subsequent decrease in cellular levels of MK in a hypoxic environment prompted us to carefully search for alternative quinones (Honaker et al., 2010; Matsoso et al., 2005). Biochemical analysis of biofilm extracts identified CoQ-like quinones that are produced by both Msmeg and Mtb. These PkQ metabolites are derived from fatty acyl units and do not require an isoprenoid biosynthetic system. These molecules are not produced under planktonic conditions, even after overexpression of their biosynthetic pathway, and require an exogenous supply of fatty acyl precursors. Therefore, a complete metabolic rerouting is likely to take place in the biofilm to produce PkQs. The mutant Msmeg strain that was defective in PkQ biosynthesis showed fragile biofilm formation with decreased ATP synthesis and accumulation of NADH. These physiological changes also indicated a disruption in the respiratory chain.
Cell-free reconstitution of a component of the intermediate electron transfer chain showed that PkQ is able to efficiently mediate electron transfer to complex III. Complementation of PkQ in the mutant strain recovered the ATP levels and also restored biofilm morphology. Hypoxia-induced reductive stress triggers ROS spillover, and respiratory quinones become crucial in such microenvironments (Korge et al., 2015; Murphy, 2009). The benzoquinone ring of PkQ provides a distinct advantage over the naphthoquinone ring system during electron transfer under oxygen-limiting conditions. Although the reduction of PkQ forms an aromatic hydroquinone, MK reduction extends the aromaticity to the second ring. The gain of aromaticity results in a larger potential energy difference between the oxidized and reduced PkQ compared with the corresponding difference between the oxidized and reduced MK. This makes MK more prone to non-catalyzed oxidation and, therefore, a less efficient electron carrier during oxidative stress (Meganathan, 2001; Schoepp-Cothenet et al., 2009).
We further explored whether these alternate electron shuttles would support mycobacterial survival during other oxygen-deficient conditions. After establishing PKS10 promotor-driven expression of GFP in biofilms, we examined GFP expression in a hypoxic planktonic culture of Msmeg. Although a 4-fold upregulation could be seen under oxygen-deficient planktonic conditions, this was comparatively lower than observed in biofilms. The apparent ambiguity in the extent of upregulation might be due to the differences in the mode of generation of hypoxia. This was also validated by gene expression analysis. Moreover, the pks10 Msmeg mutant showed a growth defect in hypoxic cultures. Our studies therefore demonstrate the crucial role of PkQ molecules during mycobacterial microaerophilic respiration. Remarkably, this biosynthetic system is conserved in several Actinobacteria and also in a few Proteobacteria. Interestingly, all of these microbes also contain several other PKS systems. The emergence of PkQ is therefore likely to have evolved after the acquisition of these secondary metabolite pathways, particularly by soil-dwelling microorganisms. Our studies reveal a crucial adaptive feature of mycobacteria during hypoxic conditions that could be pursued as a potential drug target.
Experimental Procedures
Details of the experimental procedures are described in the Supplemental Experimental Procedures.
Radiolabeled Enzymatic Assay with PKS10 Protein
The standard reaction conditions involved 100 μM starter CoA and 50 μM MCoA (including 9.12 μM of [2-14C]-MCoA [58.40 mCi/mmol]) and/or 50 μM MMCoA (including 9.12 μM of [2-14C]-MMCoA [58.40 mCi/mmol]). Reactions were carried out with 45 μg of protein at 30°C for 120 min and quenched with 5% acetic acid.
Biofilm Development
Biofilm cultures for Msmeg were grown in Sauton’s medium supplemented with 2% glucose. The plates were incubated at 37°C in a humidified incubator for 7 days. Mtb biofilms were grown in Sauton’s medium (without Tween 80) by incubation without shaking at 37°C for 5 weeks under humidified conditions. The dishes were wrapped with parafilm during incubation.
Extraction and Analysis of Type III PKS Products from Planktonic and Biofilm Cultures
Cultures were harvested and suspended in an appropriate volume of 100 mM Tris-Cl (pH 8.0) and disrupted. The resultant whole-cell lysate was acidified by addition of 6 M HCl. Low-molecular-weight molecules were then extracted by adding a double volume of ethyl acetate and left overnight on a stirrer. The organic layer was separated and evaporated to dryness, and the residual material was dissolved in a minimal volume of methanol and analyzed by liquid chromatography (LC)-MS.
ATP Estimation
Mycobacterial planktonic cultures were grown to an OD600 ranging from 0.8 to 1.2. Biofilm pellicles were harvested from 7 day old biofilms. Cells were disrupted with a bead beater using 3 pulses of 1 min each for intracellular ATP estimation. Protein estimation was done using the Pierce BCA Protein Assay Kit. ATP estimation was done using Molecular Probes’ ATP Determination Kit (A22066).
NADH/NAD+ ratio estimation
Mycobacterial planktonic cultures were grown to an optical density 600 (OD600) ranging from 0.8–1.2. Biofilm pellicles were harvested from 7-day-old biofilms. The concentrations of NAD+ and NADH were determined by the cycling assay of Bernofsky and Swan using an adaptation provided by A. Koul (Bernofsky and Swan, 1973; Koul et al., 2014). The biofilm pellicle was subjected to mild sonication to separate the cells’ extracellular matrix. Briefly, bacteria were harvested, re-suspended in 0.2 M HCl (for NAD+ extraction) or 0.2 M NaOH (for NADH extraction), heated at 55°C for 10 min, and cooled on ice. Cell debris was removed by centrifugation. The supernatant was pre-incubated for 5 min in the dark in Bicine buffer containing EDTA, ethanol, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), and phenazine methosulfate. The reaction was started by addition of alcohol dehydrogenase from Saccharomyces cerevisiae. The absorbance at 570 nm was recorded for 10 min.
Membrane Potential Estimation
Mycobacterial planktonic cultures were grown to an OD600 ranging from 0.8–1.2. Biofilm pellicles were harvested from 7-day-old biofilms. Assaying for membrane potential was performed using the BacLight bacterial membrane potential kit (Invitrogen, Life Technologies) as described previously (Rao et al., 2008). Briefly, cells were diluted to an OD600 of 0.05 in PBS, and 1 ml of cells was stained using 3 μM of DiOC2 for 15 min in the presence or absence of 25 μM carbonyl cyanide 3-chlorophenylhydrazone (CCCP). The cells were fixed using 2% paraformaldehyde and analyzed for fluorescein and Texas red dye using flow cytometry. The proton ionophore (CCCP) was used as a control to eliminate the bacterial membrane potential.
Cytochrome c Reduction Assay
An ethanolic solution of alkyl benzoquinone was used to reduce cytochrome c via cytochrome c reductase. Alcohol dehydrogenase (A3263, Sigma) was coupled to the assay to reduce alkyl benzoquinones. The reduction was monitored until saturation at 550 nm using a Cary 300 UV-visible spectrophotometer.
Mixed-Culture Biofilm Formation
Wild-type and Δpks10 cells were electroporated with GFP and RFP expressing mycobacterial expression vectors, respectively. Both strains were grown in planktonic mode to an OD600 of 0.8–1.0, and then cell density was calculated based on OD600. Mixed inoculums of both strains were inoculated in biofilm media. For imaging, biofilms were grown in Nunc Lab-Tek chambered cover-glass (catalog no. 155380), and imaging was done using a Leica DMI6000 inverted fluorescence microscope at 63× magnification, and images were processed with a Leica Application Suite (LAS) AF 2.6 platform.
Supplementary Material
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2015.10.016.
Highlights.
Mycobacterial biofilm contains physiologically diverse microenvironments
Hypoxia induces expression of the alternate electron carrier polyketide quinones
PkQs are produced by type III polyketide synthases using fatty acyl-CoA precursors
PkQs maintain cellular bioenergetics in oxygen-deficient niches
Acknowledgments
We thank Dr. Archana Singh (CSIR-IGIB, India) for her help with scanning electron microscopy. We thank Dr. Vinay Nandicoori (NII, India) and Dr. Vivek Rao (CSIR-IGIB, India) for providing mycobacterial vectors. We thank Dr. H.V. Thulsiram (CSIR-NCL, India) for his help in devising the experimental setup. We thank Prof. Yossef Av-Gay (University of British Columbia, Vancouver, BC, Canada) and Dr. Kate S. Carroll (Scripps Research Institute, Jupiter, FL) for MSH mutants. The reporter construct for the hspX promoter (phspX) was kindly provided by Dr. Jaya S. Tyagi (AIIMS, India). We acknowledge support provided by Manish Kumar for confocal imaging. We thank Rintu Kutum and Anupam K. Mondal (CSIR-IGIB, India) for help with analyzing and designing figures for the manuscript. A.A. acknowledges CSIR for an SRF fellowship. A.S. is grateful for Wellcome/DBT India Alliance fellowships. We gratefully acknowledge CSIR Grants Gencode BSC0123 and NCL-IGIB BSC0124 (to R.S.G.) and NIH Grant AI-070219 (to C.C.A.). We also acknowledge DBT for institutional support to NII and for NMR facility to ICGEB, New Delhi.
Footnotes
Author Contributions
Conceptualization, R.S.G., A.A., and P.V.; Methodology, R.S.G., A.A., P.V., and T.N.V.; Software, D.K. and D.D.; Formal Analysis, A.A. and S.K.; Investigation, A.A., P.V., M.C., A.S., Y.Y., and A.K.O.; Resources, A.K.S., C.S., H.K., C.A., R.P., C.K.E., R.B., and N.S.B.; Writing – Original Draft, R.S.G. and A.A.; Writing – Review and Editing, R.S.G., A.A., and C.A.; Supervision, R.S.G.
References
- Barry CE, 3rd, Boshoff HI, Dartois V, Dick T, Ehrt S, Flynn J, Schnappinger D, Wilkinson RJ, Young D. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat Rev Microbiol. 2009;7:845–855. doi: 10.1038/nrmicro2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernofsky C, Swan M. An improved cycling assay for nicotinamide adenine dinucleotide. Anal Biochem. 1973;53:452–458. doi: 10.1016/0003-2697(73)90094-8. [DOI] [PubMed] [Google Scholar]
- Bhaskar A, Chawla M, Mehta M, Parikh P, Chandra P, Bhave D, Kumar D, Carroll KS, Singh A. Reengineering redox sensitive GFP to measure mycothiol redox potential of Mycobacterium tuberculosis during infection. PLoS Pathog. 2014;10:e1003902. doi: 10.1371/journal.ppat.1003902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borriello G, Werner E, Roe F, Kim AM, Ehrlich GD, Stewart PS. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob Agents Chemother. 2004;48:2659–2664. doi: 10.1128/AAC.48.7.2659-2664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshoff HI, Barry CE., 3rd Tuberculosis - metabolism and respiration in the absence of growth. Nat Rev Microbiol. 2005;3:70–80. doi: 10.1038/nrmicro1065. [DOI] [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, Honoré N, Garnier T, Churcher C, Harris D, et al. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- Cook GM, Hards K, Vilchèze C, Hartman T, Berney M. Energetics of Respiration and Oxidative Phosphorylation in Mycobacteria. Microbiol Spectr. 2014;2:2. doi: 10.1128/microbiolspec.MGM2-0015-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson JF, Schiestl RH. Mitochondrial respiratory electron carriers are involved in oxidative stress during heat stress in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:8483–8489. doi: 10.1128/MCB.21.24.8483-8489.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beer D, Stoodley P, Roe F, Lewandowski Z. Effects of biofilm structures on oxygen distribution and mass transport. Biotechnol Bioeng. 1994;43:1131–1138. doi: 10.1002/bit.260431118. [DOI] [PubMed] [Google Scholar]
- Desjardin LE, Hayes LG, Sohaskey CD, Wayne LG, Eisenach KD. Microaerophilic induction of the alpha-crystallin chaperone protein homologue (hspX) mRNA of Mycobacterium tuberculosis. J Bacteriol. 2001;183:5311–5316. doi: 10.1128/JB.183.18.5311-5316.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich LE, Okegbe C, Price-Whelan A, Sakhtah H, Hunter RC, Newman DK. Bacterial community morphogenesis is intimately linked to the intracellular redox state. J Bacteriol. 2013;195:1371–1380. doi: 10.1128/JB.02273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eoh H, Rhee KY. Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2013;110:6554–6559. doi: 10.1073/pnas.1219375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabashi M, Funa N, Horinouchi S. Phenolic lipids synthesized by type III polyketide synthase confer penicillin resistance on Streptomyces griseus. J Biol Chem. 2008;283:13983–13991. doi: 10.1074/jbc.M710461200. [DOI] [PubMed] [Google Scholar]
- Gilbert P, Maira-Litran T, McBain AJ, Rickard AH, Whyte FW. The physiology and collective recalcitrance of microbial biofilm communities. Adv Microb Physiol. 2002;46:202–256. [PubMed] [Google Scholar]
- Guirado E, Schlesinger LS. Modeling the Mycobacterium tuberculosis Granuloma - the Critical Battlefield in Host Immunity and Disease. Front Immunol. 2013;4:98. doi: 10.3389/fimmu.2013.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- Harmsen M, Yang L, Pamp SJ, Tolker-Nielsen T. An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol Med Microbiol. 2010;59:253–268. doi: 10.1111/j.1574-695X.2010.00690.x. [DOI] [PubMed] [Google Scholar]
- Honaker RW, Dhiman RK, Narayanasamy P, Crick DC, Voskuil MI. DosS responds to a reduced electron transport system to induce the Mycobacterium tuberculosis DosR regulon. J Bacteriol. 2010;192:6447–6455. doi: 10.1128/JB.00978-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MS, Richards JP, Ojha AK. Targeting drug tolerance in mycobacteria: a perspective from mycobacterial biofilms. Expert Rev Anti Infect Ther. 2012;10:1055–1066. doi: 10.1586/eri.12.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kana BD, Weinstein EA, Avarbock D, Dawes SS, Rubin H, Mizrahi V. Characterization of the cydAB-encoded cytochrome bd oxidase from Mycobacterium smegmatis. J Bacteriol. 2001;183:7076–7086. doi: 10.1128/JB.183.24.7076-7086.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korge P, Calmettes G, Weiss JN. Increased reactive oxygen species production during reductive stress: The roles of mitochondrial glutathione and thioredoxin reductases. Biochim Biophys Acta. 2015;1847:514–525. doi: 10.1016/j.bbabio.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koul A, Vranckx L, Dhar N, Göhlmann HW, Özdemir E, Neefs JM, Schulz M, Lu P, Mørtz E, McKinney JD, et al. Delayed bactericidal response of Mycobacterium tuberculosis to bedaquiline involves modelling of bacterial metabolism. Nat Commun. 2014;5:3369. doi: 10.1038/ncomms4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leistikow RL, Morton RA, Bartek IL, Frimpong I, Wagner K, Voskuil MI. The Mycobacterium tuberculosis DosR regulon assists in metabolic homeostasis and enables rapid recovery from nonrespiring dormancy. J Bacteriol. 2010;192:1662–1670. doi: 10.1128/JB.00926-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidstrom ME, Konopka MC. The role of physiological heterogeneity in microbial population behavior. Nat Chem Biol. 2010;6:705–712. doi: 10.1038/nchembio.436. [DOI] [PubMed] [Google Scholar]
- Mailloux RJ, Bériault R, Lemire J, Singh R, Chénier DR, Hamel RD, Appanna VD. The tricarboxylic acid cycle, an ancient metabolic network with a novel twist. PLoS ONE. 2007;2:e690. doi: 10.1371/journal.pone.0000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manina G, Dhar N, McKinney JD. Stress and host immunity amplify Mycobacterium tuberculosis phenotypic heterogeneity and induce nongrowing metabolically active forms. Cell Host Microbe. 2015;17:32–46. doi: 10.1016/j.chom.2014.11.016. [DOI] [PubMed] [Google Scholar]
- Matsoso LG, Kana BD, Crellin PK, Lea-Smith DJ, Pelosi A, Powell D, Dawes SS, Rubin H, Coppel RL, Mizrahi V. Function of the cytochrome bc1-aa3 branch of the respiratory network in mycobacteria and network adaptation occurring in response to its disruption. J Bacteriol. 2005;187:6300–6308. doi: 10.1128/JB.187.18.6300-6308.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meganathan R. Biosynthesis of menaquinone (vitamin K2) and ubiquinone (coenzyme Q): a perspective on enzymatic mechanisms. Vitam Horm. 2001;61:173–218. doi: 10.1016/s0083-6729(01)61006-9. [DOI] [PubMed] [Google Scholar]
- Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton GL, Buchmeier N, Fahey RC. Biosynthesis and functions of mycothiol, the unique protective thiol of Actinobacteria. Microbiol Mol Biol Rev. 2008;72:471–494. doi: 10.1128/MMBR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- Ojha AK, Baughn AD, Sambandan D, Hsu T, Trivelli X, Guerardel Y, Alahari A, Kremer L, Jacobs WR, Jr, Hatfull GF. Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol Microbiol. 2008;69:164–174. doi: 10.1111/j.1365-2958.2008.06274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole RK, Cook GM. Redundancy of aerobic respiratory chains in bacteria? Routes, reasons and regulation. Adv Microb Physiol. 2000;43:165–224. doi: 10.1016/s0065-2911(00)43005-5. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan L. Revisiting the role of the granuloma in tuberculosis. Nat Rev Immunol. 2012;12:352–366. doi: 10.1038/nri3211. [DOI] [PubMed] [Google Scholar]
- Rani SA, Pitts B, Beyenal H, Veluchamy RA, Lewandowski Z, Davison WM, Buckingham-Meyer K, Stewart PS. Spatial patterns of DNA replication, protein synthesis, and oxygen concentration within bacterial biofilms reveal diverse physiological states. J Bacteriol. 2007;189:4223–4233. doi: 10.1128/JB.00107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SP, Alonso S, Rand L, Dick T, Pethe K. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2008;105:11945–11950. doi: 10.1073/pnas.0711697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou R, Miyanaga A, Ozawa H, Funa N, Katsuyama Y, Miyazono K, Tanokura M, Ohnishi Y, Horinouchi S. Structural basis for cyclization specificity of two Azotobacter type III polyketide synthases: a single amino acid substitution reverses their cyclization specificity. J Biol Chem. 2013;288:34146–34157. doi: 10.1074/jbc.M113.487272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena P, Yadav G, Mohanty D, Gokhale RS. A new family of type III polyketide synthases in Mycobacterium tuberculosis. J Biol Chem. 2003;278:44780–44790. doi: 10.1074/jbc.M306714200. [DOI] [PubMed] [Google Scholar]
- Schoepp-Cothenet B, Lieutaud C, Baymann F, Verméglio A, Friedrich T, Kramer DM, Nitschke W. Menaquinone as pool quinone in a purple bacterium. Proc Natl Acad Sci USA. 2009;106:8549–8554. doi: 10.1073/pnas.0813173106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, Schoolnik GK. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha -crystallin. Proc Natl Acad Sci USA. 2001;98:7534–7539. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Sohaskey CD, Kana BD, Dawes S, North RJ, Mizrahi V, Gennaro ML. Changes in energy metabolism of Mycobacterium tuberculosis in mouse lung and under in vitro conditions affecting aerobic respiration. Proc Natl Acad Sci USA. 2005;102:15629–15634. doi: 10.1073/pnas.0507850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- Teschler JK, Zamorano-Sánchez D, Utada AS, Warner CJ, Wong GC, Linington RG, Yildiz FH. Living in the matrix: assembly and control of Vibrio cholerae biofilms. Nat Rev Microbiol. 2015;13:255–268. doi: 10.1038/nrmicro3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaupel P, Kallinowski F, Okunieff P. Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: a review. Cancer Res. 1989;49:6449–6465. [PubMed] [Google Scholar]
- Via LE, Lin PL, Ray SM, Carrillo J, Allen SS, Eum SY, Taylor K, Klein E, Manjunatha U, Gonzales J, et al. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect Immun. 2008;76:2333–2340. doi: 10.1128/IAI.01515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol. 2013;11:157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Sambandan D, Halder R, Wang J, Batt SM, Weinrick B, Ahmad I, Yang P, Zhang Y, Kim J, et al. Identification of a small molecule with activity against drug-resistant and persistent tuberculosis. Proc Natl Acad Sci USA. 2013;110:E2510–E2517. doi: 10.1073/pnas.1309171110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb JS, Givskov M, Kjelleberg S. Bacterial biofilms: prokaryotic adventures in multicellularity. Curr Opin Microbiol. 2003;6:578–585. doi: 10.1016/j.mib.2003.10.014. [DOI] [PubMed] [Google Scholar]
- Williamson KS, Richards LA, Perez-Osorio AC, Pitts B, McInnerney K, Stewart PS, Franklin MJ. Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. J Bacteriol. 2012;194:2062–2073. doi: 10.1128/JB.00022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrano MM, Kolter R. Mycobacterial biofilms: a greasy way to hold it together. Cell. 2005;123:762–764. doi: 10.1016/j.cell.2005.11.011. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yew WW, Barer MR. Targeting persisters for tuberculosis control. Antimicrob Agents Chemother. 2012;56:2223–2230. doi: 10.1128/AAC.06288-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A, Raviglione M, Hafner R, von Reyn CF. Tuberculosis. N Engl J Med. 2013;368:745–755. doi: 10.1056/NEJMra1200894. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.