Abstract
Objective
Shared genetic background may explain phenotypic associations between depression and Type 2 diabetes (T2D). We aimed to study, on a genome-wide level, if genetic correlation and pleiotropic loci exist between depressive symptoms and T2D or glycemic traits.
Methods
We estimated single-nucleotide polymorphism (SNP)–based heritability and analyzed genetic correlation between depressive symptoms and T2D and glycemic traits with the linkage disequilibrium score regression by combining summary statistics of previously conducted meta-analyses for depressive symptoms by CHARGE consortium (N = 51,258), T2D by DIAGRAM consortium (N = 34,840 patients and 114,981 controls), fasting glucose, fasting insulin, and homeostatic model assessment of β-cell function and insulin resistance by MAGIC consortium (N = 58,074). Finally, we investigated pleiotropic loci using a bivariate genome-wide association study approach with summary statistics from genome-wide association study meta-analyses and reported loci with genome-wide significant bivariate association p value (p < 5 × 10−8). Biological annotation and function of significant pleiotropic SNPs were assessed in several databases.
Results
The SNP-based heritability ranged from 0.04 to 0.10 in each individual trait. In the linkage disequilibrium score regression analyses, depressive symptoms showed no significant genetic correlation with T2D or glycemic traits (p > 0.37). However, we identified pleiotropic genetic variations for depressive symptoms and T2D (in the IGF2BP2, CDKAL1, CDKN2B-AS, and PLEKHA1 genes), and fasting glucose (in the MADD, CDKN2B-AS, PEX16, and MTNR1B genes).
Conclusions
We found no significant overall genetic correlations between depressive symptoms, T2D, or glycemic traits suggesting major differences in underlying biology of these traits. However, several potential pleiotropic loci were identified between depressive symptoms, T2D, and fasting glucose, suggesting that previously established phenotypic associations may be partly explained by genetic variation in these specific loci.
Keywords: depression, meta-analysis, Type 2 diabetes, pleiotropy, GWAS
INTRODUCTION
Diabetes, primarily Type 2 diabetes (T2D) presents a major and increasing health burden globally, and it has been estimated that 415 million people had diabetes in 2015 (approximately 10% of the world adult population) (1). This figure is expected to exceed 700 million by 2025 (1). Many people have undiagnosed diabetes and many more are at increased risk (2). For prevention, it is important to unravel the reasons why an individual is susceptible to develop diabetes.
T2D and another growing health problem (3), depression, show bidirectional relationship (4). Based on a meta-analysis involving 172,521 participants, individuals with T2D have a 24%increased risk of incident depression compared with people without diabetes (5). On the other hand, based on a meta-analysis involving 2,411,641 participants, individuals with depression have a 41% increased risk for developing any type of diabetes and 32% increased risk for developing T2D (6). Moreover, major depressive disorder (MDD) and subclinical depressive symptoms are associated with abnormal glucose metabolism, impaired insulin secretion, and insulin resistance (7,8). However, the underlying mechanisms linking these conditions are unknown. One possibility is that these moderately heritable (9,10) conditions share a common genetic basis. Studies examining this hypothesis are still scarce and show conflicting results. A recent twin study using Swedish and Danish population registries (N = 68,606 and N = 95,403, respectively) found a moderate but significant genetic correlation between depressive disorders and diabetes among females from both cohorts ranging between 0.18 and 0.23 and among Danish male twins (genetic correlation, 0.25) suggesting significant overall shared genomic background between these disorders (11). Conversely, a recent single-nucleotide polymorphism (SNP)–based study using linkage disequilibrium score regression (LDSC) failed to show genetic correlations between MDD and T2D or quantitative glycemic traits (12). Similarly, a population-based study of 21,516 individuals showed no genetic overlap between MDD and T2D using polygenic risk score analysis, and Mendelian randomization in addition to LDSC (13).
Currently, there are no studies on genetic overlap between depressive symptoms and T2D or glycemic traits. There are several reasons why focusing on depressive symptoms rather than MDD is justified: most of the longitudinal studies investigating bidirectional associations between these traits use measures of depressive symptoms. Moreover, psychiatric patients may be reluctant to seek help or be suboptimally treated for their nonpsychiatric disorders (14). Finally, because depression in the population may be best characterized dimensionally as a continuum from depressive symptoms to subthreshold depression and finally to MDD (15), each step increasing adverse health influences (16,17), treating depression dimensionally rather than categorically could increase statistical power to detect phenotypic associations (18).
In addition to investigating overall genetic correlation between the traits, examining specific SNPs that associate with both conditions may increase our understanding in the common underlying biology of the traits (19). Methods to study these pleiotropic associations based on genome-wide meta-analyses summary data have become only recently available (20). For example, a recent study applied bivariate genome-wide association study (GWAS) analyses and identified several pleiotropic loci that associated with both C-reactive protein and lipids and suggested potential genetic interrelation between these traits (21).
Here, we report findings from a study that applied a bivariate genome-wide association analysis using summary statistics from three relatively large univariate GWAS for depressive symptoms and glycemic traits (22–24). In these studies, Dupuis and colleagues (23) identified 25 independent loci and conducted follow-up analysis leading to 16 loci that associated with fasting glucose/homeostatic model assessment of β-cell function (HOMA-β) and two that associated with fasting insulin/homeostatic model assessment of insulin resistance (HOMA-IR), and Morris and colleagues (24) found 10 new loci that associated with T2D. For depressive symptoms, one SNP in the 5q21 region reached genome-wide significance (22). In sum, we first tested in large genome-wide meta-analyses data sets (22–24) the hypothesis that a positive genetic correlation between depressive symptoms and T2D or glycemic traits exists. Second, in the same data sets, we also explored if there are potential pleiotropic SNPs that are associated with both depressive symptoms and T2D and the glycemic traits. Because of the vast number of tests in genome-wide analyses and our limited understanding on the etiology of these traits, we did not place specific hypothesis on SNPs and their direction.
METHODS
GWAS Summary Statistics
Our analyses are based on previously published univariate GWAS summary statistics. Data on T2D were contributed by DIAGRAM investigators and include 34,840 cases with T2D and 114,981 controls without T2D (www.diagram-consortium.org, accessed October 10, 2015)(24). T2D diagnosis in DIAGRAM was based on several different criteria including self-reported T2D, physician's diagnosis, registry data, and oral glucose tolerance test applying World Health Organization criteria (25). Data on glycemic traits were contributed by MAGIC investigators and downloaded from www.magicinvestigators.org (accessed November 26, 2015). The quantitative glycemic traits in MAGIC included fasting insulin, fasting glucose, HOMA-β [(20 × fasting plasma insulin)/(fasting plasma glucose − 3.5)] for assessment of β-cell function, and HOMA-IR (fasting plasma insulin × fasting plasma glucose/22.5) for estimation of the degree of insulin resistance (26) involving up to 46,186 participants without diabetes from up to 21 cohorts (23). Data on depressive symptoms were contributed by the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Depression Working Group investigators and include 51,258 individuals (22). Most cohorts used the Center for Epidemiological Studies Depression Scale 10-, 11-, or 20-item versions (27) to assess depressive symptoms, but some cohorts used the Geriatric Depression Scale, Patient Health Questionnaire, Maastricht Questionnaire, or Beck Depression Inventory-II.
Statistical Analyses
To estimate genetic correlation of depressive symptoms with T2D and quantitative glycemic traits, we combined results from univariate GWAS meta-analyses using the LDSC tool (28) according to the manual with default options. This command line tool is for estimating heritability and genetic correlation from GWAS summary statistics that relies on the fact that the GWAS effect size estimate for a given SNP incorporates the effects of all SNPs in linkage disequilibrium with that SNP. LDSC is not biased by sample overlap and has been described in detail elsewhere (12).
To simultaneously analyze two outcome variables (phenotypes) at once, we performed bivariate (or multivariate with more than two phenotypes at once) GWAS analyses. Such bivariate analysis by jointing association analysis of two traits in a GWAS allows us to identify potential pleiotropic genetic effects and offers several advantages over analyzing each trait in a separate GWAS. To identify potential pleiotropic SNPs associated with both depressive symptoms and each of the T2D and quantitative glycemic traits, we performed bivariate GWAS analyses using empirical-weighted linear-combined test statistics (eLC) (29) with aggregate data (Z test statistics) from each univariate GWAS meta-analysis (inverse-variance meta-analysis with GC controls). Briefly, eLC directly combines correlated test statistics obtained from univariate GWAS meta-analyses with a weighted sum of univariate test statistics to empirically maximize the overall association signals and to account for the phenotypical correlation between the phenotypes of interest. The eLC approach is expressed as (29):
where c is some given nonnegative constant. The weight in this new test statistics will be optimally determined by the specific data structure. For instance, when c = 0, the test statistics simply reduces into sum of squares of Tk. When c is relatively large, equal weight is assigned to each Tk. Ideally, we would like to find an optimal value of c, so the SeLC performs as a linear combination of Ti when under the H0, but under the alternative HA, more weight is given to the larger true Ti. The bona fide p value for SeLC then can be estimated by applying permutation or perturbation techniques. The variance-covariance matrix Σ of univariate test statistics uses the sample covariance matrix of the test statistics of all SNPs from univariate GWAS analyses as an approximation.
where Z1 consists of unbiased univariate test statistics of all the SNPs for the first trait on genome-wide scale, so does Z2. On the other hand, Σ can be estimated by using generalized least squares from individual-level data. The eLC method is implemented in eLX package using C++ and publicly available at https://sites.google.com/site/multivariateyihsianghsu/.
As suggested in the earlier study (21), we only performed bivariate GWAS analyses for those SNPs with nominal p values of less than.05 in univariate GWAS meta-analyses for both phenotypes analyzed. We report potential pleiotropic SNPs based on a) p value < 5 × 10−8 from the bivariate GWAS analyses and b) the bivariate p value being at least one order of magnitude smaller than the univariate p values.
Functionality of the SNPs
We examined the influence of identified potential pleiotropic SNPs on gene expression in the Brain eQTL Almanac (BRAINEAC, www.braineac.org) database and GTEx portal V6 (dbGaP Accession phs000424.v6.p1, www.gtexportal.org). The BRAINEAC comprises genotype data and gene expression data in brain regions (frontal cortex, hippocampus, white matter, putamen, cerebellar cortex, medulla, temporal cortex, hippocampus, and substantia nigra) from 134 individuals free of neurodegenerative disorders, and the GTEx database contains information on expression quantitative traits (eQTL) in 44 different tissues from 449 donors. In the BRAINEAC eQTL analysis, we set the Bonferroni-corrected significance level at 5.7 × 10−4 (0.05/[11 brain regions × 8 SNPs]).
RESULTS
Heritability Estimates and Genetic Correlation Between the Traits
Based on the LDSC analysis, we found that SNP-based heritability estimates were 0.09 [0.07, 0.12] for T2D, 0.10 [0.06, 0.15] for fasting glucose, 0.07 [0.05, 0.10] for fasting insulin, 0.07 [0.05, 0.09] for HOMA-β, 0.05 [0.03, 0.07] for HOMA-IR, and 0.04 [0.01, 0.07] for depressive symptoms. There were no significant SNP-based genetic correlations between depressive symptoms and T2D and quantitative glycemic traits; however, all correlations were negative (Table 1). T2D and all quantitative glycemic traits showed significant SNP-based genetic correlations, except for T2D and HOMA-β (Table S1, Supplemental Digital Content 1, http://links.lww.com/PSYMED/A435).
TABLE 1.
SNP-based Genetic Correlations Between Depressive Symptoms, Diabetes, and Quantitative Glycemic Traits Based on LD Score Regression on Meta-analyses Summary Statistics of DIAGRAM, MAGIC, and CHARGE Consortia
| Genetic Correlation Between Depressive Symptoms and |
rG | SE | p |
|---|---|---|---|
| T2D | −0.03 | 0.13 | .82 |
| Fasting glucose | −0.003 | 0.150 | .98 |
| Fasting insulin | −0.16 | 0.17 | .37 |
| HOMA-β | −0.10 | 0.17 | .55 |
| HOMA-IR | −0.07 | 0.18 | .70 |
Data on T2D are based on up to 149,821 participants (34,840 cases). Data on fasting glucose, fasting insulin, HOMA-β, and HOMA-IR are based on up to 46,286 participants.
SNP = single-nucleotide polymorphism; LD = linkage disequilibrium; T2D = Type 2 diabetes; HOMA-β = homeostatic model assessment of β-cell function; HOMA-IR = homeostatic model assessment of insulin resistance.
Potential Pleiotropic Loci
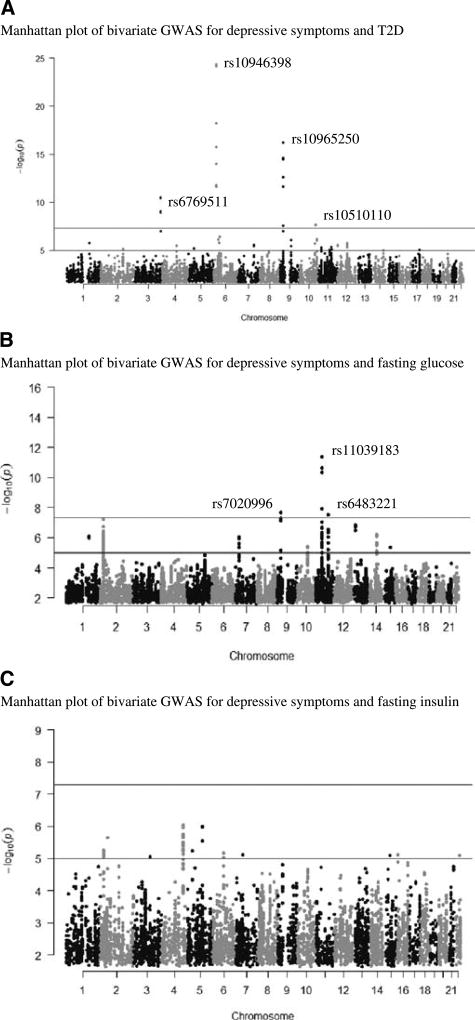
We found several SNPs showing potential pleiotropic effects between depressive symptoms and T2D and fasting glucose (Fig. 1, Tables S2 and S3, Fig. S1, Supplemental Digital Content 1, http://links.lww.com/PSYMED/A435). Bivariate GWAS analysis for depressive symptoms and T2D implicated two intronic SNPs in the insulin-like growth factor 2mRNA binding protein 2 (IGF2BP2; chr 3; rs6769511; bivariate p value = 3.32 × 10−11) and CDK5 regulatory subunit associated protein 1-like 1 (CDKAL1; chr 6; rs10946398; bivariate p value = 4.49 × 10−25) genes and two intergenic SNPs close to the CDKN2B antisense RNA 1 (CDKN2B-AS; chr 9; rs10965250; bivariate p value = 6.18 × 10−17) and pleckstrin homology domain-containing family A member 1 (PLEKHA1; chr 10; rs10510110; bivariate p value = 1.83 × 10−8) gene (Fig. 1A, Table S2, Fig. S1, Supplemental Digital Content 1, http://links.lww.com/PSYMED/A435). Of these SNPs, rs6769511 in IGFBP2 and rs10965250 in CDKN2B-AS showed an association in the same direction with depression and T2D, whereas for rs10946398 in CDKALI1 and rs10510110 in PLEKHA1, the effect was opposite. All these SNPs were associated with significant alterations in expression of several genes in brain tissues in the BRAINEAC database, as well as rs6769511 and rs10510110 with expression in other tissues in the GTEx database (Table S4, Supplemental Digital Content 1, http://links.lww.com/PSYMED/A435).
FIGURE 1.
Manhattan plots showing bivariate p values against genomic positions for associations between depressive symptoms and T2D (A), fasting glucose (B), and fasting insulin (C). Red line is indicating bivariate genome-wide significance [−log10(5 × 10−8)]. All variants here have univariate p values of less than.05. GWAS = genome-wide association study; T2D = Type 2 diabetes. Color image is available only in online version (www.psychosomaticmedicine.org).
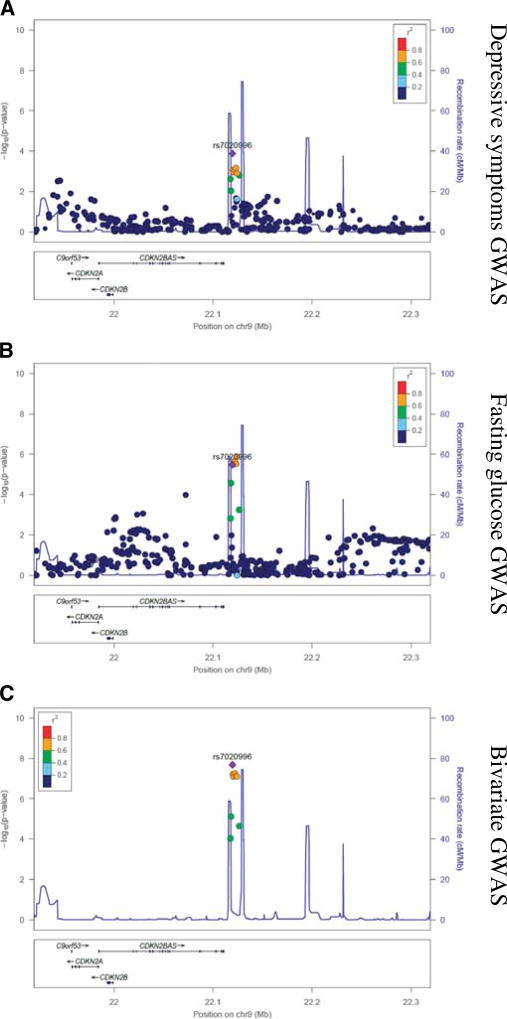
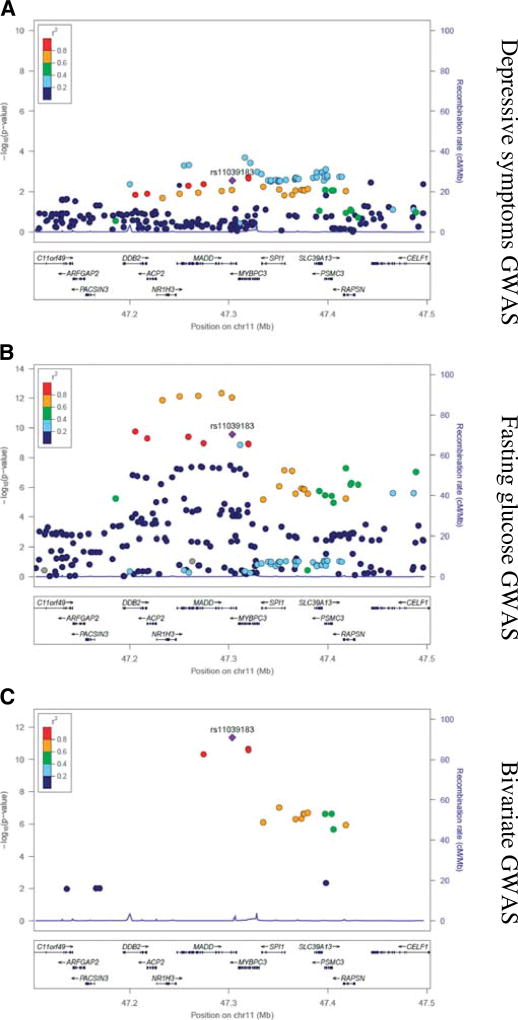
Bivariate GWAS analysis for depressive symptoms and fasting glucose implicated intronic SNPs in the MAP kinase–activating death domain protein (MADD; chr11; rs11039183; bivariate p value = 4.40 × 10−12) and peroxisomal biogenesis factor 16 (PEX16; chr 11; rs11038708; bivariate p value = 1.27 × 10−8) genes and two intergenic SNPs near CDKN2B-AS (chr 9; rs7020996; bivariate p value = 2.08 × 10−8) and melatonin receptor 1B (MTNR1B; chr 11; rs6483221; bivariate p value = 2.99 × 10−8) (Figs. 1B and 2–4; Table S3, Supplemental Digital Content 1, http://links.lww.com/PSYMED/A435). The associations of rs11039183 (MADD) and of rs7020996 (CDKN2B-AS) with depression and glucose levels were in the same direction, whereas for rs11038708 and rs10510110 in PEX16 and MTNR1B, the effect was opposite. Of these four SNPs, rs11039183 was associated with expression of several genes in the brain and other tissues in the BRAINEAC and GTEx databases (Table S4, Supplemental Digital Content 1, http://links.lww.com/PSYMED/A435).
FIGURE 2.
Regional visualization for rs7020996 of univariate and bivariate GWAS results for depressive symptoms and fasting glucose. GWAS = genome-wide association study.
FIGURE 4.
Regional visualization for rs6483221 of univariate and bivariate GWAS results for depressive symptoms and fasting glucose. GWAS = genome-wide association study.
Bivariate GWAS analyses for depressive symptoms and a) fasting insulin (Fig. 1), b) HOMA-β (Fig. S2, Supplemental Digital Content 1, http://links.lww.com/PSYMED/A435), and c) HOMA-IR (Fig. S2, Supplemental Digital Content 1, http://links.lww.com/PSYMED/A435) did not result in any genome-wide significance.
DISCUSSION
We first showed that SNP-based heritability estimate for depressive symptoms was 4% in the 51,258 individuals of the CHARGE consortium GWA meta-analyses. As shown earlier (12), SNP-based heritability estimates for T2D and quantitative glycemic traits were between 5% and 10% in the GWA meta-analyses of the DIAGRAM and MAGIC consortia. The heritability estimates are low compared with heritability estimates from Nordic twin study for depression (40%–52%) and T2D (72%–73%)(11). However, heritability estimates based on family or twin data may show inflated values due to biases related to study designs, such as assumption of size of common environmental factors (30). At the same time, heritability estimates based on SNPs may be deflated for several reasons: we may not be able to capture relevant genomic variation with the current genome-wide genotyping arrays, and we may lack power in our original GWA analyses. Consequently, our results on the heritability of depressive symptoms and T2D and quantitative glycemic traits are in line with the missing heritability gap between the SNP-based studies and twin studies.
Next, we tested, to our knowledge for the first time with GWA data, whether there are genetic correlations between depressive symptoms and T2D or quantitative glycemic traits. These correlations might explain the well-established phenotypic relationship between depression and T2D and quantitative glycemic traits (4,7,31–33). Contrary to our hypothesis, we did not find a significant overall genetic correlation between depressive symptoms and T2D or with quantitative glycemic traits by using data from more than 2.3 million SNPs, suggesting that the contribution of genetic variation to the phenotypic association is relatively small or nonexisting.
Our results are in line with previous studies suggesting lack of significant genetic overlap between MDD and T2D and glycemic traits. A recent SNP-based genetic correlation study applying GWAS meta-analyses data using LDSC showed a nonsignificant overall genetic correlation between MDD and T2D, fasting glucose, fasting insulin, HOMA-β, or HOMA-IR (12). Another recent population-based study found no genetic overlap between MDD and T2D using several different methods including LDSC (13). These studies rely on available summary statistic data of contemporary GWA studies, and their ability to detect genetic correlations is determined by the ability of the original GWA studies to detect genetic associations. Thus, future studies on genetic correlation between depressive symptoms and T2D or glycemic traits will benefit from GWA meta-analyses with even larger samples providing increased power to detect genetic correlations. Moreover, also twin studies have pointed toward the role of nongenetic factors explaining the link between depression and T2D. Mezuk and colleagues (34) showed in twins that unique environmental factors contribute significantly to the association between MDD and T2D in a study of 37,043 twins. Moreover, another recent twin study concluded that environmental factors unique to the individual but common to both depressive disorders and T2D, for example, psychosocial stress, contribute to their co-occurrence in males (11).
Although major part of the link between depression and T2D or glycemic traits may be due to nongenetic factors, some proportion may still be explained by genetic overlap, as suggested by a recent twin study showing moderate, but significant, genetic correlation between MDD and T2D in two Nordic samples, especially in females (11). Despite our limited understanding of the etiology of depression, previous research suggests that common biological pathways for both disorders may relate to functions of the hypothalamic-pituitary-adrenocortical axis, autonomic nervous system, cytokine-mediated inflammatory response, or aberrations in circadian rhythms (35,36). Moreover, it has been proposed that the reciprocal relationship between depressive symptoms and T2D may be mediated by behavioral risk factors that are partially affected by genetic makeup, such as eating habits and life-style choices (37).
To get better insight into the possible biological processes and shared genomic background linking depressive symptoms and glycemic traits, we next identified potential pleiotropic loci between the traits of interest. We observed potential pleiotropic loci for depressive symptoms and T2D in the IGF2BP2 and CDKAL1 genes and near the CDKN2B-AS and PLEKHA1 genes. Variants in the CDKALI1 and PLEKHA1 showed associations in the opposite direction with depressive symptoms and T2D. However, such findings are not uncommon: rs12720356 in the TYK2 is associated with both Crohn's disease and psoriasis, yet the G allele increases risk for Crohn's disease and decreases risk for psoriasis (38,39). Similarly, Sirota and colleagues (40) show cross-autoimmune diseases that opposite effects are frequent. These results address the fact that biological processes underlying phenotypic correlation may operate in both directions. Moreover, depression is a heterogenous disorder and it is possible that some subtypes of depression show associations with genomic variation opposite to the others: McCaffery and colleagues (41) recently tried to replicate the most significant signals in the study by Hek et al. (22) and found that of the eight top SNPs, three associated nominally with depressive symptoms in the opposite direction as in the original study in a sample of overweight/obese subjects with T2D. Of identified variants, CDKALI1 and PLEKHA1 have been implicated not only in T2D and metabolic traits (9,42) but also in neurodevelopmental traits. IGF2BP2 variants have been implicated in schizophrenia (43) and CDKN2B-AS variants in Parkinson's and Alzheimer's diseases, albeit showing only modest association with cognitive function (44). In the same study, no significant associations were found between CDKAL1 variants and Parkinson's and Alzheimer's diseases (44). However, CDKAL1 was 1 of the 226 genes associated with bipolar disorder in a gene-based meta-analyses of four studies (45), and in another study, CDKAL1 harbored an SNP with potential association with externalizing symptoms in bipolar disorder (rs17622252; p = 4.39 × 10−6) (46). Interestingly, the minor allele of our top SNP rs10510110, an intergenic variant near PLEKHA1, was associated with down-regulated FGFR2 expression in substantia nigra. FGFR2 abnormalities have been linked with aberrant glia cell function (47), and MDD patients have shown down-regulated FGFR2 in several brain regions (48). Moreover, an SNP in FGFR2 has been implicated in bipolar disorder (49). Although associated with neurodevelopmental traits, these intronic variants in the IGF2BP2 and CDKAL1 genes and intergenic variants near the CDKN2B-AS and PLEKHA1 genes are novel in relation to depressive symptoms. Interestingly, all the potentially pleiotropic variants that associated with depressive symptoms and T2D altered an expression of genes in various regions of the brain. Future research should focus on those associations and the biological relevance of those SNPs on depression.
For depressive symptoms and fasting glucose, we identified potential pleiotropic loci in the MADD and in PEX16 genes as well as close to the CDKN2B-AS and MTNR1B genes, all novel in relation to depressive symptoms. Importantly, variants in the PEX16 and MTNR1B showed associations in the opposite direction with depressive symptoms and fasting glucose. MADD has been associated with fasting glucose (23), insulin (50), and high-density lipoprotein values (51), but its variation also influences apoptosis and it plays an essential role in Ca2+-dependent neurotransmitter release (52). Recent studies reported up-regulation of PEX16 in patients with chronic fatigue syndrome (CFS) (53,54), and one study showed that allelic distribution of rs3802758 in PEX16 differed between those with CFS and healthy controls (55). This is interesting because CFS often show symptoms typical to depression such as lack of energy and sleep problems (56). Genetic variation in the PEX16 gene has also been associated with fatty acid oxidation (57). The nearest gene to rs6483221 is melatonin receptor 1B gene (MTNR1B). Genetic variation in the MTNR1B gene has been associated not only with T2D-related outcomes (58) but also with sleep (59) and recurrent MDD (60). A possibility that variation in the MTNR1B gene could affect both depression and T2D through effects on circadian regulation also exists (61,62).
Together, these data could suggest that genetic variation in the IGF2BP2, CDKAL1, CDKN2B-AS, MADD, PEX16, and MTNR1B genes may be associated with both metabolic traits and symptoms of depression. Variation in the PLEKHA1 gene could modulate expression of the FGFR2 gene, which, in turn, could influence the development of neurodevelopment diseases. The direction of the association, however, can vary and risk SNPs associated with T2D can show protection against depressive symptoms as seen for SNPs in the IGF2BP2 and in CDKN2B-AS genes. A similar situation was observed for the obesity-associated rs9939609 A-variant in the FTO gene, which has also been associated with reduced risk of MDD (63).
There are some limitations of our study. First, heterogeneity of the phenotypes may alleviate our ability to detect genomic risk loci. In addition, combining results from meta-analyses misses several SNPs, although after quality control, the number of SNPs remained between 2,351,566 and 2,390,179, which should provide adequate genomic coverage. We lacked data sets for replication, and we were not able to test these differences separately in men and women. Future studies should replicate our findings in larger data sets with higher genotypic resolution. Moreover, these analyses should be run separately in males and females. Not only may the factors that influence the association between depression and T2D or glycemic traits differ, but also women may be more vulnerable to comorbid depression and T2D (64). Despite these limitations, we showed that in the currently existing GWAS data sets, no overall genetic relation exists between depressive symptoms and glycemic traits. However, were able to identify several SNPs that showed potential pleiotropy by being associated both with depressive symptoms and T2D and glycemic traits. Although our findings should be replicated in the future studies, these genomic loci implicate biological processes that may explain the bidirectional relationship between depressive symptoms and T2D or glycemic traits. Understanding of the comorbidity may help in classifying biologically more homogenous subtypes of depression and T2D.
Supplementary Material
FIGURE 3.
Regional visualization for rs11039183 of univariate and bivariate GWAS results for depressive symptoms and fasting glucose. GWAS = genome-wide association study.
Acknowledgments
The authors thank the investigators, the staff, and the participants of each contributing cohort in CHARGE, DIAGRAM, and MAGIC consortia.
Source of Funding: K.H. has received funding from the doctoral program of psychology, learning, and education. K.R. was supported by the Academy of Finland. D.A. B. has received grant support from P30AG10161, R01AG15819, and R01AG17917. The Multi-Ethnic Study of Atherosclerosis (MESA) is one of the cohorts included in the CHARGE Depression Working Group GWAS. MESA and the MESA SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute in collaboration with MESA investigators. Support for MESA is provided by contracts HHSN268201500003I, N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079, UL1-TR-001420, UL1-TR-001881, and DK063491. Funding for SHARe genotyping was provided by National Heart, Lung, and Blood Institute contract N02-HL-64278. Genotyping was performed at Affymetrix (Santa Clara, CA) and the Broad Institute of Harvard and MIT (Boston, MA) using the Affymetrix Genome-Wide Human SNP Array 6.0. Funding for analysis of MESA data was also provided by R01 HL101161 and P60 MD 002249.
Glossary
- CFS
chronic fatigue syndrome
- GWAS
genome-wide association study
- HOMA-β
homeostatic model assessment of β-cell function
- HOMA-IR
homeostatic model assessment of insulin resistance
- LDSC
linkage disequilibrium score regression
- MDD
major depressive disorder
- SNP
single-nucleotide polymorphism
- T2D
Type 2 diabetes
Footnotes
Conflicts of Interest: No potential conflicts of interest relevant to this article were reported by contributing authors.
Contribution statement: K. H., J. L., and A.A. wrote the manuscript and researched data. K.R., L.G., and H.S. reviewed and edited the manuscript. Others contributed to the discussion and reviewed the manuscript.
References
- 1.NCD Risk Factor Collaboration (NCD-RisC) Worldwide trends in diabetes since 1980: a pooled analysis of 751 population-based studies with 4.4 million participants. Lancet. 2016;387:1513–30. doi: 10.1016/S0140-6736(16)00618-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguiree F, Brown A, Cho NH, Dahlquist G, Dodd S, Dunning T, Hirst M, Hwang C, Magliano D, Patterson C. IDF Diabetes Atlas. 6. Brussels, Belgium: International Diabetes Federation; 2013. [Google Scholar]
- 3.Ferrari AJ, Charlson FJ, Norman RE, Patten SB, Freedman G, Murray CJ, Vos T, Whiteford HA. Burden of depressive disorders by country, sex, age, and year: findings from the global burden of disease study 2010. PLoS Med. 2013;10:e1001547. doi: 10.1371/journal.pmed.1001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renn BN, Feliciano L, Segal DL. The bidirectional relationship of depression and diabetes: a systematic review. Clin Psychol Rev. 2011;31:1239–46. doi: 10.1016/j.cpr.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Nouwen A, Winkley K, Twisk J, Lloyd CE, Peyrot M, Ismail K, Pouwer F. Consortium ED in D (EDID) R. Type 2 diabetes mellitus as a risk factor for the onset of depression: a systematic review and meta-analysis. Diabetologia. 2010;53:2480–6. doi: 10.1007/s00125-010-1874-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu M, Zhang X, Lu F, Fang L. Depression and risk for diabetes: a meta-analysis. Can J Diabetes. 2015;39:266–72. doi: 10.1016/j.jcjd.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Timonen M, Rajala U, Jokelainen J, Keinänen-Kiukaanniemi S, Meyer-Rochow VB, Räsänen P. Depressive symptoms and insulin resistance in young adult males: results from the Northern Finland 1966 birth cohort. Mol Psychiatry. 2006;11:929–33. doi: 10.1038/sj.mp.4001838. [DOI] [PubMed] [Google Scholar]
- 8.Pyykkonen AJ, Raikkonen K, Tuomi T, Eriksson JG, Groop L, Isomaa B. Depressive symptoms, antidepressant medication use, and insulin resistance: the PPP-Botnia Study. Diabetes Care. 2011;34:2545–7. doi: 10.2337/dc11-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prasad R, Groop L. Genetics of Type 2 diabetes—pitfalls and possibilities. Genes (Basel) 2015;6:87–123. doi: 10.3390/genes6010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jansson M, Gatz M, Berg S, Johansson B, Malmberg B, McClearn GE, Schalling M, Pedersen NL. Gender differences in heritability of depressive symptoms in the elderly. Psychol Med. 2004;34:471–9. doi: 10.1017/s0033291703001375. [DOI] [PubMed] [Google Scholar]
- 11.Kan C, Pedersen NL, Christensen K, Bornstein SR, Licinio J, MacCabe JH, Ismail K, Rijsdijk F. Genetic overlap between Type 2 diabetes and depression in Swedish and Danish twin registries. Mol Psychiatry. 2016;21:903–9. doi: 10.1038/mp.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, Duncan L, Perry JR, Patterson N, Robinson EB, Daly MJ, Price AL, Neale BM. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke T-K, Obsteter J, Hall LS, Hayward C, Thomson PA, Smith BH, Padmanabhan S, Hocking LJ, Deary IJ, Porteous DJ, McIntosh AM. Investigating shared aetiology between Type 2 diabetes and major depressive disorder in a population based cohort. Am J Med Genet B Neuropsychiatr Genet. 2017;174:227–34. doi: 10.1002/ajmg.b.32478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lahti M, Tiihonen J, Wildgust H, Beary M, Hodgson R, Kajantie E, Osmond C, Räikkönen K, Eriksson J. Cardiovascular morbidity, mortality and pharmacotherapy in patients with schizophrenia. Psychol Med. 2012;42:2275–85. doi: 10.1017/S0033291712000396. [DOI] [PubMed] [Google Scholar]
- 15.Hankin BL, Fraley RC, Lahey BB, Waldman ID. Is depression best viewed as a continuum or discrete category? A taxometric analysis of childhood and adolescent depression in a population-based sample. J Abnorm Psychol. 2005;114:96–110. doi: 10.1037/0021-843X.114.1.96. [DOI] [PubMed] [Google Scholar]
- 16.Ayuso-Mateos JL, Nuevo R, Verdes E, Naidoo N, Chatterji S. From depressive symptoms to depressive disorders: the relevance of thresholds. Br J Psychiatry. 2010;196:365–71. doi: 10.1192/bjp.bp.109.071191. [DOI] [PubMed] [Google Scholar]
- 17.Cuijpers P, de Graaf R, van Dorsselaer S. Minor depression: risk profiles, functional disability, health care use and risk of developing major depression. J Affect Disord. 2004;79:71–9. doi: 10.1016/S0165-0327(02)00348-8. [DOI] [PubMed] [Google Scholar]
- 18.MacCallum RC, Zhang S, Preacher KJ, Rucker DD. On the practice of dichotomization of quantitative variables. Psychol Methods. 2002;7:19–40. doi: 10.1037/1082-989x.7.1.19. [DOI] [PubMed] [Google Scholar]
- 19.Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet. 2013;14:483–95. doi: 10.1038/nrg3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porter HF, O'Reilly PF. Multivariate simulation framework reveals performance of multi-trait GWAS methods. Sci Rep. 2017;7:38837. doi: 10.1038/srep38837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inflammation Working Group of the CHARGE Consortium IWG of the C, PM-IWG-XCP, LifeLines Cohort Study LC. Ligthart S, Vaez A, Hsu YH, Stolk R, Uitterlinden AG, Hofman A, Alizadeh BZ, Franco OH, Dehghan A. Bivariate genome-wide association study identifies novel pleiotropic loci for lipids and inflammation. BMC Genomics. 2016;17:443. doi: 10.1186/s12864-016-2712-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hek K, Demirkan A, Lahti J, Terracciano A, Teumer A, Cornelis MC, Amin N, Bakshis E, Baumert J, Ding J, Liu Y, Marciante K, Meirelles O, Nalls MA, Sun YV, Vogelzangs N, Yu L, Bandinelli S, Benjamin EJ, Bennett DA, Boomsma D, Cannas A, Coker LH, de Geus E, De Jager PL, Diez-Roux AV, Purcell S, Hu FB, Rimm EB, Hunter DJ, Jensen MK, Curhan G, Rice K, Penman AD, Rotter JI, Sotoodehnia N, Emeny R, Eriksson JG, Evans DA, Ferrucci L, Fornage M, Gudnason V, Hofman A, Illig T, Kardia S, Kelly-Hayes M, Koenen K, Kraft P, Kuningas M, Massaro JM, Melzer D, Mulas A, Mulder CL, Murray A, Oostra BA, Palotie A, Penninx B, Petersmann A, Pilling LC, Psaty B, Rawal R, Reiman EM, Schulz A, Shulman JM, Singleton AB, Smith AV, Sutin AR, Uitterlinden AG, Volzke H, Widen E, Yaffe K, Zonderman AB, Cucca F, Harris T, Ladwig KH, Llewellyn DJ, Raikkonen K, Tanaka T, van Duijn CM, Grabe HJ, Launer LJ, Lunetta KL, Mosley TH, Jr, Newman AB, Tiemeier H, Murabito J. A genome-wide association study of depressive symptoms. Biol Psychiatry. 2013;73:667–78. doi: 10.1016/j.biopsych.2012.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, Lindgren CM, Mägi R, Morris AP, Randall J, Johnson T, Elliott P, Rybin D, Thorleifsson G, Steinthorsdottir V, Henneman P. New genetic loci implicated in fasting glucose homeostasis and their impact on Type 2 diabetes risk. Nat Genet. 2010;42:105–16. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segrè AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A, Prokopenko I, Kang HM, Dina C, Esko T, Fraser RM, Kanoni S, Kumar A, Lagou V, Langenberg C, Luan J. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of Type 2 diabetes. Nat Genet. 2012;44:981–90. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.World Health Organization. Part 1: Diagnosis and Classification of Diabetes Mellitus. Geneva: Department of Noncommunicable Disease Surveillance; 1999. Definition, Diagnosis and Classification of Diabetes mellitus and its Complications. [Google Scholar]
- 26.Wallace TM, Levy JC, Matthews DR. Use and abuse of HOMA modeling. Diabetes Care. 2004;27:1487–95. doi: 10.2337/diacare.27.6.1487. [DOI] [PubMed] [Google Scholar]
- 27.Radloff LS. The CES-D Scale: a self-report depression scale for research in the general population. Appl Psychol Measur. 1977;1:385–401. [Google Scholar]
- 28.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Patterson N, Daly MJ, Price AL, Neale BM. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X, Hsu YH. Identifying pleiotropic effects: a two-stage approach using genome-wide association meta-analysis data. bioRxiv. 2017 [Google Scholar]
- 30.Tenesa A, Haley CS. The heritability of human disease: estimation, uses and abuses. Nat Rev Genet. 2013;14:139–49. doi: 10.1038/nrg3377. [DOI] [PubMed] [Google Scholar]
- 31.Carnethon MR. Symptoms of depression as a risk factor for incident diabetes: findings from the National Health and Nutrition Examination Epidemiologic Follow-up Study, 1971–1992. Am J Epidemiol. 2003;158:416–23. doi: 10.1093/aje/kwg172. [DOI] [PubMed] [Google Scholar]
- 32.Meurs M, Roest AM, Wolffenbuttel BH, Stolk RP, de Jonge P, Rosmalen JG. Association of depressive and anxiety disorders with diagnosed versus undiagnosed diabetes: an epidemiological study of 90,686 participants. Psychosom Med. 2016;78:233–41. doi: 10.1097/PSY.0000000000000255. [DOI] [PubMed] [Google Scholar]
- 33.Knol MJ, Twisk JW, Beekman AT, Heine RJ, Snoek FJ, Pouwer F. Depression as a risk factor for the onset of Type 2 diabetes mellitus. A meta-analysis. Diabetologia. 2006;49:837–45. doi: 10.1007/s00125-006-0159-x. [DOI] [PubMed] [Google Scholar]
- 34.Mezuk B, Heh V, Prom-Wormley E, Kendler KS, Pedersen NL. Association between major depression and Type 2 diabetes in midlife: findings from the Screening Across the Lifespan Twin Study. Psychosom Med. 2015;77:559–66. doi: 10.1097/PSY.0000000000000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moulton CD, Pickup JC, Ismail K. The link between depression and diabetes: the search for shared mechanisms. Lancet Diabetes Endocrinol. 2015;3:461–71. doi: 10.1016/S2213-8587(15)00134-5. [DOI] [PubMed] [Google Scholar]
- 36.Tully PJ, Baumeister H, Martin S, Atlantis E, Jenkins A, Januszewski A, O Loughlin P, Taylor A, Wittert GA. Elucidating the biological mechanisms linking depressive symptoms with Type 2 diabetes in men: the longitudinal effects of inflammation, microvascular dysfunction, and testosterone. Psychosom Med. 2016;78:221–32. doi: 10.1097/PSY.0000000000000263. [DOI] [PubMed] [Google Scholar]
- 37.Carter J, Swardfager W. Mood and metabolism: anhedonia as a clinical target in Type 2 diabetes. Psychoneuroendocrinology. 2016;69:123–32. doi: 10.1016/j.psyneuen.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 38.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, Anderson CA, Bis JC, Bumpstead S, Ellinghaus D, Festen EM, Georges M, Green T, Haritunians T, Jostins L, Latiano A, Mathew CG, Montgomery GW, Prescott NJ, Raychaudhuri S, Rotter JI, Schumm P, Sharma Y, Simms LA, Taylor KD, Whiteman D, Wijmenga C, Baldassano RN, Barclay M, Bayless TM, Brand S, Büning C, Cohen A, Colombel JF, Cottone M, Stronati L, Denson T, De Vos M, D'Inca R, Dubinsky M, Edwards C, Florin T, Franchimont D, Gearry R, Glas J, Van Gossum A, Guthery SL, Halfvarson J, Verspaget HW, Hugot JP, Karban A, Laukens D, Lawrance I, Lemann M, Levine A, Libioulle C, Louis E, Mowat C, Newman W, Panés J, Phillips A, Proctor DD, Regueiro M, Russell R, Rutgeerts P, Sanderson J, Sans M, Seibold F, Steinhart AH, Stokkers PC, Torkvist L, Kullak-Ublick G, Wilson D, Walters T, Targan SR, Brant SR, Rioux JD, D'Amato M, Weersma RK, Kugathasan S, Griffiths AM, Mansfield JC, Vermeire S, Duerr RH, Silverberg MS, Satsangi J, Schreiber S, Cho JH, Annese V, Hakonarson H, Daly MJ, Parkes M. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–25. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strange A, Capon F, Spencer CC, Knight J, Weale ME, Allen MH, Barton A, Band G, Bellenguez C, Bergboer JG, Blackwell JM, Bramon E, Bumpstead SJ, Casas JP, Cork MJ, Corvin A, Deloukas P, Dilthey A, Duncanson A, Edkins S, Estivill X, Fitzgerald O, Freeman C, Giardina E, Gray E, Hofer A, Hüffmeier U, Hunt SE, Irvine AD, Jankowski J, Kirby B, Langford C, Lascorz J, Leman J, Leslie S, Mallbris L, Markus HS, Mathew CG, McLean WH, McManus R, Mössner R, Moutsianas L, Naluai ÅT, Nestle FO, Novelli G, Onoufriadis A, Palmer CN, Perricone C, Pirinen M, Plomin R, Potter SC, Pujol RM, Rautanen A, Riveira-Munoz E, Ryan AW, Salmhofer W, Samuelsson L, Sawcer SJ, Schalkwijk J, Smith CH, Ståhle M, Su Z, Tazi-Ahnini R, Traupe H, Viswanathan AC, Warren RB, Weger W, Wolk K, Wood N, Worthington J, Young HS, Zeeuwen PL, Hayday A, Burden AD, Griffiths CE, Kere J, Reis A, McVean G, Evans DM, Brown MA, Barker JN, Peltonen L, Donnelly P, Trembath RC. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010;42:985–90. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sirota M, Schaub MA, Batzoglou S, Robinson WH, Butte AJ. autoimmune disease classification by inverse association with SNP alleles. PLoS Genet. 2009;5:e1000792. doi: 10.1371/journal.pgen.1000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCaffery JM, Papandonatos GD, Faulconbridge LF, Erar B, Peter I, Wagenknecht LE, Pajewski NM, Anderson A, Wadden TA, Wing RR. Look AHEAD Research Group. Genetic predictors of depressive symptoms in the Look AHEAD trial. Psychosom Med. 2015;77:982–92. doi: 10.1097/PSY.0000000000000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dorajoo R, Liu J, Boehm B. Genetics of Type 2 diabetes and clinical utility. Genes (Basel) 2015;6:372–84. doi: 10.3390/genes6020372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X, Hui L, Liu Y, Wang Z-Q, You Y, Miao L, Sun SL, Guan SL, Xiang Y, Kosten TR, Zhang XY. The Type 2 diabetes mellitus susceptibility gene IGF2BP2 is associated with schizophrenia in a Han Chinese population. J Clin Psychiatry. 2013;74:e287–92. doi: 10.4088/JCP.12m07846. [DOI] [PubMed] [Google Scholar]
- 44.Chung SJ, Kim MJ, Kim J, Ryu HS, Kim YJ, Kim SY, Lee JH. Association of Type 2 diabetes GWAS loci and the risk of Parkinson's and Alzheimer's diseases. Parkinsonism Relat Disord. 2015;21:1435–40. doi: 10.1016/j.parkreldis.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 45.Nurnberger JI, Koller DL, Jung J, Edenberg HJ, Foroud T, Guella I, Vawter MP, Kelsoe JR. Psychiatric Genomics Consortium Bipolar Group JR. Identification of pathways for bipolar disorder: a meta-analysis. JAMA Psychiatry. 2014;71:657–64. doi: 10.1001/jamapsychiatry.2014.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swaminathan S, Koller DL, Foroud T, Edenberg HJ, Xuei X, Niculescu AB, Nurnberger JI. Characteristics of Bipolar I patients grouped by externalizing disorders. J Affect Disord. 2015;178:206–14. doi: 10.1016/j.jad.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Terwisscha van Scheltinga AF, Bakker SC, Kahn RS, Kas MJ. Fibroblast growth factors in neurodevelopment and psychopathology. Neuroscientist. 2013;19:479–94. doi: 10.1177/1073858412472399. [DOI] [PubMed] [Google Scholar]
- 48.Evans SJ, Choudary PV, Neal CR, Li JZ, Vawter MP, Tomita H, Lopez JF, Thompson RC, Meng F, Stead JD, Walsh DM, Myers RM, Bunney WE, Watson SJ, Jones EG, Akil H. Dysregulation of the fibroblast growth factor system in major depression. Proc Natl Acad Sci U S A. 2004;101:15506–11. doi: 10.1073/pnas.0406788101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang T, Zeng Z, Hu Z, Zheng L, Li T, Li Y, Liu J, Li J, Feng G, He L, Shi Y. FGFR2 is associated with bipolar disorder: a large-scale case-control study of three psychiatric disorders in the Chinese Han population. World J Biol Psychiatry. 2012;13:599–604. doi: 10.3109/15622975.2011.650203. [DOI] [PubMed] [Google Scholar]
- 50.Cornes BK, Brody JA, Nikpoor N, Morrison AC, Chu H, Ahn BS, Wang S, Dauriz M, Barzilay JI, Dupuis J, Florez JC, Coresh J, Gibbs RA, Kao WHL, Liu CT, McKnight B, Muzny D, Pankow JS, Reid JG, White CC, Johnson AD, Wong TY, Psaty BM, Boerwinkle E, Rotter JI, Siscovick DS, Sladek R, Meigs JB. Association of levels of fasting glucose and insulin with rare variants at the chromosome 11p11.2-MADD locus: Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium Targeted Sequencing Study. Circ Cardiovasc Genet. 2014;7:374–82. doi: 10.1161/CIRCGENETICS.113.000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, Pramstaller PP, Penninx BW, Janssens AC, Wilson JF, Spector T, Martin NG, Pedersen NL, Kyvik KO, Kaprio J, Hofman A, Freimer NB, Jarvelin MR, Gyllensten U, Campbell H, Rudan I, Johansson A, Marroni F, Hayward C, Vitart V, Jonasson I, Pattaro C, Wright A, Hastie N, Pichler I, Hicks AA, Falchi M, Willemsen G, Hottenga JJ, de Geus EJ, Montgomery GW, Whitfield J, Magnusson P, Saharinen J, Perola M, Silander K, Isaacs A, Sijbrands EJ, Uitterlinden AG, Witteman JC, Oostra BA, Elliott P, Ruokonen A, Sabatti C, Gieger C, Meitinger T, Kronenberg F, Döring A, Wichmann HE, Smit JH, McCarthy MI, van Duijn CM, Peltonen L. ENGAGE Consortium. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miyoshi J, Takai Y. Dual role of DENN/MADD (Rab3GEP) in neurotransmission and neuroprotection. Trends Mol Med. 2004;10:476–80. doi: 10.1016/j.molmed.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 53.Kaushik N, Fear D, Richards SC, McDermott CR, Nuwaysir EF, Kellam P, Harrison TJ, Wilkinson RJ, Tyrrell DA, Holgate ST, Kerr JR. Gene expression in peripheral blood mononuclear cells from patients with chronic fatigue syndrome. J Clin Pathol. 2005;58:826–32. doi: 10.1136/jcp.2005.025718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kerr JR, Petty R, Burke B, Gough J, Fear D, Sinclair LI, Mattey DL, Richards SC, Montgomery J, Baldwin DA, Kellam P, Harrison TJ, Griffin GE, Main J, Enlander D, Nutt DJ, Holgate ST. Gene expression subtypes in patients with chronic fatigue syndrome/myalgic encephalomyelitis. J Infect Dis. 2008;197:1171–84. doi: 10.1086/533453. [DOI] [PubMed] [Google Scholar]
- 55.Shimosako N, Kerr JR. Use of single-nucleotide polymorphisms (SNPs) to distinguish gene expression subtypes of chronic fatigue syndrome/myalgic encephalomyelitis (CFS/ME) J Clin Pathol. 2014;67:1078–83. doi: 10.1136/jclinpath-2014-202597. [DOI] [PubMed] [Google Scholar]
- 56.Griffith JP, Zarrouf FA. A systematic review of chronic fatigue syndrome: don't assume it's depression. Prim Care Companion J Clin Psychiatry. 2008;10:120–8. doi: 10.4088/pcc.v10n0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lodhi IJ, Semenkovich CF. peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell Metab. 2014;19:380–92. doi: 10.1016/j.cmet.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lyssenko V, Nagorny CL, Erdos MR, Wierup N, Jonsson A, Spegel P, Bugliani M, Saxena R, Fex M, Pulizzi N, Isomaa B, Tuomi T, Nilsson P, Kuusisto J, Tuomilehto J, Boehnke M, Altshuler D, Sundler F, Eriksson JG, Jackson AU, Laakso M, Marchetti P, Watanabe RM, Mulder H, Groop L. Common variant in MTNR1B associated with increased risk of Type 2 diabetes and impaired early insulin secretion. Nat Genet. 2009;41:82–8. doi: 10.1038/ng.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Comai S, Gobbi G. Unveiling the role of melatonin MT2 receptors in sleep, anxiety and other neuropsychiatric diseases: a novel target in psychopharmacology. J Psychiatry Neurosci. 2014;39:6–21. doi: 10.1503/jpn.130009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gałecka E, Szemraj J, Florkowski A, Gałecki P, Bieńkiewicz M, Karbownik-Lewińska M, Lewiński A. Single nucleotide polymorphisms and mRNA expression for melatonin MT(2) receptor in depression. Psychiatry Res. 2011;189:472–4. doi: 10.1016/j.psychres.2011.01.021. [DOI] [PubMed] [Google Scholar]
- 61.Gale JE, Cox HI, Qian J, Block GD, Colwell CS, Matveyenko AV. Disruption of circadian rhythms accelerates development of diabetes through pancreatic beta-cell loss and dysfunction. J Biol Rhythms. 2011;26:423–33. doi: 10.1177/0748730411416341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Germain A, Kupfer DJ. Circadian rhythm disturbances in depression. Hum Psychopharmacol. 2008;23:571–85. doi: 10.1002/hup.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Samaan Z, Anand SS, Anand S, Zhang X, Desai D, Rivera M, Pare G, Thabane L, Xie C, Gerstein H, Engert JC, Craig I, Cohen-Woods S, Mohan V, Diaz R, Wang X, Liu L, Corre T, Preisig M, Kutalik Z, Bergmann S, Vollenweider P, Waeber G, Yusuf S, Meyre D. The protective effect of the obesity-associated rs9939609 A variant in fat mass– and obesity-associated gene on depression. Mol Psychiatry. 2013;18:1281–6. doi: 10.1038/mp.2012.160. [DOI] [PubMed] [Google Scholar]
- 64.Demmer RT, Gelb S, Suglia SF, Keyes KM, Aiello AE, Colombo PC, Galea S, Uddin M, Koenen KC, Kubzansky LD. Sex differences in the association between depression, anxiety, and Type 2 diabetes mellitus. Psychosom Med. 2015;77:467–77. doi: 10.1097/PSY.0000000000000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.