Abstract
Context.
The oncolytic polio-rhinovirus recombinant (PVSRIPO) has demonstrated promise in currently ongoing phase I/II clinical trials against recurrent glioblastoma and was granted breakthrough therapy designation by the Food and Drug Administration/Center for Biologics Evaluation and Research. A reliable clinical assay to document expression of the poliovirus receptor, CD155, in routinely available patient tumor samples is needed for continued clinical development of PVSRIPO oncolytic immunotherapy in primary brain tumors and beyond.
Objectives.
To validate a novel anti-CD155 antibody for immunohistochemistry and develop a robust, reliable, and specific protocol for detecting CD155 expression in glioblastoma formalin-fixed, paraffin-embedded (FFPE) tissue samples. To characterize the expression of CD155 in human glioblastoma cells as well as to evaluate the influence of CD155 expression levels on tumor cell susceptibility to PVSRIPO infection and killing.
Design.
Immunohistochemical staining on glioblastoma FFPE tissue sections and immunoblot of corresponding frozen tissues were performed. Positive controls were confirmed sites of poliovirus propagation, spinal cord anterior horn, and tonsils; negative controls were vascular smooth muscle in patient samples and FFPE sections from a confirmed CD155-negative Burkitt lymphoma line (Raji).
Results.
We succeeded in developing a reliable assay to specifically detect CD155 by immunohistochemistry in glioblastoma FFPE sections. Our data suggest widespread, virtually universal expression of CD155 in glioblastoma cells at levels commensurate with susceptibility to PVSRIPO infection and killing.
Conclusions.
Anti-CD155 antibody D3G7H achieves monospecific detection of CD155 in immunoblots of tumor homogenates and immunohistochemistry of tumor FFPE sections. Our assay has utility in defining appropriate use of PVSRIPO in oncolytic immunotherapy against malignant glioma and other cancer histotypes.
Malignant gliomas are a family of highly aggressive brain tumors, with glioblastoma representing the most frequent and most malignant type. Median survival for newly diagnosed glioblastoma with the current standard of care, including maximum safe surgical resection, radiotherapy, and concomitant chemotherapy with temozolomide, is 14.6 months.1 The modest survival improvement achieved with currently approved therapies is plagued by systemic toxicities and a poor health-related quality of life. Hence, there is a dire need for the development of new therapeutics to improve glioblastoma patient survival. The oncolytic polio-rhinovirus recombinant (PVSRIPO), a nonpathogenic human poliovirus (PV) vaccine, is showing promise in clinical trials for glioblastoma. This approach takes advantage of the marked tropism of PV for solid cancers mediated by natural ectopic overexpression of the human PV receptor (PVR), CD155, on the surface of neoplastic cells.2 Infection and killing of cancer cells with PVSRIPO initiates a broad range of proinflammatory and immunogenic events that may recruit adaptive immune-effector responses against the tumor.3
CD155, aka the PVR or nectinlike molecule 5 (Necl-5), is a cell adhesion molecule of the immunoglobulin (Ig) super-family.4 Functional CD155 is a 417-amino acid (aa) membrane-anchored glycoprotein consisting of a putative signal peptide (1–20 aa), an extracellular domain with 3 Ig-like loops (domains 1–3 [D1–3]; 21–343 aa), a transmembrane domain (344–367 aa), and a cytoplasmic domain (368–417 aa)5,6; D1 resembles an Ig-variable domain, whereas D2 and D3 are similar to Ig-constant domains (Figure 1, A). CD155 is heavily glycosylated at 8 putative glycosylation sites in D1–3.6,7 Alternative splicing of CD155 mRNA yields CD155-α/δ, membrane-bound forms that differ only in their C-terminal cytoplasmic domains; CD155-β/γ lack transmembrane domains and are secreted.8,9 Poliovirus binds to the D1 loop of the CD155 glycoprotein.10–12
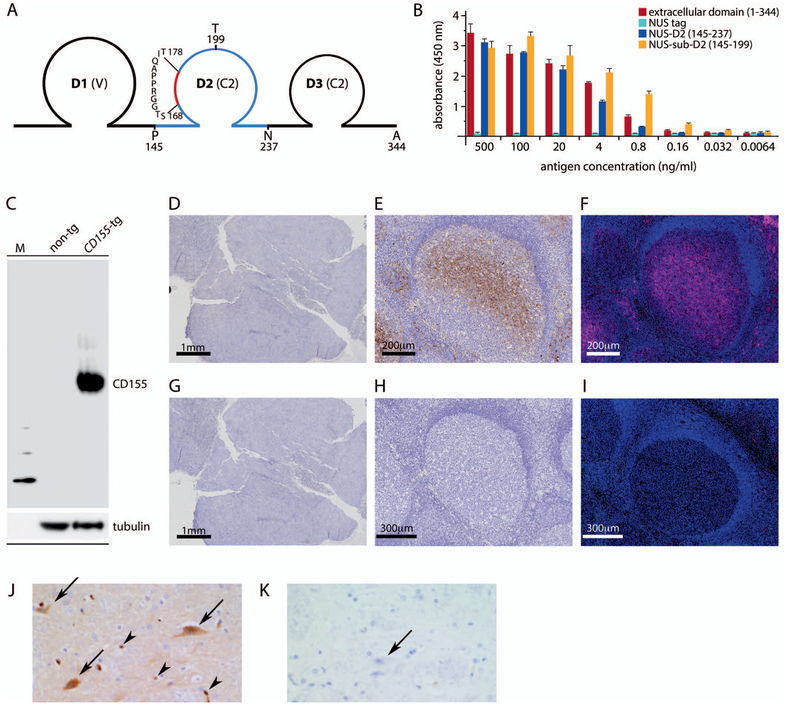
Figure 1.
Validation of anti-CD155 monoclonal antibody D3G7H in tissues with known CD155 status. A, Schematic of the CD155 extracellular domain; A344 marks the border of the transmembrane domain. Polypeptides tested for epitope mapping (domain 2 [D2; amino acids (aa) 145–237]; sub-D2 [aa 145–199]) are indicated in dark blue; a predicted B-cell epitope (aa 168–178) is indicated in red (see Materials and Methods for details). B, Epitope mapping of D3G7H. Reactivity of D3G7H against the CD155 extracellular domain, N-utilization substance (NUS)–tagged CD155 (145–237) and CD155 (145–199), or the NUS-tagged polypeptide tag alone was probed by enzyme-linked immunosorbent assay. C, CD155 immunoblot in spinal cord homogenates from wild-type mice (non-tg) and CD155-transgenic mice (CD155-tg). Note the absence of nonspecific staining. D through F, D3G7H immunohistochemistry (IHC) and immunofluorescence in Raji xenograft and human tonsil tissues. D, Raji xenograft, IHC, diaminobenzidine [DAB] stain. E, Human tonsil, IHC, DAB stain. F, Immunofluorescence, human tonsil, Opal 570. G through I, The corresponding assays with isotype-matched nonspecific IgG control. J and K, D3G7H IHC in the primate spinal cord anterior horn (DAB stain). J, CD155-positive staining in anterior horn motor neurons (arrows) as well as in the capillary endothelial cells seen in longitudinal and cross sections (arrowheads). K, There was no staining in corresponding isotype-matched nonspecific IgG controls (arrow) (original magnifications ×4 [D and G], ×10 [E, F, H, and I], and ×20 [J and K]).
Despite its role as a key pathogenic factor in paralytic poliomyelitis, CD155’s distribution in the normal primate organism remains uncertain. In humans/chimpanzees, only gastrointestinal epithelium, gastrointestinal-associated lymphatic tissues, and spinal cord are sites of significant PV propagation (implying CD155 expression).13,14 There is scant immunohistochemistry (IHC) evidence for CD155 expression in primates, except a study in the gastrointestinal tract matching CD155 expression to known sites of PV replication.15 There also is some evidence for PV presence in myeloid cells expressing CD155 (macrophages, dendritic cells)16,17 and in vascular endothelial cells16 of infected primates, although these cells likely are not significant reservoirs for PV propagation in vivo. One reason for an incomplete understanding of CD155 distribution is the lack of antibodies suitable for IHC (see Results).
Many IHC studies have yielded evidence for broad, ectopic CD155 upregulation in many cancer histotypes,3,4 albeit with immunologic probes used in assays that were not vetted in normal tissues in parallel. The present study was undertaken to establish a reliable method for unambiguous, semiquantitative analysis of CD155 expression in routinely available patient tumor tissue samples. We sought to validate our approach by including IHC for CD155 in known sites of PV replication/CD155 expression and excluding it in confirmed negative controls. Moreover, our studies address the influence of CD155 expression levels on PVSRIPO propagation and killing and document the utility of CD155 IHC as a companion diagnostic in conjunction with PVSRIPO oncolytic immunotherapy.
MATERIALS AND METHODS
Study Population, Patient Tumor Tissues, and Vertebrate Animal Tissues
De-identified archival tissue samples (62 glioblastoma and 1 giant cell glioblastoma; Table 1) were obtained with institutional review board approval from the Preston Robert Tisch Brain Tumor Center Biorepository at Duke University Medical Center, Durham, North Carolina. Cases were selected from archived formalin-fixed, paraffin-embedded (FFPE) or cryogenic tumor blocks stemming from resected tissue. The material consisted of representative FFPE glioblastoma sections, of which 34, 27, and 2 were from surgeries for newly diagnosed, recurrent, and progressive disease, respectively, at Duke University Medical Center (Table 1). The study population included 34 men and 29 women (primarily of white descent; N = 62) who were 24 to 82 years of age (average, 57 years) at the time of resection. Tissue blocks were selected by a Duke University Medical Center neuropathologist as ~80% to 100% viable tumor and more than 1 cm2 of tissue by light microscopic examination of hematoxylineosin–stained sections. Serial unstained sections were cut from these FFPE and frozen blocks and stained for CD155 by IHC or immunoblot. Raji cells (Cat. No. CCL-86, ATCC, Manassas, Virgina) were used to initiate subcutaneous tumors in athymic Balb/c mice as described previously;18 resected Raji tumor tissue was processed to obtain FFPE sections. Spinal cord homogenates were surplus tissues from euthanized wild-type and CD155-transgenic C57Bl6 mice, respectively. Institutional guidelines regarding animal experimentation were followed.
Table 1.
| Patient Characteristics | ||
|---|---|---|
| Characteristic | No. (%) of Cases (N = 63) |
|
| Sex | ||
| Male | 34 (54) | |
| Female | 29 (46) | |
| Age, y | ||
| ≤45 | 11 (17.5) | |
| >45 | 52 (82.5) | |
| Tumor status | ||
| Newly diagnosed | 34 (54) | |
| Recurrent | 27 (43) | |
| Progressed | 2 (3) | |
Construction, Expression, and Purification of CD155-Derived Polypeptides
Complementary DNAs encoding the CD155 D2 (aa 145–237) or sub-D2 (aa 145–199) were synthesized (GenScript, Piscataway, New Jersey) for insertion into the EcoRI/HinDIII sites of the pET43.1a(+) expression vector (EMD Biosciences, San Diego, California) and the sequences verified (GenScript). Individual CD155 expression constructs were transformed and expressed under control of the T7 promoter in Escherichia coli BL21 (λ DE3) (Stratagene, La Jolla, California). The bacteria were harvested by centrifugation at 8000g (10 minutes, 4°C) and the resulting pellets were resuspended in extraction buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl) containing lysozyme (Sigma-Aldrich, St Louis, Missouri). The bacterial suspension was incubated at room temperature (20 minutes) with gentle shaking for complete cell lysis to occur. The cells were further disrupted by sonication on ice and centrifuged at 16 000g (20 minutes). The insoluble cell debris was discarded and the supernatant containing the crude protein preparation was dialyzed against 1× phosphate-buffered saline (12 hours, 4°C), filtered through a 0.2-μm filter (Millipore, Billerica, Massachusetts), and used for downstream applications.
D3G7H Epitope Mapping by Enzyme-Linked Immunosorbent Assay
Different concentrations of the recombinant CD155 extracellular domain (Cat. No. CD5-H5223, Acro Biosystems, Newark, Delaware), N-utilization substance–tagged CD155 D2/sub-D2, or the N-utilization substance–tagged polypeptide alone were added to 96-well plates and incubated (12 hours, 4°C). The plates were then blocked with SuperBlock buffer (30 minutes, 37°C; Pierce, Rockford, Illinois) and incubated with (1) 0.5 μg/mL of D3G7H monoclonal antibody (mAb) (1 hour, 37°C); 2) horseradish peroxidase–conjugated anti-rabbit IgG antibody (1 hour, 37°C; Life Technologies Corporation, Carlsbad, California); or (3) 3,3’,5,5’-tetramethylbenzidine substrate solution (Pierce). The reaction was terminated with stop solution (Pierce) and the optical density was measured at 450 nm.
IHC, Histopathology, Immunofluorescence, Scoring, and Immunoblot
Serial FFPE sections (5-μm thickness) were stained for CD155 using automated IHC techniques on a Bond-maX Processing Module (Leica Microsystems, Buffalo Grove, Illinois), using the Bond Polymer Refine Detection kit (Cat. No. DS9800, Leica Microsystems). The FFPE sections were deparaffinized, hydrated with alcohol, and subjected to heat-induced epitope retrieval with Bond Epitope Retrieval Solution 1 (citrate buffer, pH 6.0, Cat. No. AR9961, Leica Microsystems). Slides were then washed with bond wash solution (Cat. No. AR9590, Leica Microsystems) and exposed to peroxide block (10 minutes). The sections were sequentially blocked with Fc receptor blocker (45 minutes; Cat. No. NB309, Innovex Biosciences, Richmond, California) and protein block (30 minutes; Cat. No. RE7102-CE, Leica Microsystems). We used the following anti-CD155 antibodies: rabbit polyclonal (aa 314–342) IHC-plus (Cat. No. LS-B10536, LSBio, Seattle, Washington), mouse mAb (Cat. No. 337602, Biolegend, San Diego, California), rabbit polyclonal (Cat. No. NBP1-02520, Novus, Littleton, Colorado); rabbit mAb (Cat. No. 13544, Cell Signaling Technology, Danvers, Massachusetts), D171,19 and D480.20 Anti-CD155 antibody was applied to the tissue sections at a concentration of 5 μg/mL (60 minutes). Sections were then treated sequentially with polymer (8 minutes), mixed 3,3’-diaminobenzidine refine (10 minutes), and hematoxylin solutions (5 minutes). CD155 expression was defined as membranous and cytoplasmic staining on tumor cells and endothelial cells. The histologic sections were scored for IHC intensity by a neuropathologist using the criteria described for evaluating human epidermal growth factor receptor 2 (HER2) expression in breast cancer,21 where scores indicate the intensity of staining as follows: 0, no staining; 1+, weak reactivity in 10% of cells or less; 2+, weak to moderate reactivity in more than 10% of cells; and 3+, strong reactivity in 30% of cells or more. CD155 immunofluorescence on tonsil FFPE sections was performed using the Opal kit (Cat. No. NEL796001KT, PerkinElmer, Shelton, Connecticut) following the manufacturer’s instructions. Briefly, the slides were deparaffinized and rehydrated in ethanol and antigen retrieval was performed in Target Antigen Retrieval buffer AR9 (Cat. No. AR900250ML, PerkinElmer) using microwave incubation. Rabbit mAb (Cat. No. 13544, Cell Signaling Technology) or rabbit (DA1E) isotype control (Cat. No. 3900, Cell Signaling Technology) antibodies were applied to sections at a concentration of 1 μg/mL (30 minutes) in a humidified chamber at room temperature. Sections were then incubated in secondary antibody working solution (10 minutes; Cat. No. ARH1001EA, PerkinElmer) at room temperature. Visualization of CD155 was accomplished using Opal 570 (1:50), after which the slides were placed in AR9 buffer and heated using microwave incubation. Nuclei were subsequently stained with 4’,6-diamidino-2-phenylindole solution (Cat. No. NEL796001KT, PerkinElmer), and the sections were coverslipped using Vectashield HardSet Antifade mounting media (Cat. No. H-1200, Vector Laboratories, Burlingame, California). The slides were scanned using the Vectra 3.0 System and image analysis was performed using the InForm image analysis software (both PerkinElmer). For immunoblot, frozen tissue sections (50-μm thickness) from each tumor were treated with the appropriate volume of T-PER Tissue Protein Extraction Reagent (Cat. No. 78510, Life Technologies) and subjected to homogenization. Total protein concentration was determined by Bradford method and 30 μg of protein (per sample) was subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis and immunoblot as described previously.22
PVSRIPO, CD155-Expressing Murine Cell Lines, and Quantitative Fluorescence-Activated Cell Sorter Analysis
Derivation, propagation and purification of PVSRIPO, and PVSRIPO infection procedures are described elsewhere.23 TRAMP-C2 cells (Cat. No. CRL-2731, ATCC) were transduced by transfection with the pSVL-H20A CD155 expression plasmid (a gift of E. Wimmer, PhD)6 and clonally selected as described before.23 CD155 numbers on the surface of transduced TRAMP-C2 cells were determined by quantitative fluorescence-activated cell sorting (qFACS), using the Quantum Simply Cellular anti-mouse IgG kit (Bangs Laboratories, Inc, Fishers, Indiana) as described previously.24 Briefly, a cocktail of uniform-sized beads, 1 blank and 4 with varying capacities to bind mouse IgG, and the cells were stained with 10 μg/mL of mouse IgG1-AF488 (isotype control Ab) and D171-AF488 (CD155-specific Ab) at 4°C (45 minutes). After washing, the beads and the cells were analyzed on a Becton Dickinson (San Jose, California) FACSCalibur instrument. Analysis of receptor density was performed by interpolation with the bead standard curves using QuickCal analysis software provided with the kit.
RESULTS
CD155 is emerging as a molecule of central interest in tumor immunology (see Discussion) and as a therapeutic target in cancer immunotherapy, for example, with the oncolytic immunotherapy agent PVSRIPO.20 Thus, developing reliable methods to determine CD155 expression in routinely available patient samples is an urgent priority.
Validating Anti-CD155 Antibody D3G7H
In the past, reliable CD155 detection by immunoblot or IHC has been hampered by the lack of suitable antibodies. The first anti-CD155 antibody generated (D171), a mouse mAb raised against fractionated HeLa cell membranes,19 led to the identification of CD155 as the PVR6 by virtue of its ability to block PV infection. D171, however, poorly recognizes denatured CD155 and exhibits profuse non-specific staining (eg, in immunoblot),25 and hence is unsuitable for IHC in FFPE materials. To address this shortcoming, we previously developed a 2-step immunoblot protocol (immunoprecipitation of CD155 from non-denatured tumor homogenates with D171 mAb, followed by immunoblot with polyclonal probes).20 This procedure circumvented the limitations of D171 and eliminated nonspecific background staining in CD155 immunoblots; however, this cumbersome and nonquantitative approach is not applicable for IHC.
To develop clinically viable IHC methods, we first conducted validation assays with a range of anti-CD155 antibodies in FFPE sections of confirmed positive and negative control tissues (Figure 1, A through K). These included assays in 2 tissues harboring confirmed sites of productive PV replication, tonsils and spinal cord,13,14 and a human-derived tissue with confirmed CD155 absence, Raji cell xenografts (the CD155 gene is transcriptionally silenced in Epstein-Barr virus–infected lymphomas).26 Our tests included antibodies D171, D480 (a rabbit polyclonal serum raised against the CD155 extracellular domain),20 and several commercially available probes (rabbit polyclonal [aa 314–342] IHC-plus, Cat. No. LS-B10536, LSBio; mAb, Cat. No. 337602, Biolegend; and rabbit polyclonal, Cat. No. NBP1–02520, Novus). Examination of IHC results in FFPE sections by the study pathologist revealed lacking specificity of all tested antibodies, suggesting their unsuitability for a robust clinical assay (data not shown).
Recently, a newly developed rabbit anti-CD155 mAb (D3G7H, Cell Signaling) has demonstrated unprecedented specificity/avidity in immunoblots from spinal cord homogenates of CD155-transgenic mice (see below). To determine the D3G7H epitope that conveys unique specificity for CD155, truncation variants of the CD155 extracellular domain corresponding to aa 145–237 and 145–199 were generated (Figure 1, A). D3G7H exhibited strong reactivity for both these CD155 fragments in an enzyme-linked immunosorbent assay (Figure 1, B). B-cell–epitope prediction analysis of the CD155 extracellular domain through the immune epitope database and analysis resource (http://www.iedb.org; accessed September 14, 2016) identified a single putative epitope at aa 168–178 in CD155 D2 (Figure 1, A). These results indicated that the epitope recognized by the D3G7H mAb is in the region spanning aa 145–199 of CD155 D2.
The remarkable specificity and avidity of the D3G7H probe for human CD155 in transgenic mouse spinal cord (Figure 1, C) was also evident in human tissues (see below). Anti-CD155 mAb D3G7H did not exhibit positive staining in FFPE sections from Raji cell xenografts (Figure 1, D and G), recapitulating the monospecificity observed in immunoblots of murine (Figure 1, C) or human tissue homogenates (see below). We observed staining for CD155 in FFPE human tonsil sections with D3G7H, both by IHC (Figure 1, E and H) and by immunofluorescence (Figure 1, F and I), in a pattern matching a prior immunofluorescence study with D171 in fresh-frozen sections from rhesus macaque Peyer patches.15 CD155 was present mainly in cells located in the germinal centers.
Poliovirus CNS infections are defined by specificity for lower motor neurons, and flaccid paralysis is pathognomonic for poliomyelitis; accordingly, studies in mice transgenic for a CD155-promoter reporter suggested CD155 gene expression to be confined to the floor plate/anterior horn/lower motor neuron compartment during embryonic development.2 To our knowledge, successful CD155 IHC in the primate spinal cord has never been reported. We performed IHC with D3G7H in FFPE spinal cord tissues from cynomolgus macaques, a World Health Organization–standard primate species for evaluating PV neurovirulence. Cynomolgus macaques develop the clinical and histopathologic hallmarks of human paralytic poliomyelitis upon exposure to neurovirulent PVs.27 Our tests revealed specific staining for CD155 in anterior horn motor neurons, consistent with the tropism and pathogenic profile of PV (Figure 1, J [arrows] and K). We also observed CD155 staining in normal CNS endothelial cells as well as in capillary endothelial cells seen in the longitudinal and cross sections (Figure 1, J, arrowheads), in line with earlier observations of PV particles in CNS endothelial cells by electron microscopy16 and more recent mechanistic studies of CD155’s involvement in transendothelial migration.28 These data suggest that (1) D3G7H recognizes an epitope in CD155 D2, possibly the putative B cell epitope at aa 168–178, (2) D3G7H is monospecific for human CD155 in immunoblot in transgenic murine tissues, (3) D3G7H is devoid of immune reactivity in a confirmed CD155-negative human-derived tissue, (4) D3G7H detects CD155 in normal primate/human tissues known to support PV infection/replication, and (5) D3G7H recognizes CD155 in cells that may not support productive PV replication (eg, vascular endothelium) but execute physiologic functions associated with CD155 expression.
CD155 IHC in Glioblastomas
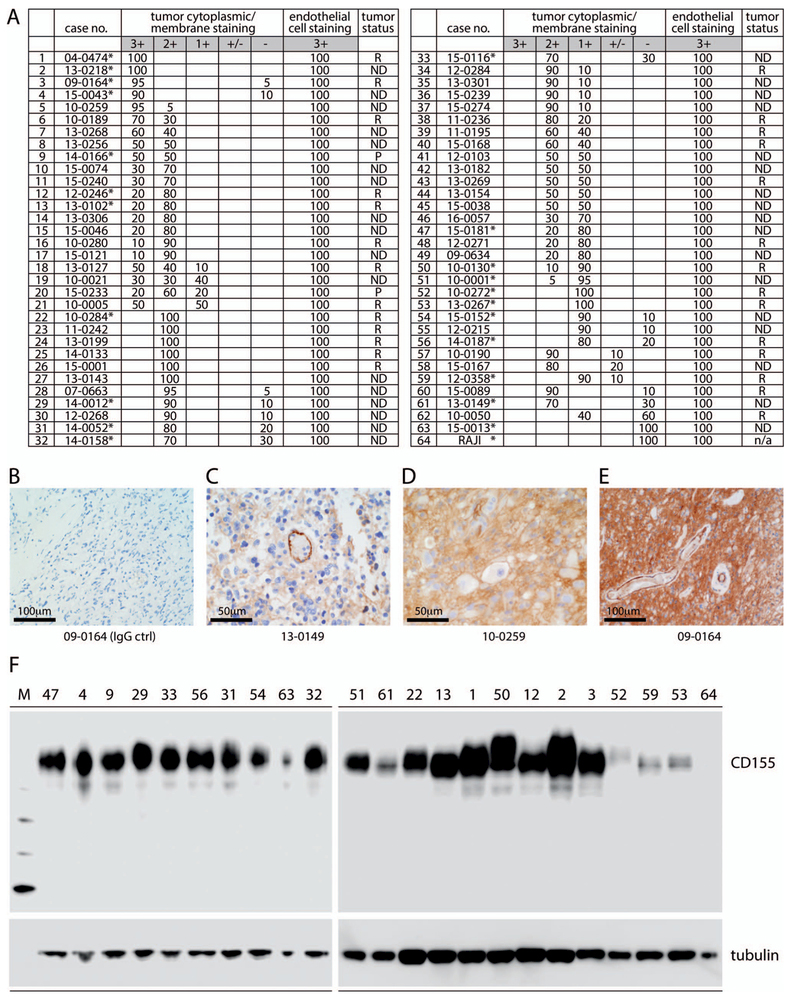
The positive validation of the anti-CD155 probe D3G7H described above qualified it for IHC tests in FFPE sections from glioblastoma patients (Figure 2, A through F). We evaluated CD155 expression in the tumor tissues of 63 patients with glioblastomas. In each case, the endothelial cells proved to exhibit 3+, dark brown stain product (Figure 2, A, C, and E) and were used to compare the tumor cell reactivity with intermediate (2+ in 5% tumor; case 10-0259; Figure 2, D) and dark brown staining (3+ in 95% tumor; case 09–0164; Figure 2, E). Each case was evaluated for percentage of positive cells (>2+ intensity) and negative cells (0–1+ reactivity). Necrotic regions were ignored. The immunoreactivity patterns in the tumors always demonstrated membranous staining (Figure 2, D); however, most tumors exhibited cells with prominent cytoplasmic staining as well (Figure 2, E). There was significant heterogeneity of D3G7H immunoreactivity within the tumors, as highlighted in Figure 2, A. Nuclear reactivity was not noted. Overall, 49 cases (78%) demonstrated more than 50% 2+ to 3+ reactivity by IHC (Figure 2, A). Of note, only one case in this small series was considered to be negative for CD155 expression.
Figure 2.
CD155 expression in glioblastoma. A, Table summarizing CD155 expression scores in 63 glioblastomas and in Raji xenograft tissue (line 64) with anti-CD155 monoclonal antibody (mAb) D3G7H immunohistochemistry (IHC) (see Materials and Methods for scoring method). Endothelial staining was scored separately. Tumor status is referred to as newly diagnosed (ND), recurrent (R), or progressive (P). Asterisks mark tumor samples that were analyzed by immunoblot in parallel (F). B through E, Examples of glioblastoma IHC with isotype-matched nonspecific IgG control (ctrl) (B) or anti-CD155 mAb D3G7H IHC (C through E) demonstrating 3+ endothelial staining (C), 2+ tumor staining (D), and 3+ tumor staining (E) in glioblastoma cells. In each case, 3+ endothelial cell staining served as the internal positive control; numbers below the panels indicate the case number (see A). F, CD155 immunoblot with anti-CD155 mAb D3G7H in select glioblastoma samples and in Raji xenograft tissue (line 64). The numbers on top correspond to line numbers for individual cases (original magnifications ×10 [B and E] and ×20 [C and D]).
CD155 Immunoblot in Glioblastoma
To supplement IHC findings, we also performed immunoblot of 23 glioblastoma samples with the anti-CD155 probe D3G7H (Figure 2, F). As in immunoblot of murine tissues (Figure 1, A), we observed monospecificity of D3G7H in human glioblastoma samples (Figure 2, F). Variable electrophoretic mobility of CD155 in distinct nonmalignant tissues or individual tumors is commonly observed and is due to tissue type–specific glycosylation. CD155 levels determined by immunoblot generally supported the IHC findings, for example, when comparing high-expressing cases (line numbers <10) with lower-expressing ones (line numbers >50). Definitive quantitative measurements of CD155 expression levels in patient tumor tissues, however, should not be based on immunoblot data. A significant presence of CD155 in abundant nonmalignant/stromal tumor components (vascular endothelium and myeloid cells) makes this unreliable, as stromal contents vary substantially individually (patient to patient) and intratumorally (sample to sample). For these reasons, we did not attempt to obtain quantitative measurements of CD155 expression in immunoblots or to correlate quantitative measurements with IHC data.
CD155 IHC as a Companion Diagnostic for PVSRIPO
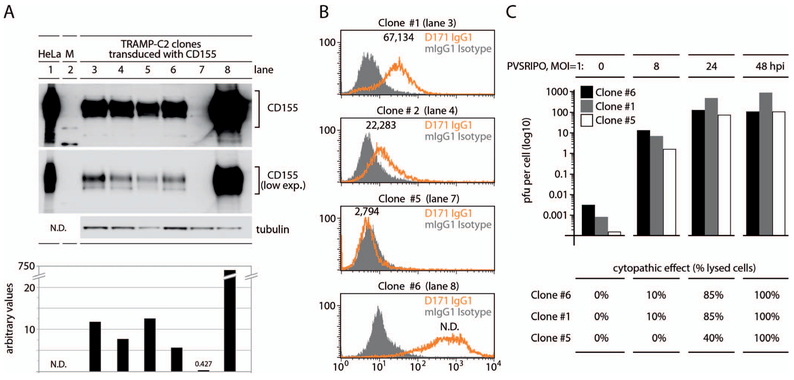
CD155 alone is sufficient for mediating PV susceptibility,6 and positive CD155 IHC correlates with known sites for PV replication (Figure 1, E, F, and J). Thus, CD155 expression suggests virtually universal susceptibility of glioblastoma to PVSRIPO.3 Because CD155 expression in glioblastoma was variable, it was important to gauge the possibility that CD155 expression levels may influence PVSRIPO tumor targeting. To test this, we carried out assays in a genetically engineered mouse prostate cancer model, TRAMP-C2,29 transduced with human CD155 (Figure 3, A through C). The mouse CD155 homolog does not function as a PVR, and thus murine cells such as TRAMP-C2 do not allow PV entry. Therefore, ectopic PVR expression in TRAMP-C2 cells enables tests to correlate CD155 expression levels with PV susceptibility. Six TRAMP-C2 clones, selected for stable transduction with CD155, exhibited inherently distinct CD155 expression levels (Figure 3, A). Immunoblot signal for the lowest-expressing clone, clone 5, was exceedingly weak and was apparent only with excessive exposure (Figure 3, A). Densitometric analysis suggested that CD155 immunoblot signal in the highest-expressing clone, clone 6, exceeded that of clone 5 ~1700-fold (Figure 3, A). Widely diverging expression levels were also evident in FACS/qFACS analyses (Figure 3, B). We carried out PVSRIPO infections of TRAMP-C2 clones 5 and 6, representing expression extremes, and of clone 1, with intermediate CD155 levels, and assessed viral propagation and cytopathogenicity (Figure 3, C). One-step growth curves of PVSRIPO revealed enhanced attachment in clone 6 versus clone 5, evident as ~15-fold elevated recovery of infectious particles from cultures lysed after a 30-minute attachment step at room temperature23 (Figure 3, C). This suggests that elevated CD155 expression levels favor virus attachment to cells, an expected finding. However, with the progress of virus propagation (upon transfer of infected cultures to 37°C), viral titers detected in infected clone 5 cultures caught up and were roughly even with those of clone 6 at 48 hours postinfection (Figure 3, C). Poliovirus/PVSRIPO causes drastic, rapid cytopathogenic effects leading to gross morphologic changes and cell detachment. Unsurprisingly, unbridled PVSRIPO propagation in clones 5 and 6 resulted in complete cytolysis 48 hours postinfection (Figure 3, C). These data suggest that even a marginal CD155 immunoblot signal (Figure 2, F, and 3, A) corresponding to marginally detectable IHC (Figure 2, A) and FACS (Figure 3, B) signals is commensurate with full susceptibility to PVSRIPO infection and tumor cell killing and, hence, oncolytic immunotherapy.
Figure 3.
CD155 expression (exp.) levels do not correlate with susceptibility to, propagation of, and cancer cell killing by polio-rhinovirus recombinant (PVSRIPO). A, CD155 immunoblot of lysates from a panel of transduced TRAMP-C2 lines; quantitation of immunoblot signal is shown below. B, Standard fluorescence-activated cell sorter (FACS)/quantitative FACS (qFACS) analyses of CD155 in TRAMP-C2 clones 1, 2, 5, 6; lane numbers corresponding to A are shown. Numbers atop the orange peaks represent CD155 receptors per cell as determined by qFACS (see Materials and Methods); receptor numbers in clone 6 were outside the linear range and could not be evaluated (N.D.). C, TRAMP-C2 clone 1, 5, and 6 cultures were infected with polio-rhinovirus recombinant (PVSRIPO) (multiplicity of infection [MOI] = 1) and lysed at the indicated intervals, and plaque-forming units (pfu) per cell were determined by standard plaque assay. The bottom panel indicates the approximate percentage of cells exhibiting cytopathogenic effects in cultures at the corresponding intervals after infection. Abbreviation: hpi, hours postinfection.
DISCUSSION
CD155 is the founding member of the nectin family of cell adhesion molecules.4 Of the nectin family members, CD155 and CD112 have gained particular attention because of their emerging roles in modulation of the innate immune response to infection, in tumor immunobiology, and as receptors for oncolytic viruses under clinical investigation (CD112 is a receptor for α-herpesviruses).30 CD112 and CD155 are ligands for the activating receptor CD226 (DNAM-1) present on natural killer cells,31 but also interact with inhibitory checkpoint molecules CD96 and T-cell immunoreceptor with Ig and ITim domains (TIGIT).32 This places CD155 at the center of a regulatory system with decisive roles in the innate defense against transformed or infected cells.32
In this context, widespread ectopic CD155 expression in solid neoplasia is of interest. The mechanisms of CD155 upregulation in cancer are poorly understood; transcriptional induction of the CD155 gene has been linked to disruption of tissue architecture with injury or malignant transformation33 or with the DNA-damage response.34 Also, the immunobiological significance of CD155 expression in neoplasia is unknown. Instances of both suppression35 and stimulation36 of immune effector functions were linked to CD155 upregulation in neoplastic cells. CD155 down-regulation occurring with DNA/retrovirus infection has been interpreted as a mechanism of immune evasion.37,38
CD155 expression in glioblastoma cells renders them susceptible to PV infection and killing, an indispensable aspect of PVSRIPO oncolytic immunotherapy.2 In our report, we provide details for a robust, reliable, and specific IHC method for detecting CD155 in FFPE tumor tissue sections based on a commercially available anti-CD155 antibody with unprecedented specificity. Our studies corroborate earlier evidence for an association of CD155 with glioblastoma,2,20 extend these initial findings to indicate virtually universal expression in glioblastoma, and confirm similar systematic analyses reaching the same conclusions in other cancer histotypes with different techniques, for example, FACS analyses in melanoma.39 Studies correlating CD155 expression levels in clonal cell populations with PVSRIPO susceptibility indicate that even marginal CD155 expression levels (determined by immunoblot or qFACS) mediate full susceptibility to viral infection and cell killing. Availability of a robust IHC assay for detecting CD155 will enable preclinical assessments of the suitability of PVSRIPO oncolytic immunotherapy in cancers other than glioblastoma.
Acknowledgments
This work was supported by the following National Institutes of Health grants: P01-5P01CA154291 (D.D.B.) and the Outstanding Investigator Award 1R35CA197264 (D.D.B.), and PHS awards CA124756 (M.G.) and CA190991 (M.G.).
Footnotes
The authors have no relevant financial interest in the products or companies described in this article.
References
- 1.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. [DOI] [PubMed] [Google Scholar]
- 2.Gromeier M, Lachmann S, Rosenfeld MR, Gutin PH, Wimmer E. Intergeneric poliovirus recombinants for the treatment of malignant glioma. Proc Natl Acad Sci USA. 2000;97(12):6803–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown MC, Dobrikova EY, Dobrikov MI, et al. Oncolytic polio virotherapy of cancer. Cancer. 2014;120(21):3277–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takai Y, Miyoshi J, Ikeda W, Ogita H. Nectins and nectin-like molecules: roles in contact inhibition of cell movement and proliferation. Nat Rev Mol Cell Biol. 2008;9(8):603–615. [DOI] [PubMed] [Google Scholar]
- 5.European Bioinformatics Institute (EMBL-EBI), SIB Swiss Institute of Bioinformatics, Protein Information Resource (PIR). Universal Protein Resource (UniProt). http://www.uniprot.org/uniprot/P15151.04.25.16.
- 6.Mendelsohn CL, Wimmer E, Racaniello VR. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 1989;56(5):855–865. [DOI] [PubMed] [Google Scholar]
- 7.He Y, Mueller S, Chipman PR, et al. Complexes of poliovirus serotypes with their common cellular receptor, CD155. J Virol. 2003;77(8):4827–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koike S, Horie H, Ise I, et al. The poliovirus receptor protein is produced both as membrane-bound and secreted forms. EMBO J. 1990;9(10):3217–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wimmer E, Harber JJ, Bibb JA, Gromeier M, Lu HH, Bernhardt G. Poliovirus receptors In: Wimmer E, ed. Cellular Receptors for Animal Viruses. Plainview, NY: Cold Spring Harbor Laboratory Press; 1993:101–128. [Google Scholar]
- 10.Belnap DM, McDermott BM Jr, Filman DJ, et al. Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc Natl Acad Sci U S A. 2000;97(1):73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernhardt G, Harber J, Zibert A, deCrombrugghe M, Wimmer E. The poliovirus receptor: identification of domains and amino acid residues critical for virus binding. Virology. 1994;203(2):344–356. [DOI] [PubMed] [Google Scholar]
- 12.He Y, Bowman VD, Mueller S, et al. Interaction of the poliovirus receptor with poliovirus. Proc Natl Acad Sci U S A. 2000;97(1):79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bodian D Emerging concept of poliomyelitis infection. Science. 1955; 122(3159):105–108. [DOI] [PubMed] [Google Scholar]
- 14.Sabin AB. Pathogenesis of poliomyelitis; reappraisal in the light of new data. Science. 1956;123(3209):1151–1157. [DOI] [PubMed] [Google Scholar]
- 15.Iwasaki A, Welker R, Mueller S, Linehan M, Nomoto A, Wimmer E. Immunofluorescence analysis of poliovirus receptor expression in Peyer’s patches of humans, primates, and CD155 transgenic mice: implications for poliovirus infection. J Infect Dis. 2002;186(5):585–592. [DOI] [PubMed] [Google Scholar]
- 16.Blinzinger K, Simon J, Magrath D, Boulger L. Poliovirus crystals within the endoplasmic reticulum of endothelial and mononuclear cells in the monkey spinal cord. Science. 1969;163(3873):1336–1337. [DOI] [PubMed] [Google Scholar]
- 17.Freistadt MS, Fleit HB, Wimmer E. Poliovirus receptor on human blood cells: a possible extraneural site of poliovirus replication. Virology. 1993;195(2):798–803. [DOI] [PubMed] [Google Scholar]
- 18.Carlson BL, Pokorny JL, Schroeder MA, Sarkaria JN. Establishment, maintenance and in vitro and in vivo applications of primary human glioblastoma multiforme (GBM) xenograft models for translational biology studies and drug discovery. Curr Protoc Pharmacol. 2011;52(14):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nobis P, Zibirre R, Meyer G, Kuhne J, Warnecke G, Koch G. Production of a monoclonal antibody against an epitope on HeLa cells that is the functional poliovirus binding site. J Gen Virol. 1985;66(pt 12):2563–2569. [DOI] [PubMed] [Google Scholar]
- 20.Merrill MK, Bernhardt G, Sampson JH, Wikstrand CJ, Bigner DD, Gromeier M. Poliovirus receptor CD155-targeted oncolysis of glioma. Neuro Oncol. 2004; 6(3):208–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25(1):118–145. [DOI] [PubMed] [Google Scholar]
- 22.Dobrikov MI, Dobrikova EY, Gromeier M. Dynamic regulation of the translation initiation helicase complex by mitogenic signal transduction to eukaryotic translation initiation factor 4G. Mol Cell Biol. 2013;33(5):937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown MC, Bryant JD, Dobrikova EY, et al. Induction of viral, 7-methylguanosine cap-independent translation and oncolysis by mitogen-activated protein kinase-interacting kinase-mediated effects on the serine/arginine-rich protein kinase. J Virol. 2014;88(22):13135–13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wikstrand CJ, McLendon RE, Friedman AH, Bigner DD. Cell surface localization and density of the tumor-associated variant of the epidermal growth factor receptor, EGFRvIII. Cancer Res. 1997;57(18):4130–4140. [PubMed] [Google Scholar]
- 25.Freistadt MS, Kaplan G, Racaniello VR. Heterogeneous expression of poliovirus receptor-related proteins in human cells and tissues. Mol Cell Biol. 1990;10(11):5700–5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solecki D, Schwarz S, Wimmer E, Lipp M, Bernhardt G. The promoters for human and monkey poliovirus receptors: requirements for basic and cell type-specific activity. J Biol Chem. 1997;272(9):5579–5586. [DOI] [PubMed] [Google Scholar]
- 27.Boulger LR, Arya SC, Ahourai P, Marsden SA. International comparison of species of monkey used for the neurovirulence test for oral poliomyelitis vaccine. J Biol Stand. 1978;6(3):233–242. [DOI] [PubMed] [Google Scholar]
- 28.Reymond N, Imbert AM, Devilard E, et al. DNAM-1 and PVR regulate monocyte migration through endothelial junctions. J Exp Med. 2004;199(10): 1331–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greenberg NM, DeMayo F, Finegold MJ, et al. Prostate cancer in a transgenic mouse. Proc Natl Acad Sci U S A. 1995;92(8):3439–3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280(5369):1618–1620. [DOI] [PubMed] [Google Scholar]
- 31.Bottino C, Castriconi R, Pende D, et al. Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J Exp Med. 2003;198(4):557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinet L, Smyth MJ. Balancing natural killer cell activation through paired receptors. Nat Rev Immunol. 2015;15(4):243–254. [DOI] [PubMed] [Google Scholar]
- 33.Erickson BM, Thompson NL, Hixson DC. Tightly regulated induction of the adhesion molecule necl-5/CD155 during rat liver regeneration and acute liver injury. Hepatology. 2006;43(2):325–334. [DOI] [PubMed] [Google Scholar]
- 34.Soriani A, Zingoni A, Cerboni C, et al. ATM-ATR-dependent up-regulation of DNAM-1 and NKG2D ligands on multiple myeloma cells by therapeutic agents results in enhanced NK-cell susceptibility and is associated with a senescent phenotype. Blood. 2009;113(15):3503–3511. [DOI] [PubMed] [Google Scholar]
- 35.Carlsten M, Norell H, Bryceson YT, et al. Primary human tumor cells expressing CD155 impair tumor targeting by down-regulating DNAM-1 on NK cells. J Immunol. 2009;183(8):4921–4930. [DOI] [PubMed] [Google Scholar]
- 36.Castriconi R, Daga A, Dondero A, et al. NK cells recognize and kill human glioblastoma cells with stem cell-like properties. J Immunol. 2009;182(6):3530–3539. [DOI] [PubMed] [Google Scholar]
- 37.Matusali G, Potesta M, Santoni A, Cerboni C, Doria M. The human immunodeficiency virus type 1 Nef and Vpu proteins downregulate the natural killer cell-activating ligand PVR. J Virol. 2012;86(8):4496–4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomasec P, Wang EC, Davison AJ, et al. Downregulation of natural killer cell-activating ligand CD155 by human cytomegalovirus UL141. Nat Immunol. 2005;6(2):181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Casado JG, Pawelec G, Morgado S, et al. Expression of adhesion molecules and ligands for activating and costimulatory receptors involved in cell-mediated cytotoxicity in a large panel of human melanoma cell lines. Cancer Immunol Immunother. 2009;58(9):1517–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]