Abstract
In this study, Saussurea bogedaensis Yu-J. Wang & Jie Chen, a new species from Bogeda Mountain in the eastern part of the Tianshan Mountains, is described and discussed based on evidence in terms of both morphological and genetic data. S. bogedaensis is morphologically similar to S. involucrata, which is distributed in the western part of the Tianshan Mountains, and it is well known because of its beauty, rarity, and medicinal value. The new species is also similar to S. orgaadayi, which is distributed in the nearby Altai Mountains. Our genetic data support the close relationships among these three species. According to their allopathic distributions, we suggest that these three species are derived from the same ancestor but that they differentiated after reaching their current range. In addition, we propose that the new species might serve as an alternative to S. involucrata in medicine due to their very high similarity. However, this species appears to be rare because we only found six mature individuals in the field despite extensive investigations.
Introduction
Saussurea involucrata is well known because of its beauty, rarity, and medicinal value in China. Its Chinese name, i.e., “snow lotus,” refers to its similar appearance to a lotus, which is a well-known ornamental plant. This species is usually found on mountains covered with snow all year around, which enhances its beauty and explains its associations with many mysterious legends. This species has been used for a long time as a traditional Chinese medicine (TCM) to treat a wide spectrum of disorders such as rheumatoid arthritis, tumor diseases, and high-altitude diseases [1–4]. TCM has been modernized and analyses have isolated and identified more than 70 compounds in S. involucrata [5,6]. In addition, this species has recently been selected as a cold-resistance model in order to exploit its genetic resources [7–9]. Partly due to its over-exploitation, S. involucrata is currently endangered and included in the list of national second-class protected plants in China [10,11], although a few methods for in vitro propagation have been reported [12–15].
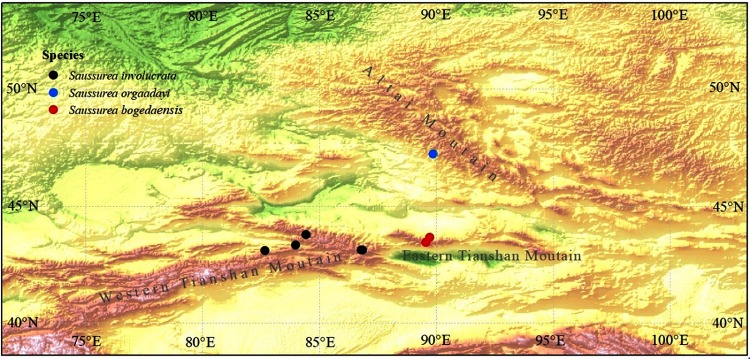
In contrast to the public popularity and significant medical value of S. involucrata, its taxonomic status has received little attention. It was considered to be widespread in the Tianshan Mountains and the nearby Altai Mountains, but recently the population in the Altai Mountains was ascribed to a new species called S. orgaadayi [16]. This species was generally recognized as S. involucrata in the local medicine market, but it can be differentiated from S. involucrata based on a number of morphological features such as the phyllary and involucre [16–19]. This unexpected taxonomic finding suggests that all of the populations cannot be treated as a single species throughout the Tianshan Mountains, which stretch 4000 km from the west to east with a width of up to 150 km in [20]. The Tianshan Mountains are divided into two parts around the Chaiwopu basin of Urumqi at a longitude of about 88° [20,21], where the western part is called Western Tianshan and the eastern part is called Bogeda Mountain (Fig 1).
Fig 1. Map showing the locations visited to obtain samples of Saussurea bogedaensis, S. orgaadayi, and S. involucrata.
To explore the possible differentiation of S. involucrata along the Tianshan Mountains from east to west, we conducted a field investigation in 2013 and found considerable differences according to the geographical regions. However, the eastern population is very small and we only found two individuals in a restricted area immediately below a peak that is covered with snow throughout the year. Thus, we made a second trip via another road in 2016. Once again, we failed to find a large population and encountered only four mature individuals and no more than 50 immature individuals.
Based on the results obtained in the present study, we propose to name the population found in the eastern part of the Tianshan Mountains as a new species called Saussurea bogedaensis Yu-J. Wang & Jie Chen. We obtained photographs in the field and determined the major differences compared with S. involucrata and S. orgaadayi. In order to determine its taxonomic status, we analyzed the genetic diversity based on the nuclear internal transcribed spacer (ITS) and three chloroplast (cp) loci for the new species and 18 other representative species of subg. Amphilaena, which includes S. involucrata and S. orgaadayi [17].
Materials and methods
Ethics statement
All the collecting locations are not in any natural conservation area and no specific permissions were required for these locations. One protected species (Saussurea involucrata) was collected with introduction letters of School of Life Sciences Lanzhou University and permission from Urumqi Forestry Bureau. The individual in this manuscript has given written informed consent (as outlined in PLOS consent form) to publish these case details.
Taxon sampling for molecular phylogeny reconstruction
In total, 44 accessions were sampled, including eight accessions of the new species (S. bogedaensis) from two populations on Bogeda Mountain, four accessions of S. orgaadayi from one population in the Altai Mountains, 15 accessions of S. involucrata from five populations in the Tianshan Mountains, 16 accessions representing the remaining species in subg. Amphilaena, and one accession comprising Jurinea multiflora as an outgroup. Fresh leaves were dried immediately after sampling with silica gel for DNA extraction. Voucher specimens were deposited in the herbarium at Lanzhou University (LZU). The detailed geographical locations of each sampled population are shown in Fig 1 and Table 1.
Table 1. Origins of materials (all these samples are from China) and GenBank accession numbers (ITS, matK, psbA-trnH, and trnK).
| Taxon | Origin | North latitude (°) | East longitude (°) | Altitude (m) | GenBank accession no. | |
|---|---|---|---|---|---|---|
| S. bracteata | Yushu, Qinghai; WYJ201607043 | 35.05681 | 93.01222 | 4644 | MF680674, MF680714, MF680754, MF680794 | |
| S. erubescens | Luqu, Gansu; SN110814017 | 34.59414 | 102.48834 | 3421 | MF680675, MF680715, MF680755, MF680795 | |
| S. wettsteiniana | Mianning, Sichuan; WYJ201607402 | 29.00106 | 102.14985 | 3381 | MF680688, MF680717, MF680757, MF680797 | |
| S. globosa | Kangding, Sichuan; W201209158 | 30.05502 | 101.95973 | 3992 | MF680676, MF680716, MF680756, MF680796 | |
| S. uniflora | Cuona, Xizang; WYJ201607254 | 27.76583 | 91.90194 | 4138 | MF680685, MF680718, MF680758, MF680798 | |
| S. nigrescens | Menyuan, Qinghai; LJQ-QLS-2008-065 | 37.40971 | 101.67202 | 2800 | MF680679, MF680719, MF680759, MF680799 | |
| S. veitchiana | Shenlongjia, Hubei; WYJ201507160 | 31.43997 | 110.30714 | 3098 | MF680686, MF680720, MF680760, MF680800 | |
| S. iodostegia | Datong; Shanxi; WYJ201507117 | 39.05578 | 113.65927 | 2514 | MF680677, MF680721, MF680761, MF680801 | |
| S. pubifolia | Jiacha, Xizang; WYJ201607272 | 29.03175 | 92.35724 | 4796 | MF680683, MF680722, MF680762, MF680802 | |
| S. velutina | Xiaojin, Sichuan; WYJ201209124 | 30.99441 | 102.82915 | 4000 | MF680687, MF680723, MF680763, MF680803 | |
| S. polycolea | Linzhi, China; LJQ07257 | 29.36866 | 94.39168 | 4680 | MF680682, MF680724, MF680764, MF680804 | |
| S. tangutica | Gansu; WYJ201607013 | 38.60685 | 99.48221 | 4096 | MF680684, MF680725, MF680765, MF680805 | |
| S. luae | Linzhi, Xizang; WYJ201607286 | 29.59022 | 94.59631 | 4121 | MF680678, MF680726, MF680766, MF680806 | |
| S. phaeantha | Gansu; WYJ201607014 | 38.60685 | 99.48221 | 4096 | MF680681, MF680727, MF680767, MF680807 | |
| S. obvallata | Cuona, Xizang; WYJ201607242 | 27.92057 | 91.84863 | 3970 | MF680680, MF680728, MF680768, MF680808 | |
| S. muliensis | Unpublished data in GenBank | --- | --- | --- | AB254665, ---, ---, --- | |
| S. involucrata | Urumqi, Xinjiang; WYJ201607025 (163) | 43.10847 | 86.84220 | 3564 | MF680689, MF680741, MF680781, MF680821 | |
| S. involucrata | Urumqi, Xinjiang; WYJ201607025 (165) | 43.10847 | 86.84220 | 3564 | MF680690, MF680742, MF680782, MF680822 | |
| S. involucrata | Urumqi, Xinjiang; WYJ201308203 (41) | 43.11985 | 86.82125 | 3768 | MF680691, MF680744, MF680784, MF680824 | |
| S. involucrata | Urumqi, Xinjiang; WYJ201308203 (42) | 43.11985 | 86.82125 | 3768 | MF680692, ---,---, --- | |
| S. involucrata | Urumqi, Xinjiang; WYJ201308203 (372) | 43.11985 | 86.82125 | 3768 | MF680693, MF680743, MF680783, MF680823 | |
| S. involucrata | Urumqi, Xinjiang; WYJ201308203 (374) | 43.11985 | 86.82125 | 3768 | MF680694, ---, ---, --- | |
| S. involucrata | Tekesi, Xinjiang; WYJ201308184 (24) | 43.09915 | 82.68382 | 3678 | MF680695, MF680738, MF680778, MF680818 | |
| S. involucrata | Tekesi, Xinjiang; WYJ201308184 (25) | 43.09915 | 82.68382 | 3678 | ---, MF680739, MF680779, MF680819 | |
| S. involucrata | Tekesi, Xinjiang; WYJ201308184 (26) | 43.09915 | 82.68382 | 3678 | MF680696, MF680740, MF680780, MF680820 | |
| S. involucrata | Dushanzi, Xinjiang; WYJ201308131 (60) | 43.77545 | 84.45615 | 2684 | ---, MF680734, MF680774, MF680814 | |
| S. involucrata | Dushanzi, Xinjiang; WYJ201308131 (61) | 43.77545 | 84.45615 | 2684 | MF680697, MF680733, MF680773, MF680813 | |
| S. involucrata | Dushanzi, Xinjiang; WYJ201308131 (63) | 43.77545 | 84.45615 | 2684 | MF680698, ---, ---, --- | |
| S. involucrata | Xinyuan, Xinjiang; WYJ201308188 (47) | 43.33469 | 84.01032 | 3543 | MF680699, MF680735, MF680775, MF680815 | |
| S. involucrata | Xinyuan, Xinjiang; WYJ201308188 (48) | 43.33469 | 84.01032 | 3543 | MF680700, MF680736, MF680776, MF680816 | |
| S. involucrata | Xinyuan, Xinjiang; WYJ201308188 (390) | 43.33469 | 84.01032 | 3543 | MF680701, MF680737, MF680777, MF680817 | |
| S. bogedaensis | Qitai, Xinjiang; WYJ201607018 (140) | 43.45321 | 89.55213 | 3471 | MF680702, MF680748, MF680788, MF680828 | |
| S. bogedaensis | Qitai, Xinjiang; WYJ201607018 (166) | 43.45321 | 89.55213 | 3471 | MF680703, MF680745, MF680785, MF680825 | |
| S. bogedaensis | Qitai, Xinjiang; WYJ201607018 (167) | 43.45321 | 89.55213 | 3471 | MF680704, MF680746, MF680786, MF680826 | |
| S. bogedaensis | Qitai, Xinjiang; WYJ201607018 (378) | 43.45321 | 89.55213 | 3471 | MF680705, MF680747, MF680787, MF680827 | |
| S. bogedaensis | Qitai, Xinjiang; WYJ201308006 (38) | 43.44370 | 89.58167 | 3386 | MF680707, MF680751, MF680790, MF680831 | |
| S. bogedaensis | Qitai, Xinjiang; WYJ201308006 (39) | 43.44370 | 89.58167 | 3386 | MF680708, MF680750, MF680791, MF680830 | |
| S. bogedaensis | Qitai, Xinjiang; WYJ201308006 (40) | 43.44370 | 89.58167 | 3386 | MF680709, MF680752, MF680792, MF680832 | |
| S. bogedaensis | Qitai, Xinjiang; WYJ201308006 (309) | 43.44370 | 89.58167 | 3386 | MF680706, MF680749, MF680789, MF680829 | |
| S. orgaadayi | Altay, Xinjiang; WYJ201308041 (11) | 47.21846 | 89.87999 | 3541 | MF680712, MF680732, MF680772, MF680812 | |
| S. orgaadayi | Altay, Xinjiang; WYJ201308041 (12) | 47.21846 | 89.87999 | 3541 | MF680713, MF680731, MF680771, MF680811 | |
| S. orgaadayi | Altay, Xinjiang; WYJ201308041 (360) | 47.21846 | 89.87999 | 3541 | MF680711, MF680730, MF680770, MF680810 | |
| S. orgaadayi | Altay, Xinjiang; WYJ201308041 (361) | 47.2184691 | 89.87999856 | 3541 | MF680710, MF680729, MF680769, MF680809 | |
| Jurinea multiflora | Tuoli, Xinjiang; WYJ201308102 | 45.73564 | 83.14712 | 1753 | MF680673, MF680753, MF680793, MF680833 | |
Morphological observations
Morphological descriptions were prepared based on examinations of the fresh and pressed specimens. Specimens deposited in E, K, PE, KUN, QTPMB, and LZU were examined to make morphological comparison with similar species, i.e., S. orgaadayi and S. involucrata. In order to determine the floral micromorphology, dry florets were boiled in distilled water for 5–10 min and photographed under a stereomicroscope (Olympus MD-90).
DNA extraction and sequencing
Total DNA was extracted from leaf tissues dried with silica gel or herbarium specimens using the modified CTAB method [22]. Four markers were employed comprising ITS, trnK, matK, and psbA-trnH. The primers [23–26] used for amplification and sequencing are listed in Table 2. PCR was performed as described in our previous study [27]. PCR products were sent to Beijing Genomics Institute (BGI) for commercial sequencing. Sequences were aligned using CLUSTALX v.2.1 [28] with the default settings and adjusted manually with Bioedit v.7.0.5 [29]. All of the sequences were registered in GenBank.
Table 2. List of the primers used in this study.
| Fragment | Primer 1 | Sequence (5′–3′) | Primer 2 | Sequence (5′–3′) |
|---|---|---|---|---|
| ITS | ITS1 | TCCTCCGCTTATTGATATGC | ITS4 | AGAAGTCGTAACAAGGTTTCCGTAGG |
| trnK | trnK(UUU) | TTAAAAGCCGAGTACTCTACC | rps16 | AAAGTGGGTTTTTATGATCC |
| trnH-psbA | psbA | GTTATGCATGAACGTAATGCTC | trnH | CGCGCATGGTGGATTCACAATCC |
| matK | matK-XF | TAATTTACGATCAATTCATTC | 5r | GTTCTAGCACAAGAAAGTCG |
Data analysis
Three datasets were constructed where one comprised the nuclear ITS sequences, the second contained the concatenated sequences of psbA-trnH, matK, and trnK, and the third of all the sequences after the incongruence length difference test that revealed little incongruence (P > 0.01) between chloroplast and ITS data [30]. MEGA v.4.0 was used to calculate the genetic distances under the Kimura two-parameter model [31]. Phylogenetic analyses were conducted using PAUP v.4.0b10 [32] and MrBayes v.3.2.1 [33]. Maximum parsimony (MP) searches were performed using heuristic search methods with tree bisection reconnection branch swapping and equal weighting for all characters. The analyses were repeated 1,000 times with a random order of sequence addition in order to sample multiple islands of the most parsimonious trees. Bootstrap tests were conducted to evaluate node support using 1,000 replicates with heuristic search settings identical to those for the original search. Bayesian inference (BI) was conducted using the different models selected by Modeltest [34] for each partition. Ten million generations were run to estimate parameters related to sequence evolution and likelihood probabilities using the Markov chain Monte Carlo method. Trees were collected every 1000 generations. Tracer v.1.5 (http://tree.bio.ed.ac.uk/software/tracer/) was used to choose a suitable burn-in period. PAUP* v.4.0b10 [32] was used to calculate a consensus tree and posterior probabilities (PP) from the sampled trees after the burn-in period.
Nomenclature
The electronic version of this article in Portable Document Format (PDF) in a work with an ISSN or ISBN will represent a published work according to the International Code of Nomenclature for algae, fungi, and plants, and hence the new names contained in the electronic publication of a PLOS ONE article are effectively published under that Code from the electronic edition alone, so there is no longer any need to provide printed copies.
In addition, new names contained in this work have been submitted to IPNI, from where they will be made available to the Global Names Index. The IPNI LSIDs can be resolved and the associated information viewed through any standard web browser by appending the LSID contained in this publication to the prefix http://ipni.org/. The online version of this work is archived and available from the following digital repositories: PubMed Central and LOCKSS.
Results
Morphological features
Figs 2 and 3 shows photographs of S. bogedaensis, including the habitat (Fig 2) and close-ups of the florets, pappus, anthers, style branches, phyllaries, and leaf margin (Fig 3). The new species could be differentiated from S. involucrata or S. orgaadayi mainly based on the shape of the phyllaries and the indumentum. In the new species, they were acuminate and covered with sericeous-villous in the upper half (Fig 4), whereas they were long, acuminate, and densely pubescent throughout the phyllaries or mostly glabrous in S. involucrata and S. orgaadayi. In addition, the three species differed in terms of their leaf, bract, and pappus features, as described in Table 3.
Fig 2. Saussurea bogedaensis in the wild.
Fig 3. Holotype of Saussurea bogedaensis (WYJ201607018).
(A) Living plant; (B) Floret; (C) Inner pappus bristle; (D, E, F) Anthers. (G) Style branches; (H, I) Phyllaries; (J) Stem leaf margin.
Fig 4.
Comparison of materials from Saussurea orgaadayi (A, D), S. involucrata (B, E), and S. bogedaensis (C, F). (A, D) from WYJ201308041; (B, E) from WYJ201607025; (C, F) from WYJ201607018.
Table 3. Comparison of Saussurea involucrata, S. orgaadayi, and S. bogedaensis.
| Features | S. involucrata | S. orgaadayi | S. bogedaensis |
|---|---|---|---|
| Distribution | Western Tianshan Mountains | Altai Mountains | Eastern Tianshan Mountains (Bogeda Mountain) |
| Petiolar remains of basal leaves | dark brown stripes up to 2–3 mm wide | yellowish brown stripes up to 1 cm wide | dark brown stripes up to 2–3 mm wide |
| Stem leaves | narrowly ovate, elliptic, or obovate, apex acute, 8–13 × 2–4cm | lanceolate, apex long acuminate 8–17 × 2–5.5 cm, |
elliptic, apex obtuse, 15–20 × 3–5 cm |
| Bracts | ovate-elliptic, apex acute 5.5–12 × 3.5–6.5 cm |
triangular-ovate, apex long acuminate 4–12 × 1.5–6.5cm |
ovate-elliptic, apex acute 5.5–12 × 3.5–6.5 cm |
| Capitula number | 10–20 | 20–30 | 15–30 |
| Involucre | hemispheric | campanulate | campanulate |
| Phyllary | triangular-ovate, apex acute or obtuse, phyllaries glabrous, rarely sparsely pubescent apically or along midvein | linear-subulate, apex long acuminate, phyllaries densely pubescent throughout | subulate to acuminate, phyllaries densely pubescent middle-upper part |
| Pappus color | dirty white | straw-colored | dirty white |
Molecular analyses
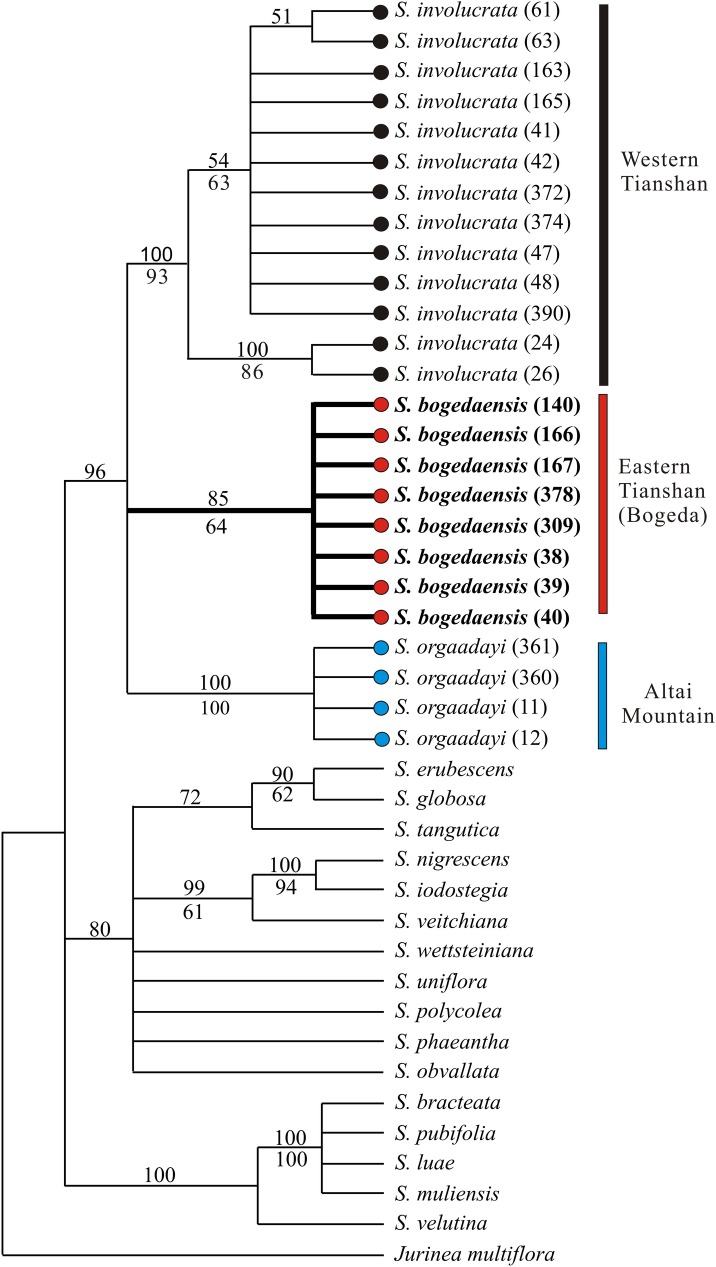
The aligned ITS data sets comprised 20 taxa with 607 positions and 69 variable characters, where 33 were parsimony informative when gaps were treated as missing. The mean pairwise distance within subg. Amphilaena was 1.4%. Those between S. bogedaensis and S. involucrata or S. orgaadayi were 0.98% or 2.0%, respectively (Table 4). Two approaches (MP and BI) obtained largely congruent tree topologies. The BI tree is shown in Fig 5 where the Bayesian PPs and MP bootstrap percentages (BPs) are denoted above or below the branches, respectively. We analyzed all three species with multiple individuals, i.e., S. bogedaensis (PP = 85%; BP = 64%), S. involucrata (PP = 100%; BP = 93%), and S. orgaadayi (PP = 100%; BP = 100%), and they were found to be monophyletic. Moreover, the three species formed a monophyletic clade (PP = 96%), whereas the other species clustered into two clades.
Table 4. Pairwise distances (%) for internal transcribed spacer (lower left) and combined plastid (upper right) sequences from 19 Saussurea species.
| CP ITS |
1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.30 | 0.10 | 0.40 | 0.20 | 0.20 | 0.30 | 0.20 | 0.20 | 0.30 | 0.20 | 0.20 | 0.20 | 0.30 | 0.20 | 0.20 | 0.50 | 0.20 | ---- | |
| 2 | 0.98 | 0.30 | 0.60 | 0.40 | 0.40 | 0.50 | 0.40 | 0.40 | 0.50 | 0.40 | 0.40 | 0.40 | 0.50 | 0.40 | 0.40 | 0.70 | 0.40 | ---- | |
| 3 | 2.00 | 1.37 | 0.40 | 0.30 | 0.30 | 0.30 | 0.30 | 0.20 | 0.30 | 0.20 | 0.20 | 0.20 | 0.30 | 0.20 | 0.30 | 0.50 | 0.20 | ---- | |
| 4 | 1.61 | 0.99 | 1.71 | 0.50 | 0.40 | 0.60 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 0.60 | 0.50 | 0.40 | 0.70 | 0.50 | ---- | |
| 5 | 1.79 | 1.16 | 1.88 | 0.50 | 0.30 | 0.20 | 0.10 | 0.10 | 0.50 | 0.10 | 0.10 | 0.10 | 0.20 | 0.30 | 0.40 | 0.70 | 0.10 | ---- | |
| 6 | 1.45 | 1.17 | 1.89 | 0.50 | 0.66 | 0.30 | 0.30 | 0.20 | 0.20 | 0.20 | 0.20 | 0.20 | 0.30 | 0.30 | 0.10 | 0.50 | 0.20 | ---- | |
| 7 | 1.84 | 1.17 | 2.24 | 1.34 | 1.51 | 1.52 | 0.20 | 0.10 | 0.50 | 0.10 | 0.10 | 0.10 | 0.00 | 0.40 | 0.50 | 0.70 | 0.10 | ---- | |
| 8 | 1.57 | 1.51 | 2.42 | 1.51 | 1.69 | 1.69 | 2.05 | 0.10 | 0.50 | 0.10 | 0.10 | 0.10 | 0.20 | 0.30 | 0.40 | 0.70 | 0.10 | ---- | |
| 9 | 2.68 | 2.00 | 3.08 | 2.34 | 2.52 | 2.53 | 0.83 | 2.90 | 0.40 | 0.00 | 0.00 | 0.00 | 0.10 | 0.30 | 0.30 | 0.60 | 0.00 | ---- | |
| 10 | 2.69 | 2.17 | 2.91 | 1.50 | 1.67 | 1.67 | 2.54 | 2.37 | 3.56 | 0.40 | 0.40 | 0.40 | 0.50 | 0.40 | 0.10 | 0.60 | 0.40 | ---- | |
| 11 | 2.15 | 1.52 | 2.20 | 1.17 | 1.35 | 1.35 | 2.05 | 1.36 | 2.90 | 2.03 | 0.00 | 0.00 | 0.10 | 0.30 | 0.30 | 0.60 | 0.00 | ---- | |
| 12 | 2.13 | 1.50 | 2.22 | 0.83 | 1.00 | 1.00 | 1.85 | 1.35 | 2.87 | 1.67 | 0.50 | 0.00 | 0.10 | 0.30 | 0.30 | 0.60 | 0.00 | ---- | |
| 13 | 2.16 | 1.50 | 2.56 | 1.84 | 2.01 | 2.01 | 0.33 | 2.03 | 1.16 | 2.69 | 2.03 | 2.01 | 0.10 | 0.30 | 0.30 | 0.60 | 0.00 | ---- | |
| 14 | 2.63 | 2.34 | 3.07 | 2.00 | 2.17 | 2.18 | 2.19 | 2.71 | 3.03 | 3.20 | 2.54 | 2.51 | 2.51 | 0.40 | 0.50 | 0.70 | 0.10 | ---- | |
| 15 | 2.47 | 1.84 | 2.35 | 1.16 | 1.33 | 1.34 | 1.86 | 2.38 | 3.21 | 2.35 | 1.69 | 1.67 | 2.70 | 2.86 | 0.30 | 0.60 | 0.30 | ---- | |
| 16 | 2.52 | 2.00 | 2.18 | 1.33 | 1.50 | 1.50 | 2.20 | 2.38 | 3.38 | 1.50 | 1.69 | 1.50 | 2.86 | 3.02 | 1.84 | 0.50 | 0.30 | ---- | |
| 17 | 2.36 | 1.84 | 2.01 | 1.16 | 1.34 | 1.34 | 2.20 | 2.21 | 3.22 | 1.33 | 1.52 | 1.34 | 2.70 | 2.86 | 1.68 | 0.16 | 0.60 | ---- | |
| 18 | 1.95 | 1.33 | 2.05 | 0.66 | 0.66 | 0.83 | 1.68 | 1.69 | 2.69 | 1.67 | 1.18 | 1.00 | 2.01 | 2.34 | 1.50 | 1.67 | 1.50 | ---- | |
| 19 | 2.33 | 1.66 | 2.74 | 2.00 | 2.18 | 2.18 | 0.50 | 2.55 | 1.33 | 3.21 | 2.38 | 2.52 | 0.83 | 2.68 | 2.87 | 3.03 | 2.87 | 2.18 |
1. S. bogedaensis, 2. S. orgaadayi, 3. S. involucrata, 4. S. obvallata, 5. S. phaeantha, 6. S. globosa, 7. S. wettsteiniana, 8. S. uniflora, 9. S. polycolea, 10. S. erubescens, 11. S. nigrescens, 12. S. iodostegia, 13. S. luae, 14. S. pubifolia, 15. S. tangutica, 16. S. muliensis, 17. S. veitchiana, 18. S. velutina, 19. S. bracteata.
Fig 5. The 50% majority rule consensus tree derived from Bayesian analysis of the nuclear internal transcribed spacer.
Posterior probabilities (PPs) and bootstrap percentages (BPs) are indicated above and below the branches, respectively.
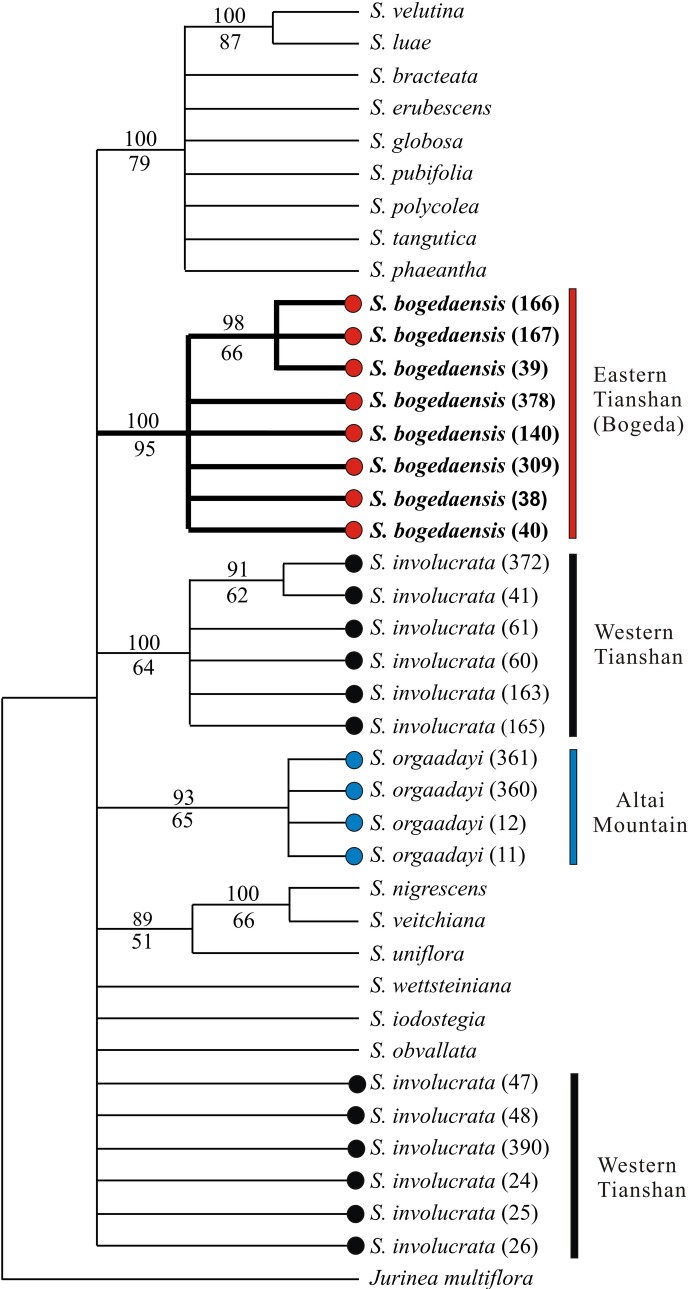
The aligned combined plastid (psbA-trnH, trnK, and matK) matrix contained 1551 characters, 49 of which were variable and 14 were phylogenetically informative. Similar to the results based on the ITS sequences, the pairwise distances of the combined cp loci between S. bogedaensis and S. involucrata or S. orgaadayi were both 0.3%, which was the smallest among the pairwise distances between S. bogedaensis and the other species (Table 4). The trees obtained by MP and BI were mainly congruent and the latter is shown in Fig 6. Both S. bogedaensis (PP = 93; BP = 65%) and S. orgaadayi (PP = 93%; BP = 65%) were resolved as monophyletic. However, those from S. involucrata failed to form a monophyletic group. Moreover, the three species did not form a monophyletic clade (Fig 6).
Fig 6. The 50% majority rule consensus tree derived from Bayesian analysis of the combined plastid dataset.
Posterior probabilities (PPs) and bootstrap percentages (BPs) are indicated above and below the branches, respectively.
The combination of ITS and plastid matrix obtained similar tree from BI and MP, and the former was shown in S1 Fig. The topology is highly similar to that from ITS, but support for a few clades, including that containing S. involucrate, is a little higher than that from ITS (PP = 100%; BP = 96%).
Discussion
As one of the four Saussurea subgenera, subg. Amphilaena is defined mainly by colored uppermost leaves or bracts surrounding the synflorescence [16,17,35]. A recent study indicated that this character might have been derived more than once and that this subgenus might be polyphyletic [36,37], but no new infrageneric system has been proposed for Saussurea or subg. Amphilaena. Thus, we tentatively ascribed the new species to subg. Amphilaena. In subg. Amphilaena, S. involucrata and S. orgaadayi were identified as similar species to the new species because of a morphological combination unique to these species, i.e, the cream-yellow bracts that aggregated below the florescence and the hollow stem at least 1.5 cm in diameter near base. Their morphological affinity was also supported by our molecular analyses. Thus, the genetic distances between the new species and S. involucrata were 0.98% based on ITS and 0.3% for cp, where were the smallest among the new species and the other sampled in-group species. Moreover, the three species resolved into a well-supported clade in the ITS phylogeny (Fig 5).
The three species are closely related in terms of both their morphology and molecular level characteristics, but they also have significant differences. First, six morphological differences were identified among the three species based on multiple individuals from at least two populations for each species. In particular, the shapes of the involucre and the abaxial indumenta are distinct in each species, whereas the other characters differ in at least two species. Second, all three species were resolved into three monophyletic clades, which were well supported and they corresponded to the morphological divisions in the ITS phylogeny. Third, all three species are geographically isolated. Thus, the Tianshan Mountains and Altai Mountains are separated by the Junggar Basin. In the Tianshan Mountains, the western and eastern parts are separated by Chaiwopu Basin (Fig 1). Both basins might be sufficiently large to impede or reduce gene flow among these regions, especially for plants that inhabit high altitude regions. Accordingly, we propose that these species might be derived from a common ancestor, but they may have differentiated after reaching their current range due to restricted gene flow.
In plants, closely related species often share the same common chemical components [38–40]. Thus, it is reasonable to hypothesize that the new species may have similar medicinal value to S. involucrata because of their very high similarity and recent differentiation. However, the population of the new species might be rather small. We found this species in two localities, which were both located in restricted areas immediately below peaks that were covered with snow all the year around, where we only found six mature individuals and 50 immature individuals. This harsh environment might at least partly explain their rarity. Thus, we suggest that exploitation of this new species should be subject to strict protection.
Taxonomic treatment
Saussurea bogedaensis Yu J. Wang & J. Chen sp. nov. [urn:lsid:ipni.org:names: 77180814–1] (Figs 2, 3, 4C and 4F) Type: China. Xinjiang: Qitai Country, Banjiegou Town, Bogeda Mountain, 43.45321°N, 89.55213°E, 3471 m, July 22, 2016, WYJ201607018 (holotype, LZU).
Diagnosis. Similar to S. involucrata or S. orgaadayi but differs in terms of acuminate and densely pubescent phyllaries in middle-upper part.
Description. Herbs 15–50 cm tall, perennial. Caudex stout, unbranched, densely covered with fibrous remains of petioles. Stem solitary, 1.5–3 cm in diam., erect, simple. Rosette and stem leaves petiolate; leaf blade narrowly ovate, elliptic, or obovate, 15–20 × 3–5 cm, both surfaces green and glandular hairy, base decurrent, margin denticulate to serrulate, apex obtuse. Uppermost stem leaves sessile, ovate to elliptic, 5.5–12 × 3.5–6.5 cm, membranous, stellate surrounding synflorescence, both surfaces pale yellow. Capitula 15–30 in a hemispheric synflorescence, 8–15 cm in diam., sessile or shortly pedunculate. Involucre broadly campanulate, 1–2.5 cm in diam. Phyllaries in four or five rows, subulate, light brown with dark margin, densely pubescent on middle-upper part, apex acuminate; outer phyllaries 25–30 × 2.5–4 mm; middle and inner phyllaries 18–23 × 1.5–3 mm. Receptacle papillose; papillae 0.5–1 mm. Corolla purple, 1.3–1.8 cm, tube 7–9 mm, limb 6–9 mm, lobes 3–5 mm. Achene straw-colored with blackish spots, cylindrical 4.8–6.7 mm. Pappus dirty white; outer bristles 0.5–3 mm; inner bristles 0.8–1.5 cm.
Distribution. The species is currently known only from two localities in Bogeda Mountain located in Qitai, Xinjiang, China.
Conservation Status. we discovered only six individuals in blossom, all without mature seeds, and no more than 50 immature ones in cliffs near the snowline of the Bogeda Mountain. We estimated the species comprise less than 500 individuals in the light of its restrict distribution. Due to its highly resembling to S. involucrata, there is risk of harvest by herb-digger and/or native shepherd. We propose that the location should be recognized as critical habitat and the species listed as ‘‘Critically Endangered” according to the IUCN red list categories and criteria [41].
Supporting information
The 50% majority rule consensus tree derived from Bayes inference of the combined sequences of nuclear ITS and all the plastid loci. Posterior probabilities and bootstrap percentages are indicated above and below the branches, respectively.
(TIF)
Acknowledgments
We are grateful to Jian-Quan Liu, Zhong-Hu Li, Yi-Xuan Kou, Fu-Shen Yang, Sonam Tso and Hiroshi Ikeda for helping with our field investigation. This study was supported by the National Natural Science Foundation of China (81274024).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the National Natural Science Foundation of China (81274024).
References
- 1.Way TD, Lee JC, Kuo DH, Fan LL, Huang CH, Lin HY, et al. (2010) Inhibition of epidermal growth factor receptor signaling by Saussurea involucrata, a rare traditional Chinese medicinal herb, in human hormone-resistant prostate cancer PC-3 cells. Journal of Agricultural and Food Chemistry 58: 3356–3365. 10.1021/jf903793p [DOI] [PubMed] [Google Scholar]
- 2.Jia Z, Wu C, Jin H, Zhang J (2014) Identification of the chemical components of Saussurea involucrata by high-resolution mass spectrometry and the mass spectral trees similarity filter technique. Rapid Commun Mass Spectrom 28: 2237–2251. 10.1002/rcm.7014 [DOI] [PubMed] [Google Scholar]
- 3.Jia JM, Wu CF, Yu H, Hu GS (2005) Anti-radiation activity of the tissue culture of Saussurea involucrata Kar. et Kir. Journal of Shenyang Pharmaceutical University 22: 444–448. [Google Scholar]
- 4.Martens S, Mithofer A (2005) Flavones and flavone synthases. Phytochemistry 66: 2399–2407. 10.1016/j.phytochem.2005.07.013 [DOI] [PubMed] [Google Scholar]
- 5.Wang XL, Gesang SL, Jiao W, Liao X, Ding LS (2007) Two new sesquiterpenoid glucosides from the aerial parts of Saussurea involucrata. Journal Of Integrative Plant Biology 49: 609–614. [Google Scholar]
- 6.Chik WI, Zhu L, Fan LL, Yi T, Zhu GY, Gou XJ, et al. (2015) Saussurea involucrata: A review of the botany, phytochemistry and ethnopharmacology of a rare traditional herbal medicine. J Ethnopharmacol 172: 44–60. 10.1016/j.jep.2015.06.033 [DOI] [PubMed] [Google Scholar]
- 7.Tang Y, Yuan H, Qin J (2012) Identification and characterization of a flavonoid-3-O-glucosyltransferase gene from Saussurea involucrata. Chinese journal of biotechnology 28: 705–714. [PubMed] [Google Scholar]
- 8.Qiu H, Zhang L, Liu C, He L, Wang A, Liu HL, et al. (2014) Cloning and characterization of a novel dehydrin gene, SiDhn2, from Saussurea involucrata Kar. et Kir. Plant Molecular Biology 84: 707–718. 10.1007/s11103-013-0164-7 [DOI] [PubMed] [Google Scholar]
- 9.Liu HL, Shen HT, Chen C, Zhou XR, Liu H, Zhu JB (2015) Identification of a putative stearoyl acyl-carrier-protein desaturase gene from Saussurea involucrata. Biologia Plantarum 59: 316–324. [Google Scholar]
- 10.Guo B, Stiles AR, Liu CZ (2013) Low-temperature preincubation enhances survival and regeneration of cryopreserved Saussurea involucrata callus. In Vitro Cellular & Developmental Biology—Plant 49: 320–325. [Google Scholar]
- 11.Fu LK, Jin JM (1992) Rare and endangered plants In: Fu LK, Jin JM, editors. China plant red data book. Shanghai, China: Science Press; pp. 234–235. [Google Scholar]
- 12.Yang L, Qin XY (2006) In Vitro Tissue Culture and Plantlet Regeneration of Saussurea involucrata Kar.et Kir. Journal of the Central University for Nationalities 15: 26–29. [Google Scholar]
- 13.Guo B, Stiles AR, Liu CZ (2012) Thidiazuron enhances shoot organogenesis from leaf explants of Saussurea involucrata Kar. et Kir. In Vitro Cellular & Developmental Biology—Plant 48: 609–612. [Google Scholar]
- 14.Guo B, Gao M, Liu CZ (2007) In vitro propagation of an endangered medicinal plant Saussurea involucrata Kar. et Kir. Plant Cell Reports 26: 261–265. 10.1007/s00299-006-0230-6 [DOI] [PubMed] [Google Scholar]
- 15.Chen Z, Jiang X, Zhou P, Cai J (2002) Comparison of the contents of total flavone between artificial cultivated and wild Saussurea involucrata. Chinese Pharmaceutical Journal 37: 98–99. [Google Scholar]
- 16.Shi Z, Raab-Straube Ev (2011) Saussurea Candolle In: Wu ZY, Raven PH, editors. Flora of China. Beijing: Science Press; pp. 56–149. [Google Scholar]
- 17.Raab-Straube EV (2017) Taxonomic revision of Saussurea subgenus Amphilaena (Compositae, Cardueae) Berlin: Botanic Garden and Botanical Museum Berlin. [Google Scholar]
- 18.Shmakov A, Chen WL, Smirnov S, Kamelin RV, Zhang SR, Liu JQ, et al. (2011) Some new or noteworthy plant species for china found in North West Xinjiang. Turczaninowia 14: 75–80. [Google Scholar]
- 19.Smirnov SV (2004) Notes on the genus Saussurea DC. (Asteraceae) in Altai. Turczaninowia 7: 11–17. [Google Scholar]
- 20.Li JY, Wang KZ, Li YP, Sun GH, Chu CH, Li LQ, et al. (2006) Geomorphological features, crustal composition and geological evolution of the Tianshan Mountains. Geol Bull China 25: 895–909. [Google Scholar]
- 21.Shen CB, Mei LF, Zhang SW, Lin L, Tang JG, Feng Z, et al. (2008) Fission-track dating evidence on space-time difference of mesozoic-cenozoic uplift of the yilianhabierga mountain and bogeda mountain. Journal of Mineralogy and Petrology 28: 63–70. [Google Scholar]
- 22.Doyle JJ (1987) A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem Bull 19: 11–15. [Google Scholar]
- 23.Berends ST, Jones JT, Mullet JE (1990) Sequence and transcriptional analysis of the barley ctDNA region upstream of psbD-psbC encoding trnK(UUU), rps16, trnQ(UUG), psbK, psbI, and trnS(GCU). Current Genetics 17: 445–454. [DOI] [PubMed] [Google Scholar]
- 24.Ford CS, Ayres KL, Toomey N, Haider N, Stahl JV, Kelly LJ, et al. (2009) Selection of candidate coding DNA barcoding regions for use on land plants. Botanical Journal Of the Linnean Society 159: 1–11. [Google Scholar]
- 25.Sang T, Crawford D, Stuessy T (1997) Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). American Journal of Botany 84: 1120 [PubMed] [Google Scholar]
- 26.White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols A guide to methods and applications. San Diego: Academic Press; pp. 315–322. [Google Scholar]
- 27.Wang YJ, Susanna A, Raab-Straube EV, Milne R, Liu JQ (2009) Island-like radiation of Saussurea (Asteraceae: Cardueae) trigged by uplifts of the Qinghai-Tibetan Plateau. Biological Journal of the Linnean Society 97: 893–903. [Google Scholar]
- 28.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- 30.Farris JS, Källersjö M, Kluge AG, Bult C (1995) Testing significance of incongruence. Cladistics 10: 315–319. [Google Scholar]
- 31.Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Molecular Biology and Evolution 24: 1596–1599. 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- 32.Swofford D (2003) PAUP*: phylogenetic analysis using parsimony (*and other methods), Version 4.0b 10 Sunderland, Massachusetts: Sinauer Associates. [Google Scholar]
- 33.Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- 34.Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- 35.Raab-Straube EV (2003) Phylogenetic relationships in Saussurea (Compositae, Cardueae) sensu lato, inferred from morphological, ITS and trnL-trnF sequence data, with a synopsis of Himalaiella gen. nov., Lipschitziella and Frolovia. Willdenowia 33: 379–402. [Google Scholar]
- 36.Ev Raab-Straube (2009) Saussurea luae (Compositae, Cardueae), a new species of snow lotus from China. Willdenowia 39: 101–106. [Google Scholar]
- 37.Kita Y, Fujikawa K, Ito M, Ohba H, Kato M (2004) Molecular phylogenetic analyses and systematics of the genus Saussurea and related genera (Asteraceae, Cardueae). Taxon 53: 679–690. [Google Scholar]
- 38.Madhavi M, Mallika G, Lokanath N, Vishnu MN, Chetty CM, Saleem TSM (2012) A review on phytochemical and pharmacological aspects of Saussurea lappa. International Journal of Review in Life Sciences 2: 24–31. [Google Scholar]
- 39.Li Y, Jia ZJ (1989) Guaianolides from Saussurea involucrata. Phytochemistry 28: 3395–3397. [Google Scholar]
- 40.Cock IE, Mpala L, Chikowe G (2010) No evidence of antiseptic properties and low toxicity of selected Aloe species. Journal of Pharmaceutical Negative Results 1: 10. [Google Scholar]
- 41.IUCN (2012) IUCN red list categories and criteria: version 3.1. Second edition. Gland: Switzerland and Cambridge.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The 50% majority rule consensus tree derived from Bayes inference of the combined sequences of nuclear ITS and all the plastid loci. Posterior probabilities and bootstrap percentages are indicated above and below the branches, respectively.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.