Abstract
Ganoderma is a large, diverse and globally-distributed genus in the Basidiomycota that includes species causing a white rot form of wood decay on a variety of tree species. For the past century, many studies of Ganoderma in North America and other regions of the world have used the name G. lucidum sensu lato for any laccate (shiny or varnished) Ganoderma species growing on hardwood trees or substrates. Molecular studies have established that G. lucidum sensu stricto (Curtis) Karst is native to Europe and some parts of China. To determine the species of the laccate Ganoderma that are present in the United States, we studied over 500 collections from recently collected samples and herbarium specimens from hardwoods, conifers, and monocots. A multilocus phylogeny using ITS, tef1α, rpb1 and rpb2 revealed three well-supported clades, similar to previously reported findings. From the U.S. collections, thirteen taxa representing twelve species were identified, including: G. curtisii, G. lucidum sensu stricto, G. martinicense, G. oregonense, G. polychromum, G. ravenelii, G. sessile, G. tsugae, G. tuberculosum, G. cf. weberianum, G. zonatum, and Tomophagus colossus (syn. G. colossus). The species G. meredithiae is synonymized with G. curtisii, and considered a physiological variant that specializes in decay of pines. The designation G. curtisii f.sp. meredithiae forma specialis nov. is proposed. Species such as G. curtisii and G. sessile, once considered as G. lucidum sensu lato, were found to be divergent from one another, and highly divergent from G. lucidum sensu stricto. Morphological characteristics such as context tissue color and features (e.g. melanoid bands), basidiospore shape and size, geographic location, and host preference were found to aid in species identification. Surprisingly, G. lucidum sensu stricto was found in the U.S., but only in geographically restricted areas of northern Utah and California. These collections appear to have resulted from the introduction of this species into the United States possibly from mushroom growers producing G. lucidum outdoors. Overall, this study clarifies the chaotic taxonomy of the laccate Ganoderma in the United States, and will help to remove ambiguities from future studies focusing on the North American species of laccate Ganoderma.
Introduction
Ganoderma is a genus of wood decay fungi in the Polyporales and Ganodermataceae containing species that are commonly observed around the world [1, 2]. The word Ganoderma is Greek from “Gano” meaning shiny and “derma” meaning skin. Ganoderma was erected as a genus in 1881 by Karsten and included only one species, G. lucidum (Curtis) Karst [3]. Previously, it was characterized as Boletus lucidus Curtis (1781) and then Polyporus lucidus (Curtis) Fr. (1821) [3]. The species P. lucidus was characterized by having a laccate pileus and stipe, which Murrill suspects was the reason for Karsten’s division because only one species was included, G. lucidum [4]. Patouillard revised Karsten’s genus Ganoderma to include all species with pigmented spores, adhering tubes and laccate crusted pilei, which resulted in a total of 48 species classified under the genus Ganoderma in his 1889 monograph [4, 5]. Until Murrill investigated Ganoderma in North America in 1902, previous work had focused solely on European species including G. lucidum, G. resinaceum Boud. (1890) and G. valesiacum Boud. (1895) [4, 6, 7].
Murrill described 17 new Ganoderma species in his treatises of North American polypores [4, 7], including G. oregonense Murrill from Picea sitchensis in Oregon, G. sessile Murrill occurring on decaying wood of deciduous trees widespread in the eastern United States (U.S.), G. tsugae Murrill from decaying trunks, stumps and roots of Tsuga canadensis in the boreal hemlock forests of the eastern U.S., G. tuberculosum Murrill occurring on dead wood in Central America and G. zonatum Murrill from decayed palm wood in Florida [4, 7]. Subsequent reports on the genus in North America were contradictory, and there was little agreement regarding the species designated by Murrill [1, 8–12]. For example, many studies focusing on North American taxa considered G. sessile as a synonym of the European species G. lucidum [8, 11]. This was based on similar fruiting body morphology and host preference for both G. sessile and G. lucidum, in which they were both laccate with reddish-brown crust, with or without a stipe, and fruiting on decaying hardwood substrates [7, 8, 11]. Similarly, G. tsugae was also synonymized with G. lucidum, despite the former being described from conifers and the latter from deciduous trees [8]. Atkinson [8] erected G. subperforatum G.F. Atk. in 1908 to include taxa that had “smooth” basidiospores, which were due to fine echinulations protruding through the perisporium (hyaline outer wall) of the double-walled basidiospore. Although Atkinson also considered G. tsugae a synonym for G. lucidum [8], Overholts accepted the species G. tsugae due to its temperate distribution, darker pileus and pure white context tissue [11]. Both Atkinson & Overholts accepted G. curtisii (Berk.) Murrill as a unique species that occurs in more southern latitudes of the U.S. [8, 11]. Although no data were reported, in 1920 in an update on Polyporaceae of North America, Murrill conceded that G. sessile was closely related to the European G. lucidum [13]. It is difficult to determine if this concession was the result of other mycologists’ scrutiny or from a scientific foundation, due to the retention of the name by Murrill in a later publication [14] and a single herbarium collection in 1926 (FLAS-F-08907). In addition, Murrill also described G. sessiliforme Murrill from dead wood in Mexico [15]. The brief description of G. sessiliforme described the fruiting body as similar to G. sessile, but with smaller dimensions. A decade later, Haddow [9] considered G. sessile as a unique taxon, and suggested Atkinson’s G. subperforatum was a synonym of G. sessile, on the basis of the “smooth” spores compared to the “rough” spores of G. lucidum, which was the original basis for G. subperforatum when erected earlier by Atkinson. Furthermore, Haddow [9] agreed with Atkinson that G. tsugae is a synonym of G. lucidum and should not be its own species, despite the previous work of Overholts [11]. Until this point, all identifications of Ganoderma taxa were based on fruiting body morphology, geography, host, and spore characters [4, 7–9, 12].
Nobles [10, 16] characterized the cultural characteristics of numerous wood-inhabiting Hymenomycetes including Ganoderma species. Her work essentially laid the foundation for culture-based identifications of the Basidiomycota [16]. Although Nobles recognized G. lucidum in her 1948 publication [10] as the correct name for the taxon from North American isolates that produce numerous broadly ovoid to elongate smooth chlamydospores (12.0–21.0 x 7.5–10.5 μm), she corrected this misnomer in 1965 by amending the name to G. sessile [16]. Nobles recognized that there were differences in cultural characteristics between G. oregonense, G. sessile, and G. tsugae [10, 16]. Clarifying further, Bazallo & Wright as well as Steyaert agreed with Haddow’s distinction between G. lucidum and G. sessile on the basis of having “smooth” spores, but they synonymized G. sessile with G. resinaceum, a previously described European taxon [12, 17].
In the monograph of “North American Polypores” [1] written in 1986, which is still the only comprehensive treatise on this group of fungi in North America, Gilbertson and Ryvarden recognize five laccate species; G. colossum (Fr.) CF. Baker, G. lucidum, G. oregonense, G. tsugae, and G. zonatum. In the comments for G. lucidum the authors discuss the taxonomic perplexity of this particular species complex, and left it somewhat unresolved. The culture morphology (e.g. presence of chlamydospores) of the North American G. lucidum sensu lato recognized by Adaskaveg & Gilbertson [6] was indistinguishable and biologically compatible with the European species G. resinaceum, where monokaryons of North American G. lucidum sensu lato could be mated with monokaryons of G. resinaceum. A similar conclusion was drawn by Bazallo & Wright as well as Steyaert, where they suggest that G. sessile and G. resinaceum are taxonomic synonyms [12, 17]. Biological species concepts are not always appropriate for designating species ranks for fungi [18]. Although intersterility groups can be successful in laboratory tests, these successful pairings may be due to geographic distributions of a shared common ancestor, and the extant taxa are in the process of allopatric speciation, or vicariance, due to a lack of gene flow because of geographical barriers (e.g. mountains, oceans, etc.) [18, 19].
Adaskaveg & Gilbertson [20] erected G. meredithiae Adask. & Gilb. as a distinct species occurring on pines in the Gulf Coast of the United States. This species was distinguished from the five species found in Gilbertson & Ryvarden’s polypore treatise [1], by its host restriction to pines, slow cultural growth rate, no chlamydospores, plumose or feathery culture morphology, and frequently lobed or branched pilocystidia [20]. Melanoid deposits in the context tissue, which Steyaert suggested was a distinguishing feature for some Ganoderma taxa, are present in G. meredithiae [12, 20]. Although this species was circumscribed, in the same paper the authors informally suggest that G. curtisii and the North American G. lucidum sensu lato were synonyms [20].
With the rise of molecular phylogenetic analyses in the late 20th century, species concept hypotheses were tested to determine the relatedness amongst the nuanced morphological variabilities of the laccate Ganoderma taxa. Moncalvo et al. [21] constructed a phylogeny of the rDNA, and found six major clades amongst the 29 samples in the analysis. Samples labeled as G. lucidum were found in five of the six clades [21]. Hong and Jung [22] found similar results, and showed that G. resinaceum from Europe, and the North American G. lucidum (which Adaskaveg and Gilbertson found to be biologically compatible in vitro) were sister taxa and were also more closely related to each other than G. lucidum sensu stricto [6, 21]. In addition, the authors showed that the North American taxa G. tsugae and G. oregonense were closely related and that these two species were relatives of the European taxon G. valesiacum Boud. From this they determined that G. carnosum Pat., G. oregonense, G. tsugae, and G. valesiacum were conspecific, and ranked them to the G. valesiacum species complex, where all taxa are restricted to temperate coniferous forests [21, 22]. However, it is hypothesized that the laccate Ganoderma species share a common tropical ancestor since more species diversity has been documented from tropical locations [21, 23].
Furthermore, early rDNA phylogenies of global collections of Ganoderma taxa showed that morphological features and cultural characteristics appeared to be highly polyphyletic [21, 22, 24]. While morphology is often polyphyletic in many groups of fungi, it is difficult to determine the validity of the polyphyletic nature of the morphological characters in the laccate Ganoderma taxa due to the use of ambiguous species names. Recent surveys of Ganoderma taxa conducted in the neotropics have revealed a diversity of Ganoderma species including several novel, phylogenetically supported species [25–28]. Most notably, Welti & Courtecuisse [28] described G. martinicense, which is a close relative of the Asian taxon G. multipileum Ding Hou. Ganoderma multipileum has recently been identified from tropical Asian locations as one of the cultivated Ganoderma species used in traditional medicine that was once labeled as G. lucidum sensu lato [29, 30]. Furthermore, the species G. sessiliforme has never been reported in the U.S., but Torres-Torres et al. [27] recently collected it in geographically restricted areas of Mexico, near the type location, along with G. sessile, which was more rare.
Finally, a recent multilocus phylogeny, using ITS, tef, rpb1, and rpb2, revealed that the global diversity of the laccate Ganoderma species included three supported major lineages [2]. These results agree with several of the earlier works focusing mostly on morphology, geography and host preference showing genetic affinity of G. resinaceum and G. sessile, but with statistical support separating the European and North American taxa [2]. Similarly, Ganoderma curtisii and G. sessile were separated with high levels of statistical support, and not considered synonyms. In addition, the phylogeny supported the similarities between G. tsugae and G. oregonense revealing that these two taxa are sister to one another, and the closest relatives of the European taxon G. lucidum sensu stricto [2]. The phylogenetic species concept using a multilocus approach is currently the most robust and accepted method for designating species ranks for the fungi [18].
The confusion surrounding the taxonomy of the North American laccate Ganoderma species has made drawing clear inferences on ecology and biology difficult or impossible. Molecular work is needed to clarify and redefine the laccate Ganoderma species both globally and regionally. Specific molecular phylogenies of local taxa would allow testing of hypotheses of various phylogeographic and ecological questions. To our knowledge a comprehensive survey of the species diversity of the genus Ganoderma in the United States using molecular phylogenetic techniques to validate species names has not been done. The objectives of this research were to I) to identify the laccate Ganoderma species present in the United States using over 500 collections, II) determine relationships between the phylogenetically supported species with a multilocus analysis, and III) identify diagnostic characters for the laccate Ganoderma species in the U.S.
Materials and methods
Fungal material examined
Basidiomata representing laccate (varnished or shiny) Ganoderma species were collected from dead and declining trees from throughout the United States by the authors or citizen scientists. In addition, selected herbarium collections from the University of Florida (Fungal Herbarium of the University of Florida: FLAS) and North Carolina State University (North Carolina State Collection Larry Grand: NCSCLG) herbaria were morphologically examined and used for this study.
Isolations from basidiomata were attempted for all fresh collections by taking small pieces (<1 mm3) of context tissue with a sterile scalpel and placing onto 2% malt extract agar (MEA) (Difco Laboratories, Franklin Lakes, NJ) according to the manufacturer’s instructions with the addition of streptomycin (100 mg/l), benomyl 95% (4 mg/l), and lactic acid (2 mL/l) to limit bacterial growth. Pure cultures were made by subculturing original isolations on MEA (without antibiotics), and grown at 28° C in the dark. Cultures were maintained on MEA slants (without antibiotics) as working stocks, and infested agar discs were submerged in sterile water for long term storage as done previously [31]. Culture collections are archived at the Center for Forest Mycology Research (CFMR) Culture Collection and Herbarium, USDA Forest Service, maintained by the Northern Research Station and housed in the Forest Products Laboratory, USDA-Forest Service in Madison, Wisconsin. Representative basidiomata collected from this study have been accessioned into the FLAS collection.
Basidiomata morphology and host substrate classification
Morphological assessments were similar to that from Zhou et al. [2]. Color standards from Ridgway [32] were used to describe macromorphological features of the basidiomata. The color and texture of the context tissue was observed visually and described. Other contextual features such as melanoid deposits or concentric zones were noted. Basidiospores were visualized on slides in 5% KOH under 1000x magnification using a Nikon Eclipse 55i light microscope (Melville, NY), and photographed with a Canon Rebel T3i (Huntington, NY). Measurements were made using ImageJ software (www.imagej.net) for at least ten basidiospores of three to five representative collections for each species, except Tomophagus colossus (Fr.) Murrill, which had measurements from two basidiomata from the same collection (255FL). The size of the basidiospore width extends the entire width at the widest point from the outer wall and the length extends from the base of the basidiospore to the length of the truncated apex. Basidiospore measurements (i.e. length, width, spore shape index, and Q-ratio) were analyzed statistically in JMP PRO 12 (SAS, Cary, NC) using analysis of variance and Tukey’s HSD means separations. In addition to basidiospores, slide mounts of the context tissue and mycelium from cultures were made to note presence or absence of chlamydospores. If chlamydospores were present, at least ten were measured under 1000x magnification as described previously. In addition to morphology, information on location and host was recorded if available. Host substrate species, when available, were identified visually by the authors, or by collectors of the various specimens. From this data, hosts were broken down into major groups including cactus, conifer, cycad, hardwood, or monocot.
DNA extraction, PCR, and sequencing
DNA was extracted from the context tissue of the basidiomata or mycelium from cultures of each accession with the Extract-N-Amp rapid DNA kit (Sigma-Aldrich, St. Louis, MO) a Qiagen DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions, or a CTAB protocol as described previously [33]. Initially, samples were identified based on morphology, host, and geographic locations, and these identifications were confirmed through sequencing the internal transcribed spacer (ITS) region of the ribosomal DNA (rDNA). ITS sequences were queried across sequences deposited in GenBank. In addition, tef1α, rpb1, and rpb2 were sequenced for representative isolates for each species. These regions were selected to make comparisons to a recently published global phylogeny of the members of the G. lucidum species complex [2].
The ITS region was amplified with primers ITS1F and ITS4b or ITS4 as done previously [34, 35]. For the tef1α, rpb1, and rpb2 loci primer pairs EF1-983/EF1-2218R [36], RPB1-2.2f/RPB1-Cr [37, 38], and fRPB2-5F/bRPB2-7R2 [39, 40] were used respectively and followed PCR protocols following Blanchette et al. [33]. For problematic samples, primers were designed from alignments made from Ganoderma sequences downloaded from GenBank for each of the loci (tef1α, rpb1, and rpb2). Multiple primer pairs were made for each locus, and tested for efficiency using a gradient PCR. The following primers were chosen and used for PCR amplification: tef1α: EF-Gano23F (5’ GGTGTCAGGCAGCTCATYGT) and EF-Gano887R (5’ CGAACTTGCARGCGATGTG), rpb1: RPB1-Gano18F (5’ GCGTGGTGAAATGGGGGGCT) and RPB1-Gano958R (5’ GCAACTGCTCGAACTCGTTG), rpb2: RPB2-Gano53F (5’ AAYTGGGGAGACCAGAA) and RPB2-Gano829R (5’ CGCCTTYAAYCGAGC). For each PCR reaction, the following reagents were used: 12.5 μl of Immomix Red Master Mix (Bioline, London, UK), 8.5μl of PCR-grade H2O, 1μl BSA (3% w/v), 1μl of each primer, and 1ng/μl of DNA template. Reactions were performed on a MJ Mini thermocycler (BioRad, Hercules, CA) with the following thermocycler conditions: cycle of 95° C for 10 min and followed by 35 cycles of 94° C for 30 sec, variable annealing temperatures of 55° C (ITS) 62 C (tef1α and rpb1) 53.2° C (rpb2) for 30 sec, and 72° C for 1 min, followed by a final extension step of 72° C for 5 min, and then 4° C. Amplicons were purified with Exo-SAP-IT (ThermoFisher, Waltham, MA) according to the manufacturer’s recommendations. Sanger sequencing was performed using both forward and reverse primers at the Interdisciplinary Center for Biotechnology Research (ICBR) at the University of Florida, Genewiz (www.genewiz.com), or the University of Minnesota Genomics Center. Forward and reverse sequences for each sample were aligned and visually edited using Geneious 10 (www.geneious.com, [41]). All sequences generated were deposited and accessioned in the GenBank sequence database.
Phylogenetic analyses
ITS, tef, rpb1, and rpb2 sequences for representative isolates generated from this study and reliable voucher sequences from Zhou et al. [2] were aligned for each locus using the MAFFT [41] plugin in Geneious 10, and visually edited to remove any ambiguities and minimize differences of each alignment that could have resulted from sequencing error. The alignments consisted of 98 individual sequences with 521 nucleotides for ITS, 58 individual sequences with 764 nucleotides for tef1α, 61 individual sequences with 615 nucleotides for rpb1, and 36 individual sequences with 570 nucleotides for rpb2. Visually edited alignments of each locus were used for independent phylogenetic analyses using the RAxML [42] plugin in Geneious 10 using a general time reversal (GTR) evolutionary model with rapid boostrapping and 1000 bootstrap replications. Trees were assessed visually to determine any incongruences between topologies across individual locus-based phylogenies. All tree topologies of the four loci were congruent, so a concatenated alignment with all four loci representing 2470 characters was used in phylogenetic analyses using RAxML and Mr. Bayes [43] plugins in Geneious 10. The RAxML analysis of the concatenated alignment used a GTR evolutionary model using rapid bootstrapping with 1000 bootsrap replications, and the Mr. Bayes analysis used a GTR evolutionary model with a gamma rate variation using four gamma categories, 1,100,000 chain length with 4 heated chains using a burn-in length of 100,000. In both analyses, Tomophagus colossus isolates UMNFL110 and TC-02 were used to root the tree as the outgroup, because it is a closely-related genus in the Ganodermataceae [2]. Alignments have been deposited on TreeBase and found under the submission ID 22399 (http://purl.org/phylo/treebase/phylows/study/TB2:S22399?x-access-code=a01554a96d36b989fd14482c059b6b82&format=html).
Ganoderma species DNA barcoding
Although ITS has been widely recognized as the fungal barcode of life [44], it is now accepted that ITS alone can underestimate the total number of species in biodiversity studies [45]. Alternative or additional DNA barcode loci should be sought after for Ganoderma species, so that rapid and accurate species diagnosis can be achieved. To determine the most appropriate barcode locus for the laccate Ganoderma species in the U.S., RAxML trees from the aforementioned analyses for each of the four loci were visually compared to the putative species tree constructed from the concatenated alignment. Scores were given for each analysis of individual loci and the concatenated (four loci) trees. They were scored on total number of well supported terminal clades and the associated summed bootstrap values for each well-supported terminal clade. These analyses were conducted as described previously using the RAxML plugin with rapid bootstrapping and 1000 bootstrap replication in Geneious 10. Alignments were deposited on TreeBase found under the submission ID 22399 (http://purl.org/phylo/treebase/phylows/study/TB2:S22399?x-access-code=a01554a96d36b989fd14482c059b6b82&format=html).
Results
Identification of laccate Ganoderma collections
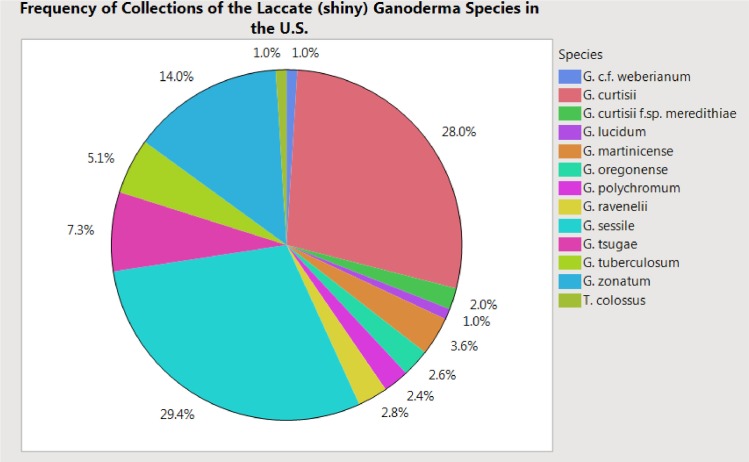
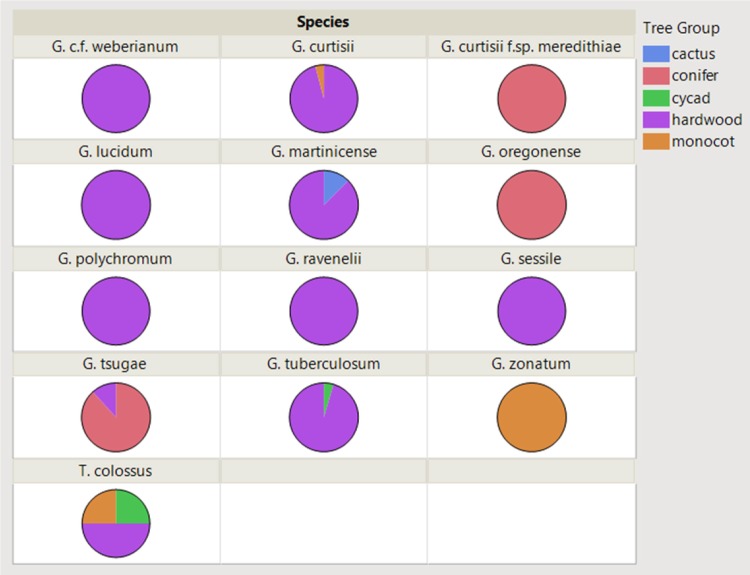
Five hundred and seven collections of laccate Ganoderma species (basidiomata and/or cultures) from our collections (n = 427) or from FLAS (n = 22) and NCSCLG (n = 58) mycological herbaria were studied (Fig 1). Collections originated from 34 U.S. states (Fig 2). Of the 22 specimens observed from FLAS, about half were collected and/or identified by Murrill, who was the taxonomic authority on all of these collections, including: G. curtisii, G. oregonense, G. tuberculosum and G. zonatum. Thirteen laccate taxa in the Ganodermataceae were identified in this U.S. survey, including G. curtisii (n = 142), G. lucidum (n = 5), G. martinicense (n = 18), G. oregonense (n = 13), G. polychromum (Copel.) Murrill (n = 12), G. ravenelii Steyaert (n = 14), G. sessile (n = 149), G. cf. weberianum (Bres. & Henn. ex. Sacc.) Steyaert (n = 5), G. tsugae (n = 37), G. tuberculosum (n = 26), G. zonatum (n = 71), Tomophagus colossus (n = 5) and G. curtisii f.sp. meredithiae (n = 10) f. sp. nov. Collections of G. curtisii f.sp. meredithiae represents collections formerly described as G. meredithiae. Here we synonymize this taxon with G. curtisii due to a lack of evidence to uphold the rank of species (see below). However, physiological differences, such as slow in vitro growth rate on MEA and an affinity to decay pine substrates, existed in collections of G. meredithiae, so the informal forma specialis designation, which is used as an informal taxonomic rank for physiological variants, is proposed here resulting with G. curtisii f.sp. meredithiae. The most commonly collected species in the eastern U.S. were G. sessile (29%), G. curtisii (28%) and G. zonatum (14%), while the more commonly encountered species in the western U.S. were G. oregonense (3%) and G. polychromum (2%) were the more commonly encountered species (Figs 1 and 2).
Fig 1. Frequency of taxa representing the laccate Ganoderma species collected in the United States.
Percentages are representative of the total collections (n = 507). Species in the legend are represented in a clockwise order on the pie chart.
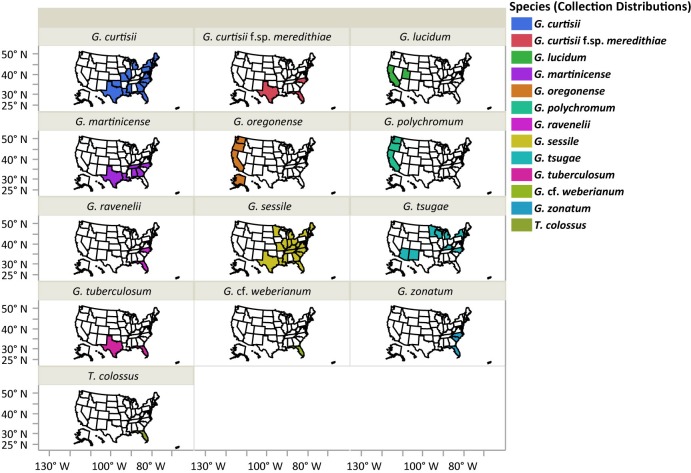
Fig 2. Distribution of collections of the laccate Ganoderma species studied.
Species are shaded in different colors in each state where a collection of that species was made.
Although our collections were heavily biased towards the eastern U.S., there were some apparent geographic distribution limitations for some of the species (Fig 2). For example, Tomophagus colossus and G. cf. weberianum were only collected in subtropical south Florida. Similarly, G. tuberculosum was only collected in subtropical locations of south Florida and south Texas. Furthermore, collections of G. oregonense and G. polychromum were restricted to the western U.S. near each type locality, while collections of G. curtisii, G. curtisii f.sp. meredithiae, G. martinicense, G. ravenelii, G. sessile, G. tsugae, and G. zonatum were generally restricted to the eastern U.S. However, there were some anomalies with respect to geographic distributions. For example, G. sessile was collected twice in California and once in Utah, suggesting it may be more widespread or these were the results of introductions. Similarly, the collections of G. lucidum sensu stricto were from California and Utah, but were restricted to small geographic regions in anthropogenic environments in northern California and northern Utah. Furthermore, there were four collections of G. tsugae from the southwestern U.S., in northern New Mexico (UMNNM13 and UMNNM46) and northern Arizona (MS182AZ and UMNAZ9).
Basidiomata morphology and host substrate affinities
Although there was some variation in basidiomata morphology within collections representing an individual species, there were generally several morphological features that were diagnostic. Diagnostic morphological features included I) stipitate vs. sessile fruiting body morphology, II) color of context tissue, III) contextual features (e.g melanized deposits), iv) presence/absence of chlamydospores. and v) shape and size of basidiospores.
Stipitate morphologies were designated based on having a stipe the same size or longer than the width of the pileus. Taxa that were generally stipitate, included: G. curtisii, G. curtisii f.sp. meredithiae, G. lucidum, and G. ravenelii. Sessile morphology of fruiting bodies were designated based on having basidiomata that lacked a stipe, or stipes, if present, were shorter than the width of the pileus (i.e. pseudostipitate). Species that generally produced sessile basidiomata included: G. martinicense, G. oregonense, G. polychromum, G. sessile, G. tsugae, G. tuberculosum, G. cf. weberianum, G. zonatum, and Tomophagus colossus. All collections of G. tuberculosum were truly sessile. Collections of G. martinicense were consistently centrally pseudostipitate, possessing a stipe that was less than half the width of the pileus. Similarly, collections of G. oregonense, G. polychromum, G. sessile, G. tsugae G. zonatum, G. cf. weberianum and T. colossus were rarely stipitate and occasionally pseudostipitate and generally the short stipe was produced laterally or off center.
All thirteen taxa, had shiny or varnished (laccate) pilei that ranged in color from yellow to orange to reddish brown with variable coloration within a species (Fig 3). For example, pilei ranged in color from yellow-orange to reddish brown often with purple hues for collections of G. curtisii. Similarly, the pileus color of collections of G. sessile ranged from deep red to orangish-red. The color of the pileus was not diagnostic for most, although T. colossus was the only species that produced basidiomata that were distinctly shiny and mustard yellow. Similar to basidiomata color, pore shape and hymenium color were not always diagnostic. This is probably due to the age and maturity of the basidiomata as well as the environment where they were fruiting. However, the color of the context tissue was diagnostic. The context tissue is the inner flesh, or area between the pileus crust and initiation of the tubes. Generally, the color of the context tissue was grouped into three major categories: I) white, II) pinkish-buff to cinnamon-buff (i.e. light brown), or III) cinnamon brown (i.e. dark brown) (Fig 4A). All thirteen species could be grouped into one of these categories; G. oregonense, G. tsugae, and T. colossus had white context tissue, G. curtisii, G. curtisii f.sp. meredithiae, G. lucidum sensu stricto, G. polychromum, G. ravenelii, G. sessile, and G. cf. weberianum had buff to light brown, and G. martinicense, G. tuberculosum, and G. zonatum had dark cinnamon brown colored context tissues.
Fig 3. In situ photos of the laccate Ganoderma species in the United States.

A) G. curtisii fruiting at the base of a dead oak tree (Quercus sp.) in Georgia (290GA), B) G. lucidum fruiting from near an oak (Quercus sp.) in California (not in collection) (photo credit: Shane Hanofee), C) G. martinicense fruiting from a southern red oak (Quercus falcata) in Georgia (230GA) (photo credit: Bill Sheehan), D) G. curtisii f. sp. meredithiae fruiting from slash pine roots in Florida (140FL), E) G. oregonense fruiting on white fir (Abies concolor) in California (no collection data) (photo credit: Arthur Grupe), F) G. polychromum fruiting on a pruning wound on a coast live oak (Quercus agrifolia) in California (331CA) (photo credit: Drew Zwart), G) G. ravenelii fruiting from the roots of an oak tree (Quercus sp.) in Florida (no collection data), H) G. sessile fruiting on the lower bole and root flare of honeylocust (Gleditsia tricanthos) in New York (276NY) (photo credit: Margery Daughtrey), I) G. tsugae fruiting on the trunk of eastern hemlock (Tsuga canadensis) in Wisconsin (342WI), J) G. tuberculosum fruiting on the root flare of pongam tree (Pongamia pinnata) in Florida (335FL), K) G. cf. weberianum near a recently removed live oak tree (Quercus virginiana) in Florida (261FL), L) G. zonatum fruiting on the trunk of an American oil palm (Elaeis oleifera) in Florida (283FL), and M) Tomophagus colossus fruiting on the cycad Macrozamia moorei in Florida (255FL) (photo credit: Michael Calonje).
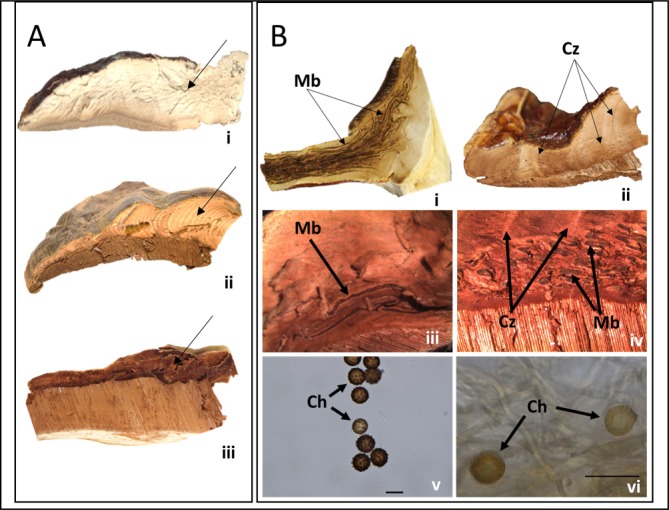
Fig 4.
Contextual colors (A) and features (B) of the laccate Ganoderma species of the U.S. (bars = 20 μm) A) broad categories of the context tissue colors, where arrows point to context tissue; i) white context tissue (324WI, G. tsugae), ii) light buff to cream context tissue (112CA, G. polychromum), and iii) dark brown context tissue (265FL, G. zonatum). B) contextual features such as melanoid bands (“Mb”), concentric growth zones (“Cz”), and contextual chlamydospores (“Ch”); i) melanoid bands embedded in context tissue of pileus and stipe (158FL, G. curtisii), ii) concentric growth zones in context tissue of the pileus (171FL, G. sessile), iii) close-up (10x) of melanoid bands in the context tissue of the pileus (NCSCLG 1804, G. curtisii f.sp. meredithiae), iv) close-up (10x) of melanoid bands and concentric growth zones in context tissue of the pileus (NCSCLG 19006, G. martinicense), v) double walled, globose contextual chlamydospore (255FL, T. colossus), and vi) double walled contextual, globose chlamydospore with striated margin (FLAS F59210, G. cf. weberianum).
In addition to the color of the context tissue, certain context tissue features such as melanoid bands, concentric growth zones, and chlamydospores were diagnostic for many species (Fig 4B). For example, G. curtisii, G. curtisii f.sp. meredithiae, and G. martinicense had melanoid shiny deposits embedded in the context tissues. Similarly, G. tuberculosum often had resinous deposits in the context tissue, but were often lighter in color relative to the context tissue. Concentric growth zones (Fig 4B) were observed consistently in collections of G. lucidum, G. martinicense, G. polychromum, G. sessile, G. tuberculosum, G. cf. weberianum, and G. zonatum. Lastly, chlamydospores found in the context tissues were consistently observed in collections of G. cf. weberianum and T. colossus. The chlamydospores produced in fruiting body context tissue of G. cf. weberianum were hyaline or pigmented in 5% KOH, double-walled, ovate to globose, and on average were 17.1 (14.1–20.1) X 12.0 (9.6–14.1) μm. In addition the outer wall of chlamydospores of G. cf. weberianum were striate (Fig 4B-vi). The chlamydospores produced in fruiting body context tissue of T. colossus were pigmented, double-walled, globose, ornamented, and on average 16.1 (15.1–17.6) μm in diameter (Fig 4B-v). When grown on MEA, G. martinicense, G. polychromum and G. sessile also constitutively produced hyaline chlamydospores, but were rarely observed in the context tissue of basidiomata (Table 1).
Table 1. Morphological assessment of the laccate Ganoderma species present in the United States.
| Taxon | Authority | Stipe | Context tissue1 | Hymenium (pores/mm) |
Chlamydospores2 | Basidiospores3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| presence | color | size | color/texture | concentric growth zones | resinous or melanoid deposits | present/absent | shape | size (length X width) | length | width | S.S.I.4 | Q-ratio5 | |||
| G. curtisii | (Berk.) Murrill 1902 | almost always present | tawny to russett with occasional mauve pigments | typically, 1.5x the diameter of the cap | pink-buff to cinnamon buff/corky | absent | present | 5–8 | absent | - | - | 10.6 (8.3–12.1) efg | 6.4 (5.4–7.5) f | 60.4 c | 1.7 cd |
| G. curtisii f.sp. meredithiae | (Adask. & Gilb) (this paper) | almost always present | tawny to russett with occasional mauve pigments | typically, 1.5x the diameter of the cap | pink-buff to cinnamon buff/corky | absent | present | 5–8 | absent | - | - | 10.8 (9.5–11.5) cd | 6.8 (6.4–7.3 def) | 62.6 bc | 1.6 cd |
| G. lucidum | (Curtis) P. Karst 1881 | almost always present | tawny to russett | long and eccentric; typically twice the diameter of the cap | pink-buff to cinnamon buff/corky | present | absent | 4–5 | absent | - | - | 10.7 (8.2–12.1) f | 7.1 (4.8–8.9) cd | 66.2 ab | 1.5 ef |
| G. martinicense | Welti & Court 2010 | present | cinnamon brown to black | short and stubby; typically less than the diameter of the cap | cinnamon brown/felty to corky | present | present | 5–6 | present and abundant in culture | hyaline to pigmented ovate to spherical or irregularly shaped with hyphal appendages protruding out (culture) | 17.1 (13.5–21.1) x 12.2 (9.2–17.3) | 11.1 (9.0–13.6) def | 6.9 (5.3–8.3) cde | 62.6 abc | 1.6 de |
| G. oregonense | Murrill 1908 | occasionally present | tawny to cinnamon brown | short and stubby; typically less than the diameter of the cap | white/spongy to corky | absent | absent | 3–4 | absent | - | - | 12.9 (11.6–14.9) b | 8.0 (6.7–9.3) b | 62.2 c | 1.6 cd |
| G. polychromum | (Copel.) Murrill 1908 | rarely present | tawny to russett | short and thin; typically less than the diameter of the cap if present | pink-buff to cinnamon-buff/corky | present | absent | 4–5 | present; rare in context and abundant in culture | elliptical to obpyriform to ovate, hyaline, smooth (culture) | 14.8 (10.3–18.3) X 9.9 (11.9–7.0) | 12.2 (10.8–13.2) cd | 6.8 (6.0–7.4) cde | 55.5 abc | 1.8 cd |
| G. ravenelii | Steyaert 1980 | almost always present | tawny to russett | typically, 1.5x the diameter of the cap | pink-buff to cinnamon-buff/corky | absent | absent | 6–7 | absent | - | - | 11.2 (9.1–13.6) cde | 5.2 (4.2–6.8) h | 46.5 d | 2.2 a |
| G. sessile | Murrill 1902 | occasionally present | tawny to russett | short and thin; typically less than the diameter of the cap if present | pink-buff to cinnamon-buff/corky | present | rarely present | 5–7 | present; rare in context and abundant in culture | elliptical to obpyriform to ovate, hyaline, smooth (culture) | 16.0 (12.0–26.0) X 11.0 (9.5–12.0) | 11.4 (9.7–14.0) cd | 6.6 (5.2–8.4) ef | 58.1 c | 1.7 c |
| G. tsugae | Murrill 1902 | occasionally present | tawny to cinnamon brown | short and stubby; typically less than the diameter of the cap | white/spongy to corky | absent | absent | 5–7 | absent | - | - | 9.9 (8.9–11.5) g | 6.5 (5.2–7.7) ef | 65.2 abc | 1.5 def |
| G. tuberculosum | Murrill 1908 | absent | - | - | cinnamon brown/felty to corky | present | present | 4–7 | absent | - | - | 10.5 (9.2–12.0) fg | 7.3 (6.2–8.6) c | 69.3 a | 1.4 f |
| G. cf. weberianum | (Bres. & Henn. ex Sacc.) Steyaert 1972 | absent | - | - | pink-buff to cinnamon-buff/corky | present | absent | 5–7 | present; context tissue and culture | ovate to globular and striate and pigmented (context tissue); elliptical to obpyriform to ovate, hyaline, smooth (culture) | 17.1 (14.1–20.1) X 12.0 (9.6–14.1) | 8.4 (7.7–9.5) h | 5.6 (4.7–7.3) gh | 67.0 abc | 1.5 ef |
| G. zonatum | Murrill 1902 | rarely present | yellow ocher to russett | short and stubby; typically less than the diameter of the cap | cinnamon brown/felty | present | absent | 4–6 | absent | - | - | 11.8 (10.3–13.7) c | 5.9 (5.0–6.6) g | 49.5 d | 2.0 b |
| T. colossus | (Fr.) Murrill 1905 | absent | - | - | white to light buff/spongy | absent | absent | 3–4 | present; abundant in context and culture | globular with abundant ornamentations pigmented (context tissue and culture) | 16.1 (15.1–17.6) | 16.1 (14.6–17.3) a | 10.4 (9.5–11.3) a | 64.8 abc | 1.5 def |
1. Sterile tissue inside fruiting body between the pileus crust and initiation of the tubes
2. Survival structures that could be produced in the fruiting body context tissue and/or in vitro culture
3. Basidiospore measurments ending in different letters are statistically different (P<0.05) with Tukey’s HSD means separation
4. The “S.S.I.” is the spore shape index, which is calculated by (width/length)*100, and is used to quantitatively describe the shape
5. The “Q-ratio” is a ratio of the length:width
The shape and size of basidiospores were diagnostic for some species. It is possible with additional measurements that do not include the inflated or truncated, hyaline apex of the basidiospores that nuanced differences in length could be elucidated as seen in Hennicke et al. (2016) [29]. However, all of our measurements included the entire spore length from base to truncated apex, which was similar and comparable to older Ganoderma literature [46]. For example, basidiospores of T. colossus were nearly one and one-half to twice as large as the other species. Although the basidiospores of T. colossus were much larger relative to the other species, the length to width ratio (Q-ratio) was similar (Q = 1.5). The basidiospore Q-ratio was distinctly larger for collections of G. ravenelii (Q = 2.2) and G. zonatum (Q = 2.0), because the spores were much more elongated than they were wide relative to the other taxa. Contrary to that, the spore shape index ((width/length)*100) of G. tuberculosum was 69.3, suggesting that 69.3% of the length can be explained by the width, which were considered broadly ovate, or “squatty”. Lastly the length of basidiospores was diagnostic for collections of G. cf. weberianum, which had basidiospores that were shorter than most (8.4(7.7–9.5) μm). Morphological descriptions are summarized in Table 1.
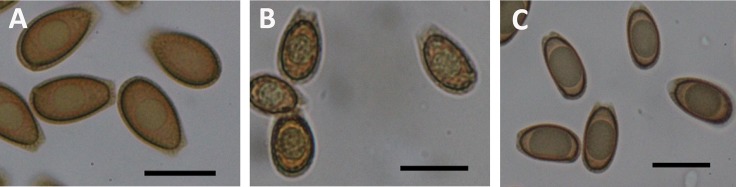
In addition to shape and size, the amount of echinulation of the basidiospores was diagnostic, and grouped into categories of “rough” or “smooth” as described previously [8, 12]. Collections of G. sessile, G. polychromum and G. cf. weberianum had “smooth” basidiospores due to fine echinulations protruding through the perisporium wall from the endosporium. The other taxa produced basidisopores that could be lumped into the “rough” basidiospore category, due to coarse echinulations protruding through the perisporium (outer, hyaline wall) from the endosporium (pigmented, inner portion of the spore) (Fig 5).
Fig 5. Examples of basidiospore morphology (bars = 10 μm).
A) “smooth” (finely echinulated) from G. sessile (287SC), B) “rough” (coarsely echinulated) basidiospores from G. curtisii (158FL), and C) elongated, elliptical basidiospores of G. zonatum (265FL).
Of the collections of laccate species of Ganoderma in the U.S. from this study, hosts were recorded for 298 collections. Of the known hosts/substrates for each collection 49 genera of plant species were identified across all collections, where the most frequent host genera were Quercus (37%), Tsuga (11%), Sabal (6%), and Acer (5%). The known host/substrate were categorized into the following broad categories: hardwood (68%), conifer (14%), monocot (17%), cycad (1%), and cactus (0.3%) (Fig 6). There were different affinities for certain groups of hosts across the different Ganoderma taxa. The following taxa were exclusively or predominately associated with hardwood host/substrate: G. curtisii, G. martinicense, G. polychromum, G. ravenelii, G. sessile, G. tuberculosum, and G. cf. weberianum. In addition, G. curtisii f.sp. meredithiae, G. oregonense, and G. tsugae were found fruiting predominately on conifers. Similarly, G. zonatum was only collected from monocots, mostly palm species, but three collections were from Bambusa vulgaris, a clumping bamboo. A species key of the laccate Ganoderma in the United States is presented using morphology, geography and host preference (Table 2).
Fig 6. Host substrate affinities for the laccate (shiny) Ganoderma species collected in the U.S., where host substrate was known (n = 298).
Table 2. Key to the laccate Ganoderma species in the United States.
Members of the laccate Ganoderma are have shiny or varnished pilei, and can be sessile, stipitate or pseudostipitate. The context tissue of the laccate Ganoderma species is corky to felty in texture, and generally white, cream to light buff, or cinnamon brown. Context tissue can have melanoid/resinous bands and/or concentric growth zones present or absent. Some taxa produce contextual chlamydospores that are double-walled, hyaline to pigmented, and ornamented or smooth. The basidiospores are double-walled, golden-brown in 5% KOH, echinulated and generally broadly ovoid to elliptical with a truncated apex at maturity. Ecologically these species are associated with a white rot type decay typically on hardwood, coniferous, or palm substrates. Disclaimer: This key is solely based on morphology, host preference and known geographic distributions of the laccate Gaonderma present in the U.S. based on this study.
| 1 | Context tissue white when fresh | 2 |
| 1 | Context tissue not as above | 4 |
| 2 | Large basidiospores measuring on average 16.1 (14.6–17.3) x 10.4 (9.5–11.3) μm; fruiting body spongy, light-weight, shiny, and yellow | T. colossus (syn. = G. colossus) |
| 2 | Basidiospores smaller than above | 3 |
| 3 | Found on conifers in the Pacific Northwest, with basidiospores measuring on average 12.9 (11.6–14.9) x 8.0 (6.7–9.3) μm | G. oregonense |
| 3 | Found on conifers, predominately Tsuga canadensis, in the boreal hemlock forests of the eastern U.S. with basidiospores measuring on average 9.9 (8.9–11.5) x 6.5 (5.2–7.7) μm | G. tsugae |
| 4 | Context tissue cream to buff | 5 |
| 4 | Context tissue dark brown (cinnamon brown) | 11 |
| 5 | Shiny, melanoid deposits present in the context tissue | 6 |
| 5 | Melanoid bands absent, concentric growth zones sometimes present in the context tissue | 7 |
| 6 | Typically a laterally stipitate fruiting body, fruiting on hardwoods in the eastern U.S with basidiospores on average measuring 10.6 (8.3–12.1) x 6.4 (5.4–7.5), and growing somewhat rapidly (approximately 6 mm/day) on malt extract agar | G. curtisii |
| 6 | Typically a laterally stipitate fruiting body, fruiting on pines in the southeastern U.S with basidiospores on average measuring 10.8 (9.5–11.5) x 6.8 (6.4–7.3), and growing slowly (less than 3 mm/day) as a dikaryotic isolate on malt extract agar | G. curtisii f.sp. meredithiae |
| 7 | Typically sessile fruiting body morphology, or if stipe present, considered a pseudostipe, where the length of the stipe is less than the width of the pileus | 8 |
| 7 | Typically laterally stipitate fruiting body with no melanoid bands and concentric growth zones sometimes present in the context tissue | 10 |
| 8 | Found in association with hardwoods in the western United States, often with conspicuous concentric growth zones present in the context tissue | G. polychromum |
| 8 | Found in the eastern United States | 9 |
| 9 | Pigmented, double-walled globose to ovoid chlamydospores found in the context tissue, and restricted to tropical locations | G. cf. weberianum |
| 9 | Widely distributed East of the Rocky Mountains predominately associating with hardwood trees/substrates, basidiospores measuring on average 11.4 (9.7–14.0) x 6.6 (5.2–8.4) μm | G. sessile |
| 10 | Concentric growth zones absent from the context tissue, present in the southeastern U.S., and basidiospores measuring on average 11.2 (9.1–13.6) x 5.2 (4.2–6.8) μm | G. ravenelii |
| 10 | Concentric growth zones present in the context tissue, and restricted to isolated populations in northern Utah and northern California | G. lucidum |
| 11 | Basidiospores elongated measuring on average 11.8 (10.3–13.7) x 5.9 (5.0–6.0) μm, and associated with monocot trees/substrates, typically palms | G. zonatum |
| 11 | Not as above | 12 |
| 12 | Typically producing a central pseudostipe that is often dark red to black, associated with hardwood trees/substrates, concentric growth zones and melanoid deposits present in context tissue, and hymenium on average having 5–6 pores/mm | G. martinicense |
| 12 | Sessile fruiting body that is orange to red when active, and dark red when mature, concentric growth zones and shiny, resinous deposits present in the context, hymenium on average having 4–7 pores/mm, restricted to tropical locations, and basidiospores measuring on average 10.5 (9.2–12.0) x 7.3 (6.2–8.6) μm | G. tuberculosum |
Multilocus-based phylogeny of laccate Ganoderma species in the United States of America
In total, 522 sequences were generated in this study representing the thirteen species, and deposited into GenBank. These sequences represented 366 ITS (MG654066-MG654431), 60 tef1 (MG754723-MG754782), 68 rpb1 (MG754783-MG754850), and 27 rpb2 (MG754851-MG754877) sequences (S1 Table). We were unable to obtain sequences from specimens that were older than five years. Representative sequences of each species were used in the phylogenetic analyses. In addition, selected voucher sequences were downloaded from GenBank (Table 3), and were selected as reference sequences, since they were used in a recent global phylogeny of members of the G. lucidum species complex [2, 28]. The only taxa that could not be validated with a voucher sequence or collections were G. polychromum, G. ravenelii, and G. cf. weberianum but these were clearly distinct genetically and generally matched the published taxonomic descriptions and type locations.
Table 3. Sample accessions, location, and GenBank Accession numbers for ITS, tef1α, rpb1, and rpb2 used in phylogenetic analysis.
| Accession | Taxon | Location | GenBank Accession Numbers1 | |||
|---|---|---|---|---|---|---|
| ITS | tef1α | rpb1 | rpb2 | |||
| WD2028 | Ganoderma boninense | Japan | KJ143905 | KJ143924 | KJ143944 | KJ143964 |
| WD2085 | Ganoderma boninense | Japan | KJ143906 | KJ143925 | KJ143945 | KJ143965 |
| 102NC | Ganoderma curtisii | NC, USA | MG654074 | MG754727 | — | MG754851 |
| 223FL | Ganoderma curtisii | FL, USA | MG654167 | — | MG754785 | MG754854 |
| 238FL | Ganoderma curtisii | FL, USA | MG654171 | — | MG754786 | — |
| CBS100131 | Ganoderma curtisii | NC, USA | JQ781848 | KJ143926 | KJ143946 | KJ143966 |
| CBS100132 | Ganoderma curtisii | NC, USA | JQ781849 | KJ143927 | KJ143947 | KJ143967 |
| UMNFL28 | Ganoderma curtisii | FL, USA | MG654097 | MG754728 | MG754788 | MG754856 |
| UMNFL6 | Ganoderma curtisii | FL, USA | MG654093 | — | — | — |
| UMNFL60 | Ganoderma curtisii | FL, USA | MG654105 | MG754729 | MG754789 | — |
| UMNGA1 | Ganoderma curtisii | GA, USA | MG654117 | MG754731 | MG754791 | MG754857 |
| UMNNC3 | Ganoderma curtisii | NC, USA | MG654130 | MG754732 | MG754794 | — |
| 124FL | Ganoderma curtisii f.sp. meredithiae | FL, USA | MG654188 | MG754734 | MG754805 | MG754861 |
| UMNFL50 | Ganoderma curtisii f.sp. meredithiae | FL, USA | MG654103 | MG754735 | MG754806 | MG754862 |
| UMNFL64 | Ganoderma curtisii f.sp. meredithiae | FL, USA | MG654106 | — | MG754807 | MG754863 |
| Wei5491 | Ganoderma flexipes | Hainan, China | JQ781850 | – | – | KJ143968 |
| Wei5494 | Ganoderma flexipes | Hainan, China | JN383979 | — | — | — |
| Cui9166 | Ganoderma lingzhi | Shandong, China | KJ143907 | JX029974 | JX029982 | JX029978 |
| Dai12479 | Ganoderma lingzhi | Anhui, China | JQ781864 | JX029975 | JX029983 | JX029979 |
| Cui9207 | Ganoderma lucidum | Yunan, China | KJ143910 | KJ143928 | KJ143949 | KJ143970 |
| K175217 | Ganoderma lucidum | United Kingdom | KJ143911 | KJ143929 | KJ143950 | KJ143971 |
| MS183CA | Ganoderma lucidum | CA, USA | MG911000 | MG754723 | — | — |
| MT26/10 | Ganoderma lucidum | Czech Republic | KJ143912 | KJ143930 | KJ143951 | — |
| Rivoire4195 | Ganoderma lucidum | France | KJ143909 | — | KJ143948 | KJ143969 |
| UMNUT1 | Ganoderma lucidum | UT, USA | MG654070 | MG754725 | MG754799 | — |
| UMNUT7 | Ganoderma lucidum | UT, USA | MG654071 | MG754726 | MG754800 | — |
| UMNUT8 | Ganoderma lucidum | UT, US | MG654072 | — | — | — |
| UMNUT9 | Ganoderma lucidum | UT, US | MG654073 | — | — | — |
| 231NC | Ganoderma martinicense | NC, USA | MG654182 | MG754736 | MG754801 | — |
| 246TX | Ganoderma martinicense | TX, USA | MG654185 | MG754737 | MG754802 | MG754858 |
| LIPSW-Mart08-44 | Ganoderma martinicense | Martinique | KF963257 | — | — | — |
| LIPSW-Mart08-55 | Ganoderma martinicense | Martinique | KF963256 | — | — | — |
| UMNAL2 | Ganoderma martinicense | AL, USA | MG654176 | — | — | — |
| UMNSC7 | Ganoderma martinicense | SC, USA | MG654177 | — | — | MG754859 |
| UMNTN1 | Ganoderma martinicense | TN, USA | MG654178 | MG754738 | MG754803 | MG754860 |
| UMNTX3 | Ganoderma martinicense | TX, USA | — | MG754739 | MG754804 | — |
| CWN04670 | Ganoderma multipileum | Tawian, China | KJ143913 | KJ143931 | KJ143952 | KJ143972 |
| Dai9447 | Ganoderma multipileum | Hainan, China | KJ143914 | KJ143932 | KJ143953 | KJ143973 |
| CBS265.88 | Ganoderma oregonense | OR, USA | JQ781875 | KJ143933 | KJ143954 | KJ143974 |
| CBS266.88 | Ganoderma oregonense | WA, USA | JQ781876 | — | KJ143955 | KJ143975 |
| UMNAK1 | Ganoderma oregonense | AK, USA | MG654190 | MG754740 | MG754808 | — |
| UMNOR1 | Ganoderma oregonense | OR, USA | MG654194 | MG754741 | MG754809 | — |
| 330OR | Ganoderma polychromum | OR, USA | MG654196 | MG754742 | — | — |
| BJ280CA | Ganoderma polychromum | CA, USA | MG910492 | — | — | — |
| BJ316CA | Ganoderma polychromum | CA, USA | MG910493 | — | — | — |
| MS343OR | Ganoderma polychromum | OR, USA | MG654197 | MG754743 | — | — |
| UMNOR3 | Ganoderma polychromum | OR, USA | MG654204 | MG754744 | MG754810 | — |
| MS187FL | Ganoderma ravenelii | FL, USA | MG654211 | MG754745 | MG754813 | MG754865 |
| UMNFL187 | Ganoderma ravenelii | FL, USA | — | — | MG754814 | — |
| UMNFL188 | Ganoderma ravenelii | FL, USA | — | MG754746 | MG754815 | — |
| CBS194.76 | Ganoderma resinaceum | Netherlands | KJ143916 | KJ143934 | KJ143956 | |
| Rivoire4150 | Ganoderma resinaceum | France | KJ143915 | — | KJ143957 | — |
| 103SC | Ganoderma sessile | SC, USA | MG654304 | — | — | — |
| 111TX | Ganoderma sessile | TX, USA | MG654306 | MG754747 | MG754816 | MG754866 |
| 113FL | Ganoderma sessile | FL, USA | MG654307 | MG754748 | — | MG754867 |
| 117TX | Ganoderma sessile | TX, USA | MG654309 | MG754749 | MG754817 | MG754868 |
| 165MO | Ganoderma sessile | MO, USA | MG654312 | — | MG754818 | — |
| 171FL | Ganoderma sessile | FL, USA | MG654316 | — | MG754819 | — |
| 228DC | Ganoderma sessile | DC, USA | MG654319 | MG754750 | MG754820 | MG754869 |
| JV1209/27 | Ganoderma sessile | AZ, USA | KF605630 | KJ143937 | KJ143959 | KJ143976 |
| NY00985711 | Ganoderma sessile | NY, US | KJ143918 | — | — | — |
| UMNCA5 | Ganoderma sessile | CA, USA | MG910998 | — | — | — |
| UMNFL10 | Ganoderma sessile | FL, USA | MG654227 | MG754753 | MG754821 | — |
| UMNFL125 | Ganoderma sessile | FL, USA | MG654239 | MG754755 | MG754825 | — |
| UMNFL19 | Ganoderma sessile | FL, USA | MG654230 | MG754754 | MG754822 | — |
| UMNKY1 | Ganoderma sessile | KY, USA | MG654257 | MG754756 | MG754827 | — |
| UMNMI22 | Ganoderma sessile | MI, USA | MG654269 | MG754757 | MG754828 | — |
| UMNMI24 | Ganoderma sessile | MI, USA | MG654271 | MG754758 | MG754829 | — |
| UMNNY14 | Ganoderma sessile | NY, US | MG654294 | — | MG754830 | — |
| UMNOH4 | Ganoderma sessile | OH, USA | MG654298 | MG754759 | MG754831 | — |
| UMNWV1 | Ganoderma sessile | WV, US | MG654302 | — | — | — |
| Cui7691 | Ganoderma sichuanense | Guangdong, China | JQ781878 | — | — | — |
| HMAS42798 (Holytype) | Ganoderma sichuanense | Sichuan, China | JQ781877 | — | — | — |
| Dai9724 | Ganoderma tropicum | Hainan, China | JQ781879 | — | — | — |
| Yuan3490 | Ganoderma tropicum | Yunnan, China | JQ781880 | KJ143938 | — | — |
| Dai1275b | Ganoderma tsugae | CT, USA | KJ143919 | KJ143939 | KJ143960 | KJ143977 |
| Dai12760 | Ganoderma tsugae | CT, USA | KJ143920 | KJ143940 | KJ143961 | KJ143978 |
| MS182AZ | Ganoderma tsugae | AZ, USA | MG910999 | MG754864 | ||
| UMNMI20 | Ganoderma tsugae | MI, USA | MG654324 | MG754764 | MG754836 | — |
| UMNMI30 | Ganoderma tsugae | MI, USA | MG654326 | MH025362 | MG754837 | MG754871 |
| UMNNC4 | Ganoderma tsugae | NC, USA | MG654329 | MG754765 | MG754838 | MG754872 |
| LIPSW-Mart08-45 | Ganoderma tuberculosum | Martinique | KF96325 | — | — | — |
| PLM684 | Ganoderma tuberculosum | FL, USA | MG654369 | MG754769 | — | — |
| UMNFL160 | Ganoderma tuberculosum | FL, USA | MG654364 | — | MG754840 | — |
| UMNFL82 | Ganoderma tuberculosum | FL, USA | — | MG754770 | — | MG754874 |
| UMNFL100 | Ganoderma cf. weberianum | FL, USA | MG654373 | MG754762 | MG754834 | — |
| UMNFL32 | Ganoderma cf. weberianum | FL, USA | MG654372 | MG754761 | MG754833 | — |
| 123FL | Ganoderma zonatum | FL, USA | MG654416 | MG754774 | MG754841 | — |
| 179NC | Ganoderma zonatum | NC, USA | MG654417 | MG754775 | MG754842 | MG754875 |
| FL-02 | Ganoderma zonatum | FL, USA | KJ143921 | KJ143941 | KJ143962 | KJ143979 |
| GAN11 | Ganoderma zonatum | FL, USA | — | MG754776 | — | MG754876 |
| UMNFL105 | Ganoderma zonatum | FL, USA | MG654408 | MG754780 | MG754847 | — |
| UMNFL137 | Ganoderma zonatum | FL, USA | MG654413 | MG754781 | MG754848 | — |
| UMNFL16 | Ganoderma zonatum | FL, USA | MG654381 | MG754777 | MG754843 | — |
| UMNFL85 | Ganoderma zonatum | FL, USA | MG654402 | MG754778 | MG754844 | MG754877 |
| UMNFL89 | Ganoderma zonatum | FL, USA | MG654403 | MG754779 | MG754845 | — |
| UMNSC4 | Ganoderma zonatum | SC, USA | MG654415 | MG754782 | MG754849 | — |
| TC-02 | Tomophagus colossus | Vietnam | KJ143923 | KJ143943 | KJ143963 | — |
| UMNFL110 | Tomophagus colossus | FL, USA | MG654429 | MG754850 | — | |
1. Accession numbers in boldface were generated from this study
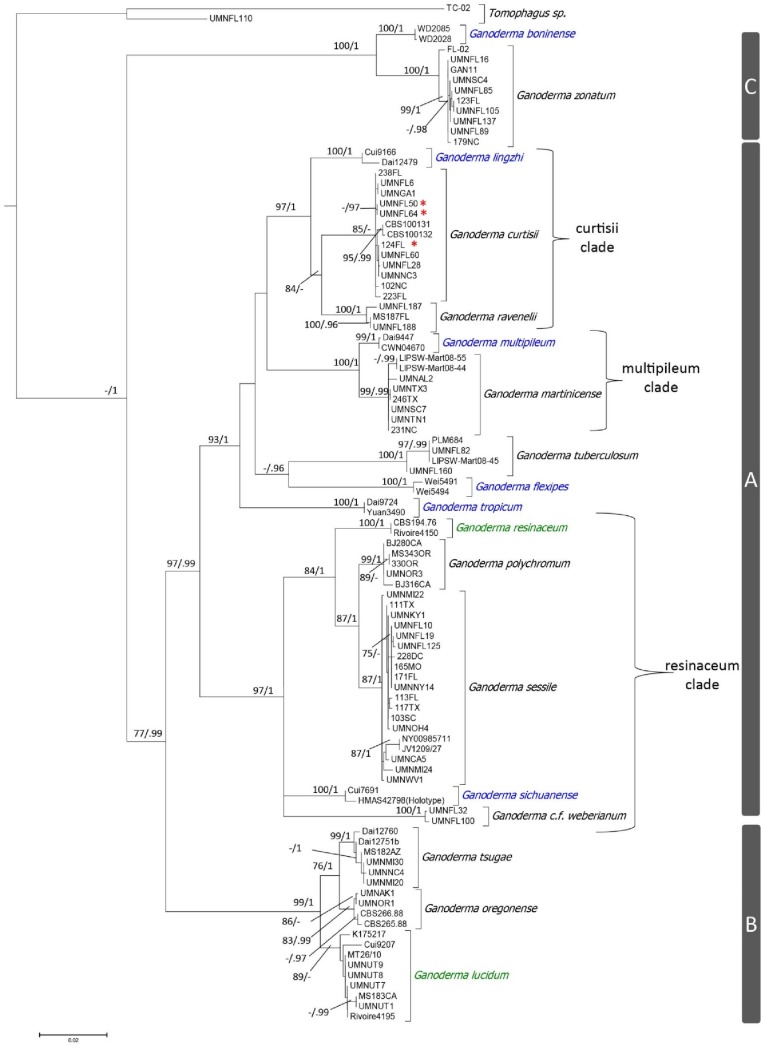
The phylogenetic analyses of the concatenated sequences with RAxML and Mr. Bayes yielded similar topologies, so the tree topology derived from the RAxML analysis was presented (Fig 7). Eighteen, well supported (ML-BS > 75%, and PP >95) terminal clades were resolved in the multilocus phylogeny using four loci representing 97 collections of the laccate Ganoderma species with Tomophagus species used as the outgroup (Fig 7). These 18 core Ganoderma terminal clades represented the following 18 taxa G. boninense Pat. 1889, G. curtisii, G. flexipes Pat. 1907, G. lingzhi Sheng H. Wu, Y. Cao & Y.C. Dai 2012, G. lucidum, G. martinicense, G. curtisii f.sp. meredithiae f.sp. nov., G. multipileum, G. oregonense, G. polychromum, G. ravenelii, G. resinaceum, G. sessile, G. sichuanense J.D. Zhao & X.Q. Zhang 1983, G. tropicum (Jungh.) Bres. 1910, G. tsugae, G. tuberculosum, G. cf. weberianum, and G. zonatum. Collections we labeled as G. meredithiae, based on the original species description, were conspecific with collections of G. curtisii, but the other 18 taxa represented well supported phylogenetic species. The 18 species in the presented phylogeny were broken into three highly supported subclades (Fig 7). The eighteen species represented a monophyletic group with high Bayesian posterior probability, but not with maximum likelihood. There did not appear to be clustering of taxa based on geographic location, but it appears there could be more genetic diversity and taxa in tropical and subtropical locations [21]. Clade labels used in Zhou et al. [2] were used in this phylogeny for consistency between studies.
Fig 7. Tree topology derived from RAxML analysis of a multilocus alignment (ITS + tef1α + rpb1 + rpb2) with 2470 characters under a GTR model with 1000 bootstrap replicates.
Statistical values shown are ML-bootstrap values above 75%, and the second value is the posterior probability (PP) where values above 95% are shown. Species native to Asia are in blue font, species native to Europe are in green font, and species native to North American are in black font. Red asterisks indicate G. curtisii f.sp. meredithiae. There are three major clades (A, B, and C), and subclades with names derived from the taxon that was described first.
Clade A was the most diverse clade, and contained thirteen of the eighteen species (Fig 7). Seven of thirteen were present in the United States. Although Clade A was highly supported as a major group separate from Clades B and C, there were also three well supported crown clades within this major clade. These clades were designated with the name based on the specific epithet of the taxon that was first described within the given crown clade. The “curtisii subclade” was highly supported, and included the North American taxa G. curtisii, and G. ravenelii, as well as the Asian taxon G. lingzhi. The “multipileum subclade” was highly supported, and included the Asian taxon G. multipileum and North American taxon G. martinicense. The “resinaceum subclade” was highly supported and included the European taxon G. resinaceum, the Asian taxon G. sichuanense, and the North American taxa G. polychromum, G. sessile, and G. cf. weberianum.
Clade B consisted of three well supported lineages including the European taxon G. lucidum sensu stricto, and the North American taxa G. oregonense and G. tsugae. Clade C consisted of two well supported lineages with the Asian taxon G. boninense and the North American G. zonatum. Lastly, Tomophagus colossus (UMNFL110) and T. cattienensis X.T. Le & Moncalvo [47] (TC-02) were used as outgroups because Tomophagus is a closely related genus in the Ganodermataceae.
Determining the optimal DNA barcode locus for Ganoderma species
The RAxML analyses of the four individual loci independently and concatenation of four loci resulted in the following cumulative bootstrap scores as described previously: ITS (761) with eight terminal clades, tef1α (1070) with twelve terminal clades, rpb1 (955) with eleven terminal clades, rpb2 (564) with eight terminal clades, and ITS + tef1α + rpb1 + rpb2 (1141) with twelve terminal clades. Samples of G. meredithiae were not resolved with any individual locus or multilocus phylogenetic analysis. Based on these data, tef1α and rpb1 resolve the laccate Ganoderma species better than the primary fungal barcode region ITS. Although there were much fewer samples with rpb2 sequences, we suspect that rpb2 would show similar or better resolution compared to tef1α and rpb1 based on previous work from Matheny et al. [40]. Furthermore, some terminal clades for tef1α, rpb1, and rpb2 had only one individual sample representative, so statistical values could not be assigned (Table 4).
Table 4. Comparisons of terminal clades and associated bootstrap scores for the laccate Ganoderma taxa present in the United States using RAxML analysis with 1,000 bootstrap replications for ITS, tef1α, rpb1, rpb2, and ITS+tef1α+ rpb1+rpb2.
| Phylogenetic Analyses Terminal Clade RAxML Bootstrap Scores | |||||
|---|---|---|---|---|---|
| Taxon | ITS | tef1α | rpb1 | rpb2 | ITS + tef1α + rpb1 + rpb2 |
| G. curtisii | 891 | 100 | 100 | 100 | 85 |
| G. curtisii f.sp. meredithiae | NR2 | NR | NR | NR | NR |
| G. lucidum | NR | 98 | 91 | 86 | 89 |
| G. martinicense | 93 | R*3 | 100 | 100 | 99 |
| G. oregonense | NR | 98 | 78 | NR | 83 |
| G. polychromum | 97 | 90 | R* | NT | 99 |
| G. ravenelii | 85 | 100 | 100 | NR* | 100 |
| G. sessile | 48 | 100 | 57 | 100 | 87 |
| G. tsugae | NR | 93 | 100 | 81 | 99 |
| G. tuberculosum | 100 | R* | 87 | NR* | 100 |
| G. cf. weberianum | 97 | 100 | 100 | NT | 100 |
| G. zonatum | 100 | 91 | 99 | 97 | 100 |
| T. colossus | 100 | 100 | 100 | NT | 100 |
| TOTAL SCORE4 | 761 | 970 | 955 | 564 | 1141 |
| NUMBER OF TERMINAL CLADES | 8 | 12 | 11 | 8 | 12 |
1. Numbers represent maximum likelihood bootstrap scores of terminal clades for each taxon from a RAxML analysis using rapid bootstrapping, a GTR evolutionary model, and 1000 boostrap replications
2. “NR” represents non-resolved terminal clades
3. “R*” represents clades where statistical value could not be computed, but lineage resolved
4. Summation of bootstrap support values for well-supported terminal clades
Discussion
The results of this study resolve problems and will help reduce confusion that is associated with the taxonomy of the G. lucidum species complex in the United States [1]. The comprehensive survey of laccate species of Ganoderma in the U.S. and the multilocus phylogeny helps resolve the species that were historically combined together as G. lucidum sensu lato. In addition to the five taxa presented in the treatise “North American Polypores” [1], eight more taxa are recognized within the United States, and include G. curtisii, G. curtisii f.sp. meredithiae, G. martinicense, G. polychromum, G. ravenelii, G. sessile, G. tuberculosum, and G. cf. weberianum. Furthermore, G. colossum, which was recognized in “North American Polypores” [1] has recently been placed back in the genus Tomophagus based on molecular data [22]. Lastly, the European taxon G. lucidum sensu stricto was found only in disturbed habitats in geographically restricted areas of northern Utah and northern California, suggesting they are likely isolated introduction events of a non-native species.
On a global scale, we found 19 well-supported terminal clades were formed with the multilocus phylogeny, including twelve species in the Ganodermataceae present in the U.S., six from Asia, and two from Europe. These 19 clades represented three highly supported clades in the genus Ganoderma and one highly supported clade in the genus Tomophagus, which was used as the outgroup. Within the core Ganoderma clades (A, B and C), Clade C that was comprised of G. boninense and G. zonatum was found to be the most basal clade. Both G. boninense and G. zonatum decay palms, which are an ancient lineage of plants [48]. We hypothesize that this relationship could explain their basal position on our phylogenetic tree, although detailed phylogenetic dating analyses are needed to address this possibility. Clade B, comprised of G. lucidum sensu stricto, G. oregonense, and G. tsugae, which includes temperate species, which suggests members of this clade share a common ancestor that was adapted for temperate climates. Finally, Clade A is the most derived and diverse clade of the laccate Ganoderma species with mostly subtropical to tropical species, including a few taxa that were geographically widespread.
Clade A was broken up into the three well-supported crown clades, the “curtisii subclade”, the “multipileum subclade”, and the “resinaceum subclade”. The “curtisii subclade” contained the three distinct species G. curtisii, G. lingzhi, and G. ravenelii. Ganoderma lingzhi is an Asian species that historically was also considered G. lucidum sensu lato, and is one of the most widely cultivated species for medicinal use [29, 49]. Although there is little information on the medicinal benefits of North American Ganoderma taxa, it is likely, given the phylogenetic placement of G. curtisii and G. ravenelii, that these species would produce similar pharmaceutically relevant compounds. All three of these species produce basidiomata that are generally laterally stipitate. Ganoderma curtisii is widespread throughout the eastern United States, and possesses melanoid deposits in the context tissue of the pileus and stipe. The basidiomata of G. meredithiae are morphologically indistinguishable from collections of G. curtisii, and based on the presented phylogeny constructed with four loci (ITS, tef1α, rpb1, and rpb2), G. meredithiae is considered to be conspecific with G. curtisii. Originally, G. meredithiae was circumscribed as a novel species based on the affinity to colonize and decay pines and slow in vitro growth rate on MEA [20]. Similar physiological differences between collections of G. meredithiae and G. curtisii were found in an in vitro growth study with our collections[50]. Furthermore, in another study, when isolates of G. meredithiae were grown on artificial media amended with pine water-soluble sapwood extracts, the linear growth was enhanced relative to the malt extract agar control. In contrast, G. curtisii was negatively affected by media amended with pine sapwood extracts relative to the MEA control [50]. Based on these physiological differences, we feel it necessary to discuss the two taxa separately, and use the forma specialis meredithiae designation for this physiological variant of G. curtisii. Based on the taxonomic code for fungi, forma specialis are used as informal classification to distinguish physiological variants of a given parasite species [51].
Ganoderma ravenelii was originally described by Steyaert [12], and was considered highly similar to G. curtisii. Type collections were from Florida, and additional specimens from South Carolina were also used to describe this species [12]. This species was originally differentiated from collections of G. curtisii by having more elongated basidiospores and lacking melanoid bands within the context tissues of the pileus and stipe [12]. Similarly, our collections studied from Florida and North Carolina lacked melanoid bands in the context tissue, and consistently had basidiospores that were more elongated than the basidiospores of G. curtisii. The average length to width ratio (Q ratio) of basidiospores of G. ravenelii was 2.2, while the Q ratio of basidiospores of G. curtisii collections was 1.7. Furthermore, this species was differentiated with high statistical support from collections of G. curtisii with all of the loci (ITS, tef1α, rpb1 and rpb2) individually. Interestingly, this species has an overlapping geographic distribution with G. curtisii, and more studies focusing on the ecology of this species are needed to understand if there are functional differences between the two species.
There were only two species included in this phylogeny from the “multipileum subclade”, which were G. multipileum and G. martinicense. This clade is sister to the “curtisii subclade”. Ganoderma multipileum is an Asian species that was historically labeled as G. lucidum sensu lato in tropical Asian locations [28]. The species G. martinicense was described from Martinique, and was suspected to be the vicariant relative of G. multipileum [28]. Our phylogeny placed these geographically distant species as sister taxa, which corroborates the hypothesis of vicariant evolution. Ganoderma martinicense is diagnosed by having dark brown context tissue, a centrally produced pseudostipe that is dark red to black, and melanoid bands as well as concentric growth zones in the context tissue. This survey reports G. martinicense from Alabama, Georgia, Louisiana, Mississippi, North Carolina, South Carolina, Tennessee, and Texas (U.S.). We suspect that G. martinicense has an even wider geographic distribution throughout the southeastern U.S. and the Caribbean. More surveys in North, Central and South America will help elucidate the geographic distribution of this species.
The “resinaceum subclade” is a diverse clade that includes species that generally produced sessile, and occasionally pseudostipitate fruiting bodies. Furthermore, all of the species within this subclade constitutively and vegetatively-produce double-walled chlamydospores with a smooth surface in vitro on MEA, which is an apomorphic characteristic [22]. The species G. resinaceum, the subclade’s name sake, is a European species that was historically thought to be a synonym for G. sessile and G. polychromum and the North American G. lucidum sensu lato [12, 17, 52]. Based on our phylogeny, European members of the “resinaceum subclade” represent a distinct lineage from the North American G. sessile and G. polychromum lineages, which are sister to one another. Previous research [53] suggests laboratory biological compatibility of monokaryotic European and North American sister species is not uncommon, and occasionally can result in dikaryotic isolates with reduced vigor, such as observed in intersterility groups of members of the European and North American Armillaria mellea species complex [53]. Furthermore, gene flow is naturally improbable with taxa that are separated by geographic barriers such as oceans and mountain ranges or taxa that do not have long distance dispersal methods [18]. Therefore, taxa that are in the process of allopatric speciation may not be intersterile because there is no mechanism that has evolved to prevent outcrossing [18].
There are three distinct North American linages representing three unique species within the resinaeum subclade, which include G. polychromum, G. sessile, and G. cf. weberianum. Ganoderma polychromum, (syn. = Polyporus polychromus Copel.), was described from California and Nevada, and the zonate context tissue and affinity for angiosperms were considered diagnostic features [7, 12]. This species, like many of the others from the U.S., has long been mislabeled as G. lucidum sensu lato. Ganoderma polychromum is the sister taxon to G. sessile, a species widespread in the eastern U.S.
Ganoderma sessile is the most taxonomically controversial of all laccate Ganoderma taxa present in the U.S. This species was originally described by Murrill in 1902, was contested by many subsequent reports and synonymized with G. lucidum sensu lato or G. resinaceum [1, 6, 8–12]. Based on the presented phylogeny, G. sessile is distinct from G. lucidum sensu stricto, as well as G. resinaceum. Ganoderma sessile is one of the most widely distributed species of the laccate Ganoderma in the eastern U.S. Based on our collections in the U.S., we suspect that G. sessiliforme Murrill, a species originally described from Mexico [15], is either a synonym of G. sessile, or the geographic limits for this species occur south of the United States [27, 54]. The other two species in the “resinaceum subclade” are the Asian species G. sichuanense and G. cf. weberianum.
Based on the holotype morphology and ITS sequence (JQ781877), G. sichuanense was incorrectly labeled and is actually part of the G. weberianum species complex [2, 55]. The holotype of G. sichuanense (HMAS 42798) is from Sichuan, China, and is a sessile fruiting body with contextual chlamydospores, which are not characteristics of the widely cultivated Asian taxon, now recognized as G. lingzhi [49]. This morphology is highly similar to collections from south Florida that we labeled as G. cf. weberianum. The species G. weberianum was originally described from Pacific Islands with likely distribution in tropical locations in Africa and the Americas [56]. Later, Bazzalo & Wright [17] described G. subamboinense var. laevisporum as a morphologically similar species to G. weberianum, but from South America, including Argentina and Cuba. Unfortunately, due to a lack of sequences for other members of the G. weberianum/subamboinense species complex, we were unable to confidently determine an appropriate name for the collections made from south Florida. However, based on our phylogeny, the North American collections of G. cf. weberianum represent a well-supported lineage distinguished from the Asian collections, including the holotype labeled as G. sichuanense. Further investigations into this species complex are warranted, and it is likely that the collections of G. cf. weberianum from the US reported in this study are an undescribed species. The ITS sequences of G. sichuanense, G. subamboinense and G. weberianum from Genbank and the G. cf. weberianum from Florida are only 2–5 bases different. We suspect that sequencing from other loci, particularly tef1α and rpb1, may help to clearly distinguish these species.
Given the large geographic distributions of G. oregonense in the western U.S. and G. tsugae in the eastern U.S., we suspect that these sister taxa are native to the U.S. Both G. oregonense and G. tsugae have distinctly white context tissue, “rough” basidiospores, and are predominately associated with decay of conifers. In the eastern U.S., G. tsugae is geographically widespread in temperate eastern hemlock forests. Similarly, G. oregonense is geographically widespread in the western U.S. in the temperate forests dominated by conifers such as Tsuga heterophylla, Pseudotsuga menziesii, Picea spp., and Abies spp. The two species can be distinguished based on geographic location and size of basidiospores. The basidiospores of G. oregonense are slightly larger (12.9±1.7 x 8.0±1.3 μm) than those of G. tsugae (9.9±1.4 x 6.5±1.2 μm). Ecologically, G. oregonense and G. tsugae appear to be highly similar, but have been evolving independently due to geological barriers (e.g. the midwestern plains and Rocky Mountain range). However, we made four collections of G. tsugae from the southwestern United States (Arizona and New Mexico) from Abies concolor or Pseudotsuga menziesii. In Gilbertson & Ryvarden’s “North American Polypores” [1] they discuss collections of G. tsugae from Arizona and southern California exclusively on Abies concolor except for one collection from Pseudotsuga menziesii. Our collections from New Mexico on Abies concolor and Pseudotsuga menziesii are the first reports of G. tsugae from New Mexico. The populations of G. tsugae in the eastern U.S. and southwestern U.S. require further study to elucidate differences between the two disjunct populations.
Clade B included G. lucidum sensu stricto, G. oregonense, and G. tsugae. All three of these species are predominately found in temperate climates and all were found in our samples from the United States, although we strongly suspect G. lucidum sensu stricto is not native to the U.S. This clade was the least resolved with ITS alone, where none of these species were statistically separated. However, the phylogeny built with tef1α resolved the three species. Collections of G. lucidum sensu stricto were only found in two small distinct geographic ranges of northern Utah and northern California. Most subclades supported a general vicariant speciation hypothesis and we suspect that given the limited geographic distributions and the presented phylogenetic anomaly of the two populations, they are the result of two independent introductions of a European taxon to the U.S. This likely occurred by the production of the introduced fungus in field sites to produce basidiomata for medicinal purposes. In a similar case, the non-laccate European species, G. adspersum (Schulzer) Donk, has been introduced to California on popular European root stocks of almonds [57]. This species is causing tree failures, and economic losses to the California almond industry in the San Joaquin Valley [57]. Morphologically, G. lucidum sensu stricto in the U.S. can be diagnosed as having a true stipe, concentric growth zones in the context tissue, possessing a highly lacquered pileus and stipe, and being found on hardwood substrates. This morphological diagnosis is similar to collections of this taxon from Europe [2]. Further collections and genetic work will continue to elucidate the ecology and phylogenetic history of this species in the U.S. If this is indeed an introduced species, isolates of G. lucidum sensu lato of unknown origin should not be used outside for medicinal mushroom production since it is likely that these exotic species can become established. Furthermore, although it has not been widely documented, successful mating tests of European and U.S. collections of some Ganoderma species suggests that hybrids are possible. Theoretically, hybrids could produce more fit Ganoderma species that could potentially displace endemic Ganoderma species in North America, and the ramifications of this are not known.
Clade C, the most basal clade in the core Ganoderma included G. boninense from Asia and G. zonatum from the U.S. Similar to the other clades, these two species are sister to one another and geographically distant [2]. Both species are primary decay agents of woody monocots, especially palms. Ganoderma boninense is a primary decay fungus associated with palms, especially oil palm (Elaeis guineensis), in southeast Asia [2, 58]. Basal stem rot, caused by G. boninense, is the most significant disease of the economically important oil palm in southeast Asia [59]. Similar to G. boninense, G. zonatum is also considered a devastating pathogen and is associated with butt rot of palm trees in the Gulf South and along the Atlantic Coast in the southeastern U.S [60]. The geographic distribution of G. zonatum is likely similar to the geographic distribution of Sabal palmetto [61]. This is presumably due to a coevolutionary relationship. The elongated spores and dark brown context tissue of G. boninense and G. zonatum are the primary morphological diagnostic traits. Our study also revealed Ganoderma zonatum in North Carolina for the first time. This was found on a planted Sabal palmetto outside its natural range, which implies the possible movement of G. zonatum through the nursery trade. Our results suggest these species share a common ancestor and evolved into distinct species vicariously. The distribution of palms in other parts of the world may show other taxa in this clade. Through our collections and surveys, G. zonatum was the primary decay fungus associated with the decay of palms and bamboo, although there were other Ganodermataceae taxa occasionally found on palms. Tomophagus colossus was collected from a decaying palm in Gainesville, FL, and a non-laccate Ganoderma species similar to G. tornatum (G. australe species complex) was collected from decaying palm stumps throughout Florida.
Although the majority of collections in this survey were from the eastern U.S., we feel these data have captured a majority of the diversity of the laccate Ganoderma species that are found in the United States. This work also serves as a baseline to evaluate future Ganoderma collections. More collecting and analyses in the western United States could yield more diversity. For example, Murrill described G. nevadense from Nevada and G. sequoiae from California, both collected on conifers [7]. In our survey, four collections from conifer substrates from New Mexico and Arizona were found to be conspecific to collections of G. tsugae from the eastern U.S., but it is possible that with more sequencing some of these southwestern populations of G. tsugae could represent unique species similar to what was described by Murrill on conifers in the southwestern U.S [7].
Diagnosing species accurately is critical for studying the biology and ecology of fungi. Diagnostic characters such as DNA sequence, context tissue features, chlamydospores and geography were important for accurate species identification within the laccate Ganoderma species present in the U.S. From the 507 samples studied, thirteen taxa and twelve species of the Ganodermataceae are present in the United States and include: G. curtisii, G. curtisii f.sp. meredithiae, G. lucidum sensu stricto, G. martinicense, G. oregonense, G. polychromum, G. ravenelii, G. sessile, G. tsugae, G. tuberculosum, G. cf. weberianum, G. zonatum, and Tomophagus colossus. This study has unraveled some of the taxonomic difficulties associated with the laccate Ganoderma species that were once all considered as G. lucidum sensu lato. For example, all FLAS and NCSCLG herbarium collections collected from the U.S. and labeled as G. lucidum were not G. lucidum sensu stricto, and have been reannotated to one of the following species: G. curtisii, G. martinicense, G. sessile and G. tuberculosum. In addition, the presented data showed the ITS region, which is the most widely-used and important fungal barcode, is diagnostic for most species and subclades, but some species, especially in Clade B are not resolved. The tef1α locus captured more genetic diversity and is better at diagnosing the laccate Ganoderma to species level. However, there are more ITS sequences in GenBank compared to tef1α, and we recommend using both ITS and tef1α to avoid missing species with fewer accessioned sequences. As genome sequencing becomes more affordable, phylogenomics will be the most robust method to understand evolutionary patterns and mechanisms of speciation, which will undoubtedly resolve the Ganodermataceae. Further studies into the evolutionary history of the laccate Ganoderma species in the United States will elucidate important ecological relationships and more detailed geographic distribution patterns. Furthermore, sequencing more loci should be the focus of future studies, and could potentially resolve more cryptic species present in the U.S.
Supporting information
(DOCX)
Acknowledgments
We are extremely grateful to Ryan Gates, Neil Dollinger, Jason Conheeney, Eric Otto and all of the many citizen scientists who submitted collection material for study as well as Drs. Guido Schnabel, Julia Kerrigan, Monica Elliott, and James Jacobs for Ganoderma cultures and collections. We are also grateful to the mycological herbaria at the University of Florida (FLAS) and North Carolina State University (NCSCLG) for allowing us to study their collections. We would also like to thank Cassondra Newman, Eric Linder, Sam Redford, Connor Lund, Camille Schlegel and Liam Genter for assistance in DNA extractions and sequencing. Also, we are grateful for two sequences from Bob Johnson and Dave Rizzo from UC Davis.
Data Availability
All relevant data are within the paper and its Supporting Information files; however, fruiting body collections have been deposited to the University of Florida Mycological Herbarium in Gainesville, FL under the accession numbers: FLAS-F-62386 through FLAS-62546. In addition, cultures have been deposited to the Center for Forest Mycological Research collection in Madison, WI. Futhermore, sequences have been deposited to GenBank, and alignments/trees are deposited to www.treebase.com under the submission accession: 22399.
Funding Statement
This project was funded in part by the F.A. Bartlett Tree Experts Company and a research grant from the International Society of Arboriculture-Florida Chapter and was also supported in part by the USDA National Institute of Food and Agriculture, Hatch project 0022081. Matthew E. Smith’s involvement in this project was made possible by the support of the Institute of Food and Agriculture Sciences at the University of Florida and by a grant from the National Institute of Food and Agriculture, USDA, under the grant #FLAPLP-005289. Finally, the herbarium studies were made possible by the Clark T. Rogerson research award from the Mycological Society of America. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The first author Andrew Loyd is employed by the F.A. Bartlett Tree Experts Company. F.A. Bartlett Tree Experts Company provided support in the form of salary for author AL, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific role of this author is articulated in the ‘author contributions’ section.
References
- 1.Gilbertson R, Ryvarden L. North American Polypores. Oslo: Abortiporus to Lindteria: Fungiflora; 1986. [Google Scholar]
- 2.Zhou L-W, Cao Y, Wu S-H, Vlasák J, Li D-W, Li M-J, et al. Global diversity of the Ganoderma lucidum complex (Ganodermataceae, Polyporales) inferred from morphology and multilocus phylogeny. Phytochemistry. 2015;114:7–15. 10.1016/j.phytochem.2014.09.023 [DOI] [PubMed] [Google Scholar]
- 3.Karsten P. Enumeratio Boletinarum et Polyporarum Fennicarum systemate novo dispositorum. Rev Mycol. 1881;3:16–8. [Google Scholar]
- 4.Murrill WA. The Polyporaceae of North America, genus I Ganoderma. Bull Torrey Bot Club. 1902;29:599–608. [Google Scholar]
- 5.Patouillard N. Le genre Ganoderma. Bull Soc Mycol Fr. 1889;5:64–80. [Google Scholar]
- 6.Adaskaveg JE. Studies of Ganoderma lucidum and Ganoderma tsugae (Delignification, Mating Systems, Root Rot, Cultural Morphology, Taxonomy). [Dissertation]: The University of Arizona; 1986.
- 7.Murrill WA. Agaricales (Polyporaceae). North American Flora. 1908;9:73–131. [Google Scholar]
- 8.Atkinson GF. Observations on Polyporus lucidus Leys and some of its Allies from Europe and North America. Botanical Gazette. 1908;46:321–38. [Google Scholar]
- 9.Haddow WR. Studies in Ganoderma. Journal of the Arnold Arboretum. 1931;12:25–46. [Google Scholar]
- 10.Nobles MK. Studies in Forest Pathology. IV. Identification of Cultures of Wood-Rotting Fungi. Canadian Journal of Research. 1948;26:281–431. [DOI] [PubMed] [Google Scholar]
- 11.Overholts LO. Comparative Studies in the Polyporaceae. Annals of the Missouri Botanical Garden. 1915;2(4):667–730. [Google Scholar]
- 12.Steyaert RL. Study of Some Ganoderma Species. Bull du Jardin Bot Nat de Belgique/ Bull van de Nat Plantentuin van Blegie. 1980;50:135–86. [Google Scholar]
- 13.Murrill WA. Corrections and additions to the Polypores of temperate North America. Mycologia. 1920;12:6–24. [Google Scholar]
- 14.Murrill WA. Index to Illustrations of Fungi, XXIII-XXXIII. Mycologia. 1922;14:332–4. [Google Scholar]
- 15.Murrill WA. Tropical polypores Lancaster, PA: Press of the New Era Printing Company; 1915. [Google Scholar]
- 16.Nobles MK. Identification of cultures of wood-inhabiting Hymenomycetes. Canadian Journal of Botany. 1965;43:1097–139. [Google Scholar]
- 17.Bazzalo ME, Wright JE. Survey of the Argentine Species of the Ganoderma lucidum Complex. Mycotaxon. 1982;16:293–325. [Google Scholar]
- 18.Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, et al. Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology. 2000;31:21–32. 10.1006/fgbi.2000.1228 [DOI] [PubMed] [Google Scholar]
- 19.Petersen RH, Hughes KW. Species and speciation in mushrooms: development of a species concept poses difficulties. Bioscience. 1999;49:440–52. [Google Scholar]
- 20.Adaskaveg J, Gilbertson R. Ganoderma meredithae, a new species on pines in the southeastern United States. Mycotaxon. 1988;31:251–7. [Google Scholar]
- 21.Moncalvo J-M, Wang H-F, Hseu R-S. Gene phylogeny of the Ganoderma lucidum complex based on ribosomal DNA sequences. Comparison with traditional taxonomic characters. Mycological Research. 1995;99:1489–99. [Google Scholar]
- 22.Hong SG, Jung HS. Phylogenetic analysis of Ganoderma based on nearly complete mitochondrial small-subunit ribosomal DNA sequences. Mycologia. 2004;96:742–55. [DOI] [PubMed] [Google Scholar]
- 23.Ryvarden LR. Genera of Polypores, nomenclatura and taxonomy, Fungiflora; Oslo Norway; 1991. p. 363. [Google Scholar]
- 24.Gottlieb AM, Ferrer E, Wright JE. rDNA analyses as an aid to the taxonomy of species of Ganoderma. Mycological Research. 2000;104:1033–45. [Google Scholar]
- 25.Ryvarden L. Studies in neotropical polypores 2: a preliminary key to neotropical species of Ganoderma with a laccate pileus. Mycologia. 2000:180–91. [Google Scholar]
- 26.Torres-Torres MG, Guzmán-Dávalos L, de Mello Gugliotta A. Ganoderma in Brazil: known species and new records. Mycotaxon. 2013;121:93–132. [Google Scholar]
- 27.Torres-Torres MG, Ryvarden L, Guzmán-Dávalos L. Ganoderma subgenus Ganoderma in Mexico. Revista Mexicana de Micología Vol. 2015;41:27–45. [Google Scholar]
- 28.Welti S, Courtecuisse R. The Ganodermataceae in the French West Indies (Guadeloupe and Martinique). Fungal Diversity. 2010;43:103–26. [Google Scholar]
- 29.Hennicke F, Cheikh-Ali Z, Liebisch T, Maciá-Vicente JG, Bode HB, Piepenbring M. Distinguishing commercially grown Ganoderma lucidum from Ganoderma lingzhi from Europe and East Asia on the basis of morphology, molecular phylogeny, and triterpenic acid profiles. Phytochemistry. 2016;127:29–37. 10.1016/j.phytochem.2016.03.012 [DOI] [PubMed] [Google Scholar]
- 30.Wang D-M, Wu S-H, Su C-H, Peng J-T, Shih Y-H, Chen L-C. Ganoderma multipileum, the correct name for 'G. lucidum' in tropical Asia. Botanical Studies. 2009;50:451–8. [Google Scholar]
- 31.Marx DH, Daniel WJ. Maintaining cultures of ectomycorrhizal and plant pathogenic fungi in sterile water cold storage. Canadian Journal of Microbiology. 1976;22:338–41. [DOI] [PubMed] [Google Scholar]
- 32.Ridgway R. Color standards and color nomenclature: The author; 1912. [Google Scholar]
- 33.Blanchette RA, Held BW, Hellmann L, Millman L, Büntgen U. Arctic driftwood reveals unexpectedly rich fungal diversity. Fungal Ecology. 2016;23:58–65. [Google Scholar]
- 34.Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes‐application to the identification of mycorrhizae and rusts. Molecular Ecology. 1993;2:113–8. [DOI] [PubMed] [Google Scholar]
- 35.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: a guide to methods and applications. 1990;18:315–22. [Google Scholar]
- 36.Rehner SA, Buckley E. A Beauveria phylogeny inferred from f nuclear ITS and EFl-a sequence: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005;97:84–98. [DOI] [PubMed] [Google Scholar]
- 37.Binder M, Larsson K-H, Matheny PB, Hibbett DS. Amylocorticiales ord. nov. and Jaapiales ord. nov.: early diverging clades of Agaricomycetidae dominated by corticioid forms. Mycologia. 2010;102:865–80. [DOI] [PubMed] [Google Scholar]
- 38.Matheny PB, Liu YJ, Ammirati JF, Hall BD. Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). American Journal of Botany. 2002;89:688–98. 10.3732/ajb.89.4.688 [DOI] [PubMed] [Google Scholar]
- 39.Liu YJ, Whelen S, Hall BD. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution. 1999;16:1799–808. 10.1093/oxfordjournals.molbev.a026092 [DOI] [PubMed] [Google Scholar]
- 40.Matheny PB, Wang Z, Binder M, Curtis JM, Lim YW, Nilsson RH, et al. Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi). Molecular Phylogenetics and Evolution. 2007;43:430–51. 10.1016/j.ympev.2006.08.024 [DOI] [PubMed] [Google Scholar]
- 41.Katoh K, Misawa K, Kuma Ki, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research. 2002;30:3059–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61:539–42. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences. 2012;109:6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gazis R, Rehner S, Chaverri P. Species delimitation in fungal endophyte diversity studies and its implications in ecological and biogeographic inferences. Molecular Ecology. 2011;20:3001–13. 10.1111/j.1365-294X.2011.05110.x [DOI] [PubMed] [Google Scholar]
- 46.Adaskaveg JE, Gilbertson RL. Basidiospores, pilocystidia, and other basidiocarp characters in several species of the Ganoderma lucidum complex. Mycologia. 1988;80:493–507. [Google Scholar]
- 47.Le XT, Le QHN, Pham ND, Dentinger BT, Moncalvo J-M. Tomophagus cattienensis sp. nov., a new Ganodermataceae species from Vietnam: Evidence from morphology and ITS DNA barcodes. Mycological Progress. 2012;11:775–80. [Google Scholar]
- 48.Byng JW, Chase MW, Christenhusz MJ, Fay MF, Judd WS, Mabberley DJ, et al. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Botanical Journal of the Linnean Society. 2016;181:1–20. [Google Scholar]
- 49.Cao Y, Wu S-H, Dai Y-C. Species clarification of the prize medicinal Ganoderma mushroom “Lingzhi”. Fungal Diversity. 2012;56:49–62. [Google Scholar]
- 50.Loyd AL, Held BW, Linder ER, Smith JA, Blanchette RA. Elucidating wood decomposition by four species of Ganoderma from the United States. Fungal Biology. 2018;122:254–63. 10.1016/j.funbio.2018.01.006 [DOI] [PubMed] [Google Scholar]
- 51.McNeill J, Barrie F, Buck W, Demoulin V, Greuter W, Hawksworth D, et al. International Code of Nomenclature for algae, fungi and plants. Regnum Vegetabile. 2012;154. [Google Scholar]
- 52.Adaskaveg JE, Gilbertson RL. Cultural studies and genetics of sexuality of Ganoderma lucidum and G. tsugae in relation to the taxonomy of the G. lucidum complex. Mycologia. 1986;5:694–705. [Google Scholar]
- 53.Anderson JB, Korhonen K, Ullrich RC. Relationships between European and North American biological species of Armillaria mellea. Experimental Mycology. 1980;4:78–86. [Google Scholar]
- 54.Gottlieb AM, Wright JE. Taxonomy of Ganoderma from southern South America: subgenus Ganoderma. Mycological Research. 1999;103:661–73. [Google Scholar]
- 55.Wang XC, Xi RJ, Li Y, Wang DM, Yao YJ. The species identity of the widely cultivated Ganoderma, 'G. lucidum' (Ling-zhi), in China. PLoS One. 2012;7:e40857 10.1371/journal.pone.0040857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steyaert R. Species of Ganoderma and related genera mainly of the Bogor and Leiden Herbaria. Persoonia-Molecular Phylogeny and Evolution of Fungi. 1972;7:55–118. [Google Scholar]
- 57.Johnson B. Ganoderma root and butt rot: an emerging threat to California almonds NPDN News; 2017. [Google Scholar]
- 58.Sariah M, Hussin M, Miller R, Holderness M. Pathogenicity of Ganoderma boninense tested by inoculation of oil palm seedlings. Plant Pathology. 1994;43:507–10. [Google Scholar]
- 59.Bivi MR, Farhana M, Khairulmazmi A, Idris A. Control of Ganoderma boninense: A causal agent of basal stem rot disease in oil palm with endophyte bacteria in vitro. Int J Agric Biol. 2010;12:833–9. [Google Scholar]
- 60.Elliott M, Broschat T. Observations and pathogenicity experiments on Ganoderma zonatum in Florida. Palms. 2001;45:62–73. [Google Scholar]
- 61.Elliott ML, Des Jardin EA, Ortiz JV, Macias T. Genetic variability of Ganoderma zonatum infecting palms in Florida. Mycologia. 2018;(in press). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files; however, fruiting body collections have been deposited to the University of Florida Mycological Herbarium in Gainesville, FL under the accession numbers: FLAS-F-62386 through FLAS-62546. In addition, cultures have been deposited to the Center for Forest Mycological Research collection in Madison, WI. Futhermore, sequences have been deposited to GenBank, and alignments/trees are deposited to www.treebase.com under the submission accession: 22399.