Abstract
Like DIP-STR markers (deletion/insertion polymorphism-short tandem repeat combinations), SNP-STR markers (single nucleotide polymorphism-STR combinations) are also valuable in forensic DNA mixture analysis. In this study, eight SNP-STRs were selected, and a stable and sensitive multiplex polymerase chain reaction (PCR) assay was developed for amplifying these SNP-STRs and the Amelogenin gender marker according to the principle of amplification refractory mutation system (ARMS). This novel multiplex set allows detection of the minor DNA contributor in a DNA mixture of any gender and cellular origin with high resolution (beyond a DNA ratio of 1:20). In addition, SNP-STR haplotype frequencies were estimated based on a survey of 350 unrelated individuals from Chinese Han population, and the combined power of discrimination (PD) and power of exclusion (PE) of the eight SNP-STRs were calculated as 0.99999999965 and 0.9996, which were obviously higher than that of the eight STR loci: 0.9999999954 and 0.9989 respectively. The results indicated that the SNP-STR compound markers have higher application value in forensic identification compared to standard autosomal STRs, especially in the analysis of imbalanced DNA mixtures.
Introduction
Mixed stains derived from different contributors are common biological evidence samples in forensic practice, and these complex biological samples generate mixed genotypes, presenting challenges in interpreting the results, especially for those imbalanced genomic mixtures [1, 2]. As the common forensic DNA analysis method, one of the limitations of the capillary electrophoresis (CE)-based polymerase chain reaction (PCR)-STR typing technique is that it does not work successfully if the proportion of the DNA quantities of the two contributors is more extreme than 1:10 [3]. Alternatively, Y-chromosome STRs can be used to detect the male component in these mixed samples when the DNA of the male contributor is present in a small amount [4]. However, compared with the autosomal STR analysis, the discriminatory power of Y-STR analysis is usually lower due to their paternal inheritance characteristics. So far, although a variety of strategies have been developed to separate different cell populations prior to analysis to reduce the challenges in mixture interpretation, including differential extraction [5, 6], filtration [7], fluorescence-activated cell sorting [8, 9], microchip-based separation [10–12], laser capture microdissection [13–16], micromanipulation [17–19], and microfluidic techniques [20], these methods are limited due to their complexity, low efficiency, high risk of sample cross-contamination, and/or lack of universality. Recently, massively parallel sequencing (MPS) is reported to be a promising technique for forensic mixture analysis, where all STR alleles of the minor contributors were detected in the sequence reads even for the 1% contributions [21]. In addition, MPS can also detect other types of markers, such as microhaplotype which can be highly informative for many forensic questions, including detection of DNA mixtures [22]. However, MPS is a complicated and costly technique. Therefore, there is still a need for the development of simple methods that allow complete DNA analysis of imbalanced mixtures irrespective of the gender of the DNA donors for those laboratories without NGS equipment.
In recent years, a simple solution to this problem based on CE detection platform is represented by detecting two types of compound genetic markers, deletion-insertion polymorphisms amplified with STRs (DIP-STR) [23–27] and single nucleotide polymorphisms amplified with STRs (SNP-STR) [28–30], which targets a genomic region unique to the minor DNA eliminating the masking effect of the major DNA. In comparison to SNP-STRs, although DIP-STRs are more sensitive markers (1:1,000 [24, 27] vs 1:40 [29, 30]) for the analysis of imbalanced DNA mixtures, there are still some disadvantages for forensic purpose. On one hand, DIP markers are significantly less frequent than SNPs in the human genome, and this greatly limits the selection of DIP-STR candidates. On the other hand, DIP markers are almost unavailable around the forensic commonly used STRs, such as the Combined DNA Index System (CODIS), Extended European Standard Set (ESS) and National Institute of Standards and Technology (NIST)-miniSTR, resulting in the results of DIP-STR typing are not comparable with that of routine STR typing. Based on these reasons, SNP-STRs may be more valuable compound genetic markers than DIP-STRs for the analysis of imbalanced DNA mixtures in forensic practice. The purpose of this study is to screen some valuable SNP-STR markers and develop a multiplex PCR assay, as well evaluate the application value in forensic identification, especially in the analysis of imbalanced DNA mixtures.
Materials and methods
DNA samples
Blood samples were collected from 350 unrelated healthy individuals of Chinese Han population in Hubei province in an anonymous way. All participants were interviewed to ensure that no individuals have common ancestry going back at least three generations. However, even so, we cannot fully exclude their distant relatedness. In addition, the peripheral blood samples of two women with singleton pregnancy (17 th and 40 th weeks respectively) and paired amniotic fluids or newborn oral swabs were also collected. Ethical approval was obtained from the medical ethics committee of Tongji Medical College of Huazhong University of Science and Technology and all individuals provided written informed consent (The informed consent for collection of the oral swabs of the female newborn was written by her mother). The control DNA 9947A (Thermo Fisher Scientific, MA, USA) was used for the multiplex assay development. Cell-free DNA of pregnant women was obtained from 2 mL of maternal plasma extracted by the QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instruction, and genomic DNA was isolated from whole blood and reference samples using the Chelex-100 method [31] and subsequently quantified with the Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, MA, USA).
Selection of SNP-STR compound marker and primer design
Genomic databases, STRBase (https://strbase.nist.gov/) and 1000 genomes (https://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/) were searched for regions containing SNP and STR polymorphisms based on the following criteria: (1) STRs are the commonly used genetic markers in forensic practice, (2) SNP and STR markers located closer than 200 bp, (3) SNP showing the minor allele frequency in Chinese Han population higher than 0.15, and (4) statistically independent SNP-STRs, such as locate on different chromosomes or on the same chromosome but the physical distance is not less than 20 Mb. According to these criteria, the first eight SNP-STRs, rs11222421-D11S4463, rs12423685-D12ATA63, rs2325399-D6S1043, rs1276598-D6S474, rs16887642-D7S820, rs9531308-D13S317, rs188010-D17S974 and rs258112-D5S2800, were selected as target compound markers in this study.
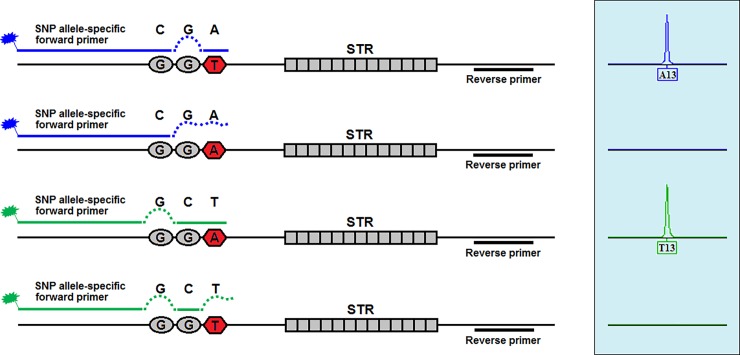
The amplification refractory mutation system (ARMS)-PCR technique [32] was used to amplify the SNP-STRs, and specificity was increased by the introduction of a deliberate mismatch at position −1, −2 or −3 of the polymorphism site. The two forward (or reverse) SNP allele-specific primers are labeled by different fluorescent dyes and the reverse (or forward) primer is located at the other flanking region of the STR which is linked to the SNP. Thus, alleles of the STR and SNP can be determined by the sizes and colors of the amplicons in one reaction respectively (Fig 1). All of primers were designed using the Primer 3 software (http://bioinfo.ut.ee/primer3/), and AutoDimer software was used to test possible primer-dimers after primer designing. All primer sequences were retested by BLAST to ensure the specificity of amplification products in the genome. In addition, we added a single G on the 5' end of the unlabeled primer within a locus-specific primer pair to promote full adenylation of PCR products amplified from that locus, and if the 5' end of the unlabeled primer was G itself, then G was not added (Table 1).
Fig 1. Schematic diagram of SNP-STR typing based on the principle of ARMS-PCR.
Table 1. Marker information, primer sequences and concentrations used in the SNP-STR multiplex assay in this study.
| SNP-STR | Chromosome | SNP allele | STR repeat | Primer sequence (5′→3′) a | Concentration (μM) | SNP-STR size (bp) | 9947A genotype |
|---|---|---|---|---|---|---|---|
| rs11222421-D11S4463 | 11q25 | A/T | TATC | F-A: 6-FAM-TTCTTATGAAATCTCTGTGTCTCCgA | 0.090 | 144–180 | T12, T13 |
| F-T: HEX-TTCTTATGAAATCTCTGTGTCTCgCT | 0.065 | ||||||
| R: GATAATTAAATACCATCTGAGCACTGAAGA | 0.090 | ||||||
| rs12423685-D12ATA63 | 12q23.3 | C/A | YAA | F: GTTGGATTTTGAGGGCCTAGGG | 1.100 | 215–242 | C13, C13 |
| R-C: 6-FAM-TCCCAGTTCTTTGGGAAGCaG | 1.100 | ||||||
| R-A: HEX-TCCCAGTTCTTTGGGAAGgTT | 0.160 | ||||||
| rs2325399-D6S1043 | 6q15 | C/G | AGAT | F: GCAGCTTACAGATGGCATATTGTGA | 0.085 | 288–343 | G12, C18 |
| R-G: 6-FAM-CATATTTTTAAGTACCCTAACAAGTAACTCAaC | 0.060 | ||||||
| R-C: HEX-CATATTTTTAAGTACCCTAACAAGTAACTCtTG | 0.085 | ||||||
| rs1276598-D6S474 | 6q21 | G/A | AGAT/GATA | F-G: 6-FAM-CATGTGTTTCTTCAGCCCCtG | 0.030 | 375–399 | A14, G18 |
| F-A: HEX-CATGTGTTTCTTCAGCCCgAA | 0.090 | ||||||
| R: GTTTGAACTTAGACTCAGCCATGC | 0.090 | ||||||
| rs16887642-D7S820 | 7q21.11 | G/A | GATA | F: GTCCTCATTGACAGAATTGCACC | 0.230 | 160–184 | G10, G11 |
| R-G: TAMRA-GTATGATAGAACACTTGTCATAGTTTAGAtC | 0.170 | ||||||
| R-A: ROX-GTATGATAGAACACTTGTCATAGTTTAGtAT | 0.230 | ||||||
| rs9531308-D13S317 | 13q31.1 | A/C | TATC | F: GACCCATCTAACGCCTATCTGT | 1.600 | 211–239 | A11, A11 |
| R-A: TAMRA-GTGGGGAAATTTGTACATTCATTAATATAgATT | 1.400 | ||||||
| R-C: ROX-GTGGGGAAATTTGTACATTCATTAATATACtTG | 1.600 | ||||||
| rs188010-D17S974 | 17p13.1 | T/C | CTAT | F: GACCCTGTCTCAGATAGATGGATAGG | 1.800 | 268–296 | T7, T10 |
| R-T: TAMRA-CCCAGAATTTAGTCTACAATTTAAAAAAGAATTtTA | 0.240 | ||||||
| R-C: ROX-CCCAGAATTTAGTCTACAATTTAAAAAAGAATTAaG | 1.800 | ||||||
| rs258112-D5S2800 | 5q11.2 | A/C | GRYW | F-A: TAMRA-ATATTACCTTCTTTATTTGATTATGTGACAaTA | 2.100 | 339–375 | C14, A23 |
| F-C: ROX-ATATTACCTTCTTTATTTGATTATGTGACtTTC | 0.330 | ||||||
| R: GTGATAGCTCAACAGGGTGACT | 2.100 | ||||||
| Amelogenin | Xp22.1–22.3, Yp11.2 | — | — | F: 6-FAM-CCCTGGGCTCTGTAAAGAATAGTG | 0.018 | 106, 112 | X, X |
| R: ATCAGAGCTTAAACTGGGAAGCTG | 0.018 |
aThe deliberately mismatched bases are indicated by lower case letters, and the added bases are underlined.
PCR amplification and genotyping
PCR amplification was performed in a total reaction volume of 20 μL containing 10 μL of Platinum® Multiplex Master Mix (Thermo Fisher Scientific, MA, USA), 2.4 μL GC Enhancer, 5.6 μL of the eight SNP-STRs primer mixture (Table 1) and 1 ng of DNA template. Thermal cycling was performed on GeneAmp 2720 (Thermo Fisher Scientific, MA, USA) under the following conditions: 95°C for 2 min; 30 cycles of 95°C for 30 s, 60°C for 90 s, 72°C for 35 s, and a final extension hold at 72°C for 10 min.
PCR products were electrophoresed on ABI 3130 Genetic Analyser (Thermo Fisher Scientific, MA, USA) following manufacturer’s protocols. Samples were prepared as a mixture of 0.3 μL GeneScan™ 500 LIZ® size standard (Thermo Fisher Scientific, MA, USA) with 8.7 μL Hi-Di™ Formamide (Thermo Fisher Scientific, MA, USA) and 1 μL PCR products. Samples were analyzed using GeneMapper ID v3.2 software (Thermo Fisher Scientific, MA, USA) after data collection.
Sensitivity testing
The control DNA 9947A (Thermo Fisher Scientific, MA, USA) was diluted with quantities of 1, 0.5, 0.25, 0.1, 0.05 and 0.03 ng, and each level of DNA was amplified with the multiplex system in duplicate.
Imbalanced DNA mixtures
Based on the typing principle of the SNP-STR markers, any two samples with different informative haplotypes can be used to construct artificially DNA mixtures. Imbalanced DNA mixtures were simulated by adding increasing quantities of a major DNA to a minor DNA, and the ratios of the minor DNA to major DNA were set from 1:10, 1:20, 1:50, 1:100, 1:500 to 1:1000, keeping the level of minor contributor at 0.05 ng. Then these mixtures were genotyped using the above-mentioned multiplex amplification conditions.
Statistical analysis
The allele frequencies and forensic parameters were evaluated using the PowerStats v1.2 software obtained from Promega [33]. Hardy–Weinberg equilibrium and pairwise linkage disequilibrium were analysed using the Arlequin v3.5 software [34]. The probability of informative genotypes (I) at a given SNP–STR marker was calculated according to Castella et al. [24]. The theoretical numbers of informative markers were also evaluated according to Castella et al. [24].
Results
Features of the selected SNP-STRs and construction of the multiplex assay
All STR loci contained in the selected eight SNP-STR compound markers are commonly used microsatellite markers: D7S820 and D13S317 are part of CODIS; D5S2800 (previous D5S2500 in NIST miniSTR 26plex and AGCU ScienTech 21-plex [35]), D6S474, D11S4463, D12ATA63 and D17S974 are part of NIST miniSTR 26plex [36]; D6S1043 is part of the commercial kits AmpFℓSTR Sinofiler™ (Thermo Fisher Scientific, MA, USA) and PowerPlex® 21 (Promega, Madison, WI, USA). Six of the eight SNP-STRs locate on different chromosomes, and the other two markers, rs1276598-D6S474 and rs2325399-D6S1043, although locate on the same chromosomal arm (6q), and their physical distance and genetic distance are about 20.8 Mb and 17.73 cM (Marshfield) respectively, the pairwise linkage disequilibrium analysis showed that the two markers were genetically independent (p = 0.1675) in the studied Chinese Han population. For the SNPs, the minor allele frequencies of all selected loci in Chinese Southern Han population is higher than 0.2 except for the rs16887642 (0.1857) according to 1000 genomes databases (https://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/).
This multiplex set was designed as a 5-dye assay, two SNP allele-specific primers for each SNP-STR marker were labeled at the 5′-end respectively with 6-FAM and HEX or TAMRA and ROX fluorescent dye for the detection by ABI 3130 Genetic Analyzer (Thermo Fisher Scientific, MA, USA). Then the eight SNP-STRs and the Amelogenin gender marker were organized by allele size ranges and assigned to each of the four dyes to achieve a single multiplex assay. After designing primers, labeling fluorescence dye and optimizing experiment conditions, a novel 9-plex fluorescent multiplex PCR system was successfully developed, and all of the eight SNP-STRs were amplified with satisfactory results (see S1 Fig). The SNP-STR markers information, primer sequences and concentrations used in our study were listed in Table 1.
Sensitivity of the multiplex PCR assay
All of the alleles could be detected from 50 pg to 1 ng of 9947A DNA when the detection threshold was set to 50 rfu, while some alleles of a few markers could not be detected at the amount of 30 pg DNA.
Genetic polymorphisms of the 8 SNP-STRs in Chinese Han population
The numbers of haplotypes observed in the studied population were 17, 15, 20, 10, 13, 14, 13 and 8 for rs11222421-D11S4463, rs12423685-D12ATA63, rs2325399-D6S1043, rs1276598-D6S474, rs16887642-D7S820, rs9531308-D13S317, rs188010-D17S974 and rs258112-D5S2800, respectively. These are significantly larger than the number of alleles for the corresponding STRs: 9, 10, 14, 7, 8, 8, 8 and 7, respectively. For the 8 SNPs, all of the minor allele frequency was higher than 0.2 except for the rs16887642 (0.1729) in Hubei Han population. The haplotype or allele frequencies and forensic statistical parameters for these SNP-STRs, STRs and SNPs in a Chinese Han population were shown in Table 2, and S1 and S2 Tables respectively.
Table 2. Haplotype frequencies and forensic statistical parameters of the 8 SNP-STRs from Hubei Han population in China (n = 350).
| rs11222421-D11S4463 | rs12423685-D12ATA63 | rs2325399-D6S1043 | rs1276598-D6S474 | rs16887642-D7S820 | rs9531308-D13S317 | rs188010-D17S974 | rs258112-D5S2800 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A9 | 0.0014 | A15 | 0.0014 | G10 | 0.0371 | G15 | 0.0057 | G8 | 0.0200 | A7 | 0.0029 | T6 | 0.0014 | A17 | 0.2857 |
| A12 | 0.0343 | A16 | 0.0186 | G11 | 0.1100 | G16 | 0.0929 | G9 | 0.0243 | A8 | 0.2829 | T7 | 0.0186 | A18 | 0.2371 |
| A13 | 0.0971 | A17 | 0.1729 | G12 | 0.1343 | G17 | 0.1143 | G9.1 | 0.0029 | A9 | 0.1186 | T8 | 0.1257 | A19 | 0.0014 |
| A14 | 0.1471 | A18 | 0.0686 | G13 | 0.1057 | G18 | 0.0243 | G10 | 0.1600 | A10 | 0.0371 | T9 | 0.2143 | A20 | 0.0886 |
| A15 | 0.1500 | A19 | 0.0086 | G13.2 | 0.0014 | G19 | 0.0014 | G11 | 0.3443 | A11 | 0.0271 | T10 | 0.0500 | A21 | 0.0014 |
| A16 | 0.0557 | A20 | 0.0029 | G14 | 0.1386 | A12 | 0.0014 | G12 | 0.2300 | A12 | 0.0086 | T11 | 0.0114 | A23 | 0.0086 |
| A17 | 0.0114 | C11 | 0.0014 | G15 | 0.0171 | A14 | 0.3614 | G13 | 0.0429 | A13 | 0.0043 | T12 | 0.0071 | C14 | 0.3743 |
| A18 | 0.0029 | C12 | 0.3600 | G19 | 0.0014 | A15 | 0.3414 | G14 | 0.0029 | A14 | 0.0014 | C8 | 0.0029 | C18 | 0.0029 |
| T9 | 0.0029 | C13 | 0.0100 | G20 | 0.0014 | A16 | 0.0543 | A8 | 0.1129 | C9 | 0.0200 | C9 | 0.0086 | ||
| T11 | 0.0043 | C14 | 0.0229 | C14 | 0.0043 | A17 | 0.0029 | A9 | 0.0400 | C10 | 0.0900 | C10 | 0.3543 | ||
| T12 | 0.0200 | C15 | 0.0029 | C16 | 0.0014 | A10 | 0.0143 | C11 | 0.2271 | C11 | 0.1743 | ||||
| T13 | 0.1400 | C16 | 0.1886 | C17 | 0.0571 | A11 | 0.0043 | C12 | 0.1343 | C12 | 0.0300 | ||||
| T14 | 0.1486 | C17 | 0.1243 | C17.3 | 0.0029 | A12 | 0.0014 | C13 | 0.0314 | C13 | 0.0014 | ||||
| T15 | 0.1143 | C18 | 0.0157 | C18 | 0.1657 | C14 | 0.0143 | ||||||||
| T16 | 0.0557 | C19 | 0.0014 | C18.2 | 0.0014 | ||||||||||
| T17 | 0.0129 | C19 | 0.1614 | ||||||||||||
| T18 | 0.0014 | C20 | 0.0471 | ||||||||||||
| C21 | 0.0071 | ||||||||||||||
| C22 | 0.0029 | ||||||||||||||
| C22.3 | 0.0014 | ||||||||||||||
| p-value | 0.6337 | 0.0097 | 0.5365 | 0.0936 | 0.6843 | 0.1113 | 0.0245 | 0.8550 | |||||||
| Hobs | 0.8743 | 0.7600 | 0.9057 | 0.7200 | 0.7971 | 0.8543 | 0.7686 | 0.7429 | |||||||
| Hexp | 0.8848 | 0.7846 | 0.8800 | 0.7286 | 0.7867 | 0.8256 | 0.7795 | 0.7151 | |||||||
| PD | 0.9731 | 0.9210 | 0.9699 | 0.8859 | 0.9245 | 0.9452 | 0.9182 | 0.8601 | |||||||
| PE | 0.7433 | 0.5270 | 0.8071 | 0.4599 | 0.5937 | 0.7033 | 0.5421 | 0.4976 | |||||||
| I | 0.3750 | 0.3181 | 0.3728 | 0.2973 | 0.2451 | 0.3747 | 0.3699 | 0.3594 | |||||||
p-value, probability of exact tests for Hardy-Weinberg disequilibrium; Hobs, observed heterozygosity; Hexp, expected heterozygosity; PD, power of discrimination; PE, power of exclusion; I, Probability of informative genotypes.
Minor DNA detection limit in DNA mixture
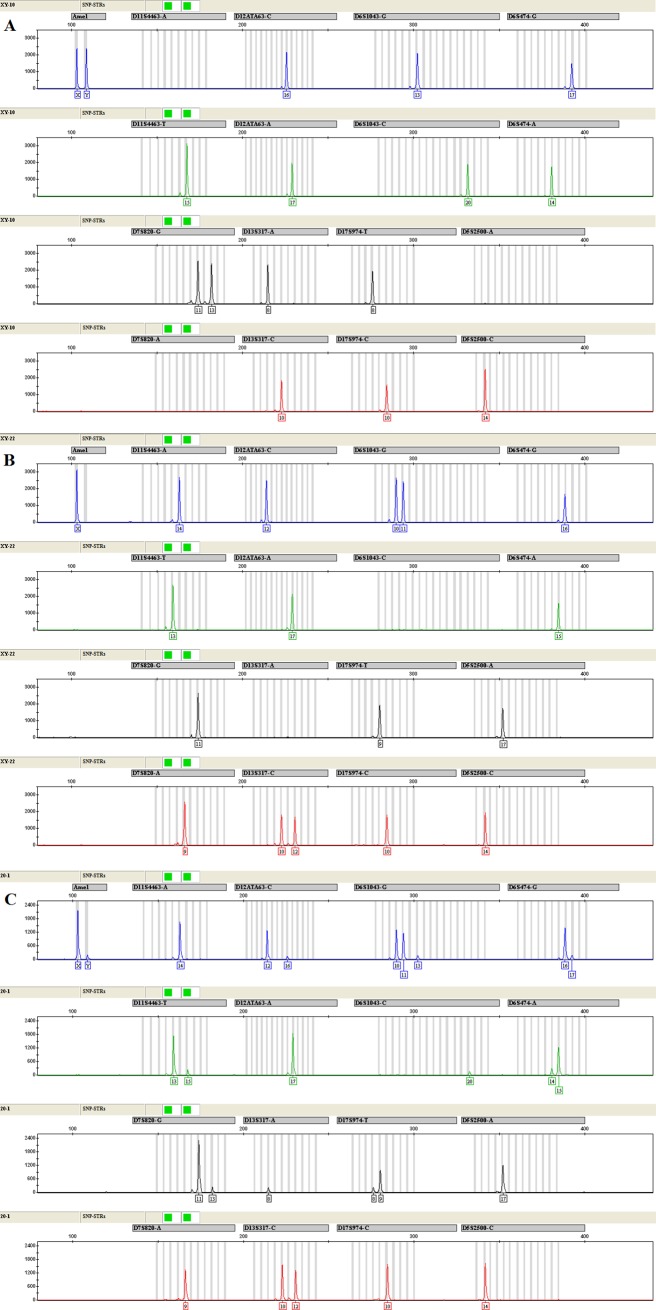
For the 9-plex fluorescent multiplex assay, all the markers were capable of discriminating the minor DNA up to 20-fold excess of major DNA (Fig 2). As shown in Table 3, when each marker was amplified separately in one reaction with three primers, different minor DNA detection limits for these markers were observed, ranged from 1:20 to 1:100 respectively.
Fig 2.
The electropherograms of the SNP-STRs multiplex assay from the sample 1 (A), sample 2 (B), and mixture mixed by samples 1 and 2 at the ratio 1:20 (C) respectively.
Table 3. The minor DNA detection limit in the artificial DNA mixtures for each marker.
| SNP-STRs | Detection ratios (minor: major) in different SNP subtypes of mixtures | Overall detection ratios (minor: major) | |||
|---|---|---|---|---|---|
| A | G | C | T | ||
| rs11222421-D11S4463 | 1:50 * | 1:100 | 1:50 | ||
| rs12423685-D12ATA63 | 1:50* | 1:20 | 1:20 | ||
| rs2325399-D6S1043 | 1:20 | 1:100 | 1:20 | ||
| rs1276598-D6S474 | 1:100 * | 1:20 | 1:20 | ||
| rs16887642-D7S820 | 1:20 | 1:20 | 1:20 | ||
| rs9531308-D13S317 | 1:50 | 1:100 * | 1:50 | ||
| rs188010-D17S974 | 1:100 | 1:50 | 1:50 | ||
| rs258112-D5S2800 | 1:100 * | 1:100 * | 1:100 | ||
* The minor DNA could be distinguished in a mixture of 1:1000 for these SNP subtypes when they were genotyped with separate SNP allele-specific primers in two reactions and 35 PCR cycles.
Analysis of SNP–STR markers’ performance
In the analysis of DNA mixtures, the informative genotypes denote the genotypes of minor DNA have the alleles that are absent in the genotypes of major DNA, and the probability of occurrence (I) is related to the allele frequency of SNP. In the present study, the I value for the current eight SNP–STR markers is reported in Table 2. As average, our markers show a probability of being informative of 0.3390.
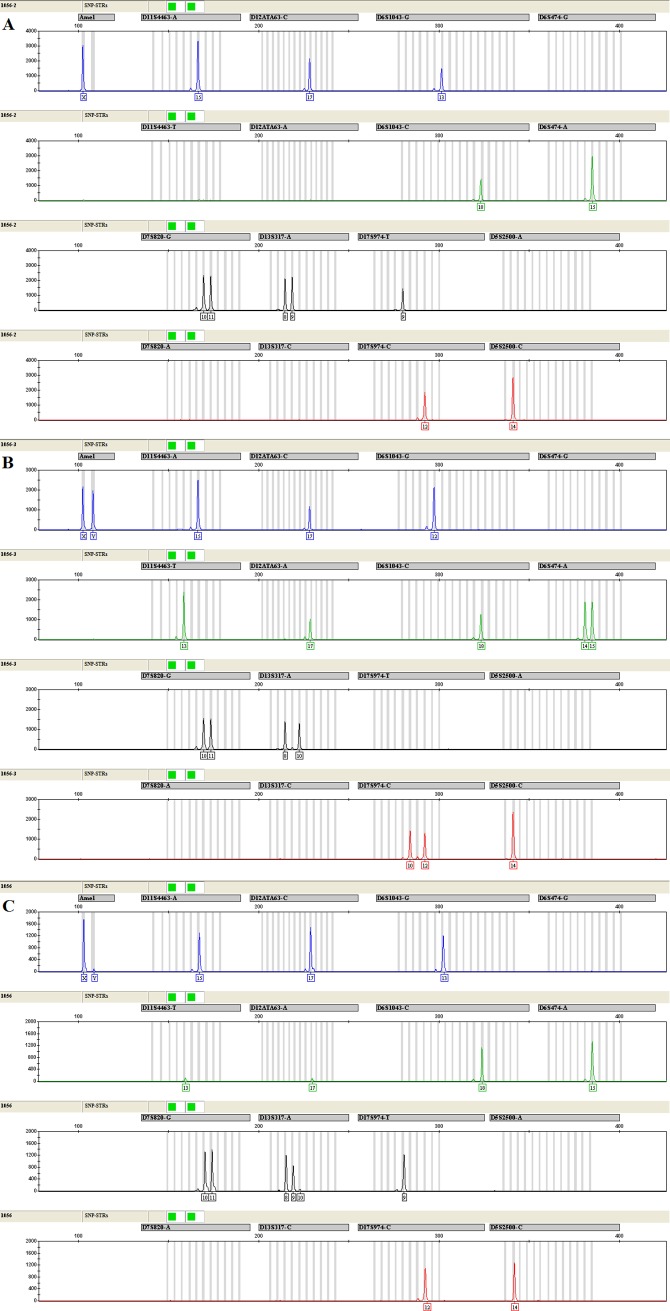
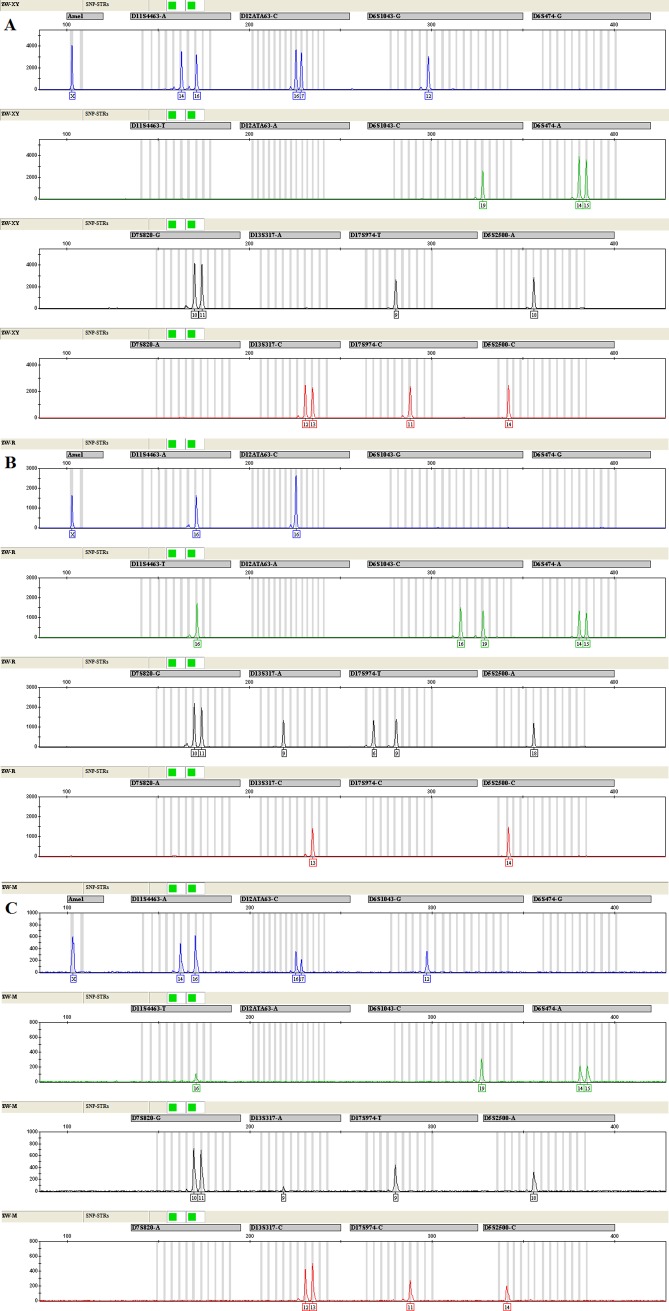
In addition, the typing results of pregnancy DNA microchimerism samples showed that the minor cell-free fetal DNA could be detected successfully for several informative markers (Figs 3 and 4).
Fig 3.
The electropherograms of the SNP-STRs multiplex assay from a woman at 17 weeks of pregnancy (A), paired amniotic fluids (B), and plasma cell-free DNA of the pregnant woman (C) respectively.
Fig 4.
The electropherograms of the SNP-STRs multiplex assay from a woman at 40 weeks of pregnancy (A), newborn oral swab (B), and plasma cell-free DNA of the pregnant woman (C) respectively.
Discussion
The previous studies showed that SNP-STRs are potentially useful and valuable markers for the analysis of unbalanced genomic mixtures [28–30]. At present study, the SNP-STRs were detected according to the principle of ARMS-PCR. Through this way, after careful design and optimization of the experimental conditions, a stable and sensitive 9-plex multiplex PCR assay was developed by us, and the detection sensitivity of this assay was up to 50 pg of DNA. In forensic practice, about 0.5–1 ng DNA is routinely recommended for typing, although 50 pg DNA is enough.
In the studied Chinese Han population, the haplotypes or genotypes distributions of all SNP-STRs, STRs and SNPs markers were in accordance with Hardy-Weinberg equilibrium after Bonferroni correction (0.05/8 = 0.00625) (Table 2, and S1 and S2 Tables). In the SNP-STR compound marker, the polymorphism of SNP locus is critical for resolving DNA mixtures. For the 8 SNPs studied, there are some differences in the allele frequency distributions among several major populations in the world (see S3 Table). For example, Africans are slightly less polymorphic at these SNPs except for the rs2325399 and rs16887642 loci. In addition, it should be noted that the SNP locus rs16887642 has very low heterozygosity in Europe and the Middle East and is fixed in the relatively unadmixed Native American samples in the human genome diversity project (HGDP), while the SNPs at the other loci have reasonable heterozygosities around the world. Therefore, this will affect the application value of the assay outside of East Asia. The combined power of discrimination and power of exclusion of the eight SNP-STR compound markers were calculated as 0.99999999965 and 0.9996 in the studied population, which were obviously higher than that of the eight STR loci: 0.9999999954 and 0.9989 respectively. The results indicated that the SNP-STR compound markers have higher application value in forensic identification compared to standard autosomal STRs.
For the analysis of DNA mixtures, the SNP-STR haplotypes of minor components (0.05 ng) in the artificially imbalanced two DNA mixtures (ratio 1:20) were successfully detected using our multiplex PCR assay. However, when each SNP-STR marker was typed separately, the detection ratios of the minor DNA increased to 1:50 and 1:100 for some SNP-STRs (Table 3). Due to the introduction of deliberate mismatch at position −1, −2 or −3 of the polymorphism site, there were different amplification specificity and efficiency for different SNP allele-specific primers [37]. Therefore, for different SNP subtypes of mixtures, each SNP-STR marker may have different minor DNA detection limits. For example, for the rs11222421-D11S4463, when the samples contained A-haplotype were used as a minor component, the detection ratio was 1:50, and when the samples contained T-haplotype were used as a minor component, the detection ratio was up to 1:100 (Table 3). It is worth noting that the minor DNA could be distinguished in a mixture of 1:1000 for six different SNP subtypes in some markers when they were genotyped with separate SNP allele-specific primers in two reactions and 35 PCR cycles (Table 3), and the other 10 subtypes were not able to do it due to the interference of non-specific amplification products derived from the major DNA. In order to avoid these influence as much as possible, therefore, it is recommended that the allele-specific primers should be separately amplified when involved in the analyses of extremely imbalanced DNA mixtures.
As shown in Table 2, for these eight SNP–STR markers studied, the maximum probability of informative genotype of each locus was 0.3750, at the rs11222421-D11S4463, and the minimum was 0.2451, at the rs16887642-D7S820. As average, our markers show a probability of being informative of 0.3390. Based on the cumulative binomial distribution of these eight SNP–STR markers each one associated to a probability of being informative of 0.3390, we found that 3.64% of the mixtures have zero informative markers, 96.36% have at least one informative marker, 81.40% have at least two informative markers, and 54.56% at least three informative markers (Table 4 column 1). In Table 4 column 2, we calculated this percentage assuming the use of 30 SNP–STR markers of allele frequencies similar to the ones already developed (I = 0.3390). The results indicate 96.92% of DNA mixtures with at least six informative markers, 84.89% with at least eight informative markers, and 59.40% with at least 10 informative markers.
Table 4. Occurrence of informative markers.
| Estimate using eight SNP-STR | Expected estimate using 30 SNP-STR |
|---|---|
| Percentage of DNA mixtures (≥N informative markers) | |
| 96.36 (≥1) | 96.92 (≥6) |
| 81.40 (≥2) | 92.56 (≥7) |
| 54.56 (≥3) | 84.89 (≥8) |
| 27.04 (≥4) | 73.58 (≥9) |
| 9.39 (≥5) | 59.40 (≥10) |
It is well known that the plasma cell-free DNA of pregnant women is a typical imbalanced DNA mixture. In order to evaluate the application value of this multiplex assay in the analysis of imbalanced DNA mixture, two cases of pregnancy DNA microchimerism samples and paired reference samples were detected. The typing results showed that the minor cell-free fetal DNA could be detected successfully only in the Amelogenin gender marker and the SNP-STR markers with smaller amplicon sizes when there were informative haplotype differences between the mother and the fetus (Figs 3 and 4). For those SNP-STR markers with larger amplicon sizes, however, even if there were informative haplotype differences between the mother and the fetus, the cell-free fetal DNA could not be detected, which is because plasma cell-free DNA molecules are mainly short DNA fragments and the fetal DNA is shorter than maternal DNA [38, 39].
Conclusions
In this study, a multiplex assay for detecting eight SNP-STRs and the Amelogenin gender marker was constructed. The SNP-STR haplotype of minor component (0.05 ng) in the artificially imbalanced two DNA mixture (ratio 1:20) can be detected successfully. In addition, the forensic efficiency of SNP–STRs is higher compared to standard autosomal STRs. Therefore, the SNP-STR compound markers should provide forensic scientists with a powerful tool for the analysis of DNA mixtures of any gender and cellular origin. Our future work is to develop more sets of SNP-STR markers and to derive an approach for the probabilistic evaluation of SNP-STR profiling results obtained from imbalanced DNA mixtures.
Supporting information
(TIF)
(DOCX)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by the National Natural Science Foundation of China (No. 81373250 to DH) and the Fundamental Research Funds for the Central Universities, HUST: No. 2016YXZD025 to DH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ladd C, Lee HC, Yang N, Bieber FR. Interpretation of complex forensic DNA mixtures. Croat Med J 2001; 42(3): 244–6. [PubMed] [Google Scholar]
- 2.Budowle B, Onorato AJ, Callaghan TF, Della Manna A, Gross AM, Guerrieri RA, et al. Mixture interpretation: defining the relevant features for guidelines for the assessment of mixed DNA profiles in forensic casework. J Forensic Sci 2009; 54(4): 810–21. 10.1111/j.1556-4029.2009.01046.x [DOI] [PubMed] [Google Scholar]
- 3.Clayton T, Buckleton J. Mixtures In: Buckleton J, Triggs C, Walsh SJ, editors. Forensic DNA Evidence Interpretation. Boca Raton: CRC Press; 2005. pp. 217–74. [Google Scholar]
- 4.Cerri N, Ricci U, Sani I, Verzeletti A, De Ferrari F. Mixed stains from sexual assault cases: autosomal or Y-chromosome short tandem repeats? Croat Med J 2003; 44(3): 289–92. [PubMed] [Google Scholar]
- 5.Gill P, Jeffreys AJ, Werrett DJ. Forensic application of DNA 'fingerprints'. Nature 1985; 318(6046): 577–9. [DOI] [PubMed] [Google Scholar]
- 6.Yoshida K, Sekiguchi K, Mizuno N, Kasai K, Sakai I, Sato H, et al. The modified method of two-step differential extraction of sperm and vaginal epithelial cell DNA from vaginal fluid mixed with semen. Forensic Sci Int 1995; 72(1): 25–33. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, Kobilinsky L, Wolosin D, Shaler R, Baum H. A physical method for separating spermatozoa from epithelial cells in sexual assault evidence. J Forensic Sci 1998; 43(1): 114–8. [PubMed] [Google Scholar]
- 8.Schoell WM, Klintschar M, Mirhashemi R, Pertl B. Separation of sperm and vaginal cells with flow cytometry for DNA typing after sexual assault. Obstet Gynecol 1999; 94(4): 623–7. [DOI] [PubMed] [Google Scholar]
- 9.Di Nunno N, Melato M, Vimercati A, Di Nunno C, Costantinides F, Vecchiotti C, et al. DNA identification of sperm cells collected and sorted by flow cytometry. Am J Forensic Med Pathol 2003; 24(3): 254–70. 10.1097/01.paf.0000070224.58005.ac [DOI] [PubMed] [Google Scholar]
- 10.Horsman KM, Barker SL, Ferrance JP, Forrest KA, Koen KA, Landers JP. Separation of sperm and epithelial cells in a microfabricated device: potential application to forensic analysis of sexual assault evidence. Anal Chem 2005; 77(3): 742–9. 10.1021/ac0486239 [DOI] [PubMed] [Google Scholar]
- 11.Bienvenue JM, Duncalf N, Marchiarullo D, Ferrance JP, Landers JP. Microchip-based cell lysis and DNA extraction from sperm cells for application to forensic analysis. J Forensic Sci 2006; 51(2): 266–73. 10.1111/j.1556-4029.2006.00054.x [DOI] [PubMed] [Google Scholar]
- 12.Norris JV, Evander M, Horsman-Hall KM, Nilsson J, Laurell T, Landers JP. Acoustic differential extraction for forensic analysis of sexual assault evidence. Anal Chem 2009; 81(15): 6089–95. 10.1021/ac900439b [DOI] [PubMed] [Google Scholar]
- 13.Di Martino D, Giuffrè G, Staiti N, Simone A, Todaro P, Saravo L. Laser microdissection and DNA typing of cells from single hair follicles. Forensic Sci Int 2004; 146 Suppl: S155–7. 10.1016/j.forsciint.2004.09.047 [DOI] [PubMed] [Google Scholar]
- 14.Anslinger K, Bayer B, Mack B, Eisenmenger W. Sex-specific fluorescent labelling of cells for laser microdissection and DNA profiling. Int J Legal Med 2007; 121(1): 54–6. 10.1007/s00414-005-0065-7 [DOI] [PubMed] [Google Scholar]
- 15.Han JP, Yang F, Xu C, Wei YL, Zhao XC, Hu L, et al. A new strategy for sperm isolation and STR typing from multi-donor sperm mixtures. Forensic Sci Int Genet 2014; 13: 239–46. 10.1016/j.fsigen.2014.08.012 [DOI] [PubMed] [Google Scholar]
- 16.Lynch L, Gamblin A, Vintiner S, Simons JL. STR profiling of epithelial cells identified by X/Y-FISH labelling and laser microdissection using standard and elevated PCR conditions. Forensic Sci Int Genet 2015; 16: 1–7. 10.1016/j.fsigen.2014.10.017 [DOI] [PubMed] [Google Scholar]
- 17.Findlay I, Taylor A, Quirke P, Frazier R, Urquhart A. DNA fingerprinting from single cells. Nature 1997; 389(6651): 555–6. 10.1038/39225 [DOI] [PubMed] [Google Scholar]
- 18.Brück S, Evers H, Heidorn F, Müller U, Kilper R, Verhoff MA. Single cells for forensic DNA analysis–from evidence material to test tube. J Forensic Sci 2011; 56(1): 176–80. 10.1111/j.1556-4029.2010.01553.x [DOI] [PubMed] [Google Scholar]
- 19.Pereira J, Neves R, Forat S, Huckenbeck W, Olek K. MtDNA typing of single-sperm cells isolated by micromanipulation. Forensic Sci Int Genet 2012; 6(2): 228–35. 10.1016/j.fsigen.2011.05.005 [DOI] [PubMed] [Google Scholar]
- 20.Geng T, Novak R, Mathies RA. Single-cell forensic short tandem repeat typing within microfluidic droplets. Anal Chem 2014; 86(1): 703–12. 10.1021/ac403137h [DOI] [PubMed] [Google Scholar]
- 21.van der Gaag KJ, de Leeuw RH, Hoogenboom J, Patel J, Storts DR, Laros JF, et al. Massively parallel sequencing of short tandem repeats-Population data and mixture analysis results for the PowerSeq™ system. Forensic Sci Int Genet 2016; 24: 86–96. 10.1016/j.fsigen.2016.05.016 [DOI] [PubMed] [Google Scholar]
- 22.Kidd KK, Speed WC. Criteria for selecting microhaplotypes: mixture detection and deconvolution. Investig Genet 2015; 6:1 10.1186/s13323-014-0018-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall D, Castella V. DIP–STR: A new marker for resolving imbalanced DNA mixtures. Forensic Sci Int Genet 2011; Suppl Ser 3: e1–2. 10.1016/j.fsigss.2011.08.06521126935 [DOI] [Google Scholar]
- 24.Castella V, Gervaix J, Hall D. DIP-STR: highly sensitive markers for the analysis of unbalanced genomic mixtures. Hum Mutat 2013; 34(4): 644–54. 10.1002/humu.22280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cereda G, Biedermann A, Hall D, Taroni F. Object-oriented Bayesian networks for evaluating DIP-STR profiling results from unbalanced DNA mixtures. Forensic Sci Int Genet 2014; 8(1): 159–69. 10.1016/j.fsigen.2013.09.001 [DOI] [PubMed] [Google Scholar]
- 26.Cereda G, Biedermann A, Hall D, Taroni F. An investigation of the potential of DIP-STR markers for DNA mixture analyses. Forensic Sci Int Genet 2014; 11: 229–40. 10.1016/j.fsigen.2014.04.001 [DOI] [PubMed] [Google Scholar]
- 27.Oldoni F, Castella V, Hall D. A novel set of DIP-STR markers for improved analysis of challenging DNA mixtures. Forensic Sci Int Genet 2015; 19: 156–64. 10.1016/j.fsigen.2015.07.012 [DOI] [PubMed] [Google Scholar]
- 28.Mountain JL, Knight A, Jobin M, Gignoux C, Miller A, Lin AA, et al. SNPSTRs: empirically derived, rapidly typed, autosomal haplotypes for inference of population history and mutational processes. Genome Res 2002; 12(11):1766–72. 10.1101/gr.238602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Schneider PM, Rothschild MA, Bai P, Liang WB, Zhang L. SNP–STR polymorphism: A sensitive compound marker for forensic genetic applications. Forensic Sci Int Genet 2013; Suppl Ser 4: e206–7. 10.1016/j.fsigss.2013.10.106 [DOI] [Google Scholar]
- 30.Wang L, He W, Mao J, Wang H, Jin B, Luo HB, et al. Development of a SNP-STRs multiplex for forensic identification. Forensic Sci Int Genet 2015; Suppl Ser 5: e598–600. 10.1016/j.fsigss.2015.09.236 [DOI] [Google Scholar]
- 31.Walsh PS, Metzger DA, Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 1991; 10(4): 506–13. [PubMed] [Google Scholar]
- 32.Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, et al. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res 1989; 17(7): 2503–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tereba A. Tools for analysis of population statistics. Profiles in DNA 1999; 3: 14–6. [Google Scholar]
- 34.Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 2010; 10(3): 564–7. 10.1111/j.1755-0998.2010.02847.x [DOI] [PubMed] [Google Scholar]
- 35.Phillips C, Parson W, Amigo J, King JL, Coble MD, Steffen CR, et al. D5S2500 is an ambiguously characterized STR: Identification and description of forensic microsatellites in the genomics age. Forensic Sci Int Genet 2016; 23: 19–24. 10.1016/j.fsigen.2016.03.002 [DOI] [PubMed] [Google Scholar]
- 36.Hill CR, Kline MC, Coble MD, Butler JM. Characterization of 26 miniSTR loci for improved analysis of degraded DNA samples. J Forensic Sci 2008; 53(1): 73–80. 10.1111/j.1556-4029.2008.00595.x [DOI] [PubMed] [Google Scholar]
- 37.Kwok S, Kellogg DE, McKinney N, Spasic D, Goda L, Levenson C, et al. Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucleic Acids Res 1990; 18(4): 999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan KC, Zhang J, Hui AB, Wong N, Lau TK, Leung TN, et al. Size distributions of maternal and fetal DNA in maternal plasma. Clin Chem 2004; 50(1): 88–92. 10.1373/clinchem.2003.024893 [DOI] [PubMed] [Google Scholar]
- 39.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Analysis of the size distributions of fetal and maternal cell-free DNA by paired-end sequencing. Clin Chem 2010; 56(8): 1279–86. 10.1373/clinchem.2010.144188 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.