Super-resolution imaging reveals subtle interplay between nanoscopic organization of membrane ligands and cellular interaction.
Abstract
Hematopoietic stem/progenitor cell (HSPC) homing occurs via cell adhesion mediated by spatiotemporally organized ligand-receptor interactions. Although molecules and biological processes involved in this multistep cellular interaction with endothelium have been studied extensively, molecular mechanisms of this process, in particular the nanoscale spatiotemporal behavior of ligand-receptor interactions and their role in the cellular interaction, remain elusive. We introduce a microfluidics-based super-resolution fluorescence imaging platform and apply the method to investigate the initial essential step in the homing, tethering, and rolling of HSPCs under external shear stress that is mediated by selectins, expressed on endothelium, with selectin ligands (that is, CD44) expressed on HSPCs. Our new method reveals transient nanoscale reorganization of CD44 clusters during cell rolling on E-selectin. We demonstrate that this mechanical force-induced reorganization is accompanied by a large structural reorganization of actin cytoskeleton. The CD44 clusters were partly disrupted by disrupting lipid rafts. The spatial reorganization of CD44 and actin cytoskeleton was not observed for the lipid raft–disrupted cells, demonstrating the essential role of the spatial clustering of CD44 on its reorganization during cell rolling. The lipid raft disruption causes faster and unstable cell rolling on E-selectin compared with the intact cells. Together, our results demonstrate that the spatial reorganization of CD44 and actin cytoskeleton is the result of concerted effect of E-selectin–ligand interactions, external shear stress, and spatial clustering of the selectin ligands, and has significant effect on the tethering/rolling step in HSPC homing. Our new experimental platform provides a foundation for characterizing complicated HSPC homing.
INTRODUCTION
Cellular interactions mediated by membrane ligands and receptors, especially in the presence of external forces, play a key role in many biologically important processes (1, 2). Often, the spatial distribution of ligands and receptors on a cell membrane governs cellular interactions and intracellular signaling (3, 4). Hematopoietic stem/progenitor cell (HSPC) homing is an important biological phenomenon, as this property can be used in treating blood disorders by bone marrow transplantation, in which transplanted HSPCs travel from peripheral blood to the bone marrow (5). The homing of HSPCs to the bone marrow is a multistep process that is regulated by highly complex spatiotemporal ligand-receptor interactions under external shear stress (6–8).
The homing process is initiated by tethering and rolling of HSPCs to endothelium mediated by the binding of selectins expressed on endothelial cells (E- and P-selectin) (9, 10) to selectin ligands expressed on HSPCs (fig. S1), which occurs against the shear stress exerted by blood flow (7, 11–13). This is followed by an integrin-mediated firm adhesion and endothelial transmigration of HSPCs. The molecular mechanisms of HSPC homing have been investigated mainly by means of gel electrophoresis, flow cytometry, confocal fluorescence microscopy, and electron microscopy. Previous studies using these conventional methods have identified the molecules that are involved in the homing process and have characterized biological processes (for example, ligand-receptor interactions, signaling) involved in these complicated multistep cellular interactions. Several studies also indicated that nanoscale clustering of selectin ligands may influence the tethering/rolling of HSPCs (14). However, because of the lack of spatial resolution and/or molecular specificity of these methods, the nanoscale molecular mechanisms of HSPC homing are still elusive. Contradictory results about the localizations of selectin ligands on the cell surface (15, 16) and their role in the cell rolling (17–19) have been reported.
Super-resolution (SR) fluorescence microscopy techniques have provided a new tool for direct visualization of subcellular structures and their dynamics in cells due to their unprecedented spatial resolution (20–22). While rapid technology development in this field enables three-dimensional (3D) SR visualization of subcellular structures with temporal resolution enough for capturing relatively fast cellular dynamics, intracellular structures and processes have currently been the main focus of the development of SR imaging techniques. SR fluorescence microscopy should be able to provide a tool not only for subcellular imaging but also for directly characterizing nanoscopic mechanisms of cellular interactions. However, because of the lack of an appropriate experimental approach, the capability of SR microscopy as a tool to study cellular interactions in the presence of external force has not been exploited.
Here, we describe a microfluidics-based SR fluorescence microscopy platform that enables the capture and characterization nanoscale architectures of selectin ligands under the presence of an external shear force and the effect of these nanoscale architectures on the macroscopic cell rolling on E-selectin (Fig. 1A). With our platform, we reveal that the transient nanoscale reorganization of selectin ligands occurs during cell rolling. We also demonstrate that the nanoscale clustering and spatial reorganization of selectin ligands occurring during cell rolling play a key role in the macroscopic cell-rolling behaviors on E-selectin.
Fig. 1. Microfluidics-based in vitro cell-rolling assay for characterizing HSPC rolling.
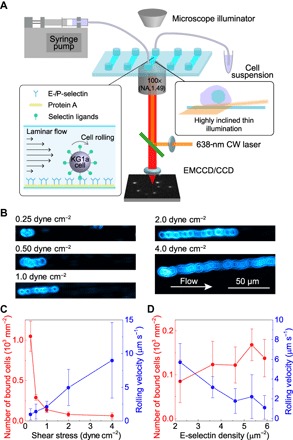
(A) Experimental configuration of the microfluidics SR microscopy platform. (B) Transmitted light microscopy images of KG1a cells rolling on E-selectin. The surface density of the E-selectin was 3.6 molecules μm−2. (C) Wall shear stress–dependent rolling velocity of KG1a cells (blue) and the number of bound cells to the E-selectin surface (red). Error bars indicate SDs of six different fields of view. (D) E-selectin density–dependent rolling velocity of KG1a cells (blue) and the number of bound cells to the E-selectin surface (red). Error bars indicate SDs of six different fields of view. CCD, charge-coupled device; EMCCD, electron multiplying CCD.
RESULTS
Macroscopic cell-rolling behavior on E-selectin under shear force
We used KG1a cells, human leukemic progenitor cell line, as a working model of HSPCs (23, 24). We deposited recombinant E-selectins with a controlled density (1.4 to 6.8 molecules μm−2) and orientation on the surface of a microfluidic chamber by a stepwise deposition of protein A and recombinant human E-selectin (Materials and Methods and fig. S2). We then perfused the KG1a cells at physiologically relevant shear stresses (0.25 to 4 dyne cm−2) over the deposited E-selectins (Materials and Methods). While the rolling velocity of the cells increased with the shear stress exerted on the cells (Fig. 1, B and C, and movies S1 to S5), the number of cells bound to the microfluidic surface increased with decreasing shear stress (Fig. 1C) (25, 26). The KG1a cells also displayed selectin density–dependent rolling behavior (that is, slower rolling velocity and larger number of surface-bound cells at higher density of E-selectins; Fig. 1D) (27). We did not observe binding when we used 10 mM EDTA to deplete Ca2+ and block the E-selectin–ligand interaction (movie S6) (25). Overall, these observations were consistent with studies documenting the in vitro rolling behavior of hematopoietic progenitor and leukocyte cells and confirmed that our microfluidics-based in vitro cell-rolling assay can mimic the cell-rolling behavior under physiologically relevant conditions.
Nanoscale reorganization of the clustering behavior of CD44 during cell rolling over E-selectin
Fluorescence images of CD44/hematopoietic cell E- and/or L-selectin ligand (HCELL), a well-known E-selectin ligand on HSPCs (23, 24, 28), were recorded using either confocal fluorescence microscopy or SR localization microscopy with a highly inclined thin illumination (HILO) configuration (see Materials and Methods for details) (29). SR images were reconstructed using CD44/HCELL molecules located within 1 μm from the surface (Materials and Methods and Supplementary Materials). After fixing the cells, we immunolabeled the CD44 molecules on the KG1a cells using Alexa Fluor (AF)–647 dye (fig. S3). We prepared two immunolabeled samples: a control sample that was fixed and stained in a cell suspension and a sample of cells that had bound and rolled on the E-selectin surface before fixing and labeling (see Methods for details). Transient behaviors caused by selectin-ligand interactions under shear stress can be captured by fixing and immunolabeling the cells inside the fluidic chamber during the rolling and binding process over E-selectins. The precision of localization was similar in both samples (fig. S4), confirming that we were able to obtain SR images of the bound and rolled cells on the E-selectin surface.
Confocal fluorescence microscopy images of CD44 showed inhomogeneous distribution of CD44 on the surface of KG1a cells (Fig. 2, A and B). The images indicate that the control and rolled cells display distinct spatial distributions of CD44. However, because of the limited spatial resolution, the confocal microscopy images do not allow us to capture nanoscale spatial organization of CD44. In contrast, SR images of CD44 revealed nanoscale architectures of CD44 on KG1a cells. A comparison of the images obtained from the control and rolled cells demonstrated an increased footprint of CD44 after rolling (Fig. 2, C and D, and fig. S5), indicating morphology changes on the cell surface caused by the E-selectin–ligand interactions. These results are consistent with the increased footprint of the cell membrane in neutrophils captured by total internal reflection fluorescence microscopy (30).
Fig. 2. SR localization microscopy imaging of CD44 on KG1a cells.
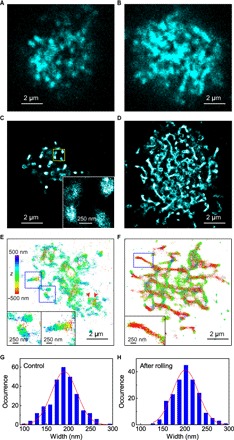
(A) Confocal fluorescence image of CD44 on a KG1a cell that was fixed and immunolabeled for CD44 using the 515 antibody followed by AF-647–conjugated secondary antibodies. (B) Confocal fluorescence image of CD44 on a KG1a cell that was fixed and immunolabeled by AF-647 dye in the microfluidic chamber after the rolling of the cell on E-selectin. (C) SR image of CD44 on a KG1a cell that was fixed and immunolabeled for CD44 using the 515 antibody followed by AF-647–conjugated secondary antibodies. The inset shows an enlarged view of the yellow region. (D) SR images of CD44 on a KG1a cell that was fixed and immunolabeled by AF-647 dye in the microfluidic chamber after the rolling of the cell on E-selectin. (E) 3D SR image of CD44 on a KG1a cell that was fixed and immunolabeled by AF-647 dye in suspension. The insets show enlarged views of the blue regions. (F) 3D SR images of CD44 on a KG1a cell that was fixed and immunolabeled by AF-647 dye in the microfluidic chamber after the rolling of the cell on E-selectin. The inset shows an enlarged view of the blue region. (G) Frequency histogram of the width of the CD44 clusters observed in the control cells. (H) Frequency histogram of the width of the CD44 clusters observed in the rolled KG1a cells.
The SR images of the control cells showed patchy nanoscale clusters of CD44 (~200 nm in size; Fig. 2, C and G, and fig. S5A). Although the clustering of CD44 on a cell surface in the length scale of several nanometers has been reported using Förster resonance energy transfer (FRET) (31), our SR imaging experiment revealed the CD44 clustering in the range of hundreds of nanometers. Three-dimensional SR localization microscopy revealed the 3D distribution of CD44 on the surfaces of KG1a cells (Fig. 2, E and F). The 3D SR images illustrated that the patchy clusters of CD44 on the control cells were located at different axial positions, even for the adjacent clusters (Fig. 2E, arrowheads). Also, the axial positions of the CD44 molecules within single clusters differed (Fig. 2E, insets). Only some of the CD44 molecules in the clusters were in close proximity to the surface. While a previous study of CD44 using immunoelectron microscopy (32) and confocal fluorescence microscopy (33) indicated that CD44 distributes to the planar cell body, our findings suggested that CD44 localizes to protruding structures of the cell surface. The observed cluster size is consistent with the length of microvilli on neutrophils (~0.3 μm) (34). Together, our results suggest that CD44 is distributed on the microvillus of KG1a cells.
The rolling caused significant reorganization of the clustering behavior of CD44, from patchy to elongated network-like structures (Fig. 2D and fig. S5B). This nanoscale reorganization of CD44 clusters was not observed when KG1a cells were incubated with recombinant E-selectin in suspension (see Materials and Methods for details; fig. S6). This result suggests that the binding of E-selectin to its ligands does not induce the structural reorganization of CD44 clusters on KG1a cells. Our results indicate instead that shear stress exerted on the cell due to the binding of surface-deposited E-selectin and CD44 molecules on the cell surface is essential for the reorganization of CD44 during cell rolling. The width of the clusters remained constant after rolling (Fig. 2H). The cell that rolled on E-selectin also exhibited 3D distribution of CD44 molecules in the elongated clusters (Fig. 2F). However, many of these network-like elongated clusters were located in close proximity to the surface (Fig. 2F, inset). This finding suggests that the reorganization of the clusters is a result of the binding of CD44 to the surface E-selectin under the presence of the shear force. This nanoscale reorganization of the clustering behavior of the selectin ligands has not been previously reported. Although the formation of tethers and slings in the length scale of several micrometers up to tens of micrometers has been observed during neutrophils rolling on selectin surfaces (35, 36), the formation of the elongated clusters of CD44 is distinct from that of the previously reported tethers and slings (see below).
Our results demonstrate that our microfluidics-based SR imaging platform can quantify both macroscopic cell-rolling and nanoscopic selectin ligand clustering behaviors. Our method revealed that nanoscale reorganization of the CD44 clusters has a significant effect on the HSPCs rolling over E-selectin (see below).
Reorganization of actin cytoskeleton during the cell rolling
The clustering of CD44 on KG1a cells points to the involvement of subcellular structures in its organization on the cell surface. Thus, we conducted two-color SR localization microscopy experiments of CD44 and actin cytoskeleton. CD44 on the KG1a cells was immunolabeled using AF-647 dye, and actin cytoskeleton was labeled by AF-488 dye–conjugated phalloidin (see Materials and Methods for details). The two-color SR image of the control cell shows strong colocalization of CD44 and actin cytoskeleton (Fig. 3A). A previous study using FRET and fluorescence after photobleaching (FRAP) techniques indicated the association of CD44 and actin cytoskeleton (31) mediated by actin-associated ERM (ezrin/radixin/moesin) proteins (37–39). Our data agree with this observation and directly demonstrate complete physical colocalization of CD44 and actin cytoskeleton on the cell surface via the ERM proteins. The result is also consistent with the 3D SR image of CD44 (Fig. 2E) that suggests the distribution of CD44 on microvillus.
Fig. 3. Two-color SR images of CD44 and actin on KG1a cells.
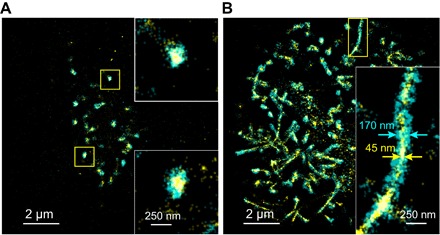
(A) SR image of CD44 (cyan) and actin cytoskeleton (yellow) on a KG1a cell that was fixed and immunolabeled by AF-647 (CD44) and labeled by AF-488 dye–conjugated phalloidin (actin). The insets show enlarged views of the yellow regions. (B) SR image of CD44 (cyan) and actin cytoskeleton (yellow) on a KG1a cell that was fixed and fluorescently labeled in the microfluidic chamber after the rolling of the cell on E-selectin. The inset shows enlarged view of the yellow region.
During rolling, they remain colocalized on the rolled cell (Fig. 3B). The actin cytoskeleton formed thin fibrous structures upon rolling on E-selectin, which displayed complete physical colocalization with the elongated CD44 clusters. Since the width of the fibrous actin cytoskeleton (~45 nm) is close to the image resolution of our SR microscopy experiments, we cannot distinguish whether they are single actin filaments or a bundle of the fibers. The elongated CD44 clusters that appeared during the cell rolling on E-selectin could be related to the membrane tethers and slings since they are believed to be formed by extending microvilli. However, the formation of the tethers requires membrane separation from the cytoskeleton (40), which is in marked contrast to the complete colocalization of the membrane-embedded CD44 and cytoskeleton observed in our experiment (Fig. 3B). Previous studies demonstrated an extension of microvilli due the selectin-ligand binding under external force, which could be responsible for the observed elongated CD44 clusters. However, the maximum length of elongated microvilli (~1 μm) (34, 41) is much shorter than that of the observed elongated clusters of CD44. Also, extended microvilli and membrane tethers exhibit a retraction upon detachment from selectins (41, 42). The elongated clusters of CD44, therefore, cannot be attributed to any previously observed membrane protrusion structures.
Spatial organization of the cortical actin cytoskeleton is one of the most important factors affecting elastic properties of cells. A recent study suggested that reorganization of the cortical actin cytoskeleton of leukocyte causes change in stiffness of the cell, which affects the interaction of the cell with endothelium during margination and demargination (43). Our observations demonstrated for the first time that the nanoscale reorganization of CD44 clusters occurring during cell rolling on E-selectins is accompanied by reorganization of the actin cytoskeleton. This finding indicates that the classical view of the initial step of HSPC homing based on simple selectin-ligand interactions cannot fully describe the cellular interaction occurring during the step. Instead, our results imply the involvement of multiple factors including change in elastic properties of the cell in the initial step of HSPC homing.
Spatial organization of lipid rafts during the cell rolling
Lipid rafts, cholesterol- and glycosphingolipid-rich microdomains, are believed to function as signaling platforms across cell membranes (14, 44, 45) by concentrating ligands for signaling in these microdomains. Thus, the clustering of CD44 could also be regulated by lipid rafts. We conducted two-color SR localization microscopy experiments of CD44 and lipid rafts. CD44 on the KG1a cells was immunolabeled using AF-647 dye, and lipid rafts were labeled by the AF-488 dye–conjugated choleratoxin B (AF-488–CtxB) subunit that binds to a glycosphingolipid, GM1 gangliosides, localized in lipid rafts (see Materials and Methods for details). Unlike the strong colocalization of CD44 and actin cytoskeleton, the two-color SR image of the control cell showed partial colocalization of CD44 and lipid rafts (Fig. 4A), suggesting that lipid rafts are not as critical as actin cytoskeleton for the nanoscale clustering of CD44 on the control cells. The partial colocalization of CD44 and lipid rafts was also observed for the rolled cell (Fig. 4B) similar to the control cell, although significant reorganization of the membrane morphology occurred during the rolling (Fig. 2). Cluster analyses (see Materials and Methods for details) (4) revealed that the size of the lipid raft microdomains does not change during the rolling (95 and 99 nm for control and rolled cells, respectively; Fig. 4C and fig. S7). A previous study described the clustering of small lipid rafts into larger domains (46), which could result in an increase in the numbers of selectin ligands including CD44 within these domains, thereby triggering outside-in signaling (46, 47). However, our result suggests the absence of merged large lipid raft domains, indicating that the CD44-dependent signaling may not require the formation of large lipid raft domains. We cannot rule out the contribution of spatial reorganization of lipid rafts to signaling because the overall spatial pattern of lipid rafts is modified during the cell rolling on E-selectin (Fig. 4, A and B) since they partially colocalized with CD44, whose spatial pattern changes significantly from a patchy to an elongated shape during rolling.
Fig. 4. Two-color SR images of CD44 and lipid rafts on KG1a cells.
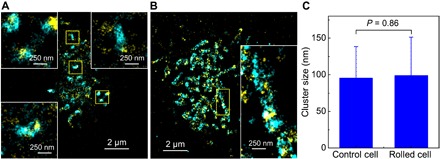
(A) SR image of CD44 (cyan) and lipid rafts (yellow) on a KG1a cell that was fixed and immunolabeled by AF-647 (CD44) and labeled by AF-488–CtxB subunit (lipid rafts). The insets show enlarged views of the yellow regions. (B) SR image of CD44 (cyan) and lipid rafts (yellow) on a KG1a cell that was fixed and fluorescently labeled in the microfluidic chamber after the rolling of the cell on E-selectin. The inset shows enlarged view of the yellow region. (C) Mean cluster sizes of lipid rafts on control and rolled cells on E-selectins. Error bars indicate SDs of the cluster sizes obtained from 12 cells for each condition.
Effects of lipid rafts on the spatial reorganization of CD44 and actin cytoskeleton during the cell rolling
We next examined how lipid rafts affect nanoscale spatial reorganization of CD44 and actin cytoskeleton during the cell rolling. To this end, we disrupted lipid raft formation using methyl-β-cyclodextrin (MβCD) (14, 48). MβCD disrupts lipid raft microdomains by extracting cholesterol from a cell membrane. The SR images of MβCD-treated KG1a cells revealed that the patchy clusters of CD44 (Fig. 2C) were moderately disrupted by MβCD treatment (Fig. 5A and figs. S4 and S8). This observation is consistent with the observed partial colocalization of CD44 and lipid rafts (Fig. 4A), suggesting that the glycolipid-enriched microdomains in lipid rafts may contribute to the spatial organization of CD44 clusters on KG1a cells to a small extent. The cluster analyses showed that while the remaining patchy clusters exhibited sizes (132 ± 61 nm; Fig. 5E and fig. S9) similar to those in the control cells (146 ± 42 nm), a nearly random distribution of CD44 was detected in the disrupted regions (cluster size, 47 ± 61 nm; Fig. 5E and fig. S9). These results are consistent with the results of the two-color SR imaging experiment of CD44 and actin cytoskeleton on the MβCD-treated KG1a cells. The two-color SR images showed that the patchy CD44 clusters usually colocalize with the actin cytoskeleton similar to the control cells, whereas the randomly distributed CD44 molecules do not colocalize with the actin cytoskeleton (Fig. 5C).
Fig. 5. Effect of lipid rafts on the nanoscale reorganization of CD44.
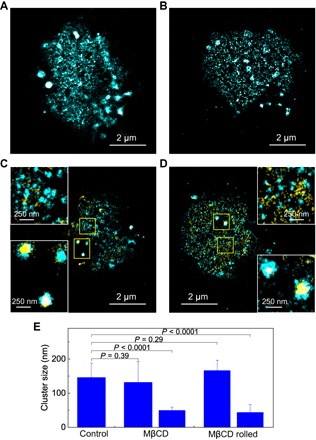
SR image of (A) CD44 on a methyl-β-cyclodextrin (MβCD)–treated KG1a cell that was fixed and immunolabeled by AF-647 dye in a suspension and (B) CD44 on an MβCD-treated KG1a cell that was fixed and fluorescently labeled in the microfluidic chamber after the rolling of the cell on E-selectin. Two-color SR images of (C) CD44 (cyan) and actin cytoskeleton (yellow) on an MβCD-treated KG1a cell that was fixed and immunolabeled by AF-647 dye in a suspension and (D) CD44 (cyan) and actin cytoskeleton (yellow) on an MβCD-treated KG1a cell that was fixed and fluorescently labeled in the microfluidic chamber after the rolling of the cell on E-selectin. The insets show enlarged view of the yellow regions. (E) Mean cluster sizes of CD44 on control cells, MβCD-treated cells, and MβCD-treated cells after rolling over E-selectin. Error bars indicate SDs of the cluster sizes obtained from 25, 19, and 12 cells for control, MβCD-treated, and MβCD-treated rolled cells, respectively.
The spatial organization of CD44 and actin cytoskeleton on the MβCD-treated KG1a cells remains unchanged during the cell rolling on E-selectins (Fig. 5, B and D), which is in marked contrast to the large spatial reorganization of CD44 and actin cytoskeleton during the cell rolling on E-selectins observed for the control KG1a cells (Fig. 3, A and B). The SR image of CD44 on the MβCD-treated KG1a cells after the rolling displayed both patchy clusters and randomly distributed CD44 (Fig. 5B). The cluster analyses demonstrated little effect of the cell rolling on the cluster sizes (158 ± 281 nm and 42 ± 22 nm for the patchy clusters and the randomly distributed CD44, respectively; Fig. 5E and fig. S9). The two-color SR image of CD44 and actin cytoskeleton on the MβCD-treated KG1a cells after the cell rolling displayed colocalization of CD44 and actin cytoskeleton in the patchy clusters (Fig. 5D), demonstrating the absence of the reorganization of actin cytoskeleton during the rolling. These results demonstrate that the intact lipid rafts and, therefore, the clustering of CD44 are essential for the nanoscale reorganization of CD44 during the cell rolling on E-selectins. We note that the MβCD treatment affects the local membrane rigidity, which may have an effect on the nanoscopic spatial pattern of CD44.
Effects of the nanoscale clustering and reorganization of CD44 on the rolling behaviors of the cell
Our microfluidics-based SR imaging experiments revealed that the spatial reorganization of CD44 and actin cytoskeleton is the result of the concerted effect of E-selectin–ligand interactions, external shear stress, and spatial clustering of the selectin ligand. Since the reorganization occurs during the cell rolling over E-selectin, we next investigated the relationship between the reorganization and rolling behaviors of the cell. We found that the rolling velocity of the MβCD-treated cells (5.7 ± 2.1 μm s−1) was much faster than that of the control cells (0.94 ± 0.39 μm s−1; Fig. 6 and movies S7 and S8). The expression level of CD44 in each cell did not change significantly following MβCD treatment (fig. S10). The footprint of CD44 on control (Fig. 2C and fig. S5A) and MβCD-treated (Fig. 5A and fig. S8) KG1a cells is similar. These results indicate that the total number of CD44 molecules that can interact with surface E-selectins is similar for both cells. We note that we conducted the cell-rolling assays at the same shear flow rate with the same surface density of E-selectin, two factors that control the rolling behaviors of the cell (Fig. 1, C and D). These results indicate that the clustering of CD44 promotes slower rolling of the KG1a cells on E-selectin.
Fig. 6. Effect of nanoscopic clustering of CD44 on the macroscopic rolling behavior of KG1a cells on E-selectin.
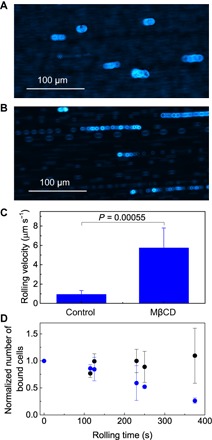
Transmitted light microscopy images of (A) control and (B) MβCD-treated KG1a cells rolling on E-selectin. The surface density of E-selectin was 2.7 molecules μm−2. (C) Mean rolling velocities of control and MβCD-treated cells. Error bars indicate SDs of six different fields of view. (D) Rolling time dependence of the number of control (black) and MβCD-treated (blue) KG1a cells tethered and rolled on E-selectin. The number is normalized by the number of cells in the first time point. Error bars indicate SDs obtained from two to four independent experiments.
The number of control cells rolled over E-selectin remained almost constant over time, whereas a significant decrease in tethered and rolled cells was observed for the MβCD-treated cells (Fig. 6D). While the spatial reorganization of CD44 occurred within several minutes during the rolling (see Materials and Methods), no spatial reorganization of CD44 was observed for the MβCD-treated cell within the same time frame. Together, these results indicate that the nanoscale reorganization of CD44 contributes to the stable rolling of the cell over E-selectin. The stable rolling may be facilitated by the increased total number of CD44 molecules that interact with the surface E-selectins, which is suggested by the increased CD44 footprint during the rolling. The change in the size and shape of the cluster may also contribute to the stable rolling.
DISCUSSION
The clustering behavior–dependent cell rolling also has important implications for the mechanism of the spatial reorganization of the CD44 clusters and actin cytoskeleton during the rolling. Since the cell-rolling experiments on the control and MβCD-treated cells were conducted at the constant flow rate (that is, wall shear stress), faster rolling velocity is a result of weaker binding of E-selectin and its ligands (that is, larger dissociation rate constant or smaller binding rate constant). Given the critical role of the external shear force for the spatial reorganization of CD44 (Fig. 2, C and D, and fig. S6), the weaker binding may not be able to exert force to the cell with enough time to reorganize CD44 and actin cytoskeleton. These results suggest the presence of a mutual effect between the spatial clustering/reorganization of CD44 and the cell-rolling behavior. The clustering causes stronger binding between E-selectin and CD44 and, thus, slow rolling. At the same time, external force is persistently exerted to the cell due to the strong binding, which causes the reorganization of CD44 clusters. The reorganized CD44 clusters further strengthen the binding and cause stable cell rolling. Our findings demonstrate that the initial step of HSPC homing is far more dynamic than previously thought and regulated by highly complex spatiotemporal interaction between the cell and E-selectin.
Our microfluidics-based SR microscopy platform enabled the capture and quantification of the nanoscale spatial reorganization of selectin ligands that result from the rolling of cells over E-selectin in the presence of external shear force. Using our approach, we also demonstrated that the nanoscopic clustering/spatial reorganization of CD44/HCELL ligand plays a key role in the macroscopic cell-rolling behaviors. These findings urge a revision of the current understanding of the molecular mechanisms used by HSPCs during the first step of the homing that is based on relatively simple selectin-ligand interactions. Our results revealed the complex and dynamic mutual effect between the spatial clustering/reorganization of the ligands and the binding/rolling behaviors of the cell on the initial tethering/rolling step of HSPC homing. Time-dependent microfluidics-based SR microscopy experiments, which can be conducted by fixing the rolling cells at different time points, will further untangle the kinetics of the nanoscale reorganization of the selectin ligands and thus will contribute to a detailed nanoscopic and molecular-level understanding of HSPC homing. Future work may also help tease out contributions of other ligands (that is, P-selectin glycoprotein ligand-1, CD43, E-selectin ligand 1, CD34, etc.) to overall cell-rolling behavior. Our platform is, in principle, compatible with selective plane illumination microscopy and thereby can be envisioned to expand the applicability of the method toward studying interactions between HSPCs and endothelium.
MATERIALS AND METHODS
Cell culturing and treatments
KG1a cells, a human acute myelogenous leukemia cell line, purchased from the American Type Culture Collection, were maintained in Iscove’s modified Dulbecco’s medium (IMDM; Gibco) supplemented with 20% fetal bovine serum, penicillin (100 U ml−1), and streptomycin (100 μg ml−1) at 37°C in a humidified atmosphere containing 5% CO2. For the disruption of actin cytoskeletons, 106 ml−1 cells were treated with cytochalasin D (CytD; 3 μg ml−1; Sigma) in a prewarmed Hanks’ balanced salt solution (HBSS; Gibco) for 30 min at 37°C. For the disruption of lipid rafts, 106 ml−1 cells were treated by a prewarmed 10 mM MβCD (Sigma) and 10 mM Hepes buffer (Sigma) in a serum/antibiotic-free IMDM at 37°C for 30 min. The viability of the cells after the treatments was confirmed by trypan blue staining.
Deposition of E-selectin
Glass coverslips (No. 1.5, ibidi GmbH) were cleaned by ultrasonication (P60H, Elma Schmidbauer GmbH) in potassium hydroxide and ethanol. A cleaned coverslip was attached to the bottom of a microfluidic chamber (channel width, 3.8 mm; channel height, 0.4 mm; sticky-Slide VI 0.4, ibidi GmbH) and incubated with protein A (10 μg ml−1; Invitrogen) overnight at room temperature. After washing off unbound protein A with HBSS, the chamber was incubated with a recombinant human E-selectin (Sino Biological), which contained the fused C-terminal polyhistidine-tagged Fc region of human immunoglobulin G1 (IgG1) at the C terminus, at 4°C at concentrations of 0.05, 0.1, 0.2, 0.3, 0.4, and 0.5 μg ml−1 for 1 hour. The chamber was then washed with HBSS and blocked with 1% casein solution in phosphate-buffered saline (PBS; Thermo) for 30 min to 1 hour at room temperature. The E-selectin–deposited chamber was used immediately for the cell-rolling assay and SR imaging experiments. The surface densities of the recombinant E-selectin were determined by labeling the surface E-selectin by anti-human CD62E (E-selectin) antibody (HAE-1f clone, BioLegend) conjugated to AF-647 dye. The microfluidic chamber with the E-selectin–coated surface was incubated with the antibody for 1 hour at room temperature. After the incubation, free antibodies were washed off with HBSS. Fluorescence images of the AF-647–conjugated antibodies were recorded using a custom-built wide-field fluorescence microscopy setup (see below). The surface densities of E-selectin were calculated by comparing the fluorescence intensities obtained from the surface and those obtained from single AF-647–conjugated antibodies.
Cell-rolling assay
The cell-rolling assay was performed at room temperature using a microfluidics chamber sticky-Slide VI 0.4 attached to the E-selectin–deposited glass coverslip. The inlet and outlet of the chamber were connected to a 0.8-mm silicon tubing (ibidi GmbH) using male Luer connectors (ibidi GmbH). The end of the inlet tube was placed in a rolling buffer [HBSS containing 1% human serum albumin (Sigma) and 1 mM CaCl2 (Sigma)], and the end of the outlet tube was connected to a programmable syringe pump (PHD ULTRA, Harvard Apparatus) using a female Luer Lock connector (ibidi GmbH). To equilibrate the flow path, we allowed the rolling buffer to flow into the chamber for 90 s prior to introducing the KG1a cells. Then, 106 ml−1 KG1a cells suspended in the rolling buffer were autoperfused for 20 s using a syringe pump. After a brief stop in flow for 10 to 20 s to allow the cells to settle down and interact with the E-selectins on the surface, we resumed flow of the cell suspension for another 20 to 30 s. Then, the chamber was washed with the rolling buffer for 90 s, during which we recorded a video of the cells rolling on the E-selectin–coated surface of the chamber. Multiple videos were recorded with a time interval of 105 to 115 s for the analysis of time-dependent rolling behaviors. The rolling experiment was conducted at wall shear stresses (W) of 0.25, 0.5, 1, 2, and 4 dyne cm−2. The cell-rolling behavior was captured by mounting the microfluidic chamber on an inverted optical microscope (CXK41, Olympus) equipped with a 20× objective (LCAch N 20X, Olympus) and by recording transmitted light images at video rate using a CCD camera (XC10, Olympus) using CellSens software (Olympus). Transmitted optical microscopy images of the KG1a cells were also recorded using an Olympus IX71 inverted optical microscope equipped with a high numerical aperture (NA) objective (UAPON 100XOTIRF, Olympus) and an EMCCD camera (iXon3 897, Andor Technology).
Analysis of the cell-rolling behavior
Each video was analyzed using TrackMate Fiji, an ImageJ plugin. The cells were localized and tracked using the LAP (Linear Assignment Problem) tracker. The displacement trajectories of each of the selected cells over time along the y axis were extracted from the tracking data, and single-cell velocities were calculated by dividing the total displacements of the rolled cells by the total number of frames, during which the cell showed continuous rolling behaviors. Mean cell velocities were calculated after applying selection criteria: The cells that were not detected in more than 75 consecutive frames were considered as passing cells and were excluded from the analysis.
Fluorescence labeling of cells
The control, CytD-treated, and MβCD-treated KG1a cells were fixed in 3% (w/v) paraformaldehyde (Electron Microscopy Sciences) and 0.2% (w/v) glutaraldehyde (Electron Microscopy Sciences) in HBSS for 20 min at room temperature. Next, the cells were blocked with 10% goat serum (Sigma) for 40 min at 37°C. The cells were incubated with purified mouse anti-human CD44 antibody (15 μg ml−1; clone 515, BD Pharmingen) diluted in 2% bovine serum albumin (BSA; Sigma) in HBSS for 40 min at 37°C, followed by AF-647–conjugated goat anti-mouse secondary antibody (5 μg ml−1; Invitrogen) diluted in 2% BSA in HBSS for 40 min at 37°C. Then, the cells were fixed again in 3% (w/v) paraformaldehyde and 0.2% (w/v) glutaraldehyde in HBSS for 10 min at room temperature. After attaching the coverslip to the microfluidic chamber sticky-Slide VI 0.4, the chamber was incubated with poly-l-ornithine (Sigma). We then mounted the fluorescently labeled cells on the surface of the chamber overnight at 4°C.
For two-color imaging of CD44 and the actin cytoskeleton, the fixed and blocked cells were incubated with purified mouse anti-human CD44 antibody (15 μg ml−1; clone 515) diluted in 2% BSA in HBSS for 40 min at 37°C, followed by AF-647–conjugated goat anti-mouse secondary antibody (5 μg ml−1) diluted in 2% BSA in HBSS for 40 min at 37°C. The immunolabeled cells were fixed again in 3% (w/v) paraformaldehyde and 0.2% (w/v) glutaraldehyde in HBSS for 10 min at room temperature. The cells were then permeabilized in 0.1% Triton X-100 (Sigma-Aldrich) in cytoskeleton buffer [10 mM MES (pH 6.1), 150 mM NaCl, 5 mM EGTA, 5 mM glucose, and 5 mM MgCl2] for 10 min at room temperature. The permeabilized cells were labeled for actin cytoskeleton with freshly prepared 0.5 μM AF-488 phalloidin (Molecular Probes-Thermo Fisher Scientific) diluted from a stock solution of 6.6 μM AF-488 phalloidin, with 1% BSA in the cytoskeleton buffer for 1 hour at room temperature. Then, the cells were fixed again in 3% (w/v) paraformaldehyde and 0.2% (w/v) glutaraldehyde in HBSS for 10 min at room temperature.
For two-color imaging of CD44 and lipid rafts, the fixed cells were incubated with AF-488–CtxB subunit (10 μg ml−1; Molecular Probes-Thermo Fisher Scientific), which binds to GM1 gangliosides localized in lipid rafts (49) on the surface of KG1a cells, with 1% BSA in PBS for 1 hour at room temperature. After blocking the cells with 10% goat serum for 40 min at 37°C, the cells were incubated with purified mouse anti-human CD44 antibody (15 μg ml−1; clone 515) diluted in 2% BSA in HBSS for 40 min at 37°C, followed by AF-647–conjugated goat anti-mouse secondary antibody (5 μg ml−1) diluted in 2% BSA in HBSS for 40 min at 37°C. Then, the cells were fixed again in 3% (w/v) paraformaldehyde and 0.2% (w/v) glutaraldehyde in HBSS for 10 min at room temperature.
To fluorescently label the control and MβCD-treated KG1a cells rolled on the E-selectin surface, either control or MβCD-treated cells were perfused into the microfluidic chamber whose surface was coated by the E-selectin (3.6 molecules μm−2) in a manner similar to the cell-rolling assay at a constant wall shear stress of 1 dyne cm−2 for 90 s. After the cell rolling, a fixation solution of 3% (w/v) paraformaldehyde and 0.2% (w/v) glutaraldehyde in HBSS was autoperfused at 1 dyne cm−2 for 3 min to arrest and fix the cells inside the chamber. This procedure was followed by a 17-min incubation of the cells in the fixation solution to completely freeze the mobility of the cell membranes and membrane proteins. The fluorescence labeling of CD44, actin cytoskeleton, and lipid rafts was conducted in a manner similar to the fluorescence labeling of KG1s cells in suspension.
SR microscopy
SR localization microscopy imaging of immunolabeled KG1a cells was performed in an imaging buffer composed of TN buffer [50 mM tris (pH 8.0) and 10 mM NaCl], oxygen scavenging system [glucose oxidase (0.5 mg ml−1; Sigma), catalase (40 μg ml−1; Sigma), 10% (w/v) glucose], and 100 mM β-mercaptoethanol (Sigma) as a reducing reagent. The imaging solution was prepared immediately before the imaging experiments. The imaging experiments were conducted on a custom-built wide-field illumination fluorescence microscope on an inverted optical microscope platform (IX71, Olympus) (50, 51). A continuous-wave diode laser operating at 638 nm (60 mW; MLD, Cobolt) was introduced into the microscope from its backside port through an achromatic convex lens (f = 300 mm; Thorlabs) that focused the beam at the back aperture of the objective lens [100×, NA = 1.49; UAPON 100XOTIRF, Olympus]. The samples were illuminated through an objective lens with a HILO configuration. The illumination power at the samples for the imaging experiments was set to 3.1 kW cm−2. The illumination area was adjusted to 10 to 15 μm in diameter such that only a single cell was illuminated in each image acquisition. The fluorescence from the sample was captured by the same objective, separated from the illumination light by a dichroic mirror (FF660-Di02-25x36, Semrock), passed an emission bandpass filter (FF01-697/58-25, Semrock), and detected by an EMCCD camera. The fluorescence images were recorded using a 150 × 150 pixel region of the EMCCD camera with 83-nm pixel size at 10-ms exposure time. The fluorescence image sequences with 10,000 frames were recorded for the reconstruction of SR localization microscopy images. The image acquisition was done using the Andor iQ3 software. The exposure of the EMCCD camera was synchronized with the sample illumination by the laser using an acousto-optic tunable filter (AA Opto Electronic).
A 3D SR imaging experiment was conducted using the illumination configuration and buffer condition in the same manner as the 2D imaging experiment. Astigmatism-based SR localization microscopy was used for the 3D SR imaging (22). A cylindrical lens with a focal length of 200 mm was inserted in front of the EMCCD camera. The calibration of the z axis positions was done by using TetraSpeck microspheres (diameter, 100 nm; Invitrogen) deposited on a cleaned coverslip. The calibration data were recorded at −500 to +500 nm with a 10-nm step size. The z axis positions in the acquisition of the calibration data were controlled by a piezo nanopositioning stage (APZ-X100 Piezo Z-Stage, Andor Technology). The stage drift in the z axis was less than 30 nm during each image acquisition (~100 s).
Two-color SR localization microscopy was conducted by introducing coaxially 638- and 488-nm (60 mW; MLD, Cobolt) lines of diode lasers into the inverted microscope in the same way as the single-color excitation with the buffer condition in the same manner as 2D imaging experiment. The samples were excited through the objective lens (100×, NA = 1.49; UAPON 100XOTIRF, Olympus) using the HILO configuration at the excitation power of 3.1 and 4.8 kW cm−2 for the 638- and 488-nm lasers, respectively. The fluorescence from the samples was captured by the same objective, separated from the illumination light by a multiband dichroic mirror (Di03-405/488/561/635-t1-25x36, Semrock), and passed through a TuCam dual-camera adaptor (Andor Technology) equipped with a filter cassette containing a dichroic mirror (FF635-Di01-25x36, Semrock) to separate the fluorescence into two channels. The separated fluorescence from the samples was detected by two EMCCD cameras (iXon3 897, Andor Technology) through emission bandpass filters (FF01-540/50-25 and FF01-697/58-25).
Analysis of SR images
The SR images were reconstructed by using either a custom-written MATLAB (MathWorks) code or Localizer software (52). The positions of the AF-647 molecules were determined by 2D Gaussian fitting of the images. We removed fluorescence spots whose width was significantly larger (>200 nm) than the point spread function (PSF) of the optical system (PSF, ~130 nm) from the analysis. The effect of the stage drift in the xy plane was corrected by reconstructing the subimages using 5000 localizations. In the 3D SR imaging, the calibration data were fitted to elliptical 2D Gaussian functions, and the z position–dependent spot widths were fitted to polynomial functions to obtain the calibration curves. In the two-color SR imaging, TetraSpeck microspheres (diameter, 100 nm) deposited on a cleaned coverslip was used to calibrate the shift between the two channels. Using fluorescence images of the TetraSpeck microspheres recorded simultaneously on the two cameras, we generated a registration map that corrects the shift between the two images and applied the registration map to the images obtained from the cell samples.
The widths of the patchy clusters of CD44 observed in the control cells were determined by manually measuring the widths of the clusters along their short axes. The widths of the elongated network-like clusters of CD44 observed in the rolled cells were determined by manually measuring the widths of the clusters at their center positions along the long axes. The cluster sizes of CD44 and lipid rafts were characterized by Ripley’s K functions. We selected cropped areas away from the edges of the images in the clustering analysis to avoid artifacts. The cluster analysis of the large and small clusters on the MβCD-treated cells was conducted by manually selecting the areas with large and small clusters.
Confocal fluorescence microscopy
The confocal fluorescence microscopy experiment of CD44 on KG1a cells was conducted using Zeiss LSM710 confocal microscope equipped with an oil immersion objective lens (100×, NA = 1.46; alpha Plan-Apochromat, Zeiss) upon excitation with a 633-nm line of HeNe laser. Fluorescence from the samples was detected using a photomultiplier tube (Hamamatsu). Fluorescence images were acquired using the ZEN 2009 software (Zeiss).
Flow cytometry
Binding specificity of the mouse anti-human CD44 (IgG1κ, clone 515) antibodies, which were used for the SR imaging experiments, to CD44 on KG1a cells was evaluated by flow cytometry. KG1a cells (106 cells ml−1) were incubated with either anti-human CD44 antibody (10 μg ml−1) or purified mouse IgG1κ isotype (10 μg ml−1; BioLegend) in HBSS at 4°C for 30 min. Subsequently, the KG1a cells were incubated with AF-488–conjugated goat anti-mouse antibody (5 μg ml−1, IgG, Invitrogen) in HBSS at 4°C for 20 min. For the secondary antibody control, KG1a cells (106 cells ml−1) were incubated with AF-488–conjugated goat anti-mouse antibody (5 μg ml−1) in HBSS at 4°C for 20 min. To assess the expression level of CD44 on the surface of KG1a cells, we fixed CytD-treated cells, MβCD-treated cells, and control cells with 1% paraformaldehyde in HBSS for 20 min at room temperature and incubated them with either mouse anti-human CD44 antibody (10 μg ml−1; IgG1κ, clone 515) in HBSS at 4°C for 30 min. The cells were then incubated with AF-488–conjugated goat anti-mouse antibody (5 μg ml−1) in HBSS at 4°C for 20 min. The fluorescence intensity was determined using a FACSCanto flow cytometer (Beckman Dickenson).
Supplementary Material
Acknowledgments
We thank M. Abadi for technical support during the 3D and two-color SR microscopy experiments. We also thank D. AbuSamra, M. Mih, K. Sakashita, A. Ali, A. Khodairi, F. Aleisa, and A. Amoodi for their contributions to this work. We thank V. Unkefer for editing the manuscript. Figure S1 was based on a figure previously created in a joint effort with E. Mikhaylova [King Abdullah University of Science and Technology (KAUST)] and modified by J. Merzaban and S. Habuchi. Funding: The research reported in this publication was supported by funding from the King Abdullah University of Science and Technology (KAUST) and the KAUST Office of Sponsored Research under Award No. CRG R2 13 MERZ KAUST 1. Author contributions: S.H. conceived the project with the help of J.S.M. and S.M.H. K.A.Z. designed the SR microscopy experiments. K.A.Z. and B.A.A. designed the cell-rolling assay. K.A.Z. conducted the experiments and analyzed the data with the support of L.I.J. J.S.M. provided the cell lines. K.A. and S.H. wrote the manuscript. All authors discussed the results. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/7/eaat5304/DC1
Fig. S1. The multistep paradigm of cell migration highlighting the main interactions that take place between the blood stem cell in flow and the endothelial cells lining the blood vessels of the bone marrow.
Fig. S2. Surface density of E-selectin on the protein A–deposited microfluidic chamber determined by immunofluorescence imaging.
Fig. S3. Flow cytometric analysis to determine the binding specificity of ligand-specific antibodies to CD44 on KG1a cells.
Fig. S4. Localization precisions of the SR localization microscopy experiments of CD44 on KG1a cells.
Fig. S5. Examples of the reconstructed SR images of CD44 on KG1a cells.
Fig. S6. SR images of CD44 on KG1a cells.
Fig. S7. Cluster analysis of the nanoscale architecture of lipid rafts on KG1a cells.
Fig. S8. Examples of the reconstructed SR images of CD44 on MβCD-treated KG1a cells.
Fig. S9. Cluster analysis of the nanoscale architecture of CD44 on KG1a cells.
Fig. S10. Expression of CD44 on untreated and MβCD-treated KG1a cells was determined by flow cytometry.
Fig. S11. Depth of the field in the SR localization microscopy imaging experiments with HILO configuration.
Movie S1. Time-lapse transmitted light microscopy images of KG1a cells perfused into the microfluidic chamber at the shear stress of 0.25 dyne cm−2.
Movie S2. Time-lapse transmitted light microscopy images of KG1a cells perfused into the microfluidic chamber at the shear stress of 0.5 dyne cm−2.
Movie S3. Time-lapse transmitted light microscopy images of KG1a cells perfused into the microfluidic chamber at the shear stress of 1.0 dyne cm−2.
Movie S4. Time-lapse transmitted light microscopy images of KG1a cells perfused into the microfluidic chamber at the shear stress of 2.0 dyne cm−2.
Movie S5. Time-lapse transmitted light microscopy images of KG1a cells perfused into the microfluidic chamber at the shear stress of 4.0 dyne cm−2.
Movie S6. Time-lapse transmitted light microscopy images of KG1a cells perfused into the microfluidic chamber in the presence of EDTA (10 mM) at the shear stress of 1.0 dyne cm−2.
Movie S7. Time-lapse transmitted light microscopy images of KG1a cells perfused into the microfluidic chamber at the shear stress of 1.0 dyne cm−2.
Movie S8. Time-lapse transmitted light microscopy images of MβCD-treated KG1a cells perfused into the microfluidic chamber at the shear stress of 1.0 dyne cm−2.
REFERENCES AND NOTES
- 1.Iskratsch T., Wolfenson H., Sheetz M. P., Appreciating force and shape—The rise of mechanotransduction in cell biology. Nat. Rev. Mol. Cell Biol. 15, 825–833 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolaczkowska E., Kubes P., Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 13, 159–175 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Lillemeier B. F., Mortelmaier M. A., Forstner M. B., Huppa J. B., Groves J. T., Davis M. M., TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat. Immunol. 11, 90–96 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williamson D. J., Owen D. M., Rossy J., Magenau A., Wehrmann M., Gooding J. J., Gaus K., Pre-existing clusters of the adaptor Lat do not participate in early T cell signaling events. Nat. Immunol. 12, 655–662 (2011). [DOI] [PubMed] [Google Scholar]
- 5.C. Magnon, P. S. Frenette, Hematopoietic stem cell trafficking, in StemBook, D. Scadden, Ed. (Harvard Stem Cell Institute, 2008). [PubMed] [Google Scholar]
- 6.Ley K., Laudanna C., Cybulsky M. I., Nourshargh S., Getting to the site of inflammation: The leukocyte adhesion cascade updated. Nat. Rev. Immunol. 7, 678–689 (2007). [DOI] [PubMed] [Google Scholar]
- 7.McEver R. P., Zhu C., Rolling cell adhesion. Annu. Rev. Cell Dev. Biol. 26, 363–396 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sundd P., Pospieszalska M. K., Ley K., Neutrophil rolling at high shear: Flattening, catch bond behavior, tethers and slings. Mol. Immunol. 55, 59–69 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lehr J. E., Pienta K. J., Preferential adhesion of prostate cancer cells to a human bone marrow endothelial cell line. J. Natl. Cancer Inst. 90, 118–123 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Schweitzer K. M., Dräger A. M., van der Valk P., Thijsen S. F. T., Zevenbergen A., Theijsmeijer A. P., van der Schoot C. E., Langenhuijsen M., Constitutive expression of E-selectin and vascular cell adhesion molecule-1 on endothelial cells of hematopoietic tissues. Am. J. Pathol. 148, 165–175 (1996). [PMC free article] [PubMed] [Google Scholar]
- 11.Frenette P. S., Subbarao S., Mazo I. B., von Andrian U. H., Wagner D. D., Endothelial selectins and vascular cell adhesion molecule-1 promote hematopoietic progenitor homing to bone marrow. Proc. Natl. Acad. Sci. U.S.A. 95, 14423–14428 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hidalgo A., Weiss L. A., Frenette P. S., Functional selectin ligands mediating human CD34+ cell interactions with bone marrow endothelium are enhanced postnatally. J. Clin. Invest. 110, 559–569 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazo I. B., Gutierrez-Ramos J.-C., Frenette P. S., Hynes R. O., Wagner D. D., von Andrian U. H., Hematopoietic progenitor cell rolling in bone marrow microvessels: Parallel contributions by endothelial selectins and vascular cell adhesion molecule 1. J. Exp. Med. 188, 465–474 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abbal C., Lambelet M., Bertaggia D., Gerbex C., Martinez M., Arcaro A., Schapira M., Spertini O., Lipid raft adhesion receptors and Syk regulate selectin-dependent rolling under flow conditions. Blood 108, 3352–3359 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Hocde S. A., Hyrien O., Waugh R. E., Cell adhesion molecule distribution relative to neutrophil surface topography assessed by TIRFM. Biophys. J. 97, 379–387 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore K. L., Patel K. D., Bruehl R. E., Li F. G., Johnson D. A., Lichenstein H. S., Cummings R. D., Bainton D. F., McEver R. P., P-selectin glycoprotein ligand-1 mediates rolling of human neutrophils on P-selectin. J. Cell Biol. 128, 661–671 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miner J. J., Xia L., Yago T., Kappelmayer J., Liu Z., Klopocki A. G., Shao B., McDaniel J. M., Setiadi H., Schmidtke D. W., McEver R. P., Separable requirements for cytoplasmic domain of PSGL-1 in leukocyte rolling and signaling under flow. Blood 112, 2035–2045 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shao B. J., Yago T., Coghill P. A., Klopocki A. G., Mehta-D’souza P., Schmidtke D. W., Rodgers W., McEver R. P., Signal-dependent slow leukocyte rolling does not require cytoskeletal anchorage of P-selectin glycoprotein ligand-1 (PSGL-1) or Integrin αLβ2. J. Biol. Chem. 287, 19585–19598 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snapp K. R., Heitzig C. E., Kansas G. S., Attachment of the PSGL-1 cytoplasmic domain to the actin cytoskeleton is essential for leukocyte rolling on P-selectin. Blood 99, 4494–4502 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Betzig E., Patterson G. H., Sougrat R., Lindwasser O. W., Olenych S., Bonifacino J. S., Davidson M. W., Lippincott-Schwartz J., Hess H. F., Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Heilemann M., van de Linde S., Schuttpelz M., Kasper R., Seefeldt B., Mukherjee A., Tinnefeld P., Sauer M., Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed. Engl. 47, 6172–6176 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Huang B., Wang W., Bates M., Zhuang X., Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.AbuSamra D. B., Al-Kilani A., Hamdan S. M., Sakashita K., Gadhoum S. Z., Merzaban J. S., Quantitative characterization of E-selectin interaction with native CD44 and P-selectin glycoprotein ligand-1 (PSGL-1) using a real time immunoprecipitation-based binding assay. J. Biol. Chem. 290, 21213–21230 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merzaban J. S., Burdick M. M., Gadhoum S. Z., Dagia N. M., Chu J. T., Fuhlbrigge R. C., Sackstein R., Analysis of glycoprotein E-selectin ligands on human and mouse marrow cells enriched for hematopoietic stem/progenitor cells. Blood 118, 1774–1783 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finger E. B., Puri K. D., Alon R., Lawrence M. B., von Andrian U. H., Springer T. A., Adhesion through L-selectin requires a threshold hydrodynamic shear. Nature 379, 266–269 (1996). [DOI] [PubMed] [Google Scholar]
- 26.Ali A. J., Abuelela A. F., Merzaban J. S., An analysis of trafficking receptors show that CD44 and P-selectin glycoprotein ligand-1 collectively control the migration of activated human T-Cells. Front. Immunol. 8, 492 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawrence M. B., Kansas G. S., Kunkel E. J., Ley K., Threshold levels of fluid shear promote leukocyte adhesion through selectins (CD62L,P,E). J. Cell Biol. 136, 717–727 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dimitroff C. J., Lee J. Y., Rafii S., Fuhlbrigge R. C., Sackstein R., CD44 is a major E-selectin ligand on human hematopoietic progenitor cells. J. Cell Biol. 153, 1277–1286 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tokunaga M., Imamoto N., Sakata-Sogawa K., Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods 5, 159–161 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Sundd P., Gutierrez E., Pospieszalska M. K., Zhang H., Groisman A., Ley K., Quantitative dynamic footprinting microscopy reveals mechanisms of neutrophil rolling. Nat. Methods 7, 821–824 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y., Yago T., Zhang N., Abdisalaam S., Alexandrakis G., Rodgers W., McEver R. P., Cytoskeletal regulation of CD44 membrane organization and interactions with E-selectin. J. Biol. Chem. 289, 35159–35171 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Andrian U. H., Hasslen S. R., Nelson R. D., Erlandsen S. L., Butcher E. C., A central role for microvillous receptor presentation in leukocyte adhesion under flow. Cell 82, 989–999 (1995). [DOI] [PubMed] [Google Scholar]
- 33.Gál I., Lesley J., Ko W., Gonda A., Stoop R., Hyman R., Mikecz K., Role of the extracellular and cytoplasmic domains of CD44 in the rolling interaction of lymphoid cells with hyaluronan under physiologic flow. J. Biol. Chem. 278, 11150–11158 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Shao J.-Y., Ting-Beall H. P., Hochmuth R. M., Static and dynamic lengths of neutrophil microvilli. Proc. Natl. Acad. Sci. U.S.A. 95, 6797–6802 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidtke D. W., Diamond S. L., Direct observation of membrane tethers formed during neutrophil attachment to platelets or P-selectin under physiological flow. J. Cell Biol. 149, 719–729 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sundd P., Gutierrez E., Koltsova E. K., Kuwano Y., Fukuda S., Pospieszalska M. K., Groisman A., Ley K., ‘Slings’ enable neutrophil rolling at high shear. Nature 488, 399–403 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Legg J. W., Isacke C. M., Identification and functional analysis of the ezrin-binding site in the hyaluronan receptor, CD44. Curr. Biol. 8, 705–708 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Yonemura S., Hirao M., Doi Y., Takahashi N., Kondo T., Tsukita S., Tsukita S., Ezrin/radixin/moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of CD44, CD43, and ICAM-2. J. Cell Biol. 140, 885–895 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thorne R. F., Legg J. W., Isacke C. M., The role of the CD44 transmembrane and cytoplasmic domains in co-ordinating adhesive and signalling events. J. Cell Sci. 117, 373–380 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Evans E., Heinrich V., Leung A., Kinoshita K., Nano- to microscale dynamics of P-selectin detachment from leukocyte interfaces. I. Membrane separation from the cytoskeleton. Biophys. J. 88, 2288–2298 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park E. Y. H., Smith M. J., Stropp E. S., Snapp K. R., DiVietro J. A., Walker W. F., Schmidtke D. W., Diamond S. L., Lawrence M. B., Comparison of PSGL-1 microbead and neutrophil rolling: Microvillus elongation stabilizes P-selectin bond clusters. Biophys. J. 82, 1835–1847 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramachandran V., Williams M., Yago T., Schmidtke D. W., McEver R. P., Dynamic alterations of membrane tethers stabilize leukocyte rolling on P-selectin. Proc. Natl. Acad. Sci. U.S.A. 101, 13519–13524 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fay M. E., Myers D. R., Kumar A., Turbyfield C. T., Byler R., Crawford K., Mannino R. G., Laohapant A., Tyburski E. A., Sakurai Y., Rosenbluth M. J., Switz N. A., Sulchek T. A., Graham M. D., Lam W. A., Cellular softening mediates leukocyte demargination and trafficking, thereby increasing clinical blood counts. Proc. Natl. Acad. Sci. U.S.A. 113, 1987–1992 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown D. A., London E., Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J. Biol. Chem. 275, 17221–17224 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Simons K., Ikonen E., Functional rafts in cell membranes. Nature 387, 569–572 (1997). [DOI] [PubMed] [Google Scholar]
- 46.Shao B. J., Yago T., Setiadi H., Wang Y., Mehta-D’souza P., Fu J. X., Crocker P. R., Rodgers W., Xia L. J., McEver R. P., O-glycans direct selectin ligands to lipid rafts on leukocytes. Proc. Natl. Acad. Sci. U.S.A. 112, 8661–8666 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yago T., Shao B. J., Miner J. J., Yao L. B., Klopocki A. G., Maeda K., Coggeshall K. M., McEver R. P., E-selectin engages PSGL-1 and CD44 through a common signaling pathway to induce integrin αLβ2-mediated slow leukocyte rolling. Blood 116, 485–494 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Setiadi H., McEver R. P., Clustering endothelial E-selectin in clathrin-coated pits and lipid rafts enhances leukocyte adhesion under flow. Blood 111, 1989–1998 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuziemko G. M., Stroh M., Stevens R. C., Cholera toxin binding affinity and specificity for gangliosides determined by surface plasmon resonance. Biochemistry 35, 6375–6384 (1996). [DOI] [PubMed] [Google Scholar]
- 50.Serag M. F., Abadi M., Habuchi S., Single-molecule diffusion and conformational dynamics by spatial integration of temporal fluctuations. Nat. Commun. 5, 5123 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Serag M. F., Habuchi S., Conserved linear dynamics of single-molecule Brownian motion. Nat. Commun. 8, 15675 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dedecker P., Duwé S., Neely R. K., Zhang J., Localizer: Fast, accurate, open-source, and modular software package for superresolution microscopy. J. Biomed. Opt. 17, 126008 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/7/eaat5304/DC1
Fig. S1. The multistep paradigm of cell migration highlighting the main interactions that take place between the blood stem cell in flow and the endothelial cells lining the blood vessels of the bone marrow.
Fig. S2. Surface density of E-selectin on the protein A–deposited microfluidic chamber determined by immunofluorescence imaging.
Fig. S3. Flow cytometric analysis to determine the binding specificity of ligand-specific antibodies to CD44 on KG1a cells.
Fig. S4. Localization precisions of the SR localization microscopy experiments of CD44 on KG1a cells.
Fig. S5. Examples of the reconstructed SR images of CD44 on KG1a cells.
Fig. S6. SR images of CD44 on KG1a cells.
Fig. S7. Cluster analysis of the nanoscale architecture of lipid rafts on KG1a cells.
Fig. S8. Examples of the reconstructed SR images of CD44 on MβCD-treated KG1a cells.
Fig. S9. Cluster analysis of the nanoscale architecture of CD44 on KG1a cells.
Fig. S10. Expression of CD44 on untreated and MβCD-treated KG1a cells was determined by flow cytometry.
Fig. S11. Depth of the field in the SR localization microscopy imaging experiments with HILO configuration.
Movie S1. Time-lapse transmitted light microscopy images of KG1a cells perfused into the microfluidic chamber at the shear stress of 0.25 dyne cm−2.
Movie S2. Time-lapse transmitted light microscopy images of KG1a cells perfused into the microfluidic chamber at the shear stress of 0.5 dyne cm−2.
Movie S3. Time-lapse transmitted light microscopy images of KG1a cells perfused into the microfluidic chamber at the shear stress of 1.0 dyne cm−2.
Movie S4. Time-lapse transmitted light microscopy images of KG1a cells perfused into the microfluidic chamber at the shear stress of 2.0 dyne cm−2.
Movie S5. Time-lapse transmitted light microscopy images of KG1a cells perfused into the microfluidic chamber at the shear stress of 4.0 dyne cm−2.
Movie S6. Time-lapse transmitted light microscopy images of KG1a cells perfused into the microfluidic chamber in the presence of EDTA (10 mM) at the shear stress of 1.0 dyne cm−2.
Movie S7. Time-lapse transmitted light microscopy images of KG1a cells perfused into the microfluidic chamber at the shear stress of 1.0 dyne cm−2.
Movie S8. Time-lapse transmitted light microscopy images of MβCD-treated KG1a cells perfused into the microfluidic chamber at the shear stress of 1.0 dyne cm−2.


