Abstract
Objective
To investigate the rich-club organization in clinically isolated syndrome (CIS) and multiple sclerosis (MS), and to characterize its relationships with physical disabilities and cognitive impairments.
Methods
We constructed high-resolution white matter (WM) structural networks in 41 CIS, 32 MS and 35 healthy controls (HCs) using diffusion MRI and deterministic tractography. Group differences in rich-club organization, global and local network metrics were investigated. The relationship between the altered network metrics, brain lesions and clinical variables including EDSS, MMSE, PASAT, disease duration were calculated. Additionally, reproducibility analysis was performed using different parcellation schemes.
Results
Compared with HCs, MS patients exhibited a decreased strength in all types of connections (rich-club: p < 0.0001; feeder: p = 0.0004; and local: p = 0.0026). CIS patients showed intermediate values between MS patients and HCs and exhibited a decreased strength in feeder and local connections (feeder: p = 0.019; and local: p = 0.031) but not in rich-club connections. Compared with CIS patients, MS patients showed significant reductions in rich-club connections (p = 0.0004). The reduced strength of rich-club and feeder connections was correlated with cognitive impairments in the MS group. These results were independent of lesion distribution and reproducible across different brain parcellation schemes.
Conclusion
The rich-club organization was disrupted in MS patients and relatively preserved in CIS. The disrupted rich-club connectivity was correlated with cognitive impairment in MS. These findings suggest that impaired rich-club connectivity is an essential feature of progressive structural network disruption, heralding the development of clinical disability in MS.
Abbreviations: MS, multiple sclerosis; CIS, clinically isolated syndrome; MRI, magnetic resonance imaging; DTI, diffusion tensor imaging; PASAT, paced auditory serial attention test; EDSS, expanded disability status scale; MMSE, mini-mental state examination
Keywords: Multiple sclerosis, Clinically isolated syndrome, Diffusion MRI, Rich-club, Brain network, Graph theory
Highlights
-
•
The rich-club organization was disrupted in MS patients and preserved in CIS.
-
•
The disrupted rich-club connectivity correlated with cognitive impairment in MS.
-
•
The rich-club results are reproducible across data analysis methods.
1. Introduction
Multiple sclerosis (MS) is an inflammatory demyelinating disease of the central nervous system and is the main cause of non-traumatic neurological disability in young adults (Compston and Coles, 2008). Complex pathological alterations including white matter demyelination, axonal injury, and primary grey matter (GM) damage were observed in MS and contribute disability progression and cognitive impairment (Reich et al., 2018). Graph theory studies have shown disrupted topological organization of structural and functional brain networks in MS and even in the early stages of this disease (clinically isolated syndrome, CIS)(Filippi et al., 2013; Fleischer et al., 2017; He et al., 2009; Liu et al., 2016; Pardini et al., 2015; Rocca et al., 2016; Schoonheim et al., 2013; Shu et al., 2016; Shu et al., 2011; Stam, 2014; Tewarie et al., 2015; Van Schependom et al., 2014), and such network changes have been associated with physical disabilities and cognitive impairments (Hawellek et al., 2011; Pardini et al., 2015; Shu et al., 2016; Stam, 2014).
When modeling the whole brain as a complex network, recent studies have demonstrated the existence of a number of highly connected and highly central neocortical hub regions (rich club), which play a key role in global information integration between different parts of the network (Bullmore and Sporns, 2009; van den Heuvel and Sporns, 2013). Emerging evidence has suggested that such central or “rich-club” regions are vulnerable to various brain disorders, such as Alzheimer's disease and schizophrenia (Crossley et al., 2014; Stam, 2014; van den Heuvel et al., 2013). Using diffusion MRI, the structural connectome can be mapped with fiber tractography techniques (Gong et al., 2009; Hagmann et al., 2007). Hub regions of structural brain networks are primarily distributed in higher-order association areas (van den Heuvel and Sporns, 2013). Damage to hub nodes in the association cortex, particularly disruptions within the default-mode network, has been reported in MS (Filippi et al., 2013; Stam, 2014). However, the pattern of alterations in the rich-club organization in MS, particularly at the CIS stage, remains largely unknown.
In the present study, we investigated the rich-club organization in CIS and MS, and characterized its relationships with physical disabilities and cognitive impairments. Additionally, the distribution of WM lesions on the rich-club organization in patients and the reproducibility of the results were examined.
2. Materials and methods
2.1. Standard protocol approvals, registrations, and patient consents
The study was approved by the local Institutional Medical Ethics Committee and all participants gave written informed consent.
2.2. Participants
We recruited 41 CIS patients (optic neuritis, n = 18; spinal cord syndrome, n = 16; brainstem syndrome n = 5; cerebellar syndrome, n = 2), 32 relapsing-remitting MS patients and 35 healthy controls (HC). All CIS patients were prospectively examined within 6 months from onset with a single clinical episode suggestive of MS (Miller et al., 2012) before steroid treatment. Fifteen CIS patients fulfilled the 2010 modified McDonald Criteria for dissemination in space, while none fulfilled for dissemination in time. Other patients (n = 26) presented with normal brain MRI. All RRMS patients were diagnosed according to the 2010 modified McDonald Criteria (Polman et al., 2011) and required to be relapse-free and without treatment with disease-modifying medications or steroids in the 2 weeks prior to MRI scanning. All of the participants were right-handed, as measured by the Edinburgh Inventory (Oldfield, 1971).
2.3. Clinical evaluation
The main demographic and clinical characteristics (Table 1), including age, gender, the Expanded Disability Status Scale (EDSS) score (Kurtzke, 1983), the disease duration, the Mini-Mental State Examination (MMSE) and the Paced Auditory Serial Addition Test (PASAT2 and PASAT3 versions) were assessed by an experienced neurologist (J.Y.). Findings of the graph analysis of brain structural and functional networks in the same cohort have been reported before (Shu et al., 2016). The current study focused on the rich-club organizations using high resolution (1024 ROIs) network analysis in CIS and MS.
Table 1.
The demographic information and clinical characteristics of all participants.
| Controls (n = 35) |
CIS (n = 41) |
MS (n = 32) |
F/T/χ2/Z value | P value | |
|---|---|---|---|---|---|
| Age (years) | 35.0 ± 11.5 | 35.7 ± 10.7 | 34.8 ± 8.3 | 0.08a | 0.92a |
| Gender (F/M) | 23/12 | 26/15 | 24/8 | 1.17c | 0.56c |
| MMSE | 29.1 ± 1.3 | 27.6 ± 1.4 | 25.9 ± 1.8 | 36.44a | <0.001a |
| PASAT2 | 46.7 ± 9.2 | 39.4 ± 7.7 | 35.4 ± 9.8 | 14.14a | <0.001a |
| PASAT3 | 53.9 ± 6.2 | 47.5 ± 7.6 | 41.0 ± 8.7 | 24.38a | <0.001a |
| Disease duration (months) | – | 2.6 ± 2.5 | 41.8 ± 28.7 | 8.73b | <0.001b |
| TWMLL (ml) | – | 4.5 ± 10.6 | 10.3 ± 10.4 | 2.33b | 0.023b |
| EDSS (range) | – | 2.0 (0–6) | 3.5 (0–6.5) | 3.35d | <0.001d |
CIS, clinically isolated syndrome; MS, multiple sclerosis; MMSE = Mini-Mental State Examination; PASAT = Paced Auditory Serial Attention Test; EDSS = Expanded Disability Status Scale; TWMLL = Total White Matter Lesion Load.
Data are shown as mean ± standard deviation, except for EDSS presented in median (range).
P value and F value were obtained using one-way analysis of variance.
P value and T value were obtained using a two-sample t-test.
P value and χ2 value were obtained using the χ2 test.
P value and Z value were obtained using the Wilcoxon rank sum test.
2.4. MRI data acquisition
The MRI data were acquired using a SIEMENS TRIO 3.0 T scanner in the Department of Radiology, Xuanwu Hospital, Capital Medical University. All participants underwent high-quality MRI scanning, which included a 3D T1-weighted magnetization prepared rapid acquisition gradient echo (MPRAGE) scan [176 sagittal slices, repetition time (TR) = 1600 ms, echo time (TE) = 2.13 ms, field of view (FOV) = 224 × 256 mm2, voxel size = 1 × 1 × 1 mm3]; a T2-weighted MRI scan (35 axial slices, TR = 5000 ms, TE = 87 ms, FOV = 256 × 256 mm2, voxel size = 1 × 1 × 4 mm3); and a diffusion MRI scan (60 axial slices, TR = 11,000 ms, TE = 98 ms, FOV = 256 × 256 mm2, voxel size = 2 × 2 × 2 mm3, 30 diffusion directions with b = 1000 s/mm2, and an additional b0 image, average = 2).
2.5. MRI data preprocessing
2.5.1. DTI data preprocessing
The preprocessing procedure for DTI data included the eddy current and motion artifact correction, diffusion tensor estimation and fractional anisotropy (FA) calculation. First, the eddy current distortions and motion artifacts in the DTI dataset were corrected by applying an affine alignment of the diffusion-weighted images to the b0 image. The b-matrix was accordingly reoriented based on the transformation matrix. Subsequently, the diffusion tensor was estimated by solving the Stejskal and Tanner equation (Basser et al., 1994), and the reconstructed tensor matrix was diagonalized to obtain 3 eigenvalues (λ1, λ2, and λ3) and their corresponding eigenvectors. The FA value of each voxel was also calculated (Basser and Pierpaoli, 1996). The preprocessing of the DTI data was performed using the FMRIB Diffusion Toolbox (FDT) in FSL (version 5.0, http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FDT).
2.5.2. Measurement of WM lesion load
Hyperintense WM lesions in MS patients were manually delineated on the T2-weighted images by an experienced radiologist blinded to the clinical details (Y.L.). The lesions were delineated at 3-month intervals in 10 patients, and the intra-rater reliability was 95.2%. A binary lesion mask for each patient was obtained by setting the values within the WM lesion borders to 1 and 0 otherwise. Then the total WM lesion load (TWMLL) for each patient was calculated.
2.6. Brain network construction
Nodes and edges are the two fundamental elements of a network. In the present study, we constructed individual high-resolution structural network using the following procedures.
2.6.1. Network node definition
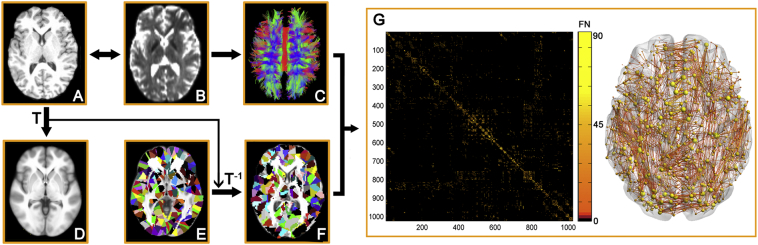
The Automated Anatomical Labeling (AAL) template (Tzourio-Mazoyer et al., 2002) was subdivided into 1024 regions of interest (ROIs) of equal size (H-1024) to define the network nodes (Zalesky et al., 2010). This procedure has been previously described (Bai et al., 2012; Zhao et al., 2015) and was performed using SPM8 software (http://www.fil.ion.ucl.ac.uk/spm/software/spm8). Briefly, the individual T1-weighted images were co-registered to the b0 images in the native DTI space. The transformed T1 images were then non-linearly transformed to the ICBM152 T1 template in the Montreal Neurological Institute (MNI) space. Inverse transformations were used to warp the H-1024 template from the MNI native DTI space. Using this procedure, we obtained 1024 cortical and subcortical ROIs, each representing a node of the network (Fig. 1).
Fig. 1.
The flowchart of structural network construction. (A) Individual T1 images and H-1024 template were used for automatic parcellation of the brain into 1024 regions, forming the nodes of the individual brain networks. (B) Streamline tractography was applied to the diffusion MRI data to reconstruct the white matter pathways. From the set of reconstructed streamlines, the streamlines that interconnected regions i and j from the set of 1024 regions were taken as an edge between nodes i and j in the structural brain network. The streamline count represents the weight of the connection and was aggregated into a structural connectivity (SC) matrix (C). (D) The matrices and 3D representations (lateral view) of the structural networks of a representative healthy subject are shown in the right panel. The nodes are located according to their centroid stereotaxic coordinates, and the edges are coded according to their connection weights. For details, see the Materials and Methods section.
2.6.2. High-resolution structural network
First, diffusion MRI tractography was performed using the Diffusion Toolkit (http://www.trackvis.org/dtk/) based on the “fiber assignment by continuous tracking” method (Mori et al., 1999). All tracts in the DTI dataset were computed by seeding each voxel with an FA >0.2. The tractography was terminated if it turned an angle >45 degrees or reached a voxel with an FA <0.2(Mori et al., 1999). As a result, all fiber pathways between the 1024 ROIs in the brain were constructed using deterministic tractography method. For each pair of ROIs i and j, we defined the number of the connected fiber streamlines (FN) with two end points located in these two regions as the weight of the edge between ROIs i and j. Therefore, for each participant, an FN-weighted 1024 × 1024 structural connectivity matrix was constructed (Fig. 1).
2.7. Network analysis
2.7.1. Rich-club organization
Based on the individual weighted structural network, we calculated the rich-club coefficient (RC) and normalized RC under a range of thresholds (k = 10–35) according to van den Heuvel and Sporns (2011). For the weighted networks, the rich-club coefficient ϕw(k) is defined as:
with E>k denoting the subset of edges between the hub nodes with a degree > k, W>k denoting the total sum weights of this subset and with Wranked denoting the ranked collection of strongest weights in the total network, with weights W representing the number of fiber streamlines of the edges. ϕ(k) was normalized relative to the ϕrandom(k) of a set of comparable random networks (n = 1000) of equal size and degree sequence, yielding a normalized RC ϕnorm(k) = ϕ(k)/ϕrandom(k). Here, threshold k is defined as the nodal degree range from 10 to 35. To identify the hub regions of the brain structural network, we constructed a structural backbone network by detecting the significant nonzero connections across all subjects with a nonparametric one-tailed sign test (p < 0.05, corrected) and identified the hub regions by sorting the nodal degree (K(i) > mean + std). Based on the categorization of the nodes of the backbone network into hub and non-hub regions, the edges of the network were classified into rich-club connections, linking hub-nodes to other hub-nodes, into feeder connections, linking hub-nodes to non-hub nodes and into local connections, linking non-hub nodes to non-hub nodes. Moreover, the connection strength of rich-club, feeder and local connections were calculated for each participant.
2.7.2. Global network organization
For the global network metrics, we quantified the network strength (Sp), global efficiency (Eglob), local efficiency (Eloc), shortest path length (Lp), clustering coefficient (Cp) and small-world parameters (λ, γ and σ). See Rubinov and Sporns (Rubinov and Sporns, 2010) and appendix e-1 for the detailed definitions of these network measures.
All network analyses were performed using in-house GRETNA software (http://www.nitrc.org/projects/gretna/)(Wang et al., 2015) and the results were visualized using BrainNet Viewer software (https://www.nitrc.org/projects/bnv/)(Xia et al., 2013).
2.8. WM lesion distribution on the rich-club organization
To investigate the WM lesion distribution on the rich-club organization in each patient, we calculated the proportion of WM lesions as the voxel number of WM lesion in each category of connections divided by the total voxel number along the fiber pathways comprising each type of connections.
2.9. Statistical analysis
2.9.1. Between group differences
Analyses were performed using SPSS software (Version 18; SPSS Inc., Chicago, USA). Kolmogorov-Smirnov tests and visual inspection of histograms were used to assess normality of the variables, and further analysis was based on normality. Demographic factor and clinical scores including age, MMSE, PASAT2 and PASAT3 among the three groups were compared using one-way analysis of variance (ANOVA). Gender distribution was compared using the Chi-square test. Disease duration and TWMLL between CIS and MS groups were compared using a two-sample t-test, and the EDSS scores were compared using the Wilcoxon rank-sum test. Group comparisons of network metrics were performed using one-way analysis of covariance (ANCOVA), and post hoc pairwise comparisons were performed using a general linear regression model when the ANCOVA model yielded significant results (p < 0.05). The effects of age and gender were removed in the statistical analysis.
2.9.2. Network-based statistic (NBS)
To localize the specific connected components in which the structural connectivity differed between each pair of groups, we used a NBS approach (Zalesky et al., 2010). For a detailed description, see appendix e-1 and Zalesky et al. (2010).
2.9.3. Relationships between network metrics and clinical scores
To investigate the relationship between the altered network metrics and clinical variables, partial correlation coefficients were calculated for the CIS and MS groups separately, while removing the effects of age and gender.
2.10. Reproducibility analysis
In addition to the high-resolution (H-1024) parcellation, we also used a low-resolution AAL (L-AAL) template with 90 brain regions to define network nodes. For each participant, the L-AAL FN-weighted WM network was constructed and the rich-club organization was investigated.
3. Results
3.1. Demographic and clinical characteristics
There were no significant differences in age or gender distribution among the three groups. As for the clinical variables, both the CIS and MS patient groups exhibited lower MMSE, PASAT2 and PASAT3 scores than the HCs, and the MS patients exhibited lower MMSE and PASAT scores than the CIS group (all p < 0.05). Additionally, the MS patients had larger TWMLL, longer disease duration and higher EDSS scores than the CIS patients (all p < 0.05) (Table 1).
3.2. Group differences in global network metrics
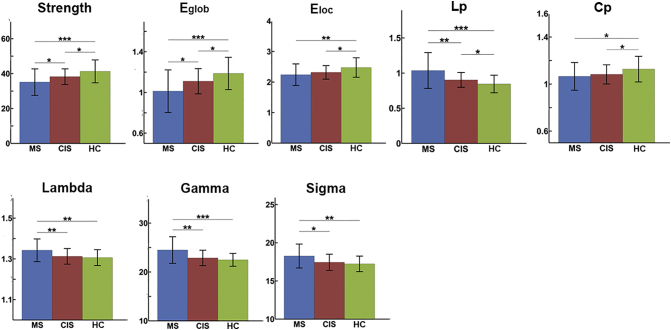
Both the patients and control subjects showed a small-world topology (lambda ≈ 1, sigma >1) of the brain structural network. Among the three groups, significant group effects in all global network metrics were observed (all p < 0.05) (Fig. S1). See appendix e-1 for the detailed descriptions of group differences in global network metrics.
Fig. S1.
Group differences in the global network metrics. The bars and error bars represent the mean values and standard deviations of the network properties in each group after removing the effects of age and gender. Significantly reduced strength, global efficiency, local efficiency, clustering and increased shortest path length of the structural networks were observed in both CIS and MS patients relative to the controls (HC). *: p < 0.05; **: p < 0.01; ***: p < 0.005.
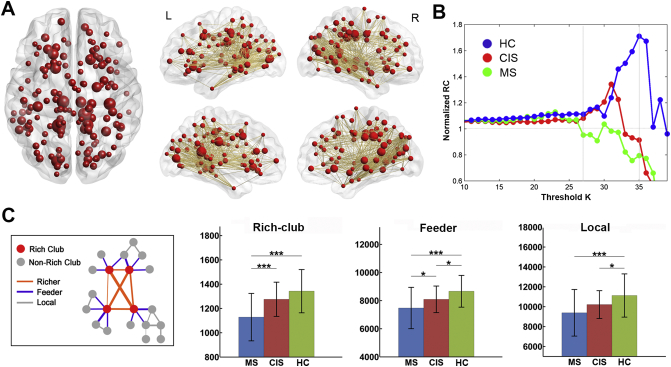
3.3. Group differences in rich-club organization
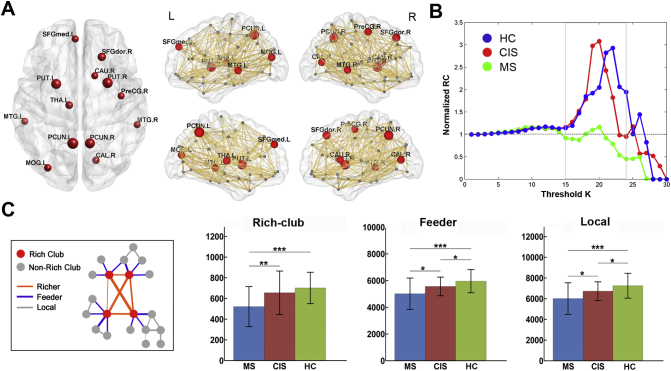
The hub regions of the structural backbone network were primarily distributed in the bilateral medial parietal, lateral temporal and prefrontal regions (Fig. 2A), which was consistent with the findings from previous studies (Shu et al., 2011; van den Heuvel and Sporns, 2011). The characteristic rich-club organization of structural networks (normalized RC > 1) was observed for both CIS patients and controls under a range of thresholds (k = 25–35). However, the MS patients showed a reduced normalized RC compared with the other groups from k = 27 to k = 35 (p < 0.05) (Fig. 2B). Moreover, significant group differences in the connection strength of the rich-club, feeder and local connections were identified (Fig. 2C). All of these connections exhibited a decreased strength in MS patients (rich-club: p < 0.0001; feeder: p = 0.0004; local: p = 0.0026). CIS patients showed intermediate values between MS patients and HCs, and exhibited a decreased strength in feeder and local connections (feeder: p = 0.019; local: p = 0.031) but not in the rich-club connections (p = 0.08). The MS group showed a significant decrease in the rich-club connections compared with the CIS group (p = 0.0004). The above results remained largely unchanged when dividing CIS into subgroups with normal or abnormal brain MRI, comparing with MS and HC. For the CIS subgroups (with normal or abnormal brain MRI), no significant differences were observed in the strength of rich-club, feeder and local connections (rich-club: p = 0.19; feeder: p = 0.35; local: p = 0.08).
Fig. 2.
Rich-club organization of high-resolution brain structural connectome. (A) Hub distribution of structural backbone network across all subjects. Network hubs are represented as nodes in red, with nodal size indicating the degree of the regions. (B) The mean normalized RC coefficient curve under a series of thresholds k for each group. (C) Group differences in the strength of the rich-club, feeder and local connections. The bars and error bars represent the mean values and standard deviations of the connection strength in each group after removing the effects of age and gender. *: p < 0.05; **: p < 0.01; ***: p < 0.005.
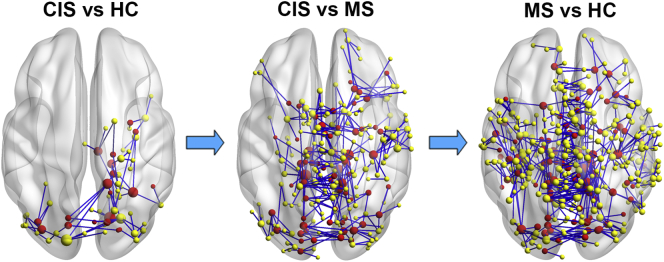
3.4. Group differences in structural connectivity
NBS analysis revealed disrupted components in structural networks for the CIS and MS patients (Fig. 3). A single connected network with 63 nodes and 66 connections was altered in CIS patients compared with the HCs (p = 0.039, corrected), comprising the bilateral occipital regions, right parahippocampal gyrus, precuneus, inferior temporal gyrus, precentral gyrus, insular and thalamus. For the MS patients, a single connected network comprising 372 nodes and 442 edges was altered (p = 0.0001, corrected). The nodes were widely distributed across the entire brain. All of the connections showed decreased values in the patients compared with the HCs. Compared with the CIS group, the MS group showed a disrupted network composed of 247 nodes and 279 edges (p < 0.0001, corrected), primarily involved in bilateral homotopic regions (such as bilateral precuneus, posterior and median cingulate gyrus, paracentral lobule, supplementary motor area, thalamus, putamen, cuneus and middle occipital gyrus) along the cortical midline and partly overlapped with the hub regions (30%).
Fig. 3.
Disrupted structural connectivity in CIS and MS patients. Connected components showing decreased structural connectivity were identified between CIS vs. controls, MS vs. CIS and MS vs. controls (HC) (p < 0.05, corrected). The nodes and connections were mapped onto the cortical surfaces using in-house BrainNet viewer software. The nodes in red represent the hub regions of the backbone network.
3.5. Relationship between network metrics and clinical variables
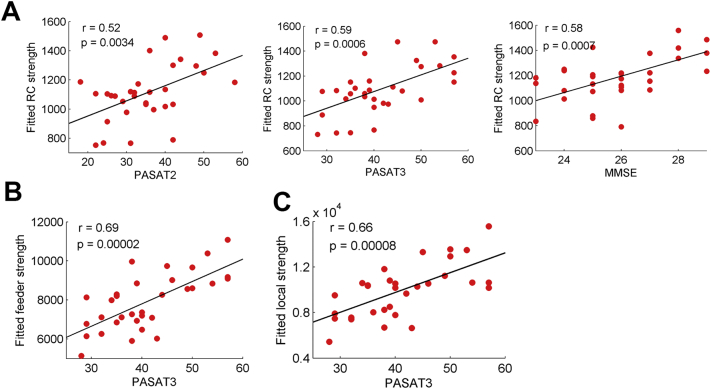
Significant clinical correlations were observed only in the MS group. First, the rich-club connection strength was correlated with PASAT2 (r = 0.52; p = 0.0034), PASAT3 (r = 0.59; p = 0.0006) and MMSE (r = 0.58; p = 0.0007) but not with EDSS (p = 0.14) (Fig. 4A). Second, the feeder and local connection strength was correlated with the PASAT3 (feeder: r = 0.69; p = 0.00002; local: r = 0.66; p = 0.00008) (Fig. 4B and C). No significant correlations were observed between network metrics and clinical variables in the CIS group.
Fig. 4.
Correlations between the rich-club metrics and clinical variables in patients with MS. (A) Plots showing the linear correlation between altered rich-club connection strength with PASAT2, PASAT3 and MMSE scores in MS patients (all p < 0.05, corrected). (B) Plots showing the linear correlation between altered feeder connection strength with PASAT3 score in MS patients. (C) Plots showing the linear correlation between altered local connection strength with PASAT3 score in MS patients. The red dots represent the adjusted values of MS patients after controlling for age and gender.
3.6. Proportion of WM lesions on the rich-club, feeder and local connections
The following mean proportions of WM lesions in the MS group were observed: rich-club (2.53%), feeder (1.90%) and local (1.55%). For the CIS group, the mean proportions of WM lesion were: rich-club (0.89%), feeder (0.75%) and local (0.60%).
3.7. Reproducibility findings
Based on the backbone of the L-AAL network across all subjects, we determined that the hub regions were primarily distributed in the bilateral precuneus, putamen, middle temporal gyrus, superior frontal gyrus, right caudate and left middle occipital gyrus (Fig. S2A), which was consistent with the findings from previous studies (Shu et al., 2011; van den Heuvel and Sporns, 2011). The characteristic rich-club organization of the L-AAL structural networks (normalized RC > 1) was observed for both CIS patients and controls under a range of thresholds (k = 15–24). However, the MS patients showed a reduced normalized RC compared with the other groups (p < 0.05) (Fig. S2B). Moreover, significant differences in the connection strength of the rich-club, feeder and local connections were identified (Fig. S2C). All connections exhibited a decreased strength in MS patients (rich-club: p < 10−4; feeder: p = 0.0003; local: p = 0.0005). CIS patients showed intermediate values of all connection strengths and a decreased strength in feeder and local connections (feeder: p = 0.031; local: p = 0.033). Between the CIS and MS patients, the most significant decreases were in the rich-club connections (p = 0.009). These results were consistent with the findings obtained from the H-1024 networks.
Fig. S2.
Reproducibility of the rich-club organization in L-AAL network. (A) Hub distribution of structural backbone network across all subjects. Network hubs are represented with nodes in red, with nodal size indicating the degree of the regions. (B) Mean normalized RC coefficient curve under a series of thresholds k for each group. (C) Group differences in the strength of the rich-club, feeder and local connections. The bars and error bars represent the mean values and standard deviations of the connection strength in each group after removing the effects of age and gender. *: p < 0.05; **: p < 0.01; ***: p < 0.005.
4. Discussion
Using diffusion MRI and graph theory approaches, we investigated the rich-club organization of brain networks in the patients with MS and CIS. Compared with the HCs, the MS patients exhibited disrupted rich-club organization with a reduced rich-club coefficient and decreased rich-club connections, and CIS patients showed intermediate levels of alterations. Importantly, the differences between CIS and MS were primarily located in the rich-club component. The network alterations, particularly the disrupted rich-club connections, were correlated with cognitive scores in MS.
Decreased connection strength in rich-club, feeder and local connections were observed in MS patients, suggesting widespread brain disconnection, including both long- and short-range connections. This is consistent with findings of disrupted global and local network efficiency in MS from different MRI modality including morphometric MRI (He et al., 2009), DTI (Shu et al., 2016) and fMRI (Rocca et al., 2016) collectively defining MS as a disconnection syndrome from a system perspective. Although disconnection were common observation in the network studies, there were inconsistencies in group difference and clinical correlation between existing studies, which might be attributable to the heterogeneity of the patient groups as well as to differences in imaging parameters and methods of analysis (Filippi et al., 2017). Further multi-center study using standardized protocol is warranted to clarify the network alterations in MS.
Various underlying pathological factors such as axonal damage, demyelination and gliosis may contribute to the network disconnection (Ciccarelli et al., 2014). Grey matter damage is another main pathological alteration in MS, the relationship between the grey matter damage and white matter disconnection is unclear and need further investigations (Calabrese et al., 2015).
The current study extended the results to the earliest stage of MS (CIS). CIS patients showed intermediate values between MS patients and controls, and exhibited a decreased strength in feeder and local connections, whereas rich-club connections were largely preserved. This interesting finding suggests that WM lesions and normal-appearing WM changes primarily affect local connections at disease onset and that rich-club disruption is not an early biomarker for MS. The difference between CIS and MS is alterations in rich-club connections, but not feeder or local connections, suggesting that rich-club disruption is the key substrate for disease progression (from CIS to MS). Rich-club components play a key role in global information integration between different parts of the network and are highly vulnerable to damage (van den Heuvel and Sporns, 2013). The important roles of rich hub regions in information integration make these areas more likely to be symptomatic, consistent with the correlation analyses in this study. Recently, the “hub overload and failure” theory was proposed as a common pathway of different neurological disorders (Crossley et al., 2014; Stam, 2014). The findings of the present study also support this hypothesis in MS: the rich-club hubs are not affected at the early stage, but subsequently become affected as the disease progresses. Furthermore, the functional consequence of rich-club damage is likely clinical disability, particularly cognitive impairment.
The NBS analysis revealed that the alterations between CIS and MS were primarily located in brain areas overlapped with rich-club connections and hub regions, such as the bilateral precuneus. These NBS findings validated the rich-club results, suggesting that rich-club damage but not disruptions in feeder or local connections is strongly associated with disease progression. Specifically deep grey matter such as thalamus alteration can be observed in both CIS and MS, implying deep grey matter changes are early and specific imaging features of MS (Liu et al., 2015).
Brain atrophy and diffusion abnormalities were correlated with clinical disability in MS in numerous previous studies (Filippi et al., 2017; Sastre-Garriga et al., 2017). However, the brain is an integrative complex network, the network metrics connecting the different brain regions are comprehensive measures to reflect the consequence of multiple pathological alterations and improving the clinical correlations (Filippi et al., 2013). Cognitive scores (PASAT2, PASAT3 and MMSE) were correlated with network metrics, particularly the strength of rich-club connections, suggesting that the rich-club connection may provide potential biomarkers for assessing and monitoring cognitive impairment in MS. No significant correlations between network metrics and clinical variables were observed in CIS, implying that the more peripheral network changes in CIS may represent a transitional phase.
To illustrate the WM lesion effects on the hub distributions in MS patients, we observed that the WM lesions were equally distributed across the rich-club, feeder and local connections. Additionally, we utilized both high and low resolutions of brain parcellation schemes to investigate the rich-club changes, and the results were consistent. Taken together, these findings imply that our rich-club results are robust: they are independent of WM lesions and that they are reproducible across different brain parcellation schemes.
Several limitations should be addressed. First, considering the cross-sectional design and relatively small sample size of the present study, the relationship between rich club connectivity and disease progression should currently be interpreted as preliminary. Future longitudinal studies with large sample size examining the alterations of the rich-club organization in MS patients are of particular interest in this regard. Second, the prevalence of CIS patients without visible brain lesions in this study is higher compared to other CIS cohorts. A possible bias selection of CIS patients with a low lesion burden might influence the results obtained, especially in the comparisons with MS patients. Third, deterministic tractography was used for the reconstruction of WM tracts, which may result in the loss of existing fibers due to “fiber crossing” problem (Mori and van Zijl, 2002). Future studies could employ more advanced tractography techniques, such as probabilistic tractography, to define the network edges. Additionally, the presence of WM lesions in MS or CIS patients could partially influence tractography results, inducing a loss or a reduction of connections measured. To overcome this limitation, some possibilities have been suggested in the previous literature (Fagerholm et al., 2015), such as the creation of an atlas of connection from controls. Finally, comprehensive neuropsychological tests (e.g., Minimal Assessment of Cognitive Function in Multiple Sclerosis, MACFIMS) for MS patients should be examined to evaluate the relationship between rich-club organization alterations and cognitive impairment in different cognitive domains.
5. Conclusion
The rich-club organization was disrupted in MS patients and relatively preserved in CIS. The disrupted rich-club connectivity was correlated with cognitive impairment in MS. These findings suggest that impaired rich-club connectivity is an essential feature of progressive structural network disruption, heralding the development of clinical disability, particularly cognitive impairment in MS.
The following are the supplementary data related to this article.
Financial disclosures
Dr. Ni Shu, Dr. Yunyun Duan, Dr. Jing Huang, Dr. Zhuoqiong Ren, Dr. Jing Ye, Dr. Huiqing Dong, Dr. Kuncheng Li and Dr. Yaou Liu report no disclosures.
Dr. Frederik Barkhof serves as a consultant for Bayer-Schering Pharma, Sanofi-Aventis, Biogen Idec, Teva, Merck Serono, Novartis, Roche, Synthon, and Jansen Research.
Author contributions
-
1
1 guarantor of integrity of the entire study: Yaou Liu.
-
2
study concepts: Yaou Liu; Frederik Barkhof.
-
3
study design: Yaou Liu; Ni Shu.
-
4
definition of intellectual content: Yaou Liu; Ni Shu.
-
5
literature research: Yaou Liu; Ni Shu.
-
6
clinical studies: Zheng Liu; Huiqiong Dong; Zhuoqiong Ren.
-
7
data acquisition: Yunyun Duan; Jing Huang; Zhuoqiong Ren; Jing Ye.
-
8
data analysis: Yaou Liu; Ni Shu; Yunyun Duan.
-
9
statistical analysis: Yaou Liu; Ni Shu.
-
10
manuscript preparation: Yaou Liu; Ni Shu.
-
11
manuscript editing: Yaou Liu; Ni Shu; Frederik Barkhof.
-
12
manuscript review: Yaou Liu; Kuncheng Li; Frederik Barkhof.
Acknowledgements
This work was supported by the ECTRIMS-MAGNMIS Fellowship from ECTRIMS (YL), the 973 program (Grant No. 2013CB837300, NS), the National Natural Science Foundation of China (Grant Nos. 81301284, 81471732,81571631, 81671761 and 30930029), the Beijing Natural Science fund (Grant No. 7133244, YL), and the Beijing Nova Program (Grant No. xx2013045, YL), the Fundamental Research Funds for the Central Universities (Grant No. 2013YB28, NS) and the Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (Grant No. ZYLX201609).
Contributor Information
Ni Shu, Email: nshu@bnu.edu.cn.
Kuncheng Li, Email: kunchengli55@gmail.com.
Yaou Liu, Email: yaouliu80@163.com.
References
- Bai F., Shu N., Yuan Y., Shi Y., Yu H., Wu D., Wang J., Xia M., He Y., Zhang Z. Topologically convergent and divergent structural connectivity patterns between patients with remitted geriatric depression and amnestic mild cognitive impairment. J. Neurosci. 2012;32:4307–4318. doi: 10.1523/JNEUROSCI.5061-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser P.J., Pierpaoli C. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. J. Magn. Reson. B. 1996;111:209–219. doi: 10.1006/jmrb.1996.0086. [DOI] [PubMed] [Google Scholar]
- Basser P.J., Mattiello J., LeBihan D. MR diffusion tensor spectroscopy and imaging. Biophys. J. 1994;66:259–267. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore E., Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- Calabrese M., Magliozzi R., Ciccarelli O., Geurts J.J., Reynolds R., Martin R. Exploring the origins of grey matter damage in multiple sclerosis. Nat. Rev. Neurosci. 2015;16:147–158. doi: 10.1038/nrn3900. [DOI] [PubMed] [Google Scholar]
- Ciccarelli O., Barkhof F., Bodini B., De Stefano N., Golay X., Nicolay K., Pelletier D., Pouwels P.J., Smith S.A., Wheeler-Kingshott C.A., Stankoff B., Yousry T., Miller D.H. Pathogenesis of multiple sclerosis: insights from molecular and metabolic imaging. Lancet Neurol. 2014;13:807–822. doi: 10.1016/S1474-4422(14)70101-2. [DOI] [PubMed] [Google Scholar]
- Compston A., Coles A. Multiple sclerosis. Lancet. 2008;372:1502–1517. doi: 10.1016/S0140-6736(08)61620-7. [DOI] [PubMed] [Google Scholar]
- Crossley N.A., Mechelli A., Scott J., Carletti F., Fox P.T., McGuire P., Bullmore E.T. The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain. 2014;137:2382–2395. doi: 10.1093/brain/awu132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerholm E.D., Hellyer P.J., Scott G., Leech R., Sharp D.J. Disconnection of network hubs and cognitive impairment after traumatic brain injury. Brain. 2015;138:1696–1709. doi: 10.1093/brain/awv075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippi M., van den Heuvel M.P., Fornito A., He Y., Hulshoff Pol H.E., Agosta F., Comi G., Rocca M.A. Assessment of system dysfunction in the brain through MRI-based connectomics. Lancet Neurol. 2013;12:1189–1199. doi: 10.1016/S1474-4422(13)70144-3. [DOI] [PubMed] [Google Scholar]
- Filippi M., Preziosa P., Rocca M.A. Brain mapping in multiple sclerosis: lessons learned about the human brain. NeuroImage. 2017 doi: 10.1016/j.neuroimage.2017.09.021. Sep 14, [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Fleischer V., Radetz A., Ciolac D., Muthuraman M., Gonzalez-Escamilla G., Zipp F., Groppa S. Graph theoretical framework of brain networks in multiple sclerosis: a review of concepts. Neuroscience. 2017 doi: 10.1016/j.neuroscience.2017.10.033. Nov 1, [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Gong G., He Y., Concha L., Lebel C., Gross D.W., Evans A.C., Beaulieu C. Mapping anatomical connectivity patterns of human cerebral cortex using in vivo diffusion tensor imaging tractography. Cereb. Cortex. 2009;19:524–536. doi: 10.1093/cercor/bhn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P., Kurant M., Gigandet X., Thiran P., Wedeen V.J., Meuli R., Thiran J.P. Mapping human whole-brain structural networks with diffusion MRI. PLoS One. 2007;2 doi: 10.1371/journal.pone.0000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawellek D.J., Hipp J.F., Lewis C.M., Corbetta M., Engel A.K. Increased functional connectivity indicates the severity of cognitive impairment in multiple sclerosis. Proc. Natl. Acad. Sci. U. S. A. 2011;108:19066–19071. doi: 10.1073/pnas.1110024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Dagher A., Chen Z., Charil A., Zijdenbos A., Worsley K., Evans A. Impaired small-world efficiency in structural cortical networks in multiple sclerosis associated with white matter lesion load. Brain. 2009;132:3366–3379. doi: 10.1093/brain/awp089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel M.P., Sporns O. Rich-club organization of the human connectome. J. Neurosci. 2011;31:15775–15786. doi: 10.1523/JNEUROSCI.3539-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel M.P., Sporns O. Network hubs in the human brain. Trends Cogn. Sci. 2013;17:683–696. doi: 10.1016/j.tics.2013.09.012. [DOI] [PubMed] [Google Scholar]
- van den Heuvel M.P., Sporns O., Collin G., Scheewe T., Mandl R.C., Cahn W., Goni J., Hulshoff Pol H.E., Kahn R.S. Abnormal rich club organization and functional brain dynamics in schizophrenia. JAMA Psychiat. 2013;70:783–792. doi: 10.1001/jamapsychiatry.2013.1328. [DOI] [PubMed] [Google Scholar]
- Kurtzke J.F. Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS) Neurology. 1983;33:1444–1452. doi: 10.1212/wnl.33.11.1444. [DOI] [PubMed] [Google Scholar]
- Liu Y., Duan Y., Huang J., Ren Z., Ye J., Dong H., Shi F.D., Barkhof F., Vrenken H., Wattjes M.P., Wang J., Li K. Multimodal quantitative MR imaging of the thalamus in multiple sclerosis and neuromyelitis optica. Radiology. 2015;277:784–792. doi: 10.1148/radiol.2015142786. [DOI] [PubMed] [Google Scholar]
- Liu Y., Wang H., Duan Y., Huang J., Ren Z., Ye J., Dong H., Shi F., Li K., Wang J. Functional brain network alterations in clinically isolated syndrome and multiple sclerosis: a graph-based connectome study. Radiology. 2016:152843. doi: 10.1148/radiol.2016152843. [DOI] [PubMed] [Google Scholar]
- Miller D.H., Chard D.T., Ciccarelli O. Clinically isolated syndromes. Lancet Neurol. 2012;11:157–169. doi: 10.1016/S1474-4422(11)70274-5. [DOI] [PubMed] [Google Scholar]
- Mori S., van Zijl P.C. Fiber tracking: principles and strategies - a technical review. NMR Biomed. 2002;15:468–480. doi: 10.1002/nbm.781. [DOI] [PubMed] [Google Scholar]
- Mori S., Crain B.J., Chacko V.P., van Zijl P.C. Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann. Neurol. 1999;45:265–269. doi: 10.1002/1531-8249(199902)45:2<265::aid-ana21>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Oldfield R.C. The assessment and analysis of handedness: the Edinburgh inventory. Neuropsychologia. 1971;9:97–113. doi: 10.1016/0028-3932(71)90067-4. [DOI] [PubMed] [Google Scholar]
- Pardini M., Yaldizli O., Sethi V., Muhlert N., Liu Z., Samson R.S., Altmann D.R., Ron M.A., Wheeler-Kingshott C.A., Miller D.H., Chard D.T. Motor network efficiency and disability in multiple sclerosis. Neurology. 2015;85:1115–1122. doi: 10.1212/WNL.0000000000001970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polman C.H., Reingold S.C., Banwell B., Clanet M., Cohen J.A., Filippi M., Fujihara K., Havrdova E., Hutchinson M., Kappos L., Lublin F.D., Montalban X., O'Connor P., Sandberg-Wollheim M., Thompson A.J., Waubant E., Weinshenker B., Wolinsky J.S. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann. Neurol. 2011;69:292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D.S., Lucchinetti C.F., Calabresi P.A. Multiple Sclerosis. N. Engl. J. Med. 2018;378:169–180. doi: 10.1056/NEJMra1401483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocca M.A., Valsasina P., Meani A., Falini A., Comi G., Filippi M. Impaired functional integration in multiple sclerosis: a graph theory study. Brain Struct. Funct. 2016;221:115–131. doi: 10.1007/s00429-014-0896-4. [DOI] [PubMed] [Google Scholar]
- Rubinov M., Sporns O. Complex network measures of brain connectivity: uses and interpretations. NeuroImage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Sastre-Garriga J., Pareto D., Rovira A. Brain atrophy in multiple sclerosis: clinical relevance and technical aspects. Neuroimaging Clin. N. Am. 2017;27:289–300. doi: 10.1016/j.nic.2017.01.002. [DOI] [PubMed] [Google Scholar]
- Schoonheim M.M., Geurts J.J., Landi D., Douw L., van der Meer M.L., Vrenken H., Polman C.H., Barkhof F., Stam C.J. Functional connectivity changes in multiple sclerosis patients: a graph analytical study of MEG resting state data. Hum. Brain Mapp. 2013;34:52–61. doi: 10.1002/hbm.21424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu N., Liu Y., Li K., Duan Y., Wang J., Yu C., Dong H., Ye J., He Y. Diffusion tensor tractography reveals disrupted topological efficiency in white matter structural networks in multiple sclerosis. Cereb. Cortex. 2011;21:2565–2577. doi: 10.1093/cercor/bhr039. [DOI] [PubMed] [Google Scholar]
- Shu N., Duan Y., Xia M., Schoonheim M.M., Huang J., Ren Z., Sun Z., Ye J., Dong H., Shi F.D., Barkhof F., Li K., Liu Y. Disrupted topological organization of structural and functional brain connectomes in clinically isolated syndrome and multiple sclerosis. Sci. Rep. 2016;6 doi: 10.1038/srep29383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam C.J. Modern network science of neurological disorders. Nat. Rev. Neurosci. 2014;15:683–695. doi: 10.1038/nrn3801. [DOI] [PubMed] [Google Scholar]
- Tewarie P., Schoonheim M.M., Schouten D.I., Polman C.H., Balk L.J., Uitdehaag B.M., Geurts J.J., Hillebrand A., Barkhof F., Stam C.J. Functional brain networks: linking thalamic atrophy to clinical disability in multiple sclerosis, a multimodal fMRI and MEG study. Hum. Brain Mapp. 2015;36:603–618. doi: 10.1002/hbm.22650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N., Landeau B., Papathanassiou D., Crivello F., Etard O., Delcroix N., Mazoyer B., Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Van Schependom J., Gielen J., Laton J., D'Hooghe M.B., De Keyser J., Nagels G. Graph theoretical analysis indicates cognitive impairment in MS stems from neural disconnection. Neuroimage Clin. 2014;4:403–410. doi: 10.1016/j.nicl.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wang X., Xia M., Liao X., Evans A., He Y. GRETNA: a graph theoretical network analysis toolbox for imaging connectomics. Front. Hum. Neurosci. 2015;9:386. doi: 10.3389/fnhum.2015.00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M., Wang J., He Y. BrainNet viewer: a network visualization tool for human brain connectomics. PLoS One. 2013;8 doi: 10.1371/journal.pone.0068910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalesky A., Fornito A., Bullmore E.T. Network-based statistic: identifying differences in brain networks. NeuroImage. 2010;53:1197–1207. doi: 10.1016/j.neuroimage.2010.06.041. [DOI] [PubMed] [Google Scholar]
- Zhao T., Cao M., Niu H., Zuo X.N., Evans A., He Y., Dong Q., Shu N. Age-related changes in the topological organization of the white matter structural connectome across the human lifespan. Hum. Brain Mapp. 2015;36:3777–3792. doi: 10.1002/hbm.22877. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.