Abstract
The precise control of assembly and packing of proteins and colloids on curved surfaces has fundamental implications in nanotechnology. In this paper, we describe dynamical simulations of the self-assembly of conical subunits around a spherocylindrical template, and a continuum theory for the bending energy of a triangular lattice with spontaneous curvature on a surface with arbitrary curvature. We find that assembly depends sensitively on mismatch between subunit spontaneous curvature and the mean curvature of the template, as well as anisotropic curvature of the template (mismatch between the two principal curvatures). Our simulations predict assembly morphologies that closely resemble those observed in experiments in which virus capsid proteins self-assemble around metal nanorods. Below a threshold curvature mismatch, our simulations identify a regime of optimal assembly leading to complete, symmetrical particles. Outside of this regime we observe defective particles, whose morphologies depend on the degree of curvature mismatch. To learn how assembly is affected by the nonuniform curvature of a spherocylinder, we also study the simpler cases of assembly around spherical and cylindrical cores. Our results show that both the intrinsic (Gaussian) and extrinsic (mean) curvatures of a template play significant roles in guiding the assembly of anisotropic subunits, providing a rich design space for the formation of nanoscale materials.
TOC image
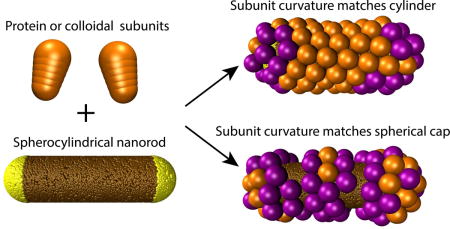
I. INTRODUCTION
Achieving molecular-scale control over the surface morphology of nanoscale particles is essential for many nano-materials and drug delivery applications. A promising route to this end is self-assembly, in which local interactions between subunits drive the emergence of particular large-scale morphologies [1–16]. However, assembly can be frustrated when the subunit-subunit interactions are incompatible with the surface geometry. For example, the densest 2D packing of spheres or discs, a six-fold coordinated hexagonal lattice, requires a minimum of 12 five-fold coordinated disks to cover a spherical surface [17, 18]. This frustration is metric in origin, arising because a hexagonal lattice is incompatible with the positive intrinsic (Gaussian) curvature of a sphere. Intrinsic curvature frustration has been the subject of intense experimental and theoretical investigation [17–45]. In contrast, there has been much less attention on extrinsic (mean) curvature, since it plays no role in the assembly of subunits that are isotropic (e.g. spheres) or live entirely in the surface tangent plane (i.e. a film of zero thickness). However, finite-thickness layers of flexible ligands undergo microphase separation or ordering on surfaces with mean curvature [5, 6, 10, 46–52]. Similarly, anisotropic subunits such as proteins or aspherical colloids have a preferred (spontaneous) mean curvature, and their assembly can be frustrated when the spontaneous curvature differs from that of the substrate on which they assemble [53].
In this article we computationally investigate assembly when both forms of geometric frustration occur simultaneously, during the assembly of anisotropic subunits on a spherocylindrical template. The inhomogeneous curvature of a spherocylinder ensures that the preferred subunit curvatures (mean and Gaussian) cannot globally match those of the template. Moreover, we show that anisotropy between the two principal curvature directions provides an additional source of frustration, even on surfaces such as cylinders for which the Gaussian curvature is zero.
Our study is motivated by recent experiments in which the capsid proteins of brome mosaic virus (BMV, a small, single-stranded RNA, icosahedral plant virus) assembled around spherocylindrical nanorods [1]. The assembly of viral capsid proteins around spherical cores has been studied for several systems [12–14, 54–57], including the assembly of BMV proteins around spherical charge-functionalized gold nanoparticles [3, 4, 58–62]. The experiments on spherical cores showed that the core size influences the protein organization, since as the core size increased, the proteins assembled larger icosahedral shells [4]. Yet, relatively little is known about viral protein assembly around nanoparticles with lower symmetry such as spherocylinders. Bancroft and coworkers [63] showed that BMV proteins form bacilliform particles around calf-thymus DNA, while Mukherjee et al. [64] showed that the capsid proteins of cowpea chlorotic mottle virus (CCMV, closely related to BMV) form tubular structures around double-stranded DNA. However the detailed arrangements of proteins in these complexes is unknown.
Zeng et al. [1] showed that BMV proteins can assemble closed shells around spherocylindrical gold nanorods whose surfaces are functionalized with negative charge. Using AFM, they visualized the arrangement of protein capsomers (pentamers or hexamers of the capsid protein) on the nanorod surfaces. While pentamer and hexamer capsomers formed partial icosahedral shells on the endcaps, particles exhibited hexagonal packing of capsomers on the cylindrical region of nanorods. Different alignments of the hexagonal lattice relative to the cylinder axis were observed, including both achiral geometries and a range of chiral angles. In addition to closed shells, the experiments identified particles with a variety of defects, several examples of which are shown in Fig. 1. The presence of defects was correlated to the nanorod dimensions.
FIG. 1.
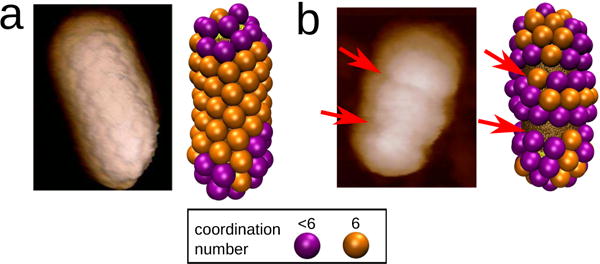
Comparison between AFM images of BMV capsid proteins assembled around gold spherocylindrical nanoparticles and simulation outcomes. The images show typical examples of (a) complete particles and (b) particle with defects, with red arrows indicating defects. Parameters for the simulation outcomes are: nanorod radius , nanorod length Lcyl = 45nm, and subunit spontaneous curvature radius Rcone = 14.0 (top) and Rcone = 9.5 (bottom). In all figures, capsomers with six neighbors are shown in orange, and capsomers with 5 or fewer neighbors in purple.
While insightful computational and theoretical studies have modeled the packing of spherical subunits on spherical [65–68] or spherocylindrical templates [69–71], spherical subunits do not possess a preferred mean curvature. In studies that did consider the effects of mean curvature, Yu et al. [53] showed that rodlike particles, with a preferred mean curvature of zero, form faceted structures on a spherical template, while other studies modeled the assembly of anisotropic subunits into icosahedral shells on spherical templates [72–78]. However, to our knowledge none have yet considered assembly around templates with inhomogeneous curvature.
In this article we examine the effect of preferred subunit curvature on assembly around templates with inhomogeneous curvature. We have developed a minimal coarse-grained model in which conical subunits with short-range lateral subunit-subunit attractions assemble on the surface of a spherocylindrical nanorod (Fig. 2). The preferred curvature of the assemblage is controlled by the subunit cone angle. Depending on the relative values of the subunit preferred curvature and the template curvature, our simulations predict a rich array of particle morphologies, including many of the structures observed in experiments such as complete shells and particles with circular vacancies, or ring defects (Fig. 1). To understand the physics underlying these results, we develop a continuum-theory description for the assembly of a triangular lattice with spontaneous curvature on a surface with arbitrary curvature. The theory shows that the assembly outcomes depend on mismatches between the mean curvatures of the subunits and nanorod, as well as the curvature anisotropy of the nanorod (i.e. difference between the two principal curvatures) even when the Gaussian curvature is zero. These results highlight the key role played by extrinsic curvature incompatibilities in the assembly of proteins and colloids on curved substrates.
FIG. 2.
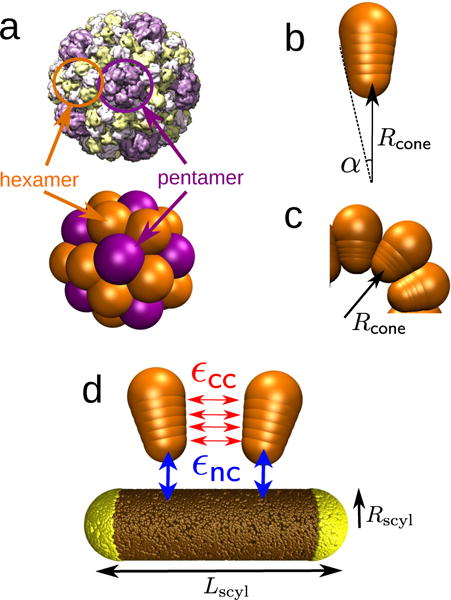
(a) Brome mosaic virus (BMV) capsid structure obtained from cyoEM (PDB ID: 3J7M) [79], compared with a capsid assembled from conical subunits in the absence of a nanoparticle. Each conical subunit represents a capsomer, and both pentamers (five neighbors, in purple) and hexamers (six neighbors, in orange) can be identified, showing icosahedral organization consistent with T=3 symmetry [22, 80]. (b,c) The cone angle α (b) sets the capsid spontaneous curvature radius Rcone (c), defined as the radial position of the capsomer bottom bead in the lowest energy capsid. (d) Capsomer model for Rcone = 9.5nm. Each of four interior beads (indices n = 1 … 4) interacts through a Morse potential of depth εcc with its counterparts in neighboring capsomers (red). The bottom capsomer bead experiences attractive interactions of strength εnc with the nanoparticle beads (blue). The equilibrium distance of the Morse potential between nanoparticle and capsomer beads, , must be considered in the template curvature. The effective template radius is then given by .
II. COMPUTATIONAL METHODS
Capsomer model
We seek a minimal, general, model capable of investigating the relationship between template curvature, subunit spontaneous curvature, and assembly morphologies. Motivated by these objectives and the limited resolution of the AFM experiments in Ref. [1], we employ a model in which the basic assembly unit corresponds to a protein capsomer (either a hexamer or pentamer). The model is sufficiently general to describe anisotropic colloidal subunits as well as proteins.
We have adapted the model from the conical subunits described in Chen et al. [69], which showed that conical subunits can self-assemble into monodisperse aggregates. Each subunit consists of a linear array of six beads with increasing radius, characterized by the cone angle α (Fig. 2b). For certain magic cone angles, the equilibrium structures exhibit icosahedral symmetry.
We modify the Chen et al. model by truncating the cones, so that in bulk (with no nanorod present) they assemble into empty shells with geometries that closely resemble viral capsids. We index the beads within a given capsomer by n = 0 … 5, with n = 0 corresponding to the bead with the smallest radius, which sits at the inner surface of an assembled capsid. Each of the four interior beads (n = 1 … 4) in a capsomer interacts with its counterpart (bead with the same index) in a nearby capsomer through a Morse potential (Eq. (9)), which drives lateral assembly of the subunits. Other pairs of beads experience excluded volume interactions (Eq. (10)).
Our subunit dimensions and aspect ratio roughly match those of BMV capsomers. The bead sizes within a subunit set the cone angle α and correspondingly the preferred capsid radius, i.e., the capsomer spontaneous curvature radius, Rcone. In an ideal shell, the innermost bead of each capsomer is located at a radial distance from the shell center Rcone, and the outer capsomer bead is located at Rout = Rcone + h, where h is the ‘height’ of a capsomer. The four interior beads are uniformly distributed between the inner and outer beads, so that the spacing between neighboring beads is h/5. The cone angle α is then determined by the criteria that beads of the same index in neighboring capsomers should be separated by a distance equal to their diameter (the equilibrium distance of their interaction potential). This requirement determines the bead radius as a function of index and cone angle as: . Specifying values for h, one bead diameter, and α thus completely determines the dimensions of the model capsomers. In our simulations we fix h = 4 nm and the diameter of the outermost bead to σcone = 2r5 = 7.7 nm, and we vary α to tune the spontaneous curvature.
To model the native BMV capsid structure, we set the cone angle α = 33.1°, so that in bulk simulations we obtain spherical shells consisting of 32 subunits and inner radius Rcone ≈ 9.5nm, in good agreement with the native BMV capsid [79]. Because we consider only a single subunit geometry, which represents both hexamer and pentamer capsomers, we find that only 40% of the assembled shells have perfect icosahedral symmetry. Robust formation of T=3 capsids with icosahedral symmetry was previously shown to require two capsomer diameters (with ‘pentamer’ subunits being smaller than those corresponding to hexamers) [67]. Moreover, the preferred curvature of individual BMV capsomers could differ from those observed in assembled capsids. Similarly, interactions with a substrate (such as the functionalized nanorod surface) could drive conformation changes that adjust the capsomer spontaneous curvature. To address these possibilities, we performed simulations over a range of cone angles α, corresponding to spontaneous curvature radii Rcone = 4.0 – 21.0nm. The inner radii of assembled capsids closely correspond to Rcone throughout this range.
Nanoparticle template
The nanoparticle is represented as a rigid body formed from many spherically symmetric pseudoatoms (or beads). In the case of spherocylinders (Fig. 2a), a central cylindrical body of length Lcyl and radius Rscyl is fused with two spherical caps also with radius Rscyl. The total length of the spherocylinder is then Lscyl = Lcyl + 2Rscyl. For studying spherical nanoparticles, we set Lcyl = 0. For cylindrical nanoparticles, we turn off the attractive interaction between the spherical caps and capsomers. We randomly distribute beads on the nanoparticle surface with a high and homogeneous density (ρ=13 beads/nm2 regardless of the nanoparticle dimensions), so that the capsomers interact with a roughly homogeneous surface. In the experimental system, positive charges on the inner surface of BMV capsomers are attracted to functionalized negative charge on the nanoparticle surface. We qualitatively represent this attractive interaction by implementing a Morse potential between the bottom bead (n = 0) of each capsomer and the nanoparticle surface beads. To account for excluded volume between capsomers and the nanoparticle, the outer three beads in each capsomer n = 3 … 5 experience a repulsive Lennard-Jones potential with nanoparticle surface beads. When considering the nanoparticle curvature, we must account for the interaction distance between the nanoparticle and capsomer beads, roughly given by the Morse potential equilibrium distance . Thus, we define an effective nanoparticle radius as
Simulations
We performed simulations in HOOMD[81], which uses GPUs to accelerate molecular dynamics simulations [82]. In our simulations, all physical units are expressed relative to the thermal energy kBT, mass m, and a reference length σ. The maximum diameter of a subunit is σcone = 7.7σ and its height (h, center-to-center distance between beads 0 and 5) is h = 4σ. By comparison with the actual height and diameter of a BMV capsomer h ≈ 5nm and d ≈ 8nm [79], we can map the length scale of our simulations into that of real viruses by setting σ ≈ 1nm. Accordingly, to facilitate comparison with experiments, we express all distances in nanometers.
Motivated by the experiments, we performed all simulations within the nanoparticle-directed assembly regime, meaning that assembly only occurs on the nanoparticle surface. To this end, we set the depth of the Morse potential for interactions between capsomer beads to εcc = 2kBT. For such weak interactions, nucleation of assembly in bulk was not observed at the simulated capsomer concentration. We set the well-depth of the Morse potential for the nanoparticle-capsomer interaction to εnc = 0.3kBT, which gives an average interaction energy of 6.65kBT/capsomer (each capsomer interacts with many nanoparticle pseudoatoms). This value promotes adsorption of capsomers onto the nanoparticle surface, resulting in a sufficiently high local capsomer concentration to drive assembly nucleation [76]. Our results are qualitatively unchanged for εnc ∈ [0.2, 0.5]kBT; for smaller adsorption energies the density of adsorbed capsomers is too low to promote rapid assembly nucleation, whereas for higher εnc capsomer adsorption becomes effectively irreversible, leading to malformed assemblies [83]. We found that achieving a homogeneous absorption free energy on the nanorod surface required slightly weakening the interaction between capsomers and nanoparticle surface beads on the spherical endcaps, εnc, sph = 0.98εnc, cyl. This reflects coupling between curvature and the finite size of the nanorod pseudoatoms.
Depending on the nanoparticle size, we considered two system sizes: for small nanoparticles (Lcyl < 45.0) we initialized 300 capsomers in a 130×130×130nm box, and for larger nanoparticles (Lcyl < 45.0) we used 500 capsomers in a 154×154×154nm box. For these system sizes the ratio of free to adsorbed capsomers was always higher than 5:1, thus ensuring results were not affected by depletion of free capsomers. The simulated capsomer concentration corresponds to approximately 220 μM. Our results were qualitatively insensitive to varying capsomer concentration provided that we remained in the nanoparticle-directed assembly regime.
We integrated the positions and orientations of capsomers using the HOOMD Brownian dynamics algorithm for rigid bodies for 1.5 × 107 timesteps, with a timestep duration of Δt = 0.005. The nanoparticle position and orientation were fixed. Based on the energy and visual inspection of particle morphologies, simulations have equilibrated (or at least reached long-lived metastable minima) by 5 × 106 timesteps.
III. CONTINUUM THEORY FOR A TRIANGULAR LATTICE WITH SPONTANEOUS CURVATURE
A. Derivation of the continuum description
In this section we elucidate the effects of mean and Gaussian curvature on assembly morphologies, by deriving a continuum approximation for the bending energy of a triangular lattice of subunits on a surface with arbitrary curvature. Our calculation is general, and extends calculations of bending energy for triangulated membrane models (e.g. Seung and Nelson [44] and Gompper and Kroll [45]) to lattices with spontaneous curvature and arbitrary template curvature. We will see that allowing for spontaneous curvature makes the relevant physics more transparent as well as increasing generality.
We consider a hexagonal lattice of subunits on a patch of surface with principal curvatures c1 and c2. We assume that particles maintain hexagonal close packing density at some neutral surface located at a distance hns above the surface, and that the long axis of each subunit points along the local surface normal. For now we neglect the presence of disclinations in the hexagonal lattice due to non-zero Gaussian curvature; we will discuss their contribution below. To make the derivation concrete, we assume the interaction potential used in our computational model. However, the derivation is easily extendable to other interaction forms and the results are generic.
We consider a reference state in which , so that all pseudoatoms pairs are separated by their equilibrium distance . We then calculate the change in the interaction energy between a pair of subunits (to quadratic order) as the curvatures vary:
| (1) |
with dn the distance between neighboring pairs of pseudoatoms with index n, and .
Distances within the shell can be calculated using the metric associated with the nanorod surface, but to make the role of curvature transparent we calculate the deviation from the reference state displacement between a pair of pseudoatoms separated by lattice vector as:
| (2) |
with σns the equilibrium separation distance at the neutral surface, hn the height of pseudoatom n above the surface, and cN the normal curvature along :
| (3) |
with θ the angle between and the principal direction associated with c1.
Substituting Eq. (2) into Eq. (1), summing over all particles and bonds, and using the hexagonal close-packing density yields a result independent of θ:
| (4) |
with A the template surface area, H = (c1 + c2)/2 the mean curvature and c0 = 2/Rcone the shell spontaneous curvature. The effective bending modulus is given by
| (5) |
Eq. (4) can also be written as
| (6) |
with K = c1c2 the Gaussian curvature. Eqs. 4 and 6 are the central result of this section.
There are several points worth noting about this result. Firstly, the energy, to quadratic order, is independent of lattice orientation, as shown by Seung and Nelson [44]. However, note that the energy does become orientation-dependent if higher order terms are considered in Eq. (1). Secondly, the first term in Eq. (6) has the standard Helfrich form for deviations between the mean and spontaneous curvatures [85], but the second term (H2 − K) arises from curvature anisotropy (c1 ≠ = c2). When this term is accounted for, the bending modulus κ is seen to be independent of surface geometry; i.e., in contrast to the suggestion in Ref. [45], the triangulated membrane model does not have a different bending modulus value on a sphere and a cylinder. Thirdly, we see that the Gaussian modulus is equal to the negative of the bending modulus, κG = −κ as found for lipid bilayers [86].
The contribution from the Gaussian curvature is constant if particles are uniformly covered, since the integrated Gaussian curvature is 4π regardless of the nanorod dimensions according to the Gauss-Bonnet theorem [87]. However, this contribution will change for particles which have vacancies on portions of the surface with non-zero K, and thus affects the relative favorability of assembling on the end caps or shaft. More interestingly, the curvature mismatch term H2 − K is zero on the spherical caps, but is nonzero on the cylindrical shaft and does depend on Rscyl.
Finally, regions with non-zero Gaussian curvature (e.g. the spherical caps) require disclinations in the hexagonal packing, which give rise to an additional energy Ud. This energy has been discussed at length for spherical packings (e.g. [17–22, 25, 30, 33, 35–45]). For our small spherical caps we can expect the total disclination energy to be proportional to Ud ∼ 12·4εcc since there are 12 five-fold coordinated subunits with 4 attractive pseudoatoms.
B. Implications of the continuum theory for assembly morphologies
To assess the relative favorability of assembly on the cylindrical or spherical portions of the nanorod, we use Eq. (6) to write the energy (per particle) due to bending and disclinations for the two surface geometries as
| (7) |
Comparing these expressions shows that there is a threshold value of the curvature ratio γnr above which assembly on the cylinder becomes more favorable than on a sphere:
| (8) |
Finally, to apply these results to our model, we set the pseudoatom height hn = nh/5, the neutral surface height hns = h/2, cone angle α = 33.1°, Morse strength εcc = 2, and Morse potential width , resulting in κ = −κG = 38.3kBT. To maintain the simplification of a harmonic interaction, we are neglecting the excluded volume repulsions from the Lennard-Jones potential (Eq. (10)), which apply only when the pseudoatom displacement is less than the equilibrium distance.
IV. RESULTS
All simulations resulted in extensive adsorption of capsomers onto the nanoparticle. To clearly illustrate the capsomer packing on the nanoparticle surface, in all figures we render only the outermost attractive bead of each capsomer (n = 4), and only show capsomers located within 5 nm of a nanoparticle surface bead. We render capsomers with six neighbors in orange, and capsomers with 5 or fewer neighbors in purple. We compute the coordination number of each capsomer from the number of neighbors within a distance , where is the equilibrium distance between the outermost attractive beads in neighboring capsomers. Due to this threshold-based definition, we never observe capsomers with 7 or more neighbors. Consistent with Luque et al. [71], we observe hexagonal packing of capsomers on the cylindrical portion of complete particles and 5-fold defects on the spherical caps.
To understand how assembly morphologies depend on mismatches in curvature, we consider results in terms of the ratio between the capsomer spontaneous curvature radius and the nanoparticle radius, . The mean curvature, defined as with c1 and c2 the principle curvatures, differs for the spherical endcaps and the cylindrical shaft. The ratio of mean curvatures on endcap and shaft is thus 1/γnr and 2/γnr respectively. Also note that the curvature anisotropy term in Eq. (6), (H2 − K), is zero on the endcap but equal to on the cylinder.
A. Spherocylinders: simulation outcomes
We first studied the assembly of conical capsomers around spherocylindrical nanoparticles of different length and radius. We focused on nanoparticles of similar dimensions to those studied in experiments, Rscyl ∼ 5 – 15 nm and Lcyl = 5 – 45 nm. Because the BMV capsid protein spontaneous curvature is not known for certain, and to understand the effect of varying γnr, we explored capsomer spontaneous curvature radii in the range Rcone = 5 – 21nm.
Within the simulated range of nanoparticle dimensions, we identify six classes of assembly outcomes (Fig. 3): incomplete III, with capsomers assembled into three isolated clusters; partial III, three partially connected domains; incomplete II, two isolated clusters; partial II, two partially connected clusters; complete capsids with capsomers homogeneously covering the entire nanoparticle surface; and incomplete, in which capsomers do not homogeneously cover the surface. We observe incomplete morphologies for very low radii of curvature (typically ≤ 5nm), suggesting that assembly is frustrated by the high substrate curvature.
FIG. 3.
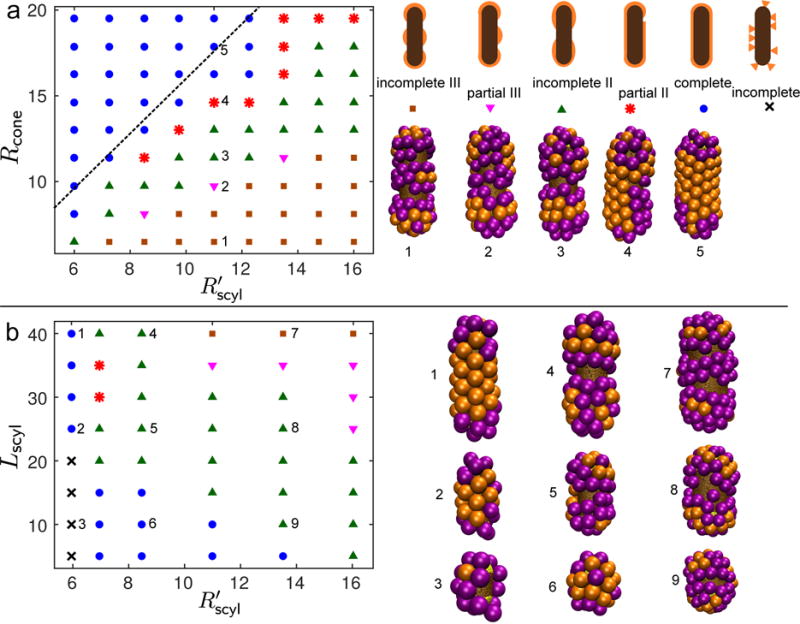
(a) Phase diagram of the most frequent simulation outcomes as a function of the capsomer spontaneous curvature radius Rcone and spherocylinder radius , for a spherocylinder length of Lcyl = 40nm. On the right, we show a schematic for each class of assembly outcome along with a representative example from the simulations. The outcomes are numbered the same as the corresponding parameters on the phase diagram. The dashed line shows the threshold above which the continuum theory (Eq. (8)) predicts that assembly is more favorable on the cylindrical shaft than on the spherical end caps. (b) Phase diagram of the most frequent outcomes as a function of the spherocylinder length Lcyl and radius Rscyl, for constant Rcone = 9.5nm. Legend as in (a). On the right we show some typical simulation snapshots for relevant spherocylinder aspect ratios. We performed 5-10 independent simulations at each parameter set. The distribution of outcomes for representative parameter values is shown in the supplemental figure [84].
Fig. 3a shows the most frequent morphology as a function of capsomer and nanoparticle radius, for fixed spherocylinder length Lcyl = 40nm. The phase diagram can be divided into two main regions: complete particles for (i.e. γnr ≳ 8/5) and incomplete particles otherwise. Among incomplete particles, for 1.2 < γnr < 1.5 we observe incomplete II particles, and for γnr < 1.2 only incomplete III particles. If we consider a cut of constant spherocylinder dimensions , as we increase Rcone the size of the capsomer domains increases and therefore the number of isolated domains that grow on the nanoparticle decreases. In this sequence, morphologies gradually change from incomplete III particles, to incomplete II and eventually complete particles, as shown in the snapshots of Fig. 3a. For parameter sets away from outcome transition boundaries, independent trajectories result in the same outcome class. However, for parameter sets near transition boundaries we observe up to three distinct outcomes at a given parameter set. The supplemental figure [84] shows the distribution of outcomes for representative parameter sets.
These results suggest that shell bending energy due to curvature mismatch and anisotropy is the key factor determining the presence and number of defects. In particular, the dashed line in Fig. 3a shows the threshold curvature ratio γnr∗ = 8/5 predicted by Eq. (8) (neglecting the contribution from disclinations, i.e. Ud = 0), above which assembly on the cylinder is more favorable than on the spherical endcaps. We see this matches the boundary between complete and incomplete particles reasonably well. Moreover, Fig. 4 plots the difference between bending energy on the cylinder and sphere, Δu = ucyl − usphere (Eq. (7)), and we see that the contours roughly track the boundaries between different assembly morphologies. We discuss in section IV B 2 how the circular ‘ring’ defects can be understood in terms of the bending energy. However, in addition to these equilibrium effects, we observed instances in which kinetics played a role in defect generation. In a few trajectories, clusters independently nucleated on both end caps, after which their growth edges met with incompatible orientations.
FIG. 4.
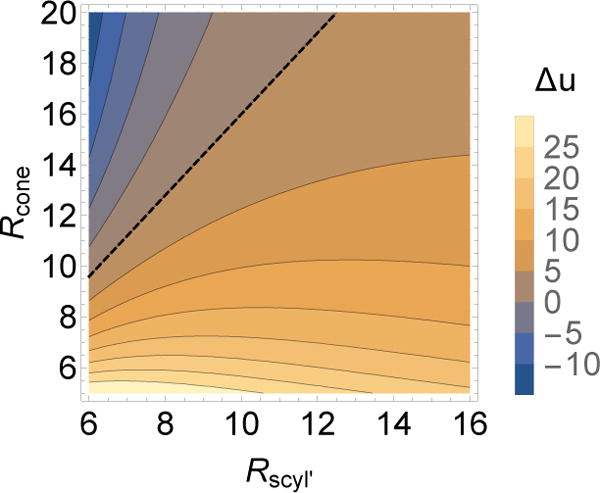
Comparison of bending energy on cylindrical and spherical surfaces. The difference in bending energy per particle predicted by the continuum theory, Δu = ucyl −usphere (Eq. (7)), is shown as a function of and Rcone for κ = 38.3kBT. The dashed line shows the threshold above which assembly is favored on the cylinder over the sphere (Eq. (8)).
Fig. 3b shows the most frequent morphology as a function of spherocylinder radius and length, for fixed capsomer radius Rcone = 9.5nm. In this case, the nanoparticle length determines the number of capsomer domains assembled. For small Lcyl, capsomers homogeneously assemble around the whole surface even for small Rcone. As we increase the nanoparticle length, two and eventually three domains are observed. The figure shows a slight coupling between radius and length; e.g. for the same Lcyl we observe incomplete II particles for and incomplete III for . The case of highest curvature exhibits a different behavior, with many vacancies and a low assembly yield for the shortest spherocylinders, presumably due to the high curvature of the template. However, as we increase the spherocylinder length we obtain complete particles. These results can be understood in terms of the relationship between the typical shell domain width and γnr discussed in section IV B 2.
B. Assembly around templates with uniform curvature
To understand how the interplay between the cylindrical and spherical portions of the nanoparticle effects assembly morphology, we performed independent sets of simulations in which assembly occured on spherical or cylindrical templates.
1. Spherical nanoparticles
We first consider capsomer assembly around a perfectly spherical nanoparticle, with a radius consistent with the T=3 capsid obtained in bulk, . We explore a range of capsomer curvature radii, 5.0 ≤ Rcone ≤ 15.5nm, so that the ratio of preferred capsid size to nanoparticle size varies from 0.5 ≲ γnr ≲ 1.75. Representative assembly outcomes are shown in Fig. 5a for cases in which the spontaneous curvature radius is smaller than, equal to, or larger than the nanoparticle size. For γnr ≈ 1 we obtain complete particles which consist of 32 capsomers, although icosahedral symmetry is observed in a lower proportion (20%) of simulations than in bulk (40%). As the preferred curvature radius deviates from the template size, assembled particles exhibit vacancies and defects. For γnr > 1 capsomers tend to form larger shells, leading to the formation of two large, flat caps whose assembly eventually stalls when the capsomers in the cap can no longer reach the nanoparticle surface. For smaller radii, γnr < 1, the capsomer curvature does not allow a well-formed assemblage to cover the entire nanoparticle surface, and instead we usually observe two small caps.
FIG. 5.
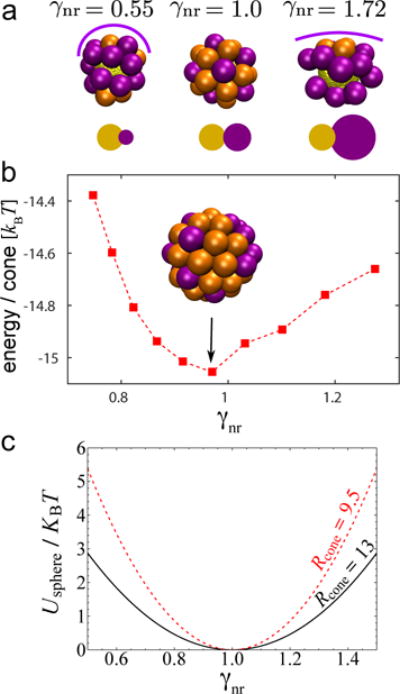
(a) Representative examples of capsomers assembled around spherical nanoparticles with radius . From left to right, Rcone = 5.0, 9.5 and 15.5nm. The line over each capsid approximately shows its preferred curvature. Below each snapshot we show a schematic of the relative size between the nanoparticle (yellow) and the protein capsid in bulk (purple). (b) Energy per capsomer assembled around spherical nanoparticles as a function of .In the simulations for (b), we kept the capsomer radius ≡ constant Rcone = 13.0nm and changed the nanoparticle size. (c) Continuum result for bending energy / capsomer on a spherical surface, usphere (Eq. (7), as a function of curvature ratio for indicated values of the spontaneous curvature radius Rcone.
In both limits (γnr > 1 and γnr < 1) the incompatibility between capsid and template curvature eventually frustrates assembly, leading to a gap between two caps. However, Yu et al. [53] showed that cylindrical rods assembling on spherical nanoparticles form ordered faceted particles. It is conceivable that the same result would be recovered in the limit γnr → ∞. In general we expect that the existence of gaps and the number of faces could depend on the degree of curvature mismatch and whether capsomer-capsomer interactions are sufficiently strong to drive assembly once a cap separates from the nanoparticle surface (recall that these simulations are performed in the limit of weak capsomer-capsomer interactions).
To quantitatively describe how the ‘quality’ of assembly depends on the curvature mismatch, we show the average energy per capsomer in Fig. 5b. For this calculation we fixed the capsomer spontaneous curvature radius at Rcone = 13nm, and then varied the nanoparticle in the range Rs ∈ [9.0, 16.0]. In this parameter regime all assembled capsids were defect-free, simplifying the interpretation of the capsomer energy. The simulation results are qualitatively consistent with the continuum theory (Eq. (6), Fig. 5c) — the energy is minimized at γnr ≈ 1, when the capsomer and nanoparticle curvatures match, and increases as the capsomer curvature radius deviates from the template size.
2. Cylindrical nanoparticles
To investigate assembly on the cylindrical portion of the nanoparticle, we performed simulations with long nanoparticles (Lcyl = 75nm), with attractions turned off between capsomers and the surface beads in the spherical caps. Thus assembly morphologies near the middle of the cylindrical region were essentially independent of end effects. As shown in Fig. 6, we observe well-formed hexagonal lattices that uniformly cover the cylindrical surface for γnr ≳ 1.4, with optimal assembly for γnr ≈ 1.5. This is roughly consistent with the behavior predicted by Eq. (6) as shown below.
FIG. 6.
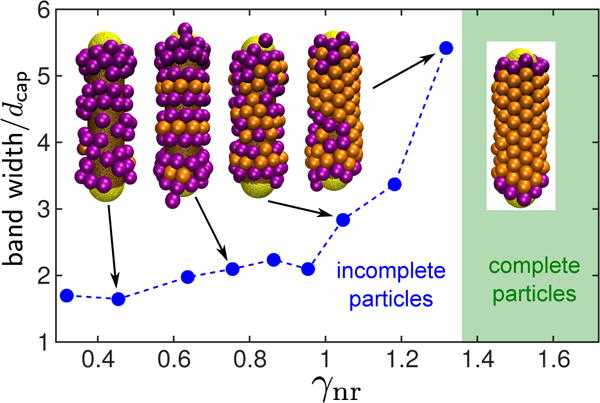
Average normalized width of clusters (in the cylinder axis direction) formed by capsomers assembled around cylinders, for increasing and constant Lcyl = 75nm.
For smaller capsomer curvature radii, we observe capsomers assembled into ‘bands’ separated by gaps, with the width of the bands depending on γnr. Capsomers at the edge of a band tilt along the cylinder axis, effectively allowing for shell curvature along the cylinder axis. To quantify the band width, we define w as as the maximum distance along the cylinder axis direction between two beads belonging to the same cluster, wl = (max(zi) − min zi) / σcone for all capsomer beads i in a given cluster l. Here we have normalized the band with by the diameter of the outer capsomer bead σcone = 7.7, so that the normalized band width roughly equals the number of capsomers per cluster in the axial direction. The normalized cylinder length is then Lcyl/σcone = 9.7.
Fig. 6 shows the average band width as a function of γnr, along with representative assembly outcomes. For a low curvature ratio γnr ≈ 0.4 – 0.5, capsomers assemble poorly on the cylinder, forming very narrow bands consisting of just 1-2 capsomers. Bands become thicker with increasing γnr, until w ≈ 5 for γnr ≈ 1.3 meaning that only two domains fit on the cylinder. The system then gradually transitions to a complete hexagonal lattice for γnr ≳ 1.4. Due to the finite length of our nanoparticle, we cannot determine whether the band width remains finite (but larger than the cylinder length), or if capsomers could assemble infinite domains with hexagonal packing. However, the energy calculations discussed next suggest that assembly is minimally frustrated for γnr ≈ 1.5, suggesting that the band width would be infinite (except for defects arising due to entropy in this effectively 1D geometry).
The increasing band width as a function of γnr can be qualitatively understood from (Eq. (4)). For small γnr both the mean curvature and curvature anisotropy terms are unfavorable because the principle curvature is zero along the cylinder axis. Tilting of capsomers near the edge of a band allows for nonzero curvature along this direction, with higher curvature for smaller bands. In particular, the typical band width for small γnr is comparable to Rcone.
We note that the bands in our simulations superficially resemble bands observed in the context of spherical colloids assembling around spherical nanoparticles in recent experiments [33] and simulations [68]. However, the physics driving band formation differs. The bands in our system arise due to curvature anisotropy and a mismatch between the extrinsic (mean) curvatures of the capsomer and template. In contrast, the spherical surfaces in Refs. [33, 68] have isotropic curvature, and the spherical colloids undergoing assembly have no spontaneous curvature. Bands arise because isotropic growth of a crystal with hexagonal order becomes frustrated by the intrinsic (Gaussian) curvature of the spherical surface.
To evaluate assembly quality, Fig.7a shows the average energy per capsomer for fixed Rcone = 13.0 and cylinder radius Rscyl = 7.5 – 15.0nm. We observe a minimum at γnr = 1.5, when capsomers form highly ordered hexagonal lattices. As the curvature ratio increases beyond γnr = 1.5, the energy increases and both visual inspection of simulation snapshots and analysis of the mean square deviations of capsomer-capsomer distances confirmed that the packings become less ordered (e.g. see Fig.7b for γnr = 2.05). This result is qualitatively consistent with the continuum theory. Minimization of the energy per particle on a cylinder (ucyl, Eq. (7)) as a function of , at fixed Rcone, gives an optimal cylinder radius corresponding to γnr = 4/3. This is qualitatively consistent with the minimum in Fig.7a, as is the variation of ucyl with γnr (Fig.7c). This result highlights the importance of the curvature anisotropy; analysis of mean curvature mismatch only would predict optimal assembly on a cylindrical surface for γnr = 2.0. Note however that if one varies Rcone at fixed the energy per particle is optimal for γnr = 2.0, since the curvature anisotropy term is independent of Rcone.
FIG. 7.
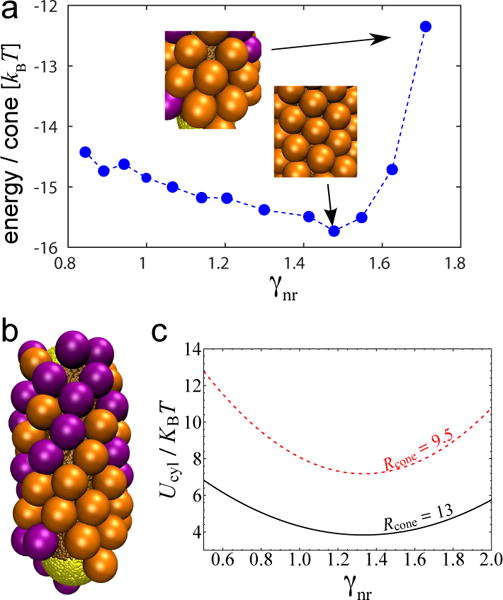
(a) Energy per capsomer assembled around cylindrical nanoparticles, for different capsomer/cylinder curvature ratios γnr. The insets show examples of ordered hexagonal packing at γnr = 1.5 and irregular hexagonal packing at γnr = 2.0. In these simulations the attractive interaction between capsomers and the spherical caps is turned off. Parameter values are Lcyl = 45.0, Rcone = 13.0 and Rscyl = 7.5–15.0nm. (b) Typical example of assembly morphology around a cylinder for γnr = 2.05. (c) Continuum result for bending energy / capsomer on a cylindrical surface, ucyl (Eq. (7), as a function of curvature ratio for indicated values of the spontaneous curvature radius.
C. Chirality
AFM imaging showed that capsid proteins form hexagonal lattices on the cylindrical region of the nanorod [1]. The particle lattice orientations relative to the nanorod were quantified by determining θ as the smallest angle between any of the lattice vectors and the long axis (Fig. 8). Following the nomenclature of Luque et al. [70, 71], there are two achiral geometries denoted as “ring” (θ = 0°) or “zigzag” (θ = 30°), while angles between 0° < θ < 30° correspond to chiral (or “skew”) configurations. The experimental particles exhibit the complete range of angles, but favor the “zigzag” configuration.
FIG. 8.
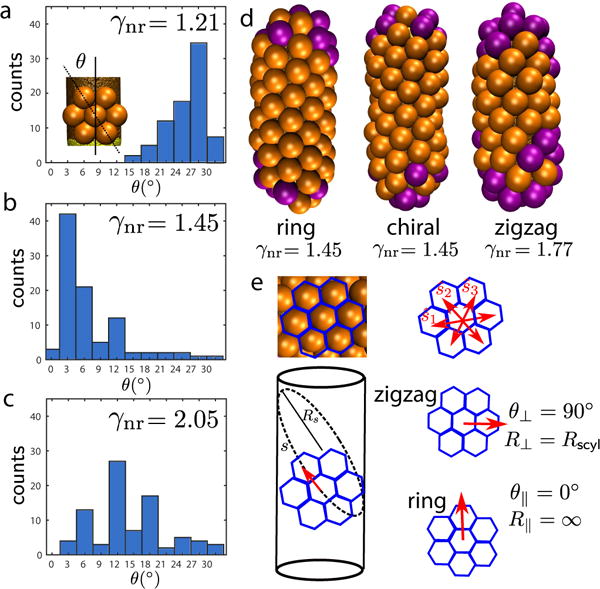
Chirality of the hexagonal lattice assembled on spherocylinders with Lcyl = 45nm and , and variable Rcone. (a) Angle distribution for γnr = 1.21. The inset shows how the chiral angle θ is defined. (b) Angle distribution for γnr = 1.45. (c) Angle distribution for γnr = 2.05. (d) From left to right, representative examples of particles with hexagonal lattice orientations that are approximately ring, chiral, and approximately zigzag. (e) Schematic of the hexagonal lattice orientation. Each capsomer interacts with six neighbors, with interaction directions given by the vectors . Capsomers experience three different interaction directions, each one along a different curve s on the cylinder, with curvature radius Rs (for simplicity only one is shown). In zigzag particles, the perpendicular orientation presents the minimum curvature radius R⊥ = Rscyl. Ring particles experience the lowest curvature radius, R║ = ∞.
Our simulation trajectories also result in a range of chiral angles (Fig. 8), with the preferred angle depending on γnr. We calculate the chiral angle θ as the minimum angle formed by the nanoparticle axis and any of the three orientation vectors of the hexagonal lattice (see Fig. 8), averaged over all the beads adsorbed onto the cylindrical face with 6 neighbors. We computed the angle distribution from 105 independent simulations at each of three parameter sets corresponding to different curvature ratios (see Fig. 8). For γnr = 1.21, adsorbed capsomers tend to form bands in which the hexagonal lattice is perpendicular to the cylinder axis (e.g. see several examples in Fig. 6). The angle distribution in Fig. 8a then exhibits a peak at θ ∼ 30°. For γnr = 1.45, we observed complete particles in which the hexagonal lattice aligns with the axis, and the angle distribution in Fig. 8b shows a clear preference for ring particles, with only a few chiral particles and generally of angle < 15°. As described in Section IV B 2, this regime corresponds to optimal hexagonal packing. For γnr = 2.05, we observe formation of both chiral and zigzag particles with disordered hexagonal packing (Fig. 8c); there is no identifiable pattern in the angle distribution.
The chiral angle distribution in simulations is most consistent with that observed in the experiments for γnr = 1.21 [1], implying a capsomer preferred curvature more closely matching the spherical end cap rather than the cylindrical region of the nanorod. A key difference in this regime is that the simulation model cannot capture the ability of the experimental capsomers to form complete particles in this regime. This difference may arise because the simulations only consider one subunit geometry while the BMV proteins form both pentameric and hexameric capsomers with different preferred curvatures.
To test whether the chiral angle is sensitive to kinetic effects (i.e. whether growth starts from the hemispherical caps or on the cylinder) we performed a set of 60 simulations for each chiral angle in which capsomers only interacted with the cylindrical surface. These simulations resulted in very similar angle distributions, suggesting the chiral angle primarily arises from energetic rather than kinetic effects. This result can be explained at least in part by the coupling between anisotropic curvature on the cylinder surface and the discrete nature of the capsomer lattice. As noted in section III the bending energy is independent of lattice orientation to quadratic order, but becomes orientation-dependent when nonlinearities are included. We have not performed a detailed analysis of the continuum theory at higher order, but the simulation results can be qualitatively understood as follows. As shown in Fig. 8d, the three lattice vectors define three curves si on the cylinder, with different curvature radii Rsi. The anisotropy in the curvatures of the three lattice directions strongly depends on the lattice orientation. Zigzag configurations allow a direction with the smallest curvature radius (for θ⊥ = 90°, R⊥ = Rscyl), while ring particles have the highest radius of curvature (for θ║ = 0°, R║= ∞). For preferred capsomer curvature radii matched to the spherical end cap (γnr ≈ 1), capsomers avoid the flat axis direction and preferentially orient perpendicular to the axis, resulting in a curvature radius along the perpendicular contour R⊥ = Rcyl ∼ Rcone. For γnr ∼ 1.5, ring orientations allow for the curvature along two directions to nearly match that of the capsomer (for θs = 30°, R30° = Rscyl/3 Rcone), although one lattice vector must then orient along the unfavorable flat axis direction.
D. Nucleation & kinetics
Although we have focused on long-time assembly morphologies thus far, the simulations also allow studying assembly dynamics. Figs. 9a,b shows the evolution of the number of capsomers adsorbed onto the cylindrical and spherical regions of the nanoparticle as a function of time for high and low curvature ratios. Note that the first adsorbed capsomers are typically isolated and randomly distributed, and hence the adsorption probability is equal for the cylindrical and spherical regions until nucleation occurs. Fig. 9c shows the density of adsorbed capsomers on each region at t = 6 × 105 as a function of γnr.
FIG. 9.
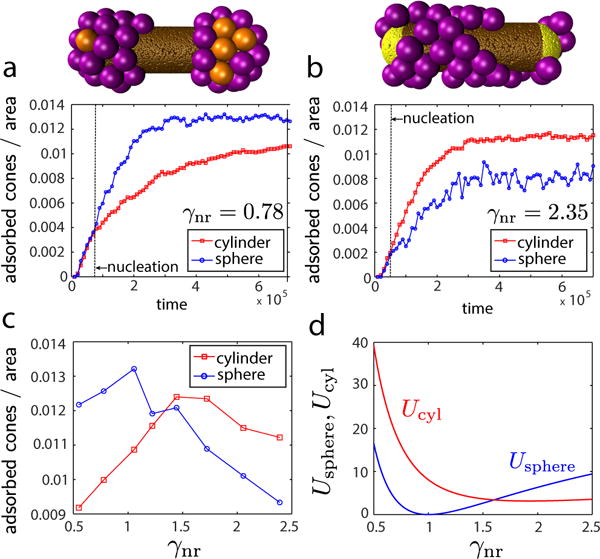
Comparison of assembly kinetics on spherical and cylindrical surfaces. (a), (b) Density of adsorbed capsomers on spherical (○ symbols) and cylindrical (□ symbols) nanoparticles as a function of time for curvature ratios below and above the threshold: (a) γnr = 0.8 and (b) γnr = 2.3. The black dashed line in each plot indicates the time point at which nucleation occurred. The image above each plot is a snapshot from a representative trajectory at (a) t = 4.5 × 104 and (b) t = 3 × 103. Each curve represents the average over ten independent simulations. (c) The average density of adsorbed capsomers on cylindrical and spherical nanoparticles as a function of γnr. (d) The bending energy per capsomer on spherical and cylindrical surfaces calculated from continuum theory (Eq. (7)) as a function of curvature ratio. In all cases the spherocylinder radius is fixed at and Rcone is varied. In the simulations Lcyl = 40.0nm.
Recall that the continuum theory predicts a threshold γnr∗ ≈ 8/5 above which there is a transition from preferential assembly on spherical surfaces to cylindrical surfaces. The dependence of the relative driving force for assembly on either surface can be estimated from the dependence of curvature energies for each geometry shown in Fig. 9d. The simulation trajectories are strongly consistent with the continuum predictions. For low γnr nucleation preferentially occurs on end caps, whereas nucleation occurs in the cylindrical region for high γnr. The crossover between these regimes, where nucleation occurs uniformly throughout the nanorod, occurs at γnr∗ (within error). Moreover, the adsorbed density is correlated to the bending energy, with a maximum in the density on the spherical region at γnr = 1 corresponding to the minimum in usphere.
Consistent with these results, experimental evidence in Zeng et al. [1] suggests both that the preferred capsomer curvature is consistent with the spherical end-caps (i.e. γnr ≈ 1) and that experimental trajectories initiate at the end-caps.
V. DISCUSSION & CONCLUSIONS
We have studied a phenomenological computational model motivated by experiments in which capsid proteins from an icosahedral virus assemble around spherocylindrical nanorods [1]. By varying a single parameter, the ratio of the preferred curvature of the subunits to the template curvature, the simulations reproduce most of the experimentally observed assembly outcomes. These results highlight the importance of the coupling between the curvatures of subunits and substrate, demonstrating that particle asymmetry adds an important control parameter for assembly confined on curved surfaces. Our subunit model is sufficiently general that it also could be realized experimentally by synthesizing conical-shaped colloidal particles.
It is well known that packing of particles with a circular cross-section on a spherical surface is frustrated due to the incompatibility of hexagonal order on surfaces with Gaussian (intrinsic) curvature. Here, we see that the assembly of anisotropic particles on a curved surface results in an additional forms of geometric frustration: when there is a mismatch between the preferred extrinsic (mean) curvature of the particles and the template, or a mismatch between curvature anisotropy of the particles and template. It was previously shown that a mismatch in mean curvatures of subunits and substrate on a spherical surface leads to line defects and faceting [53]. Here, we see a similar behavior on a cylindrical surface, with bands separated by line defects when the capsomer spontaneous curvature is sufficiently large in comparison to that of the cylindrical region of the nanorod. In contrast to a spherical surface, curvature is anisotropic on a cylinder, and our simulations show that this anisotropy can provide an additional control parameter, allowing tuning of the preferred lattice orientation on the cylinder. Thus, the class of systems described by our simple computational model could allow for designing a rich variety of particle geometries.
The simulation results suggest that measuring the width w of bands between defects could provide an estimate of the spontaneous curvature radius of the adsorbed particles (w ∼ 2Rcone), a parameter which is in general difficult to measure in bulk experiments. However, the simulations also suggest a coupling between domain width and cylinder radius, and thick cylinders may lead to underestimation of Rcone.
Finally, while the minimal model used in this work seems to capture the key physical principles dictating the assembly of anisotropic particles on curved surfaces, the model also has limitations relative to the motivating experimental system [1]. In particular, while the model reproduces the experimentally observed morphologies, different values of the subunit spontaneous curvature are required to match the experimental observation that complete particles can form on both short and long nanorods. We speculate that this discrepancy arises because, for simplicity, the model considers only a single subunit geometry, whereas BMV proteins adopt different ‘quasi-equivalent’ conformations allowing them to form pentameric or hexameric capsomers. It is likely that hexameric capsomers have a larger preferred curvature radius than pentamers, which would allow them to more readily assemble on the cylindrical region of the nanorod. However, comparison of several results between the simulations and experiments still suggests that the subunits have a spontaneous curvature which more closely matches the spherical end caps than the cylinder. In particular, the distribution of chiral angles and the fact that trajectories initiate on end-caps in this regime are both consistent with experimental observations [1].
Supplementary Material
Acknowledgments
We gratefully acknowledge Greg Grason for useful discussions, and suggestions on the approach to derive the continuum theory. This work was supported by the NIH, Award Number R01GM108021 from the National Institute Of General Medical Sciences (MFH and GRL), the Brandeis Center for Bioinspired Soft Materials, an NSF MRSEC, DMR-1420382 (GRL), and the U.S Army Research Office under award # W911NF-17-1-0329 (BD). Computational resources were provided by the NSF through XSEDE computing resources (XStream, Maverick, Bridges, Comet) through award number MCB090163, and the Brandeis HPCC which is partially supported by the Brandeis MRSEC.
VI. APPENDIX
Our model is defined by pairwise potentials describing capsomer-capsomer and capsomer-nanoparticle interactions. The capsomer-capsomer potential, Ucc, consists of two terms, . The attractive interaction between the four interior pseudoatoms of the capsomers, n = 1 … 4 is modeled by a Morse potential. Beads are attracted only to their counterparts (with the same index n) on neighboring capsomers, and the equilibrium distance of the potential depends on the bead radius, :
| (9) |
with i and j indexing capsomers, n indexing beads within a capsomer, rij,n the distance between the pair of beads with index n in capsomers i and j, and εcc = 2kBT. The potential width parameter is , and the cutoff radius is set to . The Morse potential is used for these interactions largely for historical reasons, although in principle it offers the advantage over a Lennard-Jones interaction of allowing independent control over the pseudoatom size and the width of the potential well.
All pairs of capsomer pseudoatoms that are not on the same subunit and do not mutually attract (Eq. (9)) have excluded volume interactions represented by a WCA potential [88]
| (10) |
with rij,nm the distance between bead n in capsomer i and bead m in capsomer j, εex = 1.0, and the cutoff distances
The interaction potential between the nanoparticle and capsomers, Unc, can be also split into contributions from attractions and excluded volume. The bottom bead of each capsomer experiences an attractive interaction with nanoparticle beads, described by a Morse potential:
| (11) |
with the sum running over all Nnr nanorod beads and the bottom beads of all Ncone capsomers, rij,0 the distance between nanorod pseudoatom i and the bottom bead of capsomer , and the interaction width parameter set to αnc = 3.0. The well-depth parameters are set to εnc = 0.3kBT for beads on the cylindrical portion of the NR, and εnc = 0.294kBT for beads on the spherical endcap to ensure a uniform driving force for subunit adsorption everywhere (see section II). In addition, all nanoparticle beads interact through excluded volume with the three outermost capsomer beads:
| (12) |
with rij,n the distance between nanorod pseudoatom i and bead n of capsomer j, ex = 1.0 and cutoff radius
Footnotes
CONFLICTS OF INTEREST
There are no conflicts of interest to declare.
References
- 1.C. Zeng, G. R. Lázaro, I. B. Tsvetkova, M. F. Hagan, and B. Dragnea, submitted (2018).
- 2.Glotzer SC, Solomon MJ. Nat Mater. 2007;6:557. doi: 10.1038/nmat1949. [DOI] [PubMed] [Google Scholar]
- 3.Chen C, Daniel MC, Quinkert ZT, De M, Stein B, Bowman VD, Chipman PR, Rotello VM, Kao CC, Dragnea B. Nano Lett. 2006;6:611. doi: 10.1021/nl0600878. [DOI] [PubMed] [Google Scholar]
- 4.Sun J, DuFort C, Daniel MC, Murali A, Chen C, Gopinath K, Stein B, De M, Rotello VM, Holzenburg A, Kao CC, Dragnea B. Proc Natl Acad Sci U S A. 2007;104:1354. doi: 10.1073/pnas.0610542104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh C, Ghorai PK, Horsch MA, Jackson AM, Larson RG, Stellacci F, Glotzer SC. Phys Rev Lett. 2007;99 doi: 10.1103/PhysRevLett.99.226106. [DOI] [PubMed] [Google Scholar]
- 6.Carney RP, DeVries GA, Dubois C, Kim H, Kim JY, Singh C, Ghorai PK, Tracy JB, Stiles RL, Murray RW, Glotzer SC, Stellacci F. J Am Chem Soc. 2008;130:798. doi: 10.1021/ja077383m. [DOI] [PubMed] [Google Scholar]
- 7.Van Lehn RC, Atukorale PU, Carney RP, Yang YS, Stellacci F, Irvine DJ, Alexander-Katz A. Nano Lett. 2013;13:4060. doi: 10.1021/nl401365n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sknepnek R, Vernizzi G, de la Cruz MO. Soft Matter. 2012;8:636. [Google Scholar]
- 9.Jadhao V, Thomas CK, de la Cruz MO. Proc Natl Acad Sci U S A. 2014;111:12673. doi: 10.1073/pnas.1413986111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asai M, Cacciuto A, Kumar SK. Soft Matter. 2015;11:793. doi: 10.1039/c4sm02295e. [DOI] [PubMed] [Google Scholar]
- 11.Koch AHR, Leveque G, Harms S, Jaskiewicz K, Bernhardt M, Henkel A, Sonnichsen C, Landfester K, Fytas G. Nano Lett. 2014;14:4138. doi: 10.1021/nl501783x. [DOI] [PubMed] [Google Scholar]
- 12.Allen TM, Cullis PR. Science. 2004;303:1818. doi: 10.1126/science.1095833. [DOI] [PubMed] [Google Scholar]
- 13.Mieszawska AJ, Mulder WJM, Fayad ZA, Cormode DP. Mol Pharmaceutics. 2013;10:831. doi: 10.1021/mp3005885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nune SK, Gunda P, Thallapally PK, Lin YY, Forrest ML, Berkland CJ. Expert Opinion on Drug Delivery. 2009;6:1175. doi: 10.1517/17425240903229031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manoharan VN. Science. 2015;349:1253751. doi: 10.1126/science.1253751. [DOI] [PubMed] [Google Scholar]
- 16.Yang Y, Meyer R, Hagan MF. Phys Rev Lett. 2010;104:258102. doi: 10.1103/PhysRevLett.104.258102. [DOI] [PubMed] [Google Scholar]
- 17.Thompson JJ. Philos Mag. 1904;7:237. [Google Scholar]
- 18.Kleman M. Adv Phys. 1989;38:605. [Google Scholar]
- 19.Bowick M, Nelson D, Travesset A. Phys Rev. 2000;B 62:8738. [Google Scholar]
- 20.Mermin ND. Rev Mod Phys. 1979;51:591. [Google Scholar]
- 21.Bowick M, Cacciuto A, Nelson D, Travesset A. Phys Rev Lett. 2002;89:185502. doi: 10.1103/PhysRevLett.89.185502. [DOI] [PubMed] [Google Scholar]
- 22.Caspar DLD, Klug A. Cold Spring Harbor Symp Quant Biol. 1962;27:1. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 23.Nelson D. Defects and Geometry in Condensed Matter Physics. Cambridge University Press; 2002. [Google Scholar]
- 24.Altschuler E, Williams T, Ratner E, Tipton R, Stong R, Dowla F, Wooten F. Phys Rev Lett. 1997;78:2681. [Google Scholar]
- 25.Bausch A, Bowick M, Cacciuto A, Dinsmore A, Hsu M, Nelson D, Nikolaides M, Travesset A, Weitz D. Science. 2003;299:1716. doi: 10.1126/science.1081160. [DOI] [PubMed] [Google Scholar]
- 26.Irvine WTM, Vitelli V, Chaikin PM. Nature. 2010;468:947. doi: 10.1038/nature09620. [DOI] [PubMed] [Google Scholar]
- 27.Fernández-Nieves A, Vitelli V, Utada AS, Link DR, Márquez M, Nelson DR, Weitz DA. Phys Rev Lett. 2007;99:157801. doi: 10.1103/PhysRevLett.99.157801. [DOI] [PubMed] [Google Scholar]
- 28.Lopez-Leon T, Fernandez-Nieves A, Nobili M, Blanc C. J Phys: Condens Matter. 2012;24 doi: 10.1088/0953-8984/24/28/284122. [DOI] [PubMed] [Google Scholar]
- 29.Lopez-Leon T, Fernandez-Nieves A, Nobili M, Blanc C. Phys Rev Lett. 2011;106 doi: 10.1103/PhysRevLett.106.247802. [DOI] [PubMed] [Google Scholar]
- 30.Liang HL, Schymura S, Rudquist P, Lagerwall J. Phys Rev Lett. 2011;106 doi: 10.1103/PhysRevLett.106.247801. [DOI] [PubMed] [Google Scholar]
- 31.Liang HL, Noh J, Zentel R, Rudquist P, Lagerwall JPF. Phil Trans R Soc A. 2013;371 doi: 10.1098/rsta.2012.0258. [DOI] [PubMed] [Google Scholar]
- 32.Lopez-Leon T, Koning V, Devaiah KBS, Vitelli V, Fernandez-Nieves A. Nat Phys. 2011;7:391. [Google Scholar]
- 33.Meng G, Paulose J, Nelson DR, Manoharan VN. Science. 2014;343 doi: 10.1126/science.1244827. [DOI] [PubMed] [Google Scholar]
- 34.Kusumaatmaja H, Wales DJ. Phys Rev Lett. 2013;110 doi: 10.1103/PhysRevLett.110.165502. [DOI] [PubMed] [Google Scholar]
- 35.Irvine WTM, Bowick MJ, Chaikin PM. Nat Mater. 2012;11:948. doi: 10.1038/nmat3429. [DOI] [PubMed] [Google Scholar]
- 36.Irvine WTM, Vitelli V. Soft Matter. 2012;8:10123. [Google Scholar]
- 37.Grason GM. Soft Matter. 2013;9:6761. [Google Scholar]
- 38.Azadi A, Grason GM. Phys Rev Lett. 2014;112:225502. doi: 10.1103/PhysRevLett.112.225502. [DOI] [PubMed] [Google Scholar]
- 39.Azadi A, Grason GM. Phys Rev E. 2016;94:013003. doi: 10.1103/PhysRevE.94.013003. [DOI] [PubMed] [Google Scholar]
- 40.Mbanga BL, Grason GM, Santangelo CD. Phys Rev Lett. 2012;108:017801. doi: 10.1103/PhysRevLett.108.017801. [DOI] [PubMed] [Google Scholar]
- 41.Bruss IR, Grason GM. Proc Natl Acad Sci U S A. 2012;109:10781. doi: 10.1073/pnas.1205606109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grason GM, Davidovitch B. Proc Natl Acad Sci U S A. 2013;110:12893. doi: 10.1073/pnas.1301695110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grason GM. J Chem Phys. 2016;145:110901. doi: 10.1063/1.4962629. . [DOI] [Google Scholar]
- 44.H. S. Seung and D. R. Nelson, 38 (1988).
- 45.Gompper G, Kroll D. J Phys I France. 1996;6:1305. [Google Scholar]
- 46.Tung C, Cacciuto A. J Chem Phys. 2013;139 doi: 10.1063/1.4829638. [DOI] [PubMed] [Google Scholar]
- 47.Miller WL, Bozorgui B, Klymko K, Cacciuto A. J Chem Phys. 2011;135 doi: 10.1063/1.3672104. [DOI] [PubMed] [Google Scholar]
- 48.Luedtke WD, Landman U. J Phys Chem. 1998;B 102:6566. [Google Scholar]
- 49.Luedtke WD, Landman U. J Phys Chem. 1996;100:13323. [Google Scholar]
- 50.Widmer-Cooper A, Geissler P. Nano Lett. 2014;14:57. doi: 10.1021/nl403067p. [DOI] [PubMed] [Google Scholar]
- 51.Ghorai PK, Glotzer SC. J Phys Chem. 2007;C 111:15857. [Google Scholar]
- 52.Yu M, Stellacci F. Small. 2012;8:3720. [Google Scholar]
- 53.Yu N, Ghosh A, Hagan MF. Soft Matter. 2016;12:8990. doi: 10.1039/c6sm01498d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goicochea NL, De M, Rotello VM, Mukhopadhyay S, Dragnea B. Nano Lett. 2007;7:2281. doi: 10.1021/nl070860e. [DOI] [PubMed] [Google Scholar]
- 55.Loo L, Guenther RH, Lommel SA, Franzen S. J Am Chem Soc. 2007;129:11111. doi: 10.1021/ja071896b. [DOI] [PubMed] [Google Scholar]
- 56.Kwak M, Minten IJ, Anaya DM, Musser AJ, Brasch M, Nolte RJM, Muellen K, Cornelissen JJLM, Herrmann A. J Am Chem Soc. 2010;132:7834. doi: 10.1021/ja101444j. [DOI] [PubMed] [Google Scholar]
- 57.Chang CB, Knobler CM, Gelbart WM, Mason TG. ACS Nano. 2008;2:281. doi: 10.1021/nn700385z. [DOI] [PubMed] [Google Scholar]
- 58.Chen C, Kwak ES, Stein B, Kao CC, Dragnea B. J Nanosci Nanotechno. 2005;5:2029. doi: 10.1166/jnn.2005.506. [DOI] [PubMed] [Google Scholar]
- 59.Daniel MC, Tsvetkova IB, Quinkert ZT, Murali A, De M, Rotello VM, Kao CC, Dragnea B. ACS Nano. 2010;4:3853. doi: 10.1021/nn1005073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsvetkova I, Chen C, Rana S, Kao CC, Rotello VM, Dragnea B. Soft Matter. 2012;8:4571. [Google Scholar]
- 61.Malyutin AG, Dragnea B. J Phys Chem B. 2013 doi: 10.1021/jp405603m. [DOI] [PubMed] [Google Scholar]
- 62.Cheng F, Tsvetkova IB, Khuong YL, Moore AW, Arnold RJ, Goicochea NL, Dragnea B, Mukhopadhyay S. Mol Pharm. 2013;10:51. doi: 10.1021/mp3002667. http://pubs.acs.org/doi/pdf/10.1021/mp3002667. [DOI] [PubMed] [Google Scholar]
- 63.Bancroft J. Adv Virus Res. 1970;16:99. doi: 10.1016/s0065-3527(08)60022-6. [DOI] [PubMed] [Google Scholar]
- 64.Mukherjee S, Pfeifer CM, Johnson JM, Liu J, Zlotnick A. J Am Chem Soc. 2006;128:2538. doi: 10.1021/ja056656f. [DOI] [PubMed] [Google Scholar]
- 65.Fantoni R, Salari JW, Klumperman B. Physical Review E. 2012;85:061404. doi: 10.1103/PhysRevE.85.061404. [DOI] [PubMed] [Google Scholar]
- 66.Luque A, Reguera D, Morozov A, Rudnick J, Bruinsma R. J Chem Phys. 2012;136 doi: 10.1063/1.4712304. [DOI] [PubMed] [Google Scholar]
- 67.Zandi R, Reguera D, Bruinsma RF, Gelbart WM, Rudnick J. Proc Natl Acad Sci U S A. 2004;101:15556. doi: 10.1073/pnas.0405844101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paquay S, Kusumaatmaja H, Wales DJ, Zandi R, van der Schoot P. Soft Matter. 2016;12:5708. doi: 10.1039/c6sm00489j. [DOI] [PubMed] [Google Scholar]
- 69.Chen T, Glotzer SC. Physical Review E. 2007;75:051504. doi: 10.1103/PhysRevE.75.051504. [DOI] [PubMed] [Google Scholar]
- 70.Luque A, Zandi R, Reguera D. Proc Natl Acad Sci USA. 2010;107:5323. doi: 10.1073/pnas.0915122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luque A, Reguera D. Biophysical Journal. 2010;98:2993. doi: 10.1016/j.bpj.2010.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hagan MF. Phys Rev E. 2008;77:051904. doi: 10.1103/PhysRevE.77.051904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Elrad OM, Hagan MF. Nano Lett. 2008;8:3850. doi: 10.1021/nl802269a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fejer SN, Chakrabarti D, Wales DJ. ACS Nano. 2010;4:219. doi: 10.1021/nn9013565. [DOI] [PubMed] [Google Scholar]
- 75.Williamson AJ, Wilber AW, Doye JPK, Louis AA. Soft Matter. 2011;7:3423. [Google Scholar]
- 76.Hagan MF. J Chem Phys. 2009;130:114902. doi: 10.1063/1.3086041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kusters R, Lin HK, Zandi R, Tsvetkova I, Dragnea B, van der Schoot P. J Phys Chem. 2015;B 119:1869. doi: 10.1021/jp5108125. [DOI] [PubMed] [Google Scholar]
- 78.Lin HK, van der Schoot P, Zandi R. Physical Biology. 2012;9:066004. doi: 10.1088/1478-3975/9/6/066004. [DOI] [PubMed] [Google Scholar]
- 79.Wang Z, Hryc CF, Bammes B, Afonine PV, Jakana J, CHen DH, Liu X, Baker ML, Kao C, Ludtke SJ, Schmid MF, Adams PD, Chiu W. Nature Communications. 2014;5:4808. doi: 10.1038/ncomms5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johnson JE, Speir JA. J Mol Biol. 1997;269:665. doi: 10.1006/jmbi.1997.1068. [DOI] [PubMed] [Google Scholar]
- 81.Anderson JA, Lorenz CD, Travesset A. J Comput Phys. 2008;227:5342. [Google Scholar]
- 82.Nguyen TD, Phillips CL, Anderson JA, Glotzer SC. Comput Phys Commun. 2011;182:2307. [Google Scholar]
- 83.Hagan MF. Adv Chem Phys. 2014;155:1. doi: 10.1002/9781118755815.ch01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.“Supplemental material to this manuscript,”.
- 85.Helfrich W. Zeitschrift Fur Naturforschung C-a Journal of Biosciences C. 1973;28:693. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- 86.Hu M, de Jong DH, Marrink SJ, Deserno M. Faraday Discussions. 2013;161:365. doi: 10.1039/c2fd20087b. [DOI] [PubMed] [Google Scholar]
- 87.Deserno M. Chemistry and Physics of Lipids. 2015;185:11. doi: 10.1016/j.chemphyslip.2014.05.001. membrane mechanochemistry: From the molecular to the cellular scale. [DOI] [PubMed] [Google Scholar]
- 88.Weeks JD, Chandler D, Andersen HC. J Chem Phys. 1971;54:5237. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


