Abstract
Using mass spectrometry (MS) to obtain information about a higher order structure of protein requires that a protein’s structural properties are encoded into the mass of that protein. Covalent labeling (CL) with reagents that can irreversibly modify solvent accessible amino acid side chains is an effective way to encode structural information into the mass of a protein, as this information can be read-out in a straightforward manner using standard MS-based proteomics techniques. The differential reactivity of proteins under two or more conditions can be used to distinguish protein topologies, conformations, and/or binding sites. CL-MS methods have been effectively used for the structural analysis of proteins and protein complexes, particularly for systems that are difficult to study by other more traditional biochemical techniques. This review provides an overview of the non-specific CL approaches that have been combined with MS with a particular emphasis on the reagents that are commonly used, including hydroxyl radicals, carbenes, and diethylpyrocarbonate. We describe the reagent and protein factors that affect the reactivity of amino acid side chains. We also include details about experimental design and workflow, data analysis, recent applications, and some future prospects of CL-MS methods.
Keywords: Carbenes, Covalent labeling, Diethylpyrocarbonate, Hydroxyl radicals, Mass spectrometry, Protein higher-order structure
1. Introduction to Covalent Labeling – Mass Spectrometry
A protein’s higher order structure (HOS) determines its function, and so methods that provide insight into protein structure are important for understanding protein reactivity. Traditionally, NMR spectroscopy or X-ray crystallography have been the methods of choice because of the atomic-level resolution afforded by these techniques. Recently, cryo-electron microscopy (cryo-EM) has also emerged as a very powerful tool for studying protein structure. In some cases, however, protein HOS cannot be properly studied by these techniques because of limited sample amounts, a given protein’s tendency to aggregate, or sample incompatibility. Moreover, in certain applications (e.g. protein therapeutic design) more rapid structural analysis tools are needed. Because of the inherent sensitivity, specificity, and speed of mass spectrometry (MS), methods based on this technique have emerged for the analysis of a protein’s solution HOS. These MS-based methods add to the toolbox of protein biochemists by offering approaches that give much higher structural resolution than techniques such as circular dichroism (CD) spectroscopy or fluorescence spectroscopy while at the same time being more routinely applicable and devoid of the limitations associated with NMR, X-ray crystallography, or cryo-EM.
Using MS to obtain information about a protein’s structure in solution requires that the protein’s structural properties are encoded into the mass of the protein. Three primary methods of encoding this structural information have been utilized, including hydrogen-deuterium exchange (HDX) [1–7], cross-linking [8–10] and covalent labeling [11–13]. When coupled with MS, HDX relies on the replacement of hydrogens by deuteriums on backbone amides, leading to mass increases in regions of the protein that are the least protected from this exchange. In doing so, HDX/MS provides insight into structured/unstructured and rigid/dynamic regions of a protein that can then be related to HOS. Cross-linking uses multi-functional reagents that can form new intra- or inter-molecular bonds between amino acid side chains in a protein or protein-protein complex. The choice of the cross-linking agent leads to defined distance constraints for the linked side chains that enables one to deduce HOS information. Covalent labeling (CL) methods are analogous to cross-linking methods in that they modify protein side chains, but they report on a protein’s surface structure, which can then be used to infer information about its HOS.
All three MS-based approaches typically use proteolytic digestion, liquid chromatographic separation, and mass spectrometric analysis to identify modification sites as a way to provide localized or amino acid-level structural information; however, HDX and cross-linking methods have analysis challenges that are not present in CL techniques. In HDX, the inherent reversibility and lability of the modification can lead to back-exchange and scrambling that must be minimized via the use of specialized sample handling techniques. In cross-linking experiments, branched polypeptide chains are necessarily produced, requiring custom software and often specially-designed reagents to facilitate identification of the cross-linked sites. CL methods do not suffer from these limitations. Side chain modifications are irreversible, enabling the use of well-established proteomics workflows and sample handling techniques to facilitate the identification of protein modification sites. In comparison to cross-linking methods, identifying modified peptides and pinpointing their labeled residues is more straightforward.
CL methods also offer some valuable attributes for the structural analysis of proteins. CL reactions occur on protein side chains, providing complementary information to HDX/MS, which probes the protein backbone. Because most protein complexes (e.g. protein-protein, protein-ligand) are primarily mediated by side chain interactions, it can be argued that amongst MS-based methods CL techniques are perhaps best suited for identifying interaction sites in protein complexes. A unique attribute of some CL reagents is the ability to study protein folding reactions that occur on the μsec timescale, which is a timeframe inaccessible by most techniques. The very fast reaction kinetics of reagents such as hydroxyl radicals and carbenes makes this possible. Given the advantages associated with analyzing covalently labeled proteins and peptides, as well as the distinctive features this technique offers, there has been an increasing interest in developing CL methods for studying protein HOS.
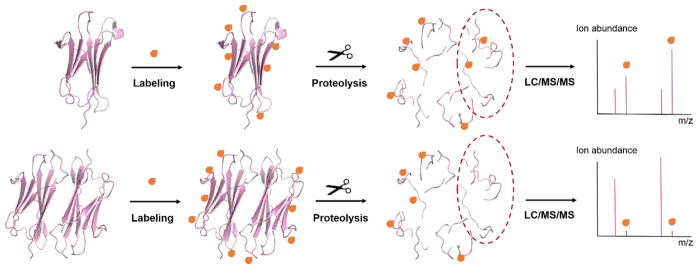
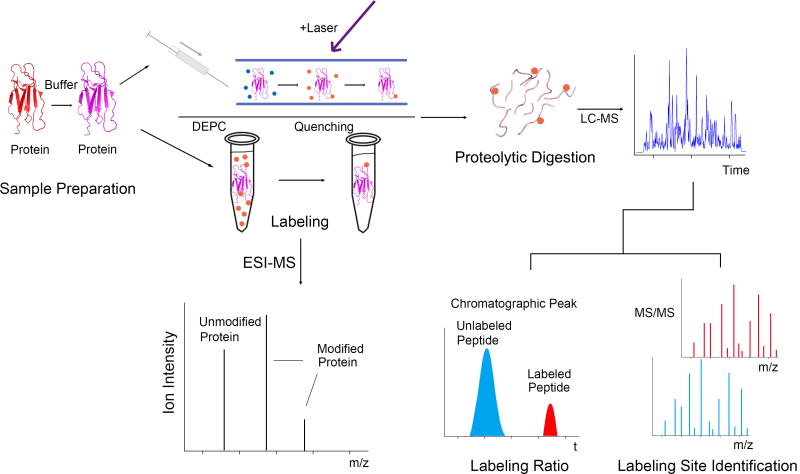
The goal of this review is to provide an overview of the most commonly used CL approaches. While there have been a large number of studies using amino acid specific labeling reagents together with MS to study protein structure [12], this review will focus on labeling reagents that are non-specific. Non-specific reagents are ones capable of modifying a wide variety of amino acid side chains simultaneously, thereby enabling broader structural coverage in a single experiment. The basic CL experiment is illustrated in Figure 1. A protein or protein complex of interest is exposed to a particular labeling reagent (see below for details), allowing solvent exposed amino acids to be modified. After halting the reaction, the modification sites on the protein are then identified using MS. Typically, this is done via proteolytic digestion and LC/MS/MS analysis to pinpoint the specific amino acid sites that have been modified, so structural information can be deduced. Structural information about a protein or protein complex is usually gathered by comparing a protein’s differential reactivity under two conditions (e.g. monomer vs. dimer to determine a binding interface as illustrated in Figure 1). A key assumption in all CL experiments is that amino acid residue will react to an extent that depends on their solvent accessibility, although solvent accessibility is not the only factor that influences the reactivity of a given amino acid residue.
Figure 1.
Scheme showing CL with MS detection. The label modifies solvent accessible amino acids. The modified protein or protein complex is subjected to proteolytic digestion and the modified peptides are analyzed using LC-MS/MS. Sites of protein conformational changes and/or protein-protein interactions can be revealed by changes in the extent of labeling at specific residues.
2. Factors Affecting Covalent Labeling Reactivity
2.1 Reagent Factors
Several reagents have been used to covalently modify amino acid side chains through oxidation or bioconjugate chemical reactions [12, 14, 15]. A portion of these reagents are used in covalent labeling – mass spectrometry (CL-MS) techniques for structural analysis of proteins and other macromolecules. In this section, we will describe how CL reactions are initiated and quenched, and the types of products that are generated.
2.1.1 Hydroxyl Radicals
Hydroxyl radicals (•OH) have been the most commonly used CL reagents when combined with MS detection. They are typically generated through radiolysis or photolysis of water or hydrogen peroxide (H2O2) [11]. The resulting •OH radicals then modify amino acid side chains on a protein’s surface via a cascade of reactions that typically begins with hydrogen abstraction and ends with formation of a new covalent bond on the side chain. High-flux X-ray or γ-ray radiation sources (e.g from a synchrotron) can be used to directly generate •OH radicals via the radiolysis of water [16]. Several excellent reviews from the Chance group have described synchrotron footprinting and its applications [17, 18]. One challenge associated with this CL approach is that a synchrotron source is necessary to directly produce •OH radicals, which limits its wide availability and applicability. In this synchrotron approach, •OH radicals are generated on a millisecond timescale, meaning protein unfolding could conceivably occur and compete with the labeling [19, 20], although adding radical scavengers to the solution can shorten the lifetime of the hydroxyl radicals and therefore shorten the reaction time frame. It is important to stress that the applicability of oxidative labeling reactions is determined not only by the radical lifetime but also the timescale of radical formation.
UV-light induced photolysis of H2O2 in aqueous solution is another common way to generate •OH radicals [21]; however, long UV irradiation times and concentrations of H2O2 up to 15% by volume are needed unless a UV laser is used [22, 23]. Laser photolysis by a Nd:YAG laser (266 nm) [22] or KrF excimer (248 nm) laser [23] was developed by Sze and co-workers and Gross and co-workers, and this approach allows H2O2 concentrations of less than 1% to be used, thereby limiting direct oxidation of sulfur groups by H2O2. Short laser pulses can shorten the time scale of radical formation to the nsec and μsec timescale [22, 23]. When used with the appropriate solution additives, this laser irradiation approach can oxidize proteins faster than they unfold, ensuring that there is no structural perturbation during labeling [24]. Gross and co-workers later coined the term “fast photochemical oxidation of protein” or FPOP for this approach.
Other reactions can also be used to generate •OH radicals for macromolecular labeling, including electron pulse radiolysis [25], metal-catalyzed oxidation (MCO) reactions [26–28], Fenton chemistry [29–31], disproportionation of peroxynitrous acid [32, 33], high voltage electrical discharge [34–36], electrochemical oxidation [37] and more recently, plasma generation [38]. Each of these •OH radical generation methods has unique aspects to their chemistry, but because they are not as widely used in CL of proteins, they will not be discussed further here. Interested readers are referred to a comprehensive review of hydroxyl radical CL by Xu and Chance [11].
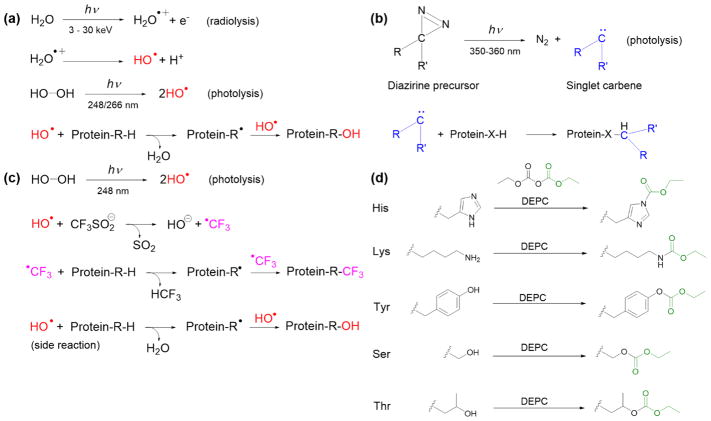
The general oxidation reactions that occur during radiolysis or UV photolysis are briefly summarized in Figure 2a. These reactions typically follow pseudo first-order kinetics [16, 24, 39] because of the excess concentration of •OH radicals. At least 14 of the 20 common amino acid side chains can be modified by •OH radicals, and over 50 different modification types on proteins can be generated [11, 13]. The most common products of aliphatic amino acids are addition of a hydroxyl group or a carbonyl group on the side chain, resulting in mass shifts of +16 Da and +14 Da, respectively. For aromatic residues, the addition of one or multiple hydroxyl groups to the aromatic ring results in mass shifts of +16. Asp and Glu residues generally undergo a −30 Da mass change due to oxidative decarboxylation, while basic side chains give rise to a series of unique oxidized products that include mass additions and side chain cleavages [11, 13]. More information about the residue-specific oxidation products can be found elsewhere [11].
Figure 2.
Modification reactions of amino acid residues used in non-specific covalent labeling with (a) hydroxyl radicals, (b) carbenes, (c) trifluoromethylation, and (d) diethylpyrocarbonate.
While •OH radicals can undergo self-quenching reactions in aqueous solution, supplemental reagents are typically added to solution to control the lifetime of the radicals or prevent unwanted side reactions. Secondary reactions from the presence of H2O2 and other oxidative species produced upon radiolysis are found to over-oxidize Met and Cys residues, thereby affecting analytical reproducibility and data interpretation regarding solvent accessibility [40]. Often, amino acids or other molecules possessing good reactivities towards •OH are selected as reaction quenchers. Addition of catalase or methionine have been found to minimize this secondary oxidation and improve quantitative protein labeling [40]. In FPOP, Gln or Phe is usually added prior to irradiation to scavenge •OH radicals and shorten their lifetimes in solution to the μsec time scale, so that oxidation can occur before any significant protein structural changes [23, 24] Even with these solution additives to control the highly reactive •OH radical chemistry, evidence has been provided that the timescale of protein oxidation can extend to tens of msec because of the formation of longer-lived secondary or higher order radicals [41].
2.1.2 Carbenes
Highly reactive singlet carbenes can be generated from the photolysis of diazirine derivatives (Figure 2b) by using near-UV wavelengths (≈350 nm) and like hydroxyl radicals, they can also be used for CL [42–44]. Carbenes can rapidly insert into any X–H bond (where X can be C, O, N, or S), and thus can potentially label any amino acid residues on a protein surface [44, 45]. Diazirine gas (CH2N2) was first used as a carbene precursor [46–49], but its poor solubility limits the extent to which proteins can be labeled with this reagent. A more useful carbene precursor is L-2-amino-4,4-azipentanoic acid (or photoleucine), which has good stability and solubility in water and has been successfully used to label proteins in several studies [44, 50, 51]. Recently, Manzi et al. reported a new aromatic diazirine precursor, 3-trifluoromethyl-3-phenyldiazirine, that has higher reaction efficiency than photoleucine [52].
The common reactions that occur during carbene production and labeling are briefly summarized in Figure 2b. The products of carbene labeling will have a substituent group of the diazirine precursor inserted into the amino acid side chains, whose mass shift is readily detected using MS. Carbene labeling is rapid, irreversible, and independent of protein concentration, implying the zero-order kinetics [44]. However, protein-dependent reactivity has been observed in some protein systems [52]. Addition of quenching agents is not necessary as carbenes are readily quenched by water. The lifetime of carbenes in aqueous solution is on the nsec time scale due to its rapid reaction with water, which allows carbene labeling to be faster than protein unfolding [44, 50]. A range of intermediates generated from aliphatic diazirines upon photolysis, such as triplet-state carbenes and diazo-mediated carbocations (diazo isomers), can result in side reactions [53]. These side reactions have been reported to be reduced somewhat by tuning functional groups on the diazirine precursor [52, 54]. Oxidation side products have also been observed after irradiation but typically at low levels [53].
2.1.3 Trifluoromethylation (CF3)
Very recently, •CF3 radicals have been introduced as labeling reagents that can be generated by pulsed laser photolysis of triflinate (Langlois’ reagent) [55]. Laser irradiation is performed in the presence of H2O2 (•OH source) and the water-soluble salt NaSO2CF3. The resulting •OH rapidly reacts with excess [SO2CF3]− to generate •CF3 (Figure 2c). The highly reactive •CF3 can insert into X-H bonds (where X is C, O, N, or S) of amino acid side chains, resulting in a mass shift of +67.987 Da. •CF3 reacts with 18 of the 20 common amino acids, including those that are relatively unreactive with hydroxyl radicals (Gly, Ala, Ser, Thr, Asp, and Glu). Because hydroxyl radicals are necessary to generate •CF3, side reactions with •OH or dissolved oxygen species during photolysis yield oxygen-incorporated products, although CF3-substitued products dominate [55]. Radical trifluoromethylation likely follows pseudo first-order kinetics because of the excess concentration of [SO2CF3]− in the reaction mixture, but this has not been verified yet. There is no need for addition of quenchers as the reactive •CF3 is readily quenched in water, having a lifetime of ~ 30 msec [56]. Using the FPOP platform along with the appropriate solution quenchers (e.g. catalase and methionine), •CF3-based CL should be faster than protein unfolding, ensuring structural integrity upon labeling [24].
2.1.4 Diethylpyrocarbonate (DEPC)
Diethylpyrocarbonate (DEPC) is a commercially-available reagent that can react with a range of nucleophilic residues. Unlike the radical reagents where specialized equipment is needed to generate the radicals, DEPC directly labels proteins when added to solution. A stock solution of the reagent is typically prepared in anhydrous acetonitrile due to its limited solubility and propensity to be hydrolyzed in water. DEPC concentrations ranging from 0.01 mM to 40 mM are readily soluble in aqueous solutions and are also useful concentrations for labeling proteins [57]. While DEPC will eventually be hydrolyzed in water, reactions between this reagent and proteins are typically quenched after a short time (10 sec – 60 sec) by adding relatively high concentrations (~20 mM) of a nucleophilic compound such as histidine [12, 58].
DEPC is a reactive electrophile that can modify nucleophilic side chains (Cys, His, Lys, Thr, Tyr, Ser) and N-termini via nucleophilic substitution reactions (Figure 2d) [12, 57–59]. Under the most relevant protein labeling conditions, the reaction follows second-order kinetics, meaning the reaction rate depends on both protein and DEPC concentrations [58]. Because DEPC reacts more slowly than radical reagents, its concentration and reaction time must be carefully controlled to prevent over-modification and preserve the structural integrity of the protein during the labeling reaction. Carbethoxylated products with a mass shift of +72.021 Da are obtained for Cys, His, Lys, Thr, Tyr, Ser, and the N-terminus. One key advantage of this labeling reagent is that it generates a single type of product, which simplifies identification of labeled sites and improves sensitivity as the signal is not distributed among numerous products. The addition of a second carbethoxyl group to His residues has been reported, but this modification is typically avoided by labeling at the low DEPC concentrations necessary to ensure the structural integrity of proteins [12]. Some modified residues, especially Ser and Thr, are subject to hydrolysis and thus label loss if they remain in solution too long, so proteolytic digestion and LC/MS/MS analysis must be performed soon after the labeling reaction is completed [59]. Moreover, DEPC label scrambling can happen in solution if free Cys residues are available. This label scrambling can be easily avoided, though, by alkylating any free thiols after disulfide reduction [60], as is typically done in most proteomics experiments.
2.2 Protein Factors
Structural analysis of proteins using CL-MS relies on the fact that in different protein conformers a certain set of residues possesses differential reactivity with the labeling reagent, and the resulting difference in modification extents of those amino acids can be used to distinguish protein topologies, conformations, and/or binding sites. The protein factors that affect the reactivity of amino acid side chains are described below.
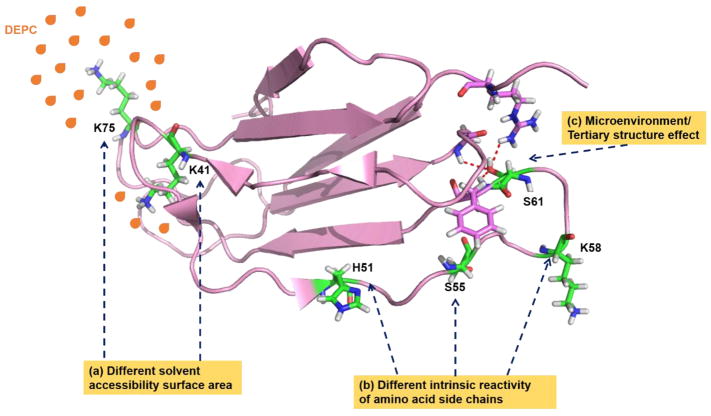
2.2.1 Solvent Accessibility
All CL reactions used to study protein structures are performed in the aqueous phase, to preserve the HOS of proteins. The implicit assumption in CL-MS is that amino acids that are exposed to solvent and accessible to the CL reagent can be modified. Meanwhile, buried residues will be modified slowly or not at all (Figure 3). However, solvent accessibility is not the sole factor that governs reactivity. Different amino acid side chains can react differently with a given labeling reagent, as will be discussed in section 2.2.2. Several groups have established that a qualitative relationship exists between solvent accessible surface area (SASA) and extent of labeling for certain types of residues [11]. For example, Chance and co-workers have found that hydroxyl radicals primarily react with surface accessible residues [11, 16, 61, 62] and that residues at protein-protein interfaces are generally protected from modification [61]. Moreover, in several proteins, oxidation rates are found to correlate fairly well with calculated SASAs of residues from known NMR and X-ray crystal structures [21, 23, 62]. A clear relationship between SASA of individual amino acid residues and carbene reactivity has not been observed, possibly due to the carbene precursor’s affinity for certain residues, and the different intrinsic reactivities of amino acid residues towards carbene labeling [44, 50]. Despite these issues, Delfino and co-workers have shown using diazirine gas as a carbene precursor that global levels of solvent accessibility for different α-lactalbumin and β-lactamase conformers are related to carbene labeling [47, 49]. This group also found a similar relationship for antibody-bound lysozyme [48]. More recently, an improved correlation between SASA and extent of carbene labels at the residue level has been demonstrated in experiments where the protein sample is flash-frozen during irradiation [53] (see section 2.2.2 for further discussion of this observation). For trifluoromethylation, Gross et al. have reported that this reagent’s reactivity patterns are consistent with SASA in a model membrane protein [55]. Our group has also demonstrated that DEPC labeling can be used to study protein topology, and DEPC modification extents for certain types of residues are found to be consistent with solvent accessibility [58]. Similarly, decreases in DEPC modification levels upon binding to transition metals are consistent with the general trend, and actually allow metal binding sites to be determined [28, 58, 63].
Figure 3.
Protein factors that affect the reactivity of CL reagents. Factors include (a) solvent accessible surface area (SASA), (b) the intrinsic reactivity of amino acid residues, and (c) primary and tertiary structure.
Differences in SASA of reactive side chains, resulting from their involvement in a conformational change and/or protein-ligand binding, will also cause amino acids to be more or less accessible to a given CL reagent and therefore, react to greater or lesser extents. Most recently, researchers have attempted to further refine and quantify the relationship between SASA and labeling extent. Yang et al. and Sharp et al. have proposed the use a protection factor (PF) to quantitatively relate hydroxyl radical reaction rates to amino acid SASA using well-characterized protein models [64, 65]. These PF are obtained by normalizing the measured labeling rate constant with the side chain’s intrinsic reactivity. More details about the applications of PF in mapping conformation changes and in modeling protein structures can be found elsewhere [13, 66].
2.2.2 Intrinsic Reactivity of Amino Acid Side Chains
Attempts to use a PF to quantify the relationship between SASA and reactivity imply that the intrinsic reactivity of a given amino acid side chain influences its reactivity (Figure 3). Thus, this factor must be considered when interpreting CL data. While •OH is highly reactive and can modify a wide range of amino acid side chains, rate constants for reactions with the 20 common amino acid residues vary over three orders of magnitude with the following reactivity order: Cys > Met > Trp > Tyr > Phe > His > Leu, Ile > Arg, Lys, Val > Ser, Thr, Pro > Gln, Glu > Asp, Asn > Ala > Gly [67]. Although every amino acid can react with hydroxyl radicals, practically speaking Gly, Ala, Ser, Thr, Asp, and Glu are often not found labeled during protein CL experiments with hydroxyl radicals. Therefore, only 14 of the 20 side chains are useful in typical labeling experiments with hydroxyl radicals, and these residues typically account for ~65% of the sequence of the average protein [11, 67]. Sharp et al. have demonstrated that normalization of a residue’s intrinsic reactivity via a PF is needed to obtain a qualitative correlation between SASA and labeling extent [65].
While CL with carbenes was initially predicted to be affected less by differences in amino acid side chain chemistry than hydroxyl radicals, studies suggest that there is a decidedly non-uniform distribution in carbene labeling of proteins [44]. Schriemer and co-workers have found that using photoleucine as a diazirine precursor leads to Glu residues being preferentially modified, which might be explained by the reagent’s affinity for negatively charged residues or the reactive carbenes favoring polar protic bond (O-H) insertion rather than C-H bond insertion [50]. Oldham et al. have also found that hydrophobic and basic residues are favored in the carbene labeling using an aryldiazirine precursor [52]. In the more recent study from Schriemer et al, surface bias is still found in the CL with carbenes generated from other different aliphatic diazirine precursors [53]. By performing carbene footprinting on flash-frozen samples to limit the diffusion of carbenes to sites of higher reactivity during irradiation, the preferred sites of labeling tend to reflect the sidechain interactions with diazirine precursor, which is influenced by the diazirine substituents rather than the intrinsic reactivity of residues [50, 52, 53]. The resulting surface biases can be avoided somewhat by tailoring the diazirine precursor or changing solution conditions to reduce the reagent’s affinity toward specific residues [50, 52].
Because •CF3 radicals have only recently emerged as CL reagents, the full details of their reactivity have not been studied enough yet. However, aromatic residues (e.g. Trp and Phe) are found to be modified at relatively high levels, indicating their good intrinsic reactivity toward •CF3 labeling [55]. For DEPC labeling, the intrinsic reactivity of side chains is governed by the nucleophilicity of residues [57–59]. Thus, the reactivity of Tyr, Ser, and Thr residues is lower than the reactivity of Cys, His, and Lys residues due to their weakly nucleophilic hydroxyl groups [58].
2.2.3 Primary and Higher Order Structure Effects
In the structure of an intact protein, the sequence context (primary structure) may contribute to the reactivity of amino acid due to electron donating and/or electron withdrawing effects. This fact appears to be true even for hydroxyl radical labeling. Sharp and co-workers have demonstrated that accurate measures of SASA for residues with poor intrinsic reactivity (e.g. Arg, Lys, Val, Thr, Ser, Pro, Glu, Gln, Asn, Asp, Ala) during hydroxyl radical CL experiments require the normalization of sequence effects by comparing label profiles of native and denatured structures [65]. For highly reactive residues (e.g. Trp, Tyr, Phe, His, Ile), the effects of primary structure on reactivity are relatively minimal [65]. In addition, the microenvironment (Figure 3) caused by the tertiary structure around an amino acid residue can influence the acid/base characteristics of side chains because of charge-charge interactions, charge-dipole interactions, dipole-dipole interactions, and/or hydrophobic effects [68–71]. For example, in dimethyl labeling proteins, Wang and co-workers found that hydrogen bonding and electrostatic interactions around lysine residues can influence the methylation reactivity of lysine residues [72]. The effects of local structural contacts of amino acid side chains, especially those with lower intrinsic reactivity, have been found to contribute to their reactivity with hydroxyl radicals [64, 65]. In CL with carbenes, the surface bias caused by the reagent’s affinity for specific protein surface sites might be explained by primary and/or tertiary effects. Sequence context and non-covalent molecular interactions (higher-order structures) may drive and steer the selectivity of diazirine precursors toward certain residues. [50, 52, 53]. However, no definitive experiments have been performed to demonstrate this hypothesis yet. For reagents like DEPC that are inherently less reactive than hydroxyl radicals and carbenes, the effect of primary or tertiary structure might be expected to be more pronounced since changes in the pKa values of a given residue lead to changes in its protonation state and thus nucleophilicity. The relatively poor correlation observed between SASA and the DEPC reactivity of Ser and Thr residues [58, 59, 73] might imply that the microenvironment around these amino acids, as formed by a protein’s tertiary structure, tunes their reactivity.
3. Using Covalent Labeling with Mass Spectrometry Detection
3.1 Principle of Experiment Design
During CL experiments only solvent exposed residues are modified and thus provide direct structural information, while the buried residues are unmodified and thus indirectly provide structural information. As illustrated in Figure 1, any structural information obtained from a CL experiment comes from comparing the labeling ratio of the protein reacted under at least two conditions, with one of the conditions usually being the protein in its native state. The resulting differential reactivity is used to deduce structural information. Another important principle of CL experiments is that the labeling reagent must be used in such a way as to not perturb a protein’s HOS. Clearly, if the probe itself changes the protein’s structure, then the resulting data will not correctly report on the protein’s structure.
There are at least three general ways that are used to assess that a protein’s structure is not perturbed during CL. The first is to use a complementary measurement such as CD, fluorescence, or an activity assay to monitor if the protein’s structure has changed during labeling [52, 58, 73, 74]. Our group has demonstrated in previous work, that while these methods are easy to implement, they tend to indicate global structural changes and lack the resolution to identify local protein structural changes [58]. A comparable approach is to measure the charge-state distributions of a protein via electrospray ionization (ESI) MS [75] as a protein will undergo a shift in its charge-state distribution upon a structural change. However, like with CD and fluorescence spectroscopy, changes in ESI-MS charge-state distributions are typically not sensitive enough to monitor local changes in HOS, so it is debatable whether these approaches are sufficient for assessing if a protein’s structure is perturbed or not from the labeling reaction.
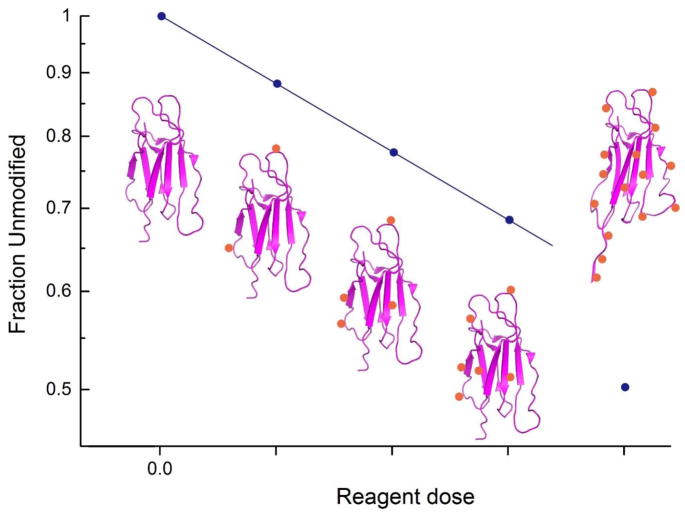
A second commonly used approach for assessing a protein’s structural integrity during CL is to measure labeling reaction kinetics (Figure 4). Chance and co-workers pioneered this approach with synchrotron-based hydroxyl radical labeling in which the unmodified fraction of a peptide or protein is monitored as a function of the reagent dose (e.g. radiolysis time). In this way, dose-response plots can be generated, and adherence to the proper reaction order kinetics indicates whether a protein’s structure has been perturbed or not [76, 77]. These plots can be generated for all modified peptides that are produced from a proteolytic digestion and provide a measure of the structural integrity throughout the entire protein. Our group has successfully used an analogous approach with DEPC labeling [58, 59, 63, 78, 79], which follows second order kinetics, and we have found this approach to offer much higher sensitivity than CD or fluorescence spectroscopy for ensuring structural integrity through all regions of a protein.
Figure 4.
Hypothetical dose-response plot, showing first-order labeling reaction kinetics. When the extent of labeling is low enough to prevent structural perturbation, reaction kinetics are monotonic (i.e. four lowest reagent doses in the plot). When over-labeling perturbs the structure of the protein, the reaction kinetics change, indicating variations in the microenvironment around one or more residue.
A third method of guaranteeing a protein’s HOS is not been perturbed by CL is to ensure labeling chemistry happens faster than structural changes can occur. As indicated in previous sections (see section 2.1), FPOP-based hydroxyl radical labeling and carbene labeling have extremely fast labeling rates that are comparable to or faster than protein folding rates [23–25, 44, 50]. The fastest protein structural changes occur in a few μsec [20, 80], and thus if hydroxyl radical or carbene labeling occurs faster, then protein modification should take place before any significant protein structural changes occur. In the FPOP experimental setup, hydroxyl radicals are generated during the nsec pulse of the laser [23], and radical scavengers that are present in solution limit the overall lifetime of the hydroxyl radicals. Calculated lifetime profiles [23] and time-resolved UV spectroscopy [25] suggest that reactive hydroxyl radical species are consumed in 0.1–1 μsec when radical scavengers are present. Such short lifetimes allow the radical-protein reactions to happen faster than protein structural changes. Further proof that proteins are modified before they can undergo structural changes comes from the Poisson distribution of modification extents, which suggest that only a single protein species is reacting [24]. Despite evidence that hydroxyl radical labeling by FPOP occurs on the μsec timescale, recent work by Konermann and coworkers suggests that the time window for FPOP-based labeling may be much longer than one millisecond as secondary and higher order radicals have longer lifetimes and are possibly not effectively quenched [41]. At this point, however, it is unclear the extent to which secondary radicals are responsible for the measured protein oxidation.
It should be noted that there are numerous examples in the literature in which nothing is done to ensure a protein’s structural integrity during CL. This approach can be successful if “single-hit” conditions are achieved [81]. In other words, as long as there is one label per protein on average, the labeling chemistry should have successfully probed the original protein structure. Any additional label, by definition, would be probing a possibly perturbed structure because the protein would now have a covalent modification that it did not have originally. While this has commonly been acknowledged, recent work by Madsen et al [82] on monoclonal antibodies (mAbs) suggests that multiple labels can be accommodated while still obtaining the desired information. It is unclear at this point how generalizable this observation is.
3.2 Experiment Design and Workflow
The goal of a CL experiment is to encode structural information into the mass of the protein. The commonly used workflow for CL experiments is sample preparation, labeling reaction, and, if necessary, quenching to consume excess reagent. After the CL experiment, the labeled protein sample can be directly analyzed by MS or LC-MS, or more typically, the labeled protein can be proteolytically digested prior to LC-MS (Figure 5). While standard proteomics techniques are typically used to analyze covalently labeled proteins to obtain the desired structural information, the following unique aspects of the CL approach must be taken into consideration.
Figure 5.
Example workflow for covalent labeling combined MS. Before MS analysis, a protein sample is prepared and labeled, during which the structural information is encoded into the mass of the protein. The labeled intact protein sample can be analyzed by ESI-MS to monitor the overall extent of modification, or more typically the labeled protein is subjected to “bottom-up” analysis via proteolytic digestion and LC/MS/MS to determine labeling ratios at each modified residue.
Unlike in typical proteomics experiments, CL-MS experiments require complete or near complete protein sequence coverage to obtain the desired structural information. Moreover, a semi-quantitative measure of labeling extents at residues throughout the protein is needed to deduce structural information in a differential experiment (see Figure 1). Like in proteomics studies, the two primary strategies for determining labeling sites are “top-down” sequencing [83–85] and “bottom-up” sequencing [83, 86, 87]. In top-down sequencing, the entire, intact protein is analyzed with MS/MS, and therefore complete sequence coverage is inherently available. In top-down sequencing, it is possible to select and apply MS/MS to the protein with only one label. This is beneficial if there is a concern that multiple labeling events could have perturbed the structure (“single-hit” principle). Despite these advantages, very limited work has been published using top-down MS to analyze proteins covalently labeled by non-specific reagents. The limited utility of top-down sequencing for CL-MS is mostly due to the relatively poor ability of top-down sequencing to localize modification sites, especially for large protein systems. Often large fragment ions containing multiple possible modification sites are produced, making it difficult to assign labeling sites to specific residues with high confidence. This difficulty effectively results in low structural resolution. In addition, semi-quantitative information that is usually required in CL-MS experiments is challenging to obtain with top-down sequencing. One study by Gross et al. compared top-down and bottom-up sequencing for analyzing covalently labeled proteins and concluded that top-down sequencing had limited sensitivity, a low dynamic range, and sometimes difficulty pinpointing modified residues in regions with numerous modified sites [88]. It should be noted that the utility of top-down sequencing for CL-MS experiments might improve with the use of UV photodissociation, as this dissociation technique has shown utility for identifying modified sites during amino-acid specific labeling experiments [89].
Bottom-up sequencing is a more commonly used strategy to analyze covalently labeled proteins. Because MS/MS is more effective on smaller peptides, this strategy provides high spatial resolution, usually pinpointing labeled sites down to 1 or 2 amino acids. Moreover, modification extents can usually be determined as low as 0.1% or lower. Bottom-up sequencing of covalently labeled proteins still suffers from the usual challenges associated with bottom-up sequencing, such as biases associated with proteolytic cleavages and peptides going undetected during LC/MS experiments. For large proteins, where sequence coverage during bottom-up sequencing can be low, using multiple enzymes for proteolytic digestions can be used to improve coverage [90].
An important consideration associated with bottom-up sequencing is the choice of dissociation techniques during the MS/MS stage of the experiment. Collision-induced dissociation (CID) remains the most commonly used dissociation technique and provides accurate information in most cases. There are examples, however, in which CID has difficulty with certain types of modifications and modified amino acid residues. For example, CID provides very poor sequence coverage for peptides containing oxidized cysteine or methionine residues [91]. The dissociation technique has also been known to lead to label misassignments for peptides in which histidines are oxidized, especially when more than one modification site exists in a peptide, as oxidation causes new dissociation pathways to emerge [92]. Sharp and co-workers reported a similar conclusion based on studies of several oxidatively modified peptides [93]. Significant neutral losses are also observed in CID of peptides modified by carbenes, making it difficult to pinpoint modification sites [50]. Another issue observed with a subset of DEPC-labeled peptides is the possibility of the label to “scramble” from one site on a peptide to another during CID, presumably due to an intramolecular nucleophilic attack during the CID process [94]. In each of the cases in which CID provided incomplete or misleading information about modification sites, electron transfer dissociation (ETD) was successfully used to correct the problem. In fact, it has been argued that ETD generally provides improved identification and quantitation of labeled sites produced during oxidative and DEPC labeling experiments [50, 94, 95].
3.3 Data Analysis and Presentation
The desired results from CL-MS experiments are measurable labeling levels at residues throughout the protein so that structural information can be obtained. The more labeled sites that are measured, the higher the structural resolution will be from the CL-MS experiment. In addition, some semi-quantitative measure of the labeling extent at each residue is desired so that any corresponding structural change can be deduced. It is rare that a particular residue is labeled in 100% of the protein molecules. More commonly, only a fraction of the protein molecules in a sample are labeled at a given site, and this labeling extent is usually provided as a labeling percentage from LC-MS measurements. Decreases in labeling percentages at given residues typically indicate a drop in solvent accessibility that could, for example, indicate a protein-protein interaction site (Figure 6). In contrast, increases in labeling percentages at a given site would suggest unfolding in that region of the protein.
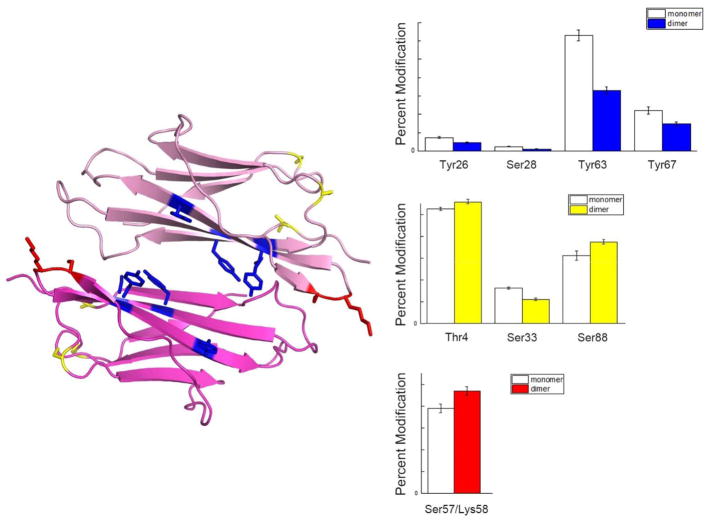
Figure 6.
An example of how changes in labeling level can be applied to predict the protein dimer interface demonstrated with β2m dimer structure extracted from β2m H13F hexamer (PDB 3CIQ). Blue colored residues indicate significant decreases in labeling level from monomer to dimer, which helps locate the dimer interface. Yellow colored residues show no significant change in labeling level, which indicates these residues are at a region away from dimer interface. The red colored residues are examples of residues with increases in labeling levels, indicating these residues becomes more solvent exposed upon dimer formation.
While residue labeling identification is achieved using tandem MS, the labeling extent is usually determined by the ion abundance or chromatographic peak area of the labeled peptide that contains the modified residue. In addition to the use of standard software packages for analyzing such data (e.g. Mascot [96, 97]), other programs such as ByOnic [51], ProtMapMS [98], a custom-designed version of Proteome Discoverer [99], and other specialized software pipelines [73] have been created to facilitate CL-MS data analysis. A key difference between these specialized software and traditional proteomics software is that they are much more efficient at finding labeling sites, especially at low levels, while also providing a measure of the labeling percentage at every identified site.
While it is important to obtain a measure of labeling extent, it should be noted that explicit quantitation is not attainable through most CL-MS experiments. There are many reasons for this fact. First off, modified and unmodified peptides are distinct chemical species that have different inherent ionization efficiencies. Moreover, a difference in chromatographic retention times of modified and unmodified peptides during reversed-phase LC gradient elutions leads to a different percentage of organic solvents from which each peptide is electrosprayed, resulting in further differences in ionization efficiencies. These ionization efficiency differences cause there to be a distinct relationship between solution concentration and ion abundance for modified and unmodified peptides. To remediate the ionization efficiency bias that occurs during LC-MS analyses, Sharp and co-workers have employed size-exclusion chromatography (SEC) instead of reversed-phase LC to separate labeled peptides. SEC ensures all peptides are electrosprayed under common solvent conditions, thereby eliminating one cause of ionization efficiency differences. A drawback of this approach is that isomeric labeled peptides co-elute, making it more difficult to quantify labeling extents for peptides with more than one modified residue. To some degree, this drawback can be overcome using ETD, as tandem mass spectra from ETD can often correctly report modification levels of each residue in an isomeric mixture [93].
4. Applications of Covalent Labeling – Mass Spectrometry Methods
The use of CL-MS methods to obtain structural information about protein topology has gained popularity because of its moderate to high structural resolution, straightforward implementation, and/or ability to probe structural changes on a μsec time scale. In this section, we describe some select examples of CL-MS applications.
4.1 Protein Folding/Unfolding
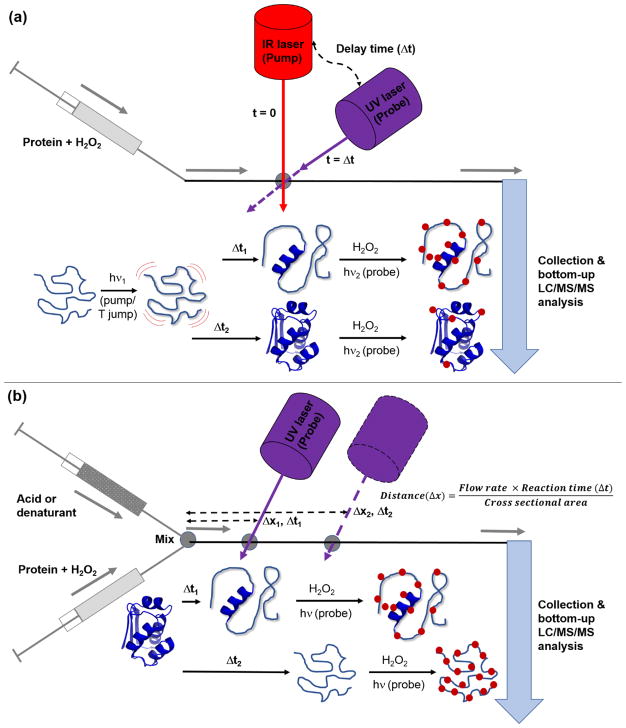
In most proteins, specific biological functions can be exerted only after folding into their appropriately folded forms. Deciphering protein folding/unfolding is necessary to understand protein-energy landscapes, the biological processes of protein misfolding disorders [100–102], and is essential for the development of therapeutics against these disorders [101]. While numerous techniques, including other MS-based techniques, have been developed to monitor conformation dynamics during protein folding/unfolding [3, 103], protein surface mapping with CL-MS can provide information with high spatial and temporal resolution, in a way that many other techniques cannot. Laser-based CL methods, such as FPOP, have proven to be useful tools for protein folding studies because proteins are labeled on the μsec time scale, which is faster than protein unfolding, due to the pulsed nature of reagent production and the rapid reaction quenching [23, 24]. Using FPOP together with denaturants such as temperature and pH, sub-msec protein folding experiments can be performed. For example, Gross and co-workers used temperature jump experiments to study the region-specific folding of the protein barstar. Barstar in aqueous buffer undergoes reversible unfolding at low temperature (~ 0 °C) [104], and ‘pump/probe’ experiments can be conducted to study the protein’s refolding (Figure 7a). Refolding of unfolded barstar was created by rapid IR laser-induced heating of a solution containing the protein (‘pump’ from ~ 0 °C to ~ room temperature), and then FPOP with a ‘probe’ UV laser was used to oxidize the protein at different times after it began refolding. By limiting the lifetime of the hydroxyl radicals in solution to around 1 μsec, ‘snapshots’ of the protein as it folded could be obtained [105–107]. Their results confirmed the presence of partially-folded intermediate conformations of barstar during its refolding (possibly a molten globule state), which was consistent with previous UV absorbance, fluorescence, and CD spectroscopic studies [108–111]. The CL-MS experiments also revealed residue-level information about conformational changes during barstar folding, which was not available from previous spectroscopic measurements. In a similar manner, the same group studied the refolding kinetics of the paramyxovirus fusion protein, which is essential in mediation of virus-host cell fusion and genetic transfer [112]. Besides temperature-triggered folding experiments, Konermann and co-workers established a platform to study short-lived protein folding/unfolding intermediates using continuous-flow mixing and pulsed laser-induced oxidative labeling [113–116]. To examine protein unfolding, a protein of interest is premixed with H2O2 in a reaction capillary and unfolding is initiated by rapid mixing with acid or denaturant [113, 115]. To study protein folding, a denatured protein is prepared by acid or denaturant, and the folding is then initiated via a pH jump or rapid denaturant dilution [114, 116]. To study conformations at different time points after initiating unfolding or folding, a pulsed laser beam is focused at different positions along the mixing capillary, and a certain amount of the resulting oxidized sample is collected for further bottom-up LC-MS/MS analysis (Figure 7b). Folding/unfolding intermediates of bacteriorhodopsin, apo- and holo-myoglobin have been studied by this method, and the in-depth residue-level information from the rapid pulsed CL, in combination with the global structural features obtained from optical spectroscopy, can provide good spatial and temporal resolution of protein folding/unfolding pathways [113–116].
Figure 7.
An illustration of the experimental setups used for studying protein folding/unfolding using laser-based CL methods. Schematic diagrams are shown for (a) temperature jump (T jump) and (b) continuous-flow mixing experiments coupled with pulsed laser-induced oxidative labeling. Details about each experiment are provided in the text.
4.2 Amyloid-Forming Proteins
Amyloids are insoluble protein aggregates that are associated with several human diseases, including Alzheimer’s and Parkinson’s [117]. Numerous efforts have been devoted to deciphering the aggregation process in vitro, but this information is challenging to obtain because amyloid proteins usually aggregate rapidly [118]. Moreover, pre-amyloid oligomers, which may in some cases be disease causing [119–121] are short-lived and can be difficult to structurally interrogate [81, 122, 123]. As compared to NMR and X-ray crystallography, CL can provide better time resolution for studying transient pre-amyloid structures, and various CL approaches have been used to gain insight into the aggregation interfaces for several amyloid proteins. This structural information can be obtained by comparing the CL ratio of residues before and during oligomer formation to help identify aggregation interfaces. As an example, Chance and coworkers applied synchrotron-based hydroxyl radical CL to Aβ40, which is the protein that aggregates in Alzheimer’s disease [124]. Protection factors were used to indicate the degree of change in solvent accessibility. Structural information of full-length Aβ40 monomer in fibrils and the core structure of two- and three-filament conformers were acquired and were compared to the previously reported structures by solid state NMR. Gross and co-workers applied FPOP in a time-dependent manner to study the pre-amyloid formation process of Aβ1–42 [125]. By plotting the fraction of modified intact Aβ1–42 as a function of time, a kinetic scheme of the aggregating process was revealed. Similarly, by plotting the labeling percentages of modified peptides over time, the regions with significantly decreased labeling ratios indicated the aggregating core structure. Moreover, because MS/MS can indicate the exact labeled residues, even more detailed structural information could be obtained. DEPC labeling has also been successfully used to study the pre-amyloid oligomers of β-2-microglobulin (β2m), which is the protein that forms amyloids in dialysis-related amyloidosis [126]. By conducting DEPC labeling at different time points after initiating amyloid formation with Cu(II), the structural changes to the monomer caused by Cu(II) binding [63], as well as structural models for the pre-amyloid dimer [78] and tetramer [79] could be determined.
4.3 Protein-Ligand Systems
CL is also well suited for studying protein-ligand complex binding sites when traditional methods, such as NMR and X-ray crystallography, are too time-consuming or fail due to limited sample amounts, protein instability, sample heterogeneity, protein molecular weight limitations, or difficulties with crystallization. In its most straightforward implementation, CL-MS can report on ligand binding sites via decreases in labeling extents at specific residues in the protein. Ligand binding protects residues on the protein surface from being labeled. Many examples of CL used for protein-ligand binding studies are available [81, 95, 127–132]. One example by Manzi et al. demonstrated that carbene-based labeling is capable of reporting the binding site of the lysozyme-NAG complex [52]. Multiple residues showed a significant decrease in labeling extent in the presence of the ligand, clearly correlating those residues to the well-studied binding pocket of the complex. Similarly, our group has applied DEPC labeling together with two other site-specific labeling reagents to study β2m binding to small molecules that inhibit its amyloid forming. The simplicity and speed of the labeling chemistry allows for ligand binding site information on a fast aggregating protein whose ligand binding site could not otherwise be determined [81]. Combining CL-MS with computational docking further refines the structural information that is acquired. CL-MS can test the pose predicted by docking [81, 131, 132] or restrain the region of protein to which the ligand is bound, but CL-MS is usually unable to show the orientation of the ligand, which is information that computational docking can provide. In one example reported by Chance and co-workers [131], a hybrid structural approach that include synchrotron-based hydroxyl radical CL, homology modeling, computational modeling, and protein ligand docking was used to predict the structure of the membrane protein Serotonin Type 4 Receptor (5-HT4R) and bound ligands. In addition to providing protein-ligand binding site information, CL has been shown to provide information about conformational changes upon ligand binding [95, 129, 130]. Gross and co-workers applied FPOP to study heme binding and conformational changes in myoglobin [129]. Several residues that are close to the heme binding site show significant differences in their ability to be modified in apo or holo form of myoglobin. In another case, the structural changes caused by the binding of the peptides melittin and mastoparan to calmodulin were compared to the known structure of the calmodulin-M13 complex [130]. The CL results indicated statistically similar patterns in labeling extents, leading to the conclusion that ligand binding changed the structure of calmodulin but the ligand bound calmodulin retained a similar structure when bound to M13, melittin, or mastoparan. It should be noted that distinguishing between ligand binding and ligand-induced conformational changes must be done carefully. For protein-ligand complexes with unknown binding sites, a decrease in labeling of a given residue (or a set of residues) might be due to protection from ligand binding or it could be due to allosteric changes that decrease the solvent accessibility of residues not directly involved in ligand binding. Further application of CL to protein-ligand binding should reveal whether such allosteric changes are likely to significantly change labeling patterns.
4.4 Structural Analysis of Protein Therapeutics
Protein therapeutics are the fastest growing pharmaceuticals on the market and have important roles in almost every field of medicine [133, 134]. Changes in HOS, e.g. protein misfolding and aggregation, can reduce drug efficacy, enhance unwanted drug actions, and/or induce immunogenicity [135, 136]. Reliable analytical tools are currently needed for characterizing the HOS of biopharmaceuticals to ensure the quality, efficacy and safety throughout a product life cycle – from manufacturing to dose administration [135, 137–140]. CL-MS techniques have the ability to probe the structure of protein therapeutics with moderate structural resolution and could be used in a semi-high throughput manner to monitor numerous therapeutic preparations [73, 141, 142]. FPOP along with MS detection has been shown by Watson and Sharp to be sensitive enough to detect subtle structural changes of expired and mishandled granulocyte colony-stimulating factor (GCSF) samples [141]. Deperalta et al. demonstrated the ability of hydroxyl radical labeling to characterize the dimer interface of an antibody dimer commonly found in formulated therapeutic IgG1 mAbs. Hydroxyl radical labeling data obtained using synchrotron radiolysis of water has shown that the Fab domain displays decreased oxidation rates and is likely involved in dimer interface [142]. Our group has also demonstrated that DEPC labeling along with MS can identify minor conformation changes in thermally-stressed human growth hormone (hGH) and IgG1 mAb samples. Heat-stressed hGH undergoes very subtle structural change that other biophysical techniques (CD, fluorescence spectroscopy, dynamic light scattering) are not sensitive enough to detect, but DEPC labeling along with MS detection can detect with good structural resolution [54]. We have also found that DEPC labeling can reveal the aggregation interface in a heat-stressed IgG1 mAb sample [73]. Epitope/paratope mapping of the specific residues involved in antibody-antigen and other protein-receptor interactions is particularly valuable for assessing epitope novelty, predicting immunogenicity, evaluating binding characteristics, and for optimizing binding [143]. Recently, Gross and co-workers used FPOP techniques to identify the interface of anti-thrombin mAb/thrombin [96], anti-vascular endothelial growth factor (VEGF) Fab-1 fragment/VEGF [97], anti-interleukin-23 (IL-23) mAb/IL-23 [90], and IL-6/IL-6 receptor interactions [144].
5. Conclusion and Outlook
5.1 Summary
In this review, we have provided an overview of CL-MS methods that use reagents that react with a wide range of amino acid residues, including hydroxyl and trifluoromethyl radicals, carbenes, and DEPC. These broadly reactive reagents are making CL-MS methods of increasing interest for the structural analysis of macromolecules. CL-MS fills in the gap among current structural characterization techniques, with higher resolution than CD and fluorescence spectroscopies, and faster measurement times than NMR and X-ray crystallography. At the same time, it retains the limited sample consumption common to most MS-based methods and can be applied to protein systems that are difficult to study by other techniques. CL-MS has very limited back exchange and label loss, allowing greater sensitivity to structural changes in some cases and better reproducibility in most cases than HDX-MS. The fact that only solvent accessible amino acid resides are modified makes the technique particularly well suited for studying protein interactions with ligands and other proteins, including aggregating proteins. Radical and carbene-based labeling can monitor protein structural changes on the μsec timescale, allowing the study of protein folding and unfolding.
While significant progress has been made with CL methods to address protein structure, some technique limitations remain. Unlike most spectroscopic methods, CL-MS relies on changing a protein in order to probe its structure. Covalent modification of amino acid residues is needed to encode a protein’s structural properties into its measured mass, and this necessarily changes the protein. Thus, every labeling procedure has to be carefully controlled and understood to ensure that accurate structural information is obtained. Our understanding of the underlying chemistry in most cases is still incomplete, and this lack of understanding can give rise to ambiguous structural information in some cases. Another limitation of CL is that not every amino acid in a protein can be probed, and this inherently limits the structural resolution obtainable by this approach. No one CL reagent has been found to react with every type of amino acid side chain. A related issue is that every amino acid residue reacts to a different extent with a given CL reagent. This fact complicates attempts to find correlations between reactivity and higher order structure. Despite these limitations, and in some cases because of these limitations, several opportunities still exist for the future development of CL-MS.
5.2 Covalent Labeling with Computational Modeling
The structural resolution provided by CL-MS is lower than that provided by NMR or X-ray crystallography because only information about the solvent accessible side chains is obtained, and this information is only a fraction of the amino acids in a protein. Even with relatively limited information, CL-MS has been successfully used to confirm or rule out proposed structural models [145, 146]. CL-MS has the potential to provide even higher resolution structural information when it is combined with computational modeling. Several groups have explored the combination of CL-MS and computational modeling techniques. For example, Gerega and Downard developed an algorithm, known as PROXIMO, that is specially designed to model protein-peptide complex structures based on oxidative labeling data from radical probe MS [147]. Similarly, Chance and co-workers have developed the ClusPro program for predicting protein complexes using the restraints from hydroxyl radical labeling to facilitate homology modeling and associated protein structural prediction for protein complexes with no NMR or X-ray crystal structure [148]. In addition, Sharp and co-workers have developed a strategy that correlates oxidative labeling levels with SASA at each modified residue, which can be utilized to assess the quality of protein structural models [65] or model protein complex structures [149]. While each of these above approaches have helped refine the protein structural information obtained by CL-MS, they have not been adopted broadly. Recent work has shown the promise of combining multiple complementary experimental methods to improve structural modeling. For example, methods like limited proteolysis [150], small-angle X-ray scattering [151], chemical crosslinking combined with native MS [152] have been combined with CL-MS and computational methods to arrive at protein structural models. It appears that CL-MS research is in the very early stages of identifying the best methods to efficiently combine constraints from CL-MS, complementary methods, and computational modeling to obtain higher resolution protein structural information.
5.3 Membrane Protein Footprinting
Another promising area in which CL-MS has the potential to have an impact is in the study of membrane proteins. Membrane proteins are inherently more difficult to study as compared to water-soluble proteins. CL-MS would appear to be a promising tool for structural analysis of membrane proteins because CL can often be done under conditions that preserve conformation and topologic information. Several groups have demonstrated the promise of CL in studying membrane proteins [55, 153–159]. Hydroxyl radical labeling has been shown to map the topology of intrinsic membrane proteins in their natural lipid environment [153], in nanodiscs [157], within amphipols [158], and even within living cells [159]. New hydrophobic-based CL labeling reagents, e.g. based on triflinate [55], might be able to further improve the information accessible by CL-MS by allowing the labeling of protein regions that are inserted into membranes themselves.
5.4 In-Cell/In-Organism Covalent Labeling
Beyond live-cell membrane protein CL [159], Jones and co-workers recently performed in-cell FPOP of live Vero cells. They demonstrated that proteins in different cellular compartments can be labeled and residue-level oxidative modification extents can be measured, and these labeling extents correlate well with the SASA of the residues [160]. It is reasonable to assume that this application of CL-MS could be expanded for studying protein interactions and conformational changes in living cells.
Highlights.
Covalent labeling - mass spectrometry can study protein structure and protein interactions
The method primarily reports on the solvent accessibility of amino acid side chains
Non-specific reagents modify many amino acid side chains, enabling broad structural coverage
Covalent labeling is used to distinguish protein topologies, conformations, and binding sites
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01 GM015092 and R43 GM116211). Catherine Tremblay and Xiao Pan are acknowledged for their valuable comments and feedback on the text of this manuscript. P.L. also acknowledges his doctoral fellowship from the Faculty of Pharmaceutical Sciences, Chulalongkorn University (Bangkok, Thailand).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Katta V, Chait BT, Carr S. Conformational changes in proteins probed by hydrogen-exchange electrospray-ionization mass spectrometry. Rapid Communications in Mass Spectrometry. 1991;5(4):214–217. doi: 10.1002/rcm.1290050415. [DOI] [PubMed] [Google Scholar]
- 2.Engen JR, Smith DL. Peer Reviewed: Investigating Protein Structure and Dynamics by Hydrogen Exchange MS. Analytical Chemistry. 2001;73(9):256 A–265 A. doi: 10.1021/ac012452f. [DOI] [PubMed] [Google Scholar]
- 3.Kaltashov IA, Eyles SJ. Studies of biomolecular conformations and conformational dynamics by mass spectrometry. Mass Spectrometry Reviews. 2002;21(1):37–71. doi: 10.1002/mas.10017. [DOI] [PubMed] [Google Scholar]
- 4.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrometry Reviews. 2006;25(1):158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 5.Konermann L, Pan J, Liu YH. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chemical Society Reviews. 2011;40(3):1224–1234. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- 6.Wei H, Mo J, Tao L, Russell RJ, Tymiak AA, Chen G, Iacob RE, Engen JR. Hydrogen/deuterium exchange mass spectrometry for probing higher order structure of protein therapeutics: methodology and applications. Drug Discovery Today. 2014;19(1):95–102. doi: 10.1016/j.drudis.2013.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pirrone GF, Iacob RE, Engen JR. Applications of Hydrogen/Deuterium Exchange MS from 2012 to 2014. Analytical Chemistry. 2015;87(1):99–118. doi: 10.1021/ac5040242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinz A. Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein–protein interactions. Mass Spectrometry Reviews. 2006;25(4):663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- 9.Holding AN. XL-MS: Protein cross-linking coupled with mass spectrometry. Methods. 2015;89:54–63. doi: 10.1016/j.ymeth.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 10.Yu C, Huang L. Cross-Linking Mass Spectrometry: An Emerging Technology for Interactomics and Structural Biology. Analytical Chemistry. 2018;90(1):144–165. doi: 10.1021/acs.analchem.7b04431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu G, Chance MR. Hydroxyl Radical-Mediated Modification of Proteins as Probes for Structural Proteomics. Chemical Reviews. 2007;107(8):3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- 12.Mendoza VL, Vachet RW. Probing protein structure by amino acid-specific covalent labeling and mass spectrometry. Mass Spectrometry Reviews. 2009;28(5):785–815. doi: 10.1002/mas.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Chance MR. Structural Mass Spectrometry of Proteins Using Hydroxyl Radical Based Protein Footprinting. Analytical Chemistry. 2011;83(19):7234–7241. doi: 10.1021/ac200567u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baslé E, Joubert N, Pucheault M. Protein Chemical Modification on Endogenous Amino Acids. Chemistry & Biology. 2010;17(3):213–227. doi: 10.1016/j.chembiol.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 15.Shannon DA, Weerapana E. Covalent protein modification: the current landscape of residue-specific electrophiles. Current Opinion in Chemical Biology. 2015;24(Supplement C):18–26. doi: 10.1016/j.cbpa.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 16.Maleknia SD, Brenowitz M, Chance MR. Millisecond Radiolytic Modification of Peptides by Synchrotron X-rays Identified by Mass Spectrometry. Analytical Chemistry. 1999;71(18):3965–3973. doi: 10.1021/ac990500e. [DOI] [PubMed] [Google Scholar]
- 17.Takamoto K, Chance MR. Radiolytic protein footprinting with mass spectrometry to probe the structure of macromolecular complexes. Annual Review of Biophysics and Biomolecular Structure. 2006;35(1):251–276. doi: 10.1146/annurev.biophys.35.040405.102050. [DOI] [PubMed] [Google Scholar]
- 18.Maleknia SD, Ralston CY, Brenowitz MD, Downard KM, Chance MR. Determination of Macromolecular Folding and Structure by Synchrotron X-Ray Radiolysis Techniques. Analytical Biochemistry. 2001;289(2):103–115. doi: 10.1006/abio.2000.4910. [DOI] [PubMed] [Google Scholar]
- 19.Maleknia SD, Wong JWH, Downard KM. Photochemical and electrophysical production of radicals on millisecond timescales to probe the structure, dynamics and interactions of proteins. Photochemical & Photobiological Sciences. 2004;3(8):741–748. doi: 10.1039/b315904c. [DOI] [PubMed] [Google Scholar]
- 20.Naganathan AN, Muñoz V. Scaling of Folding Times with Protein Size. Journal of the American Chemical Society. 2005;127(2):480–481. doi: 10.1021/ja044449u. [DOI] [PubMed] [Google Scholar]
- 21.Sharp JS, Becker JM, Hettich RL. Analysis of Protein Solvent Accessible Surfaces by Photochemical Oxidation and Mass Spectrometry. Analytical Chemistry. 2004;76(3):672–683. doi: 10.1021/ac0302004. [DOI] [PubMed] [Google Scholar]
- 22.Aye TT, Low TY, Sze SK. Nanosecond Laser-Induced Photochemical Oxidation Method for Protein Surface Mapping with Mass Spectrometry. Analytical Chemistry. 2005;77(18):5814–5822. doi: 10.1021/ac050353m. [DOI] [PubMed] [Google Scholar]
- 23.Hambly DM, Gross ML. Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale. Journal of the American Society for Mass Spectrometry. 2005;16(12):2057–2063. doi: 10.1016/j.jasms.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Gau BC, Sharp JS, Rempel DL, Gross ML. Fast Photochemical Oxidation of Protein Footprints Faster than Protein Unfolding. Analytical Chemistry. 2009;81(16):6563–6571. doi: 10.1021/ac901054w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watson C, Janik I, Zhuang T, Charvátová O, Woods RJ, Sharp JS. Pulsed Electron Beam Water Radiolysis for Submicrosecond Hydroxyl Radical Protein Footprinting. Analytical Chemistry. 2009;81(7):2496–2505. doi: 10.1021/ac802252y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bridgewater JD, Lim J, Vachet RW. Transition Metal–Peptide Binding Studied by Metal-Catalyzed Oxidation Reactions and Mass Spectrometry. Analytical Chemistry. 2006;78(7):2432–2438. doi: 10.1021/ac051983r. [DOI] [PubMed] [Google Scholar]
- 27.Bridgewater JD, Lim J, Vachet RW. Using metal-catalyzed oxidation reactions and mass spectrometry to identify amino acid residues within 10 Å of the metal in Cu-binding proteins. Journal of the American Society for Mass Spectrometry. 2006;17(11):1552–1559. doi: 10.1016/j.jasms.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 28.Srikanth R, Wilson J, Burns CS, Vachet RW. Identification of the Copper(II) Coordinating Residues in the Prion Protein by Metal-Catalyzed Oxidation Mass Spectrometry: Evidence for Multiple Isomers at Low Copper(II) Loadings. Biochemistry. 2008;47(35):9258–9268. doi: 10.1021/bi800970m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hampel KJ, Burke JM. Time-Resolved Hydroxyl-Radical Footprinting of RNA Using Fe(II)-EDTA. Methods. 2001;23(3):233–239. doi: 10.1006/meth.2000.1134. [DOI] [PubMed] [Google Scholar]
- 30.Tullius TD, Greenbaum JA. Mapping nucleic acid structure by hydroxyl radical cleavage. Current Opinion in Chemical Biology. 2005;9(2):127–134. doi: 10.1016/j.cbpa.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 31.Sharp JS, Becker JM, Hettich RL. Protein surface mapping by chemical oxidation: Structural analysis by mass spectrometry. Analytical Biochemistry. 2003;313(2):216–225. doi: 10.1016/s0003-2697(02)00612-7. [DOI] [PubMed] [Google Scholar]
- 32.Götte M, Marquet R, Isel C, Anderson VE, Keith G, Gross HJ, Ehresmann C, Ehresmann B, Heumann H. Probing the higher order structure of RNA with peroxonitrous acid. FEBS Letters. 1996;390(2):226–228. doi: 10.1016/0014-5793(96)00662-x. [DOI] [PubMed] [Google Scholar]
- 33.King PA, Jamison E, Strahs D, Anderson VE, Brenowitz M. ‘Footprinting’ proteins on DNA with peroxonitrous acid. Nucleic Acids Research. 1993;21(10):2473–2478. doi: 10.1093/nar/21.10.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maleknia SD, Chance MR, Downard KM. Electrospray-assisted modification of proteins: a radical probe of protein structure. Rapid Communications in Mass Spectrometry. 1999;13(23):2352–2358. doi: 10.1002/(SICI)1097-0231(19991215)13:23<2352::AID-RCM798>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 35.Wong JWH, Maleknia SD, Downard KM. Study of the Ribonuclease–S-Protein–Peptide Complex Using a Radical Probe and Electrospray Ionization Mass Spectrometry. Analytical Chemistry. 2003;75(7):1557–1563. doi: 10.1021/ac026400h. [DOI] [PubMed] [Google Scholar]
- 36.Wong JWH, Maleknia SD, Downard KM. Hydroxyl radical probe of the calmodulin-melittin complex interface by electrospray ionization mass spectrometry. Journal of the American Society for Mass Spectrometry. 2005;16(2):225–233. doi: 10.1016/j.jasms.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 37.McClintock C, Kertesz V, Hettich RL. Development of an Electrochemical Oxidation Method for Probing Higher Order Protein Structure with Mass Spectrometry. Analytical Chemistry. 2008;80(9):3304–3317. doi: 10.1021/ac702493a. [DOI] [PubMed] [Google Scholar]
- 38.Minkoff BB, Blatz JM, Choudhury FA, Benjamin D, Shohet JL, Sussman MR. Plasma-Generated OH Radical Production for Analyzing Three-Dimensional Structure in Protein Therapeutics. Scientific Reports. 2017;7(1):12946. doi: 10.1038/s41598-017-13371-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharp JS, Tomer KB. Analysis of the Oxidative Damage-Induced Conformational Changes of Apo- and Holocalmodulin by Dose-Dependent Protein Oxidative Surface Mapping. Biophysical Journal. 2007;92(5):1682–1692. doi: 10.1529/biophysj.106.099093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu G, Kiselar J, He Q, Chance MR. Secondary Reactions and Strategies To Improve Quantitative Protein Footprinting. Analytical Chemistry. 2005;77(10):3029–3037. doi: 10.1021/ac048282z. [DOI] [PubMed] [Google Scholar]
- 41.Vahidi S, Konermann L. Probing the Time Scale of FPOP (Fast Photochemical Oxidation of Proteins): Radical Reactions Extend Over Tens of Milliseconds. Journal of The American Society for Mass Spectrometry. 2016;27(7):1156–1164. doi: 10.1007/s13361-016-1389-x. [DOI] [PubMed] [Google Scholar]
- 42.Blencowe A, Hayes W. Development and application of diazirines in biological and synthetic macromolecular systems. Soft Matter. 2005;1(3):178–205. doi: 10.1039/b501989c. [DOI] [PubMed] [Google Scholar]
- 43.Suchanek M, Radzikowska A, Thiele C. Photo-leucine and photo-methionine allow identification of protein-protein interactions in living cells. Nature Methods. 2005;2:261. doi: 10.1038/nmeth752. [DOI] [PubMed] [Google Scholar]
- 44.Jumper CC, Schriemer DC. Mass Spectrometry of Laser-Initiated Carbene Reactions for Protein Topographic Analysis. Analytical Chemistry. 2011;83(8):2913–2920. doi: 10.1021/ac102655f. [DOI] [PubMed] [Google Scholar]
- 45.Das J. Aliphatic Diazirines as Photoaffinity Probes for Proteins: Recent Developments. Chemical Reviews. 2011;111(8):4405–4417. doi: 10.1021/cr1002722. [DOI] [PubMed] [Google Scholar]
- 46.Richards FM, Lamed R, Wynn R, Patel D, Olack G. Methylene as a possible universal footprinting reagent that will include hydrophobic surface areas: overview and feasibility: properties of diazirine as a precursor. Protein Science. 2000;9(12):2506–2517. doi: 10.1110/ps.9.12.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Craig PO, Ureta DB, Delfino JM. Probing protein conformation with a minimal photochemical reagent. Protein Science: A Publication of the Protein Society. 2002;11(6):1353–1366. doi: 10.1110/ps.4710102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gómez GE, Cauerhff A, Craig PO, Goldbaum FA, Delfino JM. Exploring protein interfaces with a general photochemical reagent. Protein Science. 2006;15(4):744–752. doi: 10.1110/ps.051960406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ureta DB, Craig PO, Gómez GE, Delfino JM. Assessing Native and Non-native Conformational States of a Protein by Methylene Carbene Labeling: The Case of Bacillus licheniformis β-Lactamase. Biochemistry. 2007;46(50):14567–14577. doi: 10.1021/bi7012867. [DOI] [PubMed] [Google Scholar]
- 50.Jumper CC, Bomgarden R, Rogers J, Etienne C, Schriemer DC. High-Resolution Mapping of Carbene-Based Protein Footprints. Analytical Chemistry. 2012;84(10):4411–4418. doi: 10.1021/ac300120z. [DOI] [PubMed] [Google Scholar]
- 51.Zhang B, Rempel DL, Gross ML. Protein Footprinting by Carbenes on a Fast Photochemical Oxidation of Proteins (FPOP) Platform. Journal of The American Society for Mass Spectrometry. 2016;27(3):552–555. doi: 10.1007/s13361-015-1313-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manzi L, Barrow AS, Scott D, Layfield R, Wright TG, Moses JE, Oldham NJ. Carbene footprinting accurately maps binding sites in protein–ligand and protein–protein interactions. Nature Communications. 2016;7:13288. doi: 10.1038/ncomms13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ziemianowicz DS, Bomgarden R, Etienne C, Schriemer DC. Amino Acid Insertion Frequencies Arising from Photoproducts Generated Using Aliphatic Diazirines. Journal of The American Society for Mass Spectrometry. 2017;28(10):2011–2021. doi: 10.1007/s13361-017-1730-z. [DOI] [PubMed] [Google Scholar]
- 54.Brunner J, Senn H, Richards FM. 3-Trifluoromethyl-3-phenyldiazirine. A new carbene generating group for photolabeling reagents. Journal of Biological Chemistry. 1980;255(8):3313–3318. [PubMed] [Google Scholar]
- 55.Cheng M, Zhang B, Cui W, Gross ML. Laser-Initiated Radical Trifluoromethylation of Peptides and Proteins: Application to Mass-Spectrometry-Based Protein Footprinting. Angewandte Chemie International Edition. 2017;56(45):14007–14010. doi: 10.1002/anie.201706697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lilie J, Behar D, Sujdak RJ, Schuler RH. Lifetime of trifluoromethyl radical in aqueous solution. The Journal of Physical Chemistry. 1972;76(18):2517–2520. [Google Scholar]
- 57.Miles EW. Methods in Enzymology. Academic Press; 1977. Modification of histidyl residues in proteins by diethylpyrocarbonate; pp. 431–442. [DOI] [PubMed] [Google Scholar]
- 58.Mendoza VL, Vachet RW. Protein Surface Mapping Using Diethylpyrocarbonate with Mass Spectrometric Detection. Analytical Chemistry. 2008;80(8):2895–2904. doi: 10.1021/ac701999b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou Y, Vachet RW. Increased Protein Structural Resolution from Diethylpyrocarbonate-based Covalent Labeling and Mass Spectrometric Detection. Journal of The American Society for Mass Spectrometry. 2012;23(4):708–717. doi: 10.1007/s13361-011-0332-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou Y, Vachet RW. Diethylpyrocarbonate Labeling for the Structural Analysis of Proteins: Label Scrambling in Solution and How to Avoid It. Journal of The American Society for Mass Spectrometry. 2012;23(5):899–907. doi: 10.1007/s13361-012-0349-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goldsmith SC, Guan JQ, Almo SC, Chance MR. Synchrotron Protein Footprinting: A Technique to Investigate Protein-Protein Interactions. Journal of Biomolecular Structure and Dynamics. 2001;19(3):405–418. doi: 10.1080/07391102.2001.10506750. [DOI] [PubMed] [Google Scholar]
- 62.Kiselar JG, Maleknia SD, Sullivan M, Downard KM, Chance MR. Hydroxyl radical probe of protein surfaces using synchrotron X-ray radiolysis and mass spectrometry. International Journal of Radiation Biology. 2002;78(2):101–114. doi: 10.1080/09553000110094805. [DOI] [PubMed] [Google Scholar]
- 63.Srikanth R, Mendoza VL, Bridgewater JD, Zhang G, Vachet RW. Copper Binding to β-2-Microglobulin and Its Pre-Amyloid Oligomers. Biochemistry. 2009;48(41):9871–9881. doi: 10.1021/bi901172y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang W, Ravikumar Krishnakumar M, Chance Mark R, Yang S. Quantitative Mapping of Protein Structure by Hydroxyl Radical Footprinting-Mediated Structural Mass Spectrometry: A Protection Factor Analysis. Biophysical Journal. 2015;108(1):107–115. doi: 10.1016/j.bpj.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xie B, Sood A, Woods RJ, Sharp JS. Quantitative Protein Topography Measurements by High Resolution Hydroxyl Radical Protein Footprinting Enable Accurate Molecular Model Selection. Scientific Reports. 2017;7(1):4552. doi: 10.1038/s41598-017-04689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang L, Chance MR. Protein footprinting comes of age: mass spectrometry for biophysical structure assessment. Molecular & Cellular Proteomics. 2017 doi: 10.1074/mcp.O116.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu G, Chance MR. Radiolytic Modification and Reactivity of Amino Acid Residues Serving as Structural Probes for Protein Footprinting. Analytical Chemistry. 2005;77(14):4549–4555. doi: 10.1021/ac050299+. [DOI] [PubMed] [Google Scholar]
- 68.Mehler EL, Fuxreiter M, Simon I, Garcia-Moreno BE. The role of hydrophobic microenvironments in modulating pKa shifts in proteins. Proteins: Structure, Function and Bioinformatics. 2002;48(2):283–292. doi: 10.1002/prot.10153. [DOI] [PubMed] [Google Scholar]
- 69.Harris TK, Turner GJ. Structural Basis of Perturbed pKa Values of Catalytic Groups in Enzyme Active Sites. IUBMB Life. 2002;53(2):85–98. doi: 10.1080/15216540211468. [DOI] [PubMed] [Google Scholar]
- 70.Isom DG, Castañeda CA, Cannon BR, García-Moreno BE. Large shifts in pKa values of lysine residues buried inside a protein. Proceedings of the National Academy of Sciences. 2011;108(13):5260–5265. doi: 10.1073/pnas.1010750108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peck MT, Ortega G, De Luca-Johnson JN, Schlessman JL, Robinson AC, García-Moreno BE. Local Backbone Flexibility as a Determinant of the Apparent pKa Values of Buried Ionizable Groups in Proteins. Biochemistry. 2017;56(40):5338–5346. doi: 10.1021/acs.biochem.7b00678. [DOI] [PubMed] [Google Scholar]
- 72.Zhou Y, Wu Y, Yao M, Liu Z, Chen J, Chen J, Tian L, Han G, Shen JR, Wang F. Probing the Lysine Proximal Microenvironments within Membrane Protein Complexes by Active Dimethyl Labeling and Mass Spectrometry. Analytical Chemistry. 2016;88(24):12060–12065. doi: 10.1021/acs.analchem.6b02502. [DOI] [PubMed] [Google Scholar]
- 73.Borotto NB, Zhou Y, Hollingsworth SR, Hale JE, Graban EM, Vaughan RC, Vachet RW. Investigating Therapeutic Protein Structure with Diethylpyrocarbonate Labeling and Mass Spectrometry. Analytical Chemistry. 2015;87(20):10627–10634. doi: 10.1021/acs.analchem.5b03180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Herrmann A, Svangård E, Claeson P, Gullbo J, Bohlin L, Göransson U. Key role of glutamic acid for the cytotoxic activity of the cyclotide cycloviolacin O2. Cellular and Molecular Life Sciences CMLS. 2006;63(2):235–245. doi: 10.1007/s00018-005-5486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tong X, Wren JC, Konermann L. Effects of Protein Concentration on the Extent of γ-Ray-Mediated Oxidative Labeling Studied by Electrospray Mass Spectrometry. Analytical Chemistry. 2007;79(16):6376–6382. doi: 10.1021/ac070724u. [DOI] [PubMed] [Google Scholar]
- 76.Kiselar JG, Janmey PA, Almo SC, Chance MR. Structural Analysis of Gelsolin Using Synchrotron Protein Footprinting. Molecular & Cellular Proteomics. 2003;2(10):1120–1132. doi: 10.1074/mcp.M300068-MCP200. [DOI] [PubMed] [Google Scholar]
- 77.Gupta S, Celestre R, Petzold CJ, Chance MR, Ralston C. Development of a microsecond X-ray protein footprinting facility at the Advanced Light Source. Journal of Synchrotron Radiation. 2014;21(4):690–699. doi: 10.1107/S1600577514007000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mendoza VL, Antwi K, Barón-Rodríguez MA, Blanco C, Vachet RW. Structure of the Preamyloid Dimer of β-2-Microglobulin from Covalent Labeling and Mass Spectrometry. Biochemistry. 2010;49(7):1522–1532. doi: 10.1021/bi901748h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mendoza VL, Barón-Rodríguez MA, Blanco C, Vachet RW. Structural Insights into the Pre-Amyloid Tetramer of β-2-Microglobulin from Covalent Labeling and Mass Spectrometry. Biochemistry. 2011;50(31):6711–6722. doi: 10.1021/bi2004894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ivankov DN, Finkelstein AV. Prediction of protein folding rates from the amino acid sequence-predicted secondary structure. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(24):8942–8944. doi: 10.1073/pnas.0402659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu T, Marcinko TM, Kiefer PA, Vachet RW. Using Covalent Labeling and Mass Spectrometry To Study Protein Binding Sites of Amyloid Inhibiting Molecules. Analytical Chemistry. 2017;89(21):11583–11591. doi: 10.1021/acs.analchem.7b02915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Madsen JA, Yin Y, Qiao J, Gill V, Renganathan K, Fu WY, Smith S, Anderson J. Covalent Labeling Denaturation Mass Spectrometry for Sensitive Localized Higher Order Structure Comparisons. Analytical Chemistry. 2016;88(4):2478–2488. doi: 10.1021/acs.analchem.5b04736. [DOI] [PubMed] [Google Scholar]
- 83.Kelleher NL, Lin HY, Valaskovic GA, Aaserud DJ, Fridriksson EK, McLafferty FW. Top Down versus Bottom Up Protein Characterization by Tandem High-Resolution Mass Spectrometry. Journal of the American Chemical Society. 1999;121(4):806–812. [Google Scholar]
- 84.McLafferty FW, Breuker K, Jin M, Han X, Infusini G, Jiang H, Kong X, Begley TP. Top-down MS, a powerful complement to the high capabilities of proteolysis proteomics. FEBS Journal. 2007;274(24):6256–6268. doi: 10.1111/j.1742-4658.2007.06147.x. [DOI] [PubMed] [Google Scholar]
- 85.Siuti N, Kelleher NL. Decoding protein modifications using top-down mass spectrometry. Nature Methods. 2007;4:817. doi: 10.1038/nmeth1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Meng F, Forbes AJ, Miller LM, Kelleher NL. Detection and localization of protein modifications by high resolution tandem mass spectrometry. Mass Spectrometry Reviews. 2005;24(2):126–134. doi: 10.1002/mas.20009. [DOI] [PubMed] [Google Scholar]
- 87.Zhang Y, Fonslow BR, Shan B, Baek MC, Yates JR. Protein Analysis by Shotgun/Bottom-up Proteomics. Chemical reviews. 2013;113(4):2343–2394. doi: 10.1021/cr3003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen J, Cui W, Giblin D, Gross ML. New Protein Footprinting: Fast Photochemical Iodination Combined with Top-down and Bottom-up Mass Spectrometry. Journal of the American Society for Mass Spectrometry. 2012;23(8):1306–1318. doi: 10.1007/s13361-012-0403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cammarata M, Lin K-Y, Pruet J, Liu H-w, Brodbelt J. Probing the Unfolding of Myoglobin and Domain C of PARP-1 with Covalent Labeling and Top-Down Ultraviolet Photodissociation Mass Spectrometry. Analytical Chemistry. 2014;86(5):2534–2542. doi: 10.1021/ac4036235. [DOI] [PubMed] [Google Scholar]
- 90.Li J, Wei H, Krystek SR, Bond D, Brender TM, Cohen D, Feiner J, Hamacher N, Harshman J, Huang RYC, Julien SH, Lin Z, Moore K, Mueller L, Noriega C, Sejwal P, Sheppard P, Stevens B, Chen G, Tymiak AA, Gross ML, Schneeweis LA. Mapping the Energetic Epitope of an Antibody/Interleukin-23 Interaction with Hydrogen/Deuterium Exchange, Fast Photochemical Oxidation of Proteins Mass Spectrometry, and Alanine Shave Mutagenesis. Analytical Chemistry. 2017;89(4):2250–2258. doi: 10.1021/acs.analchem.6b03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Srikanth R, Wilson J, Bridgewater JD, Numbers JR, Lim J, Olbris MR, Kettani A, Vachet RW. Improved Sequencing of Oxidized Cysteine and Methionine Containing Peptides Using Electron Transfer Dissociation. Journal of the American Society for Mass Spectrometry. 2007;18(8):1499–1506. doi: 10.1016/j.jasms.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 92.Srikanth R, Wilson J, Vachet RW. Correct identification of oxidized histidine residues using electron-transfer dissociation. Journal of Mass Spectrometry. 2009;44(5):755–762. doi: 10.1002/jms.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li X, Li Z, Xie B, Sharp JS. Improved Identification and Relative Quantification of Sites of Peptide and Protein Oxidation for Hydroxyl Radical Footprinting. Journal of The American Society for Mass Spectrometry. 2013;24(11):1767–1776. doi: 10.1007/s13361-013-0719-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Borotto NB, Degraan-Weber N, Zhou Y, Vachet RW. Label Scrambling During CID of Covalently Labeled Peptide Ions. Journal of The American Society for Mass Spectrometry. 2014;25(10):1739–1746. doi: 10.1007/s13361-014-0962-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li Z, Moniz H, Wang S, Ramiah A, Zhang F, Moremen KW, Linhardt RJ, Sharp JS. High Structural Resolution Hydroxyl Radical Protein Footprinting Reveals an Extended Robo1-Heparin Binding Interface. Journal of Biological Chemistry. 2015;290(17):10729–10740. doi: 10.1074/jbc.M115.648410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jones LM, Sperry JB, Carroll JA, Gross ML. Fast Photochemical Oxidation of Proteins for Epitope Mapping. Analytical Chemistry. 2011;83(20):7657–7661. doi: 10.1021/ac2007366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang Y, Wecksler AT, Molina P, Deperalta G, Gross ML. Mapping the Binding Interface of VEGF and a Monoclonal Antibody Fab-1 Fragment with Fast Photochemical Oxidation of Proteins (FPOP) and Mass Spectrometry. Journal of The American Society for Mass Spectrometry. 2017;28(5):850–858. doi: 10.1007/s13361-017-1601-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kaur P, Kiselar JG, Chance MR. Integrated Algorithms for High-Throughput Examination of Covalently Labeled Biomolecules by Structural Mass Spectrometry. Analytical Chemistry. 2009;81(19):8141–8149. doi: 10.1021/ac9013644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rinas A, Espino JA, Jones LM. An efficient quantitation strategy for hydroxyl radical-mediated protein footprinting using Proteome Discoverer. Analytical and Bioanalytical Chemistry. 2016;408(11):3021–3031. doi: 10.1007/s00216-016-9369-3. [DOI] [PubMed] [Google Scholar]
- 100.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 101.Cohen FE, Kelly JW. Therapeutic approaches to protein-misfolding diseases. Nature. 2003;426:905. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- 102.Luheshi LM, Crowther DC, Dobson CM. Protein misfolding and disease: from the test tube to the organism. Current Opinion in Chemical Biology. 2008;12(1):25–31. doi: 10.1016/j.cbpa.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 103.Konermann L, Stocks BB, Pan Y, Tong X. Mass spectrometry combined with oxidative labeling for exploring protein structure and folding. Mass Spectrometry Reviews. 2010;29(4):651–667. doi: 10.1002/mas.20256. [DOI] [PubMed] [Google Scholar]
- 104.Wong KB, Freund SMV, Fersht AR. Cold Denaturation of Barstar:1H,15N and 13C NMR Assignment and Characterisation of Residual Structure. Journal of Molecular Biology. 1996;259(4):805–818. doi: 10.1006/jmbi.1996.0359. [DOI] [PubMed] [Google Scholar]
- 105.Chen J, Rempel DL, Gross ML. Temperature Jump and Fast Photochemical Oxidation Probe Submillisecond Protein Folding. Journal of the American Chemical Society. 2010;132(44):15502–15504. doi: 10.1021/ja106518d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen J, Rempel DL, Gau BC, Gross ML. Fast Photochemical Oxidation of Proteins and Mass Spectrometry Follow Submillisecond Protein Folding at the Amino-Acid Level. Journal of the American Chemical Society. 2012;134(45):18724–18731. doi: 10.1021/ja307606f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gau BC, Chen J, Gross ML. Fast photochemical oxidation of proteins for comparing solvent-accessibility changes accompanying protein folding: Data processing and application to barstar. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2013;1834(6):1230–1238. doi: 10.1016/j.bbapap.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Khurana R, Udgaonkar JB. Equilibrium unfolding studies of barstar: Evidence for an alternative conformation which resembles a molten globule. Biochemistry. 1994;33(1):106–115. doi: 10.1021/bi00167a014. [DOI] [PubMed] [Google Scholar]
- 109.Shastry MCR, Udgaonkar JB. The Folding Mechanism of Barstar: Evidence for Multiple Pathways and Multiple Intermediates. Journal of Molecular Biology. 1995;247(5):1013–1027. doi: 10.1006/jmbi.1994.0196. [DOI] [PubMed] [Google Scholar]
- 110.Nölting B, Golbik R, Fersht AR. Submillisecond events in protein folding. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(23):10668–10672. doi: 10.1073/pnas.92.23.10668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nölting B, Golbik R, Neira JL, Soler-Gonzalez AS, Schreiber G, Fersht AR. The folding pathway of a protein at high resolution from microseconds to seconds. Proceedings of the National Academy of Sciences. 1997;94(3):826–830. doi: 10.1073/pnas.94.3.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Poor TA, Jones LM, Sood A, Leser GP, Plasencia MD, Rempel DL, Jardetzky TS, Woods RJ, Gross ML, Lamb RA. Probing the paramyxovirus fusion (F) protein-refolding event from pre- to postfusion by oxidative footprinting. Proceedings of the National Academy of Sciences. 2014;111(25):E2596–E2605. doi: 10.1073/pnas.1408983111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stocks BB, Konermann L. Structural Characterization of Short-Lived Protein Unfolding Intermediates by Laser-Induced Oxidative Labeling and Mass Spectrometry. Analytical Chemistry. 2009;81(1):20–27. doi: 10.1021/ac801888h. [DOI] [PubMed] [Google Scholar]
- 114.Pan Y, Brown L, Konermann L. Kinetic Folding Mechanism of an Integral Membrane Protein Examined by Pulsed Oxidative Labeling and Mass Spectrometry. Journal of Molecular Biology. 2011;410(1):146–158. doi: 10.1016/j.jmb.2011.04.074. [DOI] [PubMed] [Google Scholar]
- 115.Vahidi S, Stocks BB, Liaghati-Mobarhan Y, Konermann L. Mapping pH-Induced Protein Structural Changes Under Equilibrium Conditions by Pulsed Oxidative Labeling and Mass Spectrometry. Analytical Chemistry. 2012;84(21):9124–9130. doi: 10.1021/ac302393g. [DOI] [PubMed] [Google Scholar]
- 116.Vahidi S, Stocks BB, Liaghati-Mobarhan Y, Konermann L. Submillisecond Protein Folding Events Monitored by Rapid Mixing and Mass Spectrometry-Based Oxidative Labeling. Analytical Chemistry. 2013;85(18):8618–8625. doi: 10.1021/ac401148z. [DOI] [PubMed] [Google Scholar]
- 117.Hazenberg BPC. Amyloidosis: A Clinical Overview. Rheumatic Disease Clinics of North America. 2013;39(2):323–345. doi: 10.1016/j.rdc.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 118.Li H, Rahimi F, Sinha S, Maiti P, Bitan G, Murakami K. Amyloids and Protein Aggregation—Analytical Methods, Encyclopedia of Analytical Chemistry. John Wiley & Sons, Ltd; 2006. [Google Scholar]
- 119.Verma M, Vats A, Taneja V. Toxic species in amyloid disorders: Oligomers or mature fibrils. Annals of Indian Academy of Neurology. 2015;18(2):138–145. doi: 10.4103/0972-2327.144284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shankar GM, Li S, Mehta TH, Garcia-Munoz A, Shepardson NE, Smith I, Brett FM, Farrell MA, Rowan MJ, Lemere CA, Regan CM, Walsh DM, Sabatini BL, Selkoe DJ. Amyloid-β protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nature Medicine. 2008;14:837. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sengupta U, Nilson AN, Kayed R. The Role of Amyloid-β Oligomers in Toxicity, Propagation and Immunotherapy. EBioMedicine. 2016;6:42–49. doi: 10.1016/j.ebiom.2016.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bitan G. Methods in Enzymology. Academic Press; 2006. Structural Study of Metastable Amyloidogenic Protein Oligomers by Photo-Induced Cross-Linking of Unmodified Proteins; pp. 217–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Marcinko TM, Dong J, LeBlanc R, Daborowski KV, Vachet RW. Small molecule-mediated inhibition of β-2-microglobulin-based amyloid fibril formation. Journal of Biological Chemistry. 2017;292(25):10630–10638. doi: 10.1074/jbc.M116.774083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Klinger AL, Kiselar J, Ilchenko S, Komatsu H, Chance MR, Axelsen PH. A Synchrotron-Based Hydroxyl Radical Footprinting Analysis of Amyloid Fibrils and Prefibrillar Intermediates with Residue-Specific Resolution. Biochemistry. 2014;53(49):7724–7734. doi: 10.1021/bi5010409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li KS, Rempel DL, Gross ML. Conformational-Sensitive Fast Photochemical Oxidation of Proteins and Mass Spectrometry Characterize Amyloid Beta 1–42 Aggregation. Journal of the American Chemical Society. 2016;138(37):12090–12098. doi: 10.1021/jacs.6b07543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Floege J, Ketteler M. β2-Microglobulin–derived amyloidosis: An update. Kidney International. 2001;59:S164–S171. doi: 10.1046/j.1523-1755.2001.59780164.x. [DOI] [PubMed] [Google Scholar]
- 127.Guan J-Q, Vorobiev S, Almo SC, Chance MR. Mapping the G-Actin Binding Surface of Cofilin Using Synchrotron Protein Footprinting. Biochemistry. 2002;41(18):5765–5775. doi: 10.1021/bi0121104. [DOI] [PubMed] [Google Scholar]
- 128.Rashidzadeh H, Khrapunov S, Chance MR, Brenowitz M. Solution Structure and Interdomain Interactions of the Saccharomyces cerevisiae “TATA Binding Protein” (TBP) Probed by Radiolytic Protein Footprinting. Biochemistry. 2003;42(13):3655–3665. doi: 10.1021/bi027203f. [DOI] [PubMed] [Google Scholar]
- 129.Hambly D, Gross M. Laser flash photochemical oxidation to locate heme binding and conformational changes in myoglobin. International Journal of Mass Spectrometry. 2007;259(1):124–129. [Google Scholar]
- 130.Zhang H, Gau BC, Jones LM, Vidavsky I, Gross ML. Fast Photochemical Oxidation of Proteins for Comparing Structures of Protein–Ligand Complexes: The Calmodulin–Peptide Model System. Analytical Chemistry. 2011;83(1):311–318. doi: 10.1021/ac102426d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Padayatti PS, Wang L, Gupta S, Orban T, Sun W, Salom D, Jordan SR, Palczewski K, Chance MR. A Hybrid Structural Approach to Analyze Ligand Binding by the Serotonin Type 4 Receptor (5-HT4) Molecular & Cellular Proteomics: MCP. 2013;12(5):1259–1271. doi: 10.1074/mcp.M112.025536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Misra SK, Sood A, Soares PA, Pomin VH, Woods RJ, Sharp JS. Mapping of the Fondaparinux Binding Site of JR-FL gp120 by High Resolution Hydroxyl Radical Protein Footprinting and Computational Docking. bioRxiv. 2017 [Google Scholar]
- 133.Leader B, Baca QJ, Golan DE. Protein therapeutics: a summary and pharmacological classification. Nature Reviews Drug Discovery. 2008;7:21. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- 134.Ecker DM, Jones SD, Levine HL. The therapeutic monoclonal antibody market. mAbs. 2015;7(1):9–14. doi: 10.4161/19420862.2015.989042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Berkowitz SA, Engen JR, Mazzeo JR, Jones GB. Analytical tools for characterizing biopharmaceuticals and the implications for biosimilars. Nature Reviews Drug Discovery. 2012;11:527. doi: 10.1038/nrd3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Frokjaer S, Otzen DE. Protein drug stability: a formulation challenge. Nature Reviews Drug Discovery. 2005;4:298. doi: 10.1038/nrd1695. [DOI] [PubMed] [Google Scholar]
- 137.Kaltashov IA, Bobst CE, Abzalimov RR, Berkowitz SA, Houde D. Conformation and Dynamics of Biopharmaceuticals: Transition of Mass Spectrometry-Based Tools from Academe to Industry. Journal of the American Society for Mass Spectrometry. 2010;21(3):323–337. doi: 10.1016/j.jasms.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kaltashov IA, Bobst CE, Abzalimov RR, Wang G, Baykal B, Wang S. Advances and challenges in analytical characterization of biotechnology products: Mass spectrometry-based approaches to study properties and behavior of protein therapeutics. Biotechnology Advances. 2012;30(1):210–222. doi: 10.1016/j.biotechadv.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Weiss WF, Gabrielson JP, Al-Azzam W, Chen G, Davis DL, Das TK, Hayes DB, Houde D, Singh SK. Technical Decision Making With Higher Order Structure Data: Perspectives on Higher Order Structure Characterization From the Biopharmaceutical Industry. Journal of Pharmaceutical Sciences. 2016;105(12):3465–3470. doi: 10.1016/j.xphs.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 140.Zhang H, Cui W, Gross ML. Mass spectrometry for the biophysical characterization of therapeutic monoclonal antibodies. FEBS Letters. 2014;588(2):308–317. doi: 10.1016/j.febslet.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Watson C, Sharp JS. Conformational Analysis of Therapeutic Proteins by Hydroxyl Radical Protein Footprinting. The AAPS Journal. 2012;14(2):206–217. doi: 10.1208/s12248-012-9336-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Deperalta G, Alvarez M, Bechtel C, Dong K, McDonald R, Ling V. Structural analysis of a therapeutic monoclonal antibody dimer by hydroxyl radical footprinting. mAbs. 2013;5(1):86–101. doi: 10.4161/mabs.22964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Abbott WM, Damschroder MM, Lowe DC. Current approaches to fine mapping of antigen–antibody interactions. Immunology. 2014;142(4):526–535. doi: 10.1111/imm.12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li KS, Chen G, Mo J, Huang RYC, Deyanova EG, Beno BR, O’Neil SR, Tymiak AA, Gross ML. Orthogonal Mass Spectrometry-Based Footprinting for Epitope Mapping and Structural Characterization: The IL-6 Receptor upon Binding of Protein Therapeutics. Analytical Chemistry. 2017;89(14):7742–7749. doi: 10.1021/acs.analchem.7b01748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang L, Qin Y, Ilchenko S, Bohon J, Shi W, Cho MW, Takamoto K, Chance MR. Structural Analysis of a Highly Glycosylated and Unliganded gp120-Based Antigen Using Mass Spectrometry. Biochemistry. 2010;49(42):9032–9045. doi: 10.1021/bi1011332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zheng X, Wintrode PL, Chance MR. Complementary Structural Mass Spectrometry Techniques Reveal Local Dynamics in Functionally Important Regions of a Metastable Serpin. Structure. 2008;16(1):38–51. doi: 10.1016/j.str.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 147.Gerega SK, Downard KM. PROXIMO—a new docking algorithm to model protein complexes using data from radical probe mass spectrometry (RP-MS) Bioinformatics. 2006;22(14):1702–1709. doi: 10.1093/bioinformatics/btl178. [DOI] [PubMed] [Google Scholar]
- 148.Kamal JKA, Chance MR. Modeling of protein binary complexes using structural mass spectrometry data. Protein Science. 2008;17(1):79–94. doi: 10.1110/ps.073071808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Charvátová O, Foley BL, Bern MW, Sharp JS, Orlando R, Woods RJ. Quantifying Protein Interface Footprinting by Hydroxyl Radical Oxidation and Molecular Dynamics Simulation: Application to Galectin-1. Journal of the American Society for Mass Spectrometry. 2008;19(11):1692–1705. doi: 10.1016/j.jasms.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Schorzman AN, Perera L, Cutalo-Patterson JM, Pedersen LC, Pedersen LG, Kunkel TA, Tomer KB. Modeling of the DNA-binding site of yeast Pms1 by mass spectrometry. DNA repair. 2011;10(5):454–465. doi: 10.1016/j.dnarep.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Huang W, Ravikumar KM, Parisien M, Yang S. Theoretical modeling of multiprotein complexes by iSPOT: Integration of small-angle X-ray scattering, hydroxyl radical footprinting, and computational docking. Journal of structural biology. 2016;196(3):340–349. doi: 10.1016/j.jsb.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Schmidt C, Macpherson JA, Lau AM, Tan KW, Fraternali F, Politis A. Surface Accessibility and Dynamics of Macromolecular Assemblies Probed by Covalent Labeling Mass Spectrometry and Integrative Modeling. Analytical Chemistry. 2017;89(3):1459–1468. doi: 10.1021/acs.analchem.6b02875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pan Y, Piyadasa H, O’Neil JD, Konermann L. Conformational Dynamics of a Membrane Transport Protein Probed by H/D Exchange and Covalent Labeling: The Glycerol Facilitator. Journal of Molecular Biology. 2012;416(3):400–413. doi: 10.1016/j.jmb.2011.12.052. [DOI] [PubMed] [Google Scholar]
- 154.Pan Y, Ruan X, Valvano MA, Konermann L. Validation of Membrane Protein Topology Models by Oxidative Labeling and Mass Spectrometry. Journal of The American Society for Mass Spectrometry. 2012;23(5):889–898. doi: 10.1007/s13361-012-0342-x. [DOI] [PubMed] [Google Scholar]
- 155.Konermann L, Pan Y. Exploring membrane protein structural features by oxidative labeling and mass spectrometry. Expert Review of Proteomics. 2012;9(5):497–504. doi: 10.1586/epr.12.42. [DOI] [PubMed] [Google Scholar]
- 156.Bavro Vassiliy N, Gupta S, Ralston C. Oxidative footprinting in the study of structure and function of membrane proteins: current state and perspectives. Biochemical Society Transactions. 2015;43(5):983. doi: 10.1042/BST20150130. [DOI] [PubMed] [Google Scholar]
- 157.Lu Y, Zhang H, Niedzwiedzki DM, Jiang J, Blankenship RE, Gross ML. Fast Photochemical Oxidation of Proteins Maps the Topology of Intrinsic Membrane Proteins: Light-Harvesting Complex 2 in a Nanodisc. Analytical Chemistry. 2016;88(17):8827–8834. doi: 10.1021/acs.analchem.6b01945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Watkinson TG, Calabrese AN, Ault JR, Radford SE, Ashcroft AE. FPOP-LC-MS/MS Suggests Differences in Interaction Sites of Amphipols and Detergents with Outer Membrane Proteins. Journal of The American Society for Mass Spectrometry. 2017;28(1):50–55. doi: 10.1007/s13361-016-1421-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Shen G, Li S, Cui W, Liu S, Yang Y, Gross M, Li W. Membrane Protein Structure in Live Cells: Methodology for Studying Drug Interaction by Mass Spectrometry-Based Footprinting. Biochemistry. 2018 doi: 10.1021/acs.biochem.7b00874. Article ASAP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Espino JA, Mali VS, Jones LM. In Cell Footprinting Coupled with Mass Spectrometry for the Structural Analysis of Proteins in Live Cells. Analytical Chemistry. 2015;87(15):7971–7978. doi: 10.1021/acs.analchem.5b01888. [DOI] [PubMed] [Google Scholar]