Abstract
RNA-based fluorescent (RBF) biosensors have been applied to detect a variety of metabolites in vitro and in live cells. They are designed by combining the ligand sensing domain of natural riboswitches with in vitro selected fluorogenic aptamers. Different biosensor topologies have been developed to accommodate the diversity of riboswitch structures. Here we show that circular permutation of the riboswitch ligand sensing domain also gives functional biosensors, using the SAM-I riboswitch as our model. We reveal that this design can enhance fluorescence turn-on and ligand binding affinity compared to the non-permuted topology.
1. INTRODUCTION
Riboswitches have been applied for classic synthetic biology applications including inducible gene regulation and metabolic engineering [1,2]. More recently, detailed structural information of riboswitch secondary and tertiary structure has enabled their combination with in vitro selected fluorogenic aptamers to generate RNA-based fluorescent (RBF) biosensors for metabolite imaging and detection [3]. RBF biosensors are designed so that binding of a target ligand induces the riboswitch conformational change that allows a dye to bind and turn on fluorescence. In this way, the biosensor generates a fluorescent signal only in the presence of a target metabolite. To date, RBF biosensors have been applied for high-throughput screening of enzyme activity in vitro and in vivo, imaging metabolites and signals in Gram positive and Gram negative bacteria, and anaerobic imaging of bacterial signals [3].
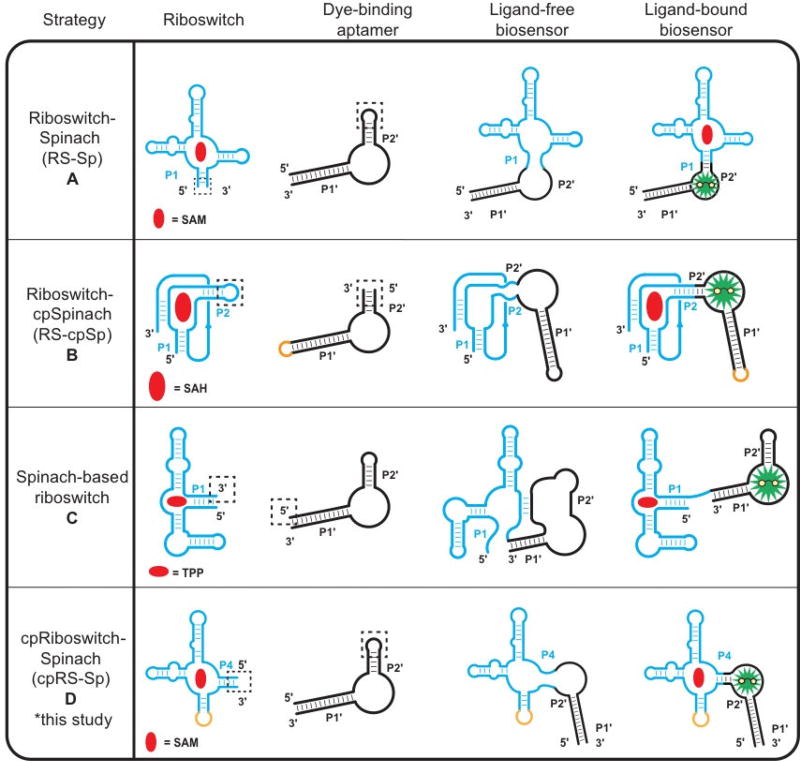
Different biosensor topologies have been explored to broaden the rational design strategy to accommodate the diversity of riboswitch and other aptamer folds. The first riboswitch-based biosensors utilized “transducer modules” consisting of randomized stem sequences to connect to the dye-binding aptamer called Spinach, which binds the profluorescent dye 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI) [4]. One improvement our group made to biosensor design was to use the natural P1 stem from native riboswitches, which simplified the transducer module design and permitted libraries of riboswitch aptamers to be screened for finding the optimal biosensor [5–7]. The conventional topology (RS-Sp, Fig. 1A) thus uses a P1-P2’ stem architecture for the transducer module, where P1 refers to the first pairing stem of the ligand-sensing riboswitch and P2’ stem refers to the second pairing stem of the dye-binding aptamer, which could be either Spinach or an improved fluorogenic aptamer variant, Spinach2 [8].
Figure 1.
Design strategies for riboswitch-based fluorescent biosensors. (a) The riboswitch–Spinach (RS-Sp) fusion strategy for biosensor development involves grafting the P1 stem of a riboswitch sensor domain to the P2’ stem of Spinach. [4–7] (b) For riboswitches with non-pairing 5’ and 3’ ends, an internal stem of the riboswitch can be fused to a circular permutation of Spinach2 to generate a Riboswitch-circularly permuted Spinach2 design (RS-cpSp). [9] (c) Spinach riboswitches can be created by replacing the gene regulatory expression platform of a riboswitch with the Spinach aptamer. [10] (d) A new biosensor strategy for any riboswitch with multiple pairing stems is to circularly permute the riboswitch and fuse it to the P2’ stem of Spinach2 to generate a circularly-permuted Riboswitch-Spinach2 (cpRS-Sp) design. Riboswitch domains (blue) and their corresponding metabolites (red) are depicted with either the Spinach/Spinach2 aptamer (black) and 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI, green). “Sp” represents either Spinach or Spinach 2, which are used in different studies.
The conventional riboswitch-Spinach fusion design is effective for many riboswitch aptamers, but is not suitable when the terminal ends of the ligand-sensing domain do not form a P1 stem. For example, the S-adenosyl-L-homocysteine (SAH) riboswitch contains a terminal pseudoknot that places the 5’ and 3’ ends far apart. To address this challenge, our group constructed a biosensor by making a circular permutation of the Spinach2 aptamer and fusing it to the P2 stem of the SAH riboswitch (RS-cpSp, Fig. 1B) [9]. An alternate strategy is to incorporate Spinach to mimic the expression platforms found in native riboswitches. The ligand-free riboswitch aptamer interferes with the Spinach aptamer folding to block DFBHI binding, but target ligand binding restores the dye-binding aptamer (Fig. 1C) [10].
Here we showcase another strategy to access new biosensor designs that uses circular permutations of a riboswitch aptamer (cpRS-Sp, Fig 1D). To our knowledge, there are no natural examples of circularly permutated riboswitches. However, circularly-permuted hammerhead ribozyme variants are found in diverse genomes [11] and some pre-tRNAs form circularized intermediates before maturation [12]. To investigate these new biosensor topologies, the SAM-I riboswitch was chosen as a model 4-way junction RNA fold to explore novel permuted architectures. This riboswitch class has been extensively characterized with structural, biochemical, and biophysical studies [13–15], which suggested that a circularly-permuted SAM-I riboswitch could be fused with a fluorogenic aptamer via different pairing stems. These constructs were generated, screened, and characterized as new biosensors utilizing circularly-permuted riboswitch topologies, then were compared to traditional biosensor architectures.
2. MATERIALS & METHODS
2.1 Reagents and oligonucleotides
DNA oligonucleotides for biosensor constructs were purchased as Ultramers from Integrated DNA Technologies (Coralville, IA) and other DNA oligonucleotides were purchased from Elim Biopharmaceuticals (Hayward, CA). S-Adenosyl-L-methionine (SAM), S-adenosyl-L homocysteine (SAH) were purchased from Sigma-Aldrich (St Louis, MO). DFHBI was synthesized following previously described protocols [16] and was stored as a 10 mM stock in DMSO at −20 °C. Chemically competent BL21 (DE3) Star cells were purchased from Life Technologies (Carlsbad, CA).
2.2 In vitro transcription procedure
DNA templates for in vitro transcription were prepared by PCR amplification using Phusion DNA polymerase (NEB) from sequence-confirmed plasmids or Ultramer oligonucleotides (for screening experiment only) using primers that added the T7 polymerase promoter sequence at the 5’ end. PCR products were purified either by a 96-well format ZR-96 DNA Clean-up kit (Zymo Research) for screening experiments or by QIAquick PCR purification kit (Qiagen) for characterization and application experiments. RNA was transcribed from DNA templates using T7 RNA polymerase in 40 mM Tris-HCl, pH 8.0, 6 mM MgCl2, 2 mM spermidine, and 10 mM DTT. RNAs were either purified by a 96-well format ZR-96 Clean & Concentrator (Zymo Research) or by denaturing (7.5 M urea) 6% PAGE. RNAs purified by PAGE were visualized by UV shadowing and extracted from gel pieces using Crush Soak buffer (10 mM Tris-HCl, pH 7.5, 200 mM NaCl and 1 mM EDTA, pH 8.0). Purified RNAs were precipitated with ethanol, dried, and then resuspended in TE buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA). Accurate RNA concentrations were determined by measuring the absorbance at 260 nm after performing a hydrolysis assay to eliminate the hypochromic effect due to RNA secondary structure [17].
2.3 General procedure for in vitro fluorescence assays
In vitro fluorescence assays were carried out in binding buffer containing 100 nM RNA, 10 µM DFHBI, 40 mM HEPES, pH 7.5, 125 mM KCl, and 3 or 10 mM MgCl2, as indicated in the figures. Other conditions, including temperature and concentration of ligand, were varied in different experiments as indicated. The RNA was renatured by heating to 72 °C for 3 min in the binding buffer then cooled to ambient temperature for 5 min prior to addition to the reaction solution. DFHBI was added to the solution containing buffer and RNA, and then ligand (or water for no ligand control) was added before fluorescence measurement. Binding reactions were performed in 100 µL volumes and were incubated at the indicated temperature in a Corning Costar 3915 96-well black plate or a Greiner Bio-One 384-well black plate in a Molecular Devices SpectraMax Paradigm Multi-Mode detection platform plate reader (Sunnyvale, CA). The fluorescence emission was measured during 30 to 60 min total with the following instrument parameters: 448 nm excitation, 506 nm emission.
2.4 Fluorescence polarization (FP) binding assays
Fluorescence polarization readings were carried out using a QuantaMaster spectrofluorometer (Photon Technology International) at excitation 646 nm, emission 662 nm. Samples were prepared in 50 ml of TBM buffer (90 mM Tris base, 89 mM boric acid, and 10 mM MgCl2, pH 7.0) containing 1 mM of a Cy5-labled SAM analog (C8-Cy5) [18] and saturation binding experiments were performed with RNA concentrations ranging from 0 to 40 µM. RNA was added successively to the sample cuvette, and concentration values were corrected for added volume. Samples were equilibrated at 30 °C for 2 min prior to each FP measurement using tubing connecting the cuvette holder to a water bath.
2.5 Binding affinity analysis of SAM biosensors
To measure the binding affinities of SAM biosensors, fluorescence assays were performed with the following conditions: binding buffer with 10 mM MgCl2, 37 °C, 100 nM RNA, 10 µM DFHBI, and ligand SAM concentrations from 10 nM to 100 µM. The fluorescence of the sample with DFHBI but no RNA was subtracted as background to determine relative fluorescence units.
2.6 In vivo fluorescence assays by flow cytometry
Preparation of cell samples for flow cytometry was carried out by inoculating 3 mL of LB/carb media with 150 µL of an overnight culture of BL21 (DE3) Star cells containing either the pET31b-RS-Spor pET31b-cpRS-Sp constructs. Cells were grown aerobically to an OD600 ~ 0.5 – 0.6, then induced with 1 mM IPTG at 37 °C for 2 h. Cell density was measured by OD600, and assuming that there are 1×109 cells/mL for for an OD600 of 1, 4×108 cells were sampled and polluted at rt for 4 min at 3,700 rcf, washed once with PBS media at pH 7.0, then resuspended in PBS media containing 100 µM DFHBI. Cellular fluorescence was measured for 30,000 cells using an Attune NxT Acoustic Focusing Cytometer (Life Technologies).
3. RESULTS
3.1 Screen of different biosensor topologies
The SAM-I riboswitch has a 4-way junction architecture. Biochemical and x-ray crystallographic information have shown that the pairing stem element P2 is involved in formation of a pseudoknot that is important for SAM binding [14]. Thus, modifications at the P2 stem were excluded from our biosensor design, but the other pairing stems P1, P3, and P4 became candidates for the transducer stem.
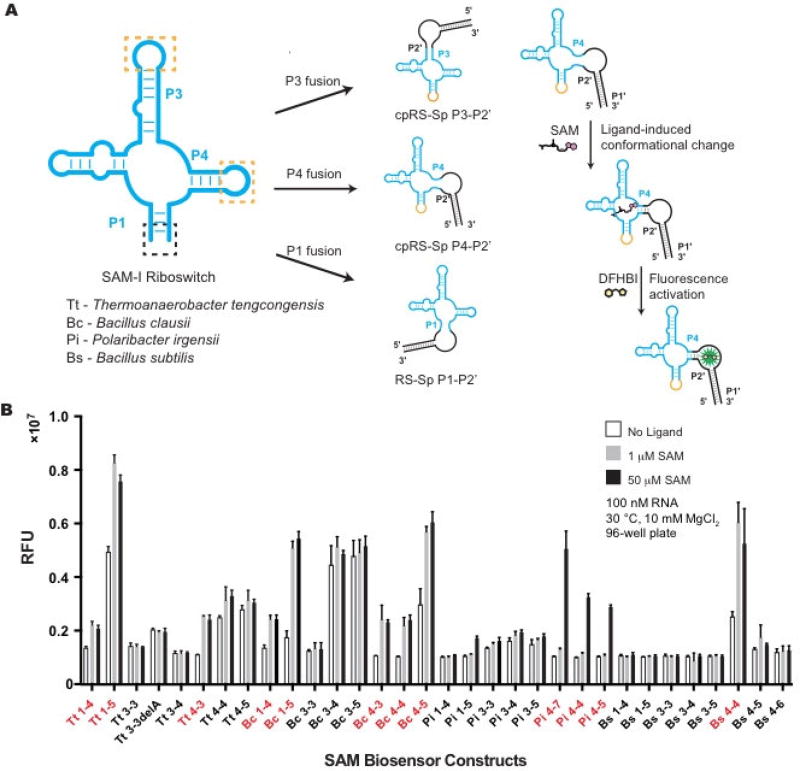
Biosensors that fuse Spinach2 to the riboswitch P1 stem are the conventional RS-Sp design. In contrast, biosensors that fuse Spinach2 to the riboswitch P3 or P4 stems would represent two new cpRS-Sp designs, which require the P1 stem to be closed by a loop sequence (GCAA, Fig. 2A). In all cases, the riboswitch or circularly permuted riboswitch was fused to the P2’ stem of Spinach2, thus the nomenclature for the different topologies is P1-P2’, P3-P2’ or P4-P2’. A small library of 32 biosensor candidates were designed based on these criteria: 1) Four SAM-I riboswitches that have been previously characterized were chosen, [13,14,19] as it has been shown that sampling riboswitches from diverse phylogeny can generate highly fluorescent and well-folded RNA biosensors [7]; 2) For each riboswitch sequence, the three biosensor topologies described above were designed; 3) Based on empirically derived rules for length of the transducer stem, [4] each P1-P2’ construct had two possible stem lengths and each P3-P2’ and P4-P2’ had three possible stem lengths.
Figure 2.
In vitro screen of cpRS-Sp biosensors derived from the SAM-I riboswitch class. (a) Design of different SAM biosensor architectures and mechanism for fluorescence turn-on is depicted for the P4-P2’ biosensor. (b) In vitro fluorescence response of SAM-I biosensors to SAM ligand. The nomenclature is based on the species origin of the sequence, the pairing stem of the SAM-I riboswitch that is fused to Spinach, and number of base pairs retained from the riboswitch stem, e.g. Bs 4-4 stands for SAM-I riboswitch from Bacillus subtilis, with Spinach fused to the P4 stem of the riboswitch that retains 4 base pairs. SAM biosensors chosen for further characterization (with greater than 1.5× fluorescence increase at 50 µM SAM) are indicated in red. Data shown are mean values ± standard deviation taken from two independent replicates.
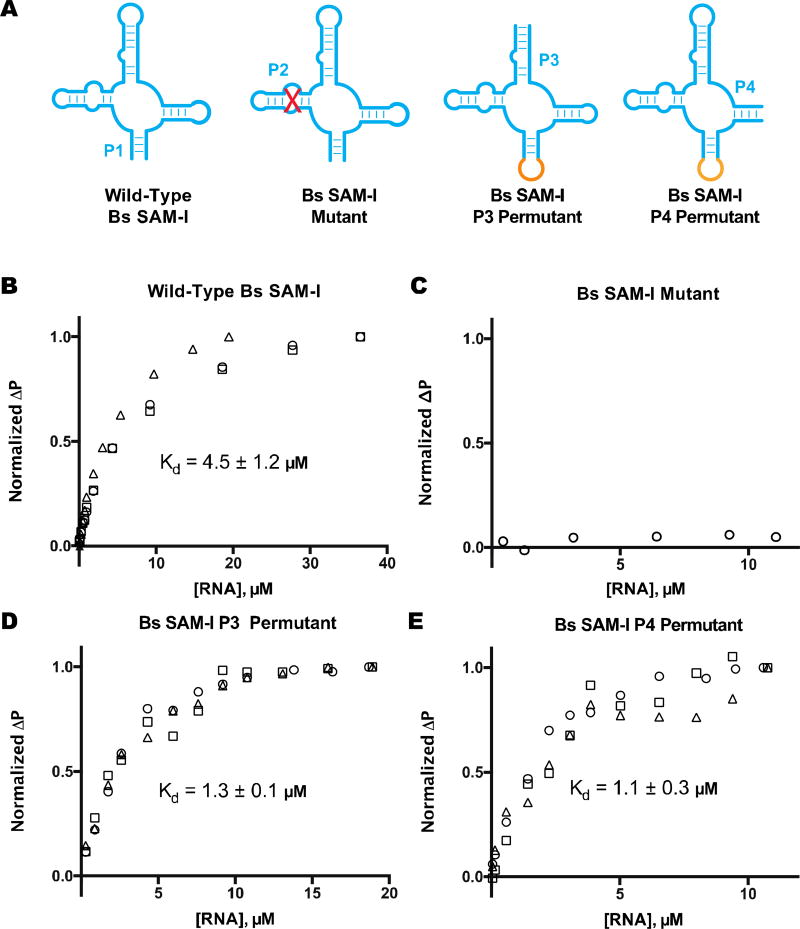
The 32 biosensors were ordered as commercial DNA oligonucleotides, amplified by PCR, synthesized by in vitro transcription, and tested for fluorescence response to ligand, all in a 96-well format. In this initial high-throughput screen, 12 candidate biosensors showed a response to SAM with greater than 1.5× fluorescence activation (Fig. 2B). Interestingly, 8 hits were cpRS-Sp P4-P2’ designs and the remaining hits were the conventional RS-Sp P1-P2’ design. None of the P3-P2‘ constructs were functional as biosensors, even though we separately verified that both P3 and P4 permutants of the Bs riboswitch aptamer retain good binding affinity to SAM (Fig. 3).
Figure 3.
Ligand binding analysis of SAM-I riboswitch aptamers. (a) Schematic of SAM-I riboswitch aptamer, mutant, and permutants. Fluorescence polarization data is shown for Cy5-labeled SAM analog with (a) wild-type Bs SAM-I (P1–7 stem), (b)Bs SAM-I mutant (P2 disruptive mutation), (c) Bs SAM-I P3 permutant (P3–5 stem), and (d) Bs SAM-I P4 permutant (P4–6 stem). Different symbols indicate independent replicates. Change in polarization (ΔP) was normalized to the maximal difference observed at the highest RNA concentration. The reported Kd value is the average of those calculated from best-fit curves with margin of error reported as SD. Data from part (a) and (b) were reproduced with permission from [18].
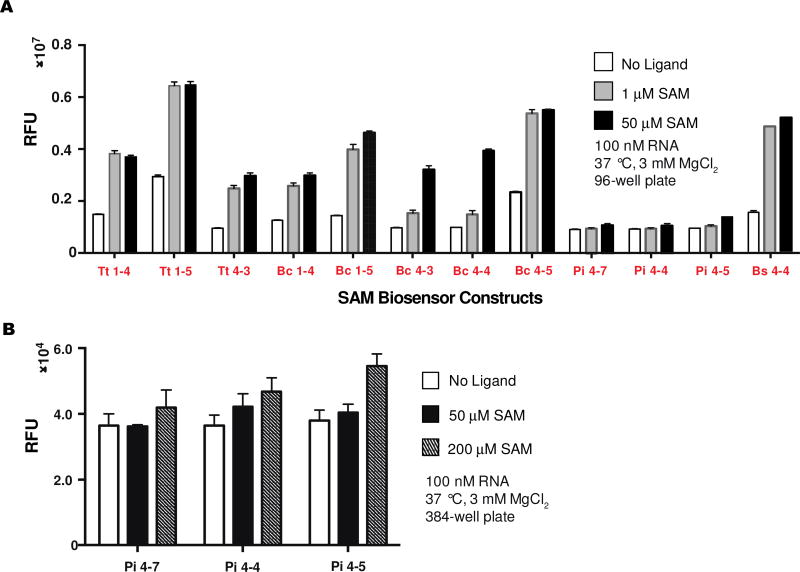
Further in vitro analysis of the 12 hits was performed at physiologically relevant conditions (37 °C and 3 mM Mg2+) to predict their performance in bacteria (Fig. 4A). Three initial hits of the CpRS-Sp P4-P2’ design with varying stem lengths derived from Polaribacter irgensii SAM-I did not give fluorescence turn-on in response to SAM in this secondary screen until higher ligand concentrations (200 µM), which is consistent with their observed poorer affinity in the initial screen (Fig. 4B). The remaining biosensors still retained activity with similar fold activations.
Figure 4.
Re-screen of cpRS-Sp biosensors at physiologically relevant conditions. (a) In vitro fluorescence response of SAM-I biosensors to SAM ligand at 37 °C and 3 mM MgCl2. (b) Pi biosensors were re-screened at higher SAM concentrations. Data shown are mean values ± standard deviation taken from two independent replicates.
3.2 Selectivity and sensitivity of cpRS-Sp biosensors for SAM
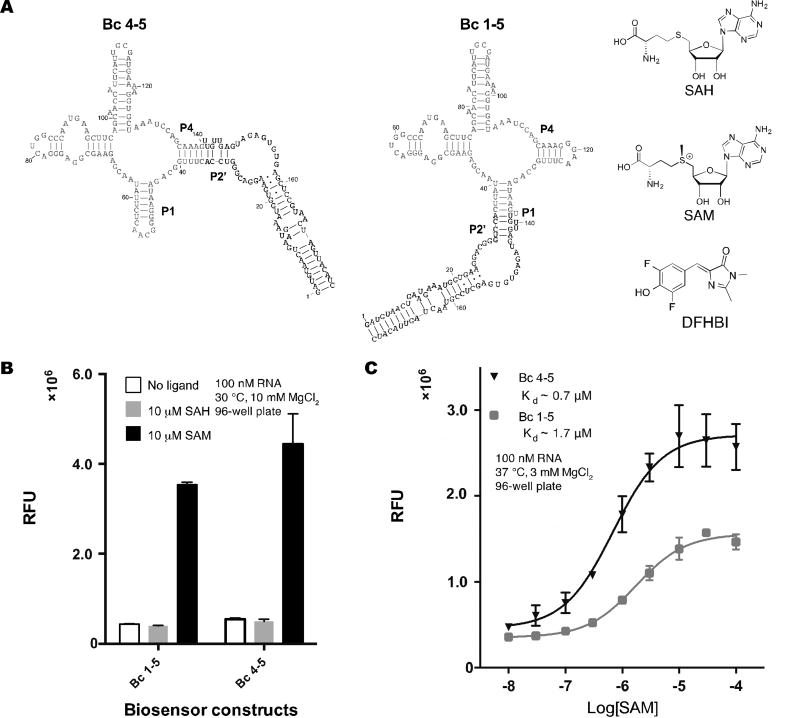
To compare cpRS-Sp and RS-Sp biosensor designs, constructs Bc 4–5 and Bc 1–5 were chosen as representative models. Both biosensors were derived from the same parent SAM-I riboswitch from Bacillus clausii and use the same base pair length in the transducer (Fig 5A). We tested their selectivity for SAM versus S-adenosyl-L-homocysteine (SAH), which differs in one methyl group from the natural ligand. Previous studies have shown that SAM-I riboswitches are 550-fold selective for SAM over SAH [20]. The two riboswitch-based biosensors also appear fully selective for SAM over SAH at equimolar concentrations. Bc 4–5 and Bc 1–5 were measured to have dissociation constants (KD) for SAM of ~0.7 and ~1.7 µM, respectively. Furthermore, Bc 4–5 had ~2-fold higher maximal fluorescence then Bc 1–5. Thus, Bc 4–5, a cpRS-Sp design, has better performance than Bc 1–5, a conventional RS-Sp design.
Figure 5.
Selectivity and sensitivity of cpRS-Sp biosensors. (a) Secondary structure models for Bc 1–5 and Bc 4–5 biosensors and chemical structures of the biosensor ligand, S-adenosyl-L-methionine (SAM), the structural analog S-adenosyl-L-homocysteine (SAH), and dye DFHBI. (b) Selectivity of biosensors for SAM over SAH. (c) In vitro analysis of biosensor binding affinities for SAM. Data shown are mean values ± standard deviation taken from at least two independent replicates.
3.3 In vivo detection of SAM with a cpRS-Sp biosensor using flow cytometry
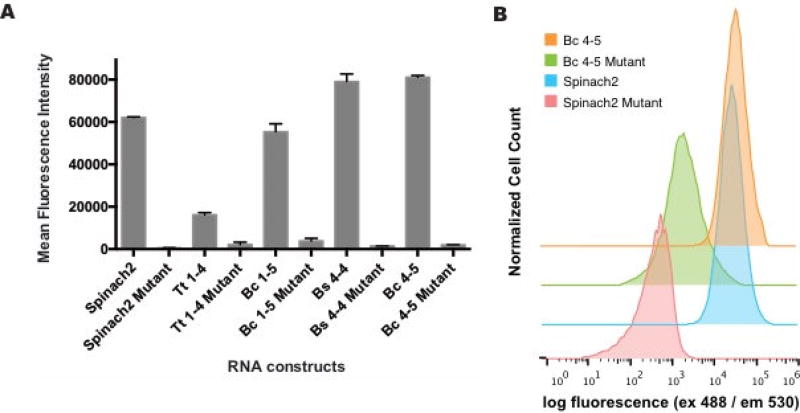
While the secondary screen at physiologically relevant conditions suggests these biosensors would function in a cellular context, we wanted to verify their activity in bacterial cells. Efforts to image metabolites such as SAM are important as it is the required cofactor for enzymatic methylation and is a key metabolite in cellular sulfur metabolism [21]. Additionally, E. coli have been shown to contain sub-millimolar levels of SAM [22] and this high abundance would allow us to measure maximal fluorescence turn-on response in cells expressing biosensor constructs. To validate that cellular fluorescence was due to ligand-dependent turn on of the biosensors, mutant biosensors were generated that carry mutations that ablate ligand binding in the SAM-I riboswitch [13]. Additionally, Spinach2 and a Spinach2 mutant [8] that cannot bind DFHBI were included to compare the maximal fluorescence of our biosensors relative to the parent dye-binding aptamer Spinach2 and the background fluorescence of biosensor mutants to the inactive Spinach2. Each of the constructs were inserted into a tRNA scaffold on a pET31b plasmid [23]. E. coli cells transformed with pET31b plasmids containing each construct were analyzed by flow cytometry. In all cases, robust fluorescence was observed for functional biosensors while their corresponding mutants showed close to background fluorescence (Fig. 6). Similar to the in vitro results, Bc 4–5 was brighter in vivo than Bc 1–5. Furthermore, two cpRS-Sp designs, Bs 4-4 and Bc 4–5, possess mean fluorescence intensity values higher than the parent Spinach2 aptamer. While their signal is saturated in this E. coli strain, these biosensors may be useful in auxotrophic strains to study biosynthetic enzyme mutants or SAM transporters. As we have seen previously [7], by surveying diverse riboswitch sequences and a limited set of transducer stem lengths, we obtained biosensors with a range of affinities.
Figure 6.
In vivo performance of SAM cpRS-Sp biosensors. (a) Live cell fluorescence measured by flow cytometry for E. coli BL21* cells expressing plasmid encoding biosensors and incubated in media containing DFHBI. (b) Mean fluorescence intensity was determined by analyzing 30,000 cells per replicate. All error bars represent standard deviation between technical replicates. (b) Representative flow histograms for cells expressing biosensors or controls.
3.4 An unexpected effect of SAM on biosensor and Spinach2 fluorescence
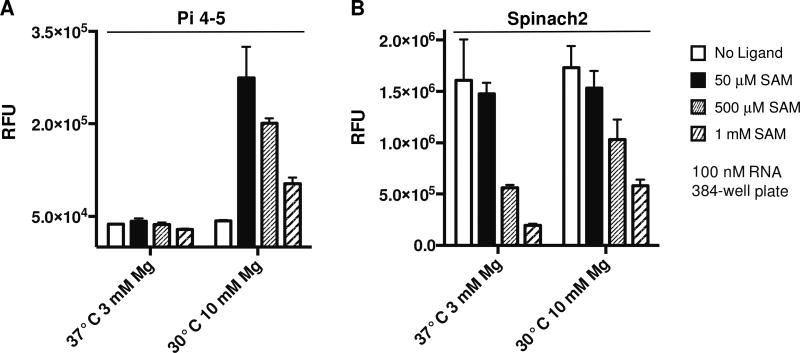
While re-investigating biosensor function at higher SAM concentrations, we observed that fluorescence turn-on was decreased at 500 µM and higher ligand concentrations (Fig. 6). These results were reminiscent of a prior observation we made, that 3 mM and higher ATP reduced Spinach2 fluorescence,[9] likely due to competitive displacement of DFHBI from the dye-binding pocket. Similarly, we found that Spinach2 fluorescence was significantly decreased at 500 µM and higher SAM. These surprising results lead us to suggest that the Spinach2 aptamer may have general affinity for adenosine-containing metabolites. Although the KD for SAM or ATP is much poorer (500-fold or higher) than for DFHBI, the intracellular concentrations of these metabolites are in the range that they can compete for binding. This explains at least in part why observed fluorescence turn-on is generally lower in vivo than in vitro. In practice, we have found that using 50 or 100 µM DFHBI gives sufficient signal for experiments in E. coli. Also, we always perform in vivo experiments with Spinach2 as a control to compare to our Spinach2-based biosensor. Biosensor function is validated for sensing changes in metabolite levels only if the same conditions result in no fluorescence change in the Spinach2 control.
4. DISCUSSION AND CONCLUSIONS
This study shows that circularly-permuted riboswitches can provide an additional scaffold for designing functional RNA-based fluorescent biosensors. In fact, our results suggest it may be possible to generate more sensitive and higher fluorescent biosensors from a parent riboswitch by surveying different transducer stem topologies. There are a number of natural riboswitches or synthetic aptamers besides the SAM-I class that possess multi-stem junctions, such as the PreQ1 or FMN riboswitches [24,25] for which this strategy could be adapted to generate biosensors for their respective ligands. Also, the ability to exploit different connections to riboswitch folds may be beneficial to make biosensors with other dye-binding aptamers besides the Spinach scaffold, such as the Mango and SRB-2 aptamers for example [26,27] or to make other RNA devices [2]. Although naturally circularly-permuted riboswitches have not yet been found, our results show that permuted aptamer domains retain or even improve on ligand binding and conformation switching, which suggests that this method may be promising for making synthetic riboswitches for gene regulation as well.
While P4-P2’ cpRS-Sp constructs were functional, biosensors utilizing a P3-P2’ topology were not. We verified at least in one example that the circularly permuted riboswitch aptamers still bind SAM with good affinities. There are two other potential explanations for this result. One is that the SAM-I riboswitch aptamer does not undergo a significant conformational change in the P3 stem upon ligand binding. A detailed study of the Bs yitJ SAM-I riboswitch discovered key differences in conformational changes with and without the expression platform [28]. The riboswitch aptamer with the expression platform displayed modest structural changes in the P3 stem upon ligand addition, but the aptamer alone showed little to no modulation in the presence of SAM. Our biosensor design incorporates only the SAM-I riboswitch aptamer, so likely does not modulate at the P3 stem efficiently to alter fluorescence output. Alternatively, even though the aptamer alone is capable of binding SAM, it is possible that the positioning of the Spinach2 aptamer at the P3 stem sterically hinders the riboswitch folding, or vice versa.
In vivo analysis by flow cytometry shows that cpRS-Sp sensors with P4-P2’ topologies, Bs 4-4 and Bc 4–5, have higher mean fluorescence intensities than the Spinach2 aptamer alone (Fig. 5A). This directly contrasts with RS-cpSp sensors for SAH, which exhibit lower fluorescence than the fluorogenic aptamer alone [9]. This suggests that the placement of circular permutation on the fluorogenic aptamer hinders maximal fluorescence more than creating a circular permutation on the riboswitch.
Additionally, there have been two previously developed biosensors for SAM that use the RS-Sp and Spinach-based riboswitch topologies (Fig. 1A, C). The first SAM biosensor fused the original Spinach aptamer to the Enterococcus faecalis SAM-III riboswitch, which is from a different riboswitch class that utilizes a 3-way junction [4,29]. The second biosensor was a Spinach-based riboswitch generated by fusing Spinach to the B. subtillis yitJ SAM-I riboswitch [5], which is one of the native riboswitches used in this study. Interestingly, the only functional biosensor construct derived from this riboswitch from our designs was Bs 4-4. The Bs 4– biosensor representing the cpRS-Sp topology has a dissociation constant of 1.0 µM, which is comparable to the reported dissociation constant of 1.2 µM for the Spinach-based riboswitch [10].
Overall, this work validates our concept that circular permutations of riboswitch aptamers can add to existing design strategies for creating RNA-based fluorescent biosensors. To our knowledge, there has not been any previous biosensor designs which utilize circular permutations on the riboswitch aptamer. Furthermore, our success with modulating biosensor fluorescence via ligand-induced structural changes in the P4 stem suggests that SAM riboswitches with this alternate topology have the potential to regulate gene expression as well.
Figure 7.
Effect of SAM on biosensor and Spinach2 fluorescence. (a) In vitro fluorescence activation of (a) biosensor Pi-4–5 and (b) Spinach2 at 30 °C, 10 mM MgCl2 and 37 °C, 3 mM MgCl2 in the presence of 0, 50, 500, and 1000 µM SAM. Data shown are mean values ± standard deviation taken from two independent replicates.
Table 1.
Candidate biosensor sequences. Bold sequences indicate the Spinach2 sequence, which flanks the riboswitch sequence. Italic and underlined indicate the part of the transducer stem derived from the native riboswitch. Blue indicates the tetraloop used for circular permutation of the P1 stem in the riboswitch.
| Name | Sequence (5’ to 3’) |
|---|---|
| Tt 1–4 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCATATCAAGAGAGGTGGAGGGACTGGCCC GATGAAACCCGGCAACCAGCCTTAGGGCATGGTGCCAATTCCTGCAGCGGTTTCGCTGAAAGATGT TGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Tt 1–5 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCATTATCAAGAGAGGTGGAGGGACTGGCC CGATGAAACCCGGCAACCAGCCTTAGGGCATGGTGCCAATTCCTGCAGCGGTTTCGCTGAAAGATG ATTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Tt 3-3 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCAGGCATGGTGCCAATTCCTGCAGCGGTTT CGCTGAAAGATGAGAGGCAACTCTTATCAAGAGAGGTGGAGGGACTGGCCCGATGAAACCCGGCA ACCAGCCTTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Tt 3-3 delA |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCAGGCTGGTGCCAATTCCTGCAGCGGTTTC GCTGAAAGATGAGAGGCAACTCTTATCAAGAGAGGTGGAGGGACTGGCCCGATGAAACCCGGCAA CCAGCCTTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Tt 3–4 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCACTGGTGCCAATTCCTGCAGCGGTTTCGC TGAAAGATGAGAGGCAACTCTTATCAAGAGAGGTGGAGGGACTGGCCCGATGAAACCCGGCAACC AGTTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Tt 4-3 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCACTGAAAGATGAGAGGCAACTCTTATCAA GAGAGGTGGAGGGACTGGCCCGATGAAACCCGGCAACCAGCCTTAGGGCATGGTGCCAATTCCTG CAGTTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Tt 4-4 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCAGCTGAAAGATGAGAGGCAACTCTTATCA AGAGAGGTGGAGGGACTGGCCCGATGAAACCCGGCAACCAGCCTTAGGGCATGGTGCCAATTCCT GCAGCTTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Tt 4–5 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCACGCTGAAAGATGAGAGGCAACTCTTATC AAGAGAGGTGGAGGGACTGGCCCGATGAAACCCGGCAACCAGCCTTAGGGCATGGTGCCAATTCC TGCAGCGTTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Bc 1–4 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCATTATAACGAGAAGCGGAGGGACTGGCC CAATGAAGCTTCAGCAACCATTCATTGCGATGAAAAGGTGCTAAATCCAGCAAAGGGAACTTTGGC AGATAATTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Bc 1–5 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCACTTATAACGAGAAGCGGAGGGACTGGC CCAATGAAGCTTCAGCAACCATTCATTGCGATGAAAAGGTGCTAAATCCAGCAAAGGGAACTTTGG CAGATAAGTTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Bc 3-3 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCAGAAAAGGTGCTAAATCCAGCAAAGGGA ACTTTGGCAGATAAGGGGCAACTCTTATAACGAGAAGCGGAGGGACTGGCCCAATGAAGCTTCAG CAACCATTCTTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Bc 3–4 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCATGAAAAGGTGCTAAATCCAGCAAAGGG AACTTTGGCAGATAAGGGGCAACTCTTATAACGAGAAGCGGAGGGACTGGCCCAATGAAGCTTCA GCAACCATTCATTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Bc 3–5 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCAATGAAAAGGTGCTAAATCCAGCAAAGG GAACTTTGGCAGATAAGGGGCAACTCTTATAACGAGAAGCGGAGGGACTGGCCCAATGAAGCTTC AGCAACCATTCATTTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Bc 4-3 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCATTGGCAGATAAGGGGCAACTCTTATAAC GAGAAGCGGAGGGACTGGCCCAATGAAGCTTCAGCAACCATTCATTGCGATGAAAAGGTGCTAAAT CCAGCAATTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Bc 4-4 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCATTTGGCAGATAAGGGGCAACTCTTATAA CGAGAAGCGGAGGGACTGGCCCAATGAAGCTTCAGCAACCATTCATTGCGATGAAAAGGTGCTAA ATCCAGCAAATTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Bc 4–5 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCACTTTGGCAGATAAGGGGCAACTCTTATA ACGAGAAGCGGAGGGACTGGCCCAATGAAGCTTCAGCAACCATTCATTGCGATGAAAAGGTGCTA AATCCAGCAAAGTTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Pi 1–4 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCATATCAAGAAAGGCGGAGGGATTAGACC CATTGAAGCCTTAGCAACCCTTTAGTAATAAAGAAGGTGCTAAATTCTACTCAATTATTCGTAATTGG ATAGATATTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Pi 1–5 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCATTATCAAGAAAGGCGGAGGGATTAGAC CCATTGAAGCCTTAGCAACCCTTTAGTAATAAAGAAGGTGCTAAATTCTACTCAATTATTCGTAATTG GATAGATAATTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Pi 3-3 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCAAAGAAGGTGCTAAATTCTACTCAATTATT CGTAATTGGATAGATAACAGCAATGTTATCAAGAAAGGCGGAGGGATTAGACCCATTGAAGCCTTA GCAACCCTTTTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Pi 3–4 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCAAAAGAAGGTGCTAAATTCTACTCAATTA TTCGTAATTGGATAGATAACAGCAATGTTATCAAGAAAGGCGGAGGGATTAGACCCATTGAAGCCT TAGCAACCCTTTTTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Pi 3–5 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCATAAAGAAGGTGCTAAATTCTACTCAATT ATTCGTAATTGGATAGATAACAGCAATGTTATCAAGAAAGGCGGAGGGATTAGACCCATTGAAGCC TTAGCAACCCTTTATTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Pi 4–7 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCATAATTGGATAGATAACAGCAATGTTATC AAGAAAGGCGGAGGGATTAGACCCATTGAAGCCTTAGCAACCCTTTAGTAATAAAGAAGGTGCTAA ATTCTACTCAATTATTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Pi 4-4 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCATTGGATAGATAACAGCAATGTTATCAAG AAAGGCGGAGGGATTAGACCCATTGAAGCCTTAGCAACCCTTTAGTAATAAAGAAGGTGCTAAATT CTACTCAATTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Pi 4–5 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCAATTGGATAGATAACAGCAATGTTATCAA GAAAGGCGGAGGGATTAGACCCATTGAAGCCTTAGCAACCCTTTAGTAATAAAGAAGGTGCTAAAT TCTACTCAATTTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Bs 1–4 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCATATCAAGAGAAGCAGAGGGACTGGCCC GACGAAGCTTCAGCAACCGGTGTAATGGCGATCAGCCATGACCAAGGTGCTAAATCCAGCAAGCTC GAACAGCTTGGAAGATATTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Bs 1–5 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCATTATCAAGAGAAGCAGAGGGACTGGCC CGACGAAGCTTCAGCAACCGGTGTAATGGCGATCAGCCATGACCAAGGTGCTAAATCCAGCAAGCT CGAACAGCTTGGAAGATAATTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Bs 3-3 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCACATGACCAAGGTGCTAAATCCAGCAAGC TCGAACAGCTTGGAAGATAAGAAGCAATTCTTATCAAGAGAAGCAGAGGGACTGGCCCGACGAAG CTTCAGCAACCGGTGTAATGTTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Bs 3–4 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCACCATGACCAAGGTGCTAAATCCAGCAAG CTCGAACAGCTTGGAAGATAAGAAGCAATTCTTATCAAGAGAAGCAGAGGGACTGGCCCGACGAA GCTTCAGCAACCGGTGTAATGGTTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Bs 3–5 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCAGCCATGACCAAGGTGCTAAATCCAGCAA GCTCGAACAGCTTGGAAGATAAGAAGCAATTCTTATCAAGAGAAGCAGAGGGACTGGCCCGACGA AGCTTCAGCAACCGGTGTAATGGCTTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Bs 4-4 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCACTTGGAAGATAAGAAGCAATTCTTATCA AGAGAAGCAGAGGGACTGGCCCGACGAAGCTTCAGCAACCGGTGTAATGGCGATCAGCCATGACC AAGGTGCTAAATCCAGCAAGTTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Bs 4–5 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCAGCTTGGAAGATAAGAAGCAATTCTTATC AAGAGAAGCAGAGGGACTGGCCCGACGAAGCTTCAGCAACCGGTGTAATGGCGATCAGCCATGAC CAAGGTGCTAAATCCAGCAAGCTTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
| Bs 4–6 |
GATGTAACTGAATGAAATGGTGAAGGACGGGTCCAAGCTTGGAAGATAAGAAGCAATTCTTAT CAAGAGAAGCAGAGGGACTGGCCCGACGAAGCTTCAGCAACCGGTGTAATGGCGATCAGCCATGA CCAAGGTGCTAAATCCAGCAAGCTTTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC |
Table 2.
Sequences used in in vivo flow cytometry analysis. Italic sequences indicates the tRNA scaffold that is appended onto the terminal ends. Bold sequences indicate the Spinach2 sequence, which flanks the riboswitch sequence. Italic and underlined indicate the part of the transducer stem derived from the native riboswitch. Blue indicates the tetraloop used for circular permutation of the P1 stem in the riboswitch. Red indicates the site of mutation(s) that abolish ligand binding to the aptamer.
| Name | Sequence (5’ to 3’) |
|---|---|
| Spinach2 |
GCCCGGATAGCTCAGTCGGTAGAGCAGCGGCCGGATGTAACTGAATGAAATGGTGAAGG ACGGGTCCAGTAGGCTGCTTCGGCAGCCTACTTGTTGAGTAGAGTGTGAGCTCCGTAAC TAGTTACATCCGGCCGCGGGTCCAGGGTTCAAGTCCCTGTTCGGGCGCCA |
| Spinach2 Mutant |
GCCCGGATAGCTCAGTCGGTAGAGCAGCGGCCGGATGTAACTGAATGAAATGGTGAAGG ACGccTCCAGTAGGCTGCTTCGGCAGCCTACTTGTTGAGTAGAGTGTGAGCTCCGTAACT AGTTACATCCGGCCGCGGGTCCAGGGTTCAAGTCCCTGTTCGGGCGCCA |
| Tt 1–4 |
GCCCGGATAGCTCAGTCGGTAGAGCAGCGGCCGGATGTAACTGAATGAAATGGTGAAGG ACGGGTCCATATCAAGAGAGGTGGAGGGACTGGCCCGATGAAACCCGGCAACCAGCCTTAG GGCATGGTGCCAATTCCTGCAGCGGTTTCGCTGAAAGATGTTGTTGAGTAGAGTGTGAGCT CCGTAACTAGTTACATCCGGCCGCGGGTCCAGGGTTCAAGTCCCTGTTCGGGCGCCA |
| Tt 1–4 Mutant |
GCCCGGATAGCTCAGTCGGTAGAGCAGCGGCCGGATGTAACTGAATGAAATGGTGAAGG ACGGGTCCATATCAgaAGAGGTGGAGGGACTGGCCCGATGAAACCCGGCAACCAGCCTTAG GGCATGGTGCCAATTCCTGCAGCGGTTTCGCTGAAAGATGTTGTTGAGTAGAGTGTGAGCT CCGTAACTAGTTACATCCGGCCGCGGGTCCAGGGTTCAAGTCCCTGTTCGGGCGCCA |
| Bc 1–5 |
GCCCGGATAGCTCAGTCGGTAGAGCAGCGGCCGGATGTAACTGAATGAAATGGTGAAGG ACGGGTCCACTTATAACGAGAAGCGGAGGGACTGGCCCAATGAAGCTTCAGCAACCATTCAT TGCGATGAAAAGGTGCTAAATCCAGCAAAGGGAACTTTGGCAGATAAGTTGTTGAGTAGAG TGTGAGCTCCGTAACTAGTTACATCCGGCCGCGGGTCCAGGGTTCAAGTCCCTGTTCGGG CGCCA |
| Bc 1–5 Mutant |
GCCCGGATAGCTCAGTCGGTAGAGCAGCGGCCGGATGTAACTGAATGAAATGGTGAAGG ACGGGTCCACTTATAAgcAGAAGCGGAGGGACTGGCCCAATGAAGCTTCAGCAACCATTCAT TGCGATGAAAAGGTGCTAAATCCAGCAAAGGGAACTTTGGCAGATAAGTTGTTGAGTAGAG TGTGAGCTCCGTAACTAGTTACATCCGGCCGCGGGTCCAGGGTTCAAGTCCCTGTTCGGG CGCCA |
| Bs 4-4 |
GCCCGGATAGCTCAGTCGGTAGAGCAGCGGCCGGATGTAACTGAATGAAATGGTGAAGG ACGGGTCCAGCTTGGAAGATAAGAAGCAATTCTTATCAAGAGAAGCAGAGGGACTGGCCCG ACGAAGCTTCAGCAACCGGTGTAATGGCGATCAGCCATGACCAAGGTGCTAAATCCAGCAAG CTTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATCCGGCCGCGGGTCCAGGGTTC AAGTCCCTGTTCGGGCGCCA |
| Bs 4-4 Mutant |
GCCCGGATAGCTCAGTCGGTAGAGCAGCGGCCGGATGTAACTGAATGAAATGGTGAAGG ACGGGTCCAGCTTGGAAGATAAGAAGCAATTCTTATCAgaAGAAGCAGAGGGACTGGCCCG ACGAAGCTTCAGCAACCGGTGTAATGGCGATCAGCCATGACCAAGGTGCTAAATCCAGCAAG CTTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATCCGGCCGCGGGTCCAGGGTTC AAGTCCCTGTTCGGGCGCCA |
| Bc 4–5 |
GCCCGGATAGCTCAGTCGGTAGAGCAGCGGCCGGATGTAACTGAATGAAATGGTGAAGG ACGGGTCCACTTTGGCAGATAAGGGGCAACTCTTATAACGAGAAGCGGAGGGACTGGCCCA ATGAAGCTTCAGCAACCATTCATTGCGATGAAAAGGTGCTAAATCCAGCAAAGTTGTTGAGT AGAGTGTGAGCTCCGTAACTAGTTACATC |
| Bc 4–5 Mutant |
GCCCGGATAGCTCAGTCGGTAGAGCAGCGGCCGGATGTAACTGAATGAAATGGTGAAGG ACGGGTCCACTTTGGCAGATAAGGGGCAACTCTTATAAgcAGAAGCGGAGGGACTGGCCCA ATGAAGCTTCAGCAACCATTCATTGCGATGAAAAGGTGCTAAATCCAGCAAAGTTGTTGAGT AGAGTGTGAGCTCCGTAACTAGTTACATC |
Circular permutations of riboswitches are a new design strategy for RBF biosensors
Circularly permuted SAM-I riboswitch improved biosensor brightness and affinity
Circularly permuted SAM-I biosensor worked in vivo
Acknowledgments
This work was supported by the National Institutes of Health (DP2 OD008677 and R01 GM124589) and Office of Naval Research (N000141712638) for J.T., Y.F.H., and M.C.H. and the Chau Hoi Shuen Foundation Women in Science program for G.J. and M.C.H. Additionally, L.T. was supported by the UC Berkeley Amgen Scholars program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Villa JK, Su Y, Contreras LM, Hammond MC. Synthetic biology of regulatory RNAs. Regulating with RNA in Bacteria and Archaea. 2017 [Google Scholar]
- 2.Chappell J, Watters KE, Takahashi MK, Lucks JB. A renaissance in RNA synthetic biology : new mechanisms, applications and tools for the future. Curr. Opin. Chem. Biol. 2016;28:47–56. doi: 10.1016/j.cbpa.2015.05.018. [DOI] [PubMed] [Google Scholar]
- 3.Hallberg ZF, Su Y, Kitto RZ, Hammond MC. Engineering and In Vivo Applications of Riboswitches. Annu. Rev. Biochem. 2017;86:515–539. doi: 10.1146/annurev-biochem-060815-014628. [DOI] [PubMed] [Google Scholar]
- 4.Paige JS, Nguyen-Duc T, Song W, Jaffrey SR. Fluorescence Imaging of Cellular Metabolites with RNA. Science (80-.) 2012;335:1194–1194. doi: 10.1126/science.1218298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kellenberger Ca, Wilson SC, Sales-Lee J, Hammond MC. RNA-based fluorescent biosensors for live cell imaging of second messengers cyclic di-GMP and cyclic AMP-GMP. J. Am. Chem. Soc. 2013;135:4906–4909. doi: 10.1021/ja311960g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kellenberger CA, Chen C, Whiteley AT, Portnoy DA, Hammond MC. RNA-Based Fluorescent Biosensors for Live Cell Imaging of Second Messenger Cyclic di-AMP. J. Am. Chem. Soc. 2015;137:6432–6435. doi: 10.1021/jacs.5b00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang XC, et al. Next-generation RNA-based fluorescent biosensors enable anaerobic detection of cyclic di-GMP. Nucleic Acids Res. 2016;44:e139–e139. doi: 10.1093/nar/gkw580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strack RL, Disney MD, Jaffrey SR. A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA. Nat. Methods. 2013;10:1219–24. doi: 10.1038/nmeth.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su Y, Hickey SF, Keyser SGL, Hammond MC. In Vitro and In Vivo Enzyme Activity Screening via RNA-Based Fluorescent Biosensors for S-Adenosyl- l -homocysteine (SAH) J. Am. Chem. Soc. 2016;138:7040–7047. doi: 10.1021/jacs.6b01621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.You M, Litke JL, Jaffrey SR. Imaging metabolite dynamics in living cells using a Spinach-based riboswitch. Proc. Natl. Acad. Sci. 2015;2015:201504354. doi: 10.1073/pnas.1504354112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammann C, Luptak A, Perreault J, de la Pena M. The ubiquitous hammerhead ribozyme. Rna. 2012;18:871–885. doi: 10.1261/rna.031401.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soma A, et al. Permuted tRNA Genes Expressed via a Circular RNA Intermediate in Cyanidioschyzon merolae. Science (80-.) 2007;318:450–453. doi: 10.1126/science.1145718. [DOI] [PubMed] [Google Scholar]
- 13.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 14.Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–5. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- 15.Heppell B, et al. Molecular insights into the ligand-controlled organization of the SAM-I riboswitch. Nat. Chem. Biol. 2011;7:384–392. doi: 10.1038/nchembio.563. [DOI] [PubMed] [Google Scholar]
- 16.Paige JS, Wu KY, Jaffrey SR. RNA Mimics of Green Fluorescent Protein. Science (80-.) 2011;333 doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson SC, Cohen DT, Wang XC, Hammond MC. A neutral pH thermal hydrolysis method for quantification of structured RNAs. RNA. 2014;20:1153–60. doi: 10.1261/rna.045856.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hickey SF, Hammond MC. Structure-guided design of fluorescent S-adenosylmethionine analogs for a high-throughput screen to target SAM-I riboswitch RNAs. Chem. Biol. 2014;21:345–356. doi: 10.1016/j.chembiol.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sudarsan N, et al. Tandem Riboswitch Architectures Exhibit Complex Gene Control Functions. Science (80-.) 2006;314 doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- 20.Montange RK, et al. Discrimination between Closely Related Cellular Metabolites by the SAM-I Riboswitch. J. Mol. Biol. 2010;396:761–772. doi: 10.1016/j.jmb.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burgess DJ. Epigenetics: Bacterial DNA methylation gets SMRT. Nat. Rev. Genet. 2013;14:4. doi: 10.1038/nrg3389. [DOI] [PubMed] [Google Scholar]
- 22.Bennett BD, et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kellenberger CA, Hallberg ZF, Hammond MC. Live cell imaging using riboswitch-Spinach tRNA fusions as metabolite-sensing fluorescent biosensors. Methods Mol Biol. 2015;1316:7–103. doi: 10.1007/978-1-4939-2730-2_8. [DOI] [PubMed] [Google Scholar]
- 24.McCown P, Liang J, Weinberg Z, Breaker R. Structural, Functional, and Taxonomic Diversity of Three PreQ1 Riboswitch Classes. Chem. Biol. 2014;21:880–889. doi: 10.1016/j.chembiol.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serganov A, Huang L, Patel DJ. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature. 2009;458:233–237. doi: 10.1038/nature07642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dolgosheina EV, et al. RNA Mango Aptamer-Fluorophore: A Bright, High-Affinity Complex for RNA Labeling and Tracking. ACS Chem. Biol. 2014;9:2412–2420. doi: 10.1021/cb500499x. [DOI] [PubMed] [Google Scholar]
- 27.Sunbul M, Jaschke A. Contact-mediated quenching for RNA imaging in bacteria with a fluorophore-binding aptamer. Angew. Chemie - Int. Ed. 2013;52:13401–13404. doi: 10.1002/anie.201306622. [DOI] [PubMed] [Google Scholar]
- 28.Lu C, et al. SAM recognition and conformational switching mechanism in the Bacillus subtilis yitJ S box/SAM-I riboswitch. J. Mol. Biol. 2010;404:803–18. doi: 10.1016/j.jmb.2010.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu C, et al. Crystal structures of the SAM-III/S(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism. Nat. Struct. Mol. Biol. 2008;15:1076–83. doi: 10.1038/nsmb.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]