Abstract
Persister cells constitute a small subpopulation of bacteria that display remarkably high antibiotic tolerance and for pathogens such as Staphylococcus aureus are suspected as culprits of chronic and recurrent infections. Persisters formed during exponential growth are characterized by low ATP levels but less is known of cells in stationary phase. By enrichment from a transposon mutant library in S. aureus we identified mutants that in this growth phase displayed enhanced persister cell formation. We found that inactivation of either sucA or sucB, encoding the subunits of the α-ketoglutarate dehydrogenase of the tricarboxylic acid cycle (TCA cycle), increased survival to lethal concentrations of ciprofloxacin by 10–100 fold as did inactivation of other TCA cycle genes or atpA encoding a subunit of the F1F0 ATPase. In S. aureus, TCA cycle activity and gene expression are de-repressed in stationary phase but single cells with low expression may be prone to form persisters. While ATP levels were not consistently affected in high persister mutants they commonly displayed reduced membrane potential, and persistence was enhanced by a protein motive force inhibitor. Our results show that persister cell formation in stationary phase does not correlate with ATP levels but is associated with low membrane potential.
Introduction
Persister cells are phenotypic variants present in the bacterial populations that in the absence of heritable mutations or antibiotic resistance genes display remarkable antibiotic tolerance. They are detected as survivors when exposed to very high antibiotic concentrations often reaching 100 times the minimal inhibitory concentration (MIC) and they are observed in both exponential and stationary growth phases for multiple bacterial species and with several antibiotics1,2. During exponential growth, persister cells (designated type II) arise at very low frequencies whereas upon entry into stationary growth phase the fraction of persister cells (type I) increases substantially3,4. Commonly, persister cells are considered to be in a state of dormancy and several mechanisms have been proposed to explain the phenomenon. In Escherichia coli and Salmonella, dormancy was previously proposed to be stochastically induced by the action of the toxin-antitoxin (TA) systems that compromise key cellular functions and are activated by ppGpp or by the SOS response5–10. However, this model was recently disproven11,12. In fact, conditions that halt transcription, translation or ATP synthesis dramatically increase persister frequency from 0.01% to up to 10–100%13. Particularly, the cellular ATP level is important as depletion of ATP with arsenate increases the number of persister cells both in E. coli and in the human pathogen, Staphylococcus aureus4,14. In the latter organism, neither the stringent response nor TA loci contribute to persistence but rather persister cells express stationary phase marker genes, such as the capsular polysaccharide operon, suggesting that they are cells that during exponential growth have prematurely entered stationary phase with low ATP levels4.
Although the central role of bacterial metabolism in bacterial persistence has been documented by several reports, a uniform picture of the contribution has not emerged. In E. coli, fumarate accumulation and perturbations of the TCA cycle as well as the electron transport chain has been linked with increased persistence while others have found that inactivation of sucB, encoding α-ketoglutarate dehydrogenase and other genes of the TCA cycle reduced persister formation15–17. Tolerance to aminoglycosides is provided by low proton motive force (PMF) as persister cells are eradicated by conditions that enhance PMF and facilitates aminoglycoside uptake18,19. The varying contributions of metabolism to persister formation may in part be explained by different experimental setups but also by the significant differences in metabolism between organisms and growth conditions13. For example, in Gram-negative bacteria which are growing under laboratory conditions, such as E. coli, ATP is produced via oxidative phosphorylation driven by products generated in the TCA cycle while the low-passage counterpart produces ATP by substrate level phosphorylation; in the Gram-positive bacteria, S. aureus, the TCA cycle is repressed during exponential growth where glucose is metabolized to acetate with ATP generated by substrate level phosphorylation20.
S. aureus is known to give rise to a diverse range of serious diseases such as endocarditis, osteomyelitis and chronic and re-occurring infections that are difficult to treat with antibiotics. The clinical relevance of persister cells has been demonstrated by infection of a chronic wound model with S. aureus that could be eradicated with an acyldepsipeptide antibiotic, ADEP4 that killed persister cells21,22. With most studies of persisters having focused on the exponential growth phase we set out to determine if the already high level of persisters observed for S. aureus in stationary growth phase may be increased even further and if so which genes are contributing. For this purpose, we have taken a transposon library approach to identify genes that either by inactivation or overproduction increase persister cell frequency in stationary phase23. During enrichment, we found that inactivation of either sucA or sucB encoding subunits of α-ketoglutarate dehydrogenase dramatically increased persister cell formation as did inactivation of other enzymes of the TCA cycle or atpA gene. Commonly, all the mutants with enhanced persister levels displayed reduced membrane potential that contributes to PMF indicating that in stationary phase membrane potential becomes critical for persister cell formation or survival.
Results
Inactivation of α-ketoglutarate dehydrogenase increases persister cell formation in stationary phase
With the aim of identifying genes that contribute to persister cell formation in stationary phase in S. aureus, we constructed a high-density transposon (Tnp) insertion library in strain Newman employing the bacteriophage-based mariner transposon delivery system that allowed both gene inactivation and gene overexpression from an outward-facing promoter in the transposon element23,24. The coverage of Tnp insertions were assessed in 10 random library isolates and the results revealed that the 10 isolates carrying insertions at 10 independent sites were evenly distributed throughout the genome indicating random Tnp insertion (see Supplementary Table S1).
Library clones with increased frequency of persister cells formation in stationary phase were identified by an enrichment strategy where the Tnp library cells were repeatedly exposed to the fluoroquinolone drug, ciprofloxacin at a lethal dose of 100 times MIC25. The survivors were collected and inoculated into fresh medium and allowed to reach stationary phase before repeated exposure to ciprofloxacin. This treatment was repeated four times in total for pooled library clones as well as for the wild type strain Newman (Fig. 1) with the starting CFU/ml indicated in Supplementary Table S2. The persister frequency of strain Newman was approximately 1% by the end of enrichment with a few samples reaching 5–10% while the percentage of persisters from the Tnp library cultures exhibited more than 1000-fold increase, from the initial 0.01% to more than 20% after the four rounds of enrichment (Fig. 1). The increase in the fraction of persisters seen for wild type cells after enrichment implies that spontaneous mutations may enhance persister formation. Genome sequencing of one randomly picked clone confirmed this notion as it carried more than 200 mutations compared to the wild type cells prior to passage (data not shown).
Figure 1.
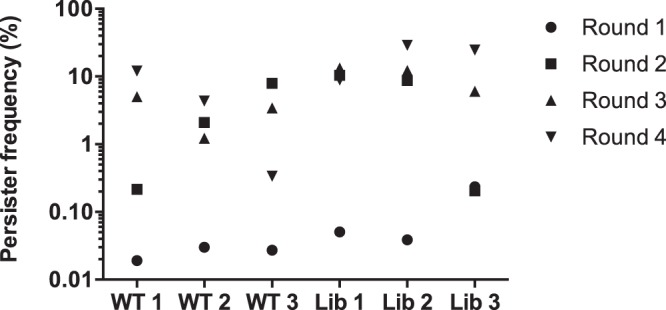
The enrichment of persisters with both S. aureus Newman wild type and transposon library. 24-hour culture of either wild type (WT 1–3) or Tnp library (Lib 1–3) was exposed to 100 × MIC of ciprofloxacin (MIC = 0.5 µg/ml) for another 24 hours before plating on TSA plates for CFU calculation. The same procedure was repeated consecutively for 4 times (Round 1–4) and the persister frequencies were recorded accordingly.
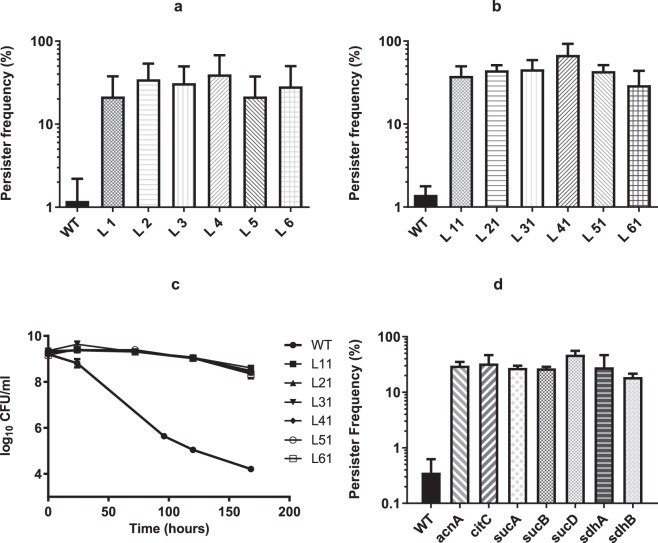
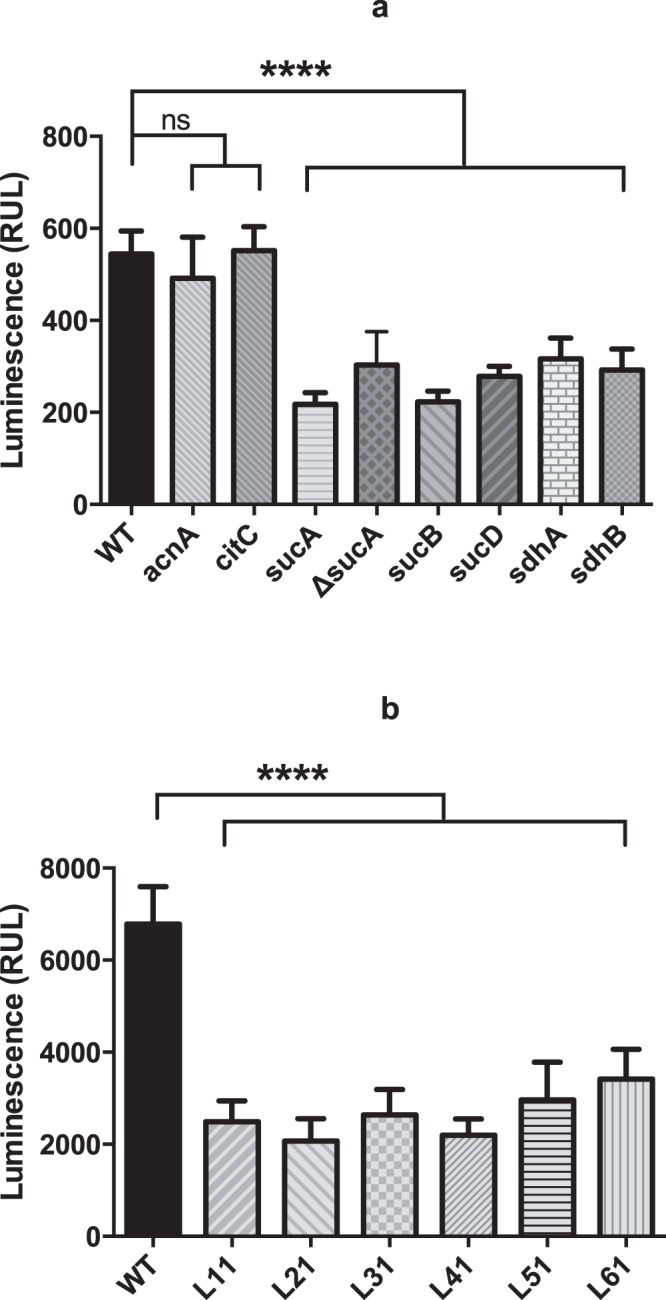
To determine if individual clones were enriched by repeated exposure of the Tnp library to high antibiotic concentrations, six colonies (named as L1-L6) were randomly picked by the end of the fourth round of enrichment to examine persister formation in individual clones. When treated with ciprofloxacin, all the six isolates exhibited persister frequencies between 20–60% compared to approximately 1% for the wild type (Fig. 2a). The transposon/chromosome junction was determined by sequencing and the result showed that in five of the six isolates (L2-L6) the transposon was inserted at one of three different locations within the sucA gene while in the last clone (L1) it was inserted in sucB. sucA and sucB encode subunits of α-ketoglutarate dehydrogenase, which is a key enzyme of the TCA cycle that catalyses the conversion of α-ketoglutarate to succinyl-CoA with the concomitant production of NADH and CO2 in an irreversible reaction26.
Figure 2.
Persister frequency in the presence of ciprofloxacin or oxacillin. Persister frequency of 6 single colonies (L1-L6) of Tnp library (a) or their corresponding transductants (L11-L61) (b) in the presence of ciprofloxacin after persister enrichment was calculated by comparing the CFU calculation of 24-hour post and before the addition of 100 × MIC of ciprofloxacin (MIC = 0.5 µg/ml) (c) Survival of the transductants (L11-L61) challenged with 100 × MIC of oxacillin (MIC = 0.25 µg/ml) was recorded by CFU counting over 7 days. (d) Persister frequency of selected TCA cycle mutants (acnA, citC, sucA, sucB, sucD, sdhA and sdhB) when challenged with 100 × MIC of ciprofloxacin (MIC = 0.5 µg/ml). One wild type colony (WT) was included as control for all the experiments and starting CFU/ml are indicated in Supplementary Table S2. Three biological triplicates were included for each sample point and error bars represent standard deviation.
To rule out the possibility that secondary mutations within the library isolates contribute to the persister phenotype, the Tnp insertion mutations were transduced for all six clones to naïve wild type Newman cells to generate transductants, L11, L21-L61, carrying the individual mutations of the original library clones. All transductants yielded more than 30% persisters in comparison to 1% for the wild type when exposed to lethal concentrations of ciprofloxacin (Fig. 2b). To corroborate this finding and confirm that the enhanced persister cell formation is indeed due to inactivation of α-ketoglutarate dehydrogenase, an unmarked sucA deletion mutant (ΔsucA) was constructed. The ΔsucA mutant yielded more than 30% persisters when exposed to 100 times MIC of ciprofloxacin (see Supplementary Fig. S1). When the survival was followed for 48 hours after addition of ciprofloxacin, ΔsucA mutant exhibited improved survival compared to wild type which displayed the typical biphasic killing curve at the observed time points (see Supplementary Fig. S2). Additionally, the enhanced persister formation was also observed when the mutations were transduced into the S. aureus strains, SA564 and RN6607, which indicates that the current findings are not confined to strain Newman (see Supplementary Fig. S3). Further, we examined if the mutants influenced persister cell formation during exponential growth and found that in this growth phase the inactivation of sucA or sucB did not have any effect (data not shown) agreeing with the notion that in S. aureus, TCA cycle activity is low during exponential growth but de-repressed upon entry into stationary phase20.
Persister cells have been observed with other antibiotics than ciprofloxacin and therefore we examined if inactivation of α-ketoglutarate dehydrogenase increased survival during treatment with lethal doses of oxacillin (Fig. 2c). Because there was no discernible differences in persister generation between wild type and library isolate transductants after a 24-hour treatment, the effect of oxacillin on bacterial survival was monitored over a 7-day period. The counting of colony forming unit (CFU/ml) showed that there was less than 1 log CFU reduction of the library isolate transductants while wild type dropped from 109 to 104 CFU/ml after the 7-day treatment with 100 × MIC of oxacillin. Furthermore, the MICs of all library transductants towards ciprofloxacin and oxacillin were identical to that of wild type cells (see Supplementary Table S3), demonstrating that the increased persister cell formation is not a result of general changes in susceptibility to antibiotics. All together, these experiments show that inactivation of α-ketoglutarate dehydrogenase increases dramatically stationary phase persister cell formation in S. aureus.
Inactivation of TCA cycle genes increases persister cell formation in stationary phase
α-ketoglutarate dehydrogenase is an enzyme of the TCA cycle and to address if TCA cycle activity in general influences persister formation, we examined persister frequency of mutants obtained from a sequence-defined transposon mutant library carrying insertional inactivations of sucD, sdhA, sdhB, acnA and citC27. Importantly, all of these mutants exhibited increased persister frequency (20–60%) paralleling that observed for the sucA and sucB mutants, when compared with wild type cells (<1%) in stationary growth phase (Fig. 2d). While it is unclear why our enrichment strategy only identified sucA and sucB mutants and not mutants lacking other TCA cycle enzymes, it may be related to the observation that after 24-hour of cultivation, the optical density at 600 nm (OD600) of sucA mutant cells was higher than observed for the other TCA cycle mutants (see Supplementary Fig. S4), indicating that this mutant may have a growth advantage. In summary, our results suggest that decreased TCA cycle activity increases S. aureus persister cell formation in stationary phase.
Increased persister cell formation is not caused by slow growth or low pH
The appearance of persister cells has been associated with arrested bacterial growth28 but for the transductants carrying sucA or sucB inactivation we only observed slight differences in cell densities upon entry into stationary phase (see Supplementary Fig. S5). Similarly, long-term survival in stationary phase was not notably different between mutants and the wild type throughout a 7-day period (see Supplementary Fig. S5). In addition, the persister assay was repeated with bacterial cultures after 48-hour cultivation to ensure that all cells had entered stationary phase and again, the persister percentage of sucA mutant remained significantly higher than for wild type cells (see Supplementary Fig. S6), suggesting that any potential differences between strains in entry into stationary phase did not influence the results.
In contrast, we observed that culture supernatants of the TCA cycle mutants displayed lower pH (see Supplementary Table S4) and accumulated more acetate than wild type cells (Fig. 3a). This observation agrees with the fact that in S. aureus, the TCA cycle is required for post-exponential catabolism of acetate that has accumulated during exponential growth20. Similarly, the accumulation of acetate was also observed for all the transductants of library isolates (L11-L61) which have mutations of either sucA or sucB within TCA cycle (Fig. 3b). To examine if the enhanced persister cell formation was related to acidification of the growth medium, TSB medium without glucose was used to achieve comparable pH of supernatants from both ΔsucA and wild type cells in stationary phase (Fig. 4a). Importantly, in the absence of glucose where the pH of both mutant and wild type cultures was similar, the persister frequency of ΔsucA cells was still enhanced with 50% of the cells producing persisters while only 1% of wild type cells (Fig. 4b). These results show that low pH is not responsible for the increased persister cell formation in the TCA cycle mutants.
Figure 3.
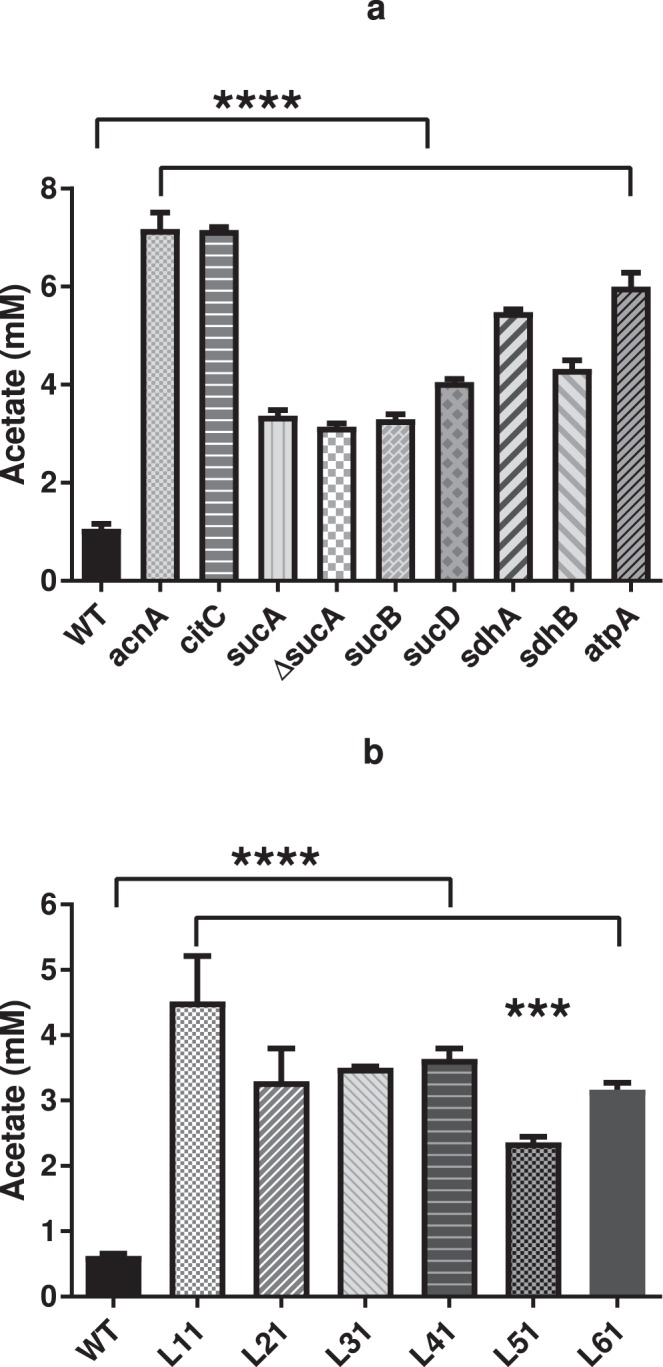
Acetate production by TCA cycle mutants (a) and transductants (L11-L61) (b). All strains were cultivated with TSB medium at 37 °C with shaking at 200 rpm and 1 ml of the culture was filtrated with 0.22 µm filter before the measurement with HPLC. The accumulation of acetate was calculated by comparing the concentration from early exponential phase (3-hour cultivation) and stationary phase (24-hour cultivation). There were 3 biological replicates for each sample and error bars represent standard deviation. The statistical comparison between wild type and mutants (a) or transductants (b) was determined with one way ANOVA followed by Dunnett’s test. The asterisks indicate significant difference at P < 0.05.
Figure 4.
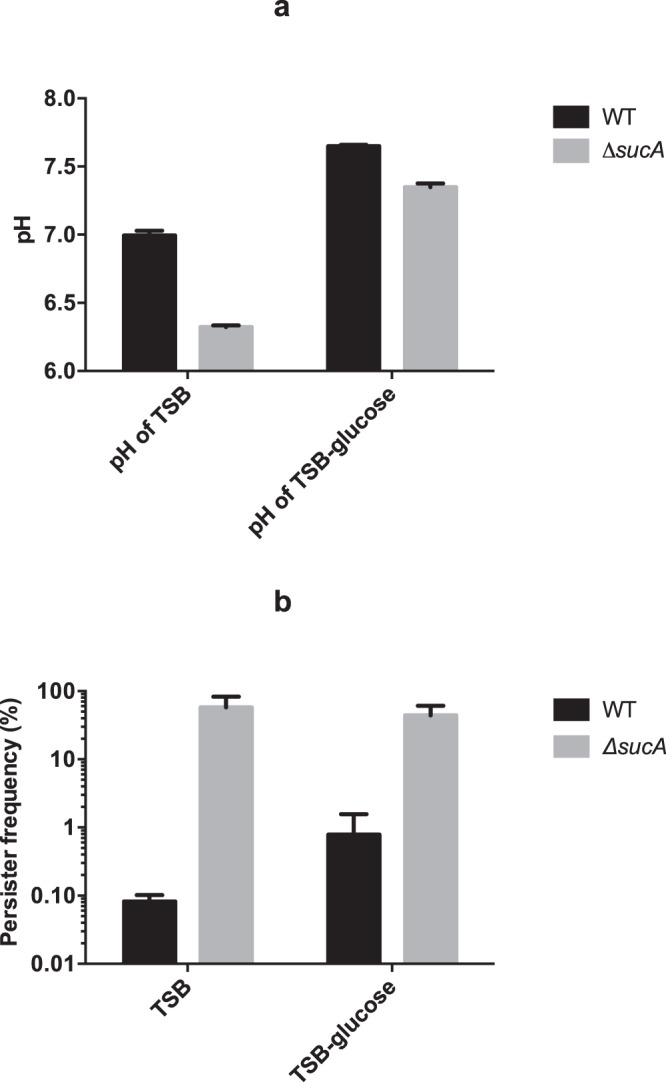
pH does not affect persister formation. (a) pH of cell-free supernatant. Wild type (WT) and ΔsucA were cultivated in TSB medium with or without glucose (TSB-glucose) for 24 hours and the pH of the supernatant was measured and compared; (b) the persister frequency of wild type (WT) and ΔsucA was calculated when cultivated with TSB medium or TSB medium without glucose (TSB-glucose). There were 3 biological replicates for each sample and error bars stand for standard deviation. Starting CFU/ml are indicated in Supplementary Table S2.
Membrane potential is associated with persister cell formation
Recently, it was demonstrated that S. aureus persisters generated during exponential growth are characterized by low ATP level4. However, when we assessed the ATP content we found that while the majority of the TCA cycle mutants (sucA, sucB, sucD, sdhA or sdhB, Fig. 5a) including the library isolate transductants (L11-L61, Fig. 5b) contained about 50% of the wild type ATP level, two mutants, namely acnA and citC, encoding aconitase and isocitrate dehydrogenase, respectively, had similar ATP levels as wild type cells (Fig. 5a) even though their ability to form persisters was remarkably higher than wild type cells (Fig. 2d). This phenomenon may be explained by the fact that some amino acids can feed into the TCA cycle at the point of α-ketoglutarate, and therefore, the inactivation of upstream enzymes, either aconitase (acnA) or isocitrate dehydrogenase (citC), may not affect the downstream metabolic activity of TCA cycle including ATP production26.
Figure 5.
ATP is not the sole determinant of cellular persistence. (a) The ATP content of TCA cycle mutants (acnA, citC, sucA, sucB, sucD, sdhA, sdhB and ΔsucA) was measured after 24 hours of cultivation; (b) the ATP levels of wild type (WT) and transductants of Tnp library isolates (L11-L61) were measured after 24-hour cultivation. There were 3 biological replicates for each sample and 3 different reads of luminescence signal for each biological replicate. The statistical comparison between wild type and TCA cycle mutants (a) or Tnp isolate transductants (b) was determined with one way ANOVA followed by Dunnett’s test. The error bars stand for standard deviation and the asterisks indicate significant difference at P < 0.05.
During exponential growth ATP is generated by substrate level phosphorylation in S. aureus, but in stationary phase oxidative phosphorylation and the ATP synthase could contribute20. Therefore, we examined a mutant lacking atpA that encodes the α-subunit of F1F0 ATP synthase. As has been observed previously for other organisms we found that there was no significant reduction in ATP level of the atpA mutant when cultivated in rich medium (Fig. 6a) indicating that also in the post-exponential growth phase substrate level phosphorylation contributed to ATP production29. Strikingly, however, the inactivation of atpA increased persister formation approximately 1000-fold compared to wild type cells (Fig. 6b) underscoring the notion that ATP content is not decisive for S. aureus persister cell formation in stationary phase.
Figure 6.
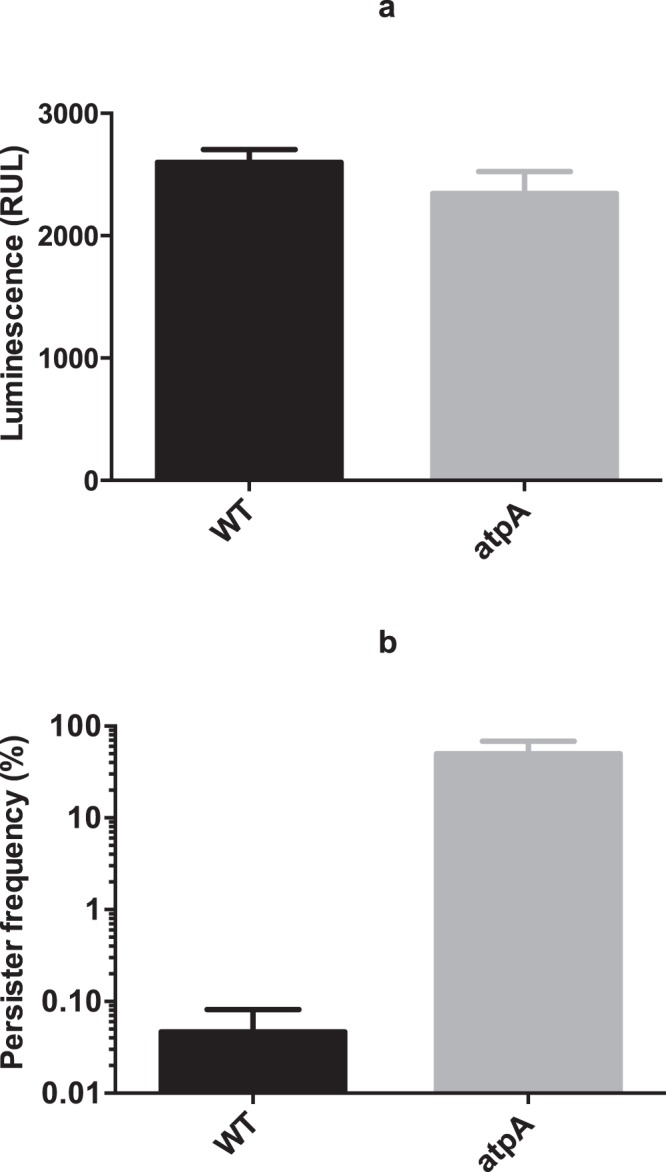
Comparison of ATP level (a) and persister frequency (b) between S. aureus Newman wild type (WT) and atpA mutant. For ATP measurement (a) 3 different reads of luminescence signal were taken for each biological replicate. Persister frequency (b) was tested with 100 × MIC of ciprofloxacin and the samples for ATP measurement were taken after 24 hours of incubation right before the addition of ciprofloxacin. There were 3 biological replicates for each sample and error bars indicate standard deviation. Starting CFU/ml are indicated in Supplementary Table S2.
Inactivation of the TCA cycle is likely to reduce the production of reducing equivalents that enters the electron transport chain and contributes to the PMF30. Therefore, we speculated that membrane potential, which contributes to PMF, may be related to persister cell formation. Indeed, when monitoring membrane potential using the fluorescent probe, DiOC2(3) and flow cytometry we consistently found that all TCA cycle mutants which exhibited increased persister cell frequencies displayed reduced membrane potential (Fig. 7a). Importantly, also the membrane potential of the atpA mutant was decreased compared to wild type cells (Fig. 7a). This result may indicate that wild type S. aureus cells in the post-exponential growth phase maintains PMF by extruding protons through the F1F0 ATP synthase with hydrolysis of ATP. The critical role of PMF for persister cell formation was lastly confirmed by addition of the PMF inhibitor, carbonyl cyanide m-chlorophenylhydrazone (CCCP) as expected it enhanced persister cell formation (Fig. 7b)31–33. Collectively, our results suggest that in stationary phase, the reduced proton motive force (PMF) characterizes S. aureus persister cells.
Figure 7.
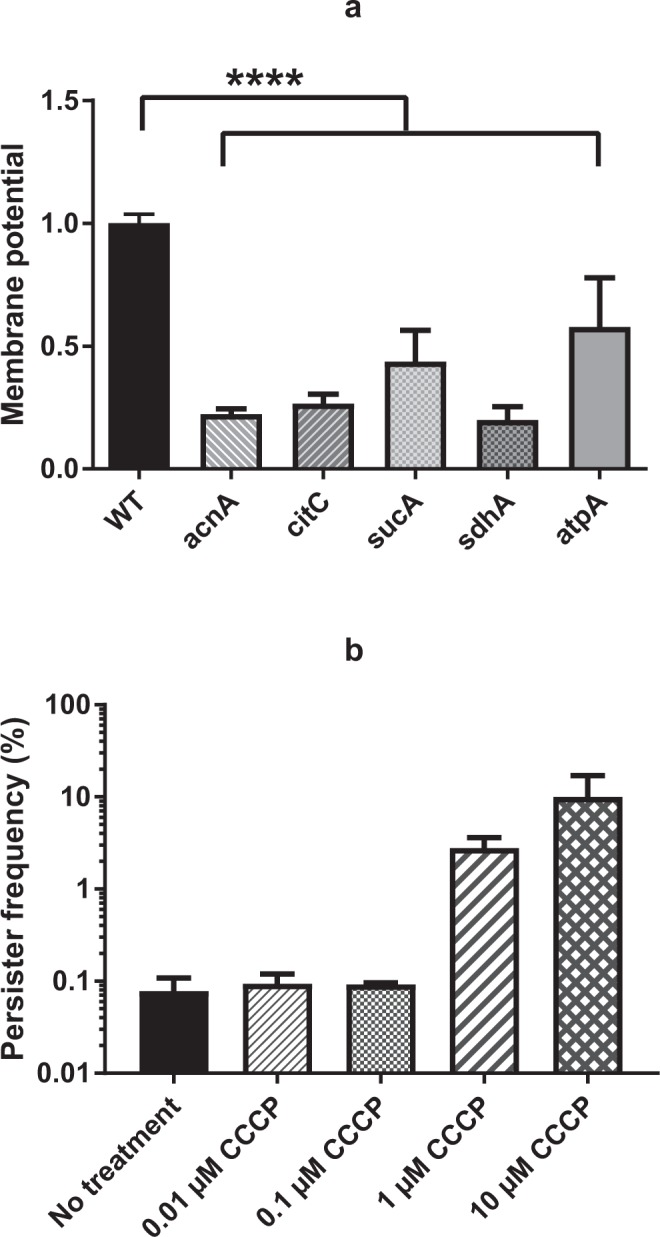
Measurement of membrane potential of selected mutants (a) and the persister formation in the presence of CCCP (b). (a) Membrane potential was measured for TCA cycle mutants (acnA, citC, sucA and sdhA) and atpA by flow cytometer after 30 minutes co-incubation with dye, DiOC2(3). (b) The persister frequency of S. aureus Newman wild type, challenged with 100 × MIC of ciproloxacin, was determined in the presence of CCCP, from 0.01 µM to 10 µM. There were biological triplicates for each sample and error bars represent standard deviation. Starting CFU/ml are indicated in Supplementary Table S2. The statistical comparison of membrane potential between wild type and selected TCA cycle mutants was determined with one way ANOVA followed by Dunnett’s test and the asterisks indicate significant difference at P < 0.05 (a).
sucA is expressed in stationary phase
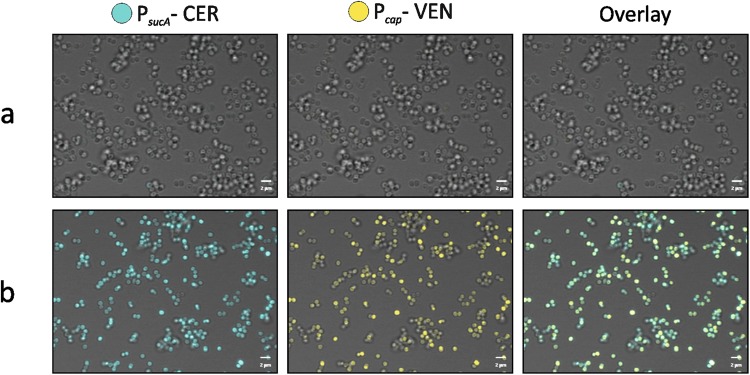
In S. aureus TCA cycle activity is de-repressed in stationary phase20. To examine if this de-repression correlates with expression of TCA cycle genes, we fused the sucA promoter to the cfp reporter gene and monitored expression in exponential and stationary phase single cells (Fig. 8). While essentially no expression was observed in exponentially growing cells, sucA was clearly expressed in stationary phase cells. Similar expression pattern was observed for the cap promoter, an established stationary phase marker that is expressed along with TCA cycle activity34. In addition, we observed that expression of sucA varied between individual cells. These results show that TCA cycle genes are expressed in stationary phase and that variation in expression of sucA and possibly of other TCA cycle genes occurs. Thus, we speculate that in wild type cells, cell to cell variation in expression of TCA cycle genes in stationary phase determines persister cell formation.
Figure 8.
Expression of sucA in S. aureus Newman in exponential or stationary phase. The fluorescence expression of both PsucA (CFP channel) and Pcap (YFP channel) in strain Newman was monitored at exponential (a) and stationary phase (b) respectively. The promoter activity of PsucA appeared blue and Pcap activity appeared yellow.
Discussion
When searching for mutants that in stationary phase gave increased persister frequencies we identified insertion mutants of sucA and sucB, which encode the subunits of the α-ketoglutarate dehydrogenase, a key enzyme of the TCA cycle that converts α-ketoglutarate to succinyl-CoA35. Importantly, also mutants lacking other genes of the TCA cycle as well as atpA encoding a subunit of the F1F0 ATP synthase displayed elevated persister cell frequencies. In contrast to our findings, Wang et al. reported that reduced activity of the electron transport chain and the TCA cycle decreased the number of persister cells when challenged with the fluoquinolone, levofloxacin36. To address the basis for this inconsistency, we examined our strains using their reported protocol. The result presented in Supplementary Fig. S7 shows that with the experimental approach by Wang et al., the sdhA (as well as sucA) mutant survived levofloxacin equally well as the wild type cells within a 5 days period. In contrast, when the persister assay was repeated with our standard protocol and using levofloxacin, both mutants (sucA and sdhA) exhibited higher persister frequencies compared to wild type cells (data not shown). Thus, we conclude that while the difference is not due to the antibiotics (ciprofloxacin vs. levofloxacin); there is a difference between the outcomes from the two protocols. We do, however, not see the same effect from an sdhA mutant in our strain background when using Wang’s persister assay protocol36, suggesting that strain choice is also important. Moreover, we have replicated our findings in different strain backgrounds (strains SA564 or RN6607, see Supplementary Fig. S3), supporting the generality of our findings. We also noted that, in Wang’s study36, the persister assay was carried out with static cultures in eppendorf tubes where the available oxygen is expected to be limited and rapidly depleted during cultivation. As an electron acceptor involved in electron transport chain, the scarcity of oxygen may restrict TCA cycle activity and oxidative phosphorylation, which could influence the outcome of their persister assay and potentially explain the differences compared to our results.
In the absence of antibiotics, a general link seems to exist between reduced TCA cycle activity and prolonged survival as inactivation of the aconitase resulted in extended stationary phase survival of S. aureus37. Also, small colony variants of S. aureus are resistant to aminoglycosides due to mutations in TCA cycle or electron transport chain genes and they display reduced membrane potential38,39. Recently, ATP content was reported to be a determining factor for whether or not exponentially growing cells become persisters4,14. However, in stationary phase cells of S. aureus we did not observe this correlation. In S. aureus, metabolism changes dramatically from exponential to stationary phase with the TCA cycle being inactive during exponential growth and ATP generated by substrate level phosphorylation whereas in stationary phase cells TCA cycle activity is de-repressed as acetate is being catabolized20,40. Consequently, persisters formed in exponential and stationary phase may be different and the genes contributing in the two phases may also vary. However, for all mutants with increased stationary phase persister cell frequencies in response to ciprofloxacin we observed a lower membrane potential than for wild type cells. Reduced membrane potential may for the TCA cycle mutants arise from reduced flow of electron donors to the electron transport chain while for the atpA mutant it indicates that in S. aureus the ATP synthase is normally working in reverse and contributes to PMF by pumping out protons fuelled by ATP hydrolysis. This may be necessary to get rid of the protons that at low pH enter the cell together with acetate which has accumulated during exponential growth. Membrane potential has previously been linked with persister cell formation but primarily in response to aminoglycosides. In Pseudomonas aeruginosa, PMF was increased by mannitol and it leads to increased killing of biofilm associated persister cells by tobramycin while in both E. coli and S. aureus, aminoglycoside uptake was stimulated by increased PMF leading to enhanced killing of persister cells18,19. Our data indicate that reduced PMF in stationary phase may enhance persister formation not only in response to aminoglycosides but also to fluoroquinolones. In our experiments, however, the high-persister phenotype with β-lactam treatment was observed over a 7 days period instead of within 24 hours as was the case for ciprofloxacin, which indicates a difference in persister mechanism between the two antibiotic classes. Since β-lactam killing relies on active cell wall synthesis, such a difference could result from increased cell turn-over in the wild type compared to the TCA cycle mutants. We speculate that mutations within TCA cycle contribute to the maintenance of S. aureus β-lactam persisters rather than their generation.
The exact mechanism by which the TCA cycle and PMF contribute to persister formation in S. aureus remains unclear. We found that expression of sucA is high and variable in stationary phase cells and correlates with the expression of cap, an established reporter of the stationary growth phase. Thus, we anticipate that in wild type populations, individual S. aureus cells with low expression of TCA cycle genes may be prone to form persisters. However, this is only in stationary phase, as in exponential phase where sucA is not expressed and TCA cycle is repressed, persister cells form at low frequency and TCA cycle inactivation does not influence this frequency. But how does reduced PMF lead to dormancy? In Salmonella enterica, the TacT toxin promotes persister formation by halting translation and its activity is modulated by the NAD+-dependent CobB sirtuin deacetylase suggesting that NAD+/NADH levels may influence persister formation9,41. However, for S. aureus, TA systems are not involved in persister cells formation indicating that alternative killing mechanisms should be sought4. Another possibility could be that the reduced membrane potential leads to a halt in cell wall synthesis as was recently shown in Bacillus subtilis42. Future analysis will be required to determine how reduced PMF leads to a persister state in S. aureus.
Methods
Strains, growth conditions and chemicals
A list of bacterial strains, plasmids and primers used in this study can be found in Supplementary Table S5. All the S. aureus strains, transposon library isolates and corresponding transductants, TCA cycle mutants and atpA mutant were all cultivated in Tryptic Soy Broth (TSB, Oxoid, Denmark) at 37 °C and 200 rpm of shaking in 12-ml centrifuge tube or plated on Tryptic Soy Agar (TSA, Oxoid, Denmark) at 37 °C of incubation unless otherwise indicated. All the plasmids and strains used for Tnp library construction were kindly provided by Timothy C. Meredith and supplemented glucose minimal medium (SGMM) was prepared as described previously23. TCA cycle mutants of sucA, sucB, sucD, sdhA, sdhB, acnA (NWMN-1263), citC and atpA were provided by bei Resources27. Ciprofloxacin, oxacillin, adenosine 5′-triphosphate disodium salt hydrate (ATP), carbonyl cyanidem-chlorophenylhydrazone (CCCP) and other mentioned chemicals were from Sigma-Aldrich (MO, USA). All restriction endonucleases and T4 ligase were from NEB BioLabs (MA, USA).
Construction of transposon library and identification of Tnp insertion sites
Transposon library was constructed in S. aureus Newman as reported previously23,24: (1) Tnp donor lysates were prepared by infecting TM43-45 with Ø11-FRT and the Tnp-plasmid packaging efficacy was determined by tittering these phage lysates on both RN4220 and TM19; (2) recipient strains were obtained by transforming plasmids pOrf5 Tnp+ or pOrf5 Tnp− into S. aureus Newman; (3) the Tnp donor lysates were gently mixed with recipient strains in SGMM and incubated at 22 °C overnight; (4) the cells were pelleted and incubated with TSB medium for 2 hours at 30 °C before plated on TSA plates containing erythromycin (5 µg/ml). Subsequently, the plates were left at room temperature until visible growth of individual Tnp insertion mutants could be observed.
The identification of the Tnp insertion site of library isolates was performed as reported by Bae et al. by PCR amplification of the transposon/chromosome junction using primers Martn-F and Martn-ermR and an annealing temperature of 63 °C. After purification, the PCR products were sequenced with primer Martn-F43.
Monitoring of Pcap and PsucA activity and image acquisition
Approximately 108 bacterial cells were harvested and re-suspended in 1 ml of pre-chilled 1x phosphate buffered saline (PBS) containing 3.7% formaldehyde. After 15 minutes of gentle mixing at room temperature, 500 µl of the fixed bacterial suspension was transferred to each well of a 24-well cell culture plate (Greiner Bio-One, Germany) lined with 12 mm diameter round coverslips. After centrifugation at 600 g for 5 minutes, coverslips were mounted onto slides with 3 µl fluorescence mounting medium (DAKO, Denmark).
Image acquisition was performed in the confocal mode of an inverted Zeiss LSM 710 NLO microscope equipped with a spectral detector and employing Zeiss Plan-Apochromat 63x/1.40 oil DIC M27 objective (Zeiss, Germany). The following excitation wavelengths, laser sources and detection spectra were used for the promoter activity experiments, gpCerulean: 405 nm/diode laser/454–516 nm; gpVenus: 514 nm/argon laser/519–621 nm. Images were exported in the different channels or overlays as 16-bit tagged image files. The exported images were assembled with CorelDRAW X7 and adjusted for brightness and contrast.
Enrichment of the mutants with enhanced persistence
1 µl of pooled wild type or Tnp library was inoculated into 2 ml TSB medium for incubation at 37 °C with shaking at 200 rpm in 12-ml centrifuge tube, after 24 hours, 1 ml of the culture was taken out to be treated by 100 × MIC of ciprofloxacin (MIC = 0.5 µg/ml) at 37 °C for another 24 hours in 12-ml centrifuge tube. The culture was washed and re-suspended with 1 ml FK buffer (9 g/l sodium chloride), and then certain dilutions were plated on TSA plates to be incubated at 37 °C for 24 hours. The plates with 100–200 colonies were eluted with FK buffer and 2 µl of the suspensions were inoculated in 2 ml fresh TSB medium which then undergone the same treatment of ciprofloxacin. This procedure was repeated for four times in a row to enrich persister cells.
Persister assay
2 µl of overnight culture of library isolates, TCA cycle mutants or wild type was inoculated into 2 ml TSB medium and incubated at 37 °C with shaking at 200 rpm for 24 hours in 12-ml centrifuge tube, then 1 ml of the culture was challenged by 100 × MIC of ciprofloxacin for another 24 hours still at 37 °C in 12-ml centrifuge tube. The persister frequency was determined by calculating the ratio between the CFU/ml counting of 24-hour post and right before the addition of ciprofloxacin by plating and incubating known dilutions of the samples on TSA plates at 37 °C for 24 hours. Aliquots of ΔsucA mutant and parent strain cultures were taken at 1, 3, 5, 24 or 48 hours post the addition of ciprofloxacin and the dilutions were plated on TSA plate at 37 °C for 24 hours for CFU determination. The transductants of Tnp library isolates were also tested with 100 × MIC of oxacillin (MIC = 0.25 µg/ml) and the CFU/ml was determined over 7 days.
To evaluate the effect of medium acidification on persister generation, TSB medium with or without glucose (fisher scientific, MA, USA) was used for the cultivation and the persister frequencies were compared between the cultures with these two media. CCCP dissolved in DMSO was added into bacterial culture right before the treatment of ciprofloxacin to test the effect of PMF on persister generation.
Quantification of cellular ATP level
The ATP level of 24-hour cultures was tested with BacTiter-GloTM Microbial Cell Viability Assay kit (Promega, WI, USA) according to manufacturer’s instruction. 100 µl suspension of bacterial culture (10-fold diluted with TSB medium) was carefully mixed with equal volume of BacTiter-GloTM Reagent and incubated at room temperature for 5 minutes before the luminescence was recorded with emission at 560 nm. The value of each sample was the average of three independent reads and there were 3 biological replicates for each individual sample.
Measurement of membrane potential
Membrane potential was assessed by BacLightTM Bacterial Membrane Potential Kit (Life Technologies, CA, USA) where 24-hour cultures were re-suspended with 0.5 ml filtered PBS, mixed with 10 µl of fluorescent membrane potential indicator dye, DiOC2(3) and incubated at room temperature for 30 minutes. The fluorescent signals were recorded by BD Biosciences Accuri C6 flow cytometer (BD Biosciences, CA, USA) counting 50,000 cells with the threshold of 16,000 at medium flow rate. The ratio between channel F3 (red fluorescence) and F1 (green fluorescence) was calculated with CFlow® (BD Accuri, CA, USA) to indicate membrane potential.
Quantification of fermentation products
The concentration of acetate in the supernatant of cultures from both early exponential (3-hour cultivation) and stationary phase (24-hour cultivation) was determined by Ultimate 3000 HPLC system (Dionex, Sunnyvale, USA) equipped with an Aminex HPX-87H column (Bio-Rad, Hercules, USA) and a Shodex RI-101 detector (Showa Denko KK, Tokyo, Japan). The column oven temperature was set as 60 °C and the mobile phase consisted of sulfuric acid (5 mM) with a flow rate of 0.5 ml/minute.
Statistical analysis
Results were represented as the mean of biological triplicates with standard deviation besides for the ATP measurements there were 3 independent luminescent reads for each biological replicate. Statistical significance was analyzed using one way ANOVA followed by Dunnett test with P < 0.05 in GraphPad Prism 7 (GraphPad software Inc., CA, USA).
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Electronic supplementary material
Acknowledgements
We would like to thank NARSA (University of Nebraska Medical Centre) and bei RESOURCES for providing mutants used in this study and TC Meredith for help with the Tnp library construction. Y. Wang, M.S. Bojer and H. Ingmer, were funded by the Danish National Research Foundation (DNRF-120); SEGeorge and C. Wolz, by the Deutsche Forschungsgemeinschaft (SFB766) and Z.H. Wang and P. R. Jensen, by the Danish Council for Strategic Research and the Danish Council for Technology and Innovation (0603-00522B).
Author Contributions
Y.W. designed and performed most of the experiments, wrote and edited the manuscript, M.S.B. designed and performed some experiments and wrote part of the manuscript, Z.H.W. and S.E.G. performed the experiments included in Figures 3 and 8 respectively, C.W. and P.R.J. conceived some experiments and wrote part of the manuscript, H.I. conceived the experiments, wrote and edited the manuscript. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-29123-0.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fisher RA, Gollan B, Helaine S. Persistent bacterial infections and persister cells. Nat Rev Microbiol. 2017;15:453–464. doi: 10.1038/nrmicro.2017.42. [DOI] [PubMed] [Google Scholar]
- 2.Van den Bergh B, Fauvart M, Michiels J. Formation, physiology, ecology, evolution and clinical importance of bacterial persisters. FEMS Microbiol Rev. 2017;41:219–251. doi: 10.1093/femsre/fux001. [DOI] [PubMed] [Google Scholar]
- 3.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 4.Conlon BP, et al. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat Microbiol. 2016;1:16051. doi: 10.1038/nmicrobiol.2016.51. [DOI] [PubMed] [Google Scholar]
- 5.Germain E, Roghanian M, Gerdes K, Maisonneuve E. Stochastic induction of persister cells by HipA through (p)ppGpp-mediated activation of mRNA endonucleases. Proc Natl Acad Sci USA. 2015;112:5171–5176. doi: 10.1073/pnas.1423536112. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Maisonneuve E, Castro-Camargo M, Gerdes K. p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell. 2013;154:1140–1150. doi: 10.1016/j.cell.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 7.Maisonneuve E, Gerdes K. Molecular mechanisms underlying bacterial persisters. Cell. 2014;157:539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 8.Helaine S, et al. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science. 2014;343:204–208. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheverton AM, et al. A Salmonella Toxin Promotes Persister Formation through Acetylation of tRNA. Mol Cell. 2016;63:86–96. doi: 10.1016/j.molcel.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorr T, Vulic M, Lewis K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010;8:e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harms, A., Fino, C., Sorensen, M. A., Semsey, S. & Gerdes, K. Prophages and Growth Dynamics Confound Experimental Results with Antibiotic-Tolerant Persister Cells. MBio8, 10.1128/mBio.01964-17 (2017). [DOI] [PMC free article] [PubMed]
- 12.Goormaghtigh, F. et al. Reassessing the Role of Type II Toxin-Antitoxin Systems in Formation of Escherichia coli Type II Persister Cells. MBio9, 10.1128/mBio.00640-18 (2018). [DOI] [PMC free article] [PubMed]
- 13.Wood TK, Knabel SJ, Kwan BW. Bacterial persister cell formation and dormancy. Appl Environ Microbiol. 2013;79:7116–7121. doi: 10.1128/AEM.02636-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shan, Y. et al. ATP-Dependent Persister Formation in Escherichia coli. MBio8, 10.1128/mBio.02267-16 (2017). [DOI] [PMC free article] [PubMed]
- 15.Kim JS, et al. Fumarate-Mediated Persistence of Escherichia coli against Antibiotics. Antimicrob Agents Chemother. 2016;60:2232–2240. doi: 10.1128/AAC.01794-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma C, et al. Energy production genes sucB and ubiF are involved in persister survival and tolerance to multiple antibiotics and stresses in Escherichia coli. FEMS Microbiol Lett. 2010;303:33–40. doi: 10.1111/j.1574-6968.2009.01857.x. [DOI] [PubMed] [Google Scholar]
- 17.Orman MA, Brynildsen MP. Inhibition of stationary phase respiration impairs persister formation in E. coli. Nat Commun. 2015;6:7983. doi: 10.1038/ncomms8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allison KR, Brynildsen MP, Collins JJ. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature. 2011;473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barraud N, Buson A, Jarolimek W, Rice SA. Mannitol enhances antibiotic sensitivity of persister bacteria in Pseudomonas aeruginosa biofilms. PLoS One. 2013;8:e84220. doi: 10.1371/journal.pone.0084220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Somerville, G. A. Staphylococcus: Genetics and Physiology. 107–118 (Caister Academic Press, 2016).
- 21.Conlon BP, et al. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature. 2013;503:365–370. doi: 10.1038/nature12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conlon BP. Staphylococcus aureus chronic and relapsing infections: Evidence of a role for persister cells: An investigation of persister cells, their formation and their role in S. aureus disease. Bioessays. 2014;36:991–996. doi: 10.1002/bies.201400080. [DOI] [PubMed] [Google Scholar]
- 23.Wang H, Claveau D, Vaillancourt JP, Roemer T, Meredith TC. High-frequency transposition for determining antibacterial mode of action. Nat Chem Biol. 2011;7:720–729. doi: 10.1038/nchembio.643. [DOI] [PubMed] [Google Scholar]
- 24.Santiago M, et al. A new platform for ultra-high density Staphylococcus aureus transposon libraries. BMC Genomics. 2015;16:252. doi: 10.1186/s12864-015-1361-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Girgis HS, Harris K, Tavazoie S. Large mutational target size for rapid emergence of bacterial persistence. Proc Natl Acad Sci USA. 2012;109:12740–12745. doi: 10.1073/pnas.1205124109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D L. Nelson, M. M. C. Lehninger Principles of Biochemistry. W.H. Freeman, 1328 (2008).
- 27.Fey PD, et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio. 2013;4:e00537–00512. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Page R, Peti W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat Chem Biol. 2016;12:208–214. doi: 10.1038/nchembio.2044. [DOI] [PubMed] [Google Scholar]
- 29.Strahl H, Hamoen LW. Membrane potential is important for bacterial cell division. Proc Natl Acad Sci USA. 2010;107:12281–12286. doi: 10.1073/pnas.1005485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchell P, Moyle J. Chemiosmotic hypothesis of oxidative phosphorylation. Nature. 1967;213:137–139. doi: 10.1038/213137a0. [DOI] [PubMed] [Google Scholar]
- 31.Heytler P. G. uncoupling of oxidative phosphorylation by carbonyl cyanide phenylhydrazones. I. Some characteristics of m-Cl-CCP action on mitochondria and chloroplasts. Biochemistry. 1963;2:357–361. doi: 10.1021/bi00902a031. [DOI] [PubMed] [Google Scholar]
- 32.Cunarro J, Weiner MW. Mechanism of action of agents which uncouple oxidative phosphorylation: direct correlation between proton-carrying and respiratory-releasing properties using rat liver mitochondria. Biochim Biophys Acta. 1975;387:234–240. doi: 10.1016/0005-2728(75)90106-1. [DOI] [PubMed] [Google Scholar]
- 33.Kwan BW, Valenta JA, Benedik MJ, Wood TK. Arrested protein synthesis increases persister-like cell formation. Antimicrob Agents Chemother. 2013;57:1468–1473. doi: 10.1128/AAC.02135-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.George SE, et al. Phenotypic heterogeneity and temporal expression of the capsular polysaccharide in Staphylococcus aureus. Mol Microbiol. 2015;98:1073–1088. doi: 10.1111/mmi.13174. [DOI] [PubMed] [Google Scholar]
- 35.Tretter L, Adam-Vizi V. Alpha-ketoglutarate dehydrogenase: a target and generator of oxidative stress. Philos Trans R Soc Lond B Biol Sci. 2005;360:2335–2345. doi: 10.1098/rstb.2005.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W, et al. Transposon Mutagenesis Identifies Novel Genes Associated with Staphylococcus aureus Persister Formation. Front Microbiol. 2015;6:1437. doi: 10.3389/fmicb.2015.01437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Somerville GA, et al. Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect Immun. 2002;70:6373–6382. doi: 10.1128/IAI.70.11.6373-6382.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kriegeskorte A, et al. Staphylococcus aureus small colony variants show common metabolic features in central metabolism irrespective of the underlying auxotrophism. Front Cell Infect Microbiol. 2014;4:141. doi: 10.3389/fcimb.2014.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vestergaard M, et al. Novel Pathways for Ameliorating the Fitness Cost of Gentamicin Resistant Small Colony Variants. Front Microbiol. 2016;7:1866. doi: 10.3389/fmicb.2016.01866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Somerville GA, Proctor RA. At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiol Mol Biol Rev. 2009;73:233–248. doi: 10.1128/MMBR.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.VanDrisse, C. M., Parks, A. R. & Escalante-Semerena, J. C. A Toxin Involved in Salmonella Persistence Regulates Its Activity by Acetylating Its Cognate Antitoxin, a Modification Reversed by CobB Sirtuin Deacetylase. MBio8, 10.1128/mBio.00708-17 (2017). [DOI] [PMC free article] [PubMed]
- 42.Muller, A. et al. Daptomycin inhibits cell envelope synthesis by interfering with fluid membrane microdomains. Proc Natl Acad Sci USA, 10.1073/pnas.1611173113 (2016). [DOI] [PMC free article] [PubMed]
- 43.Bae T, et al. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc Natl Acad Sci USA. 2004;101:12312–12317. doi: 10.1073/pnas.0404728101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.