Abstract
We examined the short-term variability, by daily to weekly sampling, of protist assemblages from March to July in surface water of the San Pedro Ocean Time-series station (eastern North Pacific), by V4 Illumina sequencing of the 18S rRNA gene. The sampling period encompassed a spring bloom followed by progression to summer conditions. Several protistan taxa displayed sharp increases and declines, with whole community Bray–Curtis dissimilarities of adjacent days being 66% in March and 40% in May. High initial abundance of parasitic Cercozoa Cryothecomonas longipes and Protaspis grandis coincided with a precipitous decline of blooming Pseudo-nitzschia diatoms, possibly suggesting their massive infection by these parasites; these cercozoans were hardly detectable afterwards. Canonical correspondence analysis indicated a limited predictability of community variability from environmental factors. This indicates that other factors are relevant in explaining changes in protist community composition at short temporal scales, such as interspecific relationships, stochastic processes, mixing with adjacent water, or advection of patches with different protist communities. Association network analysis revealed that interactions between the many parasitic OTUs and other taxa were overwhelmingly positive and suggest that although sometimes parasites may cause a crash of host populations, they may often follow their hosts and do not regularly cause enough mortality to potentially create negative correlations at the daily to weekly time scales we studied.
Introduction
Through their potentially rapid turnover (from hours to days) and metabolic versatility, microbial model systems consisting of prokaryotes and/or protists can be used to address ecological questions over multiple temporal scales and to explore community-level responses to different driving forces. Whole microbial communities can now be readily studied because the “molecular revolution” in microbial biodiversity research has been expanded from bacteria and archaea [1] to eukaryotes [2], improving our understanding of ecological microbial systems [3, 4]. In particular, time series of aquatic microbial communities have yielded key insights into the drivers of microbial dynamics in aquatic systems [5–9]. Microbial ecologists have learned that despite the obvious importance of environmental variation, such as temperature, nutrient supply, and physical mixing, that galvanize microbial communities and determine their general composition, a complex network of interrelationships (i.e., cooperation, competition, mutual dependency, predator–prey, etc.) is often itself an important regulating force of microbial communities in aquatic systems [3]. Moreover, the above cited studies have shown that microbial communities are both dynamic and resilient, leading to predictable changes in their composition at different scales: daily, seasonally, and inter-annually [3].
Marine microbial eukaryotes, or protists, are being increasingly recognized as critically important in global ecological processes and biogeochemical cycles [10–12]. Marine phytoplankton provide about half of global marine primary productivity, and a significant fraction of particulate material produced by them is exported to the deep sea by the “biological carbon pump,” providing an important contribution to global carbon sequestration [11]. Protists, including many “mixotrophic” eukaryotes, act as grazers of bacteria and other protists, providing essential links in the marine food web while remineralizing nutrients [13]. Furthermore, the discovery of multiple lineages of parasitic protists in marine planktonic communities [14–16] has raised interest regarding the roles of these abundant and diverse organisms in aquatic trophic interactions [17, 18]. The high diversity in marine protists highlights the necessity to identify the mechanisms by which this diversity is established, maintained, and modified [19].
The extensive microbial and oceanographic data at the San Pedro Ocean Time-series (SPOT) station has provided valuable insights about the monthly-to-interannual dynamics in protistan community composition, as well as their vertical structure (i.e., with depth) [8, 20–22]. However, understanding the dynamics of protists requires studying the community at all ecologically relevant timescales. The monthly timescales used in the above studies can miss interesting ecological processes or species–species interactions more evident over shorter time scales (i.e., days), because the generation time of some protists may be less than a day [23]. The few studies that examined the short time scale dynamics of protists have shown rapid shifts in community composition (1–3 weeks) in estuarine environments [24], lacustrine systems [25], and through incubation experiments [26], suggesting a continuous reassembly of protist communities. These results open new fundamental questions about protist behavior over time. Lie et al. [27] used 18S rRNA sequence-based fingerprinting to study daily and several km-scale variations in protists surrounding the SPOT location over several days, reporting that differences in the community composition between days tended to be greater than those over the space of a few kilometers [27]. More recently, our lab reported a rapid shift of bacteria, archaea, and phytoplankton community dynamics, following the same spring bloom in this study, by daily to weekly sampling [28]. This study noted particularly rapid changes in phytoplankton composition in March, characterized by the dominance of 10 different taxa over 18 days. However, Needham & Fuhrman’s study did not address heterotrophic protists that can be important agents in regulating these planktonic communities.
Here, we extend the work of Needham & Fuhrman [28], analyzing the same DNA samples, but now focusing on the entire protist community by V4 18S rRNA gene tag sequencing. Our survey, which focused on the ocean surface waters, included the end of a diatom bloom and the transition to more stable summer conditions at SPOT. Because we sampled a single location at an open ocean site, changes reflect within-community processes as well as import/export though advection and mixing, plus stochastic variation. Here we address the following questions: how variable and dynamic is the protist community on time scales from days to months? How do particular OTUs contribute to the protistan community temporal variation on scales of days to months? What is the influence of environmental factors? And finally, what kind of species–species relationships characterize protist community associations over time, including parasite–host interactions?
Materials and methods
Sample collection
Sampling occurred approximately daily from March 12 to April 1 and May 22-27, and otherwise weekly through July, in 2011. A volume of 20 L of seawater from the top meter (0–2 m) was sampled at the San Pedro Ocean Time-series location (SPOT: Lat: 33°33′ N, Long: 118°24′ W) by multiple bucket casts. Water samples were stored in a cooler until arrival (30 min) at the Wrigley Institute of Environmental Science (Catalina Island, CA, USA), where filtering began immediately. Some samples (March 15, April 29, May 24, June 22, and July 20) were collected during SPOT monthly cruises (by Niskin bottles in a rosette). Seawater was filtered through 80 µm mesh and 47 mm Type A/E glass fiber filters (~1.0 µm pore), and stored at −80°C until extraction. Several environmental parameters were analyzed (details in supplemental material).
Protist community composition
We assessed protist community composition (PCC) by targeting the V4 region of the 18S rRNA gene and using Illumina Miseq 2 x 300bp paired-end sequencing. DNA was extracted from the A/E filter with a NaCl/CTAB extraction [28]. We used the “universal” eukaryote forward primer V4F (5′-CCA GCA SCY GCG GTA ATT CC-3′) [29] and modified reverse primer V4RB (5′-ACT TTC GTT CTT GAT YRR-3′) [30] that greatly increased detection of some groups, especially Haptophyta, compared to the original V4R Stoeck primer (Table S1, Figure S1). The protocol for the library preparation is fully described in supplemental material.
Forward and reverse reads were merged using PANDAseq [31]. When mismatches occur in the overlapping region, the base with the higher quality score was chosen. Merged sequences with quality score value <0.9 were discarded [31]. Using QIIME (version 1.9.0), sequences showing one or more nucleotide mismatches in primers and/or barcodes with homopolymers higher than six were removed. Barcodes and primer sequences were trimmed. Singletons and chimeric sequences were removed using UCHIME [32] by both de novo and reference database. Sequences were clustered de novo at 98% ID with UCLUST [32] and the most abundant sequence of each Operational Taxonomic Unit (OTU) was classified with UCLUST with both SILVA database release 111 [33] and Protist Ribosomal Database [34]. These databases contain the most up to date eukaryote taxonomic classification and they place protist SSU rRNA gene sequences within a coherent ranked taxonomic framework. OTUs of metazoans or of taxa appearing in less than three samples were removed. This resulted in a total of 4259 OTUs across all samples. Only samples with >10 000 sequences were further analyzed (10,248–161,365 sequences per sample). Samples were normalized by calculating relative abundance for each OTU (i.e., proportion of all sequences in a sample).
Data Analysis
Temporal dynamics were visualized (bubble chart) with ‘bubble.pl.program’ (http://www.cmde.science.ubc.ca/hallam/bubble.php). Hierarchical agglomerative clustering of Bray–Curtis similarities was performed on the 300 most abundant protist OTUs (i.e., with most reads) (Table S2), using the group average method in PRIMER (version 6.1.18). To test the null hypothesis that there was no significant difference between the groups discriminated according to the agglomerative clustering analysis, similarities were analyzed with ANOSIM [35] in PRIMER (6.1.13). Similarity percentage (SIMPER) analysis was used to determine which taxa were important in describing differences between discriminated groups [35]. Rate of change parameter [36] was applied to monitor daily changes of PCC, averaging the degree of change (i.e., Bay–Curtis dissimilarity index) between two consecutive sampling days.
To investigate the relationships between PCC and measured environmental variables (i.e., compilation of pure bottom-up (inorganic nutrients) and other physicochemical variables), Canonical Correspondence Analysis (CCA) was performed using XLSTAT-ADA software (details in supplemental material).
Extended Local Similarity Analysis (eLSA) [37] was used for the full time series to analyze covariation between the 300 most abundant protist OTUs (Table S2). P-values were estimated using the “mixed” approach [38]. Q-value (false discovery rate) was calculated to estimate the likelihood of false positives. eLSA network of delay-shifted Spearman Correlation Cofficents (SSCC) between variables was visualized using Cytoscape v2.8.2 [39], with P < 0.01 and Q < 0.05. Because the sampling was not evenly spaced, time-lags were not considered. Network characterization (structural cohesion and robustness) were evaluated using different topological indexes: degree, betweenness centrality and closeness centrality (details in supplementary material). The module detection (i.e., clusters of taxa that interact more among themselves than with other taxa compared to a random association) was performed using AllegroMCODE [40].
Results
Environmental parameters
Seawater temperature increased gradually during the studied period, from 12°C–13°C in March to 21°C in July. Salinity ranged between 30.1 (March 16) and 33.6 (May 25) (Figure S2). Chlorophyll a was highest at the beginning of the study (10.30 µg L−1 on March 12, perhaps declining from a peak before the start of the study) and then had a smaller peak a few days later (6.95 µg L−1 on March 16). March samples displayed the highest chlorophyll a concentrations (between 1 µg L−1 and 10 µg L−1) and dropped to values lower than 1 µg L−1 for the rest of the sampling period (between 0.15 µg L−1 and 0.9 µg L−1) (Fig. 1 and S2). Both NO3− + NO2− (>2 µM with a peak of 4.5 µM) and PO43− (>0.30 µM with peak of 0.48 µM) were elevated from March 22 to March 25 (Figure S2). SiO32− fluctuated between <1 µM (detection limit) and 3.4 µM.
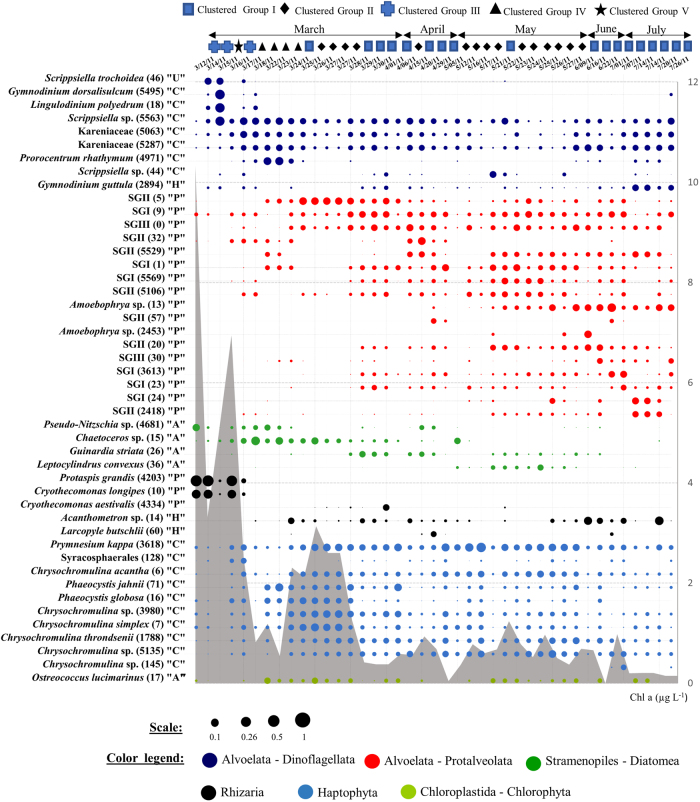
Fig. 1.
Time series dot plot showing the most abundant eukaryote OTUs, each displaying a relative abundance ≥2% in at least one sample. Note uneven sampling intervals, and symbols along the top showing community clustering (Fig. 2). Letters between quotation marks correspond to inferred trophic properties of selected OTUs based on closest known relatives: A autotroph, H heterotroph, P parasite, C with chloroplast (relatives include mixotrophs), U unassigned. The area in gray color corresponds to the temporal dynamics of chlorophyll a concentration during the studied period. To show the relative differences in sequence abundance for the displayed OTUs, the circle size is on a 0-1 scale representing proportions. SG stands for “Syndiniales Group”
OTUs dynamics
The most abundant OTUs (any that exceeded 2% of the total on a given day) belonged mainly to the Protalveolata (17 OTUs), Haptophyta (11 OTUs), and Dinoflagellata (9 OTUs) groups, as well as Diatomea and Rhizaria (Fig. 1, Table S3). The plurality (42%) of these OTUs were intermittent (present on 25–75% of sampling dates). Ephemeral (present <25% of dates) and persistent (present >75% of dates) OTUs constituted 28% and 30% of these selected OTUs, respectively.
Five taxa (Cryothecomonas longipes, Protaspis grandis, Scrippsiella trochoidea, Gymnodinium dorsalisulcum,Lingulodinium polyedrum) displayed remarkably strong dynamics early in the study, along with Pseudo-nitzschia sp. which was the most abundant phytoplankter the first day, when chlorophyll a was highest (Fig. 1). During the first four days of sampling (March 12-16), the very high abundance of two cercozoan taxa (C. longipes, P. grandis) was interrupted, in relative abundance, on March 15 by high abundance of three dinoflagellates taxa (S. trochoidea, G. dorsalisulcum, L. polyedrum). When abundant, these cercozoa taxa represented between 16% and 58% of the total community sequence counts, and then were hardly detectable for the rest of study (Fig. 1). After this period and until the end of March-early April, there were significant changes in autotrophic taxa like Prorocentrum rhathymum, Chaetoceros sp., Chrysochromulina simplex, Phaeocystis jahnii, Phaeocystis globosa, and Ostreococcus lucimarinus often with short-lived (and sometimes repeating) bursts in their relative abundances (Fig. 1). Prymnesium kappa reached 26.8% of the total community sequences on May 17 (Fig. 1). The dynamics of Syndiniales OTUs was complex, with no clear intragroup succession pattern, and multiple individual OTUs peaked in abundance starting from March 25 (reaching a relative abundance between 4% and 16%). Some Syndiniales OTUs, however, displayed interesting patterns, like Amoebophrya sp. (both OTU #13 and OTU #2453), becoming abundant later in the sampling period when temperature was ≥18°C (Fig. 1 and S2).
PCC variability over short (daily–weekly) time scales
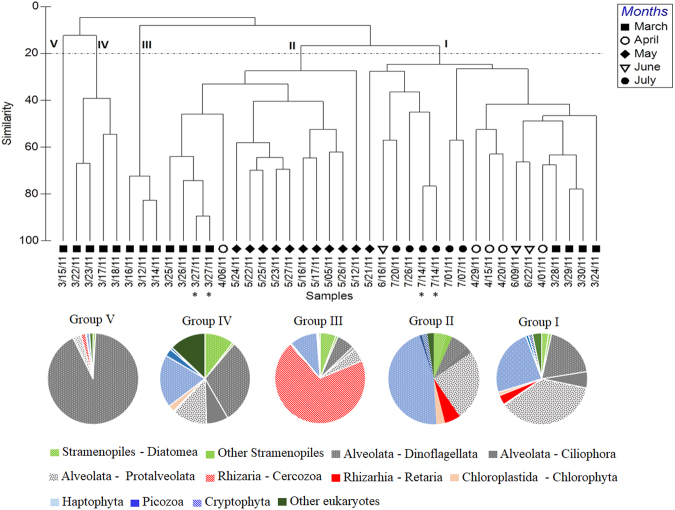
UPGMA clustering showed five distinct groups split significantly, at only 20% Bray–Curtis similarity index, indicating rapid and pronounced separation in PCC over time (Fig. 2). Cluster I, dominated by Protalveolata (37%, represented only by Syndiniales), Haptophyta (24%), and Dinoflagellata (19%) (Fig. 2) included samples from March 28 to April, also June and July. Cluster II was dominated by both Haptophyta and Protalveoalata at, cumulatively, 71% and included mainly samples from May but also March (25–27). The last three clusters (i.e., III, VI, and V) occurred exclusively in March samples (Fig. 2). Clusters III and V were very highly represented by Cercozoa (69%) and Dinoflagellata (92%), respectively (Fig. 2). The PCC exhibited a total of 10 shifts from one cluster to another. Most of these shifts (6 of 10) occurred during the March daily sampling and each appeared to last 1–3 days (Fig. 2), making this period the most dynamic for the PCC. Viewed in terms of the dominant taxa, there were 10 different such taxa from March 12 to April 1, during which dominance changed 12 times. In terms of Bray–Curtis distances, the mean rate change of the PCC was 66% (SD ± 20) per day in March, compared to 40% (SD ± 7) per day in May (Table S4). The weekly mean rate (for the study as a whole) was between 50% (April–June) and 57% (July).
Fig. 2.
Protist communities clustered into five broad community types, at >20% Bray–Curtis similarity. Dendrogram from UPGMA clustering of Bray–Curtis dissimilarity based on the 300 most abundant OTUs. * duplicate DNA extract. Pie chart plots indicate high-ranking taxonomy distribution of the clustered protist community groups. The classification is based on Adl et al. [83]
SIMPER analysis assessed the average percent contribution of individual OTUs to the dissimilarity between the five identified clusters (Table S5). On average, 12 OTUs (SD ± 2, n = 7) were identified as major contributors to differences between the detected clusters (between 60% and 89% of the dissimilarity between clusters III, V, and other discriminated clusters (I, II, and IV)) (Table S5). Overall, each OTU contributed to at least 1% of the dissimilarity between clusters. C. longipes and P. grandis drove, together, between 30% and 46% of disparity between cluster III and the other clusters, and dinoflagellates G. dorsalisulcum, L. polyedrum, and Scrippsiella sp. led to around 41% of the observed dissimilarity between clusters. Between 60% and 70% of the dissimilarity between cluster I and II was driven by more than 41 OTUs, belonging mainly to Haptophyta and Syndiniales (Table S5).
PCC in relation to environmental variables
The relative importance of physicochemical parameters in shaping PCC over time was analyzed through a direct multivariate gradient approach. A strong Spearman’s rank pairwise correlation between NO3− + NO2− and PO43− (R2 = 0.84, p < 0.001) allowed us to use PO43− as a surrogate of NO3− + NO2− to avoid overestimation of the percentage of explained variance. By CCA analysis, 31.04% of the variance of the PCC was explained by physicochemical factors (Table S6; Fig. 3). The first and second canonical axes accounted for 30.29% and 18.51% of this variance (Table S6; Fig. 3). CCA showed that almost 69% of the variance was unexplained by the model (Table S6).
Fig. 3.
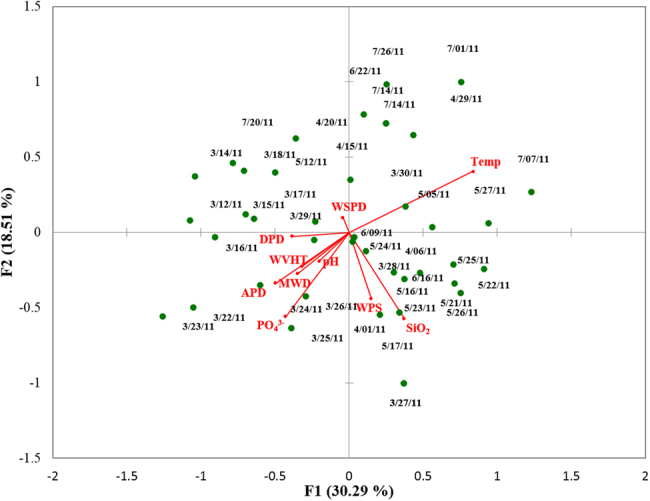
Canonical correspondence analysis of protist community composition and physicochemical parameters. Temp: Temperature (°C), PO43−: Orthophosphate (uM), SiO32−: Silicate, DPD: dominant wave period (s), MWD: direction from which DPD is coming (degree), MVHT: significant wave height (m), APD: average wave period (seconds), WPS: Wrigley precipitation sum (cm), WSPD: Wrigley (station) wind speed average (km hr−1). Wrigley refers to the weather station at Wrigley Marine Science Center on Santa Catalina Island
Variation partitioning gave more detail about the relative contribution of physical and chemical factors respectively to the observed changes in PCC. A model utilizing only PO43−, SiO32−, and pH could explain 13.52% of the variance (Table S6). Another model examining PO43− and SiO32− could statistically explain 11.29% of the variance (Table S6). The model considering only the physical variables could not significantly explain the PCC variance (p > 0.1).
Protist network
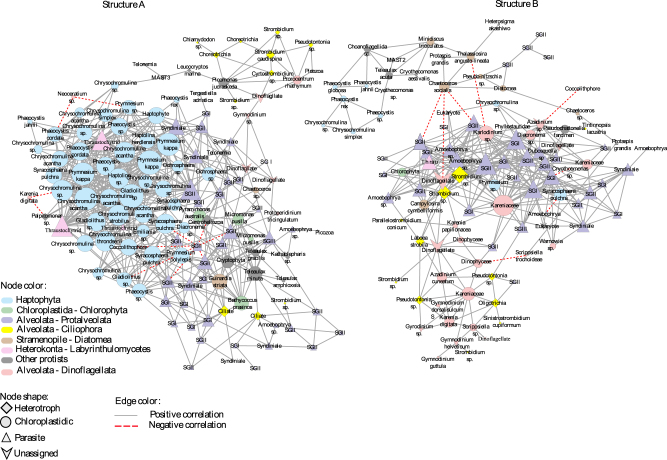
The protist network exhibited a short path length (3.7) and a power law distribution of node degree distribution (Table S7 and Figure S3), which indicates that most nodes can be reached from every other node by a small number of steps. A total of 98% of the correlations established in the network were positive (SSCC ≥ 0.6) between protist OTUs. The highest connections (SSCC ≥ 0.8) were more so within a phylum than between phyla (Figure S4). The most dynamic OTUs (C. longipes, P. grandis, G. dorsalisulcum, L. polyedrum, and Scrippsiella sp.) either were not part of the network or displayed low structural network properties (i.e., low degree, low betweenness centrality, and low closeness central) (Table S3), which suggests they did not contribute significantly to overall network cohesion, during the spring–summer period.
There were two large non-overlapping sub-networks (Fig. 4). Globally, Syndiniales, Haptophyta, and Dinoflagellata dominated in term of number of nodes (74, 55, and 30 respectively) and number of edges (434, 482, and 159, respectively), however they are unevenly distributed between the two sub-networks. Both sub-networks exhibited modular organization (i.e., strong within-group inter-correlations), with a total of 16 modules (Figures S5 and S6; Table S8). Most modules consisted of 3− to 6-node subgraphs and only three modules displayed more than seven nodes. Different members of protist groups, functionally diverse, were dispersed broadly in the different modules.
Fig. 4.
Association networks derived from the 300 most abundant protists and showing only statistically significant correlations between OTUs (Spearman correlation ≥│0.6│; p < 0.01; false discovery q < 0.05). Sub-network (a) included a total of 103 unique nodes and 348 edges (correlations) among nodes, and Sub-structural network (b) included a total of 125 unique nodes and 643 edges. Node sizes correspond to the number of neighbors (i.e., degree). Nodes colors and shapes correspond to main protist groups and their ecological behavior, respectively. Solid lines represent positive correlations and dashed lines, negative correlations
Discussion
Copy numbers and variations of rDNA sequences within individuals of single celled protists may be important for interpreting the rDNA-based surveys. While some eukaryotic organisms have intragenomic variability ([41] and references therein) that could potentially overestimate diversity from 18S surveys, we minimized such effects on our analyses. First, we did not make a point of defining or comparing diversity, per se. Second, according to Gong et al. [41] intergenomic variability reflects extremely small changes in sequences (generally <1% variation). Thus, our use of 98% OTUs should essentially avoid this problem. Variation in 18S rRNA gene copy number per cell (up to four orders of magnitude among eukaryotes) [42] limits the quantitative value of the 18S rRNA gene barcoding approach for direct comparison of relative abundance between specific different taxa, despite general correlations reported between 18S rRNA gene copy number and genome size in eukaryotes [19, 42] or cell length and biovolume in unicellular organisms [43, 44]. Most of our comparisons here—and quite notably the network analysis—depend on comparing abundance within a taxon (e.g., rank abundances over time), so they are much less influenced by 18S rRNA gene copy number [6, 22]. Nevertheless, our analyses are based primarily on relative abundance data; because cumulative protistan absolute abundance can sometimes vary greatly and our data represent relative proportions, the relative abundance of any given taxon can be influenced by major swings in the cumulative absolute abundances of other taxa.
Temporal scales of PCC
We report that the whole protist community displays fast dynamics, with several protistan taxa displaying rapid increases and declines (Fig. 1). Most previous investigations of protist temporal dynamics were mainly at monthly time scales [8, 20, 45–47], and seasonality has been reported with shifts in protist community occurring over a few months. Two studies [25, 27] have analyzed the daily temporal variability of protists, and both also reported significant changes between consecutive days, with estimated mean daily rate changes of protist community close to those obtained in our study (i.e., between 36% and 66%).
The daily shift of the PCC was more pronounced in March than in May (Fig. 2). Satellite imagery of chlorophyll a measurements made at SPOT (more detail in [28]) indicated our sampling started at the end of a strong phytoplankton bloom (peaking from March 6 to March 13 with mean chlorophyll a concentration >10 mg m−3) which seemed to have a dramatic impact on PCC during March. The high values of NO3− + NO2− (mean 1.19 µM ± 1.57, n = 15) and PO43− (mean 0.23 µM ± 0.13, n = 15) in March sampling (Figure S2), coinciding with seasonally increasing stratification and increases in day length, likely triggered the initial phytoplankton bloom.
Dense and growing bloom populations may be particularly susceptible to parasites [48–50]. Our data suggest cercozoan Protaspis and Cryothecomonas may be largely responsible for the rapid decline in the initial diatom bloom (see below) with replacement by other dominant phytoplankton (e.g., Chaetoceros sp.; Guinardia striata; Leptocylindrus convexus; Ostreococcus lucimarinus; Prorocentrum rhathymum), which themselves may also frequently be infected by parasites such as Syndiniales that we found were highly diverse and dynamic (Fig. 1). Thus, our results suggest that the post-bloom period (i.e., a few weeks after the bloom) is a strong dynamic phase for protist communities as it is for bacterial communities [28], with substantial parasitism contributing to rapid turnover of the PCC.
Parasitic protists
Among all potentially parasitic groups identified in our study (including Fungi, Labyrinthulomycetes, Apicomplexa, and parasitic Cercozoa), Syndiniales dominated in terms of sequence number and diversity. This marine group represented close to 40% of sequences obtained from our samples during the summer period. The considerable abundance and diversity of Syndiniales has previously been reported in coastal systems [45, 51] as well as in the open ocean [19]; however, their temporal dynamics are still poorly explored. Our analysis showed continual strong dynamics of Syndiniales with oscillations between low density and recovery of several OTUs, throughout the spring–summer period. This could lead to a persistent replacement of dominant hosts and parasites due to their differential susceptibility vs. infectivity. In host-parasite systems, dominant host types are expected to be eventually replaced by other hosts due to the elevated potency of their specific parasites, leading to changes in the abundance of both hosts and parasites exhibiting cycles of alternating dominance [52]. However, Guillou et al. [18] have reported that parasites within the Syndiniales are generalists and opportunists with capacity to infect very different marine hosts, from many trophic levels. This should thus lead to moderate dynamics for dominant parasitic Syndiniales, which we observed for some taxa much more than others (Fig. 1). Interestingly, by using a tritrophic configuration model (i.e., incorporation of a parasite grazer in host-parasite model), [53] demonstrated that parasite grazers lead to short-term oscillation of Amoebophrya, and thus enabled hosts-parasite coexistence. They also pointed out that other interactions such as host grazing [54] or parasite competition [55, 56] may also contribute to the observed parasitic oscillations in natural systems.
As mentioned above, we observed a marked abundance of parasitic Cercozoa (Cryothecomonas longipes and Protapsis grandis) at the beginning of the sampling period (Fig. 1), coinciding with the crash of the initial Pseudo-nitzschia diatom bloom [28]. Generally, the literature has reported co-occurrence of Cercozoa mainly with diatoms in aquatic systems [51, 57]. As is typical worldwide, diatoms are initially dominant in early spring blooms at SPOT with Pseudo-nitzschia australis usually constituting a significant portion of the total diatom assemblage [20, 58]. Although we are unaware of previous studies of parasitic Cercozoa infections of Pseudo-nitzschia in particular, the infection of diatoms by C. longipes has been reported [59, 60] and, in contrast to C. aestivalis which seems to infect specifically Guinardia striata [49], C. longipes parasitizes a large diatom host spectrum [59]. Among the 18 diatoms strains used in their study, 77% of them were infected by C. longipes. Thus, infection of Pseudo-nitzschia by parasitic Cercozoa was probably a key factor in their decline, and should be considered in studies of this harmful algal bloom organism.
Implications regarding bottom-up controls and possibly transport
Physicochemical variables explained only 31% of the PCC variation, suggesting limited short-term predictability of community variability by environmental factors at SPOT. Other studies have also reported similar findings [27, 45, 46, 61]. Among environmental parameters, temperature and nutrients seemed to be the major factors driving the short-term variability of PCC. Temperature is a fundamental determinant of physiological rates and maximal growth rates of autotrophic protists [62, 63] as well as heterotrophic ones [64–66]. Parasitic Syndiniales are also sensitive to temperature. In fact, some studies have shown disruptions in the efficiency of Amoebophrya infectivity below certain temperatures [67–69], which could explain why some Amoebophrya sp. (both OTU #13 and OTU #2453) became abundant later in the sampling period where temperature was ≥18°C. Nevertheless, because temperature determines stratification, and light and nutrients co-vary with stratification, few PCC changes can be ascribed to temperature alone.
Because 69% of variance was unexplained by physicochemical factors, other factors are relevant in explaining changes in PCC. Our study tracks the dynamics of protist community at a fixed geographic site and depth (near-surface). Changes in PCC result from responses to changing environmental conditions and interactions among the various organisms [3, 45, 46], but also potentially the advection of patches with different PCC, and mixing with adjacent water. Some of these may be considered neutral processes (i.e., random effects including dispersal); and it has been noted previously that ocean currents and mixing with adjacent water, as well as stochastic processes, could generate some variation [3, 70, 71]. This needs to be kept in mind when interpreting the data. For example, we already noted that on March 15 an increase of three dinoflagellates OTUs (S. trochoidea, G. dorsalisulcum, L. polyedrum) led them to dominate the PCC that day, but PCC was remarkably different from the days before and after. This could represent a localized patchy dinoflagellate bloom which could have been advected past the sample site by currents and/or temporarily concentrated at the surface by their own vertical migration [72–74], thus causing an abrupt and punctual shift in the PCC at the sampling site. Note that while currents and mixing prevent the certainty of tracking the same organisms over time (e.g., to estimate net growth rates, which we did not do), we can still interpret co-occurrence of organisms and clustering of communities, which would be appropriate even if samples came from different locations, Note also that Lie et al. [27] examined spatial (several km scale) and temporal (daily) variation of protists at this same sample site in May of this study, and found that temporal (daily) changes were significantly larger than spatial ones, suggesting internal community changes were dominant over advective ones when they were examined.
Protist community network
Network analysis revealed that the protist community is organized into modules (groups of inter-correlated taxa) of varying size (Figures S5, S6). Modular organization is ubiquitous in microbial systems [6, 22, 75–77] and probably reflects temporal niche arrangement and differentiation between protist taxa. Some of the identified modules displayed high and dense interconnectivity (Figures S5, S6), constituting focal points for the whole protist community interactions, and suggesting that different protist modules could be functionally linked at SPOT. In most cases, members of these modules included taxa from all trophic positions (i.e., phototrophs, mixotrophs, heterotrophs, and parasites) and diverse taxonomic groups (Figures S6), suggesting that complex parameters (resource competition, metabolic yield, and growth rate) may take part in protist module formation [78].
Taxa with positive associations have been interpreted as functional guilds of organisms performing similar or complementary functions [79, 80] or feature interactions shaped by interspecies cross-feeding [81], although sometimes they may, to a large extent, reflect similar preferred conditions. Analogously, negative associations have been suggested to reflect interactions including competition, niche partitioning, and grazing [80, 81]. Our observed interactions, including those between parasitic OTUs (i.e., Syndiniales or Chytrids) and other taxa, were overwhelmingly positive (Fig. 4), and suggest parasites are following their hosts, but not causing enough mortality to create negative correlations at the daily to weekly time scales we studied. Interestingly, the same tendency has been reported by Chow et al. [79], when they analyzed the monthly virus–bacteria network at SPOT. Hosts and parasites exert reciprocal selective pressures on each other, which may lead to rapid reciprocal adaptation. The strong positive interactions highlighted over the studied period (daily to weekly) between marine parasites and their hosts suggests that their coevolution (known as Red Queen dynamics, [51]) might be observed on small time scales in marine systems and may thus serve as a counter-example to the common notion that evolution can only be detected across extended time.
Several haptophyte OTUs exhibited more edges between them than with any other protist groups (Fig. 4), suggesting similarity in their ecological properties. Interestingly, Koid et al. [82] showed that even if haptophytes share a set of core genes for the essential metabolic and cellular pathways, there are differences in secondary pathways, such as vitamin biosynthesis potential. Thus, the high connectivity displayed by some haptophytes may reflect reduced metabolic capabilities and auxotrophy for some metabolites; which could promote dependencies with other haptophyte taxa that provide the needed biosynthesis products.
In summary, our study shows the value of frequent sampling to evaluate community responses and microbial interactions among protists, reinforcing recent ideas about rapid dynamics and the importance of parasites.
Electronic supplementary material
Supplementary figures, text, tables(DOCX 2238 kb)
Acknowledgements
We thank the USC Wrigley Institute of Environmental Science, especially Roberta Marinelli, Sean Conner, and Captain Gordon Boivin and the crew of the Miss Christie for sampling opportunities and laboratory space. This work was supported by NSF Grants 1031743 and 1136818, and grant GBMF3779 from the Gordon and Betty Moore Foundation Marine Microbiology Initiative.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0097-x) contains supplementary material, which is available to authorized users.
References
- 1.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–40. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 2.Caron DA. New accomplishments and approaches for assessing protistan diversity and ecology in natural ecosystems. Bioscience. 2009;59:287–99. [Google Scholar]
- 3.Fuhrman JA, Cram JA, Needham DM. Marine microbial dynamics and their ecological interpretation. Nat Rev Microbiol. 2015;13:133–46. doi: 10.1038/nrmicro3417. [DOI] [PubMed] [Google Scholar]
- 4.Su C, Lei L, Duan Y, Zhang KQ, Yang J. Culture-independent methods for studying environmental microorganisms: methods, application, and perspective. Appl Microbiol Biotechnol. 2012;93:993–1003. doi: 10.1007/s00253-011-3800-7. [DOI] [PubMed] [Google Scholar]
- 5.Chow CET, Sachdeva R, Cram JA, Steele JA, Needham DM, Patel A, et al. Temporal variability and coherence of euphotic zone bacterial communities over a decade in the Southern California Bight. ISME J. 2013;7:2259–73. doi: 10.1038/ismej.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cram JA, Chow CET, Sachdeva R, Needham DM, Parada AE, Steele JA, et al. Seasonal and interannual variability of the marine bacterioplankton community throughout the water column over ten years. ISME J. 2015;9:563–80. doi: 10.1038/ismej.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giovannoni SJ, Vergin KL. Seasonality in ocean microbial communities. Science. 2012;335:671–6. doi: 10.1126/science.1198078. [DOI] [PubMed] [Google Scholar]
- 8.Kim DY, Countway PD, Jones AC, Schnetzer A, Yamashita W, Tung C, et al. Monthly, seasonal and interannual variability of microbial eukaryote assemblages within and below the euphotic zone in the eastern North Pacific. ISME J. 2014;8:515–30. doi: 10.1038/ismej.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berdjeb L, Ghiglione JF, Domaizon I, Jacquet S. A two-year assessment of the main environmental factors driving the free-living bacterial community structure in Lake Bourget (France) Microb Ecol. 2011;61:941–54. doi: 10.1007/s00248-010-9767-6. [DOI] [PubMed] [Google Scholar]
- 10.Sherr EB, Sherr BF. Heterotrophic dinoflagellates: a signficant component of microzooplankton biomass and major grazers of diatoms in the sea. Mar Ecol Prog Ser. 2007;352:187–97. [Google Scholar]
- 11.Worden AZ, Follows MJ, Giovannoni SJ, Wilken S, Zimmerman AE, Keeling PJ. Rethinking the marine carbon cycle: factoring in the multifarious lifestyles of microbes. Science. 2015;347:127594–10. doi: 10.1126/science.1257594. [DOI] [PubMed] [Google Scholar]
- 12.Caron DA, Countway PD. Hypotheses on the role of the protistan rare biosphere in a changing world. Aquat Microb Ecol. 2009;57:227–38. [Google Scholar]
- 13.Fuhrman JA, Caron DA (2016). Heterotrophic planktonic microbes: virus, bacteria, archaea, and protozoa. In: Yates MV, et al. editors. Manual of Environmental Microbiology 4th ed. Washington D.C.: American Society of Microbiology. Pp. 4.2.2-1–34.
- 14.Díez B, Pedrós-Alió C, Massana R. Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Appl Environ Microbiol. 2001;67:2932–41. doi: 10.1128/AEM.67.7.2932-2941.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.López-García P, Rodríguez-Valera F, Pedrós-Alió C, Moreira D. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature. 2001;409:603–7. doi: 10.1038/35054537. [DOI] [PubMed] [Google Scholar]
- 16.Moon-van der Staay SY, Watcher RD, Vaulot D. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature. 2001;409:607–10. doi: 10.1038/35054541. [DOI] [PubMed] [Google Scholar]
- 17.Cleary AC, Durbin EG. Unexpected prevalence of parasite 18S rDNA sequences in winter among Antarctic marine protists. J Plankton Res. 2016;38:401–17. [Google Scholar]
- 18.Guillou L, Viprey M, Chambouvet A, Welsh RM, Kirkham AR, Massana R, et al. Widespread occurrence and genetic diversity of marine parasitoids belonging to Syndiniales (Alveolata) Environ Microbiol. 2008;10:3349–65. doi: 10.1111/j.1462-2920.2008.01731.x. [DOI] [PubMed] [Google Scholar]
- 19.De Vargas C, Audic S, Henry N, Decelle J, Mahé F, Logares R, et al. Eukaryotic plankton diversity in the sunlit ocean. Science. 2015;348:1261605. doi: 10.1126/science.1261605. [DOI] [PubMed] [Google Scholar]
- 20.Countway PD, Vigil P, Schnetzer A, Moorthi S, Caron DA. Seasonal analysis of protistan community structure and diversity at the USC Microbial Observatory (San Pedro Channel, Pacific Ocean) Limnol Oceanogr. 2010;55:2381–96. [Google Scholar]
- 21.Schnetzer A, Moorthi SD, Countway PD, Gast RJ, Gilg IC, Caron DA. Depth matters: microbial eukaryote diversity and community structure in the eastern North Pacific revealed through environmental gene libraries. Deep Sea Res I. 2011;58:16–26. [Google Scholar]
- 22.Steele JA, Countway PD, Xia L, Vigil PD, Beman JM, Kim DY, et al. Marine bacterial, archaeal, and protistan association networks reveal ecological linkages. ISME J. 2011;5:1414–25. doi: 10.1038/ismej.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohtsuka S, Suzaki T, Horiguchi T, Suzuki N, Not F. Marine protists: diversity and dyanmics. Tokyo: Springer; 2016. [Google Scholar]
- 24.Vigil P, Countway P, Rose J, Lonsdale D, Gobler C, Caron DA. Rapid shifts in dominant taxa among microbial eukaryotes in estuarine ecosystems. Aquat Microb Ecol. 2009;54:83–100. [Google Scholar]
- 25.Mangot JF, Domaizon I, Taib N, Marouni N, Duffaud E, Bronner G, et al. Short-term dynamics of diversity patterns: evidence of continual reassembly within lacustrine small eukaryotes. Environ Microbiol. 2013;15:1745–58. doi: 10.1111/1462-2920.12065. [DOI] [PubMed] [Google Scholar]
- 26.Kim DY, Countway PD, Gast RJ, Caron DA. Rapid shifts in protistan community structure and composition during bottle incubations. Microb Ecol. 2011;62:383–98. doi: 10.1007/s00248-011-9816-9. [DOI] [PubMed] [Google Scholar]
- 27.Lie AAY, Kim DY, Schnetzer A, Caron DA. Small-scale temporal and spatial variations in protistan community composition at the San Pedro Ocean Time-series station off the coast of southern California. Aquat Microb Ecol. 2013;70:93–110. [Google Scholar]
- 28.Needham DM, Fuhrman JA. Pronounced daily succession of phytoplankton, archaea and bacteria following a spring bloom. Nature Microbiol. 2016;1:16005. doi: 10.1038/nmicrobiol.2016.5. [DOI] [PubMed] [Google Scholar]
- 29.Stoeck T, Bass D, Nebel M, Christen R, Jones MD, Breiner HW, et al. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol. 2010;19:21–31. doi: 10.1111/j.1365-294X.2009.04480.x. [DOI] [PubMed] [Google Scholar]
- 30.Balzano S, Abs E, Leterme SC. Protist diversity along a salinity gradient in a coastal lagoon. Aquat Micro Ecol. 2015;74:263–77. [Google Scholar]
- 31.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinforma. 2012;13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–6. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guillou L, Bachar D, Audic S, Bass D, Berney C, Bittner L, et al. The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2013;41:D597–604. doi: 10.1093/nar/gks1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clarke KR. Non-parametric multivariate analyses of changes in community structure. Aust J Ecol. 1993;18:117–43. [Google Scholar]
- 36.Marzorati M, Wittebolle L, Boon N, Daffonchio D, Verstraete W. How to get more out of molecular fingerprints? Practical tools for microbial ecology. Environ Microbiol. 2008;10:1571–81. doi: 10.1111/j.1462-2920.2008.01572.x. [DOI] [PubMed] [Google Scholar]
- 37.Xia LC, Steele JA, Cram JA, Cardon ZG, Simmons SL, Vallino JJ, et al. Extended local similarity analysis (eLSA) of microbial community and other time series data with replicates. BMC Syst Biol. 2011;5:S15. doi: 10.1186/1752-0509-5-S2-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia LC, Ai D, Cram JA, Fuhrman JA, Sun F. Efficient statistical significance approximation for local similarity analysis of high-throughput time series data. Bioinformatics. 2013;29:230–7. doi: 10.1093/bioinformatics/bts668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinforma. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gong J, Dong J, Liu X, Massana R. Extremely high copy numbers and polymorphisms of the rDNA operon estimated from single cell analysis of oligotrich and peritrich ciliates. Protist. 2013;164:369–79. doi: 10.1016/j.protis.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 42.Prokopowich CD, Gregory TR, Crease TJ, Gregory TR, Crease TJ. The correlation between rDNA copy number and genome size in eukaryotes. Genome. 2003;46:48–50. doi: 10.1139/g02-103. [DOI] [PubMed] [Google Scholar]
- 43.Godhe A, Asplund ME, Harnstrom K, Saravanan V, Tyagi A, Karunasagar I. Quantification of diatom and dinoflagel-latebiomasses in coastal marine seawater samples by real-timePCR. Appl Environ Microbiol. 2008;2013:7174–82. doi: 10.1128/AEM.01298-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu F, Massana R, Not F, Marie D, Vaulot D. Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the18S rRNA gene. FEMS Microbiol Ecol. 2005;52:79–92. doi: 10.1016/j.femsec.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 45.Genitsaris S, Monchy S, Denonfoux J, Ferreira S, Kormas K, Sime-Ngando T, et al. Marine microbial community structure assessed from combined metagenomic analysis and ribosomal amplicon deep-sequencing. Mar Biol Res. 2016;12:30–42. [Google Scholar]
- 46.Genitsaris S, Monchy S, Viscogliosi E, Sime-Ngando T, Ferreira S, Christaki U. Seasonal variations of marine protist community structure based on taxon-specific traits using the eastern English Channel as a model coastal system. FEMS Microbiol Ecol. 2015;91:fiv034. doi: 10.1093/femsec/fiv034. [DOI] [PubMed] [Google Scholar]
- 47.Simon M, López-García P, Deschamps P, Moreira D, Restoux G, Bertolino P, et al. Marked seasonality and high spatial variability of protist communities in shallow freshwater systems. ISME J. 2015;9:1941–53. doi: 10.1038/ismej.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park MG, Yih W, Coats DW. Parasites and phytoplankton, with special emphasis on dinoflagellate infections. J Eukaryot Microbiol. 2004;51:145–155. doi: 10.1111/j.1550-7408.2004.tb00539.x. [DOI] [PubMed] [Google Scholar]
- 49.Peacock EE, Olson RJ, Sosik HM. Parasitic infection of the diatom Guinardia delicatula, a recurrent and ecologically important phenomenon on the New England Shelf. Mar Ecol Prog Ser. 2014;503:1–10. [Google Scholar]
- 50.Salomon PS, Imai I. Pathogens of harmful microalgae. In: Graneli E, Turner JT, editors. Ecology of harmful algae. Berlin/Hedelberg: Springer; 2006. pp. 271–82. [Google Scholar]
- 51.Christaki U, Lefèvre D, Georges C, Colombet J, Catala P, Courties C, et al. Microbial food web dynamics during spring phytoplankton blooms in the naturally iron-fertilized Kerguelen area (Southern Ocean) Biogeosciences. 2014;11:6739–6753. [Google Scholar]
- 52.Rabajante JF, Tubay JM, Uehara T, Morita S, Ebert D, Yoshimura J. Red queen dynamics in multi-host and multi-parasite interaction system. Sci Rep. 2015;2:e1501548. doi: 10.1038/srep10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arancio M, Sourisseau M, Souissi S. Processes leading to the coexistence of a host and its parasitoid inhomogeneous environments: the role of an infected dormant stage. Ecol Model. 2014;279:78–88. [Google Scholar]
- 54.Rosenheim JA, Kaya HK, Ehler LE, Marois JJ, Jaffee BA. Intraguild predation among biological-control agents: theory and evidence. Biol Control. 1995;5:303–35. [Google Scholar]
- 55.Hogarth WL, Diamond P. Interspecific competition in larvae between ento-mophagous parasitoids. Am Nat. 1984;124:552–60. [Google Scholar]
- 56.May M, Hassell MP. The dynamics of multiparasitoid–host interactions. Am Nat. 1981;117:234–61. [Google Scholar]
- 57.Georges C, Monchy S, Genitsaris S, Christaki U. Protist community composition during early phytoplankton blooms in the naturally iron-fertilized Kerguelen area (Southern Ocean) Biogeosciences. 2014;11:5847–63. [Google Scholar]
- 58.Schnetzer A, Jones BH, Schaffner RA, Cetinic I, Fitzpatrick E, Miller PE, et al. Coastal upwelling linked to toxic Pseudo-nitzschia australis blooms in Los Angeles coastal waters, 2005–7. J Plankton Res. 2013;35:1080–92. [Google Scholar]
- 59.Schnepf E, Kühn SF. Food uptake and fine structure of Cryothecomonas longipes sp. nov., a marine nanoflagellate incertae sedis feeding phagotrophically on large diatoms. Helgol Mar Res. 2000;54:18–32. [Google Scholar]
- 60.Tillmann U, Hesse KJ, Tillmann A. Large-scale parasitic infection of diatoms in the North Frisian Wadden Sea. J Sea Res. 1999;42:255–61. [Google Scholar]
- 61.Lima-Mendez G, Faust K, Henry N, Decelle J, Colin S, Carcillo F, et al. Ocean Plancton. Determinants of community structure in the global plankton interactome. Science. 2015;348:6237–1262073. doi: 10.1126/science.1262073. [DOI] [PubMed] [Google Scholar]
- 62.Eppley RW. Temperature and phytoplankton growth in the sea. Fish Bull. 1972;70:1063–85. [Google Scholar]
- 63.Goldman JC, Carpenter EJ. A kinetic approach to the effect of temperature on algal growth. Limnol Oceanogr. 1974;19:756–66. [Google Scholar]
- 64.Atkinson D, Ciotti BJ, Montagnes DJ. Protists decrease in size linearly with temperature: ca. 2.5% degrees C(−1) Proc Biol Sci. 2003;270:2605–11. doi: 10.1098/rspb.2003.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rose JM, Caron DA. Does low temperature constrain the growth rates of heterotrophic protists? Evidence and implications for algal blooms in cold waters. Limnol Oceanogr. 2007;52:886–895. [Google Scholar]
- 66.Rose JM, Feng Y, DiTullio GR, Dunbar RB, Hare CE, Lee PA, et al. Synergistic effects of iron and temperature on Antarctic phytoplankton and microzooplankton assemblages. Biogeosciences. 2009;6:3131–47. [Google Scholar]
- 67.Coats DW, Bockstahler KR. Occurrence of the parasitic dinoflagellate Amoebophrya ceratii in Chesapeake Bay populations of Gymnodinium sanguineum. J Eukaryot Microbiol. 1994;41:586–93. [Google Scholar]
- 68.Salomon PA, Janson S, Graneli E. Multiple species of the Dinophagous Dinoflagellate genus Amoebophrya infect the same host species. Environ Microbiol. 2003;5:1046–52. doi: 10.1046/j.1462-2920.2003.00511.x. [DOI] [PubMed] [Google Scholar]
- 69.Yih W, Coats DW. Infection of Gymnodinium sanguineum by the dinoflagellate Amoebophrya sp.: effect of nutrient environment on parasite generation time, reproduction and infectivity. J Eukaryot Microbiol. 2000;47:504–10. doi: 10.1111/j.1550-7408.2000.tb00082.x. [DOI] [PubMed] [Google Scholar]
- 70.Dini-Andreote F, Stegen JC, van Elsas JD, Salles JF. Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc Natl Acad Sci USA. 2015;112:E1326–E1332. doi: 10.1073/pnas.1414261112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stegen JC, Lin X, Konopka AE, Fredrickson JK. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 2012;6:1653–64. doi: 10.1038/ismej.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jephson T, Carlsson P, Fagerberg T. Dominant impact of water exchange and disruption of stratification on dinoflagellate vertical distribution. Estuar Coast Mar Sci. 2012;112:198–206. [Google Scholar]
- 73.Ji R, Franks PJS. Vertical migration of dinoflagellates: model analysis of strategies, growth, and vertical distribution patterns. Mar Ecol Prog Ser. 2007;344:49–61. [Google Scholar]
- 74.Lalli C, Parsons TR. Biological Oceanography: an introduction. 2nd ed, vol.1997. London: Butterworth-Heinemann; 1997.
- 75.Barberán A, Bates ST, Casamayor EO, Fierer N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012;6:343–351. doi: 10.1038/ismej.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Needham DM, Chow CET, Cram JA, Sachdeva R, Parada A, Fuhrman JA. Short-term observations of marine bacterial and viral communities: patterns, connections and resilience. ISME J. 2013;7:1274–85. doi: 10.1038/ismej.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Berry D, Widder S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front Microbiol. 2014;5:219. doi: 10.3389/fmicb.2014.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Freilich S, Kreimer A, Meilijson I, Gophna U, Sharan R, Ruppin E. The large scale organization of the bacterial network of ecological co-occurrence interactions. Nucleic Acids Res. 2010;38:3857–68. doi: 10.1093/nar/gkq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chow CET, Kim DY, Sachdeva R, Caron DA, Fuhrman JA. Top-down controls on bacterial community structure: microbial network analysis of bacteria, T4-like viruses and protists. ISME J. 2014;8:816–29. doi: 10.1038/ismej.2013.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eiler A, Heinrich F, Bertilsson S. Coherent dynamics and association networks among lake bacterioplankton taxa. ISME J. 2012;6:330–42. doi: 10.1038/ismej.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fuhrman JA, Steele JA. Community structure of marine bacterioplankton: patterns, networks, and relationships to function. Aquat Microb Ecol. 2008;53:69–81. [Google Scholar]
- 82.Koid AE, Liu Z, Terrado R, Jones AC, Caron DA, Heidelberg KB. Comparative transcriptome analysis of four Prymnesiophyte algae. PLoS ONE. 2014;9:e97801. doi: 10.1371/journal.pone.0097801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Adl SM, Simpson AGB, Lane CE, Lukeš J, Bass D, Bowser SS. The Revised Classification of Eukaryotes. Journal of Eukaryotic Microbiology. 2012;59:429–514. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures, text, tables(DOCX 2238 kb)