Abstract
Rhodotorula glutinis, an oleaginous red yeast, intrinsically produces several bio-products (i.e., lipids, carotenoids and enzymes) and is regarded as a potential host for biorefinery. In view of the limited available genetic engineering tools for this yeast, we have developed a useful genetic transformation method and transformed the β-carotene biosynthesis genes (crtI, crtE, crtYB and tHMG1) and cellulase genes (CBHI, CBHII, EgI, EgIII, EglA and BGS) into R. glutinis genome. The transformant P4-10-9-63Y-14B produced significantly higher β-carotene (27.13 ± 0.66 mg/g) than the wild type and also exhibited cellulase activity. Furthermore, the lipid production and salt tolerance ability of the transformants were unaffected. This is the first study to engineer the R. glutinis for simultaneous β-carotene and cellulase production. As R. glutinis can grow in sea water and can be engineered to utilize the cheaper substrates (i.e. biomass) for the production of biofuels or valuable compounds, it is a promising host for biorefinery.
Introduction
Rhodotorula glutinis is an oleaginous red yeast capable of producing several valuable compounds including microbial lipids, pigments and enzymes. R. glutinis contains up to 70% lipids in their dry weight biomass, and is non-toxic and relatively easy to grow and harvest1. It has been demonstrated that R. glutinis is a potential host for biodiesel industry due to the accumulation of polyunsaturated fatty acid triacylglycerol inside the cells, which is similar to vegetable oils2,3. Besides the high lipid production, R. glutinis can produce β-carotene, which is regarded as a valuable compound in healthcare industry and has anti-carcinogenic and antioxidant properties4,5. The Business Communications Company Research reported that β-carotene has the largest share in global carotenoids market (more than $300 million by 2018)6. So the demand of β-carotene is increasing. However, concentration of β-carotene in vegetable (e.g. carrot, 0.02 mg/g) is low and is decreased during transfer and storage7. Therefore, synthesizing β-carotene by microbes is an ideal approach. R. glutinis is a well-known β-carotene producing yeast in the industry5. Although there were many studies focused on different hosts, R. glutinis can utilize various low-cost carbon sources, making it an attractive candidate for producing lipids and β-carotene in industries5,8. Several researchers have used crude glycerol as a substrate for R. glutinis to produce microbial lipids9. Moreover, R. glutinis is successfully cultivated in brewery effluents10. Therefore, R. glutinis is a promising biorefinery host for the production of microbial lipids and β-carotene using cheap substrates as a carbon source11.
Previously, several studies have demonstrated microbial lipid and β-carotene production using wild type R. glutinis3. In order to reduce the high fermentation cost of R. glutinis, one may increase the productivity of valuable products and/or use cheaper feedstock for fermentation (i.e. biomass, waste water, effluents, etc.). Previous researchers focused on improving the carotenoid production ability of R. glutinis by optimizing the fermentation conditions (i.e. temperature, pH and dissolved oxygen) or optimizing the ratios of carbon or nitrogen sources12,13. Furthermore, R. glutinis was subjected to mutagenesis using UV irradiation or chemical mutagens to improve its carotenoid production and the mutant strain could produce higher amounts of β-carotene using sea water as a substrate14–17. Lignocellulosic biomass, which is the most available, cheap and renewable source in nature, makes it becoming an idea carbon sources for clean energy or bio-products. Lignocellulosic biomass is abundant and renewable in nature and regarded as a potential feedstock for fermentation. However, conversion of biomass into fermentable sugars requires at least three types of cellulases, including endoglucanase (EG), exoglucanase (cellobiohydrolase, CBH), and β-D-glucosidase (BGS)18,19. More recent studies revealed that some R. glutinis strains contain endoglucanases and possess wheat or rice straw degradation ability5,20,21.
So far, Agrobacterium-mediated transformation (ATMT) has been used to engineer Rhodotorula (teleomorph is Rhodosporidium) toruloides. Lin et al. conducted the genomic insertion of multiple antibiotic resistance genes by ATMT22. Johns et al. investigated four different promoters that could be induced or repressed by different carbon sources23. Moreover, Zhang et al. transformed three lipid biosynthesis genes into R. toruloides and improved the lipid production by fourfold24. A review article has pointed out the potential engineering processes in R. toruloides25.
Recently, a few studies have engineered Yarrowia lipolytica, which is an oleaginous yeast with cellulolytic ability to convert lignocellulosic substrates to lipid19,26. To produce β-carotene at a large scale, previous studies engineered Saccharomyces cerevisiae and Y. lipolytica using a synthetic biology approach27–30. However, none of the studies focused on developing a bio-refinery host to convert the lignocellulosic substrates to β-carotene and lipid. This is a first study, which focus on engineering the carotenoid pathway along with cellulolytic enzymes in R. glutinis. In this study, we used synthetic biology tools to improve the β-carotene production and install cellulolytic ability in R. glutinis by transforming β-carotene biosynthesis pathway genes (tHMG1, crtI, crtE and crtYB) and cellulase genes (CBHI, CBHII, EgIII, EgI, EglA and BGS) into its genome. Moreover, we demonstrated the unexpected link between carotenoids and cellulases that suggests the possibility to develop a cell factory to turn biomass wastes into valuable products or/and renewable energy.
Results
Establishing a heterologous gene expression platform in R. glutinis
The result of minimal inhibitory concentration (MIC) (Supplementary Table S1) showed that the R. glutinis wild type cannot grow in neither 100 nor 200 μg/ml concentration of any of the three antibiotics tested (Zeocin, G418 and Hygromycin), so either concentration can be used to select the transformants. For initial transformation, three gene expression cassettes (Table 1) including phytoene desaturase gene (crtI), cellobiohydrolase (cbhI) and G418 selection marker gene (KanMx) were integrated into the R. glutinis genome, using frozen protoplast and lithium acetate competent cells18,31. The transformants were screened using YP2D supplemented with G418 (200 μg/ml) and the wild type without expression cassette was used as a control. A total of 200 transformants were randomly selected and subcultured for 3 generations to select stable transformants (Supplementary Fig. S1). Then transformants containing both crtI and cbhI were validated by PCR using the gene specific primer pairs (Supplementary Table S2). Due to the high GC content of R. glutinis, the PCR amplification of specific genes from their genomic DNA was found to be a difficult task22,32. Hence, the genes were amplified using long-range primer pairs and modified touchdown PCR conditions and the wild type was used as a negative control. The results showed that P4-10-9 was successfully transformed and contained the crtI, cbhI and KanMx genes (Supplementary Fig. S2). This is the first study to integrate both β-carotene biosynthesis genes and cellulase genes into the R. glutinis genome by electroporation. Our gene cassettes were flanked with the specific homologous recombination (HR) region at both ends but we could not confirm the inserts in those regions (data not shown). Previous studies suggested that Rhodotorula had a low HR efficiency33,34. This suggests that the genes were integrated into the R. glutinis genome by non-homologous end joining (NHEJ). In future, we may improve the targeting frequency or HR by inhibiting or deleting the NHEJ pathway. Furthermore, R. glutinis was successfully transformed by the ATMT method to integrate the hygromycin resistance gene (Supplementary Fig. S3). Previous studies applied ATMT in some recalcitrant transformation hosts including R. glutinis22,35. However, ATMT is not suitable for the integration of multiple genes at the same time.
Table 1.
The gene expressing cassettes used in this study.
| Gene | Species | Function | Promoter | terminator | Cassette size (bp) |
|---|---|---|---|---|---|
| crtE | Xanthophyllomyces dendrorhous | Geranylgeranyl pyrophosphate synthase | ScGapDH | ScGapDH | 2,083 |
| crtI | Xanthophyllomyces dendrorhous | Phytoene desaturase | ICL | 35 S | 3,935 |
| crtYB | Xanthophyllomyces dendrorhous | Phytoene synthase/lycopene cyclase | ScPGK | ScPGK | 3,186 |
| tHMG1 | Kluyveromyces marxianus | Truncated 3-hydroxy-3-methylglutaryl-coenzyme A reductase | ScADHI | ScADHI | 2,911 |
| CBHI | Trichoderma reesei | Cellobiohydrolases | KlADHI | ScGapDH | 2,680 |
| CBHII | Trichoderma reesei | Cellobiohydrolases | KlPGK | ScPGK | 2,502 |
| EgIII | Trichoderma reesei | Endo-β-1,4-glucanases | ScGapDH | ScGapDH | 2,161 |
| EgI | Aspergillus niger | Endo-β-1,4-glucanases | ScPGK | ScPGK | 2,731 |
| EglA | Aspergillus niger | Endo-β-1,4-glucanases | Lac4 | Lac4 | 2,925 |
| BGS | Neocallimastix patriciarum | β-glucosidases | ScADHI | ScADHI | 3,691 |
| KanMx | — | Geneticin (G418) resistance | KlGapDH | ScGapDH | 1,855 |
| Sh ble | — | Zeocin resistance | ScADHI | ScADHI | 1,786 |
| hph | — | Hygromycin B resistance | ScADHI | ScADHI | 2,437 |
| Total size: | 34,890 |
Improvement of β-carotene production by installing genes into R. glutinis genome
Based on a previous study9, we tested different carbon sources to compare the β-carotene production between wild type and P4-10-9. Three different carbon sources were tested: 2% glucose, 2% galactose and 2% glycerol. The HPLC data showed that P4-10-9 produced more carotenoids than the wild type using galactose or glycerol as a carbon source, compared to glucose (Fig. 1). Similar results were obtained by Johns, Love and Aves23, who found that the ICL promoter was inhibited by glucose. It might be because the crtI gene was driven by the ICL promoter, so P4-10-9 could produce more carotenoids.
Figure 1.
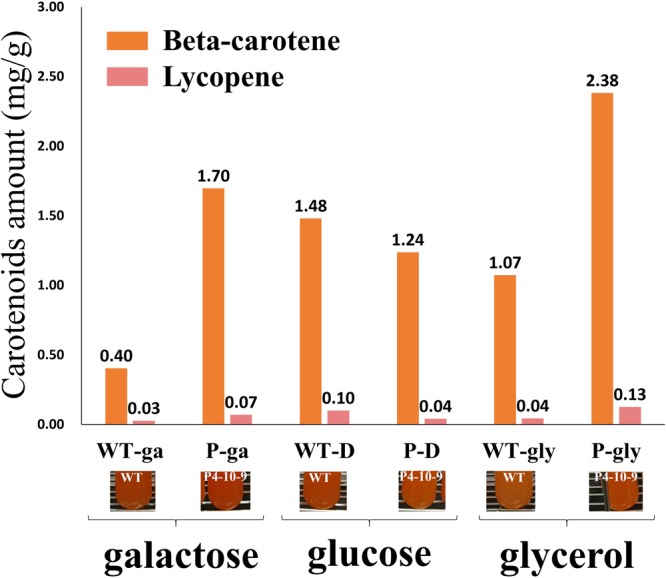
Carotenoid amounts of the P4-10-9 transformant in different carbon sources. The number at the top of a vertical bar shows the β-carotene or lycopene amounts compared to the amount in the wild type.
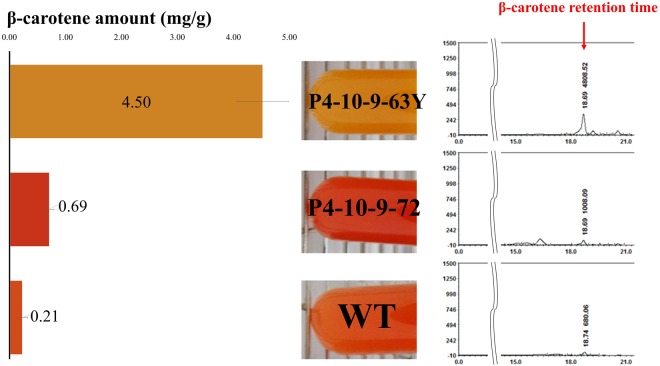
To further improve the β-carotene production, additional functional genes including crtYB and crtE were integrated into the P4-10-9 genome using electroporation, and transformants were screened using zeocin. 72 transformants were selected and subcultured for 3 generations to obtain stable transformants. The genomic integration of crtYB and crtE was confirmed by PCR using long-range gene-specific primer pairs (Supplementary Table S2). The PCR amplification (Supplementary Fig. S4) confirmed the integration of crtE in transformant 72 and it was named as P4-10-9-72. Similarly, transformant 63 was integrated only with crtYB and named as P4-10-9-63Y. To confirm the β-carotene production improvement, transformants P4-10-9-63Y and P4-10-9-72 and wild type R. glutinis were cultured in 10 ml YP2Gly for 2 weeks and total carotenoids were extracted. The HPLC data showed that both transformants produced higher β-carotene than the wild type (Fig. 2). Especially, β-carotene production of P4-10-9-63Y was improved by 20 folds (4.50 ± 0.46 mg/g), thus confirming the importance of crtYB in β-carotene biosynthesis. However, the HPLC data of P4-10-9-72 showed a small β-carotene peak at around the 18 min retention time and several new peaks were observed between 15-17 min (Fig. 2), which might be due to the production of torulene and torularhodin. Indeed, Frengova and Beshkova36 demonstrated the production of torulene and torularhodin by R. glutinis. Furthermore, transformant P4-10-9-72 showed red color and previous studies found that torulene and torularhodin showed characteristic red color37,38. Transformant P4-10-9-72 contains both crtI and crtE, which might improve the precursor availability for the torulene and torularhodin pathways39,40. The higher β-carotene producing transformant P4-10-9-63Y showed yellow color. Similarly, Bhosale and Gadre observed yellow color in R. glutinis after UV treatment, which produced increased β-carotene compared to the parent strain41. Hence, the higher β-carotene producing P4-10-9-63Y was selected as a host for further study.
Figure 2.
The amounts of β-carotene (mg/g) in P4-10-9-63Y, P4-10-9-72 and the wild type. In the HPLC data, the numbers of sample peaks represent retention times of β-carotene and peak area.
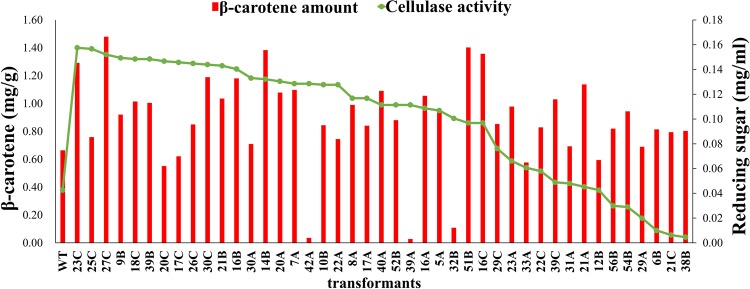
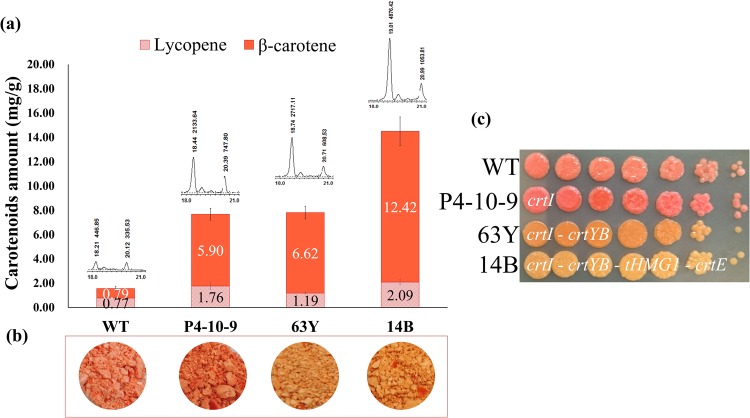
The β-carotene production of transformant P4-10-9-63Y was further improved by transforming the upstream genes (tHMG1 and crtE) of the β-carotene biosynthetic pathway. A total of 146 transformants were selected on YP2D plates with hygromycin as a selective marker and subcultured for 3 generations. The PCR results (Supplementary Fig. S5) showed that 43 transformants containing all the four β-carotene biosynthesis pathway genes and selected for functional assays. These 43 transformants could be stably subcultured on selective plates supplemented with G418, zeocin and hygromycin. The β-carotene production of 43 transformants were analyzed and almost every transformant showed improved β-carotene production (Fig. 3). We obtained transformants containing different gene arrangements (Table 2). P4-10-9, P4-10-9-63Y and P4-10-9-63Y-14B were selected for further analysis because they contained different arrangements of tHMG1, crtI, crtE and crtYB. We measured the lycopene and β-carotene amounts and observed the color of colonies of transformants compared to the wild type (Fig. 4). We reduced the growing time to avoid the decrease of β-carotene amount and used two carbon source according to Fig. 1. Therefore, we used 10 ml YP2G2Gly and cultured for 7 days. The transformant P4-10-9-63Y produced 8.4 fold higher β-carotene with a significant color change than the wild type (Table 2, Fig. 4b,c). Therefore, the bi-functional enzyme crtYB (phytoene synthase and lycopene cyclase) appears to play an important role in the carbon flux from lycopene to β-carotene. In contrast, P4-10-9-63Y-14B contains tHMG1, crtI, crtE and crtYB, which improved the conversion of HMG-CoA to β-carotene. The engineered R. glutinis strain P4-10-9-63Y-14B produced 15.7 fold β-carotene and 2.7 fold higher lycopene compared to the wild type. This is the first study that showed a significant improvement of carotenoid production in the oleaginous red yeast.
Figure 3.
Functional screening of R. glutinis 43 transformants that contained all the 10 genes.
Table 2.
R. glutinis transformants containing different β-carotene biosynthesis genes.
| R. glutinis strains | transformed carotenoid biosynthesis genes | Fold of carotenoid increase | |
|---|---|---|---|
| Lycopene | β-carotene | ||
| Wild type | — | — | — |
| P4-10-9 | crtI | 2.3 | 7.4 |
| P4-10-9-63Y | crtI - crtYB | 1.5 | 8.4 |
| P4-10-9-63Y-14B | crtI - crtYB - tHMG1 - crtE | 2.7 | 15.7 |
Figure 4.
The amounts of carotenoids (mg/g) in different R. glutinis transformants. (a) HPLC analysis. (b) The dry biomass before extraction. (c) Colony colors on YP2G2Gly agar plates.
Installation of cellulase ability in R. glutinis
To increase the value of R. glutinis as a host for biorefinery applications, the strain P4-10-9-63Y was selected as a host to be transformed with the cellulase genes to utilize the cellulosic biomass. Three types of cellulases (CBHI, CBHII, EgIII, EgI, EglA and BGS) were transformed into the R. glutinis genome according to our previous studies with other hosts18,42,43. The genomic integration of each gene was confirmed by PCR using gene specific primer pairs (Supplementary Table S2, 9–20). The PCR data (Supplementary Fig. S5) showed that several transformants contained all the cellulase genes. We first utilized mix colonies PCR to validate CBHII, EgIII, EglA and BGS genes and the results showed correct gene sizes as the positive control. Second, we tested CBHI gene by the single colony PCR and selected transformants containing the correct gene size. Then we further confirmed EgI in 43 transformants. The cellulase activity was screened using fast-cellulase screening (4% Sigmacell cellulose type 20 as the carbon source) and the data showed that almost all transformants had higher cellulase activity compared to the wild type strain (Fig. 3).
Demonstrating the biorefinery potential of the engineered R. glutinis
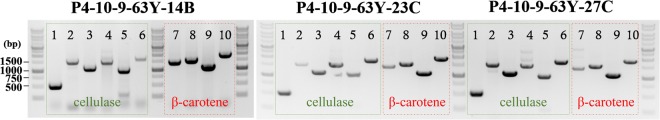
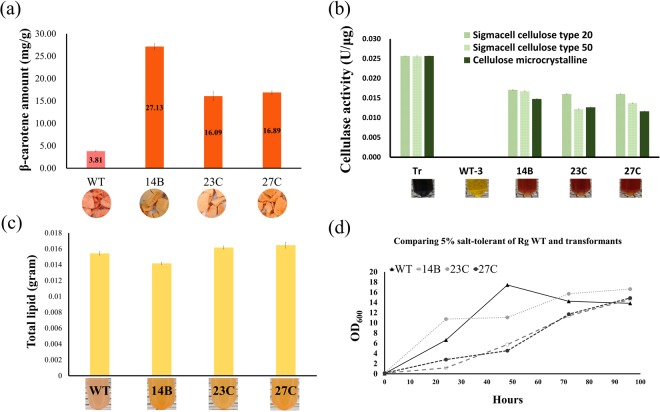
The engineered R. glutinis, P4-10-9-63Y-14B, −23C and −27C were selected based on their genotype (Fig. 5) and phenotype (Fig. 3). The four major properties, i.e., maximum β-carotene amount, total cellulase activity, total lipid and salt-tolerance of transformants, were analyzed. To analyze the maximum β-carotene amounts in selective transformants, we reduced medium from 10 ml to 7 ml because we speculated that the dissolved oxygen might improve the β-carotene amount. Therefore, the selective transformants were cultured in 7 ml YP2G2Gly for 1 week to analyze the maximum β-carotene amounts. Figure 6a showed the β-carotene amounts of selective transformants were increased compared to the wild type. The β-carotene amount of P4-10-9-63Y-14B was improved up to 27.13 ± 0.66 mg/g. To the best of our knowledge, this is the highest β-carotene amount ever produced by R. glutinis.
Figure 5.
Validation of the 10 genes in R. glutinis candidate transformants. *The numbers in the figure represent: 1: eglA (503 bp), 2: egI (1478 bp), 3: egIII (1026 bp), 4: cbhI (1441 bp), 5: cbhII (1002 bp), 6: BGS (1690 bp), 7: tHMG1 (1376 bp), 8: crtI (1489 bp), 9: crtE (1082 bp) and 10: crtYB (1821 bp). The size of amplicons were according to the designing specific primer pairs in Supplementary Table S2.
Figure 6.
Performances of R. glutinis candidate transformants. (a) Comparison of maximum β-carotene amounts in different transformants. (b) Total cellulase activity assay. (c) Total lipid weight. (d) Salt-tolerance.
To investigate the total cellulase activities of P4-10-9-63Y-14B, −23C and −27C, we tested different cellulosic substrates including Sigmacell cellulose type 20, Sigmacell cellulose type 50 and cellulose microcrystalline. The 50-fold condensed supernatants of R. glutinis wild type, 14B, 23C and 27C were harvested and the same total protein concentration was used for the total cellulase activity assay18. Figure 6b and Supplementary Fig. S6 shows that we have successfully improved the total cellulase activity of R. glutinis by transforming these cellulase genes. The results showed that the total cellulase activities of 14B (0.017 U/μg), 23 C (0.016 U/μg) and 27 C (0.016 U/μg) were only around 1.53- to 1.63-fold lower than the commercial enzyme Celluclast 1.5 L (0.026 U/μg) in Sigmacell cellulose type 20 substrate. This is the first study to engineer the cellulase genes into R. glutinis and significantly improved its total cellulase activities. Thus, the engineered R. glutinis strain might have potential to use the cellulosic substrate as a carbon source to produce a higher amount of β-carotene compared to the wild type (Supplementary Fig. S7).
In order to make sure that the insertion of additional genes did not affect the intrinsic characteristics of R. glutinis, the total lipid content and salt-tolerance of 14B, 23C and 27C was analyzed. The total lipid weight indeed showed no significant difference between the wild type and transformants (Fig. 6c); the total lipid content was around 70% lipid yield of dry weight biomass, the same as described in a previous study44. We also tested the growth of the wild type and transformants in YP2D with 5% NaCl to study their salt tolerance. The results showed that all of the strains were able to grow well in YP2D with 5% NaCl (Fig. 6d). Although the initial growth of the wild type was faster than the transformants, all the strains reached similar OD600 after 3 days culturing. We could not observe any significant growth difference between the wild type and transformants in YP2D (data not shown). Moreover, the total lipid content of both the wild type and the transformants were unaffected. So we suggest that the growth rate was not affected significantly. These results demonstrated that engineering the R. glutinis using our method did not strongly affect the lipid production and salt tolerance of R. glutinis.
Discussion
We have successfully integrated multiple gene expression cassettes including 4 β-carotene biosynthesis pathway genes and 6 cellulase genes into the genome of oleaginous red yeast R. glutinis. The transformant P4-10-9-63Y-14B produced up to 27.13 ± 0.66 mg/g of β-carotene amount. This was achieved by transforming four functional genes (tHMG1, crtI, crtE and crtYB) into the R. glutinis genome. A previous study achieved a similar result in Saccharomyces cerevisiae27. Although Li et al.30 discovered several novel β-carotene improving genes, the β-carotene amount (5.9 ± 0.1 mg/g) produced in their engineered S. cerevisiae was still as low as that in Verwaal et al. (2007). Larroude et al. applied synthetic biology tools to engineer oleaginous yeast Yarrowia lipolytica to obtain the highest β-carotene producing yeast (89.6 mg/g)29. These researchers pointed out that β-carotene may be stored and protected in lipid droplets in oleaginous yeast28. Gao et al. and Larroude et al. utilized multiple-copy insertions to improve the β-carotene production. Although there is some difficulty in analyzing the copy number in our R. glutinis transformants (i.e. RNA extraction), the strategy in this study can improve β-carotene production by 7 to 15 folds. The highest β-carotene amount (DCW) in previous studies were found in Saccharomyces cerevisiae (18 mg/g)45, Escherichia coli (72 mg/g)46 and Yarrowia lipolytica (89.6 mg/g)29. The β-carotene production of engineering strain might further improve by fed-batch cultivations in a bioreactor28,29. Our best producer transformant 14B is inferior only to E. coli and Y. lipolytica. Moreover, we might improve its β-carotene amount by a bioreactor in the future.
We improved the β-carotene production and installed cellulase ability (CBHI, CBHII, EgIII, EgI, EglA and BGS) at the same time. The cellulase from concentrated supernatants of P4-10-9-63Y-14B were only 1.53-fold lower than the commercial enzyme (Celluclast 1.5 L) purified from Trichoderma reesei. The commercial cellulases are produced by cellulolytic fungi, such as Trichoderma reesei and Aspergillus niger. Each host has its own limitation. For example, Neocallimastix patriciarum can produce highly efficient β-glucosidase but it is an obligate anaerobic rumen fungus47. Therefore, many studies engineered organisms with multiple cellulolytic properties to design an ideal cellulolytic host. Chang et al.43 engineered a cellulolytic K. marxianus with the similar cellulase genes as in this study43. The total cellulase activity of R. glutinis can be further improved by synthetic biology strategy. The functional analysis suggested that transformants also maintained its original lipid yield and salt-tolerance ability. To produce β-carotene, we need to culture for 7 days to get the highest β-carotene amounts. Our transformants can reach the same OD600 as wild type after 3 days in YP2D with 5% NaCl. So we suggested that we can produce higher β-carotene amount even in salty condition. We confirmed that the total lipid content of both the wild type and the transformants were unaffected (around 70% lipid yield), thus further supporting our statement. From these experiments, it seems that our engineering strategy is good. Several reports have described the synthesis of carotenoids, cellulases or lipid from fungi. However, in previous studies the hosts were usually engineered to produce a single product. Our goal is to turn cellulolytic wastes into β-carotene and renewable energy by a single host. In conclusion, we have upgraded the potential applications of R. glutinis for biorefinery using a synthetic biology approach and demonstrated that R. glutinis is a potential host for biorefinery.
Materials and Methods
Strains, media, and growth conditions
The oleaginous yeast R. glutinis BCRC 22360 (Bioresource Collection and Research Center, Taiwan) was kindly provided by Dr. Hong-Wei Yen (Tunghai University, Taiwan). R. glutinis was cultured in different growth media at 30 °C with 300 rpm for several days. The basic growth media contained 1% Yeast Extract (BactoDifco) and 1% Peptone (BactoDifco) as a nitrogen source and 2% dextrose (2D), 2% galactose (2 G) or 2% glycerol (2Gly) as a carbon source. For screening the R. glutinis transformants, a previously described medium was used48. The lipid inducing media (70 g/L glucose, 0.75 g/L yeast extract, 1.7 g/L yeast nitrogen base without amino acids and ammonium sulfate, and 0.1 g/L (NH4)2SO4, pH 5.6) were applied for analyzing the total lipid content in R. glutinis24.
Establishing a heterologous gene expression platform in R. glutinis
In this study, we intended to develop an efficient transformation tool for R. glutinis. R. glutinis electro-competent cells were prepared using two different methods. First, lithium acetate was used to prepare competent cells modified from previous studies for Kluyveromyces marxianus and Rhodotorula gracilis18,49. Briefly, R. glutinis cells were cultivated in 5 ml YP2D from the single colony at 30 °C with 300 rpm and then 0.2 OD cells were subcultured into 50 ml YP2D until reaching 0.6~1.4 OD. Cells were harvested at 3000 rpm for 3 min (4 °C) and washed with 5 ml ice-cold distilled H2O. Then, cells were resuspended in TMLSD buffer (10 mM Tris-HCl buffer (pH 8.0) containing 2 mM MgCl2, 100 mM lithium acetate, 270 mM sucrose, 10 mM dithiothreitol) and incubated at room temperature for 1 h. After the incubation, cells were harvested as described above and washed twice with TMS buffer (10 mM Tris-HCl buffer (pH 8.0) containing 2 mM MgCl2, 270 mM sucrose). Finally, competent cells were resuspended in TMS buffer and stored in −80 °C.
For the second method, we used the frozen protoplast protocol to engineer R. glutinis. The R. glutinis protoplast was prepared according to previous studies31,50,51 with some modifications. Briefly, single colonies of R. glutinis were inoculated in 50 mL of YP2D medium at 30 °C for 15 h. Cells were harvested at 3000 rpm for 10 min and suspended in 20 mL distilled H2O. Cells were harvested again as described above and gently resuspended in 10 mL 1 M sorbitol, followed by harvesting and suspending in 10 mL of sorbitol, sodium citrate, EDTA and β-mercaptoethanol (SCEM). Then, cells were mixed with 40 μl of lyticase solution (25,000U/ml) and incubated at 30 °C for 1 h. After the lyticase digestion cells were suspended in SCEM (109 cells/ml), 1 ml cell suspension was added 0.5 ml of lytic enzyme solution (1.5% (w/v) Zymolyase 60,000) for overnight culturing. Cells were centrifuged gently at 300 g for 5 min in round-bottom plastic tubes and suspended in 10 mL 1 M sorbitol by gently tapping the tube. Then cells were centrifuged at 300 g for 5 min, and the supernatant was discarded. This procedure was repeated to remove lyticase thoroughly. Finally, cells were suspended in 2 mL CaST solution (CaCl2, sorbitol and Tris-HCl) along with 2 mL cell-storage solution and protoplast cells were stored at −80 °C.
Electroporation
Electroporation was performed by mixing the 10–15 μl DNA with 50 μl competent cells or protoplasts and kept on ice for 15 min. Then cells were transferred to the ice-cold aluminum cuvette (0.2 cm gap Gene Pulser/MicroPulser Electroporation Cuvettes, Bio-Rad, USA) and electroporation was performed (1.2 kV or 400 V, 400 Ω and 25 μF capacitance), using a MicroPulser (Bio-Rad Laboratories, USA). After electroporation, cells were resuspended in 1 mL ice-cold YP2D and transferred into new tubes on ice for 15 min, and then incubated at 30 °C for 4 h. The cell suspension was spread onto YP2D plates containing selection markers (i.e. Kanamycin (G418), Zeocin or Hygromycin) and incubated at 30 °C for 4–5 days.
Transforming genes into the R. glutinis genome
The carotenoid biosynthetic pathway in Rhodotorula species has been well studied3,36. Generally, R. glutinis synthesizes β-carotene from the precursor acetyl-CoA (Supplementary Fig. S8). To increase the β-carotene amount in R. glutinis, the carotenogenic genes including geranylgeranyl pyrophosphate synthase (crtE), phytoene desaturase (crtI) and phytoene synthase/lycopene cyclase (crtYB) from Xanthophyllomyces dendrorhous (red yeast) were selected and integrated into the R. glutinis genome. Similarly, for improving the metabolic flux of carotenoid pathway, a tHMG1 (a truncated 3-hydroxy-3-methylglutaryl-coenzyme A reductase) gene was selected from Kluyveromyces marxianus27,52. The carotenogenic genes were in the pUC18 vector and the construction of each gene cassette was as in Chang et al.52. Briefly, the carotenogenic genes were cloned in between the yeast promoter and terminator using specific restriction enzymes. The gene cassettes were amplified using TransStart FastPfu Fly (Ultra) High-Fidelity DNA Polymerases and the primers pairs were listed in Table S2.
For the simultaneous improvement of β-carotene production and cellulose utilization ability of R. glutinis, three types of fungal cellulase genes were selected and integrated into the R. glutinis genome18,43. The cellulase genes included two cellobiohydrolases (CBHI and CBHII, from Trichoderma reesei), three endo-β-1,4-glucanases (EgIII, from T. reesei; EgI and EglA, from Aspergillus niger) and β-glucosidases (BGS, from Neocallimastix patriciarum). Each gene was fused with a secretion signal (α-factor) at the N-terminal for efficient secretion out from the cell. Each gene was flanked with independent inducible promoters and terminators (Table 1). The amplification of six cellulase gene cassettes used the same method as in the carotenogenic gene cassettes. Each cassette was in the pUC18 vector as in previous studies18,43. The gene cassettes were integrated using three selection marker genes: G418 resistance gene (neomycin phosphotransferase gene, KanMx), Zeocin resistance gene (Sh ble) and Hygromycin phosphotransferase gene (hph). The MIC of three different selection markers (i.e., Zeocin, G418 and Hygromycin) were tested on R. glutinis wild type using YP2D plates supplemented with different concentrations of antibiotics.
Transformant screening
The transformants were sub-cultured for 3 generations to select stable transformants. Then, transformants were mixed with QuickExtract ((QE), DNA Extraction Solution 1.0, Epicentre, USA) for rapid extraction of genomic DNA and used as a template for PCR verification. Each transformed functional genes were confirmed by PCR using the gene specific primer pairs (Supplementary Table S2). Finally, the transformants with integrated gene cassettes were screened for the β-carotene production and cellulase activity.
β-carotene extraction and Analytical methods
To analyze the β-carotene production in transformants, single colonies were inoculated into 5 ml medium48 and incubated at 30 °C, with 300 rpm for 7 days. Cells were harvested by centrifugation (6000 rpm, 10 min), lyophilized and suspended in 1 ml acetone. Cells were then subjected to mechanical disruption using MagNA Lyser (MagNA Lyser Instrument, Roche) at 6,000 rpm, 20 s for 3 times. For screening the transformants, 1 ml acetone was used in crude extraction. To estimate the maximum β-carotene amount of a transformant, the extraction step was repeated until colorless extracts appeared. The carotenoid extracts were analyzed using High-Performance Liquid Chromatography (HPLC) with a Nomura Chemical Develosil C30-UG Column (3 mm, ID 4.6 mm × L 250 mm- UG17346250W, Interlink Scientific Services, UK), using two buffer systems including buffer A: methanol/MTBE/water (81:15:4) and buffer B: methanol/MTBE/water (7:90:3) as mobile phases. The HPLC condition was described in previous studies52,53. The commercial free-form β-carotene and lycopene were used as the standards (Sigma–Aldrich Co. LLC, USA).
Cellulase activity assay
To screen the transformants with higher cellulase activity, a cellulase activity assay was conducted as described in previous studies18,54. Single colonies of transformants were incubated in 5 ml YP2D2G for 3 days and cells were harvested and washed twice with PBS. The washed cells were inoculated into 5 ml YP supplemented with 4% Sigmacell cellulose type 20 as the sole carbon source under 30 °C with 300 rpm for 7 days. The initial and final reducing sugar concentrations were analyzed using the Dinitrosalicylic (DNS) colorimetric method55,56. Similarly, total cellulase activity was demonstrated using different cellulosic substrates such as Sigmacell cellulose type 20, Sigmacell cellulose type 50 (SigmaAldrich, St.Louis, MO, USA) and cellulose microcrystalline (Merck, Darmstadt, Germany)54. For the cellulose degradation assay, the transformants and wild type R. glutinis were cultured in 50 ml YP2D2G for 3 days and supernatants were concentrated 50-fold using Vivaspin 20 (10,000 molecular weight cut off, PES membrane, GE Healthcare) at 4 °C. The commercial cellulases were used as a positive control, including cellulase powder from Aspergillus niger (≥0.3 units/mg solid, Sigma Aldrich C1184, USA) and cellulase liquid from Trichoderma reesei (≥700 units/g, Celluclast 1.5 L, Sigma Aldrich C2730, USA). The total protein concentration was determined by Bio-Rad Protein Assay Kit using the bovine serum albumin (BSA) as a standard. The assay reaction contained 32 μg protein, sodium acetate (50 mM, pH = 5) and cellulose substrates (2%, final concentration). The reaction mixture was incubated at 50 °C overnight. The initial and final reducing sugar amounts were analyzed by the DNS method.
Lipid content and salt tolerance analysis
The total lipid content of R. glutinis was determined using the method for Rhodosporidium toruloides with some modifications24,57. Briefly, R. glutinis wild type and transformants were cultured in 5 ml lipid-inducing medium for 3 days and cells were harvested, frozen and then freeze-dried. Then, the dry-cells were suspended in 1 ml chloroform/methanol (2:1 volumetric) and homogenized using a MagNA Lyser. The homogenized samples were then mixed with 0.2 ml ddH2O and vortexed for 15 s. The organic layer was taken using a needle and washed with 0.1 ml NaCl (0.1%, w/v) solution. The extract was repeated until the clear organic layer appeared and was dried in a hood at room temperature overnight followed by 1 h at 80 °C in a pre-weighed tube. The total lipid weight was determined. To determine the salt-tolerance of transformants, 5% NaCl was added to the YP2D media and the growth OD was measured every 24 h in 4 days.
Electronic supplementary material
Acknowledgements
The authors are thankful to Biodiversity Research Center, Academia Sinica, Taiwan for providing laboratory facility to carry out the entire research. The financial support for this study was provided by the Ministry of Science and Technology (MOST), Taiwan (Grant No. MOST 104-2621-M-039-001, MOST 105-2621-M-039 -001 -MY2, MOST 104-2311-B-039 -001 -MY3, MOST 103-2621-M-029 -007) and China Medical University Hospital (Grant No. DMR-106-120).
Author Contributions
J.-J.C., H.-W.P. and Y.-J.L. proposed the project and W.-H.L. supervised the study. J.-J.C. and H.-W.P. designed the experiments. H.-W.P. did the experiments. H.-W.P. and Y.-Y.K. conducted the analyses of carotenoids. H.-W.P. and M.A. conducted the analyses of cellulase activity. H.-W.P. and M.A. wrote the manuscript and revised by W.-H.L.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-29194-z.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jui-Jen Chang, Email: lancecjj@gmail.com.
Wen-Hsiung Li, Email: whli@uchicago.edu.
References
- 1.Li Z, et al. Overexpression of malic enzyme (ME) of Mucor circinelloides improved lipid accumulation in engineered Rhodotorula glutinis. Applied Microbiology and Biotechnology. 2013;97:4927–4936. doi: 10.1007/s00253-012-4571-5. [DOI] [PubMed] [Google Scholar]
- 2.Dai, C.-c., Tao, J., Xie, F., Dai, Y.-j. & Zhao, M. Biodiesel generation from oleaginous yeast Rhodotorula glutinis with xylose assimilating capacity. African Journal of Biotechnology6 (2007).
- 3.Kot AM, Błażejak S, Kurcz A, Gientka I, Kieliszek M. Rhodotorula glutinis—potential source of lipids, carotenoids, and enzymes for use in industries. Applied Microbiology and Biotechnology. 2016;100:6103–6117. doi: 10.1007/s00253-016-7611-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta A, Vongsvivut J, Barrow CJ, Puri M. Molecular identification of marine yeast and its spectroscopic analysis establishes unsaturated fatty acid accumulation. Journal of Bioscience and Bioengineering. 2012;114:411–417. doi: 10.1016/j.jbiosc.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Yen H-W, Liao Y-T, Liu YX. The growth of oleaginous Rhodotorula glutinis in an airlift bioreactor on crude glycerol through a non-sterile fermentation process. Bioprocess and Biosystems Engineering. 2015;38:1541–1546. doi: 10.1007/s00449-015-1396-5. [DOI] [PubMed] [Google Scholar]
- 6.Business_Communications_Company. The Global Market for Carotenoids: FOD025D (2011).
- 7.Desobry SA, Netto FM, Labuza TP. Preservation of β-Carotene from Carrots. Critical Reviews in Food Science and Nutrition. 1998;38:381–396. doi: 10.1080/10408699891274255. [DOI] [PubMed] [Google Scholar]
- 8.Mannazzu I, Landolfo S, da Silva TL, Buzzini P. Red yeasts and carotenoid production: outlining a future for non-conventional yeasts of biotechnological interest. World Journal of Microbiology and Biotechnology. 2015;31:1665–1673. doi: 10.1007/s11274-015-1927-x. [DOI] [PubMed] [Google Scholar]
- 9.Yen H-W, Yang Y-C, Yu Y-H. Using crude glycerol and thin stillage for the production of microbial lipids through the cultivation of Rhodotorula glutinis. Journal of Bioscience and Bioengineering. 2012;114:453–456. doi: 10.1016/j.jbiosc.2012.04.022. [DOI] [PubMed] [Google Scholar]
- 10.Schneider T, et al. Lipid and carotenoid production by oleaginous red yeast Rhodotorula glutinis cultivated on brewery effluents. Energy. 2013;61:34–43. doi: 10.1016/j.energy.2012.12.026. [DOI] [Google Scholar]
- 11.Karamerou EE, Theodoropoulos C, Webb C. A biorefinery approach to microbial oil production from glycerol by Rhodotorula glutinis. Biomass and Bioenergy. 2016;89:113–122. doi: 10.1016/j.biombioe.2016.01.007. [DOI] [Google Scholar]
- 12.Malisorn C, Suntornsuk W. Optimization of β-carotene production by Rhodotorula glutinis DM28 in fermented radish brine. Bioresource Technology. 2008;99:2281–2287. doi: 10.1016/j.biortech.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 13.Braunwald T, et al. Effect of different C/N ratios on carotenoid and lipid production by Rhodotorula glutinis. Applied Microbiology and Biotechnology. 2013;97:6581–6588. doi: 10.1007/s00253-013-5005-8. [DOI] [PubMed] [Google Scholar]
- 14.Bhosale P, Gadre RV. Optimization of carotenoid production from hyper-producing Rhodotorula glutinis mutant 32 by a factorial approach. Letters in Applied Microbiology. 2001;33:12–16. doi: 10.1046/j.1472-765X.2001.00940.x. [DOI] [PubMed] [Google Scholar]
- 15.Bhosale P, Gadre RV. Production of β-carotene by a Rhodotorula glutinis mutant in sea water medium. Bioresource Technology. 2001;76:53–55. doi: 10.1016/S0960-8524(00)00075-4. [DOI] [PubMed] [Google Scholar]
- 16.Bhosale PB, Gadre RV. Production of β-carotene by a mutant of Rhodotorula glutinis. Applied Microbiology and Biotechnology. 2001;55:423–427. doi: 10.1007/s002530000570. [DOI] [PubMed] [Google Scholar]
- 17.Yolmeh M, Khomeiri M. Using physical and chemical mutagens for enhanced carotenoid production from Rhodotorula glutinis (PTCC 5256). Biocatalysis and Agricultural. Biotechnology. 2016;8:158–166. [Google Scholar]
- 18.Chang J-J, et al. Assembling a cellulase cocktail and a cellodextrin transporter into a yeast host for CBP ethanol production. Biotechnology for Biofuels. 2013;6:19. doi: 10.1186/1754-6834-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei H, et al. Engineering towards a complete heterologous cellulase secretome in Yarrowia lipolytica reveals its potential for consolidated bioprocessing. Biotechnology for Biofuels. 2014;7:148. doi: 10.1186/s13068-014-0148-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herculano PN, et al. Isolation of cellulolytic fungi from waste of castor (Ricinus communis L.) Current microbiology. 2011;62:1416–1422. doi: 10.1007/s00284-011-9879-3. [DOI] [PubMed] [Google Scholar]
- 21.Yu X, Zheng Y, Dorgan KM, Chen S. Oil production by oleaginous yeasts using the hydrolysate from pretreatment of wheat straw with dilute sulfuric acid. Bioresource Technology. 2011;102:6134–6140. doi: 10.1016/j.biortech.2011.02.081. [DOI] [PubMed] [Google Scholar]
- 22.Lin X, et al. Functional integration of multiple genes into the genome of the oleaginous yeast Rhodosporidium toruloides. FEMS yeast research. 2014;14:547–555. doi: 10.1111/1567-1364.12140. [DOI] [PubMed] [Google Scholar]
- 23.Johns, A. M. B., Love, J. & Aves, S. J. Four Inducible Promoters for Controlled Gene Expression in the Oleaginous Yeast Rhodotorula toruloides. Frontiers in Microbiology 7, 10.3389/fmicb.2016.01666 (2016). [DOI] [PMC free article] [PubMed]
- 24.Zhang S, Ito M, Skerker JM, Arkin AP, Rao CV. Metabolic engineering of the oleaginous yeast Rhodosporidium toruloides IFO0880 for lipid overproduction during high-density fermentation. Applied Microbiology and Biotechnology. 2016;100:9393–9405. doi: 10.1007/s00253-016-7815-y. [DOI] [PubMed] [Google Scholar]
- 25.Park Y-K, Nicaud J-M, Ledesma-Amaro R. The Engineering Potential of Rhodosporidium toruloides as a Workhorse for Biotechnological Applications. Trends in Biotechnology. 2018;36:304–317. doi: 10.1016/j.tibtech.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 26.Guo Z, et al. Development of cellobiose-degrading ability in Yarrowia lipolytica strain by overexpression of endogenous genes. Biotechnology for Biofuels. 2015;8:109. doi: 10.1186/s13068-015-0289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verwaal R, et al. High-level production of beta-carotene in Saccharomyces cerevisiae by successive transformation with carotenogenic genes from Xanthophyllomyces dendrorhous. Applied and Environmental Microbiology. 2007;73:4342–4350. doi: 10.1128/AEM.02759-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao S, et al. Iterative integration of multiple-copy pathway genes in Yarrowia lipolytica for heterologous β-carotene production. Metabolic Engineering. 2017;41:192–201. doi: 10.1016/j.ymben.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 29.Larroude, M. et al. A synthetic biology approach to transform Yarrowia lipolytica into a competitive biotechnological producer of β‐carotene. Biotechnology and bioengineering (2017). [DOI] [PubMed]
- 30.Li J, et al. Discovery of Several Novel Targets that Enhance β-Carotene Production in Saccharomyces cerevisiae. Frontiers in Microbiology. 2017;8:1116. doi: 10.3389/fmicb.2017.01116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nickoloff, J. A. Electroporation protocols for microorganisms. Vol. 47 (Springer Science & Business Media, 1995).
- 32.Smiley JA, Kundracik M, Landfried DA, Barnes VR, Axhemi AA. Genes of the thymidine salvage pathway: thymine-7-hydroxylase from a Rhodotorula glutinis cDNA library and iso-orotate decarboxylase from Neurospora crassa. Biochimica et Biophysica Acta (BBA)-General Subjects. 2005;1723:256–264. doi: 10.1016/j.bbagen.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Koh CMJ, Liu Y, Du M, Ji L. Molecular characterization of KU70 and KU80 homologues and exploitation of a KU70-deficient mutant for improving gene deletion frequency in Rhodosporidium toruloides. BMC microbiology. 2014;14:50. doi: 10.1186/1471-2180-14-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu H, et al. Fast and efficient genetic transformation of oleaginous yeast Rhodosporidium toruloides by using electroporation. FEMS yeast research. 2017;17:fox017. doi: 10.1093/femsyr/fox017. [DOI] [PubMed] [Google Scholar]
- 35.Abbott EP, Ianiri G, Castoria R, Idnurm A. Overcoming recalcitrant transformation and gene manipulation in Pucciniomycotina yeasts. Applied Microbiology and Biotechnology. 2013;97:283–295. doi: 10.1007/s00253-012-4561-7. [DOI] [PubMed] [Google Scholar]
- 36.Frengova G, Beshkova D. Carotenoids from Rhodotorula and Phaffia: yeasts of biotechnological importance. Journal of Industrial Microbiology & Biotechnology. 2009;36:163–180. doi: 10.1007/s10295-008-0492-9. [DOI] [PubMed] [Google Scholar]
- 37.Zoz L, Carvalho JC, Soccol VT, Casagrande TC, Cardoso L. Torularhodin and torulene: bioproduction, properties and prospective applications in food and cosmetics-a review. Brazilian Archives of Biology and Technology. 2015;58:278–288. doi: 10.1590/S1516-8913201400152. [DOI] [Google Scholar]
- 38.Moran NA, Jarvik T. Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science. 2010;328:624–627. doi: 10.1126/science.1187113. [DOI] [PubMed] [Google Scholar]
- 39.Weber RW, Anke H, Davoli P. Simple method for the extraction and reversed-phase high-performance liquid chromatographic analysis of carotenoid pigments from red yeasts (Basidiomycota, Fungi) Journal of Chromatography A. 2007;1145:118–122. doi: 10.1016/j.chroma.2007.01.052. [DOI] [PubMed] [Google Scholar]
- 40.Verdoes JC, et al. Metabolic engineering of the carotenoid biosynthetic pathway in the yeast Xanthophyllomyces dendrorhous (Phaffia rhodozyma) Applied and Environmental Microbiology. 2003;69:3728–3738. doi: 10.1128/AEM.69.7.3728-3738.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhosale P, Gadre R. β-Carotene production in sugarcane molasses by a Rhodotorula glutinis mutant. Journal of Industrial Microbiology and Biotechnology. 2001;26:327–332. doi: 10.1038/sj.jim.7000138. [DOI] [PubMed] [Google Scholar]
- 42.Chang J-J, et al. PGASO: A synthetic biology tool for engineering a cellulolytic yeast. Biotechnology for Biofuels. 2012;5:53. doi: 10.1186/1754-6834-5-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang JJ, et al. Constructing a cellulosic yeast host with an efficient cellulase cocktail. Biotechnology and bioengineering. 2018;115:751–761. doi: 10.1002/bit.26507. [DOI] [PubMed] [Google Scholar]
- 44.Meng X, et al. Biodiesel production from oleaginous microorganisms. Renewable Energy. 2009;34:1–5. doi: 10.1016/j.renene.2008.04.014. [DOI] [Google Scholar]
- 45.Reyes LH, Gomez JM, Kao KC. Improving carotenoids production in yeast via adaptive laboratory evolution. Metabolic Engineering. 2014;21:26–33. doi: 10.1016/j.ymben.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 46.Nam H-K, Choi J-G, Lee J-H, Kim S-W, Oh D-K. Increase in the production of β-carotene in recombinant Escherichia coli cultured in a chemically defined medium supplemented with amino acids. Biotechnology Letters. 2013;35:265–271. doi: 10.1007/s10529-012-1072-7. [DOI] [PubMed] [Google Scholar]
- 47.Chen H-L, et al. A highly efficient β-glucosidase from the buffalo rumen fungus Neocallimastix patriciarum W5. Biotechnology for Biofuels. 2012;5:24. doi: 10.1186/1754-6834-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marova I, et al. Influence of exogenous stress factors on production of carotenoids by some strains of carotenogenic yeasts. Annals of Microbiology. 2004;54:73–86. [Google Scholar]
- 49.Takahashi S, Okada H, Abe K, Kera Y. Genetic transformation of the yeast Rhodotorula gracilis ATCC 26217 by electroporation. Applied Biochemistry and Microbiology. 2014;50:624–628. doi: 10.1134/S0003683814110040. [DOI] [Google Scholar]
- 50.Seh ML, Kenerley CM. Protoplats isolation and regeneration and nuclear staining of mycoparasitic Gliocladium species. Journal of Microbiological Methods. 1988;8:121–130. doi: 10.1016/0167-7012(88)90013-9. [DOI] [Google Scholar]
- 51.Burgers PMJ, Percival KJ. Transformation of yeast spheroplasts without cell fusion. Analytical Biochemistry. 1987;163:391–397. doi: 10.1016/0003-2697(87)90240-5. [DOI] [PubMed] [Google Scholar]
- 52.Chang J-J, et al. Integrating an algal β-carotene hydroxylase gene into a designed carotenoid-biosynthesis pathway increases carotenoid production in yeast. Bioresource Technology. 2015;184:2–8. doi: 10.1016/j.biortech.2014.11.097. [DOI] [PubMed] [Google Scholar]
- 53.Lin, Y.-J. et al. Metabolic Engineering a Yeast to Produce Astaxanthin. Bioresource Technology (2017). [DOI] [PubMed]
- 54.Worthington, C. C. Worthington enzyme manual: enzymes and related biochemicals. (Worthington Biochemical Corporation, 1988).
- 55.Zhang Y-HP, Himmel ME, Mielenz JR. Outlook for cellulase improvement: screening and selection strategies. Biotechnology advances. 2006;24:452–481. doi: 10.1016/j.biotechadv.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 56.Eveleigh DE, Mandels M, Andreotti R, Roche C. Measurement of saccharifying cellulase. Biotechnology for Biofuels. 2009;2:21. doi: 10.1186/1754-6834-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang S, et al. Engineering Rhodosporidium toruloides for increased lipid production. Biotechnology and bioengineering. 2016;113:1056–1066. doi: 10.1002/bit.25864. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.