Abstract
Marine diseases have major impacts on ecosystems and economic consequences for aquaculture and fisheries. Understanding origin, spread and risk factors of disease is crucial for management, but data in the ocean are limited compared to the terrestrial environment. Here we investigated how the marine environment drives the spread of viral disease outbreak affecting The Pacific oyster worldwide by using a spatial epidemiology framework. We collected environmental and oyster health data at 46 sites spread over an area of 300 km2 along an inshore-offshore gradient during an epizootic event and conducted risk analysis. We found that disease broke out in the intertidal farming area and spread seaward. Mortalities and virus detection were observed in oysters placed 2 km from the farming areas, but oysters of almost all sites were subclinically infected. Increasing food quantity and quality, growth rate and energy reserves of oyster were associated with a lower risk of mortality offshore whereas increasing turbidity, a proxy of the concentration of suspended particulate matter, and terrestrial inputs, inferred from fatty acid composition of oysters, were associated with a higher risk of mortality. Offshore farming and maintenance of good ecological status of coastal waters are options to limit disease risk in oysters.
Introduction
Since the mid‐1970s, disease epidemics and mass mortalities have been occurring in marine environments at a historically unprecedented rate1 and have economic consequences for fisheries and aquaculture2. Understanding where, when and why outbreaks occur and the means by which they spread is crucial for proposing effective disease management. Nevertheless, data on pathogen origin and spread in the ocean are limited3. The few cases in which information is available indicate that disease may spread at least as rapidly as any terrestrial epidemic, reflecting, among other things, the relative openness of marine systems compared to terrestrial ones4. The most emblematic case is the herpesvirus that spread through pilchard populations in Australia at a rate in excess of 10 000 km year−1 5.
The risk of disease outbreak depends on interactions between hosts, pathogens, and the environment, and any change in one or more of these components may potentially increase or decrease this risk6. Climate change related factors such as increasing temperature, rainfall anomalies and storms and acidification drive host-pathogen interactions in the marine environment and infectious disease outbreaks affecting corals, shellfish, finfish and humans6. Anthropogenic factors like enhanced terrestrial inputs from runoff, nutrient load and pollutants, and aquaculture practices have also significant implications for the emergence and spread of diseases. Ultimately, disease outbreaks in a changing ocean can be shaped by multiple factors acting on the host and the pathogen simultaneously.
Ostreid herpesvirus type 1 (OsHV-1) infecting the Pacific oyster Crassostrea gigas has gained considerable attention during the last 10 years due to economic costs associated with increased mortalities in farmed animals worldwide7,8. In addition to being socio-economically devastating, oyster diseases affect overall ecosystem productivity and health. Oysters can act as keystone species by providing ecosystem services such as, controlling phytoplankton bloom and turbidity, reducing bank erosion and providing shelters and habitats for other species9.
A great deal of work has been done on OsHV-1 risk factors but information gaps still remain7,8. One risk factor that is virtually unknown is the potential for long-distance dispersal of OsHV-1 disease. Yet, this is a fundamental prerequisite for building epidemiological models and evaluating the effectiveness of disease management scenarios. In the marine environment, distance over which viruses can be transmitted is generally limited by the interaction of hydrodynamics with viral shedding and decay rates10. Decay rates of marine viruses and persistence outside the host vary with environmental factors as seawater temperature, salinity, solar radiation, natural bacterial communities, suspended particulates and grazing11,12.
Knowledge gaps remain in the study of persistence of viruses in the marine environment in part as most experiments have been conducted at local scale whereas disease spread and risk factors in the marine environment are affected by processes that occur at broader regional scale such as hydrodynamic circulation and connectivity patterns4. Our objective is to investigate the origin and spread of a marine viral disease and to identify risk factors that affect disease dynamics at the regional scale using a spatial epidemiology framework.
Spatial epidemiology consists in the description and analysis of geographical variations in disease to identify risk factors that could explain these patterns13,14. Since the first disease maps produced during the nineteen century to identify causes of plague, yellow fever and cholera15, spatial epidemiology grew in complexity and utility, and recently benefited from advances in data availability, analytical methods, and popularization of geographic information systems (GIS)13. Spatial epidemiology is now playing an increasingly important role in our understanding of the relationship between diseases and the terrestrial environment, but the application of such analytical framework in the marine environment is still in its infancy.
Pacific oysters are well suited for spatial epidemiology because they are sessile, widely distributed in temperate waters and they survive considerable environmental fluctuations9. Also, the recent development of specific pathogen free (SPF, low Vibrio load, absence of detectable OsHV-1) oysters has enabled assessment of the infection process under natural conditions16–18. The SPF oysters are descendants of a pool of genitors produced under controlled conditions to minimize the influence of genetic and environmental parameters that could affect the host sensitivity to the disease.
In the present study, mortality and OsHV-1 DNA were monitored in SPF C. gigas deployed at 46 sites spread over a 300 km2 surface area along an inshore-offshore gradient. Seawater temperature, salinity, chlorophyll-a fluorescence, turbidity, dissolved oxygen, bacteria and vibrio concentrations and host parameters (bacteria and vibrio concentrations, energy reserves, food quality and growth rate) were regularly measured to investigate disease risk factors. Altogether, this dataset was used to quantify the influence of environmental variability on disease origin and spread in the coastal ocean.
Method
Experimental design
SPF Pacific oysters were produced under controlled conditions until they reached the age of 8 months and wet weight of 0.51 g16–18 (File S1). The oysters were screened using an OsHV-1-specific quantitative PCR assay19 at the different stages of production (3, 7 and 8 months). No OsHV-1 DNA was detected.
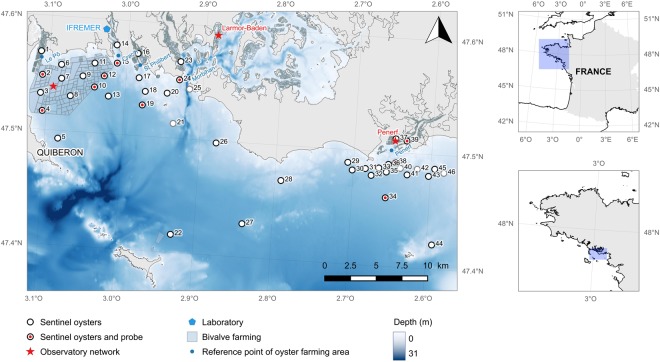
The SPF oysters (also called “sentinel oysters”) were deployed at 46 sites located along an inshore-offshore gradient in the Mor-Braz area, South Brittany (France) before the start of the OsHV-1 disease induced mortality event on 8 April 2013 for 171 d until 26 September 2013 (Fig. 1). Five sites were in intertidal oyster farming areas (1, 14, 16, 23 and 37) and 41 sites were offshore. Offshore sites were generally free of oyster farming except in the Quiberon bay where oysters are farmed on-bottom. At each site, 16 small mesh bags containing 85 individual oysters were grouped in one big mesh bag. These bags were attached to iron tables for the sites situated in the intertidal farming area or immersed vertically at 2 meters deep and attached to a mooring point for the sites in the offshore area (File S1). Seawater temperature was measured every 30 minutes during the entire experiment using SP2T recorders (NKE instrumentation, Hennebont, France) placed in ten oyster bags evenly spread throughout the study area (Fig. 1).
Figure 1.
Site map. Sentinel oysters were deployed at 46 sites represented by circles in the Mor-Braz area in southern Brittany, France. The grey circles represent the sites where the sentinel oysters were accidentally lost and where only the seawater data are available. Seawater temperature was measured every 30 minutes during the entire experiment in ten oyster bags evenly spread throughout the study area. In addition, oysters from the shellfish observatory network were monitored at two sites located in the intertidal zone (Pénerf and Larmor-Baden) and at one site in the subtidal zone (Men-er-Roué). This map was done with the open source QGIS ver. 2.18.9. (QGIS Development Team, 2016, https://qgis.org).
Sites were visited 15 times on 30 April, 15 and 27 May, 7, 14, 20 and 27 June, 4, 11, 19 and 25 July, 1, 13 and 31 August, and 26 September by means of two vessels. One boat started from the Ifremer laboratory and covered the Quiberon Bay while the other one started from the Pénerf River (site 39) up until reaching sites 27, 44 and 46 (Fig. 1). Sampling cruises were conducted at low tide slack water ±2 hours and lasted for 3 to 4 hours each.
At each visit, environmental parameters (temperature, salinity, fluorescence, turbidity and oxygen) were measured at the vicinity of the oyster bags using a multiparameter probe (MP6, NKE instrumentation). Data were acquired every 10 seconds over a period of 3–5 minutes per site. During this time, a 50-mL sterile Falcon tube (Dutsher, Issy-les-Moulineaux, France) attached to the frame of the probe was filling with seawater by means of a perforated cap for further bacterial analyses. The seawater sample was screw-capped and stored in a cooler. In a separate cooler, one small bag of oysters was placed and held at ambient seawater temperature.
Upon arrival in the laboratory at la Trinité-sur-Mer, seawater samples were plated on marine broth medium to quantify cultivable bacteria and on thiosulfate-citrate-bile salts-sucrose agar (TCBS) to quantify vibrios (File S2). Living and dead oysters were counted to evaluate survival. Living oysters were divided in three sample groups dedicated to (1) biometry, biochemical analyses and OsHV-1 DNA detection, (2) bacterial analyses and (3) monitoring of survival and detection of OsHV-1 DNA in laboratory conditions at 21 °C.
Individual shell length and wet mass were measured on a sub-sample of 25 living oysters per bag. The soft tissues of these oysters were removed from the shells, pooled together, dipped into liquid nitrogen and stored at −80 °C until laboratory analyses. The remaining shells were dried overnight at room temperature and weighted. Biochemical analyses were conducted on samples collected prior to the mortality event (27 May, 7, 14 and 20 June) whereas OsHV-1 DNA detection were conducted on samples collected prior to and during the mortality event (27 May, 20 June and 13 August). Cultivable bacteria and vibrios were quantified on a sub-sample of 10 alive oysters per bag for every sampling times17. Soft tissues were pooled, weighed, homogenized in sterile artificial seawater and plated on marine broth medium for cultivable bacteria and on TCBS for vibrios (File S2).
The remaining oysters (mean n = 42 individuals ±8) from each site were brought back to the Ifremer hatchery in Argenton where they were maintained at 21 °C for 12 d days to reveal subclinical infection i.e. asymptomatic carriers of OsHV-117 (File S3). Oysters collected on 27 May, 7, 14, 20 and 27 June, and 4, 11 and 19 July were tested. These dates corresponded (1) to times of the year when seawater temperature was lower than 16 °C (27 May and 7 June), a threshold temperature above which disease transmission was optimal and mortalities occurred, (2) to the onset of the mass mortality (14 and 20 June) and (3) to the spread of the mortality event (from 27 June onwards). The oysters were placed in 5 L jars (one for each site) filled with UV sterilized and 1 μm-filtered seawater and covered with aluminium foil. All the jars were placed in the same room with controlled air temperature. Seawater was renewed twice a day and a phytoplankton mixture was added at each water renewal (File S3). Dead animals were counted every 0.5 to 2 days. There was no cross-infection among jars (sites), and the mortality events observed under laboratory conditions were associated with OsHV-1 DNA detection (File S3).
The mooring deployed at sites 21, 25 and 42 were accidentally lost at the beginning of the experiment on 15 May. Also, the oyster bags deployed at sites 46 and 40 were lost on 7 June and 14 June respectively. Since the mooring system was still in place at site 46, new SPF oysters were added on 14 June. The data collected at these five sites were not taken into account in the risk analyses. However, the environmental and microbiological parameters of the seawater for these sites were recorded during the entire period of study.
To supplement our experimental design regarding OsHV-1 detection, the temporal evolution of OsHV-1 DNA level in three pools of three oysters collected twice a month from early May to mid-September at two sites in the intertidal zone and at one site in the subtidal in the Mor-Braz area (Fig. 1) obtained by the oyster observatory network is presented here (File S4).
Laboratory analyses
Pooled oysters (n = 25 individuals) were ground with a MM400 homogeniser (Retsch, Eragny, France) under liquid nitrogen, and the resulting powder was sub-sampled for pathogen detection and biochemical analyses (File S2). The detection and quantification of OsHV-1 DNA was carried out using a real-time PCR protocol19. Biochemical analyses consist in the quantification of energetic reserves (carbohydrates and neutral lipids) and fatty acid trophic markers. Carbohydrates were quantified by spectrophotometry and expressed in mg g−1 dry mass of tissues. Neutral lipid classes were analysed by high performance thin layer chromatography (File S4). Identified compounds were triacylglycerol (TAG) and sterols (ST). The TAG-ST ratio is a proxy for the relative contribution of lipid reserve to structure. Finally, fatty acid composition of neutral lipids was analysed by gas-chromatography. The selected fatty acid trophic markers are extensively used in ecology20 and were related to diatoms (16:1n-7/16:0 and 20:5n-3/22:6n-3); terrestrial inputs (18:2n-6 + 18:3n-3), animal tissues (carnivory, 18:1n-9/18-1n-7); freshness (polyunsaturated/saturated fatty acid, PUFA/SFA), and bacteria (sum of branched chain fatty acids and unbranched 15:0 and 17:0).
Data analyses and statistics
Analyses were conducted using SAS 9.4 (SAS institute, Carry, USA). The proportion of surviving oysters was analyzed as a function of time for each site. Survival (S) was fitted a nonlinear regression model according to the following equations:
| 1 |
where α is the mean survival before the appearance of a mortality event, d is the number of days since the deployment of oyster on the field, D0 is the number of days before the mortality event (also referred to as mortality-free time), β is the daily variation of survival of oysters during the mortality event, and δ is the duration of the mortality event (File S5).
Shell length, total body mass, shell mass and flesh mass of oysters were fitted using segmented regression models according to the following equations:
| 2 |
where b0 is the intercept, b1 and b2 are the regression coefficients before and after the mortality event respectively. The regression coefficients b1 and b2 were different for all parameters excepted for total body mass where a simple regression model was fitted to the data (File S6). When no mortality occurred, was set at the mean value (84.3 d, File S5). Each biometrical parameter was expressed on a weekly basis.
The relationship between the mortality-free time (D0) and the covariates of interest were investigated using the univariate Cox proportional hazards regression model21. These covariates consist of the environmental, microbiological, biochemical and biometrical parameters previously described. When a variable was acquired more than once between deployment and D0, the mean value and the value recorded before the onset of mortality were used, except for the biometrical parameters where the overall growth rates (the regression coefficient b1) were used. For the sites where no significant mortality was observed, D0 was censored and set at 171 d, and only covariate measures taken before 84.3 d were considered (File S5). The relationship between β, δ and the covariates were investigated using Spearman correlation coefficients.
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).
Results
Origin and spread of disease-induced mortality of oysters
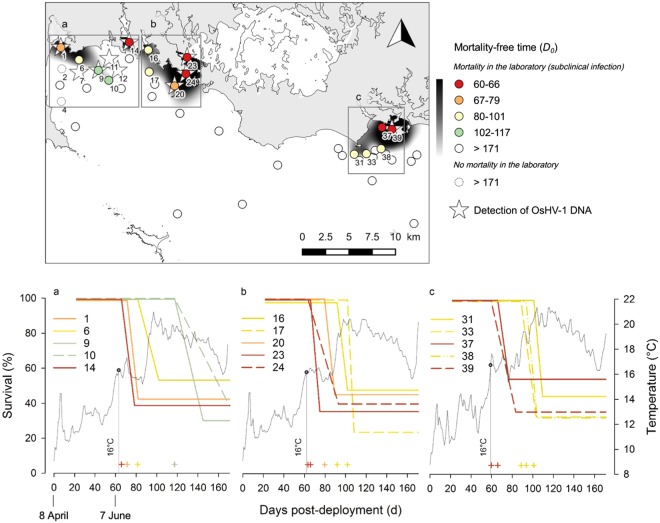
The sentinel oysters were affected by the mass mortality event at 15 sites out of 46 (Fig. 2, File S5). Mortality initiated inshore in oyster farming areas when average daily seawater temperature reached 16 °C (after 7 June, 60 d after deployment), and gradually spread seaward. The number of days before the mortality event (D0) varied from 60–66 d in inshore oyster farms (sites 14, 23 and 37) and in two adjacent sites (24 and 39), to 117 d offshore (sites 9 and 10) while seawater temperature was higher than 19 °C. The final survival of oysters at these 15 sites varied from 23.7 to 54.2% (mean value of 39.6 ± 8.5% s.d.).
Figure 2.
Survival of sentinel oysters in the field, detection of OsHV-1 DNA and subclinical infection. Top: map of the number of days before the mortality event in the field (D0). For the sites where no mortality was observed, D0 was reported as greater than 171 d. Plain circles indicate that mortality occurred at least once in the laboratory (subclinical infection) whereas dashed circles show the two sites where no mortality occurred in the laboratory. Bottom: fitted survival curves of the sentinel oysters at the sites where a mortality event was observed (left axis). The plus signs indicate D0 values for each survival curve. Daily mean seawater temperature recorded in each zone (right axis).
Overall, the spatio-temporal distribution of oyster mass mortality coincided with the general spread of OsHV-1 in oysters. OsHV-1 DNA was not detected in oyster tissues before the mortality outbreak on 27 May (49 d after deployment, averaged seawater temperature 12.8 °C), whereas it was detected on 20 June (73 d, averaged seawater temperature 16.5 °C) in 6 sites where mortalities were either already occurring (sites 1, 14, 23, 24 and 39) or upcoming (site 20, Table 1). Later on, a high level of OsHV-1 DNA (>107 cp mg−1) was detected on 13 August (127 d) in oysters from site 9 where mortality had occurred 10 days before the sampling. At the same date, low levels of OsHV-1 DNA (>104 cp mg−1) were observed in oyster samples of neighboring sites where no mortality was noticed (sites 7, 11 and 12). OsHV-1 DNA was not detected in sites 6, 10, 16, 17, 31, 33 and 38 where mortality occurred between 82–117 d, suggesting that the detection of OsHV-1 DNA was missed because of the large sampling interval. The data from the oyster observatory network confirms that the mortality (i) was associated with the presence of OsHV-1, (ii) started in oyster farms located inshore, and (iii) spread offshore (File S4).
Table 1.
Detection of OsHV-1 DNA according to site and time in oysters deployed in the Mor-Braz.
| Site | OsHV-1 DNA (cp mg−1) | ||
|---|---|---|---|
| 27 May (49d) | 20 June (73d) | 13 August (127d) | |
| 1 | nd | 2.4 × 105 | nd |
| 7 | nd | nd | 1.4 × 103 |
| 9 | nd | nd | 1.3 × 107 |
| 11 | nd | nd | 2.3 × 103 |
| 12 | nd | nd | 7.0 × 103 |
| 14 | nd | 1.8 × 105 | nd |
| 20 | nd | 8.7 × 104 | nd |
| 23 | nd | 7.6 × 103 | nd |
| 24 | nd | 2.3 × 105 | nd |
| 39 | nd | 2.6 × 104 | nd |
| All other sites (n = 36) | nd | nd | nd |
Quantification of OsHV-1 DNA was carried out in oyster tissues sampled before, during and at the end of the mortality event on 27 May, 20 June and 13 August respectively. Abbreviation: nd, not detected (<102 cp mg−1).
The relationship between oyster mortality and OsHV-1 was strengthened by the results of the challenges performed in laboratory conditions to reveal subclinical infection. When exposed during 12 days to a seawater temperature of 21 °C in the laboratory, the sentinel oysters collected on 27 May (49 d) showed no mortality (File S3). This is consistent with the lack of mortality and of OsHV-1 DNA detection in the field (Fig. 2, Table 1). Oysters collected on 7 June (60 d) exhibited significant mortality in the laboratory for 5 sites located within the inshore oyster farming areas (sites 14, 16, 23 and 37) or very close from them (site 39), suggesting that these animals were infected by OsHV-1 (File S3). Oysters deployed at these sites were the first to be hit by the mortality outbreak (Fig. 2), with the exception of oysters from site 16 (where D0 = 92 d). The oysters collected on 14 June and afterward (>67 d) showed significant mortalities in the laboratory, both for inshore farming sites and for several offshore sites located far away from the farms where no mortality was recorded on the field (e.g. sites 26, 28, 44). Interestingly, in laboratory conditions, oysters showed significant mortalities associated with OsHV-1 DNA detection (File S3) in 40 out of the 42 sites tested (95%), whereas only 15 sites (35%) were affected in the field. This reflects that (i) OsHV-1 can persist within an oyster without generating mortality in natural conditions (subclinical infection), and (ii) the spatial dispersion of the virus is much larger than the mortality pattern (Fig. 2).
Spatial and temporal variations in environmental and host parameters
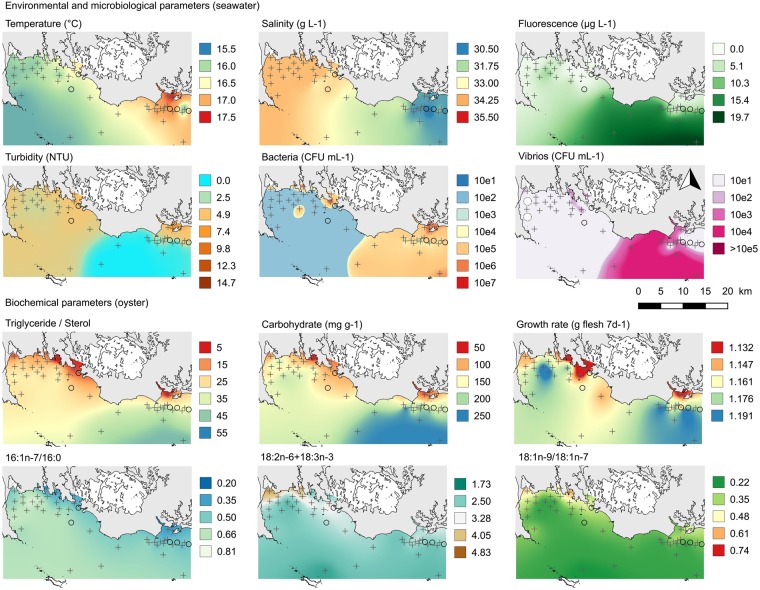
The whole dataset of physical, microbiological, biochemical and biometrical parameters was mapped (File S7), and selected variables measured on the 20 June (73 d), during the mortality event are presented (Fig. 3). From 30 April to 25 July average seawater temperature increased from 11.2 °C to 20.0 °C, and decreased thereafter. There was a thermal gradient seaward, and the Penerf area appeared slightly warmer than the Quiberon Bay (Fig. 3). Salinity varied from 29.8‰ to 35.4‰ (Fig. S7.2). Chlorophyll-a fluorescence was generally higher in the Pernerf area than in the Quiberon Bay and peaked in the eastern part of the study area on 20 June (Fig. 3). This phytoplankton patch was characterized by low salinity, low turbidity and high concentration of vibrios in seawater (Fig. 3). Turbidity was generally low, typical of late spring and summer conditions (Fig. S7.4). Bacteria and vibrio concentrations in seawater were lower and less spatially structured than in oysters (Figs S7.6–S7 and 9).
Figure 3.
Environmental and host parameters on 20 June 2013 during the mortality event. The grey circles represent the sites where the sentinel oysters were accidentally lost and where only the seawater data are available. This map was done with the open source QGIS ver. 2.18.9. (QGIS Development Team, 2016, https://qgis.org).
On average, energy reserves of oysters decreased from 38% for carbohydrates to 51% for TAG between 30 April and 14 June (Figs S7.10 and S7.11), and gradually increased seaward thereafter. The spatial pattern of oyster energy reserves mirrored chlorophyll-a fluorescence on 20 June (Fig. 3). The levels of 16:1n-7/16:0, 20:5n-3/22:6n-3 and PUFA/SFA increased between 27 May and 20 June whereas 18:2n-6 + 18:3n-3, branched +15:0 + 17:0, and 18:1n-9/18:1n-7 decreased during this period. On 20 June, some of these indicators exhibited positive (16:1n-7/16:0) or negative (18:2n-6 + 18:3n-3, branched +15:0 + 17:0, and 18:1n-9/18:1n-7) gradients from the coast to the sea (Fig. S7.12). The spatial pattern of 16:1n-7/16:0 on 14 June mirrored oyster energy reserves and chlorophyll-a fluorescence on 20 June. Oyster growth rate (shell length, total body mass, shell mass and flesh mass) were the lowest along the coast and the highest where fluorescence peaked on 20 June (Fig. S7.13).
Mortality risk factors of oysters
To better appraise the influence of the environment on the mortality event, the relationship between the mortality-free time (D0) and all environmental parameters was investigated using the Cox regression model (Table 2). Increasing bathymetry and physical distance to inshore oyster farms were associated with a lower risk of mortality (higher D0, Table 2), confirming the seaward spread of the disease outbreak. Two clusters of biogeochemical parameters were then observed: the environmental factors reducing the mortality risk (corresponding to negative estimate and higher D0), and those enhancing the mortality risk (corresponding to positive estimate and lower D0). In the first group, increasing oyster growth rate, energy reserves (TAG, TAG/ST and carbohydrates), contribution of diatoms to oyster diet (16:1n-7/16:0), food freshness (PUFA/SFA), chlorophyll-a fluorescence, oxygen content, seawater temperature and vibrio concentration in oyster were all associated with a lower risk of mortality (Table 2). In contrast, increasing turbidity, salinity and the contribution of animal (18:1n-9/18:1n-7), terrestrial (18:2n-6 + 18:3n-3) and bacterial fatty acids to the diet of oysters were associated with a higher risk of mortality (Table 2).
Table 2.
Relationship between environmental, microbiological, biochemical and biometrical parameters and the mortality-free time (D0) of oysters by univariate Cox proportional hazards models.
| N | Variable | Estimate | SE | χ2 | p | Odds ratio |
|---|---|---|---|---|---|---|
| 1 | 18:1n-9/18:1n-7last | 0.117 | 0.020 | 32.91 | <0.001 | 1.124 |
| 2 | Distance to inshore oyster farms | −0.741 | 0.134 | 30.49 | <0.001 | 0.476 |
| 3 | Turbiditymean | 0.229 | 0.042 | 30.21 | <0.001 | 1.257 |
| 4 | 16:1n-7/16:0mean | −0.180 | 0.035 | 26.78 | <0.001 | 0.835 |
| 5 | TAG/STlast | −0.685 | 0.140 | 23.91 | <0.001 | 0.504 |
| 6 | Bathymetry | −0.324 | 0.069 | 22.01 | <0.001 | 0.723 |
| 7 | PUFA/SFAlast | −0.035 | 0.008 | 20.45 | <0.001 | 0.966 |
| 8 | Flesh mass | −0.400 | 0.091 | 19.54 | <0.001 | 0.670 |
| 9 | Branched +15:0 + 17:0last | 2.512 | 0.569 | 19.47 | <0.001 | 12.326 |
| 10 | Turbiditylast | 0.373 | 0.086 | 18.91 | <0.001 | 1.451 |
| 11 | TAGlast | −0.101 | 0.024 | 18.23 | <0.001 | 0.904 |
| 12 | Fluorescencelast | −0.174 | 0.041 | 18.00 | <0.001 | 0.840 |
| 13 | Shell mass | −0.246 | 0.059 | 17.33 | <0.001 | 0.782 |
| 14 | Salinitylast | 0.544 | 0.131 | 17.18 | <0.001 | 1.722 |
| 15 | PUFA/SFAmean | −0.056 | 0.014 | 16.84 | <0.001 | 0.945 |
| 16 | Bacteria in seawaterlast | 0.000 | 0.000 | 16.75 | <0.001 | 1.000 |
| 17 | Total body mass | −0.285 | 0.070 | 16.54 | <0.001 | 0.752 |
| 18 | Fluorescencemean | −0.655 | 0.164 | 15.92 | <0.001 | 0.520 |
| 19 | 16:1n-7/16:0last | −0.103 | 0.027 | 14.21 | <0.001 | 0.902 |
| 20 | Oxygenlast | −0.083 | 0.023 | 12.85 | <0.001 | 0.920 |
| 21 | 18:2n-6 + 18:3n-3last | 1.385 | 0.395 | 12.33 | <0.001 | 3.997 |
| 22 | Carbohydrate last | −0.014 | 0.004 | 12.14 | <0.001 | 0.986 |
| 23 | 20:5n-3/22:6n-3mean | 0.032 | 0.009 | 11.61 | 0.001 | 1.032 |
| 24 | Branched + 15:0 + 17:0mean | 3.798 | 1.210 | 9.84 | 0.002 | 44.606 |
| 25 | Shell length | −0.044 | 0.014 | 9.48 | 0.002 | 0.957 |
| 26 | Temperaturemean | −0.799 | 0.269 | 8.84 | 0.003 | 0.450 |
| 27 | TAG/STmean | −0.519 | 0.191 | 7.38 | 0.007 | 0.595 |
| 28 | Vibrio in oystermean | −0.192 | 0.071 | 7.21 | 0.007 | 0.825 |
| 29 | Carbohydratemean | −0.017 | 0.007 | 6.28 | 0.012 | 0.984 |
| 30 | 18:1n-9/18:1n-7mean | 0.080 | 0.032 | 6.14 | 0.013 | 1.083 |
| 31 | TAGmean | −0.071 | 0.032 | 4.82 | 0.028 | 0.931 |
When a variable was acquired more than once between deployment and D0, mean value (mean) and the value recorded before the onset of mortality (last) were used, except for the biometrical parameters where the overall growth rates were used. Only the significant parameters are presented and sorted in ascending order of χ2. Values of 18:1n-9/18:1n-7, 16:1n-7/16:0, PUFA/SFA and 20:5n-3/22:6n-3 were multiplied by 100 and concentrations of bacteria and vibrio in seawater and oysters were divided by 1000 to obtain manageable odds ratio.
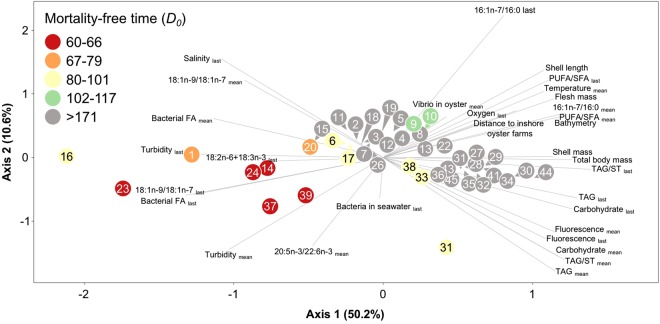
The principal component analysis of the environmental parameters associated with D0 confirms the classification in two groups. Most of the variance among sites (61%) was explained by the first two components, and all parameters associated with higher risk (lower D0) were located on the right side of the axis 1 while those related with lower risk were on the left side (Fig. 4).
Figure 4.
Principal component analysis of the parameters associated with mortality free-time of oysters (D0). Circles represent site numbers. See Table 2 for the list of parameters.
The daily survival of oysters during the mortality event (β) and the duration of the mortality event (δ), two parameters describing the dynamics of mortality at the local scale (oyster bag), were not correlated with the environmental, biochemical and biometrical parameters (Table 3).
Table 3.
Relationship between the daily survival of oysters during the mortality event (β) and the duration of the mortality event (δ) with environmental parameters.
| Parameter | Descriptor | β | Δ | ||
|---|---|---|---|---|---|
| r | p | R | p | ||
| Environment | |||||
| Bathymetry | point | 0.246 | 0.376 | 0.368 | 0.177 |
| Distance to inshore oyster farms | point | 0.164 | 0.559 | 0.161 | 0.567 |
| Temperature | mean | 0.171 | 0.541 | 0.236 | 0.398 |
| last | 0.132 | 0.639 | 0.075 | 0.791 | |
| Salinity | mean | 0.136 | 0.630 | −0.221 | 0.428 |
| last | 0.332 | 0.227 | 0.239 | 0.390 | |
| Fluorescence | mean | 0.325 | 0.237 | 0.318 | 0.248 |
| last | 0.457 | 0.087 | 0.521 | 0.056 | |
| Turbidity | mean | −0.361 | 0.187 | −0.300 | 0.277 |
| last | −0.075 | 0.791 | −0.114 | 0.685 | |
| Oxygen | mean | 0.064 | 0.820 | −0.079 | 0.781 |
| last | 0.068 | 0.810 | 0.125 | 0.657 | |
| Microbiology | |||||
| Bacteria in seawater | mean | −0.461 | 0.084 | −0.425 | 0.114 |
| last | −0.032 | 0.909 | 0.057 | 0.840 | |
| Vibrio in seawater | mean | −0.425 | 0.114 | −0.517 | 0.049 |
| last | −0.186 | 0.507 | −0.347 | 0.205 | |
| Bacteria in oyster | mean | −0.211 | 0.451 | −0.204 | 0.467 |
| last | −0.532 | 0.041 | −0.493 | 0.062 | |
| Vibrio in oyster | mean | −0.057 | 0.840 | −0.089 | 0.752 |
| last | −0.418 | 0.121 | −0.336 | 0.221 | |
| Biochemistry | |||||
| Carbohydrate | mean | 0.211 | 0.451 | 0.261 | 0.348 |
| last | 0.243 | 0.383 | 0.318 | 0.248 | |
| TAG | mean | 0.046 | 0.870 | 0.150 | 0.594 |
| last | 0.029 | 0.920 | 0.114 | 0.685 | |
| TAG/Sterol | mean | 0.071 | 0.800 | 0.182 | 0.516 |
| last | 0.029 | 0.920 | 0.118 | 0.676 | |
| 16:1n-7/16:0 | mean | 0.308 | 0.265 | 0.309 | 0.262 |
| last | 0.292 | 0.291 | 0.245 | 0.378 | |
| 20:5n-3/22:6n-3 | mean | 0.214 | 0.443 | 0.111 | 0.695 |
| last | 0.359 | 0.189 | 0.273 | 0.324 | |
| 18:2n-6 + 18:3n-3 | mean | 0.154 | 0.585 | 0.175 | 0.533 |
| last | 0.114 | 0.685 | 0.050 | 0.859 | |
| 18:1n-9/18:1n-7 | mean | 0.047 | 0.869 | 0.055 | 0.844 |
| last | −0.111 | 0.695 | −0.168 | 0.550 | |
| PUFA/SFA | mean | 0.063 | 0.825 | 0.209 | 0.454 |
| last | 0.057 | 0.840 | 0.120 | 0.671 | |
| Branched +15:0 + 17:0 | mean | −0.246 | 0.376 | −0.286 | 0.302 |
| last | −0.240 | 0.390 | −0.316 | 0.251 | |
| Biometry | |||||
| Shell length | e(b1) | 0.011 | 0.970 | 0.100 | 0.723 |
| Total body mass | e(b1) | −0.161 | 0.567 | −0.025 | 0.930 |
| Shell mass | e(b1) | −0.171 | 0.541 | −0.043 | 0.880 |
| Flesh mass | e(b1) | 0.032 | 0.910 | 0.104 | 0.713 |
Spearman correlation coefficients (r) and p-values are reported. When a variable was acquired more than once between deployment and D0, mean value (mean) and the value recorded before the onset of mortality (last) were used, except for the biometrical parameters where the overall growth rates were used.
Discussion
Oyster disease broke out in the intertidal farming area when seawater temperature reached 16 °C and gradually spread seaward. Similarly, in the Mediterranean Thau lagoon, OsHV-1 induced mortalities started within the bivalve farms and spread outward22,23. These studies, conducted in contrasting environments at different spatial scales, indicate that conditions of high-density marine aquaculture (with Pacific oysters used here as an example), provides suitable conditions for disease epizootics4. However, wild oysters coexist with the farmed animals and they probably contributed to the disease outbreak. Wild oysters are often asymptomatic carrier of the virus capable of viral shedding17.
Mortalities and OsHV-1 DNA detection were observed in oysters placed 2 km from the nearest farming areas, but laboratory challenge revealed that oysters collected at nearly all sites were subclinically infected. Virus particles likely spread seaward via water currents since there was no other known source of OsHV-1 offshore. The oyster C. gigas is the only known source of OsHV-1, and this species is restricted to intertidal and shallow subtidal areas9. Although OsHV-1 DNA can be detected in other bivalve species, their role as vectors has not been demonstrated e.g.24,25. Therefore, OsHV-1 may travel over long distances, transported by seawater currents, reflecting openness and connectivity of the studied ecosystem and persistence of infective particles in seawater4. Considering that (i) average mean and maximum velocities of seaward current in the Mor-Braz area were respectively 0.15 m.s−1 and 0.43 m.s−1 (S. Petton pers. Comm.) and (ii) herpesviruses can persist for at least 24 h in seawater26,27, viral particles could travel 13–38 km before infecting a new host which is consistent with our results.
Subclinical infection of oysters at offshore sites may reflect (i) the dilution of viral particles below a threshold value under which no mortality occurs, and (ii) the low density of susceptible host. In line with this, mortality risk of oysters injected with viral suspension increases with concentration of OsHV-1 particles28. Also, mortality risk increases with the biomass of infected oysters and decreases with seawater renewal, two parameters that influence viral particles concentration17. Moreover, mortality risk of infected oysters may also be associated with spatial variations in overall environmental and host conditions.
Increasing chlorophyll-a fluorescence, contribution of diatoms to oyster diet (16:1n-7/16:0), food freshness (PUFA/SFA), oyster growth rate and energy reserves (triglycerides and carbohydrates) were all associated with a lower risk of mortality offshore. Aquatic consumers generally show increasing growth and survival performance with increasing dietary long-chain essential fatty acid content29,30. Also, energy balance influences immune functions, and consequently disease susceptibility in terrestrial invertebrates31. Therefore, well-fed oysters with high energetic reserves may be able to mount an efficient immune response and decrease mortality risk23.
Reciprocally, contribution of terrestrial (18:2n-6 + 18:3n-3), animal (18:1n-9/18:1n-7), and bacterial (branched + 15:0 + 17:0) organic matter to the food of oysters were associated with a higher risk of mortality. These items are secondary food sources associated with reduced growth rate of oysters32. Low food quality may reduce accumulation of energy reserves which in turn alters immune response and disease susceptibility of oyster.
Increasing turbidity, an optical proxy of the concentration of suspended particulate matter (SPM), was associated with a higher risk of OsHV-1 related mortality. Several studies show that SPM facilitate disease transmission by providing microhabitats for viruses12,33. Like other marine viruses, OsHV-1 may adsorb and bind to both organic and inorganic particles, so that turbidity is a mortality risk factor for oysters. Alternatively, turbidity may increase mortality risk of oysters by attenuating the penetration of ultraviolet (UV) irradiance in the water column. Indeed, UV radiation decrease infectivity of bacteriophages and viruses infecting eukaryotic cells11,12, and they inactivate OsHV-1 in controlled conditions34. Finally, turbidity correlated positively with terrestrial inputs inferred from the fatty acid composition of oysters and negatively with chlorophyll-a fluorescence. Therefore, turbidity may reflect particles of poor nutritional quality (i.e. inorganic particles), that reduced feeding efficiency and energy intake of oysters9.
Oyster mortality initiated when seawater temperature reached 16 °C, confirming that temperature is a triggering risk factor35,36. However, at offshore sites, oyster mortalities started later when temperature was above 19 °C. This explains that the temperature measured before the onset of mortality cannot be associated with mortality risk in this study.
Apparently paradoxical results may reflect spurious correlations due to confounding factors. For instance, increasing salinity between 29.8 and 35.4‰ was associated with a higher risk of mortality whereas survival of oysters exposed to OsHV-1 at 35‰ is ∼20% higher than at 25‰ in laboratory experiment37. Also, increasing concentrations of vibrios in oysters were associated with a lower mortality risk while vibrios are involved in disease development38. Low salinity and high concentration of vibrios were confounded by distance to inshore oyster farms, food quantity and quality, growth rate of oyster and their energy reserve so that they may not be causal risk factors.
The daily survival of oysters during the mortality event (β) and the duration of the mortality event (δ), two parameters describing the dynamics of mortality at the bag scale, were not correlated with any of the studied parameters. Environmental and host parameters influence the spatio-temporal dynamics of mortality at the regional scale (D0), but not at the local scale (β and δ). Mortality risk in an oyster bag may depend on different factors acting on small spatial scales, including the biomass of susceptible hosts and the concentration of OsHV-1 particles17.
Oysters kept offshore were exposed to OsHV-1 but showed no abnormal mortality. Therefore, offshore aquaculture, a considered option to face the increasing demand for space on the intertidal zone, has an epidemiological advantage39. Nevertheless, care must be taken to (i) not recreate farming conditions (host density) that are similar to those prevailing inshore, (ii) limit the import of oysters to certified animals without OsHV-1 in order to maintain viral load to natural background and (iii) evaluate the economic and social costs of moving offshore.
However, the oysters kept offshore were subclinically infected. This reveals that the spatial distribution of the virus was much wider than that of oyster mortality. Also, these infected animals could exhibit mortality once environmental and host conditions permit virus replication, so that the risk of disease transmission can increase. The laboratory challenge supports this hypothesis by revealing asymptomatic carriers of OsHV-1, and is complementary to qPCR assays for characterizing the health status of oysters.
Overall, oyster health was associated with indicators of good ecological status of the coastal environment. Indeed, the mortality risk of oysters was maximal in inshore and intertidal farming areas with high oyster biomass, and decreases with distance to inshore oyster farms, food quantity and quality (diatoms). Reciprocally, the risk increased with turbidity, terrestrial inputs and poor food quality (terrestrial, animal and bacterial sources). Therefore, limiting the impact of disease by maintaining good ecological status of coastal waters is likely an alternative to traditional disease management strategies such as culling, vaccination, and chemotherapy that are proven inefficient for most marine invertebrates4.
Several risk factors identified in this study can be mapped using GIS and ocean color satellite remote sensing. Turbidity can be quantified in coastal waters using high resolution satellite observations, and remote sensing of colored dissolved organic matter can be used as a proxy of terrestrial inputs. By providing a spatially explicit framework in which OsHV-1 mortality risk factors would be predicted from GIS (distance to inshore oyster farms and bathymetry) and satellite remote sensing (turbidity and terrestrial inputs), our study opens original perspective in the application of Earth Observation for the sustainable management of oyster farming ecosystems.
Electronic supplementary material
Acknowledgements
We thank all the people that participated in the field work: M. Alunno-Bruscia, L. Barillé, B. Beliaeff, H. Brasselet, B. Cognie, C. Corporeau, P. Decottignies, L. Delisle, F. Descarrega, N. Djeghri, M. Doyen, Y. Epelboin, F. Enez, A. Gangnery, N. Garcia-Bueno, D. Hervio-Heath, A. Kusuma, A. Langlade, A. Le Bris, O. LeDreve, A. Lerouxel, C. Lupo, M. Miguet, J. Normand, F. Orvain, C. Quéré, J-Y. Stanisière, A. Travers, V. Turpin. We thank O. Lemarchand and N. Jeannée for their help with geostatistics, S. Petton for providing hydrodynamic model data and F. Le Roux and C. A. Burge for reviewing this MS. This work is part of the Gigassat project funded by ANR-AGROBIOSPHERE No. ANR-12-AGRO-0001-01 and the Ecoscopa project funded by the French Ministry of Ecology, Sustainable Development, Transport and Housing (Convention DPMA 2013 — Ifremer 2012/1/1210320/NYF).
Author Contributions
F.P., B.P., J.M., J.F.B., E.F. and P.G. conceived research ideas and designed methodology and collected data; F.P., M.F. and G.D. analyzed data; F.P. and P.G. led the writing of the manuscript. All authors contributed to the drafts and approved publication.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-29238-4.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Harvell CD, et al. Review: Marine ecology - Emerging marine diseases - Climate links and anthropogenic factors. Science. 1999;285:1505–1510. doi: 10.1126/science.285.5433.1505. [DOI] [PubMed] [Google Scholar]
- 2.Lafferty KD, et al. Infectious diseases affect marine fisheries and aquaculture economics. Ann Rev Mar Sci. 2015;7:471–496. doi: 10.1146/annurev-marine-010814-015646. [DOI] [PubMed] [Google Scholar]
- 3.Harvell D, et al. The rising tide of ocean diseases: unsolved problems and research priorities. Front. Ecol. Environ. 2004;2:375–382. doi: 10.1890/1540-9295(2004)002[0375:TRTOOD]2.0.CO;2. [DOI] [Google Scholar]
- 4.McCallum HI, et al. Does terrestrial epidemiology apply to marine systems? Trends Ecol. Evol. 2004;19:585–591. doi: 10.1016/j.tree.2004.08.009. [DOI] [Google Scholar]
- 5.McCallum H, Harvell D, Dobson A. Rates of spread of marine pathogens. Ecol. Lett. 2003;6:1062–1067. doi: 10.1046/j.1461-0248.2003.00545.x. [DOI] [Google Scholar]
- 6.Burge CA, et al. Climate Change Influences on Marine Infectious Diseases: Implications for Management and Society. Ann Rev Mar Sci. 2014;6:249–277. doi: 10.1146/annurev-marine-010213-135029. [DOI] [PubMed] [Google Scholar]
- 7.Pernet F, Lupo C, Bacher C, Whittington RJ. Infectious diseases in oyster aquaculture require a new integrated approach. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2016;371:20150213. doi: 10.1098/rstb.2015.0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.EFSA Oyster mortality. EFSA Journal. 2015;13:59. [Google Scholar]
- 9.Bayne, B. L. In Developments in Aquaculture and Fisheries Science Vol. Volume 41 (ed. Bayne Brian) 2–844 (Elsevier, 2017).
- 10.Murray AG. Epidemiology of the spread of viral diseases under aquaculture. Curr Opin Virol. 2013;3:74–78. doi: 10.1016/j.coviro.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Wommack KE, Colwell RR. Virioplankton: Viruses in Aquatic Ecosystems. Microbiol. Mol. Biol. Rev. 2000;64:69–114. doi: 10.1128/MMBR.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mojica KDA, Brussaard CPD. Factors affecting virus dynamics and microbial host–virus interactions in marine environments. FEMS Microbiol. Ecol. 2014;89:495–515. doi: 10.1111/1574-6941.12343. [DOI] [PubMed] [Google Scholar]
- 13.Ostfeld RS, Glass GE, Keesing F. Spatial epidemiology: an emerging (or re-emerging) discipline. Trends Ecol. Evol. 2005;20:328–336. doi: 10.1016/j.tree.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 14.Elliott P, Wartenberg D. Spatial Epidemiology: Current Approaches and Future Challenges. Environ. Health Perspect. 2004;112:998–1006. doi: 10.1289/ehp.6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walter, S. D. In Spatial epidemiology: methods and applications (eds Paul Elliot, Jon C. Wakefield, Nicola G. Best, & D. J. Briggs) 223–239 (Oxford University Press, 2000).
- 16.Petton, B. et al. Crassostrea gigas mortality in France: the usual suspect, a herpes virus, may not be the killer in this polymicrobial opportunistic disease. Front Microbiol6, 10.3389/fmicb.2015.00686 (2015). [DOI] [PMC free article] [PubMed]
- 17.Petton B, Boudry P, Alunno-Bruscia M, Pernet F. Factors influencing disease-induced mortality of Pacific oysters Crassostrea gigas. Aquaculture Env. Interact. 2015;6:205–222. doi: 10.3354/aei00125. [DOI] [Google Scholar]
- 18.Le Roux F, Wegner KM, Polz MF. Oysters and Vibrios as a Model for Disease Dynamics in Wild Animals. Trends Microbiol. 2016;24:568–580. doi: 10.1016/j.tim.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 19.Pepin JF, Riou A, Renault T. Rapid and sensitive detection of ostreid herpesvirus 1 in oyster samples by real-time PCR. J. Virol. Methods. 2008;149:269–276. doi: 10.1016/j.jviromet.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 20.Dalsgaard J, St. John M, Kattner G, Muller-Navarra D, Hagen W. Fatty acid trophic markers in the pelagic marine environment. Adv. Mar. Biol. 2003;46:225–340. doi: 10.1016/S0065-2881(03)46005-7. [DOI] [PubMed] [Google Scholar]
- 21.Cox DR. Regression Models and Life Tables. J. R. Stat. Soc. Series B Stat. Methodol. 1972;20:187–220. [Google Scholar]
- 22.Pernet F, Lagarde F, Gall PL, D’orbcastel ER. Associations between farming practices and disease mortality of oysters Crassostrea gigas in a Mediterranean lagoon. Aquaculture Env. Interact. 2014;5:99–106. doi: 10.3354/aei00096. [DOI] [Google Scholar]
- 23.Pernet F, et al. Spatial and Temporal Dynamics of Mass Mortalities in Oysters Is Influenced by Energetic Reserves and Food Quality. PLoS ONE. 2014;9:e88469. doi: 10.1371/journal.pone.0088469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burge CA, Strenge RE, Friedman CS. Detection of the oyster herpesvirus in commercial bivalve in northern California, USA: conventional and quantitative PCR. Dis. Aquat. Org. 2011;94:107–116. doi: 10.3354/dao02314. [DOI] [PubMed] [Google Scholar]
- 25.Arzul I, Renault T, Lipart C, Davison AJ. Evidence for interspecies transmission of oyster herpesvirus in marine bivalves. J. Gen. Virol. 2001;82:865–870. doi: 10.1099/0022-1317-82-4-865. [DOI] [PubMed] [Google Scholar]
- 26.Corbeil S, Williams LM, Bergfeld J, Crane MSJ. Abalone herpes virus stability in sea water and susceptibility to chemical disinfectants. Aquaculture. 2012;326-329:20–26. doi: 10.1016/j.aquaculture.2011.11.031. [DOI] [Google Scholar]
- 27.Hick P, Evans O, Looi R, English C, Whittington RJ. Stability of Ostreid herpesvirus-1 (OsHV-1) and assessment of disinfection of seawater and oyster tissues using a bioassay. Aquaculture. 2016;450:412–421. doi: 10.1016/j.aquaculture.2015.08.025. [DOI] [Google Scholar]
- 28.Paul-Pont I, Evans O, Dhand NK, Whittington RJ. Experimental infections of Pacific oyster Crassostrea gigas using the Australian ostreid herpesvirus-1 (OsHV-1) µVar strain. Dis. Aquat. Org. 2015;113:137–147. doi: 10.3354/dao02826. [DOI] [PubMed] [Google Scholar]
- 29.Delaunay F, Marty Y, Moal J, Samain J-F. The effect of monospecific algal diets on growth and fatty acid composition of Pecten maximus (L.) larvae. J. Exp. Mar. Biol. Ecol. 1993;173:163–179. doi: 10.1016/0022-0981(93)90051-O. [DOI] [Google Scholar]
- 30.Winder M, Carstensen J, Galloway AWE, Jakobsen HH, Cloern JE. The land–sea interface: A source of high-quality phytoplankton to support secondary production. Limnol. Oceanogr. 2017;62:S258–S271. doi: 10.1002/lno.10650. [DOI] [Google Scholar]
- 31.Demas GE, Adamo SA, French SS. Neuroendocrine-immune crosstalk in vertebrates and invertebrates: implications for host defence. Funct. Ecol. 2011;25:29–39. doi: 10.1111/j.1365-2435.2010.01738.x. [DOI] [Google Scholar]
- 32.Pernet F, et al. Marine diatoms sustain growth of bivalves in a Mediterranean lagoon. J. Sea Res. 2012;68:20–32. doi: 10.1016/j.seares.2011.11.004. [DOI] [Google Scholar]
- 33.Weinbauer MG, et al. Viral ecology of organic and inorganic particles in aquatic systems: avenues for further research. Aquat Microbial Ecol. 2009;57:321–341. doi: 10.3354/ame01363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schikorski D, et al. Experimental infection of Pacific oyster Crassostrea gigas spat by ostreid herpesvirus 1: demonstration of oyster spat susceptibility. Vet. Res. 2011;42:27. doi: 10.1186/1297-9716-42-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pernet F, et al. Mass mortalities of Pacific oysters Crassostrea gigas reflect infectious diseases and vary with farming practises in the Thau lagoon. Aquaculture Env. Interact. 2012;2:215–237. doi: 10.3354/aei00041. [DOI] [Google Scholar]
- 36.Renault T, Bouquet AL, Maurice J-T, Lupo C, Blachier P. Ostreid herpesvirus 1 infection among Pacific oyster (Crassostrea gigas) spat: relevance of water temperature to virus replication and circulation prior to the onset of mortality. Appl. Environ. Microbiol. 2014;80:5419–5426. doi: 10.1128/AEM.00484-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fuhrmann M, et al. Salinity influences disease-induced mortality of the oyster Crassostrea gigas and infectivity of the ostreid herpesvirus 1 (OsHV-1) Aquaculture Env. Interact. 2016;8:543–552. doi: 10.3354/aei00197. [DOI] [Google Scholar]
- 38.Lemire A, et al. Populations, not clones, are the unit of vibrio pathogenesis in naturally infected oysters. ISME J. 2015;9:1523–1531. doi: 10.1038/ismej.2014.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brenner M, et al. Bivalve aquaculture transfers in Atlantic Europe. Part B: Environmental impacts of transfer activities. Ocean Coast Manag. 2014;89:139–146. doi: 10.1016/j.ocecoaman.2013.10.009. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.