Abstract
Microorganisms frequently coexist in complex multispecies communities, where they distribute non-randomly, reflective of the social interactions that occur. It is therefore important to understand how social interactions and local spatial organization influences multispecies biofilm succession. Here the localization of species pairs was analyzed in three dimensions in a reproducible four-species biofilm model, to study the impact of spatial positioning of individual species on the temporal development of the community. We found, that as the biofilms developed, species pairs exhibited distinct intermixing patterns unique to the four-member biofilms. Higher biomass and more intermixing were found in four-species biofilms compared to biofilms with fewer species. Intriguingly, in local regions within the four member biofilms where Microbacterium oxydans was scant, both biomass and intermixing of all species were lowered, compared to regions where M. oxydans was present at typical densities. Our data suggest that Xanthomonas retroflexus and M. oxydans, both low abundant biofilm-members, intermixed continuously during the development of the four-species biofilm, hereby facilitating their own establishment. In turn, this seems to have promoted distinct spatial organization of Stenotrophomonas rhizophila and Paenibacillus amylolyticus enabling enhanced growth of all four species. Here local intermixing of bacteria advanced the temporal development of a multi-species biofilm.
Introduction
The distribution of microbes in the microenvironments of biofilms is non-random; physiological and phenotypic traits of constituent strains, as well as nutrient availability are known to be important drivers of the spatial organization of microbes [1–4]. As biofilms typically consist of multiple species [5], interspecies interactions are also expected to be key factors that determine the distribution of microbial species within biofilms, hereby influencing the diversity, stability, and activity of microbial biofilm communities [2].
Studies have highlighted the impact of spatial organization on the function and activity of biofilm communities: currently, in silico individual-based modeling is one of the most commonly used approaches in this field [6–8]. This method is powerful for exploring questions about biofilm structure and composition, and in silico-based observations have subsequently been tested experimentally: for instance, local species segregation has been shown as a way for toxin-sensitive strains to survive when co-residing with toxin-producing strains, whereas species intermixing was favored by toxin-producing strains in order to out-compete toxin-sensitive strains when competing for resources locally [9–11]. Metabolic cross-feeding and detoxification mutualisms are other examples that have been shown to promote spatial intermixing of mutualists [4, 12]. Using genetic engineering, Momeni et al. [13] verified theory predicted with in silico models, demonstrating that species intermixing was favored if strong cooperation was the main interaction occurring between two species. The aforementioned studies focused on dual-species communities, showing how relatively simple interspecies interactions impact their spatial organization. Here, we explored a more complex four-species model biofilm community to investigate if spatial positioning of individual species is impacted by species diversity and how this is reflected in the temporal development of the biofilms.
Although the revolution in meta-omics technologies has dramatically improved our understanding of the composition and activity of complex microbial communities [14–17], the spatial scale at which these technologies are applicable make them less informative when studying social interactions between individual bacteria. Indeed, spatial organization within multispecies biofilms has lately attracted more attention as recent research has revealed its crucial roles in driving community functions [2, 18]. Lee et al. [19], for example, found that a three-species biofilm was more resilient to antimicrobials compared with corresponding single-species biofilms, and the spatial organization was hypothesized to be a key factor driving this community-level protection. Such studies stress the importance and necessity of studying spatial organization at the micro-scale level in complex communities, and emphasize an urgent need for strong approaches and tools to further facilitate researches in this area.
In the current study, 16S rRNA-directed fluorescent in situ hybridization (FISH) was, in combination with confocal laser scanning microscopy (CLSM), used to visualize the spatial positioning of four different species within a well-established and reproducible biofilm model [20]. The four species; Stenotrophomonas rhizophila, Xanthomonas retroflexus, Microbacterium oxydans, and Paenibacillus amylolyticus, were originally co-isolated from soil. We present an efficient and accurate approach for three-dimensional, as opposed to two-dimensional [21], pairwise cross-correlation (PCC) analysis of the community. With this analysis we unravel the dynamics of spatial positioning and correlate this with an increased bio-volume of the community. We found that species pairs occurred in different spatial conformations when co-cultured in the four-species biofilm communities as compared to dual-species biofilms, indicating that interspecies interactions were affected by the presence of additional member species. Specifically, high levels of intermixing between X. retroflexus and M. oxydans suggested cooperative interactions between these two species. Importantly, the stable co-establishment of X. retroflexus and M. oxydans during the early development of the four-species biofilm, led to an increase in the overall bio-volume of the whole biofilm community.
Materials and methods
Bacterial strains
Four co-isolated bacteria from soil; Stenotrophomonas rhizophila, Xanthomonas retroflexus, Microbacterium oxydans, and Paenibacillus amylolyticus were used in this study to establish multispecies biofilms. Using the Nunc-TSP lid system, these bacterial strains have previously been shown to significantly increase biofilm biomass and cell numbers when co-cultured together all four, compared to when cultured as three-species, dual-species, or single-species biofilms [20].
Continuous-culture drip-flow biofilm reactor system
Biofilms were grown in a Drip Flow Biofilm Reactor (DFR) (BioSurface Technologies, Bozeman, MT) using glass microscope slides as the growth surface [22]. Microscope glass slides (VWR international, China) were placed into the DFR chambers and autoclaved. Exponentially growing cultures of the four species were adjusted to an optical density of 0.15 at 600 nm (OD600) in 1/2-strength tryptic soy broth (TSB) (Sigma, St. Louis, USA). Strains were mixed in equilibrium and a total volume of 20 ml was then inoculated into individual chambers of the DFR. Cultures were incubated statically and horizontally for 4 h at 24 °C for the initial attachment, before waste outlets were opened and the chambers were placed at a 10° angle in order to drain the waste medium. A homogeneous distribution of species along the glass surface was ensured before fresh medium was continuously dripped onto the glass slides via a Watson-Marlow 205S pump, at a flow rate of ~0.4 ml min−1 per chamber while incubation continued at 24 °C. Four-species biofilm z-stack images acquired at 0, 6, and 12 h after the attachment phase were re-used from a previous study [23]. In the current study, the four-species biofilms were cultivated up to 24 h, and two types of local regions (areas) within these biofilms were defined by visualization and imaged specifically by CLSM: (i) regions where M. oxydans was present at typical densities and (ii) aberrant regions, where few M. oxydans was present. It is specified in the text when data and discussion concerns the latter type of regions. For regions with typical densities of M. oxydans three biological replicates were included. Each biological replicate was cultivated on separate days where three z-stack images were recorded (i.e., nine z-stack images total); for regions where M. oxydans was scant, three biological replicates (three z-stack images) were collected from the four-species biofilms after 24 h incubation. Mono and dual-species biofilms were grown and sampled at corresponding time points (0, 6, 12, and 24 h), for subsequent 16S rRNA FISH and z-stack images acquisition. Three biological replicates (nine z-stack images) were acquired from the central channel following the sampling method described by Liu et al. [23]. Negative controls without inoculation of bacterial cells were included when growing both dual and four-species biofilms.
Pre-treatment of biofilms for FISH
Biofilm samples were pre-treated using the method described by Liu et al. [23] with minor modifications. Dual-species biofilms formed on glass slides from four time points (0, 6, 12, and 24 h) were dried slightly at 46 °C for 2 min and subsequently immobilized with hand-warm dissolved 0.5% (w/v) agarose. Samples were fixed in 4% (w/v) paraformaldehyde in PBS (pH 7.2) for 1 h at room temperature, followed by washing with PBS (3 × 5 min). Then, samples were transferred into 50 ml falcon tubes with 40 ml solution containing 50% ethanol in PBS and stored at −20 °C for later use. Subsequently, samples were washed in PBS (2 × 5 min), followed by incubation in permeabilisation solution containing 7 mg lysozyme (Cat. No. L6876, Sigma, St. Louis, USA) per ml in 0.1 M Tris/HCl, 5 mM EDTA (pH 7.2) for 10 min at 37 °C, and washed again with PBS (2 × 5 min). Then, the samples were treated with 1% (v/v) methanol dissolved hydrogen peroxide for 30 min and rinsed with PBS (2 × 5 min) in order to reduce background fluorescence. After this, samples were acetylated with 0.25% acetic anhydride in freshly prepared 0.1 M triethanolamine (pH 8.0) for 5 × 1 min, followed by washing in MilliQ water (3 × 5 min).
16S rRNA FISH
FISH was performed using the method described by Liu et al. [23] with minor modifications. Briefly, samples were dehydrated with a series of solution containing 50, 80, 100% ethanol for 3 min each at room temperature. Subsequently, samples were incubated with the fluorochrome-labeled oligonucleotide probes at a concentration of 5 ng each per µl hybridization buffer (0.9 M NaCl, 20 mM Tris/HCl (pH 7.2), 30% formamide, 0.01 % (w/v) SDS) at 46 °C for 2 h. After this, samples were incubated for 2 × 10 min at 48 ℃ in washing buffer containing 20 mM Tris/HCl (pH 7.5), 5 mM EDTA, 108 mM NaCl and 0.01% (w/v) SDS. After washing, the samples were submerged into 4 °C dH2O for a couple of seconds and air-dried completely at room temperature. The specimens were mounted in VECTASHIELD Antifade Medium (Cat. No. H-1000, Vector laboratories, California, USA), covered with coverslips, sealed with colorless nail polish and stored at 4 °C for later use. HPLC-purified oligonucleotide (FISH) probes were synthesized commercially by TAG Copenhagen A/S (Frederiksberg, Denmark) and 5′ labeled with different fluorochromes: sulfoindocyanine dye indocarbocyanine (Cy3), indodicarbocyanine (Cy5) or 6-carboxyfluorescein (FAM). The nucleotide sequences of the species specific FISH probes are specified in Liu et al. [23]. The probe tracking for P. amylolyticus was labeled with Cy3 or Cy5, in order to distinguish two species co-cultured. The maximum excitation wavelengths for FAM, Cy3 and Cy5 were 495, 550, and 649 nm, respectively. The maximum emission wavelengths for FAM, Cy3 and Cy5 were 520 (~480–675), 570 (~525–700), and 670 (~625–800) nm, respectively. In addition, unlabeled competitor probe (COM437) was introduced to distinguish S. rhizophila and X. retroflexus due to their high 16S rRNA sequence similarity [24]. The specificity of each oligonucleotide probe used in this study has been tested in a previous study [23].
Image processing and pixel quantification analyses
Microscopy and image acquisition was performed by CLSM using a TCS SP5-X (Leica) with ×40 oil-immersion objective (NA = 1.3) using the Quick-LUT function to set the pixel saturation limits. Standard images were acquired with a zoom setting of 1.0 corresponding to physical dimensions of 387.5 × 387.5 μm2. For each sample, three separate and representative image stacks were acquired from the center of the biofilm not close to the edges of the glass slide, covering a total area of approximately 4.5 × 105 μm2, which was more than the minimum of 1 × 105 μm2 required for representative data [25]. The sampling area of each section was transformed into a digital image containing 512 × 512 pixels. Images were segmented, hereby transforming each pixel for each channel into binary data (presence–absence) using the Li’s auto-thresholding method available in Fiji [26, 27]. The number of present pixels showed Gaussian distribution from top to bottom layers of z-stack images, and we removed the bottom layers from where the decreased pixels were detected. Bio-volume was defined as the voxel size (0.145 µm3) multiplied by the number of pixels in all images of a stack. The threshold for each stack, set by Fiji, was also applied for the following 3D images reconstruction by IMARIS (Bitplane AG, Belfast, UK) in order to make the data produced by different software comparable [27]. Reproducibility of the biofilms was determined according to Jackson et al. [28]. Statistical analyses (One-way ANOVA, Tukey HSD post-hoc test) were done using the R language [29].
3D Pairwise cross-correlation (PCC) analyses
The pre-processed z-stack images were further treated by the following algorithm for calculating pairwise cross-correlations between all pairs of species in all images (see also Fig. 1): (1) a virtual 3D cube of a set size was created, the distance (in μm) was calculated from the center of the cube to all other pixels in the cube, and the distances were binned to the nearest integer. (2) N pixels in the image were randomly chosen. (3.1) For 1 to N the cube was centered on the focal pixel (n). (3.2) If part of the cube extended beyond the image it was cut off. (3.3) For each distance bin (d), it was counted how many times the focal pixel and the pixels in the bin were of different colors (i.e., from different channels); we named this Hits (Hdn). (3.4) it was then counted how many possible hits there could have been; we named this Total (Tdn). (4) Hits and Totals were then summed across the random pixels for each distance bin, such that and . (5) A hit probability (P) was then calculated for each distance bin: . (6) The pairwise cross-correlation (PCC) was then this probability normalized by the overall densities of the two channels (Di and Dj), where and and , such that PCCd=1 equals random spatial positioning, PCCd>1 equals aggregated spatial positioning, and PCCd<1 equals segregated spatial positioning. (7) The entire analysis was run 10 times to estimate analytic variability. The final PCC was then the mean from these 10 runs. The analytic approach with a virtual 3D cube was borrowed with permission from the iMage GitHub repository (www.github.com/rmnppt/iMage) and the CC calculation is similar to the 2D co-aggregation one implemented in daime [21]. Pixel quantification and PCC analyses were evaluated with in-house R scripts [29], which are freely available at GitHub (http://www.github.com/Russel88/RCon3D version 0.9). All z-stack images captured from three biological replicates for each sample were used for these calculations, with standard errors estimated from the biological replicates. See Supplemental Figure S1 for PCC calculations on simulated images with channels of different densities placed randomly in the 3D space.
Fig. 1.
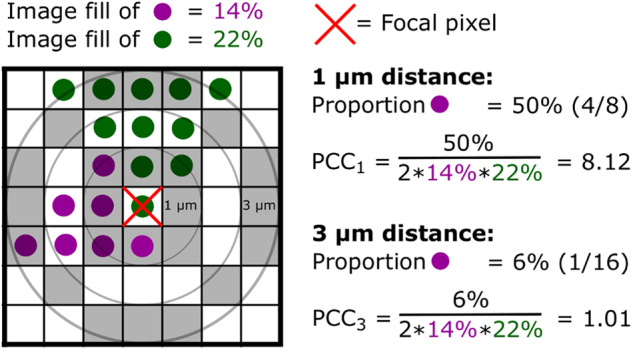
Schematic diagram of the 3D pairwise cross-correlation analysis. The diagram is shown in two dimensions, but the actual analysis is conducted in three dimensions. PCC at a certain distance is calculated as the amount of pixels that are of a different color than the focal pixel, divided by the total amount of pixels at that distance (gray zones); this proportion is then normalized to the overall image fill of the two colors, such that random positioning will give a PCC equal to one as in daime [21]. In practice, results are aggregated across many randomly chosen focal pixels, and pixels are aggregated in distance bins
Viable cell number counting
Enumeration of viable cells was performed using the spread plate method following procedures described by Jackson et al. [28] with minor modifications. Four and three-species (excluding M. oxydans) biofilms were collected for viable cell counting after 12 and 24 h incubation in the DFR. Biofilms were detached from the glass slide by applying 300 µl PBS (pH=7.2), followed by scraping of all visible biomass using aseptic NuncTM cell scrapers (Thermo Fisher Scientific) into centrifuge tubes containing 700 µl PBS. The samples were then homogenized using a vortex mixer (IKA® MS 3 basic) for 30 s. at 2500 rpm, and serially diluted, generally to 1 × 10–7. A 100 µl aliquot of each serial dilution was spread, in triplicate, onto Congo red agar plates (TSA with 40 µg/ml Congo Red and 20 µg/ml Coomassie). In addition, Congo red agar supplemented with kanamycin (50 µg/ml) was used to distinguish S. rhizophila and X. retroflexus, because their colony morphology is similar, but S. rhizophila is sensitive to kanamycin whereas X. retroflexus is resistant. Otherwise the colony morphologies of the different species of bacteria used in this study are distinguishable on Congo red agar. Agar plates were incubated at 24 °C and the numbers of colonies for each species were counted after 48 h. The assay was performed in triplicates.
Results
The biomass production of individual species in dual and four-species biofilms differed depending on which other species were present
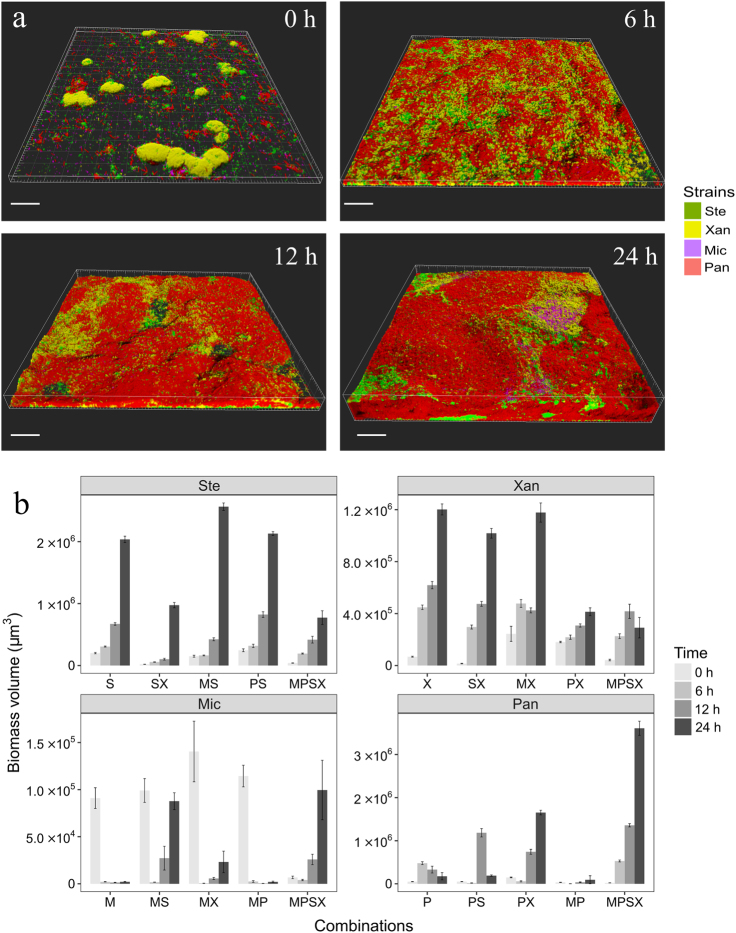
Six dual-species and one four-species biofilms were grown for 24 h in order to investigate if biomass production and spatial positioning of individual species was affected when co-cultured with the other species. In dual-species biofilms, both Xanthomonas retroflexus and Stenotrophomonas rhizophila dominated relative to Microbacterium oxydans (Supplementary Figure S2h, S2l, and S3a). Similar bio-volumes of X. retroflexus and S. rhizophila where found when the two were co-cultured for up to 24 h where they segregated locally within the biofilm (Supplementary Figure S2d and S3a). Interestingly, the growth of Paenibacillus amylolyticus was highly promoted when co-cultured with X. retroflexus or S. rhizophila for up to 12 h compared to when grown with M. oxydans (Fig. 2; Supplementary Figure S2o, s and w). However, P. amylolyticus displayed very different growth and spatial organization after 24 h of co-cultivation with X. retroflexus or S. rhizophila: P. amylolyticus was almost outgrown by S. rhizophila (Fig. 2; Supplementary Figure S2p; Supplementary Figure S3a), but not by X. retroflexus (Supplementary Figure S2t and S3a). M. oxydans and P. amylolyticus was the only pair of species that did not form a dense biofilm when co-cultured for up to 24 h (Supplementary Figure S2x). Mono-species biofilms were also grown for up to 24 h, and when comparing the bio-volume of mono-species with those in dual–species biofilms (Supplementary Figure S3a) it was found that both M. oxydans and P. amylolyticus are poor biofilm formers on their own, yet, the growth of P. amylolyticus was strongly facilitated by X. retroflexus when co-cultured (Two-sample t-test: P < 0.001) and the growth of M. oxydans, even though present at relatively low abundance, was intensively promoted by either S. rhizophila or X. retroflexus compared to when grown alone (Two-sample t-test: P < 0.01). Overall fewer negative interactions occurred as time progressed (Supplemental Figure S3b).
Fig. 2.
Spatiotemporal development of the four-species biofilm. a Confocal images showing the four-species biofilms after 0, 6, 12, and 24 h of growth under flow conditions. Ste; S. rhizophila (green), Xan; X. retroflexus (yellow), Mic; M. oxydans (purple) and Pan; P. amylolyticus (red). Magnification ×40. Scale bar: 50 µm. b Bio-volumes of species in single, dual, and four-species biofilms at 0, 6, 12, and 24 h. Capital letters; S, X, M, and P represent S. rhizophila, X. retroflexus, M. oxydans, and P. amylolyticus, respectively. Error bars represent standard error of the mean of three biological replicates
We have previously described how biofilms formed by the four species led to significantly increased bio-volumes compared to any combinations of single and three-species biofilms [23]. In accordance, distinct spatial distributions of individual species were observed and the bio-volume of the mature four-species biofilm was larger, compared with the dual-species biofilms (Supplementary Figure S2 and S3a). Image pixel quantifications confirmed that the bio-volume production was significantly increased when the four species were co-cultured for up to 24 h, compared to the most productive dual-species biofilm community, which consisted of S. rhizophila and M. oxydans (One-way ANOVA, Tukey HSD post-hoc test: P < 0.001; Supplementary Figure S3a).
Distinct pairwise cross-correlations were found in dual- and four-species biofilms as time progressed
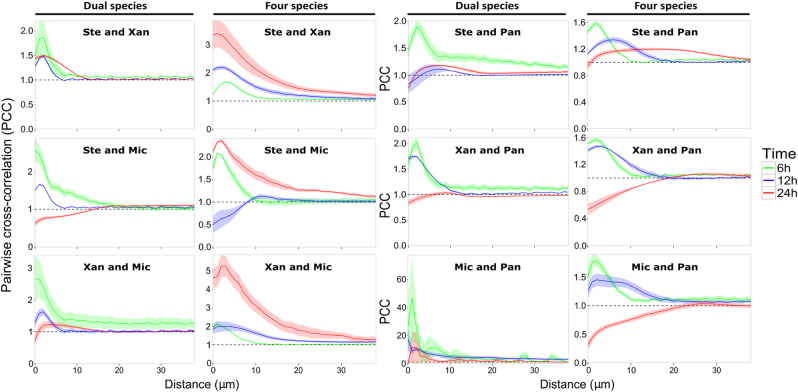
An optimized 3D pairwise cross-correlation (PCC) analysis was developed to further study the spatial organization of cells within the biofilms (see Materials & methods for further information). PCC analyses were performed on the pre-processed stacked images generated by CLSM and the output indicates whether two species tend to intermix (PCC > 1), are randomly distributed (PCC = 1), or segregate (PCC < 1) from each other (Fig. 3). Generally, the maximum PCC values of species pairs in dual-species biofilms decreased over time, showing that less intermixing and more segregation occurred in mature dual-species biofilms. Likewise, PCC values of species pairs including P. amylolyticus also decreased over time when grown in the four-species biofilm. In contrast, the other three species obtained higher PCC values in the mature four-species biofilm compared to the early time points, indicating that the pairwise intermixing of these three species varied as a response to P. amylolyticus. Specifically, the PCC value of species pair X. retroflexus and M. oxydans increased as the four-species biofilm developed, exhibiting the highest PCC value at around 3 µm between-species distance in the mature community among all six species pairs, demonstrating that these two species were intermixing consistently.
Fig. 3.
Pairwise cross-correlation analyses of dual and four-species biofilms. The mean PCC value (continuous line) and the standard error of the mean (shaded parts) are plotted against distances spaced at intervals of 1 µm. The dashed horizontal line (PCC value of 1) corresponds to random positioning. Ste; S. rhizophila, Xan; X. retroflexus, Mic; M. oxydans and Pan; P. amylolyticus. PCC values of the six species pairs were calculated from nine stacked images (three biological replicates) obtained from dual and four-species biofilms after 6 (green), 12 (blue), and 24 (red) hours of growth under flow conditions. PCC values of the 24 h four-species biofilms were calculated based on images of local regions with typical densities of M. oxydans
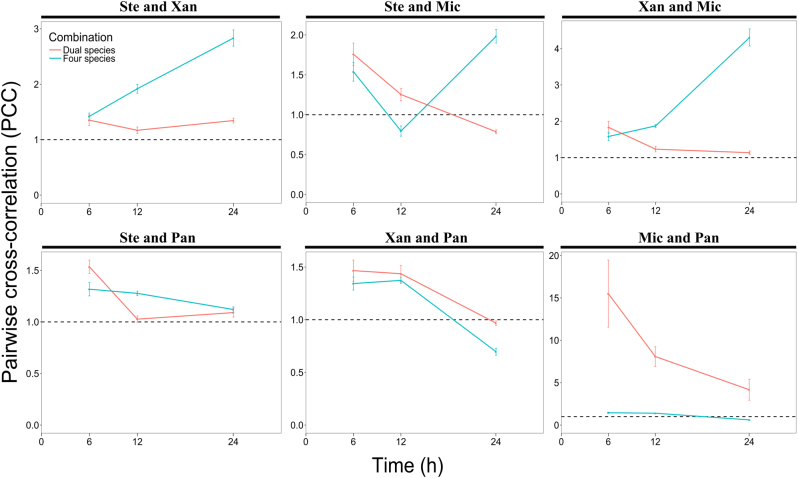
The distances at which the PCC values peaked varied considerably among the species pairs in dual and four-species biofilms (Fig. 3). Some species pairs reached the highest PCC value at distances <5 µm, and others at around 10 µm, and some at even larger distances. To better elucidate the PCC analyses shown in Fig. 3, we plotted the mean of the PCC value of each species pair in dual and four-species biofilms from 0 to 10 µm for each time point (Fig. 4). We chose this interval (0 to 10 µm) as we judged it unlikely that interactions would occur beyond a distance of 10 µm. This analysis confirmed that species pair X. retroflexus and M. oxydans became more intermixed with time when co-cultured in the four-species biofilm, but not in the dual-species biofilm. This was in accord with observations from CLSM images (Supplementary Figure S2j, k and l; Supplementary Figure S4).
Fig. 4.
Mean of pairwise cross-correlation values of dual and four-species biofilms from 0 to 10 µm as a function of time (6, 12, and 24 h of growth under flow conditions). The red and blue solid lines represent PCC values of dual and four-species biofilms, respectively. The dashed horizontal line (PCC value of 1) corresponds to random positioning. Ste; S. rhizophila, Xan; X. retroflexus, Mic; M. oxydans and Pan; P. amylolyticus. Error bars represent standard error of the mean of the average of 10 PCC values represented in the 0–10 µm interval (n = 3)
Spatial positioning of species pairs in mature dual and four-species biofilms
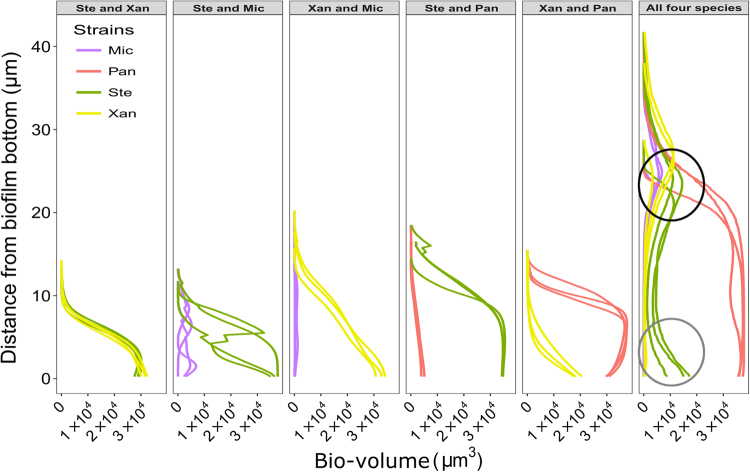
Next, we quantified pixels of each species in each layer of the pre-processed stacked CLSM images from mature dual and four-species biofilms. This showed that the number of pixels of S. rhizophila and X. retroflexus were equivalent in all layers in the mature dual-species biofilms (Fig. 5), which was partly supported by the relatively high PCC value found between these two species (Fig. 3). Similar spatial patterns were observed in all other dual-species biofilms, where one species was evenly or heterogeneously distributed in lower abundances throughout the entire biofilm that was dominated by the other species (Fig. 5). These types of distribution patterns indicate that there was a higher probability that the dominating species was surrounded by itself in all layers, demonstrating that all the four species pairs intermixed at a relatively low level (Fig. 3).
Fig. 5.
Distribution of species throughout the mature (24 h of growth under flow conditions) dual- and four-species biofilms. The bio-volume of pixels from each species is plotted against the thickness of biofilms. The four-species biofilm is more than twice as thick as the dual-species biofilms. The black circle emphasizes data showing that S. rhizophila (Ste), X. retroflexus (Xan), and M. oxydans (Mic) reside in the top layers of the four-species biofilm in relatively high cell numbers simultaneously. The gray circle emphasizes data showing a local bio-volume peak of S. rhizophila (Ste) in the bottom layers of the four-species biofilm. Each curve represents individual biological replicates that are the average of three technical replicates
In the mature four-species biofilm, P. amylolyticus dominated throughout the whole community, and the other three species had relatively low pixel counts (Fig. 5). This was consistent with the low intermixing level of species pairs including P. amylolyticus, as shown in Fig. 3. The bio-volumes of S. rhizophila, X. retroflexus, and M. oxydans all peaked in the top layer of the four-species biofilm at around 25 µm from the glass slide (Fig. 5; top black circle), indicating that these three species grew better in the top part of the biofilm. Species pairs of the three species showed relatively high levels of intermixing compared to species pairs that included P. amylolyticus (Figs. 3 and 4). In addition, more bio-volume of S. rhizophila was found in the bottom layers compared to X. retroflexus and M. oxydans (Fig. 5; gray circle), which is in agreement with the lower PCC values of species pairs S. rhizophila & X. retroflexus and S. rhizophila & M. oxydans compared to X. retroflexus & M. oxydans (Figs. 3 and 4).
Species intermixing promoted stable co-residence of X. retroflexus and M. oxydans in the four-species biofilm
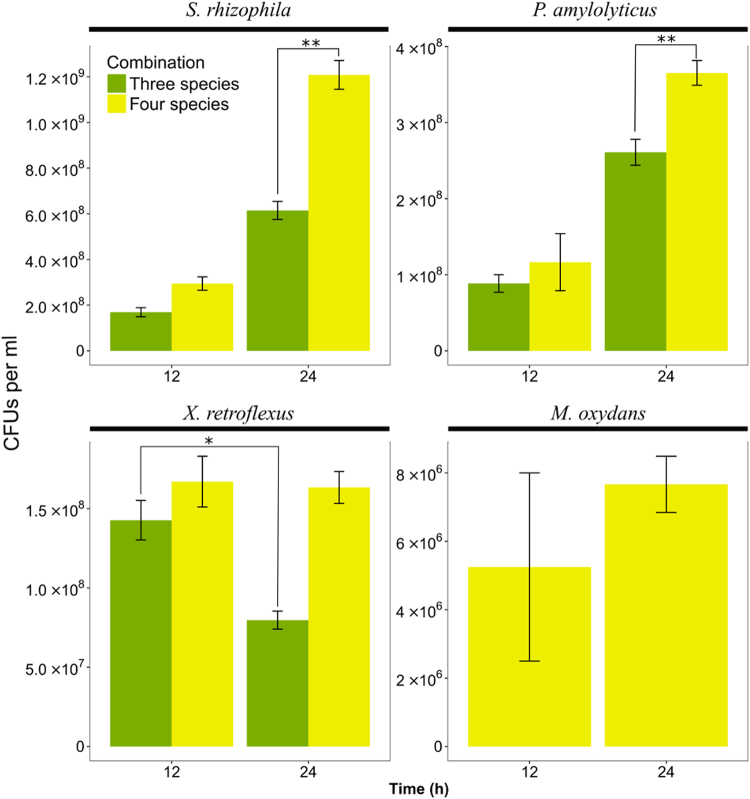
The PCC analyses suggested a high level of intermixing by X. retroflexus and M. oxydans in the mature four-species biofilms (Fig. 3). In addition, 3D images and pixel quantification showed that P. amylolyticus overall was dominating the mature four-species biofilm (Figs. 2a and 5). The intermixing of X. retroflexus and M. oxydans may therefore have occurred due to the overgrowth of P. amylolyticus, as only limited space was available to the community. To test this hypothesis, the four-species biofilms and three-species biofilms without M. oxydans were grown in the DFR for 12 and 24 h. Then, the biofilms were collected and CFUs of each species were enumerated (Fig. 6). The cell number of X. retroflexus decreased significantly from 12 to 24 h in the absence of M. oxydans (Two-sample t-test: P < 0.05). The cell number of both X. retroflexus and M. oxydans were, on the other hand, stably maintained in the four-species community from 12 to 24 h (Two-sample t-test: P > 0.05). Therefore, the presence of M. oxydans indeed had a positive effect on the growth fitness of X. retroflexus, facilitating the stable establishment of the latter species in the four-species community, and the above hypothesis was rejected. Meanwhile, it is very likely that the local intermixing of these two species enabled a mutual facilitation, i.e., a stable maintenance of M. oxydans in the presence of X. retroflexus in the four-species biofilm. Also, S. rhizophila and P. amylolyticus exhibited significantly higher numbers of cells in the mature four-species biofilm compared to those in the mature three-species biofilms that were cultivated without M. oxydans (Two-sample t-test: P < 0.05). This suggests that species intermixing promoted growth synergy of the four-species community.
Fig. 6.
CFUs per ml of each strain in the triple-species (green, excluding M. oxydans) and four-species (yellow) biofilms at two time points (12 and 24 h of growth under flow conditions). Error bars represent standard error of the mean from three biological replicates. *P < 0.05; **P < 0.01 (Two-sample t-test)
Distinct intermixing patterns and a decrease of bio-volumes of all members in local regions of mature four-species biofilms where M. oxydans was scant
To further verify that intermixing of X. retroflexus and M. oxydans lead to an increase in the biomass of all members of the mature four-species biofilms, we analyzed images of the four-species biofilm where very few M. oxydans were present (Supplementary Figure S5), and compared these to images where it was present at typical densities (Fig. 2a). Based on the z-stack images from these regions PCC values and bio-volumes were calculated: Locally M. oxydans intermixed with X. retroflexus and to a less degree S. rhizophila (Fig. 7a), while segregating apart from P. amylolyticus within 20 µm between-species distance (Supplementary Figure S6). Interestingly, the local variance in cell densities of M. oxydans had a relatively moderate influence on the intermixing of the other species pairs. In local regions with few M. oxydans, S. rhizophila was vaguely less likely to intermix with X. retroflexus but marginally more with P. amylolyticus, while X. retroflexus and P. amylolyticus were slightly less likely to segregate (Supplementary Figure S6). Most strikingly, however, we found that within the mature four-species biofilms, the bio-volumes of all members were significantly lower in local regions where the presence of M. oxydans was scant (Fig. 7b).
Fig. 7.
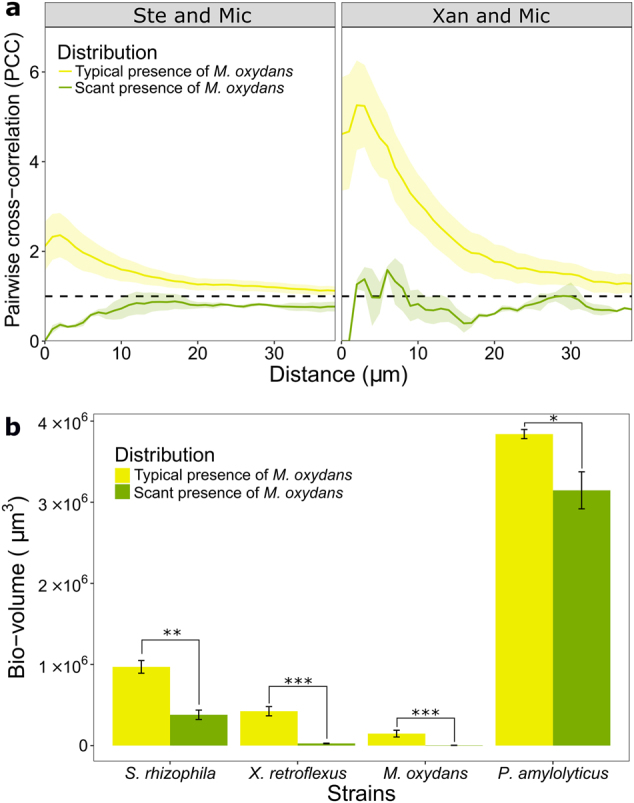
Reduced bio-volumes of all members in local regions within four-species biofilms where the presence of M. oxydans was scant. a PCC values of M. oxydans (Mic) with S. rhizophila (Ste) or X. retroflexus (Xan) in regions where the local presence of Mic was typical or scant in four-species biofilms after 24 h of growth under flow conditions. The mean PCC values (continuous line) and the standard error of the mean (SEM, shaded part) are plotted against distances spaced at intervals of 1 µm. The dashed horizontal line (PCC value of 1) corresponds to random positioning. b Bio-volumes of each species in the 24 h four-species biofilms in regions where the local presence of Mic was typical or scant. Error bars represent standard error of the mean from three biological replicates. *P < 0.05; **P < 0.01; ***P < 0.001 (Two-sample t-test)
Discussion
Microorganisms typically live in dense multispecies biofilms with distinct patterns of spatial organization, which are believed to be of central importance for driving community functions and the activity of the individual species. Oral plaques are examples of multispecies biofilms where co-aggregation seems to drive further community assembly [30, 31]. Here, cell-cell communication [32], oxygen protection [33], and horizontal gene transfer [34] have been shown to occur between co-aggregating species. For instance, Fusobacterium nucleatum is known for its roles in physically connecting aerobes and anaerobes that otherwise do not associate by co-aggregation, which hereby promotes the growth of both F. nucleatum and the rest of the community [32, 33]. Schillinger et al. [35] illustrated the usefulness of the two dimensional PCC analysis in vivo by finding high PCC values between synergistic partners F. nucleatum and Tannerella forsythia at short distances. Analogously, we find that the level of intermixing between X. retroflexus and M. oxydans increased as succession proceeded, hereby promoting growth synergy locally in the four-species community compared to regions scant with M. oxydans and three-species community without M. oxydans. These findings demonstrate the important roles that microbial co-localization plays on the activity of whole communities, emphasizing the necessity and significance of exploring spatial organization of multispecies biofilms.
In the present study, an efficient protocol was developed for PCC analyses in 3D. The analytic approach is borrowed from daime (2D based), but with an extra dimension and the possibility of processing many large z-stack images in parallel. Here, where two and four-species biofilm communities were investigated, we found distinct distribution patterns of member species in both horizontal and vertical directions. Similar distribution patterns have been observed in multispecies communities in natural environments and it is thus believed to be a universal phenomenon [36]. Accordingly, the PCC analysis developed in this study provides an alternative quantitative approach for investigating spatial organization of multispecies communities in 3D, as opposed to 2D. Performing pixel quantifications on each layer of the z-stack biofilm images confirmed the findings of the 3D PCC analyses, hereby supporting the strength of the developed method.
We show that the 3D PCC values of species pair X. retroflexus and M. oxydans increased over time at within 10 µm between-species distances in the four-species biofilms, but not in dual-species biofilms. X. retroflexus and M. oxydans was also the pair of species that intermixed to the highest degree in the mature biofilm compared to the other five species pairs. Experimental and computational research supports that both competitive and cooperative interactions may lead to varying levels of interspecies intermixing. Theory predicts that microbial toxins will be most strongly favored when competition for resources is localized and competing species are moderately intermixing [10, 37, 38]. In the present study, interactions that drive the spatial organization of X. retroflexus and M. oxydans in the four-species biofilm are unlikely to be interference competition, because these two strains previously were shown not to inhibit the growth of each other in an antagonistic assay [23]. In addition, it was established that M. oxydans was not able to stably reside in the three-species biofilm in the absence of X. retroflexus, supporting that these two species mutually facilitated the stable establishment of each other during maturation of the four-species biofilm. Collectively, our findings support that cooperative interactions are the main driving force, which results in the intermixing of the two species when co-cultured in the four-species community. In accordance, Momeni et al. [39] have shown that two engineered obligatory cooperative yeasts intermixed when they had large fitness benefits from being spatially colocalized with each other.
Studies of relatively simple and synthetic multispecies communities have showed that micro-scale spatial organization is required for stable community development as it balances competitive and cooperative interactions [40]. It has been found that species frequently modify their gene expression profiles dependent on which other species are present, in order to adapt to the local environment [50]. This is very likely to be reflected in the relative spatial localization of species pairs residing in structured communities, as was found in the current study where biofilm succession was investigated. In addition, the species pair X. retroflexus and M. oxydans only displayed increasing level of intermixing during the succession of the four-species biofilm, implying that different interactions occurred between these two species in the complex four-species biofilms compared to the dual-species ones. Our findings show that species interactions are affected by surrounding microbes and further highlight the importance of investigating complex microbial communities as an entirety and, in the future, apply this narrative to in situ studies, including medical biofilms. Focusing only on one or two dominant species might lead to a misconception of the actual activities occurring in microbial communities, where hundreds of species can co-reside.
Conventionally, interactions between two species have been studied by comparing the productivity of each species growing on its own, with the productivity of each of the two species in a 1:1 co-culture [41, 42]. This type of analysis was also conducted here (Supplemental Figure S3b), however, was of little help when trying to unravel the interactions occurring in four-species biofilms. Another issue with this analysis was that interactions changed as the dual-species biofilms developed. The only two constant influences over time were that X. retroflexus affected S. rhizophila negatively and P. amylolyticus affected X. retroflexus negatively. This approach should be used with caution when studying communities that are composed of more than two species, because the growth fitness of one species is most likely affected by multiple of the co-residing species and can therefore obscure what actual influences one species has on the growth of another. This is an important notion, as the emergences of such community-intrinsic properties seem common in complex multispecies communities [43–46]. Observations in the current study showed that P. amylolyticus was only able to stably establish in biofilms in the presence of X. retroflexus where it, in the four-species biofilm, exhibited the largest bio-volume and highest cell numbers. However, P. amylolyticus and X. retroflexus were in the present study not found to intermix. In addition, the growth of S. rhizophila and X. retroflexus was hampered by the presence of P. amylolyticus in the four-species biofilm compared to mono- and dual-species biofilms. One exception to this was the dual-species biofilm composed of M. oxydans and P. amylolyticus. Overall this suggests that P. amylolyticus acted exploitatively towards other member species. Therefore, we propose that positive interactions occurred between X. retroflexus and M. oxydans were facilitated by, or resulting in, high levels of intermixing and that this might be a response that enabled these strains to cope with P. amylolyticus as the four-species community developed.
Collectively, the findings presented here suggest that the interactions between X. retroflexus and M. oxydans, both low abundant species (in terms of bio-volume), enabled intermixing which facilitated growth synergy of all members locally in the four-species biofilms, compared both to local regions in the four-species biofilm where M. oxydans was scant, and biofilms with fewer species. In accordance, others have illustrated that low abundant species can have large effects on community growth and function [47, 48]. Yet, few studies have focused on this important aspect. Our observations underline the need for advanced and sensitive techniques that allow detection and quantification of low abundant species when studying microbial interactions. Recently Mark Welch et al. [36] succeeded in doing combinatorial labeling and spectral imaging (CLASI) FISH of 15 different bacterial genera residing in oral biofilms. The 3D PCC analysis and general approach presented here is compatible with techniques like CLASI FISH and will enable researchers to investigate spatiotemporal dynamics in highly complex microbial communities that, in addition, will provide empirical data for improved mathematical models that can help overcome some of the challenges encountered when researching community function and dynamics of complex communities [49].
Electronic supplementary material
Acknowledgements
This study was partly funded by grants from The Danish Council for Independent Research; ref no: DFF-1323–00235 (SIMICOM), from the Villum Foundation (ref no. 10098 and 11735) and from the China Scholarship Council.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Søren J. Sørensen, Email: sjs@bio.ku.dk
Jonas S. Madsen, Email: jsmadsen@bio.ku.dk
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0112-2) contains supplementary material, which is available to authorized users.
References
- 1.Hansen SK, Rainey PB, Haagensen JAJ, Molin S. Evolution of species interactions in a biofilm community. Nature. 2007;445:533–6. doi: 10.1038/nature05514. [DOI] [PubMed] [Google Scholar]
- 2.Liu W, Røder HL, Madsen JS, Bjarnsholt T, Sørensen SJ, Burmølle M. Interspecific bacterial interactions are reflected in multispecies biofilm spatial organization. Front Microbiol. 2016 10.3389/fmicb.2016.01366. [DOI] [PMC free article] [PubMed]
- 3.Nadell CD, Drescher K, Foster KR. Spatial structure, cooperation and competition in biofilms. Nat Rev Microbiol. 2016;14:589–600. doi: 10.1038/nrmicro.2016.84. [DOI] [PubMed] [Google Scholar]
- 4.Estrela S, Brown SP. Metabolic and demographic feedbacks shape the emergent spatial structure and function of microbial communities. PLoS Comput. Biol. 2013 10.1371/journal.pcbi.1003398. [DOI] [PMC free article] [PubMed]
- 5.Bao K, Belibasakis GN, Selevsek N, Grossmann J, Bostanci N. Proteomic profiling of host-biofilm interactions in an oral infection model resembling the periodontal pocket. Sci Rep. 2015 doi: 10.1038/srep15999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hellweger FL, Clegg RJ, Clark JR, Plugge CM, Kreft JU. Advancing microbial sciences by individual-based modelling. Nat Rev Microbiol. 2016;14:461–71. doi: 10.1038/nrmicro.2016.62. [DOI] [PubMed] [Google Scholar]
- 7.Kreft J, Picioreanu C, Wimpenny JWT, Van Loosdrecht MCM. Individual-based modelling of biofilms. Microbiology. 2001;147:2897–912. doi: 10.1099/00221287-147-11-2897. [DOI] [PubMed] [Google Scholar]
- 8.Xavier JB, Picioreanu C, Van Loosdrecht MCM. A framework for multidimensional modelling of activity and structure of multispecies biofilms. Environ Microbiol. 2005;7:1085–103. doi: 10.1111/j.1462-2920.2005.00787.x. [DOI] [PubMed] [Google Scholar]
- 9.Borenstein DB, Ringel P, Basler M, Wingreen NS. Established microbial colonies can survive type VI secretion assault. PLoS Comput Biol. 2015;11:1–16. doi: 10.1371/journal.pcbi.1004520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bucci V, Nadell CD, Xavier JB. The evolution of bacteriocin production in bacterial biofilms. Am Nat. 2011;178:162–73. doi: 10.1086/662668. [DOI] [PubMed] [Google Scholar]
- 11.Tait K, Sutherland IW. Antagonistic interactions amongst bacteriocin-producing enteric bacteria in dual species biofilms. J Appl Microbiol. 2002;93:345–52. doi: 10.1046/j.1365-2672.2002.01692.x. [DOI] [PubMed] [Google Scholar]
- 12.Mitri S, Xavier JB, Foster KR. Social evolution in multispecies biofilms. Proc Natl Acad Sci USA. 2011;108:10839–46. doi: 10.1073/pnas.1100292108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Momeni B, Brileya KA, Fields MW, Shou WY. Strong inter-population cooperation leads to partner intermixing in microbial communities. Elife. 2013 10.7554/eLife.00230. [DOI] [PMC free article] [PubMed]
- 14.Hua ZS, Han YJ, Chen LX, Liu J, Hu M, Li SJ, et al. Ecological roles of dominant and rare prokaryotes in acid mine drainage revealed by metagenomics and metatranscriptomics. ISME J. 2015;9:1280–94. doi: 10.1038/ismej.2014.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Z, Klatt CG, Wood JM, Rusch DB, Ludwig M, Wittekindt N, et al. Metatranscriptomic analyses of chlorophototrophs of a hot-spring microbial mat. ISME J. 2011;5:1279–90. doi: 10.1038/ismej.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura Y, Yamamoto N, Kino Y, Yamamoto N, Kamei S, Mori H, et al. Establishment of a multi-species biofilm model and metatranscriptomic analysis of biofilm and planktonic cell communities. Appl Microbiol Biotechnol. 2016;100:7263–79. doi: 10.1007/s00253-016-7532-6. [DOI] [PubMed] [Google Scholar]
- 17.Vikram A, Lipus D, Bibby K. Metatranscriptome analysis of active microbial communities in produced water samples from the Marcellus Shale. Microb Ecol. 2016;72:571–81. doi: 10.1007/s00248-016-0811-z. [DOI] [PubMed] [Google Scholar]
- 18.Pandin C, Le Coq D, Canette A, Aymerich S, Briandet R. Should the biofilm mode of life be taken into consideration for microbial biocontrol agents? Microb Biotechnol. 2017 10.1111/1751-7915.12693. [DOI] [PMC free article] [PubMed]
- 19.Lee KWK, Periasamy S, Mukherjee M, Xie C, Kjelleberg S, Rice SA. Biofilm development and enhanced stress resistance of a model, mixed-species community biofilm. ISME J. 2014;8:894–907. doi: 10.1038/ismej.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren D, Madsen JS, Sørensen SJ, Burmølle M. High prevalence of biofilm synergy among bacterial soil isolates in cocultures indicates bacterial interspecific cooperation. ISME J. 2015;9:81–89. doi: 10.1038/ismej.2014.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daims H, Lücker S, Wagner M. Daime, a novel image analysis program for microbial ecology and biofilm research. Environ Microbiol. 2006;8:200–13. doi: 10.1111/j.1462-2920.2005.00880.x. [DOI] [PubMed] [Google Scholar]
- 22.Goeres DM, Hamilton MA, Beck Na, Buckingham-Meyer K, Hilyard JD, Loetterle LR, et al. A method for growing a biofilm under low shear at the air-liquid interface using the drip flow biofilm reactor. Nat Protoc. 2009;4:783–8. doi: 10.1038/nprot.2009.59. [DOI] [PubMed] [Google Scholar]
- 23.Liu W, Russel J, Røder HL, Madsen JS, Burmølle M, Sørensen SJ. Low-abundant species facilitates specific spatial organization that promotes multispecies biofilm formation. Environ Microbiol. 2017 10.1111/1462-2920.13816. [DOI] [PubMed]
- 24.Hugenholtz P, Tyson GW, Blackall LL. Design and evaluation of 16S rRNA-targeted oligonucleotide probes for fluorescence in situ hybridization. Methods Mol Biol. 2002;179:29–42. doi: 10.1385/1-59259-238-4:029. [DOI] [PubMed] [Google Scholar]
- 25.Korber DR, Lawrence JR, Hendry MJ, Caldwell DE. Analysis of spatial variability within mot+and mot- Pseudomonas fluorescens biofilms using representative elements. Biofouling. 1993;7:339–58. doi: 10.1080/08927019309386264. [DOI] [Google Scholar]
- 26.Li CH, Tam PKS. An iterative algorithm for minimum cross entropy thresholding. Pattern Recogn Lett. 1998;18:771–6. doi: 10.1016/S0167-8655(97)00051-2. [DOI] [Google Scholar]
- 27.Schindelin J, Arganda-carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson G, Beyenal H, Rees WM, Lewandowski Z. Growing reproducible biofilms with respect to structure and viable cell counts. J Microbiol Methods. 2001;47:1–10. doi: 10.1016/S0167-7012(01)00280-9. [DOI] [PubMed] [Google Scholar]
- 29.R development Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2016. [Google Scholar]
- 30.Kolenbrander PE, Palmer RJ, Jr, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 2010;8:471–80. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- 31.Rickard AH, Gilbert P, High NJ, Kolenbrander PE, Handley PS. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 2003;11:94–100. doi: 10.1016/S0966-842X(02)00034-3. [DOI] [PubMed] [Google Scholar]
- 32.Kolenbrander P. Oral microbial communities: biofilms, interactions, and genetic systems. Annu Rev Microbiol. 2000;54:413–37. doi: 10.1146/annurev.micro.54.1.413. [DOI] [PubMed] [Google Scholar]
- 33.Bradshaw AJ, Marsh PD, Watson GK, Allison C. Role of Fusobacterium nucleatum and coaggregation in anaerobe survival in planktonic and biofilm oral microbial communities during aeration. Infect Immun. 1998;66:4729–32. doi: 10.1128/iai.66.10.4729-4732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei H, Håvarstein LS. Fratricide is essential for efficient gene transfer between Pneumococci in biofilms. Appl Environ Microbiol. 2012;78:5897–905. doi: 10.1128/AEM.01343-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schillinger C, Petrich A, Lux R, Riep B, Kikhney J, Friedmann A, et al. Co-localized or randomly distributed? pair cross correlation of in vivo grown subgingival biofilm bacteria quantified by digital image analysis. PLoS ONE. 2012 10.1371/journal.pone.0037583. [DOI] [PMC free article] [PubMed]
- 36.Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci USA. 2016;113:791–800. doi: 10.1073/pnas.1522149113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inglis RF, Gardner A, Cornelis P, Buckling A. Spite and virulence in the bacterium Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2009;106:5703–7. doi: 10.1073/pnas.0810850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gardner A, West SA. Spite and the scale of competition. J Evol Biol. 2004;17:1195–203. doi: 10.1111/j.1420-9101.2004.00775.x. [DOI] [PubMed] [Google Scholar]
- 39.Momeni B, Waite AJ, Shou WY. Spatial self-organization favors heterotypic cooperation over cheating. Elife. 2013 10.7554/eLife.00960. [DOI] [PMC free article] [PubMed]
- 40.Kim HJ, Boedicker JQ, Choi JW, Ismagilov RF. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc Natl Acad Sci USA. 2008;105:18188–93. doi: 10.1073/pnas.0807935105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reisner A, Höller BM, Molin S, Zechner L, Ho BM, Zechner EL. Synergistic effects in mixed Escherichia colibiofilms: conjugative plasmid transfer drives biofilm expansion. J Bacteriol. 2006;188:3582–8. doi: 10.1128/JB.188.10.3582-3588.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vetsigian K, Jajoo R, Kishony R. Structure and evolution of streptomyces interaction networks in soil and in silico. PLoS Biol. 2011 10.1371/journal.pbio.1001184. [DOI] [PMC free article] [PubMed]
- 43.Levine JM, Bascompte J, Adler PB, Allesina S. Beyond pairwise mechanisms of species coexistence in complex communities. Nature. 2017;546:56–64. doi: 10.1038/nature22898. [DOI] [PubMed] [Google Scholar]
- 44.Madsen JS, Røder HL, Russel J, Sørensen H, Burmølle M, Sørensen SJ. Coexistence facilitates interspecific biofilm formation in complex microbial communities. Environ Microbiol. 2016;18:2565–74. doi: 10.1111/1462-2920.13335. [DOI] [PubMed] [Google Scholar]
- 45.Madsen JS, Sørensen SJ, Burmølle M. Bacterial social interactions and the emergence of community-intrinsic properties. Curr Opin Microbiol. 2018;42:104–9. doi: 10.1016/j.mib.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 46.Friedman J, Higgins LM, Gore J. Community structure follows simple assembly rules in microbial microcosms. Nature Ecol. Evol. 2017 10.1038/s41559-017-0109. [DOI] [PubMed]
- 47.Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10:497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kindaichi T, Ito T, Okabe S. Ecophysiological interaction between nitrifying bacteria and heterotrophic bacteria in autotrophic nitrifying biofilms as determined by microautoradiography-fluorescence in situ hybridization. Appl Environ Microbiol. 2004;70:1641–50. doi: 10.1128/AEM.70.3.1641-1650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Widder S, Allen RJ, Pfeiffer T, Curtis TP, Wiuf C, Sloan WT, et al. Challenges in microbial ecology: building predictive understanding of community function and dynamics. ISME J. 2016;10:2557–68. doi: 10.1038/ismej.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hansen LBS, Ren D, Burmølle M, Sørensen SJ. Distinct gene expression profile of Xanthomonas retroflexus engaged in synergistic multispecies biofilm formation. ISME J. 2017;11:300–3. doi: 10.1038/ismej.2016.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.