Abstract
The advent of high-throughput ‘omics approaches coupled with computational analyses to reconstruct individual genomes from metagenomes provides a basis for species-resolved functional studies. Here, a mutual information approach was applied to build a gene association network of a commensal consortium, in which a unicellular cyanobacterium Thermosynechococcus elongatus BP1 supported the heterotrophic growth of Meiothermus ruber strain A. Specifically, we used the context likelihood of relatedness (CLR) algorithm to generate a gene association network from 25 transcriptomic datasets representing distinct growth conditions. The resulting interspecies network revealed a number of linkages between genes in each species. While many of the linkages were supported by the existing knowledge of phototroph-heterotroph interactions and the metabolism of these two species several new interactions were inferred as well. These include linkages between amino acid synthesis and uptake genes, as well as carbohydrate and vitamin metabolism, terpenoid metabolism and cell adhesion genes. Further topological examination and functional analysis of specific gene associations suggested that the interactions are likely to center around the exchange of energetically costly metabolites between T. elongatus and M. ruber. Both the approach and conclusions derived from this work are widely applicable to microbial communities for identification of the interactions between species and characterization of community functioning as a whole.
Introduction
In nature, microbial species exist in complex communities, which feature cell-cell communication, coordination of metabolism, and division of labor across community members [1–5]. As individual behaviors of neighboring cells cascade into a cumulative response, understanding the community structure-function relationship rests on delineating interspecies interactions. While studies have used species co-occurrence to infer potential connectivity between organisms [6–8], coexistence of organisms does not provide specific evidence of their interactions, as the latter requires information at the phenotypic and functional level. Because community responses often appear idiosyncratic and involve multi-level regulatory mechanisms, obtaining a molecular-scale resolution requires an appropriate computational and experimental framework capable of integrating quantitative ‘omics data derived from a variety of different experimental conditions. While it is difficult to apply this approach to microbial communities in nature, technological developments in high-throughput cultivation and analysis [9, 10] make such approaches tractable in the laboratory by using constructed microbial consortia. One such approach involves modeling metabolic interactions between bacterial species. Genome-scale metabolic network-based flux balance analysis (FBA) [11] enables predicting flux distributions and metabolite exchanges and within and across species, which can serve as ab initio hypotheses for experimental validation [12, 13]. Reliable prediction of FBA requires the reconstruction of high-quality metabolic networks, which is a challenging task requiring an iterative process of network refinement and validation [14]. Instead of quantitative FBA, which requires high accuracy in stoichiometry, one may alternatively analyze topological metabolic networks to understand cross-species metabolic interactions [15]. This analysis, described as ‘reverse ecology’ in the literature [16], can infer some key elements of the ecological interactions among species and populations from genomic-based data.
Following the reverse ecology approach, we sought to use a high-throughput method that can facilitate mining and identification of interspecies molecular events in microbial communities from meta-omics data. Building on previous success with mutual information (MI) methods to characterize regulatory responses in single species [17–19], we tested the applicability of the Context Likelihood of Relatedness (CLR) algorithm [20] to reconstruct multi-organism gene-association network from transcriptomic data and predict interactions between organisms from coordinated changes in gene expression. Networks made using MI are able to link genes that are co-expressed across a range of conditions based on the information that one can gather about one gene (variable) compared to another. Networks based on MI that link co-expressed genes can therefore also link genes with similar or coordinated functions [21], allowing prediction of gene function [22]. We previously demonstrated that this method is especially useful for linking genes in biological systems where genomic context is ineffective due to the localization of functionally and structurally related genes at disparate genome locations [18, 23]. We hypothesized that network-based MI analysis would identify genes that are functionally linked (e.g., involved in division of labor or exchange of nutrients) but are found in separate organisms and are thus physically separated. Network inference can aid in grouping these genes and highlighting possible points of interaction between organisms in microbial communities. Previous studies have examined gene co-expression networks of cyanobacterial species but did not infer links across species as we do here [24]. Other studies that have created networks that link genes across species have focused on host/pathogen interactions [25, 26]. However, the studies presented here differ in that many of the interactions we describe are likely complementary in nature and lead to beneficial interactions between each species, in contrast to an infection system where interactions are usually not beneficial to either organism.
To test the applicability of network-based MI analysis for identification of putative inter-species interactions, we carried out a transcriptional gene co-expression network reconstruction using data from a model phototroph-heterotroph binary system, previously developed and characterized in our laboratory [27], which consisted of a unicellular cyanobacterium Thermosynechococcus elongatus strain BP1 (hereafter, T. elongatus) and a Gram-positive chemoheterotrophic bacterium Meiothermus ruber strain A (hereafter, M. ruber). Building on the results of our previous study, which identified clusters of co-expressed genes in both organisms as a function of light and O2 availability [27], we expanded the transcriptomic dataset to 20 additional conditions (Supplemental Table 1) This enabled us to apply a network-based analysis which allowed to us link pairs of genes across species based on MI scores. This approach can provide far more detailed information regarding correlations in gene expression across species and identify individual genes that show statistically significant co-expression, thus allowing for a greater dissection of instances of metabolite exchange and coordination across species. An additional study was carried out by our group on this dataset performing genome-scale metabolic network modeling of T. elongatus-M. ruber binary consortium [28]. Their main goal was to evaluate various alternative approaches of data-model integration for community metabolic network reconstruction. Nevertheless, they predicted transfer of organic nitrogen and carbon sources (such as amino acids and vitamins) by T. elongatus to M. ruber. This commensal provider-consumer relationship predicted by the FBA was aligned with experimental understanding, but the details in the predicted fluxes and exchanged metabolites lack full accuracy. While further network refinement through a labor-intensive manual curation would lead to improved predictions, the difficulty arises in obtaining key data required for model validation such as metabolite exchanges across the two species.
As an alternative approach to examine interactions and exchanges that were not covered by the previous FBA study, we used the high-throughput approach described here to determine where specific genes of T. elongatus and M. ruber are coordinated. The resulting gene co-expression network provides a high level view of both intra- and inter-species regulatory interactions and allows identification of a significant number of cross-species links. This approach identified several links between amino acid synthesis, transport, vitamin metabolism and nitrogen metabolism genes. Gene linkages suggest the transfer of amino acids from T. elongatus to M. ruber and that these amino acids may be a source of organic nitrogen for M. ruber. B12 synthesis and scavenging genes were linked in T. elongatus and M. ruber, respectively and the position of certain uncharacterized genes of M. ruber in the network suggests their role in sugar or amino acid metabolism for this heterotroph. The resulting network is reflective of specific interaction mechanisms which can be inferred based on the functional identities of the interconnected genes. Ability to infer interactions in defined co-culture systems demonstrates that this approach could be applied to more complex communities including diverse environmental systems or even microbiomes of the human body.
Materials and methods
Experimental design and cultivation conditions
The batch and continuous cultivation of axenic T. elongatus in cultures was carried out using modified BG-11 medium as described previously [29]. For the cultivation of axenic M. ruber the BG-11 medium was supplemented with 1% yeast extract. Turbidostat co-cultivation of both species in photobioreactors [9] was carried out at a steady state with a 5.5 L working volume of modified BG-11 agitated at 250RPM and held at a constant 52°C, pH 7.5, incident irradiance of 1995 µmol photons m−2 s−1 and sparged at 4 L min−1 with N2 gas supplemented to 1% with CO2. The relative abundance of T. elongatus and M. ruber at this steady state was 90.2 ± 2.0 and 9.1 ± 1.5% cell-counts, respectively.
To construct a gene association network, we generated a large RNA-seq dataset from 25 distinct culturing conditions that examined M. ruber and T. elongatus. These conditions were all obtained by deriving inoculum from the steady state photobioreactor for batch assays or by transitioning the photobioreactor to new steady states. Specifics of the 25 growth conditions used in this study are shown in Supplemental Table 1. Five of the conditions used (Conditions 21–25) involved transitioning the photobioreactor to new steady states and were generated in a previous study examining T. elongatus/M. ruber growth, details on the conditions can be found in that study [27]. Briefly, we examined the response to three incident irradiance levels 197, 1190, and 1995 µmol photons m−2 s−1 and the response to three partial pressures of O2, 0, 0.3, and 0.59. A subset of these conditions, (conditions 22–25 in Supplemental Table 1) were examined with two biological replicates. To generate the remaining 20 conditions, 400 mL of co-culture cell suspension was harvested from the steady state photobioreactor for batch analysis. Cells were collected by centrifugation, 8000 r.p.m. for 4 min and washed twice in 10 ml of the media corresponding to the specific batch ‘perturbation’ to be performed and resuspended in 200 ml of that media. If the perturbation did not involve a change in media (i.e., temperature or irradiance) then the pellet was washed and incubated in the media used for the ‘Standard’ condition, see Supplemental Table 1. Cultures were incubated under each respective perturbation condition for 4 h before harvesting by centrifugation as above. Supernatants were removed and cell pellets were flash frozen in liquid N2 and stored at −80 °C.
Metatranscriptome sequencing and quantitation
RNA was extracted using the SV Total RNA Isolation System (Promega, Madison, WI) followed by genomic DNA removal and purification using the TURBO DNA-free kit (Life Technologies, Carlsbad, CA). The integrity of the RNA samples was assessed on an Agilent 2100 Bioanalyzer and only RNA samples with an RNA Integrity Number between 7 and 10 were used for library construction. cDNA libraries were synthesized using the Ovation Universal RNA seq system (NuGEN, San Carlos, CA), which also included targeted rRNA depletion using specific InDA-C probes designed to recognize both 16 S and 23 S rRNA from both T. elongatus and M. ruber. The constructed cDNA libraries were examined using the Agilent 2100 Bioanalyzer to confirm proper size and construction and were sequenced on an Illumina NextSeq 500 using 75 bp single end reads. Reads from all samples were aligned to the genomes of Thermosynechococcus elongatus BP-1 (NCBI: NC_004113) and Meiothermus ruber strain A (NCBI: NZ_JXOP01000000.1) using the Burrows Wheeler Aligner (BWA) with the default settings [30]. Gene counts were determined using HT-seq [31] and were normalized with DESeq2 [32]. Gene counts were normalized in a species-specific manner to ensure that changes in abundance under select conditions in one organism will not skew the transcript levels. We obtained an average of 58,741,485 (±13,108,960) reads per sample with multiplexing on the NextSeq. On average 32,514,216 (±8,673,206) aligned to T. elongatus and 1,125,333 (±425,756) aligned to M. ruber (Supplemental Table 2). This corresponds to 27-fold coverage of the M. ruber genome and 940-fold coverage of T. elongates, with this substantial difference in coverage due to low cell density of M. ruber in the co-culture. All RNA-seq fastq files were analyzed with FastQC. No adapter sequences were found in the reads and the quality of all basepair positions in all reads never dropped below a Phred score of 28.
Construction of gene co-expression network
After the data normalization described above, genes with a count of zero in all conditions were removed from the data set leaving a total of 2633 and 2214 genes from the M. ruber and T. elongatus datasets, respectively. Biological replicates for a subset of conditions were averaged by mean. Networks were then inferred from the expression data using CLR [20] along with resampling methods previously described [18]. Briefly, the CLR program computes the mutual information between all gene pairs in the dataset. Mutual information is a measure of the mutual dependence between two variables and is the amount of information in bits needed for one variable to predict the behavior of the other. Once a mutual information matrix is calculated, CLR computes the Z-scores of all gene pairs, which is defined as a number of standard deviations the mutual information score of gene x and gene y is above the average mutual information scores of all gene pairs including either gene x or gene y. This approach is superior to simply taking the mutual information score as it does not highlight or discriminate against genes which may have generally higher or lower mutual information scores with all other genes, respectively. Supplemental Table 3 shows the expression levels and functions of all genes examined with CLR. Unweighted gene co-expression networks were calculated for an interspecies dataset including expression levels from genes of both organisms, as well as individual networks of M. ruber genes only and T. elongatus genes only all using a Z-score of 3.0. When counting the number of edges linking each functional pathway in M. ruber and T. elongatus we used larger cross-species network of 7303 edges using a Z-score cutoff of 2.5. When comparing degree values for genes in networks of two different sizes a normalized degree value was used that divided each degree value by the total number of nodes in that network. In the case of interspecies networks degree values for nodes representing M. ruber genes were divided by total number of nodes representing M. ruber genes only, not the total number of nodes in the network, the same is true for degree values of nodes representing T. elongatus genes. We next used Pearson correlation coefficient to assign a positive or negative value to the correlation of the genes that were found to have edges by CLR. We chose to use CLR to determine edges, rather than Pearson, as mutual information is able to identify edges that may be missed by correlation coefficient, such as those between a gene changing its expression exponentially and one changing is expression linearly. CLR was also ranked higher than Pearson correlation coefficient in a test of accuracy in a previous study [33]. However, CLR does not report whether an edge results from positive or negative correlation of the gene pair while Pearson does. For these reasons CLR was used to determine edges and Pearson correlation coefficient data was used to determine positive and negative correlation for those edges already identified by CLR. Viewing of networks and determination of centrality measurements were done using Cytoscape [34]. A force-directed layout was used for all networks. Functional enrichment on certain gene groups was also carried out using Fisher’s exact test and annotation data obtained from KEGG.
Results and discussion
Topological features of the co-culture transcriptional network reveal functional inter-species linkages
The applicability of the CLR approach for community network reconstruction was tested using the T. elongatus-M. ruber binary culture previously developed by our group [27]. This photoautotroph-heterotroph association is based on a commensal relationship, in which M. ruber is dependent on T. elongatus for the production of organic carbon, reduced nitrogen sources and vitamin B12. Previous studies suggested that the cyanobacterium responds to co-cultivation under varying irradiance and O2 levels through changes in the expression level of core genes involved in photosynthesis, carbon uptake/fixation, vitamin biosynthesis and scavenging of reactive oxygen [27]. To identify the putative interactions and pathways leading to the exchange of metabolites and coordination of metabolism between T. elongatus and M. ruber, we reconstructed and analyzed a binary culture network built from transcriptomic data collected during co-cultivation of both organisms under 25 separate conditions (Supplemental Tables 1, 2). The edges within the network were then cross-referenced with Pearson correlation coefficient data to determine whether gene pairs in the network were positively or negatively correlated in their expression (Fig. 1). Pearson correlation was only used to determine the direction of the correlation, the magnitudes of Pearson correlation were not used.
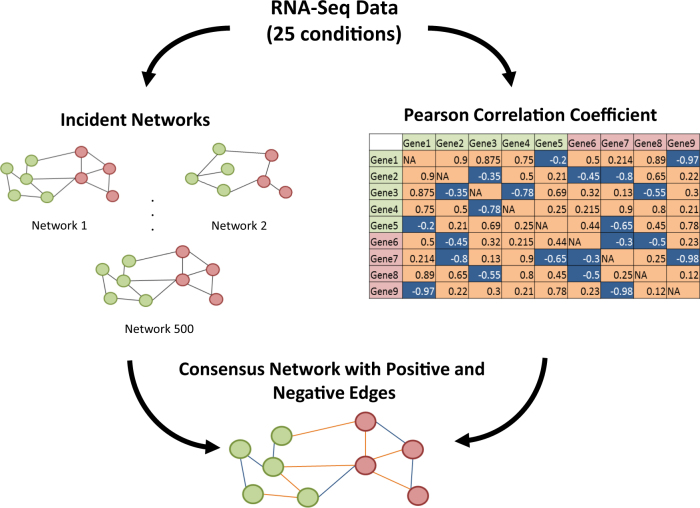
Fig. 1.
Methodology of Cross-Species Network. Both T. elongatus and M. ruber were cultured under 25 different environmental conditions and RNA-seq data was collected. Transcriptomic data was used to build 500 incident networks with random removals of 20% of the data for each incident network. These 500 incident networks were then combined into a single consensus network that averaged mutual information scores for all gene pairs. Transcriptomic data was also used to generate a Pearson correlation coefficient dataset. Both data types were combined to assign positive (orange) or negative (blue) values to the edges in the CLR network
The reconstructed network of 4408 (2344 M. ruber and 2064 T. elongatus) genes was inferred using a Z-score cutoff of 3.0 and contained 38,778 edges, of which 1396 (3.6%) were interspecies edges that connected genes in both organisms (Supplemental Fig. 1). As edges between species point to transcriptional-level coordination of cellular functions and, possibly, metabolite exchange between the organisms, we next created a subnetwork consisting only of edges that connected genes from both T. elongatus and M. ruber (Supplemental Fig. 1). Using all of the 1396 edges delineated from the cross-species subnetwork, we assigned each edge a positive or negative value based on its Pearson correlation coefficient with the corresponding node (i.e., gene) pair. This resulted in 743 positive edges (i.e., the expression of the gene pairs comprising these edges has a positive correlation) and 653 negative edges, (i.e., the expression of the gene pairs comprising these edges has a negative correlation) (Supplemental Table 4). We also inferred networks consisting of T. elongatus genes only and M. ruber genes only also using a Z-score cutoff of 3.0 (Supplemental Fig. 2).
To elucidate the linkages between network topological features and gene functionality, we next calculated the degree value (i.e., the number of connecting edges per node) of all genes in the inter-species network and grouped them into functional categories. We next compared the average degree value of a particular functional category in the cross-species subnetwork (based on the average degree values of genes within that function) with the degree value of that same functional category in a single-species network. We chose to use degree as it has been used in several previous studies as a metric of centrality with success [35–37]. As previous studies have shown that genes of high degree and centrality are those that are particularly important to bacterial metabolism [38–40], we reasoned that an increase in the degree value among genes in the cross-species subnetwork suggests that a particular function is essential to cross-species metabolism (Fig. 2a). M. ruber functions with increased degree values in the cross-species subnetwork included cell motility and surface adherence functions, as well as amino acids, defense systems, regulation of gene expression, and terpenoid/polyketide metabolism. Notably, many of these functions overlapped with T. elongatus, where increased degree values in the cross-species subnetwork also included defense and invasion response functions, as well as nitrogen, glycan and terpenoid/polyketides metabolism genes.
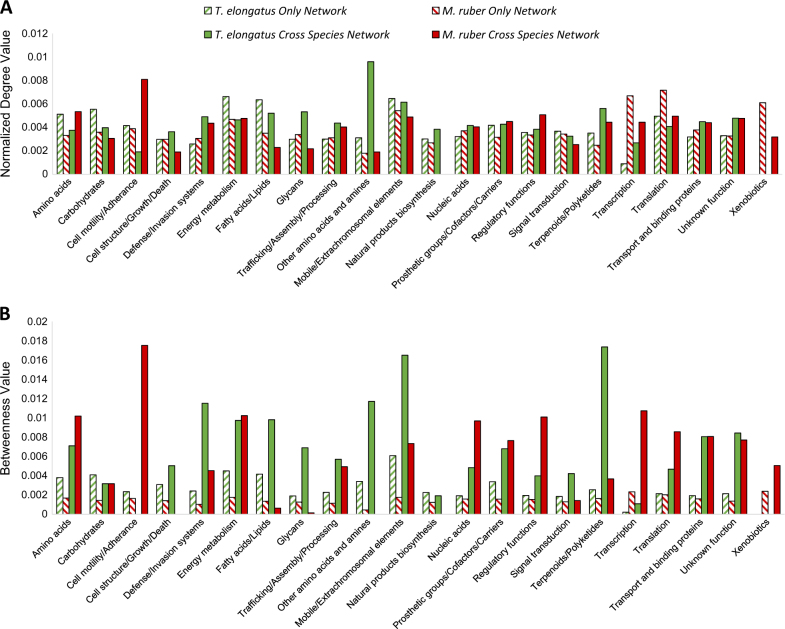
Fig. 2.
Degree values of T. elongatus and M. ruber functions. a Average normalized degree values for genes in each of the functions shown on the x-axis were calculated for a network comprised only of M. ruber genes (red hatched bars), comprised only of T. elongatus genes (hatched green bars) or for M. ruber (filled red bars) or T. elongatus (filled green bars) genes in the cross-species network. Degree values are shown on the y-axis and are normalized to the total nodes in the network. b Same as a but using betweenness as a measure of centrality rather than degree
M. ruber genes displaying increased degree values offer some interesting biological insight into microbial co-culture dependencies. For instance, our previous studies [27, 41] collected confocal micrographs that qualitatively showed tight physical associations between cyanobacteria and heterotrophs. While this related result does not directly account for the importance of cell adhesion in M. ruber, combined with results we see here, that cell adherence genes occupy central positions in a cross-species subnetwork, it does supply corroborating evidence that heterotrophic cells are likely more successful at scavenging carbon, nitrogen, O2, exopolymers and released organic substrates when physically attached to their cyanobacterial partners. Similarly, the increased M. ruber degree values for amino acid metabolism are likely related to the fact that heterotrophic cells cannot assimilate inorganic nitrogen in this experiment and rely on proteins, peptides or direct amino acid exchanges from T. elongatus. The relevance for terpenoid/polyketide metabolism—shared between both species—was less intuitive and suggested the possibility of as yet unexplored interactions between these two species. One interpretation relates to the fact that many of the experimental conditions were performed under high incident irradiance (up to 1995 µmol photons m−2 s−1) which likely presented an environmental stimulus for expressing photoprotective pigments such as carotenoids that relate to the broad functional class of terpenoid/polyketide metabolism. At the same time, M. ruber is known to produce a number of carotenoids [42]. The synthesis of related molecules from T. elongatus and M. ruber may lead to specific exchanges centered on these metabolites and to a central position for genes involved in this synthesis in a cross-species network. The increase in centrality for glycan related genes in both T. elongatus and M. ruber is also of interest. Other studies have found a role for surface glycans in cyanobacteria in recognizing symbiotic partners [43, 44]. While these studies have been mainly limited to interactions of cyanobacteria and plants, the central position of glycans here may suggest that these molecules have a role in the recognition of other kinds of partners, such as M. ruber.
The network topology was also mined for biological insight by quantifying betweenness as a measure of centrality in addition to degree (Fig. 2b). Nearly all of the functions for both species that we found to be increased using degree as a centrality measure were also found to be increased when using betweenness. Notably, we also found that cell adherence genes of M. ruber showed increased centrality in a cross-species network compared to the single-organism network pointing at a potential role of cell–cell interactions in the co-culture. Similar changes in betweenness were displayed by the terpenoid and glycan genes of T. elongatus in a cross-species network compared to the same genes in a T. elongatus-only network thus underscoring the essential role of pigment and cell wall biosynthesis in photosynthetic metabolism. There were also differences when using betweenness compared to degree. Certain functions that were not of higher centrality in the cross-species network by degree were higher by betweenness. These include energy metabolism (both species), fatty acids and lipids (T. elongatus), extrachromosomal functions (both species), as well as transcription, translation and transport (both species). While betweenness and degree are both measures of centrality they can point to different aspects of interspecies regulation. Genes of high degree in the cross-species network are those that are connected to a large number of genes from the opposite species. The position of these genes as hubs means that they are likely strongly involved with a single process in the opposite species, having edges with several genes in the same proximity within the network. Other studies have also shown that networks are particularly sensitive to the removal of such genes, biological networks tend to be robust against perturbation but removal of hub genes of high degree can have a major impact on network topology [45, 46]. Therefore, those processes that have the highest degree value increases in the cross-species network are those that are especially important to co-cultivation. In the case of M. ruber these include both adherence genes and defense/invasion responsive genes, in the case of T. elongatus they include amine metabolism and glycan metabolism genes. M. ruber is completely dependent on T. elongatus for several metabolites so it is biological intuitive that adherence genes, which allow M. ruber to physical associate with T. elongatus cells, their source of nitrogen and carbon, are of critical importance. In the same way, M. ruber respiration reduces the partial pressure of O2, thus increasing T. elongatus’ selectivity of CO2 during growth. Therefore, genes of T. elongatus that promote the growth of M. ruber, glycan and amine genes providing needed carbon and nitrogen, are of high degree.
In contrast genes of high betweenness occupy positions linking two larger groups of genes suggesting that they act to link multiple processes in the opposite species, having edges with two distinct clusters which may be enriched for different processes. While several of the processes that had high degree also had high betweenness in the cross-species network there were certain functions that only had high betweenness values suggesting their centrality is mainly a result of linking two larger groups of genes in the opposite species. For M. ruber this includes energy metabolism and transcription/translation. For T. elongatus this includes fatty acids and terpenoids/polyketides among others. Genes in these groups may act to link distinct groups of genes in the opposite species and thus may be part of exchanges of metabolites or possibly division of labor processes.
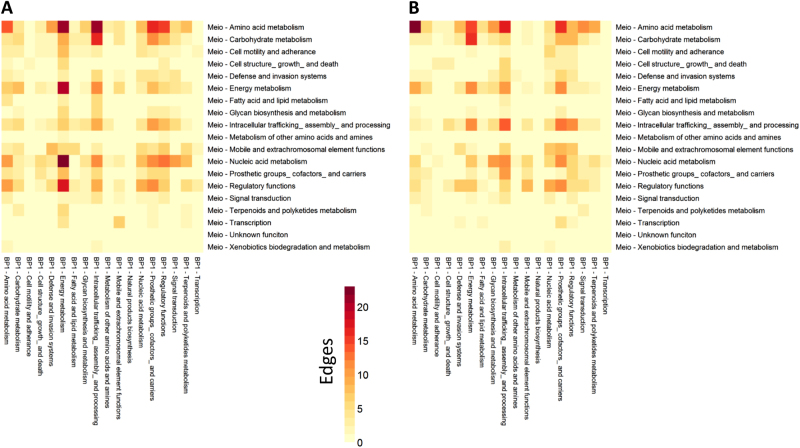
We next looked in more detail at which particular functions were linked in the cross-species network and whether these linkages came from positive or negative edges, as determined by Pearson correlation coefficient. A large number of edges linked transport and translation genes of M. ruber and T. elongatus and for ease of viewing these have been grouped into separate figures (Supplemental Figs. 3, 4). Most of these edges centered on ribosomal proteins of M. ruber having positive and negative edges with a number of functions of T. elongatus suggesting that growth of M. ruber (ribosomal proteins being a proxy for growth) is linked to metabolic activity of T. elongatus. Of the remaining edges, functional enrichment of M. ruber and T. elongatus network nodes revealed that amino acid metabolism genes of M. ruber had a large number of positive cross-species edges with T. elongatus energy metabolism and intracellular trafficking functions (Fig. 3a). A similar correlation pattern was also observed, although to a lesser extent, for the nucleic acid metabolism genes of M. ruber which had a higher number of positive edges with energy metabolism and prosthetic/cofactor biosynthesis genes of the cyanobacterium. For T. elongatus, the highest number of positive interspecies edges was observed for energy metabolism genes which were linked to the amino acid metabolism, energy metabolism, nucleic acid metabolism and regulatory functions of M. ruber. Among negative edges (Fig. 3b), M. ruber amino acid metabolism genes represented by far the largest group that displayed negative correlations with T. elongatus genes in the cross-species network. Notably, the highest number of T. elongatus negative edges was also found across amino acid metabolism, as well as co-cofactor biosynthesis genes, which likely points at the uptake of these excreted cyanobacterial metabolites by the heterotroph. Negative correlations were also found for intracellular trafficking genes of both organisms as well energy metabolism and carbohydrate transport genes of M. ruber further pointing at the metabolic dependence of the heterotroph on the organic carbon excretion by the cyanobacterium.
Fig. 3.
Number of edges between functions of T. elongatus and M. ruber. a A network with a Z-score cutoff of 2.5 was used to increase total edges and gain a more detailed view of interactions. The number of positive edges for each function in T. elongatus (x-axis) and M. ruber (y-axis) is indicated, darker colors indicate more edges. Meio refers to M. ruber and BP1 to T. elongatus. b Identical to a but showing negative edges for each function in T. elongatus (x-axis) and M. ruber (y-axis), darker colors indicate more edges
Gene-level resolution allows identification of putative interaction mechanisms
To identify the specific pathways driving the co-culture growth, we next examined the specific genes of T. elongatus and M. ruber genes that were connected by edges in the inter-species network. As our primary objective was to validate the applicability of transcriptional context-based networks to delineate interactions between community members, we focused on metabolite cross-feeding that we had previously predicted to underlie the commensal growth of the T. elongatus-M. ruber co-culture [27]. Consistent with the experimental constraints, the co-culture network contained a number of edges that linked genes that are likely to participate in the exchange of reduced vitamins, nitrogen species and organic carbon compounds.
Vitamin B12 is an important cofactor which, among other functions [47], is required for methionine synthesis. Since M. ruber is auxotrophic for B12, an exogenous supply of this co-factor is required for growth, which during co-cultivation can be provided by the B12 prototroph T. elongatus [27]. Notably, genes encoding putative B12 scavenging proteins in M. ruber (btuF; SY28_RS12145) were inversely correlated to B12 biosynthesis genes in T. elongatus (cobQ, tll1716. When plotting the expression profile across conditions for btuF and cobQ, a clear negative relationship can be observed based on Pearson correlation coefficient of −0.65 (Supplemental Fig. 5). B12 exchange appeared to also affect other metabolic functions in each species. Specifically, the M. ruber metH gene (SY28_RS08890) which encodes methionine synthase, a B12-dependent enzyme, also had a large number of edges with T. elongatus genes. Of 14 edges, 8 were negative and linked to T. elongatus genes involved in fatty acid synthesis (for example, fabI, accD, tll1693, tlr1643), while the positive edges connected to two putative transporters (tll1329, tlr1438) though the specific molecule they may transport is unknown.
Given the inability of M. ruber to carry out assimilatory nitrate reduction, the organism is dependent on T. elongatus for a source of reduced nitrogen when only NaNO3− is used as the sole nitrogen source [27]. Collective evidence from this study suggests that amino acids or small peptides could be acting as an organic nitrogen source that is being transferred from T. elongatus to M. ruber. First, amino acid and nitrogen metabolism genes of both T. elongatus and M. ruber show higher degree values in the cross-species network compared to axenic M. ruber or T. elongatus networks (Fig. 2a), in addition four of six edges between amino acid metabolism genes in M. ruber and T. elongatus are negative edges. Negative edges between similar metabolism processes suggests that one species carries out the process, the resulting metabolites are shared and the partner species has no need to carry out the same process and thus downregulates genes involved in that process. This is also apparent with the large number of amino acid synthesis genes of M. ruber having negative edges with amino acid synthesis genes of T. elongatus (Fig. 3b).
Further evidence can be found in the ammonium transporter (amt; SY28_RS01750) of M. ruber, which had both negative and positive edges with several T. elongatus genes (Fig. 4a). Genes with negative edges included, among others, three amino acid metabolism genes (aguA, aguB, thrCII; tlr0111, tlr0112, tlr0982) and a sulfur metabolism gene (sir; tlr0339). Genes with positive edges were mainly unknown in function though a cytochrome C gene (coxC; tll2009) was positively associated with ammonium transport. In addition, there were three peptide transporter genes in M. ruber that had positive edges with amino acid synthesis genes of T. elongatus (Fig. 4a). The fact that only negative edges exist between the ammonium transporter of M. ruber and amino acid metabolism genes of T. elongatus suggest that an organic nitrogen source in the form of amino acids is being provided by T. elongatus and it is less energetically favorable for M. ruber to assimilate ammonia to synthesize amino acids under these conditions. This is further supported by the presence of positive edges between peptide transporters of M. ruber and amino acid synthesis genes of T. elongatus (Fig. 4a). As these nitrogen sources are being produced by T. elongatus, M. ruber upregulates genes involved in their transport and assimilation. In addition to this data specific edges existed that crossed species for genes coding for proteins that synthesize energetically costly amino acids with branched side chains such as tryptophan and phenylalanine. These include pheA2 (SY28_RS10870) and trpE (SY28_RS04715) in M. ruber and aroE (tll0590) in T. elongatus. Finally, two of the peptide transporters in M. ruber that have edges with T. elongatus genes (SY28_RS06075 and dppC; SY28_RS10470) are expressed at lower levels (1.5 and 2.3-fold, respectively) when an alternative nitrogen source, NH4, is present in one of the experimental conditions used to generate the network.
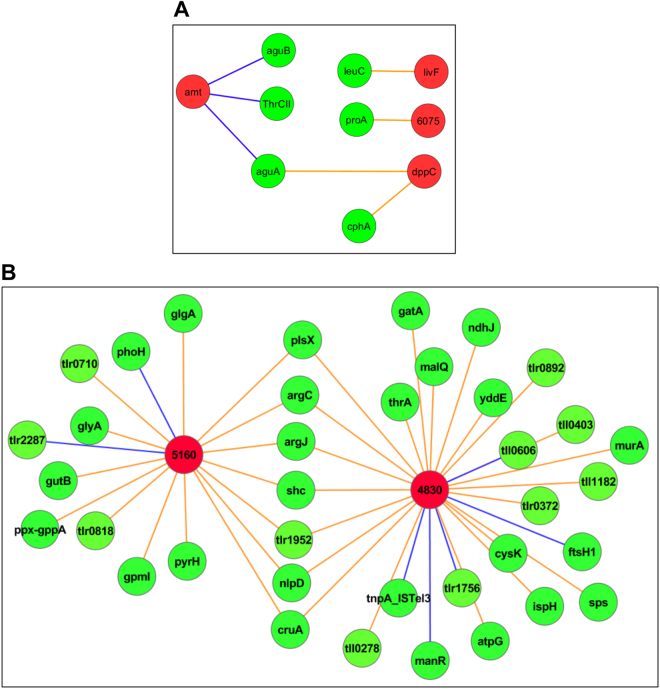
Fig. 4.
Specific edges between nitrogen metabolism and unknown genes of M. ruber and T. elongatus. a Ammonium transporters of M. ruber (amt) show negative edges to amino acid synthesis genes of T. elongatus (green circles) while M. ruber peptide transporters had positive edges to amino acid synthesis genes of T. elongatus. b Uncharacterized genes of M. ruber (red circles) show a large number of edges indicating positive correlation (direct edges, orange lines) and negative correlation (inverse edges, blue lines). Many of these edges are to the same T. elongatus genes
We also observed that two uncharacterized genes of M. ruber (SY28_RS04830, SY28_RS05160) each had several edges with T. elongatus genes (27 and 17, respectively); in several instances, both of these genes had edges to the same T. elongatus genes (Fig. 4b). When examining these 37 T. elongatus genes that had an edge with one or both of the uncharacterized M. ruber genes, carbohydrate and amino acid metabolism functions were significantly enriched (p-value of 0.006 and 0.012, respectively) as were terpenoid metabolism functions (p-value < 0.015) (Supplemental Table 5). While the role of these M. ruber genes is unknown, their position in the network suggests their importance to cross species interactions, likely centering on sugar or amino acid metabolism or transport. This notion is also supported by the increase in relative expression levels of these genes (compared to the mean across all conditions) when M. ruber was cultured with exogenous carbon sources (i.e., glucose and lactate, data not shown). Under the majority of experimental conditions, no exogenously added organic carbon was available for M. ruber and carbon must be obtained from T. elongatus. The annotated function of these M. ruber genes is unknown, though SY28_RS05160 does contain the DUF4900 domain. The linking of these M. ruber genes with T. elongatus carbon genes combined with the larger number of negative (compared to positive) edges linking carbohydrate processes of M. ruber with energy metabolism genes of T. elongatus (Fig. 3) can begin to reveal how carbon sources are shared between these species. Organic carbon is only produced by T. elongatus as CO2 is the only carbon source added to the co-culture. M. ruber can gain access to this organic carbon either through uptake after death and cell lysis of T. elongatus or through collection from excreted carbon sources from living T. elongatus, possibly aided by the tight physical association seen with M. ruber to T. elongatus cells. The ultimate collection strategy is likely a combination of both but the tight coordination between energy metabolism genes of T. elongatus and carbohydrate genes of M. ruber suggest that M. ruber has access to a not insignificant amount of carbon excreted by still living and metabolically active T. elongatus.
While functional gene association and correlative linkages within the inter-species CLR network are reflective of specific interactions, they can also point to mechanisms of co-regulation and/or co-acclimation to growth in the presence of a partner. In particular, this is reflected in the large diversity of T. elongatus genes with high degree values in the cross-species network. For instance, T. elongatus photosystem genes also had a large number of negative edges with energy metabolism genes of M. ruber, specifically NADH dehydrogenase genes (nuoG, ndh, nuoD, nuoC; SY28_RS08610, SY28_RS13475, SY28_RS13910, SY28_RS13915) and positive edges with ribosomal protein genes (rplM, rplE; SY28_RS06500, SY28_RS06635).
Network analysis of M. ruber and T. elongatus also allowed us to further the outcomes of our previous study [27], which identified instances of metabolic cross-talk and coordination between the species from clusters of co-expressed genes. For instance, while sugar synthesis genes of T. elongatus were generally co-clustered with carbon uptake and metabolism genes of M. ruber, the network analysis identified gene-specific linkages (rather than broad clusters of genes) between these two organisms (Fig. 4b) which included uncharacterized genes of M. ruber (SY28_RS04830 and SY28_RS05160) that have a number of links to sugar synthesis genes of T. elongatus. The network linkages also point to a putative role for these M. ruber genes in sugar synthesis or uptake (collecting sugars after they have been synthesized by T. elongatus), a new observation as currently there is nothing known regarding the function of these proteins. Furthermore, while the clustering analysis grouped T. elongatus B12 synthesis with M. ruber B12 scavenging genes [27], the network analysis identified a specific edge between cobQ of T. elongatus and btuF of M. ruber. The network approach also allowed us to link metH from M. ruber with several T. elongatus genes of various functions (pglS, petL, accD, fabI, tnpA_ISTel2, ycf50, mrcB, era), pointing to potentially new interactions among these organisms driven by B12 metabolism. Finally, network inference allowed us to identify gene-specific interactions facilitating the exchange of organic nitrogen in the co-culture. As M. ruber lacks the ability to use nitrate and depends on T. elongatus to provide fixed organic nitrogen, the nitrogen and amino acid metabolism genes of M. ruber were found in same clusters as amino acid synthesis genes of T. elongatus. Network analysis not only identified edges between specific genes and pathways but also provided specific hypotheses regarding the metabolic co-regulation between the cyanobacterium and the heterotroph. Specifically, the negative correlation of ammonium uptake genes of M. ruber (amt, SY28_RS01750) with amino acid synthesis genes of T. elongatus (aguA, aguB, thrCII) seen in the current study suggested that amino acids may be a source of organic nitrogen for M. ruber.
Metabolic dependency of M. ruber on T. elongatus revealed by gene co-expression network analysis in this work is also well supported by the FBA results reported by Henry et al. [28] carried out on the same dataset. However, much like the clustering analysis above we also identify new instances of interaction in our network study that were absent in the FBA analysis. In an autotrophic growth medium, for example, the FBA model predicted the T. elongatus’ provision of amino acids such as proline, leucine, histidine, and glycine, and isoleucine. The network presented here identifies a positive edge between a branched amino acid uptake gene of M. ruber (livF; SY28_RS00355) and an isoleucine synthesis gene of T. elongatus (leuC; tlr0909). In addition, another peptide uptake gene of M. ruber (SY28_RS06075) has a positive edge with a proline synthesis gene of T. elongatus (proA; tlr0764). The metabolic model also predicted the provision of cofactors involved in energy generation such as NADP + , a precursor of NAD + (i.e., nicotinamide mononucleotide or NMN). This gene association network presented here also predicts several edges between NAM salvage genes of M. ruber (pncB, nrtR, pnuC; SY28_RS02380, SY28_RS02390, SY28_RS02375) and central metabolic genes of T. elongatus. In addition, citrate (an intermediate in the TCA cycle, which also contributes to energy generation, as well as a source of organic nitrogen) was predicted by the FBA model to be provided by T. elongatus and the gene co-expression network identifies links between citrate metabolism genes of T. elongatus (fumC, gltA; tll1534, tlr2393) and genes of M. ruber. Finally, there were certain metabolites such as vitamin B12 (i.e., cobalamin) that are believed to be provided from T. elongatus to M. ruber, but was not predicted by the FBA model. This gap might be caused by incomplete reconstruction of metabolic pathways involved in B12 synthesis and transport in the community network model and might be resolved in principle by further network refinement. In contrast, the gene co-expression network analysis in this work was able to correctly predict the B12-associated metabolism between the two species by identifying putative B12 scavenging proteins in M. ruber to be inversely correlated to B12 biosynthesis genes in T. elongatus. This prediction can serve as a new observation that can be incorporated back into metabolic network refinement for improved FBA predictions. In addition, neither the FBA model, nor the earlier clustering study by Bernstein, et. al., predicted interactions between genes synthesizing terpenoids and polyketides of either species, while the gene co-expression network highlighted both of these processes as highly important in a cross-species network. This emphasizes that a mutual information network approach is critical for analyzing interactions in systems with poorly annotated metabolism and/or where genome reconstructions are not yet available. Gene co-expression networks can also act as crucial companion datasets to FBA analyses as both approaches identify putative interactions that might be missed by the other. Furthermore, the ability to infer interactions in multi-species system demonstrates the potential of this approach to uncover new structure-function relationships in environmental, plant, and human microbiomes. In that regard the ability to incorporate dynamic data from multiple time points can significantly expand the utility of the multi-species network approach and provide high-level view of how interactions change as a function of external inputs and shifts in environment variables.
Conclusions
Microbial communities are important drivers of ecology and understanding the interactions and coordination between species in them is crucial to understanding ecosystems. These studies demonstrate how network inference can be used to highlight potential points of interaction and exchange of nutrients between phototrophic and heterotrophic species growing in co-culture (Fig. 5). Specific genes involved with carbon and nitrogen reduction and acquisition in both species are significantly co-expressed, identifying where these exchanges and interactions might be taking place at the single gene level. These experiments are among the first to build a co-expression network linking genes across species in a phototroph-heterotroph system and the knowledge gained will be critical not only in understanding interactions specific to these two species but also in building paradigms that describe fundamental coordination of metabolism and exchange of nutrients that are common to a large number of bacterial communities.
Fig. 5.
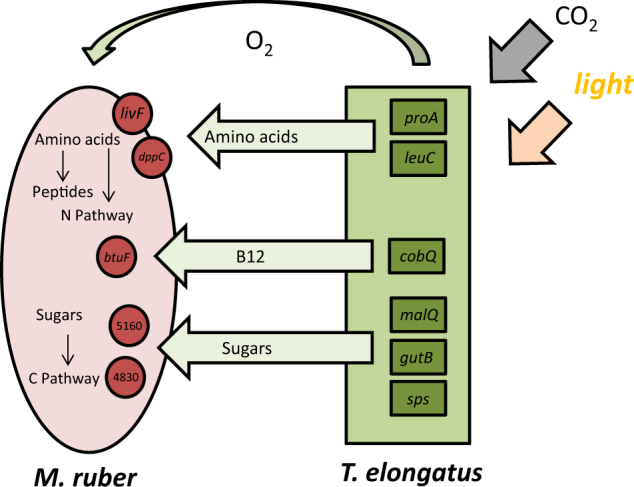
Conceptual model of putative interactions and coordination between T. elongatus and M. ruber. Genes with specific edges in M. ruber and T. elongatus are shown, as well as the putative metabolite and nutrient transfers they are involved in. Amino acid synthesis genes of T. elongatus are linked to peptide transporters of M. ruber as are B12 synthesis genes in T. elongatus and B12 scavenging genes of M. ruber. Certain uncharacterized genes of M. ruber had a larger number of edges to a variety of T. elongatus genes, specifically those involved in sugar synthesis, suggested that these uncharacterized heterotroph genes may be involved in sugar metabolism or transport in M. ruber
Availability of data and materials
All transcriptome data generated in the current study have been deposited into GEO (Accession: GSE113134).
Electronic supplementary material
Supplemental Figure 1: Cross-Species Network
Supplemental Figure 2: Singe Species Network
Supplemental Figure 3: Positive Edges Of Transport and Translation Genes
Supplemental Figure 4: Negative Edges Of Transport and Translation Genes
Supplemental Figure 5: Pearson correlation of btuB and cobQ
Supplementary Table 1: Culture Conditions
Supplementary Table 2: RNA-seq alignment statistics
Supplementary Table 3: Gene expression levels
Supplementary Table 4: Pearson Correlation Coeffecients of Edges
Supplementary Table 5: Functional enrichment of T. elongatus genes linked to uncharacterized M. ruber genes SY28_RS04830 and SY28_RS05160
Acknowledgements
The authors wish to acknowledge Drs. Margie Romine, William Nelson, and Jim Fredrickson for help with the functional genome annotation and manuscript. PNNL is operated for the DOE by Battelle Memorial Institute under Contract DE-AC05-76RLO 1830. The research was supported by the Genomic Science Program (GSP), Office of Biological and Environmental Research (BER), U.S. Department of Energy (DOE), and is a contribution of the PNNL Foundational Scientific Focus Area (FSFA). A significant portion of the research was performed using the Environmental Molecular Sciences Laboratory (EMSL), a national scientific user facility sponsored by DOE BER and located at PNNL. HCB is grateful for the support of the Linus Pauling Distinguished Postdoctoral Fellowship program at PNNL.
Author contributions
RSM performed the experiments, analyzed the data and wrote the manuscript, CCO analyzed the data and contributed to the manuscript preparation, EAH contributed to the experiments, MC contributed to the experiments, HCB contributed to the experiments and manuscript preparation, JM contributed to the experimental design and manuscript preparation, ASB developed the project concept, contributed to the experimental design, and wrote the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0145-6) contains supplementary material, which is available to authorized users.
References
- 1.Di Cagno R, De Angelis M, Calasso M, Gobbetti M. Proteomics of the bacterial cross-talk by quorum sensing. J Proteom. 2011;74:19–34. doi: 10.1016/j.jprot.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Little AE, Robinson CJ, Peterson SB, Raffa KF, Handelsman J. Rules of engagement: interspecies interactions that regulate microbial communities. Annu Rev Microbiol. 2008;62:375–401. doi: 10.1146/annurev.micro.030608.101423. [DOI] [PubMed] [Google Scholar]
- 3.Nadell CD, Xavier JB, Foster KR. The sociobiology of biofilms. FEMS Microbiol Rev. 2009;33:206–24. doi: 10.1111/j.1574-6976.2008.00150.x. [DOI] [PubMed] [Google Scholar]
- 4.Pande S, Shitut S, Freund L, Westermann M, Bertels F, Colesie C, et al. Metabolic cross-feeding via intercellular nanotubes among bacteria. Nat Commun. 2015;6:6238. doi: 10.1038/ncomms7238. [DOI] [PubMed] [Google Scholar]
- 5.Benomar S, Ranava D, Cardenas ML, Trably E, Rafrafi Y, Ducret A, et al. Nutritional stress induces exchange of cell material and energetic coupling between bacterial species. Nat Commun. 2015;6:6283. doi: 10.1038/ncomms7283. [DOI] [PubMed] [Google Scholar]
- 6.Cazelles K, Araujo M, Mouquet M, Gravel D. A theory for species co-occurance in interaction networks. Theor Ecol. 2016;9:39–48. doi: 10.1007/s12080-015-0281-9. [DOI] [Google Scholar]
- 7.Harris D. Inferring species interactions from co-occurance data with Markov networks. Cold Spring Harbor Laboratory. 2015.
- 8.Madeo D, Comolli LR, Mocenni C. Emergence of microbial networks as response to hostile environments. Front Microbiol. 2014;5:407. doi: 10.3389/fmicb.2014.00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melnicki MR, Pinchuk GE, Hill EA, Kucek LA, Stolyar SM, Fredrickson JK, et al. Feedback-controlled LED photobioreactor for photophysiological studies of cyanobacteria. Bioresour Technol. 2013;134:127–33. doi: 10.1016/j.biortech.2013.01.079. [DOI] [PubMed] [Google Scholar]
- 10.Wittebolle L, Marzorati M, Clement L, Balloi A, Daffonchio D, Heylen K, et al. Initial community evenness favours functionality under selective stress. Nature. 2009;458:623–6. doi: 10.1038/nature07840. [DOI] [PubMed] [Google Scholar]
- 11.Orth JD, Thiele I, Palsson BO. What is flux balance analysis? Nat Biotechnol. 2010;28:245–8. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biggs MB, Medlock GL, Kolling GL, Papin JA. Metabolic network modeling of microbial communities. Wiley Interdiscip Rev Syst Biol Med. 2015;7:317–34. doi: 10.1002/wsbm.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardona C, Weisenhorn P, Henry C, Gilbert JA. Network-based metabolic analysis and microbial community modeling. Curr Opin Microbiol. 2016;31:124–31. doi: 10.1016/j.mib.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Thiele I, Palsson BO. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc. 2010;5:93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borenstein E, Feldman MW. Topological signatures of species interactions in metabolic networks. J Comput Biol. 2009;16:191–200. doi: 10.1089/cmb.2008.06TT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levy R, Borenstein E. Reverse ecology: from systems to environments and back. Adv Exp Med Biol. 2012;751:329–45. doi: 10.1007/978-1-4614-3567-9_15. [DOI] [PubMed] [Google Scholar]
- 17.Ishchukov I, Wu Y, Van Puyvelde S, Vanderleyden J, Marchal K. Inferring the relation between transcriptional and posttranscriptional regulation from expression compendia. BMC Microbiol. 2014;14:14. doi: 10.1186/1471-2180-14-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McClure RS, Overall CC, Mcdermott JE, Hill EA, Markillie LM, McCue LA, et al. Network analysis of transcriptomics expands regulatory landscapes in Synechococcus sp. PCC 7002. Nucleic Acids Res. 2016;44(18):8810–8825. doi: 10.1093/nar/gkw737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Netotea S, Sundell D, Street NR, Hvidsten TR. ComPlEx: conservation and divergence of co-expression networks in A. thaliana, Populus and O. sativa. BMC Genom. 2014;15:106. doi: 10.1186/1471-2164-15-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faith JJ, Hayete B, Thaden JT, Mogno I, Wierzbowski J, Cottarel G, et al. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Dam S, Vosa U, Van Der Graaf A, Franke L, De Magalhaes JP. Gene co-expression analysis for functional classification and gene-disease predictions. Brief Bioinform. 2017;p.1–18. [DOI] [PMC free article] [PubMed]
- 22.Wolfe CJ, Kohane IS, Butte AJ. Systematic survey reveals general applicability of “guilt-by-association” within gene coexpression networks. BMC Bioinforma. 2005;6:227. doi: 10.1186/1471-2105-6-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDermott JE, Oehmen CS, McCue LA, Hill E, Choi DM, Stockel J, et al. A model of cyclic transcriptomic behavior in the cyanobacterium Cyanothece sp. ATCC 51142. Mol Biosyst. 2011;7:2407–18. doi: 10.1039/c1mb05006k. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, Wu G, Chen L, Zhang W. Cross-species transcriptional network analysis reveals conservation and variation in response to metal stress in cyanobacteria. BMC Genom. 2013;14:112. doi: 10.1186/1471-2164-14-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Musungu BM, Bhatnagar D, Brown RL, Payne GA, G OB, Fakhoury AM, et al. A Network Approach of Gene Co-expression in the Zea mays/Aspergillus flavus Pathosystem to Map Host/Pathogen Interaction Pathways. Front Genet. 2016;7:206. doi: 10.3389/fgene.2016.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tierney L, Linde J, Muller S, Brunke S, Molina JC, Hube B, et al. An Interspecies Regulatory Network Inferred from Simultaneous RNA-seq of Candida albicans Invading Innate Immune Cells. Front Microbiol. 2012;3:85. doi: 10.3389/fmicb.2012.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernstein HC, Mcclure R, Thiel V, Sadler N, Kim YM, Chrisler WB, et. al. Indirect interspecies regulation; transcriptional and physiological responses of a cyanobacterium to heterotrophic partnership. mSystems. 2017;2. [DOI] [PMC free article] [PubMed]
- 28.Henry CS, Bernstein HC, Weisenhorn P, Taylor RC, Lee JY, Zucker J, et al. Microbial community metabolic modeling: a community data-driven network reconstruction. J Cell Physiol. 2016;231:2339–45. doi: 10.1002/jcp.25428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwai M, Katoh H, Katayama M, Ikeuchi M. Improved genetic transformation of the thermophilic cyanobacterium, Thermosynechococcus elongatus BP-1. Plant Cell Physiol. 2004;45:171–5. doi: 10.1093/pcp/pch015. [DOI] [PubMed] [Google Scholar]
- 30.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marbach D, Costello JC, Kuffner R, Vega NM, Prill RJ, Camacho DM, et al. Wisdom of crowds for robust gene network inference. Nat Methods. 2012;9:796–804. doi: 10.1038/nmeth.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khuri S, Wuchty S. Essentiality and centrality in protein interaction networks revisited. BMC Bioinforma. 2015;16:109. doi: 10.1186/s12859-015-0536-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pang K, Sheng H, Ma X. Understanding gene essentiality by finely characterizing hubs in the yeast protein interaction network. Biochem Biophys Res Commun. 2010;401:112–6. doi: 10.1016/j.bbrc.2010.09.021. [DOI] [PubMed] [Google Scholar]
- 37.Yang L, Wang J, Wang H, Lv Y, Zuo Y, Jiang W. Characterization of essential genes by topological properties in the perturbation sensitivity network. Biochem Biophys Res Commun. 2014;448:473–9. doi: 10.1016/j.bbrc.2014.04.136. [DOI] [PubMed] [Google Scholar]
- 38.McDermott JE, Diamond DL, Corley C, Rasmussen AL, Katze MG, Waters KM. Topological analysis of protein co-abundance networks identifies novel host targets important for HCV infection and pathogenesis. BMC Syst Biol. 2012;6:28. doi: 10.1186/1752-0509-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDermott JE, Taylor RC, Yoon H, Heffron F. Bottlenecks and hubs in inferred networks are important for virulence in Salmonella typhimurium. J Comput Biol. 2009;16:169–80. doi: 10.1089/cmb.2008.04TT. [DOI] [PubMed] [Google Scholar]
- 40.Song HS, McClure RS, Bernstein HC, Overall CC, Hill EA, Beliaev AS. Integrated in silico analyses of regulatory and metabolic networks of Synechococcus sp. PCC 7002 reveal relationships between gene centrality and essentiality. Life. 2015;5:1127–40. doi: 10.3390/life5021127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beliaev AS, Romine MF, Serres M, Bernstein HC, Linggi BE, Markillie LM, et al. Inference of interactions in cyanobacterial-heterotrophic co-cultures via transcriptome sequencing. ISME J. 2014;8:2243–55. doi: 10.1038/ismej.2014.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burgess ML, Barrow KD, Gao C, Heard GM, Glenn D. Carotenoid glycoside esters from the thermophilic bacterium meiothermus ruber. J Nat Prod. 1999;62:859–63. doi: 10.1021/np980573d. [DOI] [PubMed] [Google Scholar]
- 43.Hirsch AM. Role of lectins (and rhizobial exopolysaccharides) in legume nodulation. Curr Opin Plant Biol. 1999;2:320–6. doi: 10.1016/S1369-5266(99)80056-9. [DOI] [PubMed] [Google Scholar]
- 44.Lerouge I, Vanderleyden J. O-antigen structural variation: mechanisms and possible roles in animal/plant-microbe interactions. FEMS Microbiol Rev. 2002;26:17–47. doi: 10.1111/j.1574-6976.2002.tb00597.x. [DOI] [PubMed] [Google Scholar]
- 45.Levy SF, Siegal ML. Network hubs buffer environmental variation in Saccharomyces cerevisiae. PLoS Biol. 2008;6:e264. doi: 10.1371/journal.pbio.0060264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zotenko E, Mestre J, O’leary DP, Przytycka TM. Why do hubs in the yeast protein interaction network tend to be essential: reexamining the connection between the network topology and essentiality. PLoS Comput Biol. 2008;4:e1000140. doi: 10.1371/journal.pcbi.1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Romine MF, Rodionov DA, Maezato Y, Anderson LN, Nandhikonda P, Rodionova IA, et al. Elucidation of roles for vitamin B12 in regulation of folate, ubiquinone, and methionine metabolism. Proc Natl Acad Sci USA. 2017;114:E1205–14. doi: 10.1073/pnas.1612360114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Cross-Species Network
Supplemental Figure 2: Singe Species Network
Supplemental Figure 3: Positive Edges Of Transport and Translation Genes
Supplemental Figure 4: Negative Edges Of Transport and Translation Genes
Supplemental Figure 5: Pearson correlation of btuB and cobQ
Supplementary Table 1: Culture Conditions
Supplementary Table 2: RNA-seq alignment statistics
Supplementary Table 3: Gene expression levels
Supplementary Table 4: Pearson Correlation Coeffecients of Edges
Supplementary Table 5: Functional enrichment of T. elongatus genes linked to uncharacterized M. ruber genes SY28_RS04830 and SY28_RS05160
Data Availability Statement
All transcriptome data generated in the current study have been deposited into GEO (Accession: GSE113134).