Abstract
Paenibacillus polymyxa (formerly known as Bacillus polymyxa) has been extensively studied for agricultural applications as a plant-growth-promoting rhizobacterium and is also an important biocontrol agent. Our team has developed the P. polymyxa strain HY96-2 from the tomato rhizosphere as the first microbial biopesticide based on P. polymyxa for controlling plant diseases around the world, leading to the commercialization of this microbial biopesticide in China. However, further research is essential for understanding its precise biocontrol mechanisms. In this paper, we report the complete genome sequence of HY96-2 and the results of a comparative genomic analysis between different P. polymyxa strains. The complete genome size of HY96-2 was found to be 5.75 Mb and 5207 coding sequences were predicted. HY96-2 was compared with seven other P. polymyxa strains for which complete genome sequences have been published, using phylogenetic tree, pan-genome, and nucleic acid co-linearity analysis. In addition, the genes and gene clusters involved in biofilm formation, antibiotic synthesis, and systemic resistance inducer production were compared between strain HY96-2 and two other strains, namely, SC2 and E681. The results revealed that all three of the P. polymyxa strains have the ability to control plant diseases via the mechanisms of colonization (biofilm formation), antagonism (antibiotic production), and induced resistance (systemic resistance inducer production). However, the variation of the corresponding genes or gene clusters between the three strains may lead to different antimicrobial spectra and biocontrol efficacies. Two possible pathways of biofilm formation in P. polymyxa were reported for the first time after searching the KEGG database. This study provides a scientific basis for the further optimization of the field applications and quality standards of industrial microbial biopesticides based on HY96-2. It may also serve as a reference for studying the differences in antimicrobial spectra and biocontrol capability between different biocontrol agents.
Keywords: genome sequencing, Paenibacillus polymyxa, comparative genomic analysis, biocontrol mechanism, biofilm, antibiotics, induced resistance
Introduction
Paenibacillus polymyxa is a multifunctional Gram-positive (G+) bacterium that has been reported to have applications in agriculture (Hao and Chen, 2017), medicine (Galea et al., 2017; Yu et al., 2017), and industry (Lal and Tabacchioni, 2009). P. polymyxa has been in the list of substances reported under the Toxic Substance Control Act (TSCA) by the United States Environmental Protection Agency (EPA)1. It means P. polymyxa is a safe and commercially available microbe. The application of P. polymyxa in agriculture includes two main aspects: the biocontrol of plant diseases and the promotion of plant growth. P. polymyxa is an important biocontrol agent that has been reported to suppress a large variety of fungal and bacterial plant diseases, such as those caused by the fungi Fusarium oxysporum and Botrytis cinerea and the bacteria Xanthomonas campestris and Ralstonia solanacearum (Xu et al., 2006; Kim et al., 2010; Fan et al., 2012; Weselowski et al., 2016). P. polymyxa can withstand various adverse conditions during the biopesticide manufacturing process, especially drying, and exhibits a long and resilient shelf life owing to its endogenous spore formation. Therefore, P. polymyxa is an important strain for the production of microbial biopesticides2,3. In 2004, P. polymyxa strain HY96-2 from the tomato rhizosphere was developed as the first microbial biopesticide based on P. polymyxa and registered in China, for the control of soil-borne diseases caused by R. solanacearum on tomatoes and F. oxysporum on watermelons, as well as leaf diseases caused by B. cinerea and Pseudomonas syringae on cucumbers2. In addition, other studies have demonstrated that HY96-2 inhibits the growth of many fungal and bacterial pathogens, such as Colletotrichum gloeosporioides, Rhizoctonia solani, and Erwinia carotovora (Fan et al., 2012). Although microbial biopesticides derived from P. polymyxa HY96-2 have been manufactured and sold in 24 provinces across China, further research into its precise biocontrol mechanism, especially at the molecular level, is still needed.
Genomes are a very useful resource for understanding the mechanism of biocontrol agents. Kim et al. (2017) determined the key genes responsible for the production of antimicrobial agents and volatile organic compounds, indoleacetic acid (IAA) synthesis, siderophore secretion, phosphate transporter, and phosphonate cluster biosynthesis in Paenibacillus yonginensis DCY84T by genome sequencing and confirmed the ability of this strain to induce plant resistance and protect plant growth by a combination of physiological experiments. By comparing the genomes of Bacillus amyloliquefaciens FZB42 with B. subtilis, Chen et al. (2007) discovered that over 8.5% of the genome of B. amyloliquefaciens FZB42 is involved in antibiotic and siderophore synthesis, whereas Stein (2005) estimated that not more than 4–5% of the average B. subtilis genome is devoted to antibiotic production. Therefore, this genomic comparison demonstrated that B. amyloliquefaciens FZB42 is capable of protecting plants from diseases at the molecular level via the production of antibiotics.
Furthermore, despite its importance as a biocontrol agent, there have only been a small number of comprehensive studies into the biocontrol mechanism of P. polymyxa using genomic comparisons or other molecular methods. To further understand the biocontrol mechanism of HY96-2 at the molecular level, its genome was completely sequenced and compared with those of other P. polymyxa strains. Thus far, the complete genomes of seven P. polymyxa strains have been published, including the strains SC2 (Ma et al., 2011), E681 (Kim et al., 2010), YC0136 (Liu et al., 2017a), M-1 (Niu et al., 2011), SQR-21 (Li et al., 2014), CR1 (Eastman et al., 2014b), and YC0573 (Liu et al., 2017b). The details of these strains are presented in Table 1. Compared with other P. polymyxa strains, SC2 and E681 have been studied in greater detail and found to inhibit the growth of many plant pathogens. SC2 was isolated from the rhizosphere of pepper plants in Guizhou Province, China, and demonstrated to inhibit plant pathogenic fungi, such as F. oxysporum, B. cinerea, Pseudoperonospora cubensis, and plant pathogenic bacteria, the species of which were not published (Zhu et al., 2008). E681, isolated from the rhizosphere of barley plants in South Korea, was reported to possess inhibitory activity against plant pathogenic fungi such as F. oxysporum, B. cinerea, and R. solani (Ryu et al., 2006) and the plant pathogenic bacterium P. syringae (Kwon et al., 2016).
Table 1.
Sources and functions of the studied P. polymyxa strains (the complete genomes of which have been published).
| Name | Accession number(s) | Origin | Activity | Reference |
|---|---|---|---|---|
| HY96-2 | CP025957 | Tomato rhizosphere (Nanchang, Jiangxi, China) | As biocontrol agent against fungi (F. oxysporum, C. gloeosporioides, B. cinerea, R. solani, Alternaria solani, Penicillium italicum, Curvularia lunata, etc.) and Gram-negative (G-) bacteria (R. solanacearum, P. syringae, E. carotovora, etc.). | Industrial microbial biopesticides that have been commercially registered in China; Fan et al., 2012 |
| SC2 | CP002213- CP002214 | Pepper rhizosphere (Guizhou, China) | As biocontrol agent against fungi (F. oxysporum, B. cinerea, and P. cubensis) and bacteria (species not stated), and as plant-growth-promoting rhizobacterium (PGPR) to dissolve phosphorus and potassium. | Zhu et al., 2008; Ma et al., 2011 |
| E681 | CP000154 | Winter barley rhizosphere (South Korea) | As biocontrol agent against fungi (F. oxysporum, B. cinerea, B. allii, R. solani, Pythium sp., Aspergillus sp., etc.), and bacteria (P. syringae, E. coli, etc.), and as PGPR. | Ryu et al., 2006; Kim et al., 2010; Kwon et al., 2016 |
| YC0136 | CP017967 | Tobacco rhizosphere (Guizhou, China) | As biocontrol agent against fungus Phytophthora parasitica var. nicotianae and as PGPR. | Liu et al., 2017a |
| M-1 | HE577054- HE577055 | Wheat rhizosphere (China) | As biocontrol agent against fungus R. solani and as PGPR. | Niu et al., 2011 |
| SQR-21 | CP006872 | Melon rhizosphere (China) | As biocontrol agent against fungus F. oxysporum and as PGPR. | Yaoyao et al., 2017 |
| CR1 | CP006941 | Degraded corn straw (Southern Ontario, Canada) | As biocontrol agent to antagonize fungi (R. solani, Cylindrocarpon destructans, etc.) and bacteria (P. syringae, X. campestris, etc.), and as PGPR, as well as biodegradation agent. | Eastman et al., 2014b; Weselowski et al., 2016 |
| YC0573 | CP017968 | Tobacco rhizosphere (Guizhou, China) | As biocontrol agent to antagonize fungus P. parasitica var. nicotianae and as PGPR. | Liu et al., 2017b |
Colonization (Bianciotto et al., 2001; Bais et al., 2004; Sang and Kim, 2014), antagonism (Lugtenberg and Kamilova, 2009; Sharma et al., 2009), and induced resistance (Ongena et al., 2007; Shi et al., 2017) are the three main reported mechanisms of biological control. Biofilm formation indicates that a biocontrol agent possesses good colonization ability (Ongena and Jacques, 2008; Sang and Kim, 2014). Besides the key genes involved in biofilm formation, quorum sensing also plays an important role (Miller and Bassler, 2001). The types and amounts of antibiotics generated by a biocontrol agent affect their antimicrobial spectra and biocontrol efficacies. It has been reported that P. polymyxa secretes antifungal and antibacterial metabolites, which mainly include fusaricidins, polymyxins, and other antibiotics (Jeong et al., 2011; Shaheen et al., 2011). Biocontrol agents can also generate and release systemic resistance inducers including volatile organic compounds (mainly consisting of 2,3-butanediol, methanethiol, isoprene, butyl acetate, n-hexadecane, etc.) into the surrounding environment, both of which have been shown to have a positive effect on plant protection (Ryu et al., 2004; Lee et al., 2012; Shi et al., 2017).
To the best of our knowledge, no genomic comparison has yet been applied to analyze the biocontrol mechanism of P. polymyxa. Thus, in this study the complete genome of P. polymyxa strain HY96-2 was sequenced and compared with those of seven other strains. To elucidate the differences in the biocontrol mechanisms between strain HY96-2 and other agriculturally undeveloped P. polymyxa strains, the main genes (or gene clusters) of HY96-2 involved in biofilm formation, antibiotic synthesis, and systemic resistance inducer production were analyzed in comparison with the strains SC2 and E681. This study provides a scientific basis for the further optimization of the field applications and product quality standards of the microbial biopesticide derived from P. polymyxa HY96-2.
Materials and Methods
Strains and Genomic DNA Preparation
P. polymyxa strain HY96-2 was isolated from the rhizosphere of tomato plants in the suburbs of Nanchang, Jiangxi Province, China, and preserved in the China General Microbiological Culture Collection Center (CGMCC No. 0829). HY96-2 was cultured at 30°C in Luria–Bertani broth. Genomic DNA was purified from overnight liquid cultures (OD600 nm ≈ 0.7) using the cetyltrimethylammonium bromide method (Watanabe et al., 2010). A TBS-380 fluorometer (Turner BioSystems, United States) or NanoDrop 2500 (Thermo Scientific, United States) was applied to ensure the DNA quality (≥10 μg, without degradation, OD260/OD280 ≈ 1.8–2.0).
Sequencing and Assembly
The whole genome was sequenced using the PacBio RS II platform with a 10-kb library. Reads were assembled using HGAP (version 2.3.0, SMRT Analysis) (Chin et al., 2013). The assembly data for the complete genome have been deposited in GenBank with the accession number CP025957.
Genome Components and Genome Annotation
Coding DNA sequence (CDS) prediction was performed using Glimmer 3.02 (Delcher et al., 1999). A circular map of the genome was obtained using Circos version 0.64 (Krzywinski et al., 2009). Genomic islands (GIs) were predicted using the GI prediction method IslandViewer 4 (Dhillon et al., 2015). tRNA and rRNA were predicted using the tRNAscan-SEv1.3.1 (Lowe and Eddy, 1997) and barrnap 0.7 software4, respectively. Clustered regularly interspaced short palindromic repeat sequences (CRISPRs) were found using CRISPRFinder (Grissa et al., 2007). Functional annotation was based on BLASTP searches (BLAST 2.2.28+) against the NCBI non-redundant (NR) database, gene database, string database, and gene ontology (GO) database. Based on the string database, the BLASTP comparison was used to perform the Clusters of Orthologous Groups of proteins (COG) annotation, according to which the protein function could be classified (Tatusov et al., 2001). The BLAST algorithm was used to compare the predicted genes with the KEGG database, and the corresponding genes involved in specific biological pathways could be obtained according to the KEGG Orthology (KO) numbers obtained from the alignment. GO was annotated with blast2go (Conesa et al., 2005).
Genome Comparison
The complete genome sequences of seven P. polymyxa strains (SC2, E681, YC0136, M-1, SQR-21, CR1, and YC0573) and the related species strain B. subtilis 168 (AL009126) and B. amyloliquefaciens FZB42 (CP00560) examined in this study were obtained from GenBank. Phylogenetic analysis was conducted for P. polymyxa strains, B. subtilis, and B. amyloliquefaciens inferred by analyzing homologous gene. The single-copy homologous genes of each strain were selected for multiple sequence alignment and quality control comparison. Multiple sequence alignment was conducted by MAFFT software5. Quality control comparison was conducted by Gblocks software6. Then the phylogenetic tree was constructed by RAxML software with maximum likelihood method based on single gene or multiple genes (Stamatakis, 2014). Pan-genome analysis and nucleic acid co-linearity were conducted for the seven P. polymyxa strains and HY96-2. The pan-genome analysis was performed using the OrthoMCL software (Chen et al., 2006). The nucleic acid co-linearity was determined using the MUMmer 3.0 software (Kurtz et al., 2004).
The genes or gene clusters involved in biofilm formation, antibiotic synthesis, and systemic resistance inducer production were compared between strain HY96-2 and strains SC2 and E681. According to the biofilm formation pathways retrieved from the KEGG database7, the possible pathways for biofilm formation in P. polymyxa were analyzed. In this study, the genes analyzed for biofilm formation in P. polymyxa were selected according to Molinatto et al. (2017). The gene clusters for secondary metabolites (containing antibiotics) in P. polymyxa were annotated using the antiSMASH database version 4.0.2 and the other antibiotics were selected based on previous studies (Walsh, 2004; Ma et al., 2011). The resistance inducers of P. polymyxa in this study were selected according to Lee et al. (2012) and Shi et al. (2017). The key genes involved in resistance inducer synthesis were searched for in the KEGG database. BLAST was used to compare the identities in the genes or gene clusters between HY96-2, SC2, and E681.
Results
Genome Features
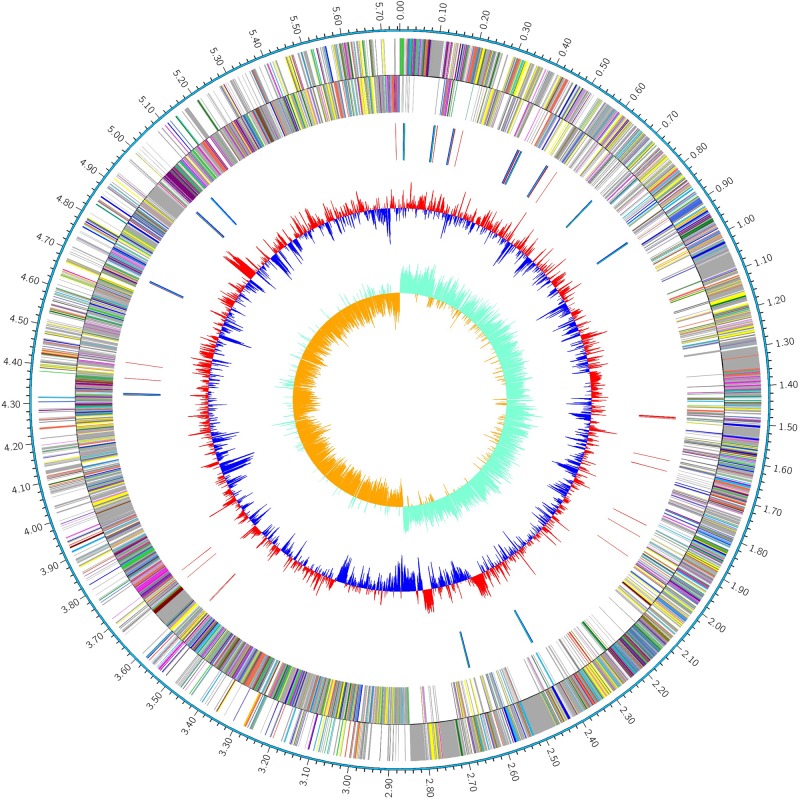
The complete genome of P. polymyxa strain HY96-2 was 5.75 Mb (Figure 1), in which the average GC content of the chromosome was 45.61%. A total of 5207 CDSs were predicted. Furthermore, the HY96-2 genome contained 42 rRNA and 110 tRNA genes. The general features are shown in Table 2. However, no plasmids were found using Webcutter version 2.0 or PlasmidFinder version 1.3.
FIGURE 1.
Circular map of the genome of the P. polymyxa strain HY96-2. The distribution of the circle from the outside to the inside indicates the genome size, forward CDS, reverse CDS, repeat sequences, tRNA (blue), rRNA (purple), GC ratio (red and blue indicate regions where the GC ratio is higher than average and lower than average, respectively), and GC skew (cyan and orange indicate regions where the G content is greater than and less than the C content, respectively).
Table 2.
General features of the genome of the P. polymyxa strain HY96-2.
| Attribute | Value |
|---|---|
| Genome size (bp) | 5745779 |
| G+C ratio (%) | 45.61 |
| Protein-coding genes | 5207 |
| Gene total length (bp) | 4958220 |
| rRNA | 42 |
| tRNA | 110 |
| Genes assigned to COGs | 2605 |
| Genomic islands | 16 |
| CRISPR | 1 |
Genomic Islands and CRISPR Prediction
Genomic islands often carry genes important for genome evolution and adaptation to surrounding environment such as those involved in pathogenesis and antibiotic resistance. Therefore, GI prediction has gradually become an increasingly important aspect of microbial genome analysis (Lu and Leong, 2016). The GIs predicted in HY96-2 are listed in Supplementary Table S1; a total of 16 GIs were predicted in the chromosome. The CRISPRs containing multiple short and repeated sequences can confer resistance to exogenous genetic elements such as phages and plasmids (Didovyk et al., 2016; Ran, 2017). A putative CRISPR was detected in the HY96-2 genome by CRISPRFinder (Supplementary Table S2).
Genome Annotation
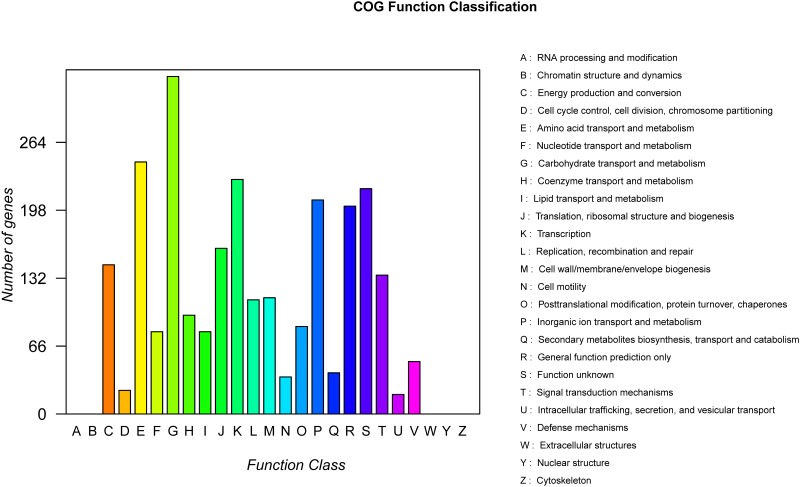
According to the results of the COG annotation, 2605 proteins were classified into 20 COG families (Supplementary Table S3). The largest group of genes was involved in carbohydrate transport and metabolism (328 genes, 6.30%) (Figure 2). In total, KEGG orthologs were found for 2352 proteins by BLAST. In addition, the GO annotation results revealed that the largest group of genes was involved in the biological process domain. Among these genes, the content of genes related to the metabolic process category was highest (Supplementary Figure S1).
FIGURE 2.
Distribution of genes across COG functional categories in the chromosome of P. polymyxa strain HY96-2.
Genome Comparison
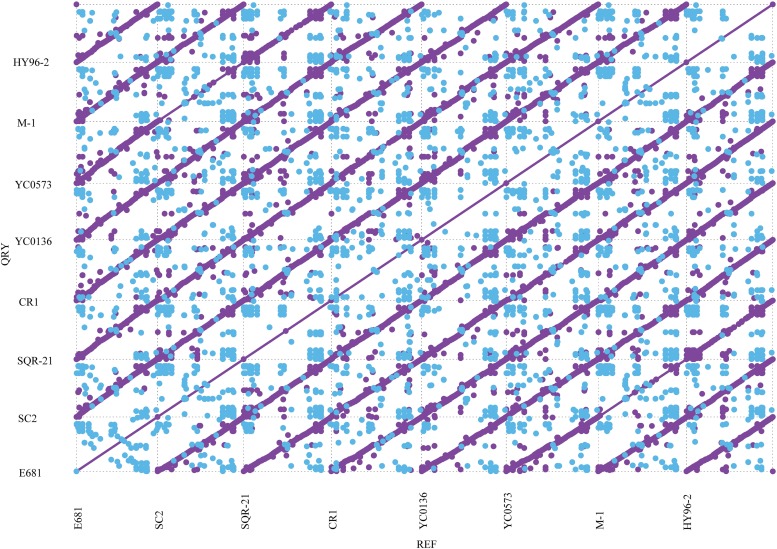
The phylogenetic tree indicated that HY96-2 is closest to P. polymyxa SQR-21 (Figure 3). However, P. polymyxa strains M-1 and SC2 were found to exhibit low homologies with strain HY96-2. We compared the 16S rRNA sequence of each P. polymyxa strain to calculate the percentage of homology (Supplementary Table S4). The percentage of homology between strain HY96-2 to strains M-1 and SC2 were 97.87 and 97.82%. We also performed a pan-genome analysis to compare HY96-2 and the seven other P. polymyxa strains, namely, SC2, E681, YC0136, M-1, SQR-21, CR1, and YC0573. As shown in Figure 4, 3382 gene families were found to be involved in the core genome shared by all of the studied strains. In addition, the number of gene families unique to strain HY96-2 was 448, corresponding to 468 genes, which was the highest among all of the analyzed strains (Supplementary Table S5). The annotation revealed that these specific genes encoded a large number of transcriptional regulators, helicase domain proteins, hypothetical proteins, aminotransferases, transposases, drug resistance transporters, and chloramphenicol resistance proteins, etc. The nucleic acid co-linearity results showed that strain HY96-2 has high co-linearity with the seven other P. polymyxa strains (Figure 5).
FIGURE 3.
Phylogenetic tree for P. polymyxa HY96-2 and the seven other P. polymyxa strains, B. subtilis 168, and B. amyloliquefaciens FZB42 based on homologous genes.
FIGURE 4.
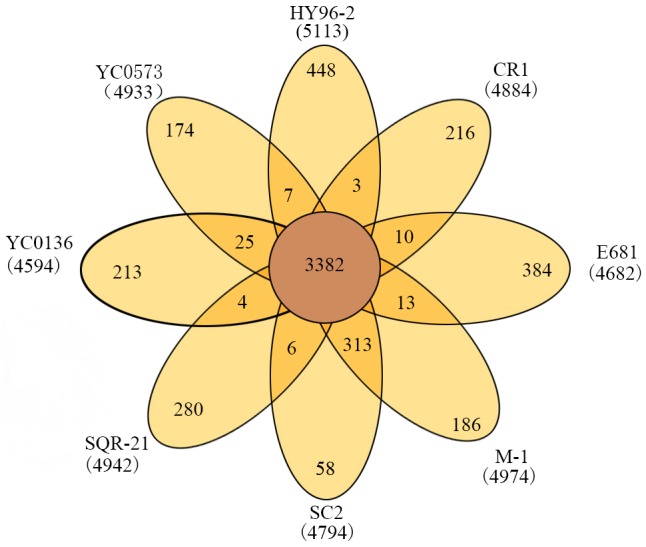
Venn diagram showing numbers of specific and shared gene families among the eight different P. polymyxa strains.
FIGURE 5.
Co-linearity results for strain HY96-2 and the seven other P. polymyxa strains. The purple dots represent the forward alignment of the two genome sequences, and the blue dots represent the reverse alignment of the corresponding segments.
Comparison of Genes Involved in Biofilm Formation
Based on the studies of Yang et al. (2014) and Molinatto et al. (2017), we searched the genomes of strains HY96-2, SC2, and E681 for genes related to biofilm formation and compared their similarities using BLAST. These genes are as follows: the key quorum-sensing gene luxS, which affects biofilm formation (Yang et al., 2014); the flagellar motility-related genes, motA, motB, and flgM (Domka et al., 2007; Hu et al., 2011); and the Bacillus-based biofilm formation pathway genes, kinB, spo0A, spo0F, degU, and degS, etc. (Grossman et al., 1992). The results showed that 14 genes involved in biofilm formation were certainly found in the genomes of these three strains. The identities of these genes between HY96-2 and SC2 exceeded 93%, which were higher than those between HY96-2 and E681. In particular, the identities of the genes motA, sfp, and kinB between strain HY96-2 and E681 were below 90% (Table 3).
Table 3.
Comparison of core genes involved in biofilm formation in strain HY96-2, as well as in strains SC2 and E681 (sequence similarity is expressed as a percentage of the nucleotide identity).
| Gene name | Locus tag | Product | Identity (%) | |||
|---|---|---|---|---|---|---|
| HY96-2 | SC2 | E681 | SC2 | E681 | ||
| luxS | C1A50_RS02845 | PPSC2_RS31750 | PPE_RS02675 | S-ribosylhomocysteine lyase | 97.19 | 91.99 |
| Efp | C1A50_RS14785 | PPSC2_RS43880 | PPE_RS13870 | Elongation factor P | 98.57 | 93.19 |
| degU | C1A50_RS22485 | PPSC2_RS51810 | PPE_RS21415 | DNA-binding response regulator | 95.99 | 93.64 |
| degS | C1A50_RS22490 | PPSC2_RS51815 | PPE_RS21420 | Histidine kinase | 98.71 | 96.30 |
| flgM | C1A50_RS22450 | PPSC2_RS51780 | PPE_RS21385 | Flagellar biosynthesis anti-sigma factor FlgM | 97.10 | 94.57 |
| motA | C1A50_RS00510 | PPSC2_RS29245 | PPE_RS00510 | Flagellar motor protein MotA | 98.24 | 88.18 |
| motB | C1A50_RS00505 | PPSC2_RS29240 | PPE_RS00505 | Flagellar motor protein MotB | 97.22 | 91.18 |
| sacB | C1A50_RS03600 | PPSC2_RS32495 | PPE_RS03320 | Levansucrase | 95.00 | 93.60 |
| sfp | C1A50_RS07585 | PPSC2_RS36260 | PPE_RS07145 | 4′-phosphopantetheinyl transferase | 93.02 | 86.32 |
| sigD | C1A50_RS09600 | PPSC2_RS38730 | PPE_RS09310 | RNA polymerase sigma factor SigD | 96.58 | 94.04 |
| spo0A | C1A50_RS14655 | PPSC2_RS43750 | PPE_RS13740 | Sporulation transcription factor spo0A | 97.39 | 93.66 |
| spo0F | C1A50_RS00680 | PPSC2_RS29415 | PPE_RS00675 | Sporulation initiation phosphotransferase F | 99.46 | 95.39 |
| kinB | C1A50_RS19570 | PPSC2_RS48955 | PPE_RS18645 | Two-component sensor histidine kinase | 96.95 | 89.75 |
| hfq | C1A50_RS13605 | PPSC2_RS42680 | PPE_RS12655 | RNA chaperone Hfq | 97.53 | 93.09 |
To date, no studies have been reported on the biofilm formation pathway in P. polymyxa. We explored the possible pathways responsible for biofilm formation in P. polymyxa by combining genomes and the information from KEGG database. An important biofilm formation pathway in Bacillus is as follows: (1) the sensor histidine kinase KinB (or KinA, KinC, or KinD) phosphorylates its own conserved histidine residue in response to an environmental stimulus; (2) the phosphate group is transferred from the activated KinB to Spo0F; (3) because Spo0F lacks an effector domain, the phosphate group of Spo0F-P is then transferred to the intermediary molecule Spo0B; (4) finally, the receptor domain of the effector Spo0A is phosphorylated by the phosphate group of Spo0B-P and subsequently triggers biofilm formation (Fabert et al., 1999; Jiang et al., 2000). However, no spo0B gene was detected in the genomes of the P. polymyxa strains. By querying the KEGG database, another pathway involved in sporulation in B. subtilis was found, which revealed that Spo0F-P can directly transfer the phosphate group to the effector Spo0A without Spo0B acting as a mediator, and when a high concentration of Spo0A-P accumulates it will trigger the sporulation. Therefore, we propose that a biofilm formation pathway exists in P. polymyxa in which KinB autophosphorylates in response to an environmental stimulus and then subsequently phosphorylates the Spo0F response regulator, whereupon the phosphate group is directly transferred to Spo0A to trigger biofilm formation (Figure 6A). In addition, degS and degU genes were detected in the genome of the three P. polymyxa strains. It is therefore presumed that another possible biofilm formation pathway in P. polymyxa involves activation of the sensor histidine kinase DegS by the external stimulus to phosphorylate the response regulation protein DegU, thereby triggering biofilm formation (Figure 6B). This biofilm formation pathway has already been reported in B. subtilis (Stanley and Lazazzera, 2005).
FIGURE 6.
Possible pathways of biofilm formation in P. polymyxa. (A) Pathway A. (B) Pathway B.
Comparison of Genes/Gene Clusters Involved in Antibiotic Synthesis
The annotation of gene clusters related to secondary metabolite synthesis was performed using the antiSMASH database. For strain HY96-2, 15 gene clusters related to secondary metabolites were retrieved, accounting for 15.77% of the genome; the corresponding values for strains SC2 and E681 were 12 (11.89%) and 11 (9.85%), respectively. Among these three strains, HY96-2 exhibited a greater variety and higher content of secondary metabolites in the genome. The comparison of genes/gene clusters involved in antibiotic synthesis showed that the identities between strains HY96-2 and SC2 were significantly higher than those between strains HY96-2 and E681 (Table 4).
Table 4.
Comparison of gene clusters and core genes involved in antibiotic biosynthesis in strain HY96-2, as well as in strains SC2 and E681.
| Antibiotic name | Activity | Locus tag | Type | Identity (%) | |||
|---|---|---|---|---|---|---|---|
| HY96-2 | SC2 | E681 | SC2 | E681 | |||
| Fusaricidin | Broad-spectrum antifungal activity, especially against Fusarium sp.; also suppresses G+ bacteria (Kajimura and Kaneda, 1995, 1996; Liu et al., 2011) | C1A50_RS00290- C1A50_RS00500 | PPSC2_RS29015- PPSC2_RS29230 | PPE_RS00290- PPE_RS00500 | NRPS | 81.54 | 61.53 |
| Paenilarvins | Broad-spectrum antifungal activity (Sood et al., 2014) | C1A50_RS04650- C1A50_RS04845 | PPSC2_RS33500- PPSC2_RS33695 | – | TransATPKS– NRPS | 81.88 | – |
| Polymyxin | Broad-spectrum antibacterial activity, especially against G- bacteria (Falagas and Kasiakou, 2005; Gounani et al., 2018) | C1A50_RS21330- C1A50_RS21525 | PPSC2_RS50695- PPSC2_RS50860 | PPE_RS20180- PPE_RS20350 | NRPS | 63.21 | 54.94 |
| Polyketide | Suppresses G- bacteria (Chakraborty et al., 2018) | C1A50_RS00805 | PPSC2_RS29535 | PPE_RS00800 | PKS | 94.93 | 90.29 |
| Tridecaptin | Suppresses G- bacteria (Cochrane et al., 2015) | C1A50_RS10840- C1A50_RS11035 | PPSC2_RS40010- PPSC2_RS40205 | – | NRPS | 31.82 | – |
| Bacitracin | Suppresses G+ bacteria (Konzl et al., 1997) | C1A50_RS24385 | PPSC2_RS53660 | PPE_RS23255 | NRPS | 94.38 | 90.62 |
| Kalimantacin | Suppresses G+ bacteria (Thistlethwaite et al., 2017) | C1A50_RS15500- C1A50_RS15755 | PPSC2_RS44500- PPSC2_RS44755 | – | TransATPKS– Otherks-NRPS | 68.75 | – |
| Mersacidin | Suppresses G+ bacteria (Sahl and Bierbaum, 1998) | C1A50_RS07480- C1A50_RS07590 | PPSC2_RS36150- PPSC2_RS36265 | PPE_RS06970- PPE_RS07075 | Lantipeptide | 83.54 | 72.92 |
Two gene clusters involved in fungicide synthesis (fusaricidins and paenilarvins) as well as six genes or gene clusters involved in bactericide synthesis (polymyxin, tridecaptin, mersacidin, kalimantacin, polyketide, and bacitracin) were found in the genomes of strains HY96-2 and SC2. However, no gene clusters for the biosynthesis of paenilarvins, tridecaptin, or kalimantacin were detected in the E681 genome. For the main antimicrobial agents in P. polymyxa, fusaricidins and polymyxin, the comparison revealed that the synthetic gene clusters in strains SC2 and E681 did not exhibit very high similarities with those in strain HY96-2, but the similarities of the corresponding synthetic gene clusters between SC2 and HY96-2 were still higher than those between E681 and HY96-2. The identity of the fusaricidin synthetic gene clusters between SC2 and HY96-2 was 81.54%, whereas that between E681 and HY96-2 was only 61.53%. Similarly, the identity of the polymyxin synthetic gene clusters between SC2 and HY96-2 was 63.21%, whereas that between E681 and HY96-2 was only 54.94%. These variations in the genes and gene clusters involved in antibiotic synthesis between the three strains may explain the differences in their target profiles and efficiency against plant diseases.
Comparison of Genes Involved in Resistance Inducer Synthesis
Key genes involved in the synthesis of resistance inducers in P. polymyxa were retrieved from the KEGG database, and the identities of these genes between HY96-2 and SC2 and E681 were also compared. The results revealed that the key genes for 2,3-butanediol, methanethiol, and isoprene were all found in the genomes of HY96-2, SC2, and E681, with sequence identities exceeding 86%. The identities of the corresponding genes between HY96-2 and SC2 (91.33–97.05%) were significantly higher than those between HY96-2 and E681 (86.83–92.05%) (Table 5). This indicates that the three strains can be expected to possess similar abilities to induce plant resistance, although their induction efficiencies may differ owing to the variation in the corresponding genes.
Table 5.
Comparison of genes involved in the synthesis of resistance inducers in strain HY96-2, as well as in strains SC2 and E681.
| Resistance inducer | Gene | Locus tag | Identity (%) | |||
|---|---|---|---|---|---|---|
| HY96-2 | SC2 | E681 | SC2 | E681 | ||
| 2,3-Butanediol | alsD | C1A50_RS10210 | PPSC2_RS39445 | PPE_RS09950 | 95.18 | 90.76 |
| ilvN | C1A50_RS07135 | PPSC2_RS35800 | PPE_RS06620 | 95.47 | 91.98 | |
| Methanethiol | mtnE | C1A50_RS13950 | PPSC2_RS43025 | PPE_RS13035 | 93.23 | 89.85 |
| metH | C1A50_RS13320 | PPSC2_RS42420 | PPE_RS12455 | 95.47 | 89.74 | |
| metE | C1A50_RS23245 | PPSC2_RS52550 | PPE_RS22160 | 94.52 | 88.18 | |
| mmuM | C1A50_RS24195 | PPSC2_RS53470 | PPE_RS23100 | 94.30 | 87.13 | |
| Isoprene | idi | C1A50_RS22765 | PPSC2_RS52085 | PPE_RS21690 | 92.55 | 86.83 |
| lytB | C1A50_RS07370 | PPSC2_RS36040 | PPE_RS06865 | 91.33 | 86.83 | |
| gcpE | C1A50_RS18800 | PPSC2_RS48225 | PPE_RS17845 | 97.05 | 91.24 | |
| ispF | C1A50_RS21940 | PPSC2_RS51265 | PPE_RS20840 | 94.76 | 89.10 | |
| ispE | C1A50_RS00170 | PPSC2_RS28895 | PPE_RS00170 | 96.14 | 92.05 | |
Discussion
Microbial biopesticides have attracted increasing attention over recent years owing to their effectiveness, environmental friendliness, and safety toward humans and livestock (Berg, 2009). The development of microbial biopesticides in China is also proceeding rapidly and the number of registered and commercially available ones is growing sharply, in line with the Chinese government’s policy of “two reductions” (i.e., reducing the amounts of chemical pesticides and fertilizers used). As an important biocontrol agent, P. polymyxa is a relatively novel species and the number of registered products based on this species is growing rapidly8. Microbial biopesticides based on P. polymyxa have also been produced industrially. However, only a few comprehensive studies of the biocontrol mechanism of P. polymyxa at the molecular level have been published to date (Eastman et al., 2014a; Xie et al., 2016). The previous reports concerning the complete genome of P. polymyxa predominantly focused on the general features of the genome and analysis of the effect of this species on promoting growth, but rarely involved the analysis of the biocontrol function and mechanism (Kim et al., 2010; Ma et al., 2011; Niu et al., 2011; Eastman et al., 2014b; Li et al., 2014; Liu et al., 2017a,b). In this study we have presented the complete genome of the industrial P. polymyxa strain HY96-2, which consists of a 5.75 Mb chromosome. The genome of HY96-2 was compared with those of seven other P. polymyxa strains (SC2, E681, YC0136, M-1, SQR-21, CR1, and YC0573). In particular, the biocontrol-related genes and gene clusters involved in the formation of biofilms, antibiotics, and systemic resistance inducers were compared between HY96-2 and the SC2 and E681 strains. The results of this comparison revealed that the genome of strain HY96-2 exhibits some degree of variation. And this finding possibly explained the relationship between genes relating to the biocontrol mechanism and the biocontrol target and efficacy.
According to the phylogenic tree, the homology between strains HY96-2 and E681 is greater than that between strains HY96-2 and SC2. However, comparison of the genes involved in biocontrol mechanism and 16s rRNA (Supplementary Table S4) suggested that strain HY96-2 is more similar to SC2 than E681. SC2 contains a large 0.51 Mb plasmid, whereas HY96-2 and E681 do not. This could be the main reason why the phylogenetic tree suggested that strain HY96-2 is closer to E681 than SC2. Strain SC2 was isolated from rhizosphere of pepper in Guizhou Province, China (latitude 24°37′–29°13′N), and strain HY96-2 was obtained from rhizosphere of tomato in Nanchang, Jiangxi Province, China (latitude 28°16′–28°58′N). Strain E681 was isolated from rhizosphere of winter barley in South Korea (latitude 33°–43°N). The biggest difference between these three strains were the regions and hosts where they were isolated. Strains HY96-2 and SC2 were all isolated from Solanaceae plants, and their geographic locations were the nearest, and they demonstrated the highest homology by comparing the genes related to biocontrol mechanism and 16s rRNA within the three strains. Thus, we speculated the host environment might be one of the reasons of the variation of strains HY96-2, SC2, and E681. Pan-genome analysis revealed that the number of specific gene families in strain HY96-2 was 448, corresponding to 468 genes, which was the highest among the eight P. polymyxa strains studied. These unique genes of strain HY96-2 were found to encode transcriptional regulators, helicase domain proteins, aminotransferases, transposases, etc. These functional genes might affect biofilm formation and the production of antibiotics and systemic resistance inducers. Thus, the variation and specialization of these genes could possibly result in the differences in biocontrol targets and efficacy between strain HY96-2 and the other P. polymyxa strains.
Biofilm formation is an important trait that has been linked to the colonization ability of biocontrol microorganisms (Ongena and Jacques, 2008). Comparison of the main genes involved in biofilm formation revealed that all 14 of the target genes could be found in strains HY96-2, SC2, and E681, and the similarities of these genes between strains SC2 and HY96-2 were much higher than those between strains HY96-2 and E681. In addition, quorum sensing has been reported to significantly affect biofilm formation in bacteria (Miller and Bassler, 2001). For B. subtilis, which can be considered a representative G+ biocontrol bacterium, it has been reported that quorum sensing is mainly regulated by the comA, comP, comX, and comQ genes (Schneider et al., 2002). These B. subtilis quorum-sensing-related genes were deficient in the eight P. polymyxa strains studied, although another important quorum-sensing regulatory gene, luxS, was observed. luxS is a key regulatory gene in quorum sensing mediated by autoinducer 2 (AI-2) (Federle, 2009; Ma et al., 2017), which significantly affects biofilm formation in P. polymyxa (this conclusion was obtained in other study by our group, which has not yet been published). In addition, luxS was also found to be a key regulatory gene involved in quorum sensing in P. polymyxa CR1 (Eastman et al., 2014a). From the above results, it is possible to speculate that all three strains, HY96-2, SC2, and E681, could prevent disease by forming biofilms, but the biofilm formation ability of the three strains might differ owing to the variations in the genes involved in biofilm formation observed in their genomes.
In B. subtilis, Spo0A is a key transcriptional regulatory protein that controls the expression of over 100 genes, including those involved in biofilm formation and sporulation. DegU is also a global regulator in B. subtilis and controls multiple cellular processes such as competence, motility, and hydrolase secretion, without which biofilm formation would be unsuccessful (Kobayashi, 2007). As confirmed using the KEGG database, the genes related to the biofilm formation pathways regulated by these two key genes were found in all of the three tested P. polymyxa strains. Therefore, a possible pathway for biofilm formation in P. polymyxa can be preliminarily proposed as follows: in response to an environmental stimulus, KinB phosphorylates the Spo0F response regulator, which subsequently transfers the phosphate group directly to Spo0A to trigger biofilm formation. The main differences in the pathways mentioned above between P. polymyxa and B. subtilis are that the mediator Spo0B, which transfers the phosphate group between Spo0F and Spo0A, is present in B. subtilis but absent from P. polymyxa, and that the phosphate group can be directly transferred from Spo0F to Spo0A in P. polymyxa. Moreover, another possible pathway for biofilm formation in P. polymyxa was deduced, wherein the histidine kinase sensor DegS phosphorylates the response regulator protein DegU to form the biofilm in response to an external stimulus. To date, there have been no reports of the biofilm formation pathway in P. polymyxa. Thus, these two possible pathways for biofilm formation in P. polymyxa were first deduced and revealed in this paper and found to possess high similarity with those reported in B. subtilis.
Comparison of the genes and gene clusters involved in antibiotic synthesis revealed more significant differences between the three P. polymyxa strains than those involved in biofilm formation. Strains HY96-2 and SC2 exhibited relatively high homology, whereas strains HY96-2 and E681 had relatively low homology in terms of the genes and gene clusters involved in antibiotic production. The variations in these genes and gene clusters between the three strains might be responsible for the differences in the biocontrol targets and efficacies.
For the control of fungi, as the main antifungal metabolites in P. polymyxa, gene clusters for fusaricidins were found in all three of the tested P. polymyxa strains. To date, at least ten members of the fusaricidin family have been isolated from P. polymyxa, including fusaricidins A–D, LI-F03, LI-F04, LI-F05, LI-F06, LI-F07, and LI-F08 (Liu et al., 2011). Fusaricidins display excellent antifungal activities against many plant pathogenic fungi, especially F. oxysporum; fusaricidin B is particularly effective against Candida albicans and Saccharomyces cerevisiae. Fusaricidins also exhibit excellent germicidal activity against G+ bacteria such as Staphylococcus aureus (Kajimura and Kaneda, 1995, 1996; Liu et al., 2011). The gene cluster for fusaricidin synthesis in strain HY96-2 showed 81.54% identity with that in strain SC2, which was higher than that for strain E681 (61.53% identity). Fusaricidin A was the main fusaricidin found in strain HY96-2 (Liu et al., 2011). Choi et al. (2008) discovered that the fusA gene in E681 plays an important role in fusaricidin biosynthesis, and the inactivation of this gene led to the complete loss of antifungal activity against F. oxysporum; moreover, fusA can produce more than one kind of fusaricidin. Though Mikkola et al. (2017) mention that fusaricidin A and B possess toxicity to mammalian cells at a certain concentration, the results of animal and environmental toxicology tests of the products with living cells of P. polymyxa HY96-2 showed that the animal and environmental toxicities of the products were slight and low, respectively, at the lowest level of toxicity in the corresponding test (Supplementary Figures S2, S3). Thus, we considered that as long as fusaricidins were not purified and developed as pesticide directly, microbial pesticides with living cells of P. polymyxa are environmentally friendly and safe toward animal. In addition, the gene clusters for paenilarvins, a class of iturin-like compounds with broad-spectrum antifungal activity, were also found in strains HY96-2 and SC2 but not in E681. The identity of the paenilarvin biosynthetic gene clusters between HY96-2 and SC2 was 81.88%. All three strains were reported to suppress the plant pathogenic fungus F. oxysporum and B. cinerea. Besides F. oxysporum and B. cinerea, the reported control targets of the three strains were different (Table 1). Strains HY96-2 and E681 were also found to suppress R. solani. The different biocontrol targets of these three strains may be attributable to the variation in their gene clusters responsible for the synthesis of antifungal metabolites.
For the control of bacteria, many differences were also observed between the three strains in terms of the genes or gene clusters involved in the synthesis of antibacterial metabolites. Using the antiSMASH database, six genes or gene clusters related to antibiotic synthesis were found in HY96-2 and SC2, including those involved in the biosynthesis of polymyxin, tridecaptin, polyketide, mersacidin, kalimantacin, and bacitracin. In contrast, only four were detected in strain E681, as the genes or gene clusters for tridecaptin and kalimantacin synthesis were absent. Among the six antibiotics, polymyxin, tridecaptin, and polyketide are able to suppress G- bacteria (Cochrane et al., 2015; Chakraborty et al., 2018; Gounani et al., 2018), whereas mersacidin, kalimantacin, and bacitracin show activity against G+ bacteria (Konzl et al., 1997; Sahl and Bierbaum, 1998; Thistlethwaite et al., 2017). The identities of the genes or gene clusters related to the six antibiotics were clearly higher between strains HY96-2 and SC2 than between strains HY96-2 and E681. All three of the P. polymyxa strains were found to contain polymyxin-related gene clusters. Polymyxin is another key antibiotic in P. polymyxa and possesses broad-spectrum antibacterial activity, especially against G- bacteria (Falagas and Kasiakou, 2005). However, the polymyxin-related gene clusters in strains SC2 and E681 displayed low identities with that in strain HY96-2 of only 63.21 and 54.94%, respectively. Bacterial plant diseases are mainly caused by G- bacteria, such as those belonging to the genera Ralstonia (Liu et al., 2017c), Erwinia (Bell et al., 2004), Pseudomonas (Wang et al., 2018), and Xanthomonas (Islam et al., 2018). Strain HY96-2 has been found to effectively control many G- bacteria plant diseases in the field (Table 1), such as bacterial wilt, a soil-borne disease caused by R. solanacearum that has been referred to as “plant cancer,” and angular leaf spot on cucumbers, a leaf disease caused by P. syringae. In the field, the biocontrol efficacy of 109 CFU/g P. polymyxa wettable powder (PPWP, developed with strain HY96-2) on tomato bacterial wilt was found to reach 91.03% with a dosage of 10.8 kg/hectare. Moreover, the biocontrol efficacy of PPWP against angular leaf spot on cucumbers reached 81.08% with 300-fold dilution, which was significantly higher than that obtained for a chemical pesticide (Supplementary Table S6). In contrast to strain HY96-2, neither strain E681 nor strain SC2 has found application as a bactericide. Strain E681 was only reported to inhibit the growth of the G- plant pathogenic bacterium P. syringae and the G- human pathogenic bacterium Escherichia coli (Table 1), while the detailed and definitive antibacterial spectrum of strain SC2 has not been reported. It can be deduced that the variations of the genes and gene clusters involved in antibacterial metabolite synthesis between these three strains might be responsible for the differences in their antibacterial spectra and control efficacy.
Comparison of the key genes involved in systemic resistance inducer production revealed that all three of the tested strains contained the key genes related to volatile organic compounds (2,3-butanediol, methanethiol, and isoprene). The genes responsible for these inducers in strains SC2 and HY96-2 exhibited 91.33–97.05% identity, which is higher than the 86.83–92.05% identity observed between strains E681 and HY96-2. It can be deduced that the three strains can induce similar systemic resistance in plants but with varying effectiveness owing to the variations in the related genes.
In summary, in this study we determined the complete genome sequence of P. polymyxa strain HY96-2 and performed a comparative genomic analysis between various P. polymyxa strains. Based on this comparison, especially in terms of biofilm formation, antibiotic synthesis, and systemic resistance inducer production, the biocontrol mechanisms of P. polymyxa strain HY96-2 were determined at the molecular level. These mechanisms can be deduced as follows: (1) biofilm formation to prevent infection of the plant; (2) synthesis of fusaricidins and paenilarvins to protect against fungal plant pathogens, and the secretion of polymyxin, tridecaptin, and polyketide antibiotics with activity against G- bacteria plant pathogens and the production of mersacidin, kalimantacin, and bacitracin with activity against G+ bacteria plant pathogens; (3) induction of the systemic resistance of plants via inducing volatile organic compounds, including 2,3-butanediol, methanethiol, and isoprene, etc. The differences between the various P. polymyxa strains in terms of the control targets and efficacies might be attributable to the variations in the genes or gene clusters responsible for these three aspects of the biocontrol mechanism. The goal of this study was to provide a scientific basis for the further optimization of microbial biopesticides based on P. polymyxa strain HY96-2 in terms of field application and quality standards. For example, to develop a biopesticide based on strain HY96-2 for specifically controlling plant diseases caused by G- bacteria, the levels of polymyxin in the product could be increased by altering the fermentation conditions or genetically modifying the producing strain. This study may also serve as a reference for future investigations into the differences in biocontrol targets and efficacy between different biocontrol agents.
Author Contributions
YCL, YC, and YGL experimental design and authorship. YCL, YC, JY, ZZ, and QL experiments and data analysis. YCL, YC, YGL, and DZ manuscript revision. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This study was supported by the National Natural Science Fund (31501693) and the National Key Research and Development Program of China (2017YFD0201107-2-3).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01520/full#supplementary-material
References
- Bais H. P., Fall R., Vivanco J. M. (2004). Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 134 307–319. 10.1104/pp.103.028712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell K. S., Sebaihia M., Pritchard L., Holden M. T. G., Hyman L. J., Holeva M. C., et al. (2004). Genome sequence of the enterobacterial phytopathogen Erwinia carotovora subsp atroseptica and characterization of virulence factors. Proc. Natl. Acad. Sci. U.S.A. 101 11105–11110. 10.1073/pnas.0402424101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg G. (2009). Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 84 11–18. 10.1007/s00253-009-2092-7 [DOI] [PubMed] [Google Scholar]
- Bianciotto V., Andreotti S., Balestrini R., Bonfante P., Perotto S. (2001). Mucoid mutants of the biocontrol strain Pseudomonas fluorescens CHA0 show increased ability in biofilm formation on mycorrhizal and nonmycorrhizal carrot roots. Mol. Plant Microbe Interact. 14 255–260. 10.1094/MPMI.2001.14.2.255 [DOI] [PubMed] [Google Scholar]
- Chakraborty K., Thilakan B., Kizhakkekalam V. K. (2018). Antibacterial aryl-crowned polyketide from Bacillus subtilis associated with seaweed Anthophycus longifolius. J. Appl. Microbiol. 124 108–125. 10.1111/jam.13627 [DOI] [PubMed] [Google Scholar]
- Chen F., Mackey A. J., Stoeckert C. J., Roos D. S. (2006). OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 34 D363–D368. 10.1093/nar/gkj123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. H., Koumoutsi A., Scholz R., Eisenreich A., Schneider K., Heinemeyer I., et al. (2007). Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 25 1007–1014. 10.1038/nbt1325 [DOI] [PubMed] [Google Scholar]
- Chin C. S., Alexander D. H., Marks P., Klammer A. A., Drake J., Heiner C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10 563–571. 10.1038/nmeth.2474 [DOI] [PubMed] [Google Scholar]
- Choi S. K., Park S. Y., Kim R., Lee C. H., Kim J. F., Park S. H. (2008). Identification and functional analysis of the fusaricidin biosynthetic gene of Paenibacillus polymyxa E681. Biochem. Biophys. Res. Commun. 365 89–95. 10.1016/j.bbrc.2007.10.147 [DOI] [PubMed] [Google Scholar]
- Cochrane S. A., Lohans C. T., van Belkum M. J., Bels M. A., Vederas J. C. (2015). Studies on tridecaptin B-1, a lipopeptide with activity against multidrug resistant Gram-negative bacteria. Org. Biomol. Chem. 13 6073–6081. 10.1039/c5ob00780a [DOI] [PubMed] [Google Scholar]
- Conesa A., Gotz S., Garcia-Gomez J. M., Terol J., Talon M., Robles M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21 3674–3676. 10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- Delcher A. L., Harmon D., Kasif S., White O., Salzberg S. L. (1999). Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27 4636–4641. 10.1093/nar/27.23.4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon B. K., Laird M. R., Shay J. A., Winsor G. L., Lo R., Nizam F., et al. (2015). IslandViewer 3: more flexible, interactive genomic island discovery, visualization and analysis. Nucleic Acids Res. 43 W104–W108. 10.1093/nar/gkv401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didovyk A., Borek B., Tsimring L., Hasty J. (2016). Transcriptional regulation with CRISPR-Cas9: principles, advances, and applications. Curr. Opin. Biotechnol. 40 177–184. 10.1016/j.copbio.2016.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domka J., Lee J., Bansal T., Wood T. K. (2007). Temporal gene-expression in Escherichia coli K-12 biofilms. Environ. Microbiol. 9 332–346. 10.1111/j.1462-2920.2006.01143.x [DOI] [PubMed] [Google Scholar]
- Eastman A. W., Heinrichs D. E., Yuan Z. C. (2014a). Comparative and genetic analysis of the four sequenced Paenibacillus polymyxa genomes reveals a diverse metabolism and conservation of genes relevant to plant-growth promotion and competitiveness. BMC Genomics 15:22. 10.1186/1471-2164-15-851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman A. W., Weselowski B., Nathoo N., Yuan Z. C. (2014b). Complete genome sequence of Paenibacillus polymyxa CR1, a plant growth-promoting bacterium isolated from the corn rhizosphere exhibiting potential for biocontrol, biomass degradation, and biofuel production. Genome Announc. 2 e01218–13. 10.1128/genomeA.01218-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabert C., Feher V. A., Hoch J. A. (1999). Two component signal transduction in Bacillus subtilis: How one organism sees its world. J. Bacteriol. 181 1975–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falagas M. E., Kasiakou S. K. (2005). Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 40 1333–1341. 10.1086/429323 [DOI] [PubMed] [Google Scholar]
- Fan L., Zhang D. J., Liu Z. H., Tao L. M., Luo Y. C. (2012). Antifungal lipopeptide produced by Paenibacillus polymyxa HY96-2. Nat. Prod. Res. Dev. 24 729–735. 10.1007/s00253-013-5157-6 [DOI] [PubMed] [Google Scholar]
- Federle M. J. (2009). “Contributions to microbiology,” in Autoinducer-2-Based Chemical Communication in Bacteria: Complexities of Interspecies Signaling, eds Collin M., Schuch R. (Basel: Karger; ), 18–32. 10.1159/000219371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galea C. A., Han M., Zhu Y., Roberts K., Wang J., Thompson P. E., et al. (2017). Characterization of the polymyxin D synthetase biosynthetic cluster and product profile of Paenibacillus polymyxa ATCC 10401. J. Nat. Prod. 80 1264–1274. 10.1021/acs.jnatprod.6b00807 [DOI] [PubMed] [Google Scholar]
- Gounani Z., Asadollahi M. A., Meyer R. L., Arpanaei A. (2018). Loading of polymyxin B onto anionic mesoporous silica nanoparticles retains antibacterial activity and enhances biocompatibility. Int. J. Pharm. 537 148–161. 10.1016/j.ijpharm.2017.12.039 [DOI] [PubMed] [Google Scholar]
- Grissa I., Vergnaud G., Pourcel C. (2007). CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35 W52–W57. 10.1093/nar/gkm360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman A. D., Lewis T., Levin N., Vivo R. D. (1992). Suppressors of a spo0A missense mutation and their effects on sporulation in Bacillus subtilis. Biochimie 74 679–688. 10.1016/0300-9084(92)90140-A [DOI] [PubMed] [Google Scholar]
- Hao T. Y., Chen S. F. (2017). Colonization of wheat, maize and cucumber by Paenibacillus polymyxa WLY78. PLoS One 12:e0169980. 10.1371/journal.pone.0169980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. E., Xia Y., Xiong Y., Li X. F., Su X. Y. (2011). Inhibition of biofilm formation by the antisense peptide nucleic acids targeted at the motA gene in Pseudomonas aeruginosa PAO1 strain. World J. Microbiol. Biotechnol. 27 1981–1987. 10.1007/s11274-011-0658-x [DOI] [Google Scholar]
- Islam M. T., Lee B. R., Das P. R., La V. H., Jung H. I., Kim T. H. (2018). Characterization of p-Coumaric acid-induced soluble and cell wall-bound phenolic metabolites in relation to disease resistance to Xanthomonas campestris pv. campestris in Chinese cabbage. Plant Physiol. Biochem. 125 172–177. 10.1016/j.plaphy.2018.02.012 [DOI] [PubMed] [Google Scholar]
- Jeong H., Park S. Y., Chung W. H., Kim S. H., Kim N., Park S. H., et al. (2011). Draft genome sequence of the Paenibacillus polymyxa type strain (ATCC 842T), a plant growth-promoting bacterium. J. Bacteriol. 193 5026–5027. 10.1128/JB.05447-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Shao W. L., Perego M., Hoch J. A. (2000). Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 38 535–542. 10.1046/j.1365-2958.2000.02148.x [DOI] [PubMed] [Google Scholar]
- Kajimura Y., Kaneda M. (1995). Fusaricidin A, a new depsipeptide antibiotic produced by Bacillus polymyxa KT-8. Taxonomy, fermentation, isolation, structure elucidation and biological activity. J. Antibiot. 49 129–135. 10.7164/antibiotics.49.129 [DOI] [PubMed] [Google Scholar]
- Kajimura Y., Kaneda M. (1996). Fusaricidins B, C and D, new depsipeptide antibiotics produced by Bacillus polymyxa KT-8: isolation, structure elucidation and biological activity. J. Antibiot. 50 220–228. 10.7164/antibiotics.50.220 [DOI] [PubMed] [Google Scholar]
- Kim J. F., Jeong H., Park S. Y., Kim S. B., Park Y. K., Choi S. K., et al. (2010). Genome sequence of the polymyxin-producing plant-probiotic rhizobacterium Paenibacillus polymyxa E681. J. Bacteriol. 192 6103–6104. 10.1128/JB.00983-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. J., Sukweenadhi J., Seok J. W., Kang C. H., Choi E. S., Subramaniyam S., et al. (2017). Complete genome sequence of Paenibacillus yonginensis DCY84T, a novel plant Symbiont that promotes growth via induced systemic resistance. Stand. Genomic Sci. 12:63. 10.1186/s40793-017-0277-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K. (2007). Bacillus subtilis pellicle formation proceeds through genetically defined morphological changes. J. Bacteriol. 189 4920–4931. 10.1128/JB.00157-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konzl D., Klensl A., Schbrgendorfer K., Marahiel M. A. (1997). The bacitracin biosynthesis operon of Bacillus licheniformis ATCC 10716: Molecular characterization of three multi-modular peptide synthetases. Chem. Biol. 4 927–937. 10.1016/S1074-5521(97)90301-X [DOI] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., Phillippy A., Delcher A. L., Smoot M., Shumway M., Antonescu C., et al. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5:R12. 10.1186/gb-2004-5-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y. S., Lee D. Y., Rakwal R., Baek S. B., Lee J. H., Kwak Y. S., et al. (2016). Proteomic analyses of the interaction between the plant-growth promoting rhizobacterium Paenibacillus polymyxa E681 and Arabidopsis thaliana. Proteomics 16 122–135. 10.1002/pmic.201500196 [DOI] [PubMed] [Google Scholar]
- Lal S., Tabacchioni S. (2009). Ecology and biotechnological potential of Paenibacillus polymyxa: a minireview. Indian J. Microbiol. 49 2–10. 10.1007/s12088-009-0008-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B., Farag M. A., Park H. B., Kloepper J. W., Lee S. H., Ryu C. M. (2012). Induced resistance by a long-chain bacterial volatile: elicitation of plant systemic defense by a C13 volatile produced by Paenibacillus polymyxa. PLoS One 7:e48744. 10.1371/journal.pone.0048744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. Q., Yang D. Q., Qiu M. H., Shao J. H., Guo R., Shen B., et al. (2014). Complete genome sequence of Paenibacillus polymyxa SQR-21, a plant growth-promoting rhizobacterium with antifungal activity and rhizosphere colonization ability. Genome Announc. 2 e00281–14. 10.1128/genomeA.00281-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Liu K., Li Y. H., Wang C. Q., Hou Q. H., Xu W. F., et al. (2017a). Complete genome sequence of Paenibacillus polymyxa YC0136, a plant growth-promoting rhizobacterium isolated from tobacco rhizosphere. Genome Announc. 5 e01635–16. 10.1128/genomeA.01635-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Wang C. Q., Li Y. H., Liu K., Hou Q. H., Xu W. F., et al. (2017b). Complete genome sequence of Paenibacillus polymyxa YC0573, a plant growth-promoting rhizobacterium with antimicrobial activity. Genome Announc. 5 e01636–16. 10.1128/genomeA.01636-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Tang Y. M., Qin X. Y., Yang L., Jiang G. F., Li S. L., et al. (2017c). Genome sequencing of Ralstonia solanacearum CQPS-1, a phylotype I strain collected from a highland area with continuous cropping of tobacco. Front. Microbiol. 8:974. 10.3389/fmicb.2017.00974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. H., Fan L., Zhang D. J., Li Y. G. (2011). Antifungal depsipeptide compounds from Paenibacillus polymyxa HY96-2. Chem. Nat. Compd. 47 496–497. 10.1007/s10600-011-9978-1 [DOI] [Google Scholar]
- Lowe T. M., Eddy S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25 955–964. 10.1093/nar/25.5.955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B. X., Leong H. W. (2016). Computational methods for predicting genomic islands in microbial genomes. Comput. Struct. Biotechnol. J. 14 200–206. 10.1016/j.csbj.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg B., Kamilova F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63 541–556. 10.1146/annurev.micro.62.081307.162918 [DOI] [PubMed] [Google Scholar]
- Ma M. C., Wang C. C., Ding Y. Q., Li L., Shen D. L., Jiang X., et al. (2011). Complete genome sequence of Paenibacillus polymyxa SC2, a strain of plant growth-promoting Rhizobacterium with broad-spectrum antimicrobial activity. J. Bacteriol. 193 311–312. 10.1128/JB.01234-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y. P., Hao L., Ke H., Liang Z. L., Ma J. Y., Liu Z. X., et al. (2017). LuxS/AI-2 in Streptococcus agalactiae reveals a key role in acid tolerance and virulence. Res. Vet. Sci. 115 501–507. 10.1016/j.rvsc.2017.07.032 [DOI] [PubMed] [Google Scholar]
- Mikkola R., Andersson M. A., Grigoriev P., Heinonen M., Salkinoja-Salonen M. S. (2017). The toxic mode of action of cyclic lipodepsipeptide fusaricidins, produced by Paenibacillus polymyxa, toward mammalian cells. J. Appl. Microbiol. 123 436–449. 10.1111/jam.13498 [DOI] [PubMed] [Google Scholar]
- Miller M. B., Bassler B. L. (2001). Quorum sensing in bacteria. Annu. Rev. Microbiol. 55 165–199. 10.1146/annurev.micro.55.1.165 [DOI] [PubMed] [Google Scholar]
- Molinatto G., Franzil L., Steels S., Puopolo G., Pertot I., Ongena M. (2017). Key impact of an uncommon plasmid on Bacillus amyloliquefaciens subsp. plantarum S499 developmental traits and lipopeptide production. Front. Microbiol. 8:17. 10.3389/fmicb.2017.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu B., Rueckert C., Blom J., Wang Q., Borriss R. (2011). The genome of the plant growth-promoting rhizobacterium Paenibacillus polymyxa M-1 contains nine sites dedicated to nonribosomal synthesis of lipopeptides and polyketides. J. Bacteriol. 193 5862–5863. 10.1128/JB.05806-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongena M., Jacques P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16 115–125. 10.1016/j.tim.2007.12.009 [DOI] [PubMed] [Google Scholar]
- Ongena M., Jourdan E., Adam A., Paquot M., Brans A., Joris B., et al. (2007). Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ. Microbiol. 9 1084–1090. 10.1111/j.1462-2920.2006.01202.x [DOI] [PubMed] [Google Scholar]
- Ran F. A. (2017). Adaptation of CRISPR nucleases for eukaryotic applications. Anal. Biochem. 532 90–94. 10.1016/j.ab.2016.10.018 [DOI] [PubMed] [Google Scholar]
- Ryu C. M., Farag M. A., Hu C. H., Reddy M. S., Kloepper J. W., Pare P. W. (2004). Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 134 1017–1026. 10.1104/pp.103.026583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu C. M., Kim J., Choi O., Kim S. H., Park C. S. (2006). Improvement of biological control capacity of Paenibacillus polymyxa E681 by seed pelleting on sesame. Biol. Control 39 282–289. 10.1016/j.biocontrol.2006.04.014 [DOI] [Google Scholar]
- Sahl H. G., Bierbaum G. (1998). Lantibiotics: biosynthesis and biological activities of uniquely modified peptides from Gram-positive bacteria. Annu. Rev. Microbiol. 52 41–79. 10.1146/annurev.micro.52.1.41 [DOI] [PubMed] [Google Scholar]
- Sang M. K., Kim K. D. (2014). Biocontrol activity and root colonization by Pseudomonas corrugata strains CCR04 and CCR80 against Phytophthora blight of pepper. Biocontrol 59 437–448. 10.1007/s10526-014-9584-9 [DOI] [Google Scholar]
- Schneider K. B., Palmer T. M., Grossman A. D. (2002). Characterization of comQ and comX, two genes required for production of ComX pheromone in Bacillus subtilis. J. Bacteriol. 184 410–419. 10.1128/jb.184.2.410-419.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen M., Li J. R., Ross A. C., Vederas J. C., Jensen S. E. (2011). Paenibacillus polymyxa PKB1 produces variants of polymyxin B-type antibiotics. Chem. Biol. 18 1640–1648. 10.1016/j.chembiol.2011.09.017 [DOI] [PubMed] [Google Scholar]
- Sharma R. R., Singh D., Singh R. (2009). Biological control of postharvest diseases of fruits and vegetables by microbial antagonists: a review. Biol. Control 50 205–221. 10.1016/j.biocontrol.2009.05.001 [DOI] [Google Scholar]
- Shi Y., Niu K. J., Huang B. R., Liu W. H., Ma H. L. (2017). Transcriptional responses of creeping bentgrass to 2,3-butanediol, a bacterial volatile compound (BVC) analogue. Molecules 22:E1318. 10.3390/molecules22081318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood S., Steinmetz H., Beims H., Mohr K. I., Stadler M., Djukic M., et al. (2014). Paenilarvins: Iturin family lipopeptides from the honey bee pathogen Paenibacillus larvae. Chembiochem 15 1947–1955. 10.1002/cbic.201402139 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley N. R., Lazazzera B. A. (2005). Defining the genetic differences between wild and domestic strains of Bacillus subtilis that affect poly-gamma-DL-glutamic acid production and biofilm formation. Mol. Microbiol. 57 1143–1158. 10.1111/j.1365-2958.2005.04746.x [DOI] [PubMed] [Google Scholar]
- Stein T. (2005). Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol. Microbiol. 56 845–857. 10.1111/j.1365-2958.2005.04587.x [DOI] [PubMed] [Google Scholar]
- Tatusov R. L., Natale D. A., Garkavtsev L. V., Tatusova T. A., Shankavaram U. T., Rao B. S., et al. (2001). The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29 22–28. 10.1093/nar/29.1.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thistlethwaite I. R. G., Bull F. M., Cui C., Walker P. D., Gao S. S., Wang L., et al. (2017). Elucidation of the relative and absolute stereochemistry of the kalimantacin/batumin antibiotics. Chem. Sci. 8 6196–6201. 10.1039/c7sc01670k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C. T. (2004). Polyketide and nonribosomal peptide antibiotics modularity and versatility. Science 303 1805–1810. 10.1126/science.1094318 [DOI] [PubMed] [Google Scholar]
- Wang R. L., Li Q., He S. S., Liu Y., Wang M. T., Jiang G. (2018). Modeling and mapping the current and future distribution of Pseudomonas syringae pv. actinidiae under climate change in China. PLoS One 13:e0192153. 10.1371/journal.pone.0192153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Lee K., Goto K., Kumagai S., Sugita-Konishi Y., Hara-Kudo Y. (2010). Rapid and effective DNA extraction method with bead grinding for a large amount of fungal DNA. J. Food Prot. 73 1077–1084. 10.4315/0362-028X-73.6.1077 [DOI] [PubMed] [Google Scholar]
- Weselowski B., Nathoo N., Eastman A. W., MacDonald J., Yuan Z. C. (2016). Isolation, identification and characterization of Paenibacillus polymyxa CR1 with potentials for biopesticide, biofertilization, biomass degradation and biofuel production. BMC Microbiol. 16:244. 10.1186/s12866-016-0860-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J. B., Shi H. W., Du Z. L., Wang T. S., Liu X. M., Chen S. F. (2016). Comparative genomic and functional analysis reveal conservation of plant growth promoting traits in Paenibacillus polymyxa and its closely related species. Sci. Rep. 6:21329. 10.1038/srep21329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Wang W., Wei H. G., Shen G. M., Li Y. G. (2006). Effect of Paenibacillus polymyxa HY96-2 on bacterial wilt of tomato. Chin. J. Biol. Control 22 216–220. 10.3969/j.issn.2095-039X.2006.03.012 [DOI] [Google Scholar]
- Yang Y., Zhou M. X., Hou H. Y., Zhu J., Yao F. H., Zhang X. J., et al. (2014). Quorum-sensing gene luxS regulates flagella expression and Shiga-like toxin production in F18ab Escherichia coli. Can. J. Microbiol. 60 355–361. 10.1139/cjm-2014-0178 [DOI] [PubMed] [Google Scholar]
- Yaoyao E., Yuan J., Yang F., Wang L., Ma J. H., Li J., et al. (2017). PGPR strain Paenibacillus polymyxa SQR-21 potentially benefits watermelon growth by re-shaping root protein expression. AMB Express 7:104. 10.1186/s13568-017-0403-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z. L., Zhu Y. Y., Qin W. R., Yin J. H., Qiu J. P. (2017). Oxidative stress induced by polymyxin E is involved in rapid killing of Paenibacillus polymyxa. Biomed. Res. Int. 2017:5437139. 10.1155/2017/5437139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Yao L. T., Tian F., Du B. H., Ding Y. Q. (2008). Screening and study on biological characteristics of antagonistic bacteria against Fusarium solani. Biotechnol. Bull. 1 156–159. 18396832 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.