Abstract
The type III NAD–dependent histone deacetylase Sirt1 plays important roles in a variety of pathobiological functions through targeting either the acetylated histones or transcription factors. However, the molecular mechanisms underlying how the Sirt1 functions are regulated remain vague. Herein we identified that the Janus kinase 1 (JAK1) interacts with Sirt1 and catalyzes its phosphorylation at the tyrosine residues of 280 and 301, both of which are highly conserved and located in the histone deacetylase catalytic domain of Sirt1. IL-6 stimulation enhanced Sirt1 interaction with JAK1 and JAK1-mediated Sirt1 phosphorylation. Interestingly, JAK1-mediated Sirt1 phosphorylation did not alter Sirt1 deacetylase catalytic activity, but instead it is required for Sirt1 interaction with the downstream transcription factor STAT3. JAK1-mediated phosphorylation enhanced Sirt1 suppression of STAT3 acetylation and transcriptional activity. As a consequence, Sirt1 activation attenuates IL-6 activity in protecting cancer cells from chemotherapeutic drug–induced apoptosis. Our studies identify JAK1 as a previously unappreciated tyrosine kinase of Sirt1 and reveal a novel negative feedback of the JAK1-STAT3 pathway.
Keywords: Janus kinase (JAK), signal transduction, sirtuin 1 (SIRT1), STAT3, post-translational modification (PTM), JAK1, negative feedback
Introduction
Sirt1, the yeast orthologue of yeast Sir2 (silent information regulator 2), is a type III NADH (NAD)–dependent histone deacetylase that plays important roles in a variety of biological functions including development, aging, immunity, metabolism, and stress response (1–3). In addition to the acetylated histone substrates, Sirt1 has been discovered to deacetylate a substantial number of transcription factors, including NF-κB (4), AP-1 (5), p53 (6), PGC-1α (7), c-Myc (8), MEF2 (9), FoxOs (10), and STAT3 (signal transducer and activator of transcription 3) (11), as well as many other nonhistone substrates to achieve its diverse biological and pathological functions. The biological functions of Sirt1 are regulated at multiple levels, such as the availability of NAD, mRNA transcription, microRNA-mediated suppression, and ubiquitination-induced protein degradation (12). It has been also shown that Sirt1 can be phosphorylated by several serine/threonine kinases, including AMPK (13, 14), JNK (15), and HIPK2 (16), which either inhibit or potentiate Sirt1 functions. A systemic kinome analysis by mass spectrum has identified Sirt1 as a substrate for tyrosine phosphorylation in human cancer cells (17), which has been also shown in https://www.phosphosite.org3 (37). However, the tyrosine kinases that catalyze Sirt1 phosphorylation and its functional consequences in cancer development and progression have not been identified.
The JAK-STAT pathway is the principal signaling mechanism for many cytokines and growth factors (18). The binding of cytokines and growth factors with their cell surface receptors leads to the activation of JAK kinases, including JAK1–3 to phosphorylate the downstream transcription factors STAT1-6, which is required for the transcription of STAT target genes. This activation process stimulates cell proliferation, differentiation, cell migration, and apoptosis, which are critical for a variety of biological functions including hematopoiesis, immune development, mammary gland development and lactation, adipogenesis, and other processes (19, 20). The mutations that constitutively activate or fail to regulate JAK signaling result in inflammatory disease, erythrocytosis, gigantism, and tumorigenesis (20). On the other hand, the JAK/STAT activation is negatively regulated by the SOCS (suppressors of cytokine signaling) family E3 ubiquitin ligase-induced degradation (21) and sumoylation catalyzed by the PIAS (protein inhibitors of activated stats) family proteins and PTPs (protein tyrosine phosphatases) (20). In addition, it has been shown that acetylation of STAT3 by the histone acetyltransferase p300 plays a positive role in regulating the STAT3 transcriptional activation which is reversed by the histone deacetylase Sirt1 (11). However, how the Sirt1-mediated deacetylation of STAT3 is regulated remain to be studied.
In the current study, we have demonstrated that JAK1 is a tyrosine kinase of Sirt1. Interestingly, whereas the tyrosine residues phosphorylated by JAK1 locate in the highly conserved histone deacetylase catalytic domain, Sirt1 deacetylase activity was unaltered by JAK1-mediated phosphorylation. Rather, JAK1-induced tyrosine phosphorylation facilitates Sirt1 interaction with STAT3 to suppress STAT3 transcription activity. Therefore, our study defines a previously unappreciated negative feedback of JAK1-STAT3 pathway.
Results
Sirt1 interacts with JAK1
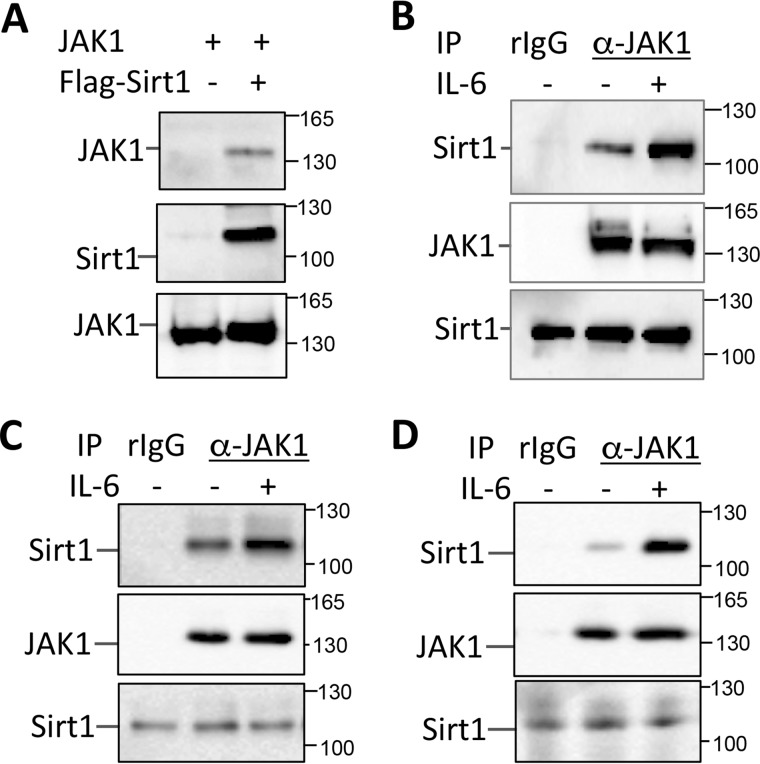
We have shown that both JAK1 and STAT3 are in an interactome of Sirt1 by mass spectrum analysis (22), together with the fact that Sirt1 has been shown as a negative regulator of STAT3 (11); these evidences suggest a possible role of Sirt1 in regulating the JAK1-STAT pathway. To test this hypothesis, we validated the interaction of Sirt1 with JAK1 in the transiently transfected HEK293 cells. Indeed, JAK1 was detected in the anti-Sirt1 immunoprecipitate from the lysate of HEK293 cells when both JAK1 and Sirt1 were co-transfected, but not with JAK1 expression alone, implying an interaction between JAK1 and Sirt1 (Fig. 1A). Further analysis detected the endogenous Sirt1 proteins in the immunoprecipitate by anti-JAK1, but not the normal rabbit IgG control antibody, in MCF-10 cells (Fig. 1B), a human breast epithelial cell line that responds to IL-6–induced activation of JAK1-STAT3 pathway (23). More importantly, stimulation of MCF-10 cells with IL-6 significantly enhanced Sirt1-JAK1 interaction (Fig. 1B). A similar result was further confirmed when human colon cancer cell HCT116 and melanoma cells MM.1s were used (Fig. 1, C and D). These results indicate that Sirt1 is an interacting protein of JAK1 and their interaction is regulated by IL-6 stimulation.
Figure 1.
JAK1 interacts Sirt1. A, JAK1 expression plasmid was co-transfected with or without FLAG-Sirt1 into HEK293 cells. The interaction of Sirt1 with JAK1 in the transfected cells was analyzed by co-immunoprecipitation with anti-FLAG antibody and by Western blotting with JAK1 antibody (top panel). The same membrane was reprobed with anti-Sirt1 (middle panel). JAK1 expression in the whole cell lysate was determined by Western blotting (bottom panel). B–D, MCF-10 (B), HCT116 (C), and MM.1s (D) cells were stimulated with or without 10 ng/ml IL-6 and lysed with RIPA buffer. The interaction of Sirt1 and JAK1 was determined by immunoprecipitation of JAK1 using normal rabbit IgG (rIgG) as a control and by Western blotting with anti-Sirt1 antibody (top panels). The same membrane was stripped and reblotted with anti-JAK1 (middle panels). Sirt1 expression in the whole cell lysate was determined by Western blotting (bottom panels).
JAK1 is a tyrosine kinase of Sirt1
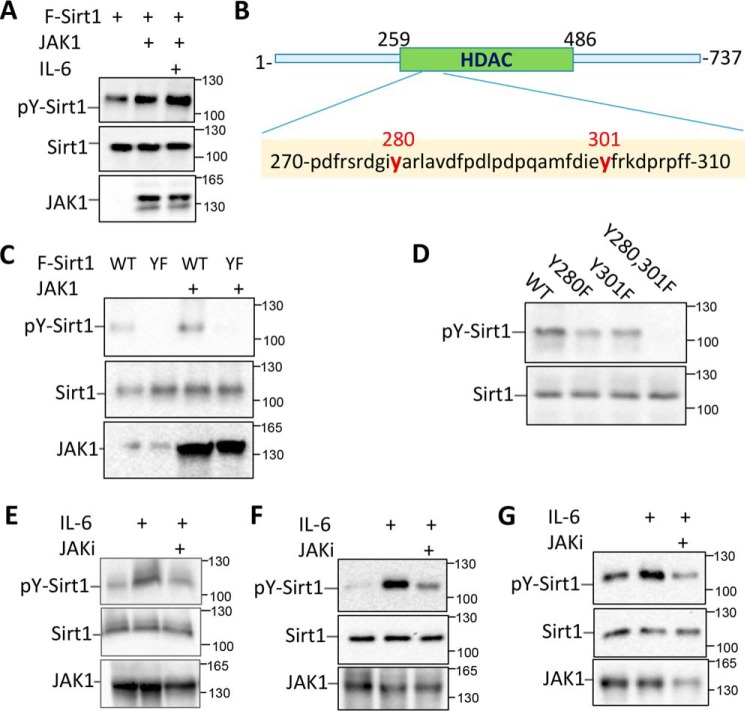
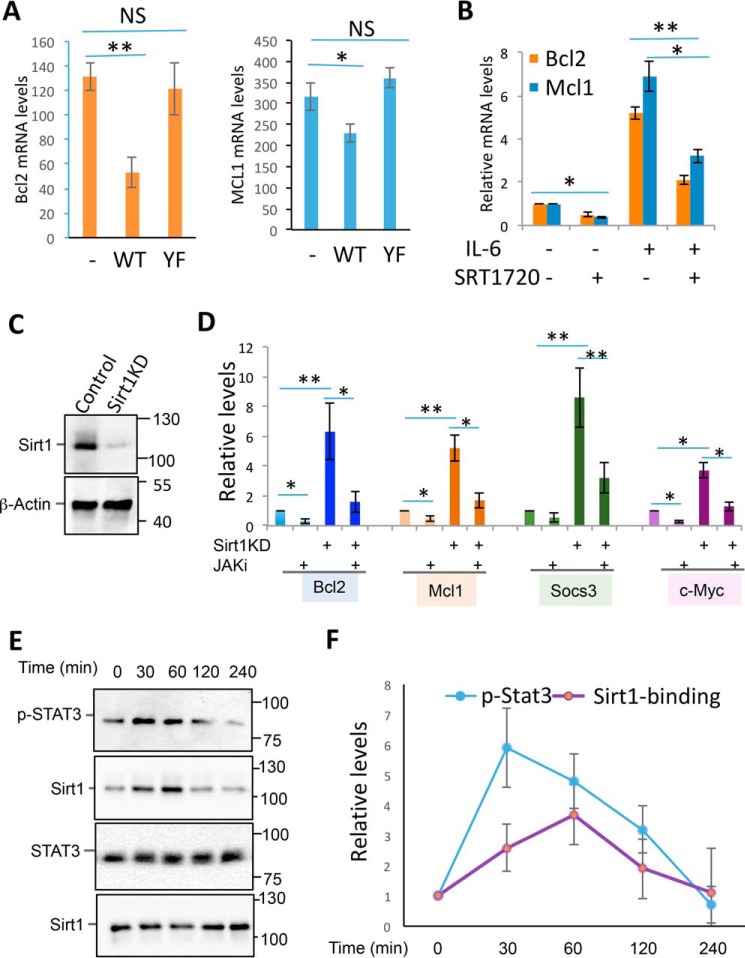
As JAK1 is a tyrosine kinase, we then reasoned whether JAK1 catalyzes Sirt1 phosphorylation. Indeed, the anti-phosphotyrosine antibody detected a 110 kDa band in the anti-Sirt1 immunoprecipitate, suggesting that Sirt1 is phosphorylated in MCF-10 cells. Stimulation of the cells with IL-6, which promoted Sirt1-JAK1 interaction, dramatically enhanced Sirt1 phosphorylation (Fig. 2A), suggesting that JAK1 is a tyrosine kinase of Sirt1. A systemic kinome analysis by mass spectrum has identified two tyrosine residues (Tyr-280 and Tyr-301) of Sirt1 are phosphorylated in human cancer cells (17), which also has been shown in https://www.phosphosite.org3 (37). Notably, both tyrosine residues are conserved from yeast to human and locate in the histone deacetylase catalytic domain of Sirt1 (Fig. 2B). We then speculated that these two tyrosine residues are potential targets of JAK1. As expected, mutation of these two tyrosine residues to phenylalanine abolished Sirt1 phosphorylation even when JAK1 was co-expressed (Fig. 2C), indicating that the two tyrosine residues are the predominant phosphorylation sites of JAK1. Further analysis indicates that mutation of either Tyr-280 or Tyr-301 alone partially abolished Sirt1 phosphorylation (Fig. 2D), confirming that the tyrosine residues of both 280 and 301 are substrates of JAK1. More importantly, IL-6 stimulation, which enhanced JAK1-Sirt1 interaction, dramatically enhanced Sirt1 tyrosine phosphorylation, and this IL-6–mediated enhancement is largely diminished by the treatment with a JAK1-specific inhibitor in MCF-10 cells (Fig. 2E). Similarly, a significant increase in Sirt1 phosphorylation was detected in both colon cancer cell HCT116 and melanoma cell MM.1s when treated with IL-6, and JAK1-specific inhibitor largely abolished the IL-6–induced Sirt1 tyrosine phosphorylation (Fig. 2, F and G). Collectively, our results indicate that JAK1 is a tyrosine kinase of Sirt1 and catalyzes its phosphorylation at the 280 and 301 tyrosine residue.
Figure 2.
JAK1 is a tyrosine kinase of Sirt1. A, MCF-10 cells were transfected with Sirt1 and JAK1 as indicated. The Sirt1 proteins were immunoprecipitated with anti-Sirt1 Abs. Sirt1 protein phosphorylation was detected by anti-phosphotyrosine antibody (4G10) (top panel). The same membrane was reprobed with anti-Sirt1 (middle panel) and the expression levels of JAK1 in the whole cell lysate were detected by Western blotting (bottom panel). B, Sirt1 structure and the locations of tyrosine residues 280 and 301. C and D, JAK1 expression plasmid was transfected into MCF-10 cells with either FLAG-Sirt1 or Flag-Sirt1/YF mutant (C), or with FLAG-Sirt1/Tyr-280, Y301F, or Y280F,Y301F (D). Sirt1 phosphorylation was determined as in (A). E–G, MCF-10 (E), HCT116 (F), and MM.1s (G) cells were stimulated with or without IL-6 or further with the JAK1 inhibitor PF-04965842 (1 μm). Cells were then lysed with RIPA buffer. Sirt1 phosphorylation was determined as in (A).
JAK1-mediated phosphorylation enhances Sirt1 interaction with STAT3
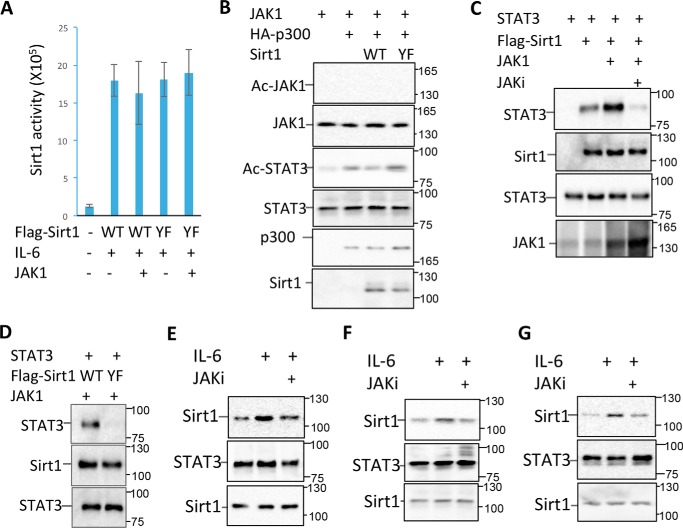
Because both the tyrosine residues locate in the HDAC domain of Sirt1 (Fig. 2B), we speculated the possibility that the JAK1-mediated tyrosine phosphorylation may regulate the catalytic activity of Sirt1. However, neither JAK1 expression nor mutation of the two tyrosine residues altered the Sirt1 deacetylase activity (Fig. 3A). We then speculated whether Sirt1 functions as a deacetylase of JAK1 to regulate the cytokine signaling. However, we were unable to detect JAK1 acetylation by Western blot analysis in the anti-JAK1 immunoprecipitates with anti–acetyl-lysine antibodies, even when the cells were co-expressed with p300 acetyltransferase and treated with IL-6 (Fig. 3B). As a positive control, STAT3 acetylation was enhanced by p300 expression. Interestingly, in contrast to the fact that WT Sirt1 inhibited STAT3 acetylation, Sirt1YF expression showed no effect of p300-mediated STAT3 acetylation (Fig. 3B). These results largely excluded the possibility that Sirt1 regulates JAK1 acetylation.
Figure 3.
Sirt1 tyrosine phosphorylation enhances its interaction with STAT3. A, JAK1 expression plasmid was transfected into MCF-10 cells with either FLAG-Sirt1 or FLAG-Sirt1/YF mutant. Two days after the transfected cells were treated with or without IL-6 for 30 min and lysed. Sirt1 was immunoprecipitated with anti-FLAG antibody, the deacetylase activity of Sirt1 was determined. Error bars represent data from three independent experiments. B, JAK1 and STAT3 were co-expressed with p300 and Sirt1 or its YF mutant in the MCF-10 cells as indicated. JAK1 acetylation was determined by immunoprecipitation of JAK1 and Western blotting with anti–acetyl-lysine Abs (top panel). The same membrane was reprobed with anti-JAK1 (second panel). STAT3 acetylation was determined by immunoprecipitation of STAT3 and by Western blotting with anti–acetyl-lysine Abs (third panel). The same membrane was reprobed with anti-STAT3 (fourth panel). The expression levels of p300 and Sirt1 in the whole cell lysates were analyzed by Western blotting (bottom two panels). C, STAT3, JAK1, and Sirt1 expression plasmids were transfected into MCF-10 cells as indicated. The transfected cells were treated with the JAK inhibitor PF-04965842 and then collected. STAT3 interaction with Sirt1 was determined by immunoprecipitation with anti-FLAG antibody and Western blotting with anti-STAT3 (top panel). The same membrane was reprobed with anti-Sirt1 (second panel). The expression levels of STAT3 (third panel) and JAK1 (bottom panel) in the whole cell lysates were determined by Western blotting. D, MCF-10 cells were transfected with STAT3, JAK1, and Sirt1 or its YF mutant. The interaction of STAT3 with Sirt1 in the lysate of treated cells was determined as in (C). E–G, MCF-10 (E), HCT116 (F), and MM.1s (G) cells were stimulated with or without IL-6 or further with JAK1 inhibitor PF-04965842 (1 μm), and then lysed with RIPA buffer. Sirt1 interaction with STAT3 was determined as in (C).
It has been shown that Sirt1 functions as a negative regulator of the STAT3 transcriptional activity (11). We then asked whether JAK1-mediated phosphorylation is involved in this regulatory pathway. Interestingly, JAK1 expression significantly enhanced Sirt1-STAT3 interaction, and this enhanced interaction was largely abolished by the JAK1 inhibitor PF-04965842 treatment (Fig. 3C), indicating that JAK1-mediated phosphorylation promotes Sirt1-STAT3 interaction. To support this notion, mutation of the tyrosine residues of 280 and 301 largely diminished the interaction of Sirt1 with STAT3 (Fig. 3D). We then speculated whether IL-6 treatment, which induces both Sirt1 and STAT3 tyrosine phosphorylation, enhances Sirt1 interaction with STAT3. Indeed, IL-6 treatment significantly enhanced Sirt1-STAT3 interaction in all three human cancer cell lines tested, including MCF-10, HCT116, and MM.1s. Treatment of cells with the JAK1 inhibitor largely diminished STAT3-Sirt1 interaction (Fig. 3, E–G). Collectively, our data indicate that JAK1-mediated phosphorylation of Sirt1 at the tyrosine residues of 280 and 301 promotes Sirt1-JAK1 interaction upon cytokine stimulation in cancer cells.
JAK1-mediated Sirt1 tyrosine phosphorylation inhibits STAT3 promoter-binding activity
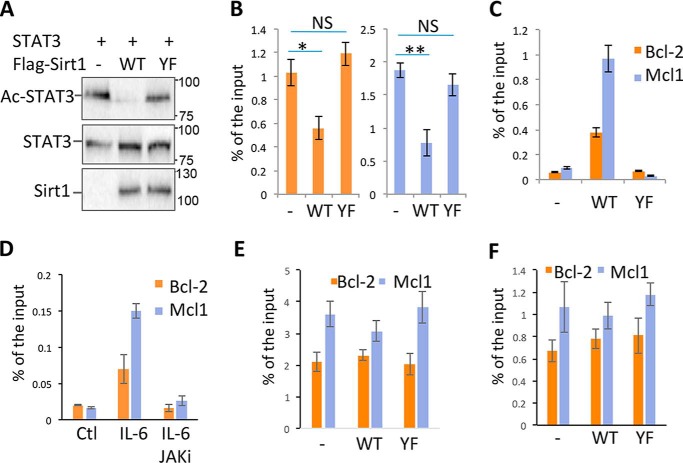
Sirt1 has been shown as a deacetylase of STAT3 to suppress its transcriptional activity (11). We reasoned whether JAK1-mediated Sirt1 phosphorylation to enhance Sirt1 interaction with STAT3 functions as a negative feedback to suppress JAK1-STAT3 signaling. Indeed, in contrast to the fact that expression of Sirt1 inhibited STAT3 acetylation, mutation of the tyrosine residues of 280 and 301 totally abolished Sirt1 activity in suppressing STAT3 acetylation (Fig. 4A), implying that JAK1-mediated tyrosine phosphorylation of Sirt1 is required to suppress STAT3 transcription activity. To support this notion, expression of Sirt1 significantly inhibited the binding activity of STAT3 onto the promoters of both Bcl-2 and Mcl1 genes. In contrast, the tyrosine to phenylalanine (YF)4 mutation of Sirt1, which abolishes its interaction with STAT3, largely diminished Sirt1 suppressive activity on STAT3 promoter binding (Fig. 4B). These results clearly indicate that JAK1-mediated Sirt1 tyrosine phosphorylation at the residues of 280 and 301 provides a docking site for Sirt1 binding to STAT3 and inhibits STAT3 transcriptional activation.
Figure 4.
JAK1-mediated Sirt1 phosphorylation inhibits STAT3 transcription activity. A, MCF-10 cells were transfected with STAT3 and FLAG-Sirt1 or its YF mutant. STAT3 acetylation was determined by immunoprecipitation with anti-STAT3 and by Western blotting with anti–acetyl-lysine antibody (top panel). The same membrane was reprobed with anti-STAT3 (middle panel), and the expression levels of Sirt1 in the whole cell lysate were analyzed by Western blotting (bottom panel). B and C, MCF-10 cells were transfected with Sirt1 or its YF mutant. The transfected cells were stimulated with IL-6 for 6 h and the binding of STAT3 (B) and Sirt1 (C) onto Bcl2 or Mcl1 promoter was determined by ChIP. D, MCF-10 cells were treated with IL-6 or together with JAK1 inhibitor PF-04965842; the binding of Sirt1 onto Bcl2 or Mcl1 promoter was determined by ChIP. E and F, MCF-10 cells were transfected with Sirt1 or its YF mutant. The transfected cells were stimulated with IL-6 for 6 h and the levels of H3K8Ac (E) and H4K16Ac (F) at the Bcl2 or Mcl1 promoter was determined by ChIP. The Student's t test was used for the statistical analysis. NS, not significantly different; *, p < 0.05; and **, p < 0.01. Error bars represent mean ± S.D.
We then speculated whether Sirt1 is recruited onto the promoter of STAT3 target genes through its interaction with STAT3. ChIP analysis detected Sirt1 binding onto the promoter regions of both Bcl-2 and Mcl1, which was totally diminished by the mutation of tyrosine 280 and 301 (Fig. 4C). Because Sirt1 interaction with STAT3 relies on JAK1-mediated tyrosine phosphorylation, these results indicate that Sirt1 is recruited onto the promoter of STAT3 target genes, and this recruitment is regulated by JAK1-mediated tyrosine phosphorylation of Sirt1. To further support this conclusion, we found that IL-6 treatment significantly enhanced Sirt1 recruitment onto both Bcl-2 and Mcl1 promoter, and this enhanced recruitment was largely blocked by JAK1-specific inhibitor treatment (Fig. 4D).
Recent studies have shown that, instead of directly deacetylating its binding transcription factors, Sirt1 modulates gene transcription though suppressing histone acetylation with a preference for deacetylating histone H4 lysine 16 (H4K16Ac) and H3 lysine 9 (H3K9Ac) (24–26). However, ChIP analysis with antibodies (Abs) specific for either H4K16Ac or H3K9Ac did not detect any changes in the levels of H4K16Ac and H3K9Ac at Bcl-2 and Mcl1 promoters in MCF-10 cells by the expression of either Sirt1 or Sirt1/YF mutation (Fig. 4, E and F). Collectively, our data suggest that JAK1-mediated Sirt1 phosphorylation enhances Sirt1-STAT3 interaction to inhibit STAT3 acetylation and STAT3 promoter–binding activity.
Sirt1 suppresses STAT3 target gene expression
Next, we analyzed the effect of Sirt1 on the transcription of STAT3 target genes. As indicated in Fig. 5A, expression of Sirt1 significantly inhibited the expression of STAT3 target genes, both Bcl2 and Mcl1, in MCF-10 cells, but Sirt1 with mutations of the tyrosine residues failed to suppress STAT3 target gene expression. In addition, treatment of MCF-10 cells with the Sirt1-specific activator SRT1720 dramatically suppressed STAT3 target gene expression even after IL-6 stimulation (Fig. 5B). Conversely, knockdown of the endogenous Sirt1 resulted in a significant increase in the expression of STAT3 target genes including Bcl-2, Mcl1, Socs3, and c-Myc. This enhanced expression of STAT3 target genes was largely diminished by JAK1-specific inhibitor (Fig. 5, C and D). These results indicate that JAK1-mediated tyrosine phosphorylation as a negative feedback to suppress STAT3 transcriptional activity.
Figure 5.
JAK1-mediated Sirt1 phosphorylation inhibits STAT3 target gene transcription. A, MCF-10 cells were transfected with Sirt1 or its YF mutant. Two days after, the expression levels of STAT3 target genes, Bcl2 and Mcl1, were analyzed by real-time RT-PCR. B, MCF-10 cells were stimulated with IL-6 (10 ng/ml) and SRT1720 (1 μm) as indicated for overnight; the levels of Bcl2 and Mcl1 mRNA were determined by real-time RT-PCR. C and D, MCF-10 cells were transfected with control or Sirt1-specific siRNA followed by treatment with IL-6 or together with JAK1 inhibitor. The levels of STAT3 target genes including Bcl2, Mcl1, Socs3, and c-Myc were determined by real-time PCR. E and F, MCF-10 cells were treated with IL-6 for different amounts of time. The levels of phospho-STAT3 and STAT3-Sirt1 interaction were analyzed. Representative images from three independent experiments are shown (E) and the quantitation of phospho-STAT3 and STAT3-Sirt1 are indicated (F). Error bars represent data from five (A, B, and D) or three (F) independent experiments. The Student's t test was used for the statistical analysis. NS, not significantly different; *, p < 0.05; and **, p < 0.01.
Because JAK1 catalyzes the phosphorylation of both Sirt1 and STAT3, it is conceptually difficult to understand if STAT3 activation and Sirt1-mediated STAT3 inhibition of Stat3 occur with an identical kinetics. We then dynamically analyzed STAT3 phosphorylation and Sirt1-STAT3 in MCF-10 cells upon IL-6 stimulation. Indeed, as indicated in Fig. 5, E and F, there is a delay in Sirt1-STAT3 interaction comparing to STAT3 phosphorylation, indicating that STAT3 transcriptional activation occurs earlier than Sirt1-mediated suppression.
Sirt1 diminishes the protective activity of IL-6 from cisplatin-induced MCF-10 cell apoptosis
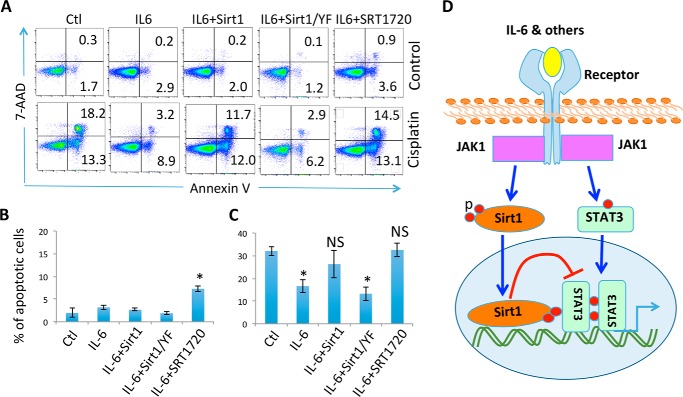
IL-6 has been shown to protect chemotherapy drug–induced cancer cell death and drug resistance of a variety of types of tumors (27). Because Sirt1 suppresses IL-6–induced anti-apoptotic gene transcription through deacetylating STAT3, we asked whether Sirt1 attenuates IL-6 protective activity from cisplatin-induced MCF-10 cell apoptosis. Consistent with previous studies (28), cisplatin treatment at 4 μg/ml significantly increased the percentage of annexin V–positive apoptotic cells, and further addition of IL-6 largely protected MCF-10 from cisplatin-induced apoptosis. Importantly, ectopic Sirt1 expression largely diminished the protective activity of IL-6. In contrast, Sirt1/YF mutant had no effect on cisplatin-induced cell death (Fig. 6, A–C). In addition, treatment with the Sirt1-specific activator SRT1720 abolished IL-6 protective functions from cisplatin-induced apoptosis of MCF-10 cells (Fig. 6, A and C). These results indicate that Sirt1 activation inhibits IL-6 functions in human breast epithelial cells.
Figure 6.
Sirt1 diminishes IL-6 protection activity. A and B, MCF-10 cells were transfected with Sirt1 or its YF mutant. Two days after, cells were treated with or without cisplatin (4 μm), IL-6 (10 ng/ml), and Sirt1 activator SRT1720 (1 μm) for 24 h. Cell apoptosis was analyzed by annexin V and 7-AAD levels and flow cytometry. A–C, representative data (A) and the average cell death (B, without cisplatin; C, with cisplatin) from five independent experiments are shown. The Student's t test was used for the statistical analysis. NS, not significantly different; *, p < 0.05; and **, p < 0.01. D, a proposed model of JAK1-mediated Sirt1 phosphorylation in suppressing STAT3 transcriptional activity.
Based on the results of the current studies, we propose a model for JAK1-mediated Sirt1 phosphorylation as a negative feedback in suppressing the IL-6–JAK1–STAT3 pathway: JAK1 functions as a tyrosine kinase of Sirt1 to catalyze its phosphorylation at the residues of Tyr-280 and Tyr-301. This phosphorylation event is required for the optimal interaction of Sirt1 with the JAK1 downstream transcription factor STAT3 to suppress STAT3 transcriptional activity (Fig. 6D). The STAT3 activation and Sirt1-mediated STAT3 inhibition of STAT3 appears to occur with a different kinetics because STAT3 phosphorylation peaks earlier than its interaction with Sirt1.
Discussion
Our studies define a previously unappreciated negative feedback in regulating the cytokine-JAK1-STAT3 pathway. This conclusion is supported by the following observations. First, Sirt1 interacts with JAK1 and their interaction is regulated by IL-6. Second, JAK1 catalyzes Sirt1 phosphorylation at the tyrosine residues 280 and 301. Third, although both tyrosine residues phosphorylated by JAK1 are located in the histone deacetylase catalytic domain of Sirt1, this phosphorylation appears not to alter Sirt1 deacetylase activity. Rather, the phosphorylation of 280 and 301 provides a docking site for JAK1 downstream transcription factor STAT3 to interact with, and as a consequence, Sirt1 deacetylates STAT3 to inhibit its transcription activity. Therefore, Sirt1 activation diminishes IL-6 protective functions from chemotherapeutic drug–induced cancer cell death.
JAK1-STAT3 has been reported to play an important role in a variety of biological and pathological conditions including gluconeogenesis, oxidative stress, immune regulation, inflammation, and tumorigenesis (19, 20). Our studies suggest JAK1-mediated Sirt1 phosphorylation as a possible common molecular feedback for the JAK1-STAT3 signaling pathways. In addition to this Sirt1-mediated deacetylation event, previous studies have defined that the posttranslational modification, either by ubiquitin catalyzed by the SOCS family E3 ubiquitin ligase (21, 29) or by the small ubiquitin-like modifier (SUMO) catalyzed by PIAS family proteins, plays critical roles in negatively regulating the JAK-STAT pathways (30). Therefore, the cytokine-induced JAK1-STAT3 activation appears to be controlled by multiple negative regulations. Our results indicate that the dynamics of JAK1-mediated STAT3 phosphorylation and Sirt1-STAT3 interaction are not identical. There is a delay in Sirt1-STAT3 interaction comparing to STAT3 phosphorylation, implying that Sirt1-mediated negative regulation occurs following STAT3 activation. It will be interesting to further determine the molecular mechanisms underlying how this different dynamic is regulated.
The role of Sirt1 in tumorigenesis has been at best controversial (3). On the one hand, it deacetylates and thus inactivates p53 (6, 31), yet on the other hand its overexpression attenuates tumor formation because Sirt1 acts to preserve genome integrity and serves as a haploinsufficient tumor suppressor protein in mice (32). Our studies here demonstrate that Sirt1 diminishes IL-6–induced cancer cell survival through suppressing the downstream transcription factor STAT3. Persistent JAK-STAT3 activation is found in many types of tumors including up to 50% of lung adenocarcinomas and breast cancers (33). Future studies will be interesting to characterize the Sirt1 role in cancer development and progression and its association with the elevated JAK-STAT3 activation.
Materials and methods
Cells and reagents
HEK293, MCF-10, HCT116, and MM.1s cells were cultivated in DMEM supplemented with 10% FBS. Human recombinant IL-6 was purchased from BioLegend (San Diego, CA) and the JAK1 inhibitor was from Sigma-Aldrich. Antibodies specific to each protein were purchased from the indicated companies including Sirt1, STAT3, and JAK1 (Abcam, Cambridge, MA), anti-phosphotyrosine (4G10), GAPDH, and FLAG (Sigma-Aldrich). Camptothecin and cisplatin were purchased from Calbiochem. Human recombinant IL-6 was purchased from PeproTECH (Rocky Hill, NJ). JAK1-specific inhibitor PF-04965842 was purchased from (Sigma-Aldrich).
Plasmid DNA
FLAG-tagged human Sirt1 expression plasmid was used as reported (34). Y280F,Y301F mutations were generated by PCR using a Phusion Site-Directed Mutagenesis Kit (Thermo Fisher). STAT3 and JAK1 expression plasmids were purchased from Addgene (Cambridge, MA).
Gene transfection, co-immunoprecipitation, and Western blotting
HEK293 or MCF-10 cells were plated in the 60-mm dishes and transfected with the indicated plasmids using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. Forty-eight h after transfection, cells were lysed in RIPA lysis buffer, which was freshly added with protease inhibitor mixture (Sigma-Aldrich). After precleaning with protein G beads (GE Healthcare), the cell lysates were immunoprecipitated with the indicated antibodies (1 μg) and incubated at 4 °C for 2 h, followed by the addition of 30 μl of protein G-Sepharose beads at 4 °C for an additional 4 h or overnight. Immunoprecipitates were washed four times with RIPA lysis buffer and then were boiled at 95 °C in 20 μl of Laemmli 2× buffer. Samples were subjected to an 8–12% SDS-PAGE gel and electrotransferred onto polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA). After blocking with 4% milk dissolved in Tris-buffered saline with Tween-20 (TBST), membranes were probed with the appropriate primary antibodies, and then washed twice with TBST and incubated with horseradish peroxidase–conjugated secondary antibodies. After washing twice with TBST, membranes were treated with enhanced chemiluminescence detection system (ECL) (GE Healthcare) and exposed. If necessary, membranes were stripped with stripping buffer (Bio-Rad), washed, blocked, and then reprobed with other antibodies.
In vitro Sirt1 activity assay
MCF-10 cells were plated in the 60-mm dishes and transfected with FLAG-Sirt1 or its YF mutant and JAK1 using Lipofectamine 2000 (Invitrogen). Two days after transfection, Sirt1 proteins in the lysates of transfected cells were immunoprecipitated with anti-FLAG antibodies. The histone deacetylase activity of beads-bound Sirt1 protein was measured using a fluorometric Sirt1 Activity Assay kit (Abcam) following a protocol provided by the manufacture.
Real-time quantitative PCR
Total RNA was extracted from cells with TRIzol reagent per the manufacturer's instructions (Ambion, Grand Island, NY). cDNA was synthesized using qScript cDNA Synthesis Kit (Quanta Biosciences, Gaithersburg, MD). Real-time qPCR was performed using iQ5 and the SYBR Green detection system (Bio-Rad). Data were normalized to the expression of β-actin in each sample as reported (35). Primers are shown in Table S1.
ChIP assay
MCF-10 cells were transfected with Sirt1 or Sirt1 mutant plasmid, cross-linked with 1% formaldehyde, and lysed with SDS lysis buffer. Cell lysates were sonicated, and 5% of cell lysate was removed and used to determine the total amount of target DNA in input. Remaining cell lysates were diluted in ChIP dilution buffer. Immunoprecipitation was performed with each of the indicated antibodies (4 μg) at 4 °C overnight. Immune complexes were then mixed with salmon sperm DNA/protein agarose 50% slurry at 4 °C for 1 h. The immunoprecipitates were then washed sequentially with low-salt buffer, high-salt buffer, LiCl wash buffer, and Tris-EDTA. DNA-protein complexes were eluted with elution buffer, and cross-linking was reversed. Genomic DNA was extracted using phenol/chloroform, and ethanol-precipitated DNA was resuspended in Tris-EDTA. PCR was performed with specific primers (purchased from Qiagen) according to the manufacturer's protocol (EMD Millipore).
Cell apoptosis analysis
Cancer cell apoptosis was analyzed as reported (36). Briefly, MCF-10 cells were transfected with Sirt1 or its YF mutant, or with control empty vector. Two days after transfection, cells were treated with or without IL-6 (10 ng/ml) and cisplatin (4 μg/ml) for an additional 24 h. The cell apoptosis was determined by staining with annexin V and 7-AAD, and analyzed by flow cytometry.
Author contributions
W. W., Y. X., J. W., H. Y., and B. G. data curation; W. W. and B. G. formal analysis; W. W., F. L., Y. X., J. W., Y. Z., H. Y., and B. G. investigation; W. W. and F. L. methodology; Y. X., J. W., Y. Z., and B. G. validation; Y. Z. and D. F. writing-original draft; B. G. and D. F. project administration; G. Y. and D. F. conceptualization; G. Y. and D. F. writing-review and editing; D. F. funding acquisition.
Supplementary Material
Acknowledgments
We thank Fang lab members for critical reading of the manuscript and constructive suggestions during our research.
This work was supported by National Institutes of Health Grants R01 AI079056, AI108634, and AR006634 (to D. F.) and the National Natural Science Foundation of China Grant 81600784 (to Y. Z.) The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- YF
- tyrosine to phenylalanine mutation
- Abs
- antibodies
- TBST
- TBS with Tween 20.
References
- 1. Fang Y., and Nicholl M. B. (2011) Sirtuin 1 in malignant transformation: Friend or foe? Cancer Lett. 306, 10–14 10.1016/j.canlet.2011.02.019 [DOI] [PubMed] [Google Scholar]
- 2. Kong S., McBurney M. W., and Fang D. (2012) Sirtuin 1 in immune regulation and autoimmunity. Immunol. Cell Biol. 90, 6–13 10.1038/icb.2011.102 [DOI] [PubMed] [Google Scholar]
- 3. Lin Z., and Fang D. (2013) The roles of SIRT1 in cancer. Genes Cancer 4, 97–104 10.1177/1947601912475079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yeung F., Hoberg J. E., Ramsey C. S., Keller M. D., Jones D. R., Frye R. A., and Mayo M. W. (2004) Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 23, 2369–2380 10.1038/sj.emboj.7600244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang J., Lee S. M., Shannon S., Gao B., Chen W., Chen A., Divekar R., McBurney M. W., Braley-Mullen H., Zaghouani H., and Fang D. (2009) The type III histone deacetylase Sirt1 is essential for maintenance of T cell tolerance in mice. J. Clin. Invest. 119, 3048–3058 10.1172/JCI38902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Luo J., Nikolaev A. Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., and Gu W. (2001) Negative control of p53 by Sir2α promotes cell survival under stress. Cell 107, 137–148 10.1016/S0092-8674(01)00524-4 [DOI] [PubMed] [Google Scholar]
- 7. Nemoto S., Fergusson M. M., and Finkel T. (2005) SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1α. J. Biol. Chem. 280, 16456–16460 10.1074/jbc.M501485200 [DOI] [PubMed] [Google Scholar]
- 8. Yuan J., Minter-Dykhouse K., and Lou Z. (2009) A c-Myc-SIRT1 feedback loop regulates cell growth and transformation. J. Cell Biol. 185, 203–211 10.1083/jcb.200809167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao X., Sternsdorf T., Bolger T. A., Evans R. M., and Yao T. P. (2005) Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol. Cell. Biol. 25, 8456–8464 10.1128/MCB.25.19.8456-8464.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., Hu L. S., Cheng H. L., Jedrychowski M. P., Gygi S. P., Sinclair D. A., Alt F. W., and Greenberg M. E. (2004) Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303, 2011–2015 10.1126/science.1094637 [DOI] [PubMed] [Google Scholar]
- 11. Nie Y., Erion D. M., Yuan Z., Dietrich M., Shulman G. I., Horvath T. L., and Gao Q. (2009) STAT3 inhibition of gluconeogenesis is down-regulated by SirT1. Nat. Cell Biol. 11, 492–500 10.1038/ncb1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang T., and Kraus W. L. (2010) SIRT1-dependent regulation of chromatin and transcription: linking NAD(+) metabolism and signaling to the control of cellular functions. Biochim. Biophys. Acta 1804, 1666–1675 10.1016/j.bbapap.2009.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee C. W., Wong L. L., Tse E. Y., Liu H. F., Leong V. Y., Lee J. M., Hardie D. G., Ng I. O., and Ching Y. P. (2012) AMPK promotes p53 acetylation via phosphorylation and inactivation of SIRT1 in liver cancer cells. Cancer Res. 72, 4394–4404 10.1158/0008-5472.CAN-12-0429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lau A. W., Liu P., Inuzuka H., and Gao D. (2014) SIRT1 phosphorylation by AMP-activated protein kinase regulates p53 acetylation. Am. J. Cancer Res. 4, 245–255 [PMC free article] [PubMed] [Google Scholar]
- 15. Gao Z., Zhang J., Kheterpal I., Kennedy N., Davis R. J., and Ye J. (2011) Sirtuin 1 (SIRT1) protein degradation in response to persistent c-Jun N-terminal kinase 1 (JNK1) activation contributes to hepatic steatosis in obesity. J. Biol. Chem. 286, 22227–22234 10.1074/jbc.M111.228874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Conrad E., Polonio-Vallon T., Meister M., Matt S., Bitomsky N., Herbel C., Liebl M., Greiner V., Kriznik B., Schumacher S., Krieghoff-Henning E., and Hofmann T. G. (2016) HIPK2 restricts SIRT1 activity upon severe DNA damage by a phosphorylation-controlled mechanism. Cell Death Differ. 23, 110–122 10.1038/cdd.2015.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., et al. (2007) Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203 10.1016/j.cell.2007.11.025 [DOI] [PubMed] [Google Scholar]
- 18. Stark G. R., and Darnell J. E. Jr. (2012) The JAK-STAT pathway at twenty. Immunity 36, 503–514 10.1016/j.immuni.2012.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Villarino A. V., Kanno Y., and O'Shea J. J. (2017) Mechanisms and consequences of Jak-STAT signaling in the immune system. Nat. Immunol. 18, 374–384 10.1038/ni.3691 [DOI] [PubMed] [Google Scholar]
- 20. Rawlings J. S., Rosler K. M., and Harrison D. A. (2004) The JAK/STAT signaling pathway. J. Cell Sci. 117, 1281–1283 10.1242/jcs.00963 [DOI] [PubMed] [Google Scholar]
- 21. Starr R., Willson T. A., Viney E. M., Murray L. J., Rayner J. R., Jenkins B. J., Gonda T. J., Alexander W. S., Metcalf D., Nicola N. A., and Hilton D. J. (1997) A family of cytokine-inducible inhibitors of signalling. Nature 387, 917–921 10.1038/43206 [DOI] [PubMed] [Google Scholar]
- 22. Lin Z., Yang H., Kong Q., Li J., Lee S. M., Gao B., Dong H., Wei J., Song J., Zhang D. D., and Fang D. (2012) USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol. Cell 46, 484–494 10.1016/j.molcel.2012.03.024 [DOI] [PubMed] [Google Scholar]
- 23. Sullivan N. J., Sasser A. K., Axel A. E., Vesuna F., Raman V., Ramirez N., Oberyszyn T. M., and Hall B. M. (2009) Interleukin-6 induces an epithelial-mesenchymal transition phenotype in human breast cancer cells. Oncogene 28, 2940–2947 10.1038/onc.2009.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vaquero A., Scher M., Erdjument-Bromage H., Tempst P., Serrano L., and Reinberg D. (2007) SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature 450, 440–444 10.1038/nature06268 [DOI] [PubMed] [Google Scholar]
- 25. Chen C. W., Koche R. P., Sinha A. U., Deshpande A. J., Zhu N., Eng R., Doench J. G., Xu H., Chu S. H., Qi J., Wang X., Delaney C., Bernt K. M., Root D. E., Hahn W. C., Bradner J. E., and Armstrong S. A. (2015) DOT1L inhibits SIRT1-mediated epigenetic silencing to maintain leukemic gene expression in MLL-rearranged leukemia. Nat. Med. 21, 335–343 10.1038/nm.3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kong S., Kim S. J., Sandal B., Lee S. M., Gao B., Zhang D. D., and Fang D. (2011) The type III histone deacetylase Sirt1 protein suppresses p300-mediated histone H3 lysine 56 acetylation at Bclaf1 promoter to inhibit T cell activation. J. Biol. Chem. 286, 16967–16975 10.1074/jbc.M111.218206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Conze D., Weiss L., Regen P. S., Bhushan A., Weaver D., Johnson P., and Rincón M. (2001) Autocrine production of interleukin 6 causes multidrug resistance in breast cancer cells. Cancer Res. 61, 8851–8858 [PubMed] [Google Scholar]
- 28. Yan H. Q., Huang X. B., Ke S. Z., Jiang Y. N., Zhang Y. H., Wang Y. N., Li J., and Gao F. G. (2014) Interleukin 6 augments lung cancer chemotherapeutic resistance via ataxia-telangiectasia mutated/NF-κB pathway activation. Cancer Sci. 105, 1220–1227 10.1111/cas.12478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Minamoto S., Ikegame K., Ueno K., Narazaki M., Naka T., Yamamoto H., Matsumoto T., Saito H., Hosoe S., and Kishimoto T. (1997) Cloning and functional analysis of new members of STAT induced STAT inhibitor (SSI) family: SSI-2 and SSI-3. Biochem. Biophys. Res. Commun. 237, 79–83 10.1006/bbrc.1997.7080 [DOI] [PubMed] [Google Scholar]
- 30. Liu B., Liao J., Rao X., Kushner S. A., Chung C. D., Chang D. D., and Shuai K. (1998) Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl. Acad. Sci. U.S.A. 95, 10626–10631 10.1073/pnas.95.18.10626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vaziri H., Dessain S. K., Ng Eaton E., Imai S. I., Frye R. A., Pandita T. K., Guarente L., and Weinberg R. A. (2001) hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107, 149–159 10.1016/S0092-8674(01)00527-X [DOI] [PubMed] [Google Scholar]
- 32. Wang R. H., Sengupta K., Li C., Kim H. S., Cao L., Xiao C., Kim S., Xu X., Zheng Y., Chilton B., Jia R., Zheng Z. M., Appella E., Wang X. W., Ried T., and Deng C. X. (2008) Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell 14, 312–323 10.1016/j.ccr.2008.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gao S. P., Mark K. G., Leslie K., Pao W., Motoi N., Gerald W. L., Travis W. D., Bornmann W., Veach D., Clarkson B., and Bromberg J. F. (2007) Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J. Clin. Invest. 117, 3846–3856 10.1172/JCI31871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gao B., Kong Q., Kemp K., Zhao Y. S., and Fang D. (2012) Analysis of sirtuin 1 expression reveals a molecular explanation of IL-2-mediated reversal of T-cell tolerance. Proc. Natl. Acad. Sci. U.S.A. 109, 899–904 10.1073/pnas.1118462109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee S. M., Gao B., and Fang D. (2008) FoxP3 maintains Treg unresponsiveness by selectively inhibiting the promoter DNA-binding activity of AP-1. Blood 111, 3599–3606 10.1182/blood-2007-09-115014 [DOI] [PubMed] [Google Scholar]
- 36. Chen A., Gao B., Zhang J., McEwen T., Ye S. Q., Zhang D., and Fang D. (2009) The HECT-type E3 ubiquitin ligase AIP2 inhibits activation-induced T-cell death by catalyzing EGR2 ubiquitination. Mol. Cell. Biol. 29, 5348–5356 10.1128/MCB.00407-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hornbeck P. V., Zhang B., Murray B., Kornhauser J. M., Latham V., Skrzypek E. (2015) PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 10.1093/nar/gku1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.