Abstract
Innate immune cells express danger-associated molecular pattern (DAMP) receptors, T-cell costimulation/coinhibition receptors, and major histocompatibility complex II (MHC-II). We have recently proposed that endothelial cells can serve as innate immune cells, but the molecular mechanisms involved still await discovery. Here, we investigated whether human aortic endothelial cells (HAECs) could be transdifferentiated into innate immune cells by exposing them to hyperlipidemia–up-regulated DAMP molecules, i.e. lysophospholipids. Performing RNA-seq analysis of lysophospholipid-treated HAECs, we found that lysophosphatidylcholine (LPC) and lysophosphatidylinositol (LPI) regulate largely distinct gene programs as revealed by principal component analysis. Metabolically, LPC up-regulated genes that are involved in cholesterol biosynthesis, presumably through sterol regulatory element–binding protein 2 (SREBP2). By contrast, LPI up-regulated gene transcripts critical for the metabolism of glucose, lipids, and amino acids. Of note, we found that LPC and LPI both induce adhesion molecules, cytokines, and chemokines, which are all classic markers of endothelial cell activation, in HAECs. Moreover, LPC and LPI shared the ability to transdifferentiate HAECs into innate immune cells, including induction of potent DAMP receptors, such as CD36 molecule, T-cell costimulation/coinhibition receptors, and MHC-II proteins. The induction of these innate-immunity signatures by lysophospholipids correlated with their ability to induce up-regulation of cytosolic calcium and mitochondrial reactive oxygen species. In conclusion, lysophospholipids such as LPC and LPI induce innate immune cell transdifferentiation in HAECs. The concept of prolonged endothelial activation, discovered here, is relevant for designing new strategies for managing cardiovascular diseases.
Keywords: inflammation, lysophospholipid, metabolism, endothelial cell, atherosclerosis, immunometabolism, lysophosphatidylcholine, lysophosphatidylinositol, RNA-Seq, transdifferentiation
Introduction
Cardiovascular disease (CVD)2 is a leading cause of death in well developed countries. As a chronic autoimmune inflammatory disease, atherosclerosis is fueled by both the innate and adaptive immune responses, which mediate initiation, progression, and ultimate thrombotic complications (1). We and others have reported that hyperlipidemia, together with other CVD risk factors, such as hyperglycemia, chronic kidney disease, obesity, and hyperhomocysteinemia, promote atherosclerosis development via several mechanisms. These mechanisms include endothelial cell (EC) activation and injury (2–6), monocyte recruitment and differentiation (7, 8), decreased function of regulatory T cells (Tregs) (9–11) and transdifferentiation of Tregs into antigen-presenting cell–like Tregs (12), impaired vascular repair ability of bone marrow–derived progenitor cells (13–15), increased migration and proliferation of vascular smooth muscle cells (16, 17), and high fat–induced adipocyte hypertrophy and metabolic healthy obesity (18). However, the underlying mechanisms of how hyperlipidemia promotes prolonged EC activation response during atherosclerosis development remain poorly defined.
Lysophospholipids are a group of bioactive, proinflammatory lipid molecules, which include lysophosphatidylcholine (LPC), lysophosphatidylinositol (LPI), and others. As our newly proposed conditional danger-associated molecular patterns (DAMPs) (19, 20), lysophospholipids contribute to aortic EC activation (4–6) and development of atherosclerosis (21). It has been suggested that LPC-induced EC activation is an initiation step of atherogenesis (4). During early hyperlipidemia, LPCs are induced and activate ECs to produce adhesion molecules, such as intercellular adhesion molecule-1 (ICAM1), which mediates the adhesion and migration of leukocyte into the aortic artery intima (22). However, whether other members of the lysophospholipids, such as LPI, could activate aortic ECs in a manner differentially from LPC and additional lysophospholipid-regulated gene targets in EC remain unknown.
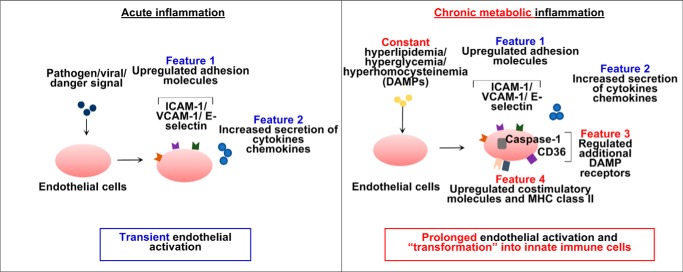
As we reported, during acute inflammation elicited by pathogen infection, ECs are transiently activated and express adhesion molecules, cytokines, and chemokines, which mediate leukocyte recruitment into the aorta (23). In addition, we have proposed a new concept of physiological EC activation for facilitation of patrolling immune surveillance cell trans-EC migration (5). During chronic metabolic inflammation, however, constant stimulation by cardiovascular stressors causes prolonged pathogenic EC activation, which is responsible for the prolonged pathogenic monocyte recruitment into the aorta, contributing to the development of CVD. We have recently reported that caspase-1 and NLR family pyrin domain–containing 3 (NLRP3) inflammasomes play a critical role in this process as deficiencies in caspase-1/inflammasomes decrease EC activation, aortic monocyte recruitment, and early atherosclerosis-induced hyperlipidemia (2) and hyperhomocysteinemia (24). These findings strengthen our recently proposed new concept that ECs are innate immune cells (25). More specifically, we have proposed that, in response to inflammatory stimuli and vascular stressors during CVD development, ECs could be transdifferentiated into antigen-presenting cells by expressing major histocompatibility complex class II (MHC-II) molecules and T-cell costimulation/coinhibition molecules, which present endothelial cell antigens to activate T cells through MHC-II (signal 1) and T-cell costimulation (signal 2). ECs could also up-regulate the expressions of DAMP receptors, proinflammatory cytokines, and chemokines, which can modulate the activities of immune cells (signal 3). Nevertheless, it remains poorly defined whether conditional DAMPs and chronic CVD stressors, such as lysophospholipids, could induce prolonged/sustained EC activation in a manner different from acute pathogen-induced EC activation.
ECs from different tissues are heterogeneous with respect to their protein and surface marker expressions (25). The heterogeneity of ECs contributes to their diversity in function at different vascular sites. More specifically, ECs are highly flexible and may be induced to stem cell–like properties via endothelial–mesenchymal transition (EndMT) (26). During EndMT, a mesenchymal phenotype is acquired by mature and progenitor ECs, which can give rise to other cell types. EndMT was found to be common in atherosclerotic lesions and to be associated with plaque instability. Mechanistically, transforming growth factor-β and Notch pathways have been shown to be important in regulating endothelial plasticity (25). However, it remains unknown whether lysophospholipids are involved in the process of EndMT.
In response to bacterial and viral infections, innate immune cells, such as macrophages, undergo extensive metabolic reprogramming. These metabolic changes not only provide energy and biosynthesis power but also directly regulate macrophage effector functions. In lipopolysaccharide (LPS)-stimulated macrophages, the increased mitochondrial oxidation of succinate drives mitochondrial reactive oxygen species (mtROS) production. As a result, LPS-stimulated macrophages shift their mitochondrial metabolism from ATP production toward mtROS production that promotes a proinflammatory state (27). In human aortic ECs, we have reported that lysophospholipid-induced mtROS are crucial in mediating adhesion molecule gene expression as well as site-specific histone 3 lysine 14 acetylation (6), a long-term epigenetic reprogramming of EC activation (4). Nevertheless, the roles of lysophospholipids in the specific transcriptional regulation of endothelial metabolic pathways remain to be elucidated.
Thus, despite significant progress, including our new concept of ECs as innate immune cells (25), several important knowledge gaps exist. First, the global transcriptomic targets of lysophospholipids, especially LPI, in ECs is unknown. Second, whether lysophospholipids could induce “professional” innate immune cell signatures in ECs is unclear. Third, whether lysophospholipids are involved in the process of EndMT remains unknown. Fourth, the role of lysophospholipids in EC metabolic reprogramming into prolonged and sustained activation status is uncharacterized. In this study, we hypothesized that lysophospholipids, as conditional DAMPs, could induce innate immune transdifferentiation of ECs, which might contribute to sustained EC activation and vascular pathology. Using RNA sequencing (Seq), we determined the transcriptomic effects of LPI and LPC treatment in human aortic ECs (HAECs). We found that LPC and LPI engage with distinct cellular metabolic pathways in HAECs. More importantly, LPI, similar to LPC, not only induces the transcriptional up-regulation of transient EC activation marker genes, including cytokines, chemokines, and EC adhesion molecules, but also contributes to the transcriptional induction of additional DAMP receptors, such as CD36 and caspase-1, T-cell costimulation and coinhibition receptors, and MHC-II molecules. Thus, a new system of innate immune transdifferentiation of EC, induced by lysophospholipids, has been uncovered. Our in-depth analysis of the roles of lysophospholipids in EC activation as well as our new concept of characterizing prolonged and sustained EC activation provides novel therapeutic potential for vascular inflammation and other CVDs.
Results
LPI induces both acute and sustained endothelial inflammation
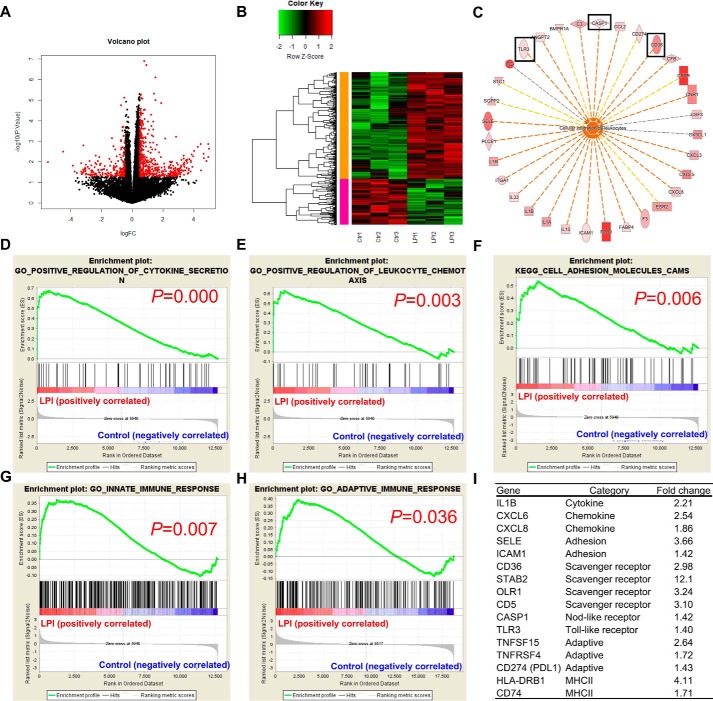
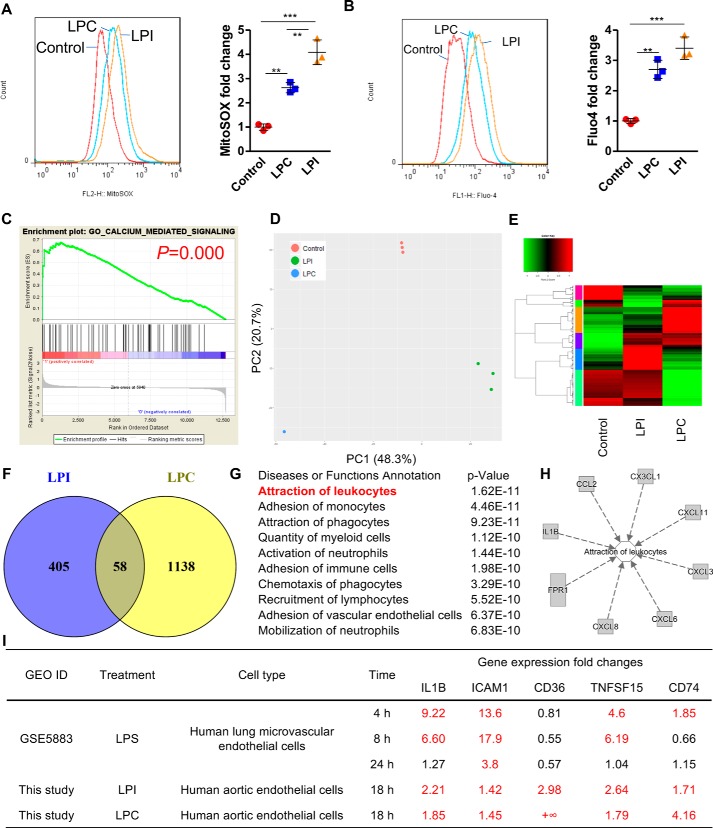
To examine the global gene expression changes in ECs after LPI stimulation, we performed RNA-Seq analysis in HAECs treated with either vehicle control or LPI (10 μm) for 18 h. The results showed that LPI regulated gene expression extensively in HAECs with more up-regulated genes (437 genes) than down-regulated genes (215 genes), indicating that LPI acts mostly by promoting gene transcription in HAECs (Fig. 1, A and B). Next, we performed Ingenuity Pathway Analysis (IPA) on LPI–up-regulated genes and found that the “cellular infiltration by leukocytes” pathway was the most significantly enriched pathway (Fig. S1A), suggesting that LPI promoted EC activation, which is an initiation step for leukocyte infiltration into the aorta and early atherogenesis. Further examination of the LPI–up-regulated genes in this pathway reveals that LPI not only promoted the up-regulation of cytokine (IL1A, IL1B, IL15, and IL33), chemokine (CCL2, CX3CL1, CXCL3, CXCL5, and CXCL8), and adhesion molecule (ICAM1 and SELE) gene expression changes but also increased gene expression changes related to the DAMPs (CASP1, TLR3, and CD36) (Fig. 1C). Furthermore, IPA predicted TLR3 as one of the top upstream regulators of LPI-induced genes (Fig. S1B), and as many as 22 LPI–up-regulated genes might be induced indirectly through the up-regulation of TLR3 (Fig. S1C). IPA also predicted extracellular signal-regulated kinase 1/2, which leads to AP-1 transcription factor activation, as a top upstream regulator of LPI-promoted transcriptome. Moreover, gene set enrichment analysis (GSEA) confirms that LPI treatment is associated with the gene signatures of “positive regulation of cytokine secretion,” “positive regulation of leukocyte chemotaxis,” “cell adhesion molecules,” and “innate immune response” (Fig. 1, D–G). These results indicate that LPI not only provokes “classic” acute endothelial cell activation process, as judged by increased expressions of adhesion molecules, cytokine, and chemokines, but also activates a previously uncharacterized “innate immune” EC activation process. We classified this as the first key feature of ECs as innate immune cells, characterized by engagement with amplification of the gene expression of additional DAMP receptors, which allow ECs to continuously receive the stimulation from DAMPs in blood circulation and tissues and sustain the EC activation (20, 25, 28). Of note, the gene signature of “adaptive immune response (functioning as antigen-presenting cells to activate T-cell responses)” was also positively correlated with the LPI treatment group (Fig. 1H), indicating that LPI also promotes “adaptive immune” EC activation, which we classified as the second key features of ECs' antigen-presenting function as innate immune cells. Examination of gene expression -fold changes further consolidated these findings with several scavenger receptors (CD36, STAB2, OLR1, and CD5), DAMPs (CASP1 and TLR3), T-cell costimulation and coinhibition receptors (TNFSF15, tumor necrosis factor receptor superfamily member 4 (TNFRSF4), and CD274), and MHC-II molecules (HLA-DRB1 and CD74) also highly up-regulated in HAECs after LPI treatment (Fig. 1I). Taken together, these results indicate that LPI induces both the classic EC activation process for acute vascular inflammation and unique innate immune and adaptive immune EC activations for prolonged inflammatory process.
Figure 1.
RNA-Seq analysis reveals that LPI induces both transient and sustained endothelial cell activation. Human aortic ECs were treated with either vehicle control (Ctr) or LPI (10 μm) for 18 h, and RNA-Seq experiments were performed. n = 3 in each group. A, volcano plot showing log(-fold change (FC)) and −log10(p value) of control versus LPI treatment. Red genes indicate genes significantly changed by more than 1.4-fold by LPI. B, heat map of genes that are significantly changed by more than 1.4-fold by LPI in ECs. C, the LPC–up-regulated genes from the top regulated pathway, cellular infiltration by leukocytes, are shown. Genes that are related to innate immunity are boxed. D–H, GSEA of the gene signatures that are significantly enriched in the LPI-treated EC group. I, representative gene expression changes in different categories corresponding to the GSEA plots.
LPI extensively reprograms endothelial cell glucose, amino acid, mitochondrial, and lipid metabolism
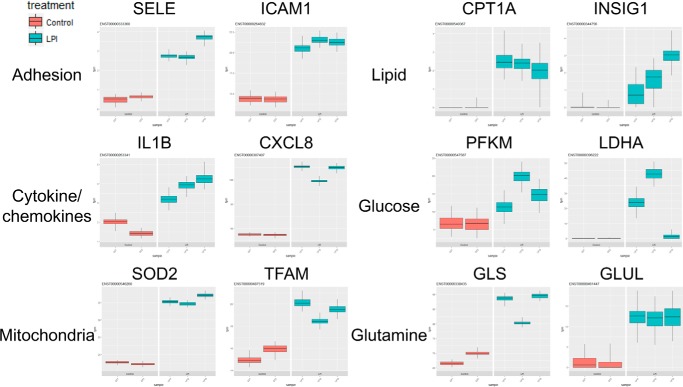
During innate immune cell activation induced by proinflammatory stimuli, such as LPS, macrophages undergo an extensive cellular metabolic reprogramming process, which is crucial for both meeting its synthetic and energetic needs and inducing the inflammatory response (29, 30). We hypothesized that LPI treatment also drives metabolic gene expression changes during innate immune reprogramming of ECs. We found that besides the induction of adhesion molecules, cytokines, and chemokines LPI treatment also strongly induced the transcripts of several metabolic genes, which were related to the metabolism of glucose, amino acids, and lipids and mitochondrial genes (Fig. 2). These molecules included the master mitochondrial transcription factor mitochondrial transcription factor A (TFAM), mitochondrial reactive oxygen species scavenger superoxide dismutase 2 (SOD2), rate-limiting glycolysis enzymes (phosphofructokinase, muscle (PFKM) and lactate dehydrogenase A (LDHA)) that drive the first and final steps of glycolysis, enzymes that carry out the conversion between glutamine and glutamate (glutaminase (GLS) and glutamate-ammonia ligase (GLUL)), and critical cholesterol and fatty acid metabolism enzymes (carnitine palmitoyltransferase 1A (CPT1A) and insulin-induced gene 1 (INSIG1)). These results indicate that LPI globally reprograms EC metabolism, presumably required for the innate reprogramming of EC activation, similarly to that reported for macrophages (29, 30).
Figure 2.
LPI reprograms endothelial cell metabolism extensively besides up-regulating adhesion molecules and cytokines/chemokines in human aortic endothelial cells. Human aortic ECs were treated with either vehicle control or LPI (10 μm) for 18 h, and RNA-Seq experiments were performed. The transcript level in units of transcripts per million (tpm) of the genes related to different categories, including EC adhesion molecules, cytokines/chemokines, and metabolic regulators, are shown. Box and whisker plot is shown for each sample (n = 100 bootstrap replicates from Kallisto). Lower and upper whiskers (error bars) indicate 25th and 75th percentiles, respectively.
LPC induces both acute activation and prolonged innate reprogramming in HAECs
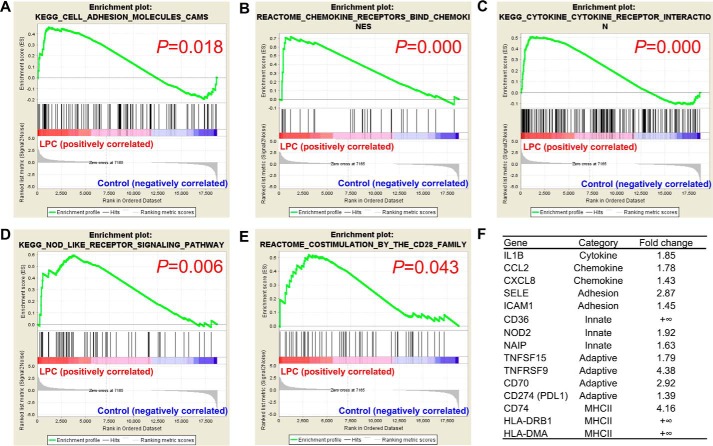
Next, we hypothesized that other lysophospholipids could also induce innate reprogramming of EC for sustained inflammatory response. We performed RNA-Seq analysis in HAECs treated with either vehicle control or LPC (10 μm) for 18 h, the same experimental conditions as those used for LPI treatment of HAECs. Similarly, we found that LPC induced the enrichment of gene transcription signatures of cell adhesion molecules, “chemokine receptors bind chemokines,” and “cytokine–cytokine receptor interaction” (Fig. 3, A–C). More importantly, the gene modulation patterns of LPC also positively correlated with the gene signatures of the innate “NOD-like receptor signaling pathway” and adaptive “costimulation by the CD28 family” (Fig. 3, D and E). Further examination of selected genes from these signatures revealed that LPC similarly promoted the gene expression of cytokine (IL1B), chemokines (CCL2 and CXCL8), adhesion molecules (ICAM1 and SELE), DAMPs (CD36, NOD2, and NAIP), costimulatory/coinhibitory molecules (TNFSF15, TNFRSF9, CD70, and CD274), and MHC class II molecules (CD74, HLA-DRB1, and HLA-DMA) (Fig. 3F). These results expanded our previous findings and suggested that innate reprogramming of EC could be a common shared feature of DAMP lysophospholipids.
Figure 3.
RNA-Seq analysis reveals that LPC induces both acute and sustained endothelial cell activation. Human aortic ECs were treated with either vehicle control or LPC (10 μm) for 18 h, and RNA-Seq experiments were performed. n = 3 in each group. A–E, GSEA of the gene signatures that are significantly enriched in the LPC-treated EC group. F, representative gene expression changes in different categories corresponding to the GSEA plots.
LPC positively regulates genes downstream of master regulator of lipid metabolism, sterol regulatory element–binding protein 2 (SREBP2)
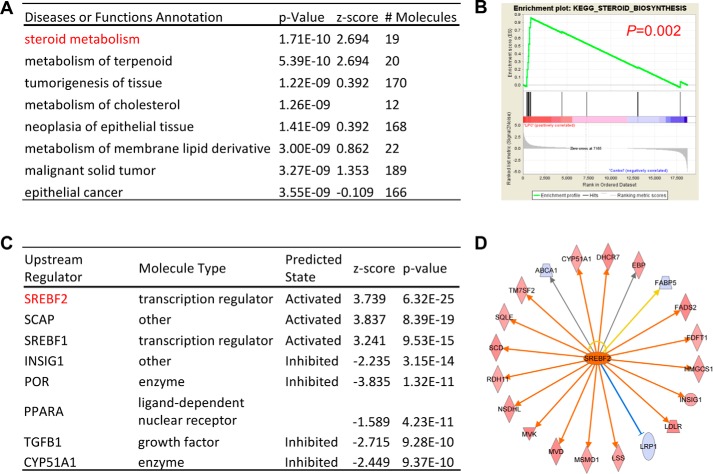
To examine whether LPC could drive metabolism changes during EC activation, we performed IPA pathway enrichment profiling on LPC-regulated genes. Strikingly, IPA results showed that “steroid metabolism” is the most significantly enriched pathway in LPC-treated HAECs (Fig. 4A). GSEA of “steroid biosynthesis” also showed that the LPC treatment group strongly correlates with the gene signature of steroid biosynthesis (Fig. 4B). Furthermore, IPA predicted sterol regulatory element–binding transcription factor 2 (SREBF2), which encodes the protein SREBP2, to be the upstream transcription factor of LPC-regulated genes in HAECs (Fig. 4C). Other protein members related to the regulation of SREBP2 (SCAP and INSIG1) as well as its other family member SREBP1 were also predicted to be the upstream regulators. SREBP2 is a master transcription factor and a proinflammatory M1 macrophage polarization marker (31), important for the regulation of cholesterol metabolism. In addition, we found that as many as 20 SREBP2-regulated genes are significantly changed by LPC in HAECs, including nine of 13 critical cholesterol biosynthesis enzymes (Fig. 4D). These results indicate that LPC drives EC activation and inflammation via inducing unique cholesterol metabolism pathways compared with LPI that are potentially mediated by SREBP2.
Figure 4.
LPC positively regulates genes downstream of master regulator of lipid metabolism SREBP2. HAECs were challenged with LPC (10 μm) for 18 h and RNA-Seq with IPA was performed. A, top regulated diseases or functions annotation of the genes that are significantly changed by LPC in HAECs. B, gene set enrichment analysis from the top LPI-regulated pathway “steroid biosynthesis” was shown. C, top upstream regulator analysis of the genes that are changed by LPC in HAECs predicted by the IPA. D, the 20 SREBP2-regulated genes that are significantly changed by LPC in HAECs. The red genes are induced by LPC and the blue genes are decreased by LPC.
LPI and LPC similarly activate mitochondrial reactive oxygen species (ROS), cytosolic calcium, and acute EC activation marker genes but regulate largely distinct gene programs in human aortic endothelial cells
Because LPI and LPC have similar structures (21), we hypothesized that LPC could activate similar cellular signaling pathways as LPI. Using a flow cytometry method with specific mtROS and cellular calcium probes, we found that both LPC and LPI acute treatments (1 h) strongly up-regulate both mitochondrial ROS production and cytosolic calcium concentrations (Fig. 5, A and B). The induction of cytosolic calcium correlated with the induction of calcium signaling–dependent genes (Fig. 5C), suggesting that cytosolic calcium influx–mitochondrial ROS signaling and endoplasmic reticulum–mitochondrial tethering pathways (32) might partially mediate lysophospholipid-induced inflammatory gene expression as we reported previously (4). Although LPI and LPC both induced cytosolic calcium influx–mitochondrial ROS signaling pathways, they regulate largely distinct gene expression changes in HAECs as determined by principal component analysis (58). Although the PC2 axis, which explains 20.7% of the variance among the data, separated the control group from the LPI and LPC groups, the LPI and LPC treatment groups were in the opposite directions from the control group in the PC1 axis, which showed the largest variations and explained 48.3% of the variances among the groups (Fig. 5D). Differential gene regulation by LPC and LPI was also evident from the differential gene expression heat map as there was a small overlap between LPC-induced and LPI-induced genes (Fig. 5, E and F, purple group). Furthermore, IPA enrichment profiling of the genes commonly induced by LPI and LPC revealed that “attraction of leukocytes” was the most enriched gene signature, suggesting that LPC and LPI could commonly induce acute EC activation, presumably through induction of mitochondrial ROS–cytosolic calcium influx signaling pathways (Fig. 5, G and H). Moreover, by analyzing published microarray data from LPS-induced endothelial cell activation (Gene Expression Omnibus (GEO) accession number GSE5883), we found that although LPS acutely induced cytokine IL1B, adhesion molecule ICAM1, scavenger receptor CD36, T-cell costimulatory molecule TNFSF15, and MHC class II molecule CD74, it could only up-regulate ICAM1 expression after 24 h. By contrast, both LPC and LPI could sustain the up-regulation of these genes in HAECs (Fig. 5I). These results indicate that during acute inflammation LPS could transiently induce EC activation by up-regulation of adhesion molecules, such as ICAM1. In comparison, lysophospholipids could induce prolonged endothelial activation and transform ECs into innate immune cells by up-regulating additional DAMP receptors, costimulatory molecules, and MHC class II molecules (Fig. 6).
Figure 5.
LPI and LPC similarly activate mitochondrial ROS, cytosolic calcium, and acute EC activation marker genes but regulate largely distinct gene programs in human aortic endothelial cells. A and B, HAECs were treated with either LPC (10 μm) or LPI (10 μm) for 1 h. Flow cytometry analysis with mitochondrial ROS (A) and cytosolic calcium (B) probes was performed afterward. C, GSEA of the LPI–up-regulated genes in the “calcium-mediated signaling” collection. D, principal component analysis showing the global transcription profile relationship among control, LPI, and LPC. E, heat map showing the similarities and differences between LPC-regulated and LPI-regulated genes. F, Venn diagram showing the number of LPC and LPI co-up-regulated genes. G, top enriched pathways of the LPI and LPC co-up-regulated genes (58 genes from F) as determined by Ingenuity Pathway Analysis. H, the LPC and LPI co-up-regulated genes from their top regulated pathway, attraction of leukocytes (in G), are shown. I, comparison of LPS- and lysophospholipid-induced endothelial activation. Red numbers indicate gene expression -fold changes that are greater than 1.4-fold. For all panels, data are expressed as mean ± S.D. (error bars). **, p < 0.01; ***, p < 0.001.
Figure 6.
A new working model. Left, during acute inflammation, a danger signal from pathogen or virus infection induces transient endothelial cell activation as characterized by two features, up-regulated adhesion molecule expression and increased secretion of cytokines and chemokines. Right, in the process of chronic metabolic inflammation during cardiovascular disease development, constant stimulation from endogenous metabolic DAMPs, such as lysophospholipids (during hyperlipidemia), glucose (during hyperglycemia), and homocysteine (during hyperhomocysteinemia), transform endothelial cells into innate immune cells and induce prolonged endothelial cell activation. The innate reprogramming of endothelial cells is characterized not only by up-regulation of cytokine/chemokines and adhesion molecule gene expression but also by up-regulation of additional DAMP receptors, such as caspase-1 and CD36, as well as up-regulation of costimulatory molecules and MHC class II molecules.
LPI and LPC commonly induce proinflammatory cytokines and chemokines that are downstream of NF-κB transcription factor
To strengthen the conclusion of innate immune transdifferentiation of HAECs by lysophospholipids, we analyzed the expression profile of a comprehensive list of inflammation-modulating cytokines (Table S1). Although LPI and LPC commonly induced the gene expression of a few proinflammatory cytokines, including IL1A, IL1B, and TNFSF15, they also differentially regulated other cytokine gene expression. LPC highly induced the expression of CD70 and IL32, whereas LPI highly up-regulated CSF2, IL16, and LTB. We also examined the gene expression of all the chemokine family members after LPC/LPI stimulation. We found that both LPC and LPI commonly up-regulated a few α-chemokine (CXC) and δ-chemokine (CX3C) family members, including CXCL3, CXCL6, CXCL8, CXCL11, and CX3CL1, whereas they largely regulate distinct β-chemokines (CC) (Table S2). Next, we hypothesized that LPI and LPC could activate certain transcription factor pathways, leading to the up-regulation of cytokine and chemokine gene expressions. To examine this, we analyzed the predicated transcription factors for LPI (Fig. S2A) and LPC (Fig. S2B) using IPA. IPA listed interferon regulatory factor 1 (IRF1), nuclear factor NF-κB p65 subunit (RELA), signal transducer and activator of transcription 3 (STAT3), and NF-κB inhibitor ϵ (NFKBIE) as top upstream transcription factors for LPI-regulated transcriptomes and listed SREBF2, SREBF1, NADH (NAD)-dependent deacetylase silent information regulator 2 (sirtuin 2 (SIRT2)), and tumor protein p53 (TP53) as top upstream transcription factors for LPC-regulated transcriptomes in HAECs. Strikingly, there is an overlap of nearly half (nine of 20) of the predicted transcription factors for both LPI and LPC, many of which are related to the NF-κB transcription factor pathway (Fig. S2C). Further examination of the genes downstream of RELA revealed that LPI and LPC commonly regulated genes downstream of RELA transcription factor, such as inflammatory cytokines IL1A and IL1B, chemokines CCL2 and CXCL8, and adhesion molecules ICAM1 and SELE (Fig. S2C). These results indicated that NF-κB might be involved in the up-regulation of EC activation marker genes induced by LPC and LPI.
LPC and LPI differentially regulate T-cell costimulation and coinhibition receptors as well as mesenchymal regulators/markers in EndMT
To further confirm immune transdifferentiation of HAECs by lysophospholipids, we examined the gene expression changes of a comprehensive list of T-cell costimulation and coinhibition receptors after LPC and LPI stimulation in HAECs (33). We found that LPC up-regulated CD28, TNFRSF9 (CD137), and TNFRSF18 (Table S3). Although LPI also up-regulated CD28 gene expression, LPI differentially up-regulated B- and T-lymphocyte attenuator (BTLA; CD272), TNFRSF4, hepatitis A virus cellular receptor 1 (HAVCR1), leukocyte-associated immunoglobulin-like receptor 1 (LAIR1), and CD274. The induction of different T-cell costimulation and coinhibition receptors suggested that LPC and LPI stimulation might lead to distinct immune transdifferentiation programs of HAECs. Finally, we examined the hypothesis that lysophospholipids might be involved in the EndMT process. To test this hypothesis, we examined the gene expression changes of previously characterized EndMT regulators and mesenchymal markers in HAECs (34). The results showed that LPC up-regulated gene expression of caldesmon 1 (CALD1), Snail superfamily of C2H2-type zinc finger transcription factor 2 (SNAI2), WNT gene family secreted signaling protein 11 (WNT11), disintegrin and metalloproteinase domain–containing protein 12 (ADAM12), basic smooth muscle protein calponin 1 (CNN1), matrix metallopeptidase 9 (MMP9), and S100 calcium-binding protein A4 (S100A4), whereas LPI induced nidogen-2 (NID2) and Notch receptor family 3 (NOTCH3) (Tables S4 and S5). Thus, LPC and LPI might be involved in the process of EndMT, but they regulated distinct regulators/markers in this process.
Discussion
Anti-inflammatory strategies are promising therapeutic approaches for prevention and treatment of atherosclerotic CVD (35). The majority of lysophospholipids serve as conditional DAMPs (19, 20), and they are considered to play a causative role in atherogenesis. For this reason, inhibitors of the key enzymes responsible for the generation of lysophospholipids were developed as potential anti-inflammatory therapies against atherosclerosis (35). Despite the beneficial effects of these inhibitors in atherosclerotic animal models (36, 37) and human phase 2 clinical trials (38, 39), human phase 3 clinical trials failed to show their therapeutic efficacy (40–42). One possibility that may explain these failures is that lysophospholipids might also be important under normal physiological conditions, and blocking their physiological effects could be detrimental to the host. To consolidate this argument, we recently reported that LPC-induced mtROS, uncoupled from ATP synthesis, determine EC activation for both physiological recruitment of patrolling cells and pathological recruitment of inflammatory cells (4, 5, 43). Thus, targeting the pathogenic downstream effects of LPC, instead of blocking the production of LPC itself, may lead to better therapeutics. In our current study, we found that different lysophospholipid species, including LPC and LPI, regulated largely distinct gene signatures in HAECs. In terms of cellular metabolism regulation, whereas LPC was involved in cholesterol biosynthesis potentially mediated by SREBP2, LPI broadly regulated critical enzymes involved in cellular glucose, lipid, and amino acid metabolism. Nevertheless, LPC and LPI could both induce EC activation marker genes, such as adhesion molecules, cytokines, and chemokines. More importantly, they could both induce innate immune reprogramming/transdifferentiation of ECs into innate immune cells by up-regulating additional DAMP receptors, such as CD36, to receive additional DAMPs for prolonged/sustained activation and MHC-II molecules, such as HLA-DRB1, to present endothelial cell–specific and non-EC–specific self-antigen epitopes to activate T cells (12, 44). Similar to the findings in the present study on cell transdifferentiation, we also reported recently that in response to environmental stimulations, including DAMPs, hypoxia, and master gene mutations, cell identification could be plastic. First, transcription factor GATA-binding protein 3 (GATA3), histone deacetylase 6 (HDAC6), and B-cell lymphoma 6 (BCL6) regulate forkhead box P3 (FOXP3)+ regulatory T cell (Treg) plasticity and determine Treg conversion into either novel antigen-presenting cell–like or type 1 T-helper cell Tregs (12), which further corroborates our recently proposed new working model that pathological conditions reshape physiological Tregs into pathological Tregs (45). Second, thrombus leukocytes exhibit more endothelial cell–specific angiogenic markers than do peripheral blood leukocytes in acute coronary syndrome patients, suggesting a possibility of transdifferentiation of leukocytes into angiogenic endothelial cells (46). Third, in patients with lymphomas, Tregs can function in four different formats (47). Collectively, our data presented here also indicated that targeting the innate immune system in ECs, such as by targeting CD36, could lead to novel anti-inflammatory therapies against hyperlipidemia-induced endothelial cell activation and atherosclerotic CVD.
When there is ample oxygen in the environment, cancer cells utilize mainly glycolysis rather than mitochondrial oxidative phosphorylation, an effect termed the “Warburg effect” (48). The Warburg effect is beneficial for tumor cells as it supports macromolecule biosynthesis. Quiescent ECs also display the Warburg effect, but very little is known about the roles of cellular metabolism during hyperlipidemia stimuli–induced EC activation (49). Although there was a report indicating that LPC shares the same receptor, G protein–coupled receptor 55 (GPR55), with LPI (50), our detailed analyses of the differential EC transcriptomes induced by LPC and LPI emphasize strongly that LPI and LPC may use different receptor-initiated signaling pathways, which correlated with another report that LPI species 20:4 is the most potent GPR55 agonist (51), emphasizing the high specificity of LPI on GPR55. Here, by using transcriptomic profiling, we demonstrated for the first time that in ECs LPI induced global reprogramming of cellular metabolism by inducing specific isoforms of metabolic gene transcripts, such as LDHA. Notably, a large portion of these metabolic enzymes are regulated at the transcript level, suggesting that LPI uses transcription and alternative splicing mechanisms to reprogram cellular metabolism. Further studies are needed to elucidate the specific molecules that mediate LPI-regulated metabolic pathways.
Lipid biosynthesis is regulated by SREBPs and liver X receptor transcription factors. SREBPs are master transcription factors that control the expression of a range of enzymes required for cholesterol, fatty acid, triacylglycerol, and phospholipid syntheses. Cholesterol and its derivatives are established to be upstream regulators of SREBPs by directly binding to the protein and preventing its translocation to the nucleus (52). Recent studies suggested that maturation of nuclear, transcriptionally active SREBP protein is also controlled by phospholipids (53). In addition, proatherogenic oscillatory flow, acting as a DAMP, activates SREBP2 and induces NLRP3 inflammasomes in ECs (54). However, to our knowledge, our results indicate, for the first time, that SREBP might be regulated by conditional DAMP lysophospholipids. Up-regulation of SREBP2-regulated genes has been found to be a proinflammatory M1 macrophage polarization marker (31), suggesting that proinflammatory immunometabolism remodeling can be the same in different cell types (55, 56). Future studies are needed to determine whether LPC could directly bind to and regulate SREBP as cholesterol does. Remarkably, LPI does not regulate SREBP-dependent genes, indicating the specificity of LPC in regulating cellular lipid homeostasis.
LPC and LPI both induce comparable levels of mitochondrial ROS and cytosolic calcium influx, which correlates with their common induction of EC adhesion molecules, cytokines, chemokines, DAMP receptors, and adaptive immune molecules, such as T-cell costimulation/coinhibition receptors and MHC class II molecules. These results indicate that the calcium influx–mitochondrial ROS pathway might mediate lysophospholipid-induced EC activation. By contrast, LPC and LPI differentially regulate metabolic pathways in ECs, suggesting that calcium influx/mitochondrial ROS are not responsible for these physiological responses. Thus, as we reported with the mitochondrial ROS inhibitor MitoTempol (4), targeting the mitochondrial ROS signaling–histone 3 lysine 14 acetylation pathway (6) might be superior to targeting lysophospholipids themselves as therapies because it does not interfere with the normal physiological metabolism of ECs but specifically targets the innate immune response in ECs.
Taken together, we propose a new system of characterizing prolonged and sustained EC activation and innate immune transdifferentiation (Fig. 6) based on the following differentiation criteria: 1) induction of DAMP receptors and 2) induction of T-cell costimulation/coinhibition receptors and MHC-II molecules. During acute inflammation induced by pathogens, such as bacteria and viruses, ECs are transiently activated by up-regulation of adhesion molecules and secretion of cytokines and chemokines. When ECs are equipped to sense and are activated by chronic metabolic stressors/DAMPs, such as lysophospholipids, additional DAMP receptors, T-cell costimulation/coinhibition receptors, and MHC-II molecules are also up-regulated in addition to the induction of classic EC activation marker genes. By doing this, ECs are transdifferentiated into innate immune cells, which can keep receiving stimulations from additional DAMPs and mediate long-term and consistent pathogenic leukocyte recruitment to the aorta. Moreover, activated ECs with up-regulated T-cell costimulation/coinhibition receptors and MHC class II can activate/modulate encountered T cells and enhance T-cell participation in vascular inflammation and atherogenesis. Our new working model of EC activation provides novel insight into how ECs are differentially activated and innate immune transdifferentiated during CVD development. Our findings are significant for future design of novel anti-inflammatory/immunosuppressive therapies against CVD.
Experimental procedures
Chemicals and antibodies
All chemicals were from Sigma-Aldrich unless otherwise indicated. LPC (16:0) and LPI (16:0) were purchased from Avanti Polar Lipids, Inc. (855675P; Alabaster, AL). For mtROS measurement, MitoSOX Red Mitochondrial Superoxide Indicator (M36008, Life Technologies) was used. Fluo-4 (Life Technologies) was purchased for the measurement of cytosolic calcium.
Human samples
All experiment procedures were performed in accordance with protocols approved by the Institutional Review Board at Temple University, which conforms to National Institutes of Health guidelines. All studies involving human subjects abide by the Declaration of Helsinki principles.
Cell culture
HAECs (CC2535, Lonza, Walkersville, MD) were cultured in medium M199 (HyClone Laboratories, Logan, UT) supplemented with 15% fetal bovine serum (FBS; HyClone Laboratories), 50 μg/ml endothelial cell growth supplement (BD Biosciences), 50 μg/ml heparin, and 1% penicillin, streptomycin, and amphotericin (Invitrogen). HAECs were grown on 0.2% gelatin-coated flasks, plates, or dishes, and experiments were performed at passage 9.
Fluorescence-activated cell sorting (FACS)
For mtROS and cytosolic calcium measurement, after staining with MitoSOX and Fluo-4, HAECs were incubated at 37 °C for 10 min and washed with PBS twice afterward. After LPC and LPI treatment, cells were washed once with ice-cold PBS, and trypsin-EDTA was added to detach cells. Trypsinization was terminated by adding FACS buffer (2% FBS in PBS), and cells were collected by centrifugation. After resuspension in 0.2 ml of FACS buffer, samples were subjected to flow cytometry analysis where fluorescence emissions were measured using a FACSCalibur machine (BD Biosciences).
RNA-Seq of LPC
RNA-Seq of LPC was performed by BGI (Shenzhen, China). Total RNAs were extracted from samples, and then mRNAs and noncoding RNAs were enriched by removing rRNA from the total RNA with a kit. Using the fragmentation buffer, the mRNAs were fragmented into short fragments (about 200–500 nucleotides), then the first-strand cDNA was synthesized by random hexamer primer using the fragments as templates, and dTTP was substituted by dUTP during the synthesis of the second strand. Short fragments were purified and resolved with elution buffer for end repair and single nucleotide A (adenine) addition. After that, the short fragments were connected with adaptors, and then the second strand was degraded finally using uracil N-glycosylase (2). After agarose gel electrophoresis, the suitable fragments were selected for PCR amplification as templates. During the quality control steps, an Agilent 2100 Bioanalyzer and ABI StepOnePlus Real-Time PCR System (Thermo Fisher) were used for quantification and qualification of the sample library. At last, the library was sequenced with an Illumina HiSeq4000 using the PE100 strategy. Primary sequencing data produced by the Illumina Hiseq4000, called raw reads, were filtered into clean reads by removing adaptor-containing and low-quality reads by BGI in-house software. A reference annotation–based assembly method was used to reconstruct the transcripts by TopHat (v2.0.10) and Cufflinks (v2.1.1), and background noise was reduced using fragments per kilobase million and coverage threshold. Data were deposited in the ArrayExpress database (59) (http://www.ebi.ac.uk/arrayexpress)3 under accession number E-MTAB-6604.
RNA-Seq of LPI
RNA-Seq of LPI was performed by Novogene (Beijing). Briefly, mRNA from HAECs was purified from total RNA using poly(T) oligo–attached magnetic beads. The mRNA was first fragmented randomly by addition of fragmentation buffer. Then first-strand cDNA was synthesized using random hexamer primer and Moloney murine leukemia virus reverse transcriptase (RNase H−). Second-strand cDNA synthesis was subsequently performed using DNA polymerase I and RNase H. Double-stranded cDNA was purified using AMPure XP beads (Beckman Coulter, Beverly, MA). Remaining overhangs of the purified double-stranded cDNA were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3′-ends of DNA fragments, NEBNext Adaptor with a hairpin loop structure was ligated to prepare for hybridization. To select cDNA fragments of preferentially 150–200 bp in length, the library fragments were purified with the AMPure XP system (Beckman Coulter). Finally, the final library was obtained by PCR amplification and purification of PCR products by AMPure XP beads. After library construction, the library was diluted to 1.5 ng/μl using the preliminary quantitative result by Qubit2.0, and the insert size was detected using Agilent 2100. Quantitative PCR was used to accurately quantify the library effective concentration (>2 nm) to ensure library quality. Libraries were fed into Illumina HiSeq machines after pooling according to activity and expected data volume. Data were deposited in the ArrayExpress database (59) under accession number E-MTAB-6605.
Sequencing data analysis
Data analysis was carried out using the statistical computing environment R, the Bioconductor suite of packages for R, and RStudio (57). Raw data were background-subtracted, variance-stabilized, and normalized by robust spline normalization. Differentially expressed genes were identified by linear modeling and Bayesian statistics using the Limma package. Probe sets that were differentially regulated (≥1.4-fold, p value <0.05) after controlling for multiple testing using the Benjamini-Hochberg method were used for hierarchical clustering and heat map generation in R. Clusters of coregulated genes were identified by Pearson correlation using the hclust function of the stats package in R. Pathway analysis was performed using IPA (Qiagen Bioinformatics) and GSEA (http://software.broadinstitute.org/gsea/index.jsp).3 Briefly, IPA is a web-based software application that goes beyond pathway analysis by identifying key upstream regulators to explain expression patterns and predicting downstream effects on biological and disease processes. GSEA is a computational method that determines whether an a priori defined set of genes shows statistically significant, concordant differences between two biological states. GSEA does not focus on only significantly/highly changed genes but examines all the genes that belong to a certain biological process instead.
Statistical analysis
Data are expressed as the mean ± S.D. throughout. For comparisons between two groups, two-tailed Student's t test was used for evaluation of statistical significance, or when the data were not normally distributed, a nonparametric Mann–Whitney U test was used. For comparisons across multiple groups, one-way analysis of variance with Bonferroni post-test adjustment was used, or when the data were not normally distributed, the data were analyzed using one-way analysis of variance with the Kruskal–Wallis test followed by pairwise comparison using the Dunn test. For linear regression tests, simple linear regression analyses were performed using GraphPad Prism to determine the coefficient of determination and p value. Data shown are representative of two to three independent experiments. NS, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Author contributions
X. L. and X.-F. Y. conceptualization; X. L. and L. W. data curation; X. L. software; X. L. and X.-F. Y. formal analysis; X. L. validation; X. L. investigation; X. L. methodology; X. L. writing-original draft; X. L. and X.-F. Y. writing-review and editing; P. F., Y. S., X. J., and H. W. resources; H. W. and X.-F. Y. supervision; X.-F. Y. funding acquisition.
Supplementary Material
This work was supported by Grants HL131460, HL132399, HL138749, HL130233, HL110764, and HL117654 from the NHLBI, National Institutes of Health and Grants DK104116 and DK113775 from the NIDDK, National Institutes of Health (to X.-F. Y. and H. W.) and American Heart Association Scientist Development Grant 17SDG33671051 (to P. F.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2 and Tables S1–S5.
Microarray data are available in the ArrayExpress database under accession numbers E-MTAB-6604 and E-MTAB-6605.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party-hosted site.
- CVD
- cardiovascular disease
- DAMP
- danger-associated molecular pattern
- MHC
- major histocompatibility complex
- HAEC
- human aortic endothelial cell
- Seq
- sequencing
- LPC
- lysophosphatidylcholine
- LPI
- lysophosphatidylinositol
- SREBP
- sterol regulatory element–binding protein
- EC
- endothelial cell
- Treg
- regulatory T cell
- ICAM1
- intercellular adhesion molecule-1
- NLRP3
- NLR family pyrin domain–containing 3
- EndMT
- endothelial–mesenchymal transition
- LPS
- lipopolysaccharide
- mtROS
- mitochondrial reactive oxygen species
- IPA
- Ingenuity Pathway Analysis
- SELE
- E-selectin
- GSEA
- gene set enrichment analysis
- NOD
- nucleotide oligomerization domain
- NLR
- NOD-like receptor
- NAIP
- NLR family apoptosis inhibitory protein
- SREBF
- sterol regulatory element–binding transcription factor
- SCAP
- SREBP cleavage–activating protein
- INSIG1
- insulin-induced gene 1
- ROS
- reactive oxygen species
- LTB
- lymphotoxin-β
- TNFRSF
- tumor necrosis factor receptor superfamily member
- GPR55
- G protein–coupled receptor 55
- LDHA
- lactate dehydrogenase A.
References
- 1. Packard R. R., Lichtman A. H., and Libby P. (2009) Innate and adaptive immunity in atherosclerosis. Semin. Immunopathol. 31, 5–22 10.1007/s00281-009-0153-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yin Y., Li X., Sha X., Xi H., Li Y. F., Shao Y., Mai J., Virtue A., Lopez-Pastrana J., Meng S., Tilley D. G., Monroy M. A., Choi E. T., Thomas C. J., Jiang X., et al. (2015) Early hyperlipidemia promotes endothelial activation via a caspase-1-sirtuin 1 pathway. Arterioscler. Thromb. Vasc. Biol. 35, 804–816 10.1161/ATVBAHA.115.305282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shao Y., Cheng Z., Li X., Chernaya V., Wang H., and Yang X. F. (2014) Immunosuppressive/anti-inflammatory cytokines directly and indirectly inhibit endothelial dysfunction—a novel mechanism for maintaining vascular function. J. Hematol. Oncol. 7, 80 10.1186/s13045-014-0080-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li X., Fang P., Li Y., Kuo Y. M., Andrews A. J., Nanayakkara G., Johnson C., Fu H., Shan H., Du F., Hoffman N. E., Yu D., Eguchi S., Madesh M., Koch W. J., et al. (2016) Mitochondrial reactive oxygen species mediate lysophosphatidylcholine-induced endothelial cell activation. Arterioscler. Thromb. Vasc. Biol. 36, 1090–1100 10.1161/ATVBAHA.115.306964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li X., Fang P., Yang W. Y., Chan K., Lavallee M., Xu K., Gao T., Wang H., and Yang X. (2017) Mitochondrial ROS, uncoupled from ATP synthesis, determine endothelial activation for both physiological recruitment of patrolling cells and pathological recruitment of inflammatory cells. Can. J. Physiol. Pharmacol. 95, 247–252 10.1139/cjpp-2016-0515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li X., Shao Y., Sha X., Fang P., Kuo Y. M., Andrews A. J., Li Y., Yang W. Y., Maddaloni M., Pascual D. W., Luo J. J., Jiang X., Wang H., and Yang X. (2018) IL-35 (interleukin-35) suppresses endothelial cell activation by inhibiting mitochondrial reactive oxygen species-mediated site-specific acetylation of H3K14 (histone 3 lysine 14). Arterioscler. Thromb. Vasc. Biol. 38, 599–609 10.1161/ATVBAHA.117.310626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Combadière C., Potteaux S., Rodero M., Simon T., Pezard A., Esposito B., Merval R., Proudfoot A., Tedgui A., and Mallat Z. (2008) Combined inhibition of CCL2, CX3CR1, and CCR5 abrogates Ly6C(hi) and Ly6C(lo) monocytosis and almost abolishes atherosclerosis in hypercholesterolemic mice. Circulation 117, 1649–1657 10.1161/CIRCULATIONAHA.107.745091 [DOI] [PubMed] [Google Scholar]
- 8. Fang P., Zhang D., Cheng Z., Yan C., Jiang X., Kruger W. D., Meng S., Arning E., Bottiglieri T., Choi E. T., Han Y., Yang X. F., and Wang H. (2014) Hyperhomocysteinemia potentiates hyperglycemia-induced inflammatory monocyte differentiation and atherosclerosis. Diabetes 63, 4275–4290 10.2337/db14-0809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ait-Oufella H., Salomon B. L., Potteaux S., Robertson A. K., Gourdy P., Zoll J., Merval R., Esposito B., Cohen J. L., Fisson S., Flavell R. A., Hansson G. K., Klatzmann D., Tedgui A., and Mallat Z. (2006) Natural regulatory T cells control the development of atherosclerosis in mice. Nat. med. 12, 178–180 10.1038/nm1343 [DOI] [PubMed] [Google Scholar]
- 10. Xiong Z., Yan Y., Song J., Fang P., Yin Y., Yang Y., Cowan A., Wang H., and Yang X. F. (2009) Expression of TCTP antisense in CD25(high) regulatory T cells aggravates cuff-injured vascular inflammation. Atherosclerosis 203, 401–408 10.1016/j.atherosclerosis.2008.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li X., Fang P., Yang W. Y., Wang H., and Yang X. (2018) IL-35, as a newly proposed homeostasis-associated molecular pattern, plays three major functions including anti-inflammatory initiator, effector, and blocker in cardiovascular diseases. Cytokine, in press 10.1016/j.cyto.2017.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu K., Yang W. Y., Nanayakkara G. K., Shao Y., Yang F., Hu W., Choi E. T., Wang H., and Yang X. (2018) GATA3, HDAC6 and BCL6 regulate FOXP3+ Treg plasticity and determine Treg conversion into either novel antigen-presenting cell-like Treg or Th1-Treg. Front. Immunol. 9, 45 10.3389/fimmu.2018.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Du F., Zhou J., Gong R., Huang X., Pansuria M., Virtue A., Li X., Wang H., and Yang X. F. (2012) Endothelial progenitor cells in atherosclerosis. Front. Biosci. 17, 2327–2349 10.2741/4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li Y. F., Ren L. N., Guo G., Cannella L. A., Chernaya V., Samuel S., Liu S. X., Wang H., and Yang X. F. (2015) Endothelial progenitor cells in ischemic stroke: an exploration from hypothesis to therapy. J. Hematol. Oncol. 8, 33 10.1186/s13045-015-0130-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li Y. F., Huang X., Li X., Gong R., Yin Y., Nelson J., Gao E., Zhang H., Hoffman N. E., Houser S. R., Madesh M., Tilley D. G., Choi E. T., Jiang X., Huang C. X., et al. (2016) Caspase-1 mediates hyperlipidemia-weakened progenitor cell vessel repair. Front. Biosci. 21, 178–191 10.2741/4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Monroy M. A., Fang J., Li S., Ferrer L., Birkenbach M. P., Lee I. J., Wang H., Yang X. F., and Choi E. T. (2015) Chronic kidney disease alters vascular smooth muscle cell phenotype. Front. Biosci. 20, 784–795 10.2741/4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ferrer L. M., Monroy A. M., Lopez-Pastrana J., Nanayakkara G., Cueto R., Li Y. F., Li X., Wang H., Yang X. F., and Choi E. T. (2016) Caspase-1 plays a critical role in accelerating chronic kidney disease-promoted neointimal hyperplasia in the carotid artery. J. Cardiovasc. Transl. Res. 9, 135–144 10.1007/s12265-016-9683-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Virtue A., Johnson C., Lopez-Pastraña J., Shao Y., Fu H., Li X., Li Y. F., Yin Y., Mai J., Rizzo V., Tordoff M., Bagi Z., Shan H., Jiang X., Wang H., et al. (2017) MicroRNA-155 deficiency leads to decreased atherosclerosis, increased white adipose tissue obesity, and non-alcoholic fatty liver disease: a novel mouse model of obesity paradox. J. Biol. Chem. 292, 1267–1287 10.1074/jbc.M116.739839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang X., Li Y. F., Nanayakkara G., Shao Y., Liang B., Cole L., Yang W. Y., Li X., Cueto R., Yu J., Wang H., and Yang X. F. (2016) Lysophospholipid receptors, as novel conditional danger receptors and homeostatic receptors modulate inflammation—novel paradigm and therapeutic potential. J. Cardiovasc. Transl. Res. 9, 343–359 10.1007/s12265-016-9700-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shao Y., Nanayakkara G., Cheng J., Cueto R., Yang W. Y., Park J. Y., Wang H., and Yang X. (2018) Lysophospholipids and their receptors serve as conditional DAMPs and DAMP receptors in tissue oxidative and inflammatory injury. Antioxid. Redox Signal., in press 10.1089/ars.2017.7069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Y. F., Li R. S., Samuel S. B., Cueto R., Li X. Y., Wang H., and Yang X. F. (2016) Lysophospholipids and their G protein-coupled receptors in atherosclerosis. Front. Biosci. 21, 70–88 10.2741/4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vestweber D. (2015) How leukocytes cross the vascular endothelium. Nat. Rev. Immunol. 15, 692–704 10.1038/nri3908 [DOI] [PubMed] [Google Scholar]
- 23. Sha X., Meng S., Li X., Xi H., Maddaloni M., Pascual D. W., Shan H., Jiang X., Wang H., and Yang X. F. (2015) Interleukin-35 inhibits endothelial cell activation by suppressing MAPK-AP-1 pathway. J. Biol. Chem. 290, 19307–19318 10.1074/jbc.M115.663286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xi H., Zhang Y., Xu Y., Yang W. Y., Jiang X., Sha X., Cheng X., Wang J., Qin X., Yu J., Ji Y., Yang X., and Wang H. (2016) Caspase-1 inflammasome activation mediates homocysteine-induced pyrop-apoptosis in endothelial cells. Circ. Res. 118, 1525–1539 10.1161/CIRCRESAHA.116.308501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mai J., Virtue A., Shen J., Wang H., and Yang X. F. (2013) An evolving new paradigm: endothelial cells–conditional innate immune cells. J. Hematol. Oncol. 6, 61 10.1186/1756-8722-6-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Piera-Velazquez S., Li Z., and Jimenez S. A. (2011) Role of endothelial-mesenchymal transition (EndoMT) in the pathogenesis of fibrotic disorders. Am. J. Pathol. 179, 1074–1080 10.1016/j.ajpath.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mills E. L., Kelly B., Logan A., Costa A. S. H., Varma M., Bryant C. E., Tourlomousis P., Däbritz J. H. M., Gottlieb E., Latorre I., Corr S. C., McManus G., Ryan D., Jacobs H. T., Szibor M., et al. (2016) Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell 167, 457–470.e413 10.1016/j.cell.2016.08.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yin Y., Pastrana J. L., Li X., Huang X., Mallilankaraman K., Choi E. T., Madesh M., Wang H., and Yang X. F. (2013) Inflammasomes: sensors of metabolic stresses for vascular inflammation. Front. Biosci. 18, 638–649 10.2741/4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Langston P. K., Shibata M., and Horng T. (2017) Metabolism supports macrophage activation. Front. Immunol. 8, 61 10.3389/fimmu.2017.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Van den Bossche J., O'Neill L. A., and Menon D. (2017) Macrophage immunometabolism: where are we (going)? Trends Immunol. 38, 395–406 10.1016/j.it.2017.03.001 [DOI] [PubMed] [Google Scholar]
- 31. Dushkin M. I. (2012) Macrophage/foam cell is an attribute of inflammation: mechanisms of formation and functional role. Biochemistry 77, 327–338 10.1134/S0006297912040025 [DOI] [PubMed] [Google Scholar]
- 32. Filadi R., Theurey P., and Pizzo P. (2017) The endoplasmic reticulum-mitochondria coupling in health and disease: molecules, functions and significance. Cell Calcium 62, 1–15 10.1016/j.ceca.2017.01.003 [DOI] [PubMed] [Google Scholar]
- 33. Chen L., and Flies D. B. (2013) Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat. Rev. Immunol. 13, 227–242 10.1038/nri3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Evrard S. M., Lecce L., Michelis K. C., Nomura-Kitabayashi A., Pandey G., Purushothaman K. R., d'Escamard V., Li J. R., Hadri L., Fujitani K., Moreno P. R., Benard L., Rimmele P., Cohain A., Mecham B., et al. (2016) Endothelial to mesenchymal transition is common in atherosclerotic lesions and is associated with plaque instability. Nat. Commun. 7, 11853 10.1038/ncomms11853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bäck M., and Hansson G. K. (2015) Anti-inflammatory therapies for atherosclerosis. Nat. Rev. Cardiol. 12, 199–211 10.1038/nrcardio.2015.5 [DOI] [PubMed] [Google Scholar]
- 36. Fraser H., Hislop C., Christie R. M., Rick H. L., Reidy C. A., Chouinard M. L., Eacho P. I., Gould K. E., and Trias J. (2009) Varespladib (A-002), a secretory phospholipase A2 inhibitor, reduces atherosclerosis and aneurysm formation in ApoE−/− mice. J. Cardiovasc. Pharmacol. 53, 60–65 10.1097/FJC.0b013e318195bfbc [DOI] [PubMed] [Google Scholar]
- 37. Wilensky R. L., Shi Y., Mohler E. R. 3rd, Hamamdzic D., Burgert M. E., Li J., Postle A., Fenning R. S., Bollinger J. G., Hoffman B. E., Pelchovitz D. J., Yang J., Mirabile R. C., Webb C. L., Zhang L., et al. (2008) Inhibition of lipoprotein-associated phospholipase A2 reduces complex coronary atherosclerotic plaque development. Nat. Med. 14, 1059–1066 10.1038/nm.1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rosenson R. S., Hislop C., Elliott M., Stasiv Y., Goulder M., and Waters D. (2010) Effects of varespladib methyl on biomarkers and major cardiovascular events in acute coronary syndrome patients. J. Am. Coll. Cardiol. 56, 1079–1088 10.1016/j.jacc.2010.06.015 [DOI] [PubMed] [Google Scholar]
- 39. Serruys P. W., García-García H. M., Buszman P., Erne P., Verheye S., Aschermann M., Duckers H., Bleie O., Dudek D., Botker H. E., von Birgelen C., D'Amico D., Hutchinson T., Zambanini A., Mastik F., et al. (2008) Effects of the direct lipoprotein-associated phospholipase A2 inhibitor darapladib on human coronary atherosclerotic plaque. Circulation 118, 1172–1182 10.1161/CIRCULATIONAHA.108.771899 [DOI] [PubMed] [Google Scholar]
- 40. Nicholls S. J., Kastelein J. J., Schwartz G. G., Bash D., Rosenson R. S., Cavender M. A., Brennan D. M., Koenig W., Jukema J. W., Nambi V., Wright R. S., Menon V., Lincoff A. M., Nissen S. E., and VISTA-16 Investigators (2014) Varespladib and cardiovascular events in patients with an acute coronary syndrome: the VISTA-16 randomized clinical trial. JAMA 311, 252–262 10.1001/jama.2013.282836 [DOI] [PubMed] [Google Scholar]
- 41. STABILITY Investigators, White H. D., Held C., Stewart R., Tarka E., Brown R., Davies R. Y., Budaj A., Harrington R. A., Steg P. G., Ardissino D., Armstrong P. W., Avezum A., Aylward P. E., Bryce A., et al. (2014) Darapladib for preventing ischemic events in stable coronary heart disease. N. Engl. J. Med. 370, 1702–1711 10.1056/NEJMoa1315878 [DOI] [PubMed] [Google Scholar]
- 42. O'Donoghue M. L., Braunwald E., White H. D., Steen D. P., Lukas M. A., Tarka E., Steg P. G., Hochman J. S., Bode C., Maggioni A. P., Im K., Shannon J. B., Davies R. Y., Murphy S. A., Crugnale S. E., et al. (2014) Effect of darapladib on major coronary events after an acute coronary syndrome: the SOLID-TIMI 52 randomized clinical trial. JAMA 312, 1006–1015 10.1001/jama.2014.11061 [DOI] [PubMed] [Google Scholar]
- 43. Cheng J., Nanayakkara G., Shao Y., Cueto R., Wang L., Yang W. Y., Tian Y., Wang H., and Yang X. (2017) Mitochondrial proton leak plays a critical role in pathogenesis of cardiovascular diseases. Adv. Exp. Med. Biol. 982, 359–370 10.1007/978-3-319-55330-6_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang X. F., Mirkovic D., Zhang S., Zhang Q. E., Yan Y., Xiong Z., Yang F., Chen I. H., Li L., and Wang H. (2006) Processing sites are different in the generation of HLA-A2.1-restricted, T cell reactive tumor antigen epitopes and viral epitopes. Int. J. Immunopathol. Pharmacol. 19, 853–870 10.1177/039463200601900415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang W. Y., Shao Y., Lopez-Pastrana J., Mai J., Wang H., and Yang X. F. (2015) Pathological conditions re-shape physiological Tregs into pathological Tregs. Burns Trauma 3, 1 10.1186/s41038-015-0001-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fu H., Vadalia N., Xue E. R., Johnson C., Wang L., Yang W. Y., Sanchez C., Nelson J., Chen Q., Choi E. T., Ma J. X., Yu J., Wang H., and Yang X. (2017) Thrombus leukocytes exhibit more endothelial cell-specific angiogenic markers than peripheral blood leukocytes do in acute coronary syndrome patients, suggesting a possibility of trans-differentiation: a comprehensive database mining study. J. Hematol. Oncol. 10, 74 10.1186/s13045-017-0440-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ke X., Wang J., Li L., Chen I. H., Wang H., and Yang X. F. (2008) Roles of CD4+CD25(high) FOXP3+ Tregs in lymphomas and tumors are complex. Front. Biosci. 13, 3986–4001 10.2741/2986 [DOI] [PubMed] [Google Scholar]
- 48. Vander Heiden M. G., Cantley L. C., and Thompson C. B. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jang C., and Arany Z. (2013) Metabolism: sweet enticements to move. Nature 500, 409–411 10.1038/nature12549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Drzazga A., Sowinska A., Krzeminska A., Rytczak P., Koziolkiewicz M., and Gendaszewska-Darmach E. (2017) Lysophosphatidylcholine elicits intracellular calcium signaling in a GPR55-dependent manner. Biochem. Biophys. Res. Commun. 489, 242–247 10.1016/j.bbrc.2017.05.145 [DOI] [PubMed] [Google Scholar]
- 51. Yamashita A., Oka S., Tanikawa T., Hayashi Y., Nemoto-Sasaki Y., and Sugiura T. (2013) The actions and metabolism of lysophosphatidylinositol, an endogenous agonist for GPR55. Prostaglandins Other Lipid Mediat. 107, 103–116 10.1016/j.prostaglandins.2013.05.004 [DOI] [PubMed] [Google Scholar]
- 52. Brown M. S., and Goldstein J. L. (1997) The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89, 331–340 10.1016/S0092-8674(00)80213-5 [DOI] [PubMed] [Google Scholar]
- 53. Walker A. K., Jacobs R. L., Watts J. L., Rottiers V., Jiang K., Finnegan D. M., Shioda T., Hansen M., Yang F., Niebergall L. J., Vance D. E., Tzoneva M., Hart A. C., and Näär A. M. (2011) A conserved SREBP-1/phosphatidylcholine feedback circuit regulates lipogenesis in metazoans. Cell 147, 840–852 10.1016/j.cell.2011.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xiao H., Lu M., Lin T. Y., Chen Z., Chen G., Wang W. C., Marin T., Shentu T. P., Wen L., Gongol B., Sun W., Liang X., Chen J., Huang H. D., Pedra J. H., et al. (2013) Sterol regulatory element binding protein 2 activation of NLRP3 inflammasome in endothelium mediates hemodynamic-induced atherosclerosis susceptibility. Circulation 128, 632–642 10.1161/CIRCULATIONAHA.113.002714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gaber T., Strehl C., and Buttgereit F. (2017) Metabolic regulation of inflammation. Nat. Rev. Rheumatol. 13, 267–279 10.1038/nrrheum.2017.37 [DOI] [PubMed] [Google Scholar]
- 56. Man K., Kutyavin V. I., and Chawla A. (2017) Tissue immunometabolism: development, physiology, and pathobiology. Cell Metab. 25, 11–26 10.1016/j.cmet.2016.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Beiting D. P., Hidano S., Baggs J. E., Geskes J. M., Fang Q., Wherry E. J., Hunter C. A., Roos D. S., and Cherry S. (2015) The orphan nuclear receptor TLX is an enhancer of STAT1-mediated transcription and immunity to Toxoplasma gondii. PLoS Biol. 13, e1002200 10.1371/journal.pbio.1002200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ringnér M. (2008) What is principal component analysis? Nat. Biotechnol. 26, 303–304 10.1038/nbt0308-303 [DOI] [PubMed] [Google Scholar]
- 59. Kolesnikov N., Hastings E., Keays M., Melnichuk O., Tang Y. A., Williams E., Dylag M., Kurbatova N., Brandizi M., Burdett T., Megy K., Pilicheva E., Rustici G., Tikhonov A., Parkinson H., et al. (2015) ArrayExpress update-simplifying data submissions. Nucleic Acids Res. 43, D1113–D1116 10.1093/nar/gku1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.