Abstract
In the yeast Saccharomyces cerevisiae, genomic instability in rDNA repeat sequences is an underlying cause of cell aging and is suppressed by the chromatin-silencing factor Sir2. In humans, rDNA instability is observed in cancers and premature aging syndromes, but its underlying mechanisms and functional consequences remain unclear. Here, we uncovered a pivotal role of sirtuin 7 (SIRT7), a mammalian Sir2 homolog, in guarding against rDNA instability and show that this function of SIRT7 protects against senescence in primary human cells. We found that, mechanistically, SIRT7 is required for association of SNF2H (also called SMARCA5, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin, subfamily A, member 5), a component of the nucleolar heterochromatin-silencing complex NoRC, with rDNA sequences. Defective rDNA–heterochromatin silencing in SIRT7-deficient cells unleashed rDNA instability, with excision and loss of rDNA gene copies, which in turn induced acute senescence. Mounting evidence indicates that accumulation of senescent cells significantly contributes to tissue dysfunction in aging-related pathologies. Our findings identify rDNA instability as a driver of mammalian cellular senescence and implicate SIRT7-dependent heterochromatin silencing in protecting against this process.
Keywords: sirtuin, senescence, heterochromatin, genomic instability, nucleolus, aging, epigenetics, rDNA, SIRT7, sirtuin 7
Introduction
Cellular senescence is a state of permanent cell cycle arrest that is induced by diverse types of stress (1–3). Replicative senescence of primary mammalian cells occurs as a result of exhaustive cell division, during which erosion of telomere sequences eventually triggers senescence as a DNA damage response (4). Senescence can also be induced by cellular stress associated with oncogene activation, DNA damage, or chromatin deregulation and can have tumor-suppressive effects (4). However, senescent cells also have profound deleterious effects that enhance tumor malignancy or contribute to tissue dysfunction in aging and disease. Indeed, senescent cells undergo dramatic alterations in metabolic and gene expression profiles with acquisition of a “senescence-associated secretory phenotype” (SASP)5 (1, 5). Through the SASP, even relatively low levels of senescent cells (<20%) can have far-ranging noncell-autonomous effects that influence tissue function (2).
The evolutionarily conserved sirtuin family of proteins is an important group of lysine deacetylase and deacylase enzymes (6, 7). The founding member of this family, Saccharomyces cerevisiae silent information regulator-2 (Sir2), promotes chromatin silencing through histone deacetylation at telomeres, mating-type loci, and ribosomal DNA (rDNA) genes (8, 9). Interest in Sir2 greatly increased with the finding that it protects against yeast cell senescence by preventing genomic instability at rDNA genes, a large family of gene repeats that is highly prone to recombination (9, 10). As the most abundant and highly transcribed gene family in eukaryotes, rDNA sequences pose a significant challenge for the replication machinery (11). They are particularly susceptible to replication stress, and stalled replication forks at rDNA give rise to DNA double strand breaks (DSBs). Recombinational repair of rDNA DSBs can lead to deletions or expansions via unequal sister chromatid exchanges (12).
In the human genome, rDNA genes comprise ∼350 copies distributed in large clusters (13). As in yeast, mammalian rDNA genes are prone to instability, and recombination among repeats can lead to expansions, contractions, or translocations (11). Thus, maintaining rDNA stability is a serious challenge for genome integrity, and rDNA instability is a potential driving force of genomic instability in cancer. Indeed, rDNA sequences are “hot spots” for DNA DSBs and recombinational instability in adult solid tumors (14, 15). rDNA instability is also observed in cells from patients with Bloom syndrome and ataxia telangiectasia, genetic syndromes associated with cancer susceptibility, genomic instability, and signs of premature aging (16). Moreover, rDNA replication stress is a potent driver of functional decline in old hematopoietic stem cells (17). Thus, elucidating the factors and mechanisms that guard against rDNA instability should provide important insights into aging and cancer pathways.
Roughly half of rDNA copies in human cells are packaged in transcriptionally active chromatin, and high levels of transcription from these genes are important for ribosome biogenesis and protein translation requirements of cells, particularly in high metabolic states (13). The other ∼50% of rDNA genes are packaged in heterochromatin and are transcriptionally silent. A central regulator of rDNA heterochromatin silencing is the nucleolar remodeling complex (NoRC), which consists of SNF2H/SMARCA5, a SWI/SNF-like nucleosome remodeling factor, and TIP5/BAZ2A (18). Notably, recent work has shown that defective rDNA chromatin silencing in cells lacking TIP5 leads to increased rDNA instability with loss of rDNA gene copies (19). However, very little is known about other upstream regulators of rDNA stability in mammalian cells.
In mammals, there are seven Sir2 family members (called “sirtuins”), SIRT1–SIRT7, and a growing body of work has implicated these enzymes in protecting against diverse aging-related pathologic states from cancer to metabolic and neurodegenerative diseases (6, 7). SIRT7 is the only mammalian sirtuin that is concentrated in nucleoli, subnuclear compartments where rDNA genes are located, and early studies of SIRT7 function examined whether the nucleolus might function like yeast Sir2 in rDNA silencing (20, 21). These studies found that SIRT7 binds rDNA regulatory sequences. Surprisingly, however, SIRT7 was found to stimulate rather than repress rDNA transcription (through stabilization of RNA PolI via deacetylation of the PolI subunit PAF53 (20, 21)), apparently having the opposite effect on rDNA silencing as yeast Sir2. SIRT7 also has been shown to have nonnucleolar functions, through which it regulates tumor-suppressive and stress response pathways, via deacetylation of histone H3K18Ac or several nonhistone proteins (22–27). Recent work has also implicated SIRT7 in various aspects of DNA DSB repair and DNA damage signaling (28, 29). However, no studies have examined potential effects of SIRT7 on nucleolar DSBs at rDNA loci.
Several reports have now implicated SIRT7 in regulation of mammalian aging. Decreased SIRT7 expression is observed in certain tissues with aging (23, 30), and loss of SIRT7 in mice leads to shortened lifespan and aging-associated pathologies, including cardiomyopathy, lipodystrophy, kyphosis, hepatic steatosis, reduced IGF-1 levels, and loss of hematopoietic stem cell–regenerative potential (24, 28, 31). However, much remains to be learned about the underlying molecular mechanisms through which SIRT7 influences aging pathology. Here, we report a novel role of human SIRT7 in protecting against cellular senescence by maintaining heterochromatin silencing and genomic stability at ribosomal DNA gene clusters. Our findings provide the first demonstration that rDNA instability has a causal role in triggering acute senescence of primary human cells and show that SIRT7-dependent heterochromatin silencing is a key mechanism protecting against this process.
Results
SIRT7 protects against cellular senescence and is localized at silent rDNA repeats
Given the aging-related phenotypes observed in SIRT7-deficient mice, we investigated a potential role of SIRT7 in protecting against senescence of primary human cells. We found that shRNA depletion of SIRT7 from WI38 primary human fibroblasts led to substantial accumulation of senescent cells within days after SIRT7 depletion (Fig. 1A). Levels of the senescence marker p16 were also increased in the SIRT7-deficient cell cultures (data not shown). Notably, the acute onset of senescence occurred much earlier than replicative senescence induced by telomere shortening, and a similar increase in senescent cells was observed when the WI38 cells were first immortalized by expression of hTERT telomerase subunit, which counteracts telomere shortening (Fig. 1B). These data indicate that SIRT7 protects against a novel trigger of cellular senescence that is independent of telomere shortening.
Figure 1.
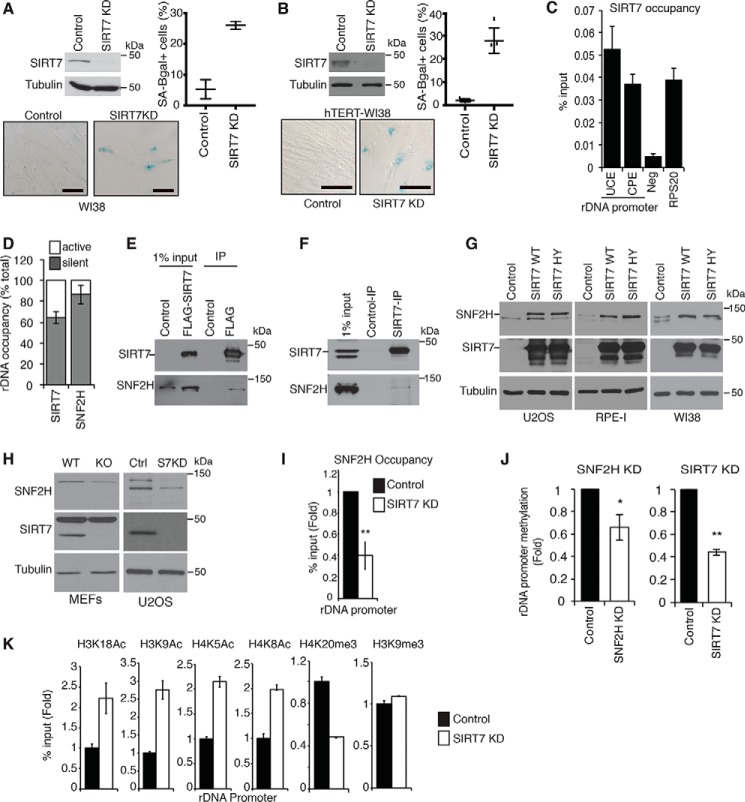
SIRT7 maintains rDNA heterochromatin and guards against cellular senescence. A and B, SA-β-gal assays showing increased senescence following SIRT7 KD in WI38 (A) or hTERT-immortalized WI38 (B) cells. Scale bars, 100 μm. Data are average of two to five fields ±S.D. (error bars). Immunoblots of SIRT7 protein and representative SA-β-gal images are shown. C, ChIP-qPCR showing SIRT7 occupancy at rDNA promoter regions upstream promoter element (UCE) and core promoter element (CPE). Neg, intergenic negative control; RPS20, positive control. D, ChIP-CHOP analysis showing percentage of rDNA-bound SNF2H or SIRT7 present at silent versus active rDNA copies. E and F, immunoblots showing coimmunoprecipitation of SNF2H with FLAG-SIRT7 expressed in U2OS cells (E) or endogenous SIRT7 IPs from 293T cells (F). Anti-FLAG antibodies were used for negative control IPs. G, immunoblots showing increased SNF2H levels in the indicated cell lines overexpressing WT or catalytically inactive (H187Y) SIRT7 proteins. H, immunoblots showing decreased SNF2H protein levels in SIRT7 KO MEFs and SIRT7 KD U2OS cells. I, ChIP-qPCR showing decreased SNF2H occupancy at rDNA promoters in SIRT7 KD U2OS cells. J, decreased DNA methylation at rDNA promoters in SIRT7 KD or SNF2H KD cells. K, ChIP-qPCR showing changes in histone acetylation and methylation marks at rDNA promoters in SIRT7 KD cells. In I and J, * indicates p < 0.05, and ** indicates, p < 0.01 (one-tailed Student's t test). In all graphs, data show averages of three technical replicates ±S.E. (error bars) and are representative of two to four biological replicates.
Because SIRT7 is enriched in nucleoli where rDNA genes are located, we hypothesized that SIRT7 might function like yeast Sir2 in guarding against senescence by preventing rDNA instability. To test this model, we first explored SIRT7 association with rDNA regulatory sequences using chromatin immunoprecipitation (ChIP) assays in U2OS cells (which allowed more robust ChIP DNA purification than primary cells). These analyses revealed SIRT7 binding to multiple rDNA promoter elements, consistent with previous reports (Fig. 1C) (20, 32). The association of SIRT7 with rDNA promoters was previously linked to the function of SIRT7 in stimulating RNA PolI activity, which occurs at transcriptionally active rDNA genes (20, 32). By contrast, we sought to determine whether a fraction of the rDNA-bound SIRT7 protein might occur in the context of rDNA silencing at heterochromatic rDNA gene clusters. Because traditional ChIP assays cannot distinguish between silent versus active rDNA, we used a modified protocol, termed “ChIP-CHOP” (32), to quantify SIRT7 occupancy specifically at silent rDNA (Fig. 1D). Digestion of ChIP DNA within rDNA genes by the SmaI methylation–sensitive restriction enzyme selectively cleaves within active, but not silent, rDNA copies, and subsequent PCR with primers that bridge the SmaI site amplifies only silent rDNA, whereas downstream primers detect total rDNA. This assay revealed that over 60% of rDNA-bound SIRT7 is present at silent rDNA (Fig. 1D). As a control, the NoRC protein SNF2H was almost exclusively detected at silent rDNA, as previously shown, consistent with its known function in maintaining rDNA heterochromatin structure (19). Together, these data suggested that, in addition to its activity in stimulating PolI transcription at active rDNA genes, SIRT7 has a novel function at silent rDNA clusters.
SIRT7 acts as a scaffold to stabilize SNF2H protein at rDNA promoters for chromatin silencing
Previous proteomic studies have identified components of the NoRC complex as potential SIRT7-interacting proteins, but the physiologic relevance of these interactions was not known (33, 34). To confirm these interactions, we carried out coimmunoprecipitation assays of both FLAG-tagged and endogenous SIRT7 proteins. This analysis revealed specific binding of SIRT7 to both the SNF2H and TIP5 subunits of NoRC (Fig. 1, E and F, and data not shown). Intriguingly, these experiments also revealed that overexpression of SIRT7 leads to increased SNF2H protein levels (Fig. 1, E and G). To verify the generality of this finding, we also confirmed this effect in two primary cell lines (RPE-I and WI38) (Fig. 1G). These observations suggested that SIRT7 might function as a scaffold that stabilizes SNF2H protein. To test this possibility, we asked whether the catalytic activity of SIRT7 is required for increasing SNF2H levels. Overexpression of the catalytically inactive H187Y SIRT7 protein (22) had the same effect on SNF2H levels as wildtype (WT) SIRT7 (Fig. 1G). Moreover, cycloheximide chase experiments revealed a longer half-life of SNF2H protein in SIRT7-overexpressing cells (Fig. S1). Together, these data suggest that interaction of SIRT7 with SNF2H enhances SNF2H stability, and increasing SIRT7 expression is sufficient to increase cellular levels of SNF2H protein.
We next asked whether, conversely, a decrease in SIRT7 can reduce SNF2H levels. We found that depletion of SIRT7 (by shRNA depletion in human cells or gene-targeted knockout in mouse embryonic fibroblasts) led to modest decreases in SNF2H protein (Fig. 1H). However, the magnitude of this effect was quite variable, possibly due to cell type–specific compensatory mechanisms. Alternatively, it is possible that the stabilizing effect of SIRT7 on SNF2H protein is only limiting in nucleoli, which could be masked by nonnucleolar SNF2H complexes. We therefore asked whether SIRT7-deficient cells show decreased SNF2H levels specifically in nucleoli or rDNA loci. First, quantitative SILAC analysis of nucleolar protein extracts from SIRT7-deficient or control cells revealed that SIRT7 depletion leads to decreased nucleolar levels of SNF2H (data not shown). We then examined SNF2H levels specifically at rDNA promoters by ChIP analysis, which revealed a dramatic loss of SNF2H occupancy in SIRT7-depleted cells (Fig. 1I). Similar results were observed for TIP5 (data not shown). Together, these observations identify a novel, nonenzymatic scaffolding function of SIRT7 that is required for maintaining SNF2H levels at silent rDNA genes.
SNF2H functions as a component of NoRC, which promotes heterochromatin silencing at rDNA by recruiting histone-modifying and DNA methylation enzymes. Thus, inactivation of NoRC by RNAi leads to changes in histone marks and DNA methylation patterns that reflect heterochromatin loss (19). We asked whether previously reported NoRC-dependent changes in DNA methylation and histone modifications are also observed in SIRT7-deficient cells. shRNA depletion of either SNF2H or SIRT7 led to a significant decrease in DNA methylation levels at rDNA promoters (Fig. 1J). Curiously, rDNA promoter methylation levels were lower in SIRT7-depleted cells than in SNF2H-depleted cells. This result could reflect different thresholds required for SIRT7 versus SNF2H protein or, alternatively, could indicate that SIRT7 influences rDNA methylation through additional mechanisms. SIRT7-deficient cells also showed an increase in multiple histone acetylation marks associated with active rDNA loci (Fig. 1K). Conversely, analysis of histone methylation marks associated with silent heterochromatin revealed a substantial decrease in rDNA promoter levels of H4K20me3, but not H3K9me3 (Fig. 1K), a pattern that is also observed in NoRC-deficient cells (19). The mechanistic underpinnings that distinguish H4K20me3 from H3K9me3 in this setting are still unclear, but our findings indicate that SIRT7 knockdown phenocopies multiple aspects of chromatin deregulation in NoRC-deficient cells.
SIRT7 protects against endogenous DSBs, rDNA instability, and nucleolar fragmentation
Chromatin silencing by yeast Sir2 represses both rDNA transcription and instability (35). In mammalian systems, much work on rDNA chromatin silencing has focused on rDNA transcription, but relatively little is understood about the factors that govern rDNA stability. Recently, inactivation of NoRC by RNAi depletion of its TIP5 subunit was shown to cause a loss of rDNA copies, implicating NoRC as one of the first known factors to control rDNA instability in human cells (19). When we assayed for rDNA copy number loss in SIRT7-depleted cells (36), we detected dramatically increased rDNA instability with an ∼50% reduction in rDNA copy number, levels comparable with what we observed in NoRC-deficient cells (generated by shRNA depletion of SNF2H) (Fig. 2A). One consequence of rDNA recombination is thought to be an increase in nucleolar fragmentation, which has been associated with rDNA silencing defects from yeast to humans (37–39). Accordingly, SIRT7-deficient mouse embryonic fibroblasts (MEFs) and WI38 cells exhibited significant disruption in nucleolar integrity (Fig. 2B and data not shown). These data suggest that, like Sir2 in yeast, SIRT7 links maintenance of rDNA heterochromatin to rDNA stabilization.
Figure 2.
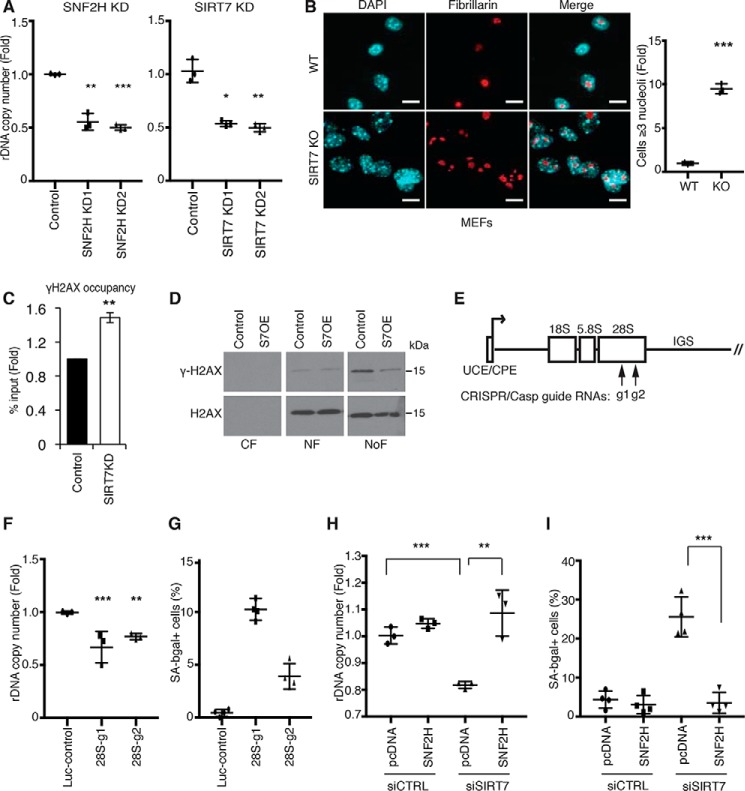
rDNA instability is a driver of mammalian cellular senescence and is regulated by SIRT7. A, decreased rDNA copy number in SIRT7 KD or SNF2H KD cells. Data show two biological replicates. B, representative confocal microscopy images showing nucleolar fragmentation in SIRT7 KO MEFs. Nucleoli are stained with anti-fibrillarin; nuclear DNA is stained with DAPI. Scale bars, 15 μm. C, ChIP-qPCR showing increased γ-H2AX levels at rDNA promoters in SIRT7 KD cells. Data show the average of three biological replicates. D, immunoblots showing γ-H2AX levels in cytoplasmic (CF), nuclear (NF), and nucleolar (NoF) biochemical fractions of U2OS cells overexpressing SIRT7 (S7OE) or vector control. E, schematic of a human rDNA repeat. Arrows indicate target locations of the g1 and g2 CRISPR/Cas9 guide RNAs. F and G, decreased rDNA copy number (F) and increased senescence (G) in clonal cell lines following rDNA break induction with g1, g2, or luciferase control guide RNAs. Error bars indicate S.D. *, p < 0.05; **, p < 0.01 (one-tailed Student's t test). H and I, reversal of rDNA copy number loss (H) and cellular senescence (I) by overexpression of SNF2H in cells depleted of SIRT7 by siRNA transfection. Error bars indicate S.D. **, p < 0.01; ***, p < 0.001(one-tailed Student's t test). In all other panels, error bars indicate S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (one-tailed Student's t test). Graphs are representative of two (A, F, and H) or three (B) biologic replicates.
A large body of work in yeast has provided evidence that rDNA instability results from recombinational repair of DNA DSBs that arise in rDNA due to replication stress (35). Moreover, rDNA sequences are thought to be the most fragile part of the genome in both yeast and mammalian cells (11, 14). In addition to being a hot spot for translocations in human cancers, recent work indicates that rDNA sequences are hot spots for phosphorylated γ-H2AX, a chromatin marker of DNA DSBs (40). These DSB hot spots colocalize with signs of active transcription, consistent with the model that an open chromatin state may be permissive for rDNA damage. We hypothesized that SIRT7-dependent rDNA silencing may be important for protecting against baseline rDNA damage that could otherwise arise as a result of replication stress. Consistent with this model, we found that, in SIRT7-deficient cells, γ-H2AX levels at rDNA are significantly increased (Fig. 2C). In biochemical fractionation studies, baseline γ-H2AX levels were considerably higher in nucleoli than throughout the nucleoplasm, consistent with the increased fragility of rDNA sequences, and overexpression of SIRT7 in these cells preferentially reduced nucleolar γ-H2AX levels (Fig. 2D). Notably, this analysis considerably underestimates the relative concentration of endogenous DSBs at rDNA because rDNA sequences comprise less than 0.5% of the human genome (15). Together, our results identify a novel function of SIRT7 in protecting against baseline endogenous rDNA damage and rDNA instability in mammalian cells.
rDNA instability is a trigger of mammalian cellular senescence and is regulated by SIRT7
Our data suggest that SIRT7 functions like yeast Sir2 in protecting against cellular senescence by preventing rDNA instability. However, although rDNA instability is a key driver of senescence in yeast, it has not yet been implicated in mammalian cellular senescence. We therefore set up a system to induce rDNA instability and directly test its effects on senescence. We used CRISPR/Cas9 technology to generate DNA DSBs specifically at rDNA sequences using guide RNAs homologous to 28S rDNA sequences or negative control luciferase sequence guide RNAs (which lack genomic targets) (Fig. 2E). rDNA DSB induction led to increased rDNA sequence loss and cellular senescence, similar to what we observed in cells lacking SIRT7 (Fig. 2, F and G).
Next, we sought to test directly whether rDNA silencing defects and instability are underlying causes of the senescence observed in SIRT7-deficient cells. Strikingly, we found that overexpression of SNF2H in SIRT7-depleted cells is sufficient to completely reverse both the rDNA instability and increased senescence in these cells (Fig. 2, H and I). This observation provides direct evidence that cellular senescence induced by SIRT7 loss is due to defective SNF2H-dependent rDNA silencing rather than other functions of SIRT7. We note that in these experiments SIRT7 levels were depleted transiently by siRNA knockdown, which led to a more modest change in rDNA copy number than was observed with stable shRNA knockdown of SIRT7 (Fig. 1A). Nonetheless, this level of instability was sufficient to induce acute senescence. Together, our data demonstrate that rDNA instability can be a driving force in provoking human cellular senescence and that SIRT7 plays a crucial role in preventing rDNA instability and senescence through rDNA chromatin silencing.
Discussion
Senescent cells accumulate in aging organisms and in the context of numerous aging-related chronic disease processes from osteoarthritis to diabetes (5). A growing body of work has shown that genetic or pharmacologic clearance of senescent cells in mice can extend lifespan and prevent or reverse a wide range of aging-related pathologies (41, 42). Thus, elucidating the upstream triggers and regulatory factors that control cellular senescence should provide important insights into basic mechanisms of aging and aging-related pathology. In this study, we have identified rDNA instability as an underlying trigger of senescence in mammalian cells and implicated SIRT7 as an important regulator of this process. Notably, by using CRISPR/Cas9 targeted cleavage to generate rDNA deletions, we also provide the first demonstration that rDNA instability has a direct causal role in provoking mammalian cellular senescence. These findings fit well with models of aging mechanisms that have been proposed based on rDNA-driven senescence in yeast, collectively called the “rDNA theory of aging” (35). In this theory, rDNA sequences are more susceptible to DSBs and instability than other genomic regions and thus can serve as a sensor that induces senescence to prevent accumulation of whole-genome damage. Our data support such models, showing that a majority of baseline endogenous DSBs that accumulate in cells occur at rDNA in nucleoli, and SIRT7 is both necessary and sufficient to protect against accumulation of such rDNA DSBs.
Our findings show that, in human cells, rDNA-driven senescence occurs acutely, within days of SIRT7 depletion or targeted induction of rDNA DSBs, and is observed at levels (∼25%) that are sufficient to induce tissue dysfunction through the SASP (2). These observations suggest that even transient increases in rDNA instability, which could be induced by conditions of stress, might contribute to human pathologies by increasing senescent cell burden. Indeed, SIRT7 has previously been shown to translocate dynamically out of nucleoli in response to stress (21), and a resulting decrease in local SIRT7 concentrations at rDNA could contribute to induction of senescence in such contexts.
Senescence has been widely associated with epigenetic changes that can establish and maintain the senescence-specific epigenetic landscape. SIRT7-dependent regulation of rDNA copy number adds another layer to the epigenetic changes that can lead to senescence. Studies in Drosophila have demonstrated that rDNA copy number has a profound effect on genome-wide gene expression, also known as “rDNA-sensitive genes” (43, 44), which postulates rDNA copy number as a major modulator of the genome. An interesting question for future studies is whether there is a conserved role for rDNA copy number changes in human cells, which could be examined in cells lacking SIRT7 or following targeted rDNA DSBs.
On a mechanistic level, our findings in this study reveal that SIRT7 has a novel nonenzymatic scaffolding function in targeting or stabilizing SNF2H at rDNA sequences. However, we cannot exclude that SIRT7 might also influence rDNA chromatin silencing by additional mechanisms. For example, SIRT7 inactivation leads to hyperacetylation of histone acetylation sites, including the known SIRT7 substrate H3K18Ac, and it is possible that SIRT7 also directly deacetylates this residue at rDNA. In addition, during preparation of our manuscript, a study from Bober and co-workers (45) suggested that SIRT7 interacts with and recruits DNA methyltransferase DNMT1 to rDNA promoters. Because NoRC recruits DNMT1 (and histone-modifying enzymes) to rDNA, our data are consistent with the model that the effects of SIRT7 on DNMT1 are mediated by SNF2H in the context of NoRC. However, our observation that SIRT7-deficient cells exhibit a greater loss of rDNA methylation than SNF2H-deficient cells is consistent with the possibility that SIRT7 interacts independently with SNF2H and DNMT1 and that the coordinated interactions provide stronger effects than each interaction alone.
rDNA stability may contribute to a number of human pathologies associated with aging. In a study of adult solid tumors, rDNA rearrangements were the most common chromosomal alterations detected and were present in over 50% of the samples analyzed (15). In addition, growing evidence suggests that rDNA instability may be linked to aging-associated disorders. Cells from patients with Werner syndrome progeria exhibit an abnormal number of palindromic rDNA repeats (46), and cells from Bloom syndrome and ataxia telangiectasia patients exhibit rDNA copy number variability and instability (16). Furthermore, age-related disorders such as Hodgkin's, Parkinson's, and Alzheimer's diseases show rDNA rearrangements, nucleolar disruption, and changes in rDNA chromatin marks, respectively (47–49). Thus, our data implicating SIRT7 in rDNA silencing and stabilization suggest potential roles for SIRT7 in the pathology of these and other human disease processes.
Experimental procedures
Cell culture and transfections
Human U2OS, 293T, WI38, hTERT-WI38, and hTERT-RPE-I were cultured in Eagle's minimum essential medium supplemented with 10% FBS and penicillin/streptomycin. Stable retroviral and lentiviral knockdown or expression was performed as described previously (50). shRNA target sequences for SIRT7 knockdown (KD) were described previously (22). SNF2H shRNA target sequence was 5′-gtactaatcttcagtcaaa-3′. rDNA guide RNA targets were caaagcatcgcgaaggcccg (28S1) and atgaagcgcgggtaaacggc (28S2). siRNA transfections were performed with Dharmafect reagent (Dharmacon) according to the manufacturer's instructions. SIRT7 and control double-stranded siRNAs were purchased from Dharmacon as described previously (22).
ChIP and quantitative real-time PCR
ChIPs were performed in U2OS cells as described previously (51) with the following modifications for SIRT7 and SNF2H ChIPs. Briefly, cells were cross-linked with 1% formaldehyde for 25 min at room temperature, quenched with 1.25 m glycine, and washed in ice-cold PBS, and pellets were resuspended in lysis buffer (20 mm Tris, pH 8.0, 85 mm KCl, 0.5% Nonidet P-40). Nuclear pellets were obtained by centrifugation and resuspended in 50 mm Tris, pH 8.0, 10 mm EDTA, 1% SDS. Cellular lysates were sonicated and diluted in dilution buffer (10 mm Tris, pH 7.5, 140 mm NaCl, 1 mm EDTA, 0.5 mm EGTA, 1.1% Triton, 0.01% SDS). Magnetic bead–antibody complexes (coupled for >2 h at 4 °C) were added to sonicated lysates and rotated overnight at 4 °C. Rabbit anti-mouse IgG was used as a negative control. Beads were washed three times in radioimmune precipitation assay buffer (10 mm Tris, pH 7.5, 140 mm NaCl, 1 mm EDTA, 0.5 mm EGTA, 1% Triton, 0.1% SDS, 0.1% sodium deoxycholate), and DNA was eluted and reverse cross-linked at 65 °C for 2 h using elution buffer (20 mm Tris, pH 7.5, 5 mm EDTA, 50 mm NaCl, 1% SDS, 5 μg/ml proteinase K). DNA was purified using a PCR purification kit (Qiagen).
ChIP-associated DNA was analyzed by qPCR on a LightCycler 480 II (Roche Applied Science) using SYBR Green Master Mix (Roche Applied Science). For γ-H2AX ChIPs, -fold enrichment over control was calculated as the percentage of input normalized to total H2AX. Purified DNA was diluted 10× before PCR to avoid saturation of repetitive sequences.
qPCR rDNA copy number quantification
DNA was isolated from U2OS cells 5–10 days after lentiviral knockdown of SIRT7 or SNF2H using a DNeasy Blood and Tissue kit (Qiagen). Purified DNA was quantified, and 10 ng was used for PCR. PCR was performed on a LightCycler 480 II using SYBR Green Master Mix. rDNA copy number was quantified by amplification of the 18S gene normalized to the tRNALys gene. Primer sequences are listed on Table S1.
Coimmunoprecipitation
Cells pellets were resuspended and sonicated in Buffer A (50 mm Tris-HCl, pH 7.4, 250 mm NaCl, 0.5% Triton X-100, 10% glycerol), and lysates were incubated overnight at 4 °C with anti-FLAG-agarose beads (Sigma). Following five washes in Buffer A, the FLAG IPs were pelleted and eluted in Laemmli buffer.
Endogenous IP was performed similarly but under higher salt concentration (50 mm Tris-HCl, pH 7.4, 300 mm NaCl, 0.5% Triton X-100, 10% glycerol). Lysates were incubated with primary antibodies overnight, and Protein A/G-agarose beads (Thermo Scientific) were added postincubation for 2 h.
Nucleolar fractionation
Nucleolar fractionation was performed as described previously (52).
Mass spectrometry
Light and heavy labeled cell extracts (53) were combined, and nucleolar material was isolated using a sucrose gradient. Sucrose was removed by SDS-PAGE, and proteins were processed by in-gel digestion with trypsin prior to analysis by LC and tandem MS (LC-MS/MS). Briefly, samples were electrophoresed 1 cm on an SDS-polyacrylamide gel, and lanes were diced into small pieces, treated with 10 mm DTT for 30 min at 60 °C and then with 55 mm iodoacetamide for 60 min in darkness at room temperature, dehydrated with acetonitrile (ACN), and rehydrated in 50 mm ammonium bicarbonate containing 12 ng/μl trypsin (Promega). Samples were digested overnight at room temperature and stopped with 50% ACN, 5% formic acid. Peptides were extracted by dehydration in ACN, and salts were removed using a C18 Stage Tip (Thermo Scientific). LC-MS/MS was performed on an Orbitrap Fusion mass spectrometer (Thermo Scientific) using data-dependent acquisition with a 4-h HPLC gradient. Protein identification and SILAC quantification were performed using MaxQuant version 1.3.0.5 with default settings (54). Proteins quantified with a minimum of four peptides were considered for further analysis.
Senescence-associated β-gal assay
SA-β-gal activity was detected with a Senescence Cells Histochemical Staining kit (Sigma-Aldrich) according to the manufacturer's instructions. Cells were imaged with a Leica DM5000B microscope.
DNA methylation
DNA was isolated from hTERT-RPE-I cells 5–10 days following transduction of SIRT7 or SNF2H shRNA using a DNeasy Blood and Tissue kit. Purified DNA (250 ng) was digested overnight with SmaI and purified using a PCR Purification kit. SmaI-resistant rDNA was normalized to total rDNA calculated by amplification of undigested DNA.
ChIP-CHOP
ChIP was done as described above, and CHOP was done as described previously (32) with some modifications. Briefly, 10 μl of chromatin immunoprecipitated or input DNA and 2 ng of pBlueScript plasmid were mixed and digested overnight with SmaI. The restriction enzyme was heat-inactivated, and DNA was purified using a PCR Purification kit. DNA methylation was detected by qPCR quantification using 2 μl of purified SmaI-digested DNA on a LightCycler 480 II using SYBR Green Master Mix.
Immunofluorescence
Cells were grown in glass coverslips, fixed with 4% paraformaldehyde, washed with 1× PBS, permeabilized with 0.1% Triton X-100 for 10 min, blocked with 2% BSA in PBS, and immunostained with primary antibodies (Table S2). Coverslips were mounted with ProLong Gold Antifade reagent with DAPI (Invitrogen) and imaged with a Zeiss LSM700 confocal laser-scanning microscope.
Author contributions
S. P. and K. F. C. conceptualization; S. P., L. T., S. M. C., M. A.-I., and W. Z. data curation; S. P. formal analysis; S. P. and K. F. C. funding acquisition; S. P., M. A.-I., L. T., S. M. C., W. Z., and T.-M. L. investigation; S. P., M. A.-I., T.-M. L., and K. F. C. methodology; S. P. writing-original draft; S. P. and K. F. C. project administration; S. P. and K. F. C. writing-review and editing.
Supplementary Material
Acknowledgments
We thank members of the laboratories of K. F. C. and O. Gozani for useful discussions.
This work was supported in part by a grant from the NIA, National Institutes of Health; a Department of Veterans Affairs merit award; and the Paul F. Glenn Laboratories (to K. F. C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
This article contains Tables S1 and S2.
- SASP
- senescence-associated secretory phenotype
- Sir2
- silent information regulator-2
- rDNA
- ribosomal DNA
- SIRT7
- sirtuin 7
- DSB
- double strand break
- NoRC
- nucleolar remodeling complex
- PolI
- polymerase I
- H3K18Ac
- acetylated H3 Lys-18
- hTERT
- human telomerase reverse transcriptase
- SILAC
- stable isotope labeling by amino acids in cell culture
- H4K20me3
- trimethylated H4 Lys-20
- H3K9me3
- trimethylated H3 Lys-9
- MEF
- mouse embryonic fibroblast
- KD
- knockdown
- qPCR
- quantitative PCR
- ACN
- acetonitrile
- SA
- senescence-associated.
References
- 1. Coppé J. P., Desprez P. Y., Krtolica A., and Campisi J. (2010) The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu. Rev. Pathol. 5, 99–118 10.1146/annurev-pathol-121808-102144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Campisi J., and d'Adda di Fagagna F. (2007) Cellular senescence: when bad things happen to good cells. Nat. Rev.. Mol. Cell Biol. 8, 729–740 10.1038/nrm2233 [DOI] [PubMed] [Google Scholar]
- 3. van Deursen J. M. (2014) The role of senescent cells in ageing. Nature 509, 439–446 10.1038/nature13193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Magalhães J. P., and Passos J. F. (2018) Stress, cell senescence and organismal ageing. Mech. Ageing Dev. 170, 2–9 10.1016/j.mad.2017.07.001 [DOI] [PubMed] [Google Scholar]
- 5. He S., and Sharpless N. E. (2017) Senescence in health and disease. Cell 169, 1000–1011 10.1016/j.cell.2017.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Toiber D., Sebastian C., and Mostoslavsky R. (2011) Characterization of nuclear sirtuins: molecular mechanisms and physiological relevance. Handb. Exp. Pharmacol. 206, 189–224 10.1007/978-3-642-21631-2_9 [DOI] [PubMed] [Google Scholar]
- 7. Haigis M. C., and Sinclair D. A. (2010) Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 5, 253–295 10.1146/annurev.pathol.4.110807.092250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Smith J. S., and Boeke J. D. (1997) An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11, 241–254 10.1101/gad.11.2.241 [DOI] [PubMed] [Google Scholar]
- 9. Kaeberlein M., McVey M., and Guarente L. (1999) The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13, 2570–2580 10.1101/gad.13.19.2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sinclair D. A., and Guarente L. (1997) Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 91, 1033–1042 10.1016/S0092-8674(00)80493-6 [DOI] [PubMed] [Google Scholar]
- 11. Ganley A. R., and Kobayashi T. (2014) Ribosomal DNA and cellular senescence: new evidence supporting the connection between rDNA and aging. FEMS Yeast Res. 14, 49–59 10.1111/1567-1364.12133 [DOI] [PubMed] [Google Scholar]
- 12. Kobayashi T., Horiuchi T., Tongaonkar P., Vu L., and Nomura M. (2004) SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell 117, 441–453 10.1016/S0092-8674(04)00414-3 [DOI] [PubMed] [Google Scholar]
- 13. McStay B., and Grummt I. (2008) The epigenetics of rRNA genes: from molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 24, 131–157 10.1146/annurev.cellbio.24.110707.175259 [DOI] [PubMed] [Google Scholar]
- 14. Tchurikov N. A., Fedoseeva D. M., Sosin D. V., Snezhkina A. V., Melnikova N. V., Kudryavtseva A. V., Kravatsky Y. V., and Kretova O. V. (2015) Hot spots of DNA double-strand breaks and genomic contacts of human rDNA units are involved in epigenetic regulation. J. Mol. Cell Biol. 7, 366–382 10.1093/jmcb/mju038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stults D. M., Killen M. W., Williamson E. P., Hourigan J. S., Vargas H. D., Arnold S. M., Moscow J. A., and Pierce A. J. (2009) Human rRNA gene clusters are recombinational hotspots in cancer. Cancer Res. 69, 9096–9104 10.1158/0008-5472.CAN-09-2680 [DOI] [PubMed] [Google Scholar]
- 16. Killen M. W., Stults D. M., Adachi N., Hanakahi L., and Pierce A. J. (2009) Loss of Bloom syndrome protein destabilizes human gene cluster architecture. Hum. Mol. Genet. 18, 3417–3428 10.1093/hmg/ddp282 [DOI] [PubMed] [Google Scholar]
- 17. Flach J., Bakker S. T., Mohrin M., Conroy P. C., Pietras E. M., Reynaud D., Alvarez S., Diolaiti M. E., Ugarte F., Forsberg E. C., Le Beau M. M., Stohr B. A., Méndez J., Morrison C. G., and Passegué E. (2014) Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature 512, 198–202 10.1038/nature13619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Santoro R., Li J., and Grummt I. (2002) The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 32, 393–396 10.1038/ng1010 [DOI] [PubMed] [Google Scholar]
- 19. Guetg C., Lienemann P., Sirri V., Grummt I., Hernandez-Verdun D., Hottiger M. O., Fussenegger M., and Santoro R. (2010) The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 29, 2135–2146 10.1038/emboj.2010.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ford E., Voit R., Liszt G., Magin C., Grummt I., and Guarente L. (2006) Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 20, 1075–1080 10.1101/gad.1399706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen S., Seiler J., Santiago-Reichelt M., Felbel K., Grummt I., and Voit R. (2013) Repression of RNA polymerase I upon stress is caused by inhibition of RNA-dependent deacetylation of PAF53 by SIRT7. Mol. Cell 52, 303–313 10.1016/j.molcel.2013.10.010 [DOI] [PubMed] [Google Scholar]
- 22. Barber M. F., Michishita-Kioi E., Xi Y., Tasselli L., Kioi M., Moqtaderi Z., Tennen R. I., Paredes S., Young N. L., Chen K., Struhl K., Garcia B. A., Gozani O., Li W., and Chua K. F. (2012) SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 487, 114–118 10.1038/nature11043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mohrin M., Shin J., Liu Y., Brown K., Luo H., Xi Y., Haynes C. M., and Chen D. (2015) Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science 347, 1374–1377 10.1126/science.aaa2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shin J., He M., Liu Y., Paredes S., Villanova L., Brown K., Qiu X., Nabavi N., Mohrin M., Wojnoonski K., Li P., Cheng H. L., Murphy A. J., Valenzuela D. M., Luo H., et al. (2013) SIRT7 represses Myc activity to suppress ER stress and prevent fatty liver disease. Cell Rep. 5, 654–665 10.1016/j.celrep.2013.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ryu D., Jo Y. S., Lo Sasso G., Stein S., Zhang H., Perino A., Lee J. U., Zeviani M., Romand R., Hottiger M. O., Schoonjans K., and Auwerx J. (2014) A SIRT7-dependent acetylation switch of GABPbeta1 controls mitochondrial function. Cell Metab. 20, 856–869 10.1016/j.cmet.2014.08.001 [DOI] [PubMed] [Google Scholar]
- 26. Li Z., Bridges B., Olson J., and Weinman S. A. (2017) The interaction between acetylation and serine-574 phosphorylation regulates the apoptotic function of FOXO3. Oncogene 36, 1887–1898 10.1038/onc.2016.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu J., Qin B., Wu F., Qin S., Nowsheen S., Shan S., Zayas J., Pei H., Lou Z., and Wang L. (2017) Regulation of serine-threonine kinase Akt activation by NAD+-dependent deacetylase SIRT7. Cell Rep. 18, 1229–1240 10.1016/j.celrep.2017.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vazquez B. N., Thackray J. K., Simonet N. G., Kane-Goldsmith N., Martinez-Redondo P., Nguyen T., Bunting S., Vaquero A., Tischfield J. A., and Serrano L. (2016) SIRT7 promotes genome integrity and modulates non-homologous end joining DNA repair. EMBO J. 35, 1488–1503 10.15252/embj.201593499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li L., Shi L., Yang S., Yan R., Zhang D., Yang J., He L., Li W., Yi X., Sun L., Liang J., Cheng Z., Shi L., Shang Y., and Yu W. (2016) SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat. Commun. 7, 12235 10.1038/ncomms12235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wronska A., Lawniczak A., Wierzbicki P. M., and Kmiec Z. (2016) Age-related changes in sirtuin 7 expression in calorie-restricted and refed rats. Gerontology 62, 304–310 10.1159/000441603 [DOI] [PubMed] [Google Scholar]
- 31. Vakhrusheva O., Smolka C., Gajawada P., Kostin S., Boettger T., Kubin T., Braun T., and Bober E. (2008) Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ. Res. 102, 703–710 10.1161/CIRCRESAHA.107.164558 [DOI] [PubMed] [Google Scholar]
- 32. Santoro R. (2014) Analysis of chromatin composition of repetitive sequences: the ChIP-Chop assay. Methods Mol. Biol. 1094, 319–328 10.1007/978-1-62703-706-8_25 [DOI] [PubMed] [Google Scholar]
- 33. Tsai Y. C., Greco T. M., Boonmee A., Miteva Y., and Cristea I. M. (2012) Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription. Mol. Cell. Proteomics 11, 60–76 10.1074/mcp.A111.015156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee N., Kim D. K., Kim E. S., Park S. J., Kwon J. H., Shin J., Park S. M., Moon Y. H., Wang H. J., Gho Y. S., and Choi K. Y. (2014) Comparative interactomes of SIRT6 and SIRT7: implication of functional links to aging. Proteomics 14, 1610–1622 10.1002/pmic.201400001 [DOI] [PubMed] [Google Scholar]
- 35. Kobayashi T. (2014) Ribosomal RNA gene repeats, their stability and cellular senescence. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 90, 119–129 10.2183/pjab.90.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Paredes S., and Maggert K. A. (2009) Expression of I-CreI endonuclease generates deletions within the rDNA of Drosophila. Genetics 181, 1661–1671 10.1534/genetics.108.099093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Espada J., Ballestar E., Santoro R., Fraga M. F., Villar-Garea A., Németh A., Lopez-Serra L., Ropero S., Aranda A., Orozco H., Moreno V., Juarranz A., Stockert J. C., Längst G., Grummt I., et al. (2007) Epigenetic disruption of ribosomal RNA genes and nucleolar architecture in DNA methyltransferase 1 (Dnmt1) deficient cells. Nucleic Acids Res. 35, 2191–2198 10.1093/nar/gkm118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Peng J. C., and Karpen G. H. (2007) H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat. Cell Biol. 9, 25–35 10.1038/ncb1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sinclair D. A., Mills K., and Guarente L. (1997) Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science 277, 1313–1316 10.1126/science.277.5330.1313 [DOI] [PubMed] [Google Scholar]
- 40. Tchurikov N. A., Yudkin D. V., Gorbacheva M. A., Kulemzina A. I., Grischenko I. V., Fedoseeva D. M., Sosin D. V., Kravatsky Y. V., and Kretova O. V. (2016) Hot spots of DNA double-strand breaks in human rDNA units are produced in vivo. Sci. Rep. 6, 25866 10.1038/srep25866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Childs B. G., Gluscevic M., Baker D. J., Laberge R. M., Marquess D., Dananberg J., and van Deursen J. M. (2017) Senescent cells: an emerging target for diseases of ageing. Nat. Rev. Drug Discov. 16, 718–735 10.1038/nrd.2017.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhu Y., Tchkonia T., Pirtskhalava T., Gower A. C., Ding H., Giorgadze N., Palmer A. K., Ikeno Y., Hubbard G. B., Lenburg M., O'Hara S. P., LaRusso N. F., Miller J. D., Roos C. M., Verzosa G. C., et al. (2015) The Achilles' heel of senescent cells: from transcriptome to senolytic drugs. Aging Cell 14, 644–658 10.1111/acel.12344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Paredes S., Branco A. T., Hartl D. L., Maggert K. A., and Lemos B. (2011) Ribosomal DNA deletions modulate genome-wide gene expression: “rDNA-sensitive” genes and natural variation. PLoS Genet. 7, e1001376 10.1371/journal.pgen.1001376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Paredes S., and Maggert K. A. (2009) Ribosomal DNA contributes to global chromatin regulation. Proc. Natl. Acad. Sci. U.S.A. 106, 17829–17834 10.1073/pnas.0906811106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ianni A., Hoelper S., Krueger M., Braun T., and Bober E. (2017) Sirt7 stabilizes rDNA heterochromatin through recruitment of DNMT1 and Sirt1. Biochem. Biophys. Res. Commun. 492, 434–440 10.1016/j.bbrc.2017.08.081 [DOI] [PubMed] [Google Scholar]
- 46. Caburet S., Conti C., Schurra C., Lebofsky R., Edelstein S. J., and Bensimon A. (2005) Human ribosomal RNA gene arrays display a broad range of palindromic structures. Genome Res. 15, 1079–1085 10.1101/gr.3970105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. MacLeod R. A., Spitzer D., Bar-Am I., Sylvester J. E., Kaufmann M., Wernich A., and Drexler H. G. (2000) Karyotypic dissection of Hodgkin's disease cell lines reveals ectopic subtelomeres and ribosomal DNA at sites of multiple jumping translocations and genomic amplification. Leukemia 14, 1803–1814 10.1038/sj.leu.2401894 [DOI] [PubMed] [Google Scholar]
- 48. Rieker C., Engblom D., Kreiner G., Domanskyi A., Schober A., Stotz S., Neumann M., Yuan X., Grummt I., Schütz G., and Parlato R. (2011) Nucleolar disruption in dopaminergic neurons leads to oxidative damage and parkinsonism through repression of mammalian target of rapamycin signaling. J. Neurosci. 31, 453–460 10.1523/JNEUROSCI.0590-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pietrzak M., Rempala G., Nelson P. T., Zheng J. J., and Hetman M. (2011) Epigenetic silencing of nucleolar rRNA genes in Alzheimer's disease. PLoS One 6, e22585 10.1371/journal.pone.0022585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tasselli L., Xi Y., Zheng W., Tennen R. I., Odrowaz Z., Simeoni F., Li W., and Chua K. F. (2016) SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat. Struct. Mol. Biol. 23, 434–440 10.1038/nsmb.3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dahl J. A., and Collas P. (2007) Q2ChIP, a quick and quantitative chromatin immunoprecipitation assay, unravels epigenetic dynamics of developmentally regulated genes in human carcinoma cells. Stem Cells 25, 1037–1046 10.1634/stemcells.2006-0430 [DOI] [PubMed] [Google Scholar]
- 52. Chamousset D., Mamane S., Boisvert F. M., and Trinkle-Mulcahy L. (2010) Efficient extraction of nucleolar proteins for interactome analyses. Proteomics 10, 3045–3050 10.1002/pmic.201000162 [DOI] [PubMed] [Google Scholar]
- 53. Ong S. E., and Mann M. (2006) A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 1, 2650–2660 10.1038/nprot.2006.427 [DOI] [PubMed] [Google Scholar]
- 54. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.