Abstract
The U1 small nuclear ribonucleoprotein 70 kDa (U1-70K) and other RNA-binding proteins (RBPs) are mislocalized to cytoplasmic neurofibrillary Tau aggregates in Alzheimer's disease (AD), yet the co-aggregation mechanisms are incompletely understood. U1-70K harbors two disordered low–complexity domains (LC1 and LC2) that are necessary for aggregation in AD brain extracts. The LC1 domain contains highly repetitive basic (Arg/Lys) and acidic (Asp/Glu) residues, referred to as a basic-acidic dipeptide (BAD) domain. We report here that this domain shares many of the properties of the Gln/Asn-rich LC domains in RBPs that also aggregate in neurodegenerative disease. These properties included self-assembly into oligomers and localization to nuclear granules. Co-immunoprecipitations of recombinant U1-70K and deletions lacking the LC domain(s) followed by quantitative proteomic analyses were used to resolve functional classes of U1-70K-interacting proteins that depend on the BAD domain for their interaction. Within this interaction network, we identified a class of RBPs with BAD domains nearly identical to that found in U1-70K. Two members of this class, LUC7L3 and RBM25, required their respective BAD domains for reciprocal interactions with U1-70K and nuclear granule localization. Strikingly, a significant proportion of RBPs with BAD domains had elevated insolubility in the AD brain proteome. Furthermore, we show that the BAD domain of U1-70K can interact with Tau from AD brains but not from other tauopathies. These findings highlight a mechanistic role for BAD domains in stabilizing RBP interactions and in potentially mediating co-aggregation with the pathological AD–specific Tau isoforms.
Keywords: protein–protein interaction, protein aggregation, systems biology, Tau protein (Tau), neurodegeneration, mass spectrometry (MS), proteomics, RNA-binding protein, intrinsically disordered protein, RNA processing
Introduction
The molecular processes that contribute to neurodegenerative diseases are not well-understood. Recent observations suggest that numerous neurodegenerative diseases are promoted by the accumulation of RNA-binding protein (RBP)4 aggregates (1–3). This includes Alzheimer's disease (AD), where pathological RNA–protein aggregates are often, but not exclusively, associated with Tau neurofibrillary tangles in brain (4). For example, U1 small nuclear ribonucleoprotein 70 kDa (U1-70K) and other core components of the spliceosome complex co-aggregate with Tau in both sporadic and familial human cases of AD, but not other tauopathies (5–7). Furthermore, RNA-sequencing analysis from AD and control brains revealed a significant accumulation of unspliced pre-mRNA disease–related transcripts in AD consistent with a loss of U1-spliceosome function (7, 8). Currently, our knowledge of the specific mechanisms underlying U1-70K and Tau co-aggregation in AD is limited. This has proved to be a barrier to developing cellular models that would further our understanding of U1-70K and related RBP aggregation events in the pathogenesis of AD.
Supporting evidence indicates that a select group of RBPs are poised for aggregation because they self-assemble to form structures, including RNA granules (9), which are membrane-free organelles composed of RNA and RBPs (1, 9–12). RNA granules form via liquid–liquid phase separation (LLPS), which is driven by a dynamic network of multivalent interactions between structurally disordered low complexity (LC) domains (13–15) that have limited diversity in their amino acid composition (16). LLPS allows specific RBPs to concentrate and separate, leading to the formation of higher-order structures, including oligomers, granules, and ultimately aggregates (16–18). Notably, several RBPs that aggregate in neurodegenerative disease contain LC domains, including TDP-43 and fused in sarcoma (FUS) (19–21). The LC domains found in TDP-43 and FUS mediate self-association, are necessary for RNA granule formation, and polymerize into amyloid-like aggregates (9, 22, 23). Mutations within the LC domains of TDP-43 and FUS cause amyotrophic lateral sclerosis (ALS) and increased RNA granule stability, highlighting a critical role for LC domains in disease pathogenesis (24–26).
We recently reported that human AD brain homogenates induce the aggregation of soluble U1-70K from control brain as well as recombinant U1-70K, making U1-70K detergent-insoluble (5). The C terminus of U1-70K, which contains two LC domains (LC1 and LC2), is necessary for this aggregation (5). Furthermore, the LC1 domain (residues 231–308) of U1-70K is sufficient for robust aggregation, and through cross-linking studies it was found to directly interact with insoluble U1-70K in AD brain homogenates (5). Collectively, these observations led to a hypothesis that pathological aggregation of RBPs in neurodegenerative diseases, including U1-70K, is driven by LC domains. However, unlike the prion-like Gln/Asn-rich LC domains of TDP-43 and FUS, the LC1 domain of U1-70K contains highly repetitive basic (Arg/Lys) and acidic (Asp/Glu) residues that we refer to as a Basic-Acidic Dipeptide (BAD) domain. These structurally unique motifs were originally described by Perutz (27), who proposed their ability to self-assemble and form higher-order structures termed polar zippers. Currently, the physiological role of BAD domains in U1-70K and other RNA-binding proteins is unclear, and understanding their role in protein–protein interactions may shed light on the mechanisms underlying RBP aggregation and their association with Tau in AD (7, 28).
Here, we report that the BAD domain of U1-70K shares many of the properties of the Gln/Asn-rich LC domains found in TDP-43 and FUS, despite having a vastly different amino acid composition. These properties include the ability to self-assemble with homotypic selectivity into high-molecular weight oligomers and to associate with nuclear granules in cells. Coupling quantitative proteomics and network-based systems biology, we mapped classes of U1-70K–interacting proteins that show a dramatic reduction in their association with U1-70K in the absence of the BAD domain. This revealed a class of functionally and structurally similar RBPs that also contains BAD domains analogous to those in U1-70K. Furthermore, global analysis of the AD detergent–insoluble proteome revealed elevated levels of RBPs with BAD domains within AD brain compared with controls. Finally, we show that the BAD domain of U1-70K can interact with Tau from AD brain, but not other tauopathies. Collectively, our findings support a mechanistic role for BAD domains in stabilizing RBP interactions and in potentially mediating co-aggregation with pathological Tau isoforms specific to AD.
Results
BAD domain of U1-70K is necessary and sufficient for self-association
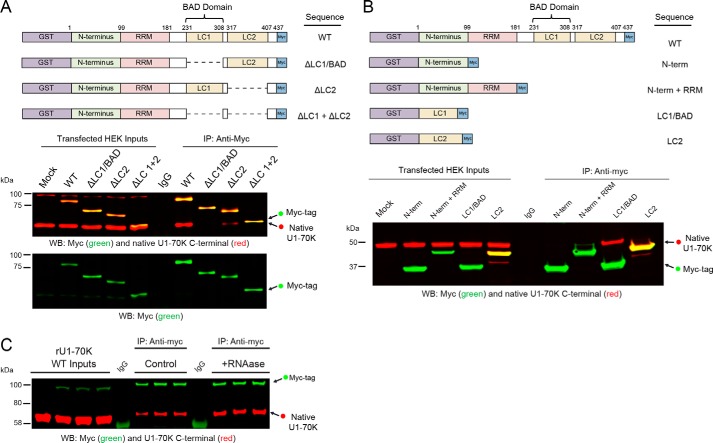
The U1-70K LC1 domain (residues 231–308), which we also refer to as the LC1/BAD domain, contains highly repetitive basic (Arg/Lys) and acidic (Asp/Glu) residues. This domain is necessary and sufficient for robust aggregation in AD brain homogenates (5). However, whether this domain is required for endogenous U1-70K self-association under physiological conditions is not known. To test this question, we overexpressed full-length recombinant GST-fused, Myc-tagged U1-70K (rU1-70K) in HEK293 cells with serial deletions lacking one or both LC domains followed by co-immunoprecipitation (co-IP) and Western blot analysis (Fig. 1A). Full-length rU1-70K (WT) and the ΔLC2 mutant co-IP endogenous U1-70K (∼55 kDa), while rU1-70K variants that lack the LC1/BAD domain (ΔLC1 and ΔLC1 + 2), show a significant decrease in the ability to co-IP native U1-70K. Together, these observations indicate that the LC1 domain is necessary for U1-70K self-association.
Figure 1.
LC1/BAD domain of U1-70K is necessary and sufficient for self-association. A, full-length (WT) recombinant GST-fused, Myc-tagged U1-70K (rU1-70K) and variants lacking one or both LC domains (ΔLC1, ΔLC2, and ΔLC1 + ΔLC2) were overexpressed in HEK293 cells and immunoprecipitated (IP) with anti-Myc antibodies. IP with a nonspecific IgG was also performed from mock-transfected cells as a negative control. Western blotting for recombinant Myc-tagged proteins (green) and native U1-70K (red) are shown for both the inputs and co-IPs (A, bottom panels). B, full-length WT and rU1-70K truncations, including the N terminus (1–99 residues) alone, the N terminus and RRM (1–181 residues), LC1/BAD alone (231–308 residues), and the LC2 domain alone (residues 317–407). IP with a nonspecific IgG was also performed from mock-transfected cells as a negative control. Western blotting for recombinant Myc-tagged proteins (green) and native U1-70K (red) are shown for both the inputs and co-IPs. C, WT rU1-70K was immunoprecipitated from untreated and RNase (50 ng/μl)-treated lysates followed by Western blotting for the Myc tag recombinant protein (green) and native U1-70K (red). IP with a nonspecific IgG was also performed from mock-transfected cells as a negative control.
To determine whether the LC1 domain is sufficient for self-association, co-IPs were performed following the overexpression of specific rU1-70K domains (Fig. 1B), including the N-terminal domain alone (residues 1–99), the N terminus, and RNA recognition motif (residues 1–181), the LC1/BAD domain alone, and the LC2 domain alone. Only the LC1/BAD domain was sufficient for self-association with native U1-70K. Furthermore, this interaction was likely not influenced by the presence of RNA, as treatment of the lysates with RNase prior to co-IP did not impair the ability of rU1-70K to interact with native U1-70K (Fig. 1C). Thus, our findings show that the LC1/BAD domain is necessary and sufficient for U1-70K self-association and that this interaction is predominantly RNA-independent.
U1-70K BAD domain oligomerizes in vitro
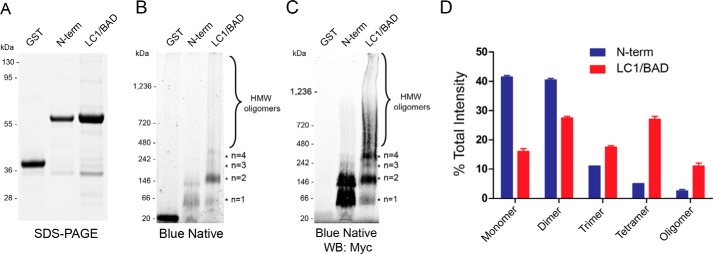
Although our results show that the LC1/BAD domain of rU1-70K is sufficient to interact with native U1-70K in cells, it is unclear whether this interaction is direct or facilitated by indirect interactions with additional RBPs. To determine whether the LC1/BAD domain of U1-70K can directly self-associate, we performed blue native gel-PAGE (BN-PAGE) of the GST-purified LC1/BAD domain (residues 231–310) and N-terminal domain (residues 1–99) of rU1-70K; the latter was unable to interact with native U1-70K (Fig. 1B). In contrast to SDS-PAGE, which resolves proteins under denaturing conditions, BN-PAGE is used to determine native protein complex masses, including high molecular weight oligomeric states, and to identify physiological protein–protein interactions (29). Under the denaturing conditions of SDS-PAGE (Fig. 2A), both the LC1 and N-terminal domain have equivalent molecular masses (∼65 kDa) compared with purified GST (∼20 kDa). However, under native conditions (Fig. 2B), the BAD domain formed dimers, trimers, tetramers, and high-molecular mass oligomers in the megadalton range (>1,236 kDa). In contrast, the N-terminal domain mainly existed in the monomeric and dimeric state with some evidence of lower abundance high-molecular mass oligomers; GST alone was almost exclusively monomeric (Fig. 2B). These complexes were more evident following the transfer to a membrane and Western blot analysis with Myc antibodies (Fig. 2C). Notably, a higher proportion of the LC1/BAD domain formed dimers (n = 2) and tetramers (n = 4) compared with trimers (n = 3), suggesting that dimer intermediates are favored over trimer intermediates for tetramer formation (Fig. 2D). These in vitro findings demonstrate that the LC1/BAD domain can directly self-associate to form oligomers, including high molecular weight species, which implicates direct BAD–BAD domain interactions as a mechanism of U1-70K self-association.
Figure 2.
LC1/BAD domain of U1-70K directly self-interacts and oligomerizes in vitro. A, SDS-PAGE of GST alone, purified N-terminal domain (N-term), and purified LC1/BAD domain of rU1-70K. Both the LC1/BAD and N-term domains have equivalent molecular masses (∼65 kDa), whereas GST alone is (∼20 kDa). B, BN-PAGE of GST alone, the N-terminal domain, and the LC1 domain of rU1-70K, respectively. The LC1/BAD domain formed higher molecular weight species (*) consistent with dimers (∼130 kDa), trimers (∼195 kDa), tetramers (∼260 kDa), and high-molecular weight (HMW) oligomers (>400 kDa). C, Western blotting detection of blue native complexes using Myc antibodies. D, densitometry of monomeric, dimeric, trimeric, and high-molecular weight species of the N-term domain (blue) and LC1/BAD domain (red) of rU1-70K. Each form is represented as the fraction of total signal intensity in each sample analyzed in technical replicate (n = 2). Error bars represent standard deviation.
U1-70K BAD domain is necessary and sufficient for robust nuclear granule localization
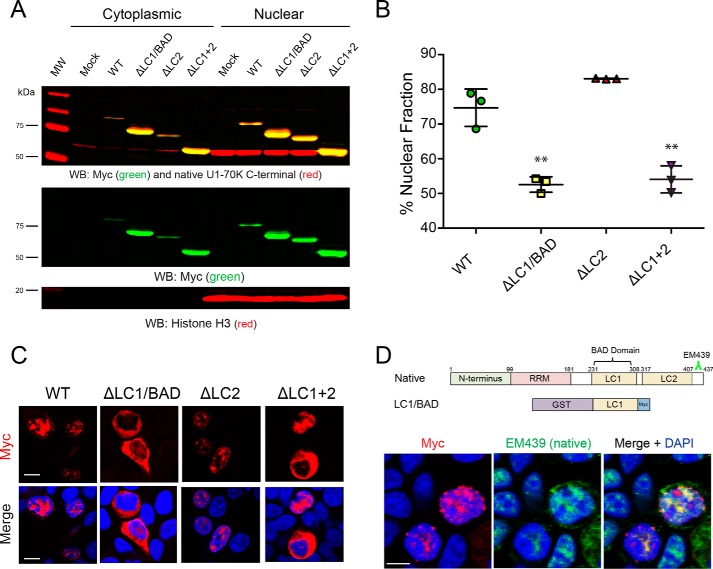
To explore whether the LC1/BAD domain influences nuclear localization and granule formation in cells, full-length rU1-70K or variants lacking one or both LC domains (Fig. 1A) were overexpressed in HEK293 cells followed by subcellular biochemical fractionation into nuclear and cytoplasmic pools (Fig. 3, A and B). The rU1-70K variants containing an LC1/BAD domain (WT and ΔLC2) partitioned mainly to the nuclear fraction (∼75% nuclear), but variants lacking the LC1 domain (ΔLC1 and ΔLC1 + 2) were equally distributed between the nucleus and the cytoplasm, indicating a significant impairment of nuclear localization. These biochemical findings are further supported by immunocytochemistry, which shows that rU1-70K variants lacking the LC1/BAD domain display diffuse patterns of localization in both the nucleus and cytoplasm compared with WT and ΔLC2 proteins (Fig. 3C). Given that the LC1/BAD domain in isolation can directly self-associate and oligomerize (Fig. 2), we sought to determine whether the LC1/BAD domain is necessary for RNA granule localization in cells (Fig. 3C). As expected, full-length rU1-70K protein localized to nuclear granules, in agreement with previous studies (30–32). Although the nuclear granule localization of variants lacking the LC1/BAD domain was diminished, the ΔLC2 mutant retained nuclear granule localization, supporting the requirement for the LC1/BAD domain in subnuclear granule localization. Furthermore, the LC1/BAD domain alone co-localizes with native U1-70K in nuclear granules (Fig. 3D), consistent with the ability of the LC1/BAD domain to interact with native U1-70K from cell lysates (Fig. 1B). Collectively, these results demonstrate that the LC1/BAD domain is important for U1-70K subcellular nuclear localization and suggest a role for LC1/BAD-mediated intermolecular interactions as a mechanism of nuclear granule formation.
Figure 3.
LC1/BAD domain of U1-70K is necessary and sufficient for nuclear granule localization. A, WT full-length rU1-70K or variants lacking one or both LC domains were overexpressed in HEK293 cells. The cells were then fractionated into nuclear and cytoplasmic pools followed by Western blot (WB) analysis for both recombinant Myc-tagged proteins (green) and native U1-70K (red). Western blottings for histone H3 (bottom panel) were used as a positive control in the nuclear fraction. B, densitometry analysis was performed to calculate the levels of cytoplasmic and nuclear WT rU1-70K and variants, and the percent nuclear intensity for each rU1-70K protein is reported. Each experiment was performed in biological triplicate (n = 3) with error bars representing the standard deviation (S.D.). Both the ΔLC1 and ΔLC1 + 2 rU1-70K fragments were significantly less nuclear than full-length rU1-70K (**, p value <0.01 by ANOVA compared with WT). C, immunocytochemistry for WT rU1-70K and deletion variants that lacked the LC1/BAD, LC2, or both LC domains was performed and visualized by confocal microscopy. Scale bar equates to 10 μm. D, overexpression of the rU1-70K LC1/BAD domain alone (red) resulted in nuclear granule association and sequestration of native U1-70K (green). The EM439 antibody detects an extreme C-terminal epitope not present in the LC1/BAD rU1-70K protein, which allows discrimination between the recombination protein and native U1-70K. DAPI-stained nuclei are shown in blue. Scale bar equates to 10 μm.
Protein–protein interaction network analysis resolves functionally distinct classes of U1-70K–interacting proteins
To further assess the physiological function of the LC domains of U1-70K, we performed co-IPs of full-length rU1-70K and various rU1-70K variants lacking either or both LC domains from HEK293 cells. Co-interacting proteins were identified by LC coupled to tandem MS (LC-MS/MS). Each co-IP was performed in biological quadruplicate (n = 4), and an equal number of mock IPs were performed using a nonspecific immunoglobulin (IgG) as a negative control. Protein abundance was determined by peptide ion–intensity measurements across LC-MS/MS runs using the label-free quantification (LFQ) algorithm in MaxQuant (33). In total, 45,223 peptides mapping to 3,458 protein groups were identified. To limit the number of nonspecific interactors, proteins with less than a 1.5-fold enrichment over IgG were not considered. This resulted in the final quantification of high-confidence interactors falling into 716 protein groups mapping to 713 unique gene symbols (Table S1).
The Weighted co-expression network analysis (WGCNA) is typically used for large-scale transcriptome and proteome datasets to categorize gene products into biologically meaningful complexes, molecular functions, and cellular pathways (34). Here, we sought to leverage co-enrichment patterns to better classify protein–protein interactions (PPIs) across WT and rU1-70K deletions to determine whether specific classes of proteins selectively favor interactions with the LC domains. In WGCNA, correlation coefficients between each protein pair in the dataset are calculated, and groups of highly correlated proteins are segregated into modules (35).
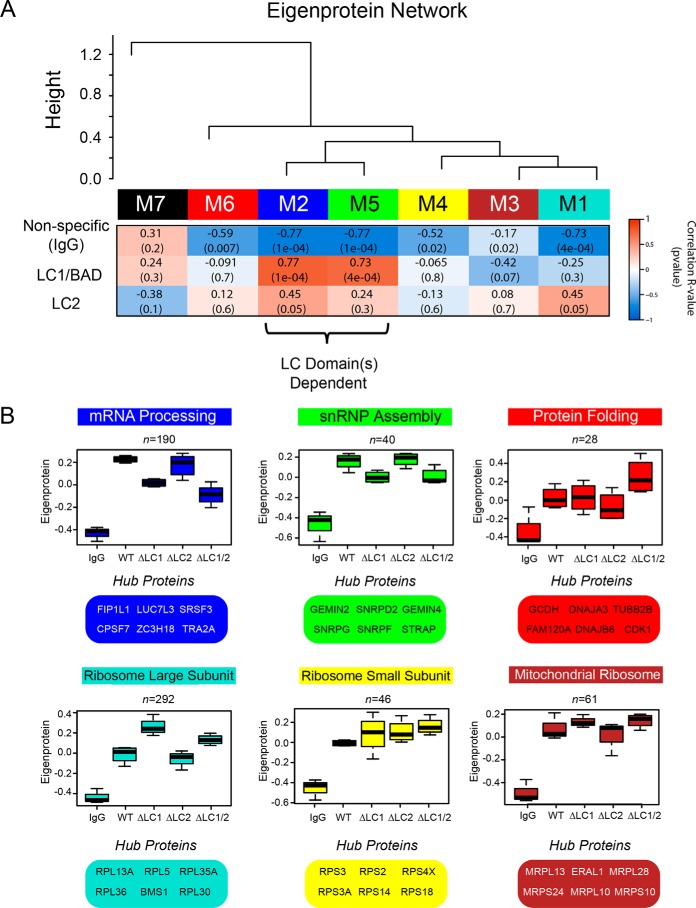
In our dataset, a total of seven modules were defined (Fig. 4A). These modules range from 292 proteins in turquoise to 19 in black (Table S1). The premise of co-expression, or in this case protein co-enrichment analysis, is that the strong correlation between two or more proteins is indicative of a physical interaction, functional relationship, and/or co-regulation. We therefore hypothesized that following a co-IP for rU1-70K, specific modules would reflect biologically relevant PPIs and thus highlight distinct complexes. As expected, modules were significantly enriched for biologically meaningful gene ontologies (GO) as well as established cellular functions and/or organelles as determined by GO-Elite (Table 1).
Figure 4.
Correlation network analysis resolves distinct modules of U1-70K-interacting proteins that differ in their association with the LC1/BAD domain. A, WGCNA-clustered proteins that were measured across all co-IP samples (n = 716) into modules (M1–M7) that represent classes of proteins defined by their correlation to each other across the five co-IP conditions analyzed (IgG, WT rU1-70K and deletions ΔLC1, ΔLC2, and ΔLC1 + ΔLC2). Listed in the heat map are bicor (R) correlations and p values defining the relationship between module eigenprotein level and rU1-70K LC domains (red is positively and blue is negatively correlated, respectively). B, eigenproteins, which correspond to the first principal component of a given module and serve as a summary abundance profile for all proteins within a module, are shown for six modules generated by WGCNA. Box plots with error bars beyond the 25th and 75th percentiles are shown for all five groups (IgG, WT, ΔLC1, ΔLC2, and ΔLC1 + 2). For each module, hub proteins are also enumerated below. Hub proteins are defined by high kME scores, which is a measure of how well a given protein matches that of the module eigenprotein, with high scores approaching one, signifying a high correlation (Table S1).
Table 1.
U1-70K protein–protein interaction network generates modules enriched with specific gene ontology (GO) terms
| Module color | Ontology type | Top GO terms | Fisher exact p value |
|---|---|---|---|
| Turquoise (n = 292) | Biological process | ncRNA processing | 2.07E-11 |
| rRNA metabolic process | 4.19E-10 | ||
| Endocrine pancreas development | 9.99E-09 | ||
| Molecular function | Nucleic acid binding | 1.8E-08 | |
| Structural constituent of ribosome | 1.28E-20 | ||
| Protein–DNA loading ATPase activity | 0.00153 | ||
| Cellular component | Cytosolic large ribosomal subunit | 2.66E-17 | |
| Nucleolus | 2.16E-10 | ||
| Intracellular | 1.58E-05 | ||
| Blue (n = 190) | Biological process | mRNA processing | 1.01E-44 |
| RNA splicing | 6.82E-41 | ||
| mRNA export from nucleus | 2.14E-16 | ||
| Molecular function | RNA binding | 1.97E-08 | |
| RS domain binding | 6.57E-06 | ||
| Nucleotide binding | 2.49E-05 | ||
| Cellular component | Nuclear speck | 3.55E-14 | |
| Spliceosomal complex | 1.29E-12 | ||
| Nucleus | 3.26E-15 | ||
| Brown (n = 61) | Biological process | Translation | 1.82E-15 |
| Molecular function | Structural constituent of ribosome | 1.28E-20 | |
| Translation regulator activity | 0.002275 | ||
| Cellular component | Mitochondrion | 9.67E-34 | |
| Ribosome | 1.91E-28 | ||
| Mitochondrial large ribosomal subunit | 1.51E-11 | ||
| Yellow (n = 46) | Biological process | Endocrine pancreas development | 9.99E-09 |
| Viral transcription | 9.99E-09 | ||
| Viral infectious cycle | 4.61E-08 | ||
| Molecular function | Structural constituent of ribosome | 1.28E-20 | |
| mRNA binding | 0.005748 | ||
| Cellular component | Cytosolic small ribosomal subunit | 1.78E-44 | |
| Ribosome | 1.91E-28 | ||
| Intracellular | 1.58E-05 | ||
| Green (n = 40) | Biological process | Spliceosomal snRNP assembly | 2.67E-20 |
| Regulation of cyclin-dependent protein kinase activity | 1.4E-06 | ||
| Spliceosome assembly | 5.74E-06 | ||
| Molecular function | snRNA binding | 6.37E-08 | |
| Cellular component | Small nuclear ribonucleoprotein complex | 5.26E-09 | |
| U12-type spliceosomal complex | 4.85E-08 | ||
| Cajal body | 1.05E-07 | ||
| Red (n = 20) | Biological process | Protein folding | 3.65E-06 |
| Response to abiotic stimulus | 2.37E-05 | ||
| Oxidation–reduction process | 0.000248 | ||
| Molecular function | Heat-shock protein binding | 7.28E-07 | |
| Purine ribonucleoside triphosphate binding | 3.21E-05 | ||
| Catalytic activity | 0.000225 | ||
| Cellular component | Microtubule | 0.000258 | |
| Membrane | 2.47E-05 | ||
| Intrinsic to membrane | 0.016518 | ||
Each module has an abundance profile for all member proteins across the rU1-70K co-IP conditions, termed the eigenprotein (Fig. 4, A and B). Notably, six (M1–M6) of the seven modules showed a significantly higher level of co-enrichment in the WT and rU1-70K deletions compared with the IgG negative controls, indicative of specific interactions for members of these modules (Fig. 4, A and B). In contrast, M7 (Fig. 4A, black) had essentially equivalent levels across all conditions and was the only module with positive correlation to the nonspecific IgG. Therefore, it was considered nonspecific and not considered for further analysis (Fig. 4A). The protein interactors with reduced affinity for rU1-70K following LC1/BAD domain deletion (e.g. ΔLC1 and ΔLC 1/2) include members of both the mRNA processing (blue) and snRNP assembly (green) modules (Table 1). Therefore, the LC1/BAD domain-dependent interactors are defined by membership in these modules. In contrast, modules enriched with large ribosomal subunit components (turquoise) and mitochondrial ribosome subunits (brown) display increased levels following deletion of the LC1/BAD domain or both LC domains, suggesting that the LC1/BAD domain negatively regulates their interactions with U1-70K (Table 1 and Fig. 4, A and B). Finally, the modules enriched with proteins involved in protein folding (Fig. 4, A and B, red) and the small ribosomal subunit (yellow) show little difference across the co-IP conditions, suggesting that these protein interactions are mainly with the N terminus and/or RRM domain of U1-70K. The hub proteins, with the highest correlation to the module abundance profile (i.e. eigenprotein), are highlighted in Fig. 4B. These findings demonstrate that a weighted PPI network analysis of the U1-70K interactome successfully resolves biologically and structurally distinct complexes.
Confirmation of U1-70K BAD domain–dependent interacting proteins
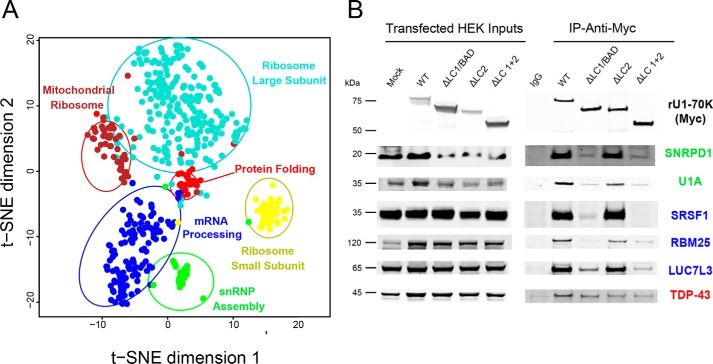
To validate and extend the module assignments, we performed both in silico and biochemical analysis. First, to visualize the relationships among modules with an independent clustering method, the T-distributed stochastic neighbor embedding (tSNE) algorithm was used to map the relatedness of proteins of top module members. The tSNE analysis largely agreed with and confirmed module assignments, whereby the majority of proteins clustered with their own module members as assigned by WGCNA (Fig. 5A). The tSNE analysis also allows for visualization of module relatedness, with similar modules in close proximity to each other and dissimilar modules further apart. For example, modules involved in translation cluster together (Fig. 5A, brown and turquoise), whereas those involved in mRNA processing (blue) and snRNP assembly (green) formed a separate cluster.
Figure 5.
Confirmation of U1-70K-interacting partners that favor interactions via the LC1/BAD domain. A, to visualize the relationship between modules and validate the WGNCA results, the tSNE algorithm was used to map the relatedness of proteins with a kME score of 0.5 or greater, which is a measure of intramodular connectivity, defined as the Pearson correlation between the expression pattern of a protein and the module eigenprotein. The tSNE analysis overlaid with module assignments determined by WGCNA allows for visualization of module relatedness, with the distance between proteins representing similarity of co-enrichment, with the more similar clusters of proteins (related modules) in closer proximity to each other compared with dissimilar modules. Proteins with similar co-enrichment across the rU1-70K co-IPs are highly correlated to one another and are related to modules with distinct biological functions (Table 1). B, Western blot analysis for select interactors of the snRNP assembly (SNRPD1 and U1A), mRNA processing (SRSF1, RBM25, and LUC7L3), and protein folding (TDP-43) modules following co-IP across the five experimental conditions (IgG, WT rU1-70K, and deletions ΔLC1, ΔLC2, and ΔLC1 + ΔLC2).
To experimentally validate the module assignments, co-IP of WT rU1-70K and deletion variants was performed followed by Western blotting for interactors of the snRNP assembly module (SNRPD1 and U1A), mRNA -processing module (SRSF1, RBM25, and LUC7L3), and the protein-folding module (TDP-43) (Fig. 5B). Members of both the mRNA processing and snRNP assembly modules have higher abundance in the WT and ΔLC2 co-IPs compared with that of the IgG, ΔLC1, and ΔLC1 + 2 IP samples. This mirrored the pattern observed for the blue and green eigenprotein values, confirming the proteomic findings (Fig. 4B). In contrast, TDP-43 shows a similar level of interaction across WT rU1-70K and the various deletions, consistent with TDP-43 being an N-terminal interactor of U1-70K and not influenced by the absence of the disordered LC domains.
mRNA processing module is enriched with RNA-binding proteins containing BAD domains
U1-70K–interacting proteins that map to the mRNA processing and snRNP assembly modules are related by their affinity for the LC1/BAD domain, yet they contain RBPs with distinct biological functions (Table 1). For example, all U1 snRNP components and assembly factors, including U1A, U1C, Sm proteins, and the SMN complex, are enriched in the green module. The SMN complex is responsible for loading the Sm proteins onto the snRNA scaffold, a critical step in U1 snRNP assembly (36). This module also contains the components of the 7SK snRNP, which regulates snRNA transcription and is present in Cajal bodies, a site of U1 snRNP maturation (37, 38). In contrast, the mRNA processing module (blue) is enriched with proteins associated with RNA splicing, polyadenylation, mRNA export, and “nuclear specks,” the latter being an analogous term for splicing speckles (39). However, it is their respective association with rU1-70K LC deletion variants that discriminates the mRNA processing and snRNP module members. For example, proteins involved in snRNP assembly are less influenced by the loss of the LC2 domain, yet associations of members of the mRNA-processing module are affected, suggesting that proteins involved in granule/speckle assembly interact in part via both LC domains of U1-70K, whereas core spliceosome assembly factors do not favor interactions with the LC2 domain (40, 41).
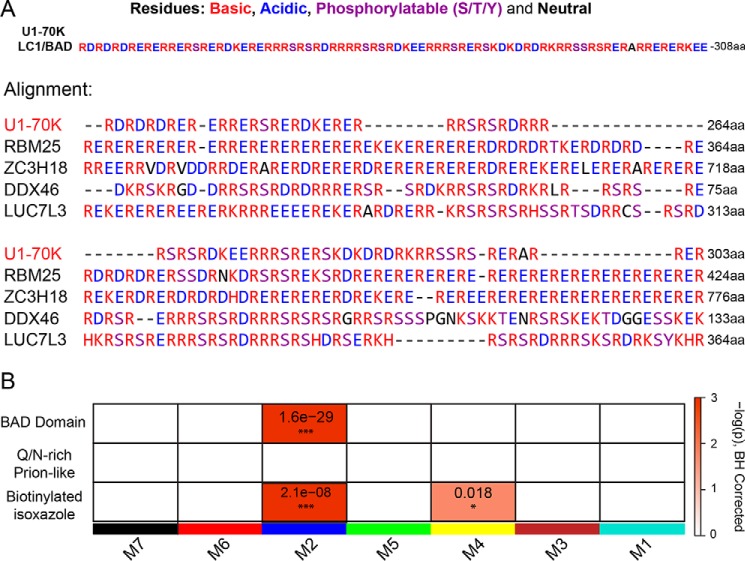
Our observations also reveal that several members of the mRNA-processing module (blue) contain stretches of highly repetitive basic (Arg/Lys) and acidic (Asp/Glu) dipeptides, analogous to the LC1/BAD domain of U1-70K (Fig. 6A). To examine the relationship between this sequence similarity and U1-70K–interacting proteins, a list of LC1-like (i.e. BAD domain) proteins was created using the Uniprot protein Basic Local Alignment Search Tool (BLAST) feature. Many proteins (n = 255) in the proteome were determined to have significant sequence overlap to the BAD domain of U1-70K. These include other members of the mRNA-processing module such as RBM25, ZC3H18, DDX46, and LUC7L3 among others. Although not identical in length, sequence alignment highlights the similar stretches of highly repetitive basic (Arg/Lys) and acidic (Asp/Glu) dipeptides across these distinct proteins (Fig. 6A). Indeed, a one-tailed Fisher's exact test revealed that the mRNA-processing module is significantly enriched with proteins harboring BAD domains (Fig. 6B). In contrast, a similar analysis comparing disordered RNA-binding proteins containing prion-like (Gln/Asn-rich) domains (42), including TDP-43 and FUS, showed no enrichment in any of the modules of U1-70K–interacting proteins. Notably, the mRNA-processing module also has a significant over-representation of nuclear proteins that selectively precipitate after treatment with biotinylated isoxazole (43). Many of these proteins participate in RNA granule assembly and form hydrogels in vitro (9, 44). Collectively, these results suggest that structurally similar BAD domains, analogous to the U1-70K LC1/BAD domain, engage in protein–protein interactions, which are essential for nuclear granule assembly and mRNA processing.
Figure 6.
mRNA-processing module is enriched with structurally similar RNA-binding proteins containing BAD domains. A, LC1/BAD domain of U1-70K (residues 231–308) contains highly repetitive dipeptide repeats of basic (Arg/Lys) and acidic (Asp/Glu) residues. A list of 255 proteins that shared greater than 20% similarity to the LC1/BAD domain of U1-70K (E-values less than 0.005) was created using the Uniprot protein BLAST feature. Using Clustal Omega, an alignment was performed on the U1-70K LC1/BAD domain and the four most structurally similar proteins to highlight the repetitive basic and acidic residues in the sequence. Residues that can be phosphorylated (Ser/Thr/Tyr) are also highlighted as they can be negatively charged after modification. B, one-tailed Fisher's exact test was used to assess structural overlap of LC1-like BAD proteins from BLAST analysis with module membership for U1-70K-interacting partners (upper panel). The same analysis was repeated using Gln/Asn-rich prion-like RNA-binding proteins (middle panel) or RNA-binding proteins that were precipitated from nuclear extracts using biotin–isoxazole compound (bottom panel). Benjamini-Hochberg multiple comparison corrected p values for the module enrichment are highlighted. Significance is demonstrated by the color scales, which go from 0 (white) to ≥3 (red), representing −log(p).
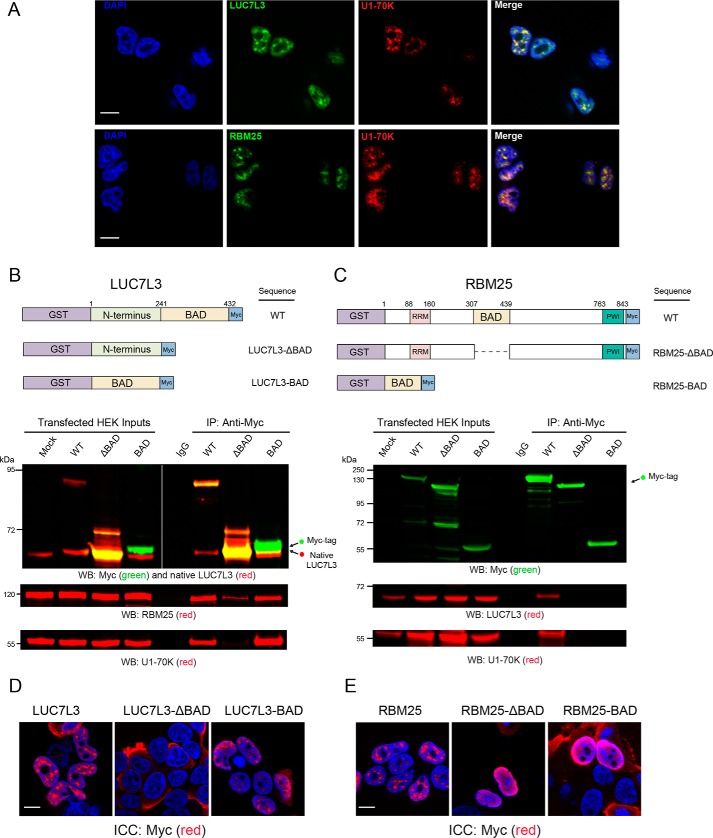
BAD domains in LUC7L3 and RBM25 are necessary for reciprocal interactions with U1-70K and nuclear granule localization
Based on their related structural and functional roles in nuclear speckle assembly and affinity for the BAD domain of U1-70K, we tested whether members of the mRNA-processing module co-localize with U1-70K in cells. Both RBM25 and LUC7L3 contain BAD domains analogous to the LC1/BAD domain of U1-70K with similar E-values of 5.5E-27 and 1.8E-26, respectively, and amino acid overlap of 55.1 and 44.2%, respectively (Fig. 6A). As expected, all three proteins localize to nuclear granules (30, 45, 46), where U1-70K shows strong co-localization with LUC7L3 and RBM25 (Fig. 7A). Our current findings support a model where the LC1/BAD domain is necessary and sufficient for U1-70K self-association and nuclear granule association. By extension, we hypothesized that the BAD domains in LUC7L3 and RBM25 would similarly be important in mediating interactions with U1-70K and other structurally similar proteins. To test this possibility, we overexpressed full-length recombinant GST-fused, Myc-tagged rRBM25 or rLUC7L3 in HEK293 cells, and their respective deletion variants lacking the BAD domains (ΔBAD) as well as the BAD domains alone followed by IP and Western blot analysis (Fig. 7B). The full-length rLUC7L3 and the BAD domain were each able to co-IP endogenous LUC7L3, mirroring the self-association observed for U1-70K (Fig. 1A). Unfortunately, the rLUC7L3-ΔBAD domain protein migrated at a similar molecular weight to endogenous LUC7L3, and thus, we were unable to determine whether the BAD domain is necessary for self-association in cells. Full-length rLUC7L3 and the BAD domain interact with endogenous U1-70K and RBM25 (Fig. 7B), whereas the rLUC7L3-ΔBAD variant does not (Fig. 7B). Similarly, full-length rRBM25 interacts with both endogenous U1-70K and LUC7L3, whereas the rRBM25-ΔBAD variant does not (Fig. 7C). In contrast, the BAD domain of rRBM25 is not sufficient to interact with U1-70K or LUC7L3, perhaps due to misprocessing, post-translation modifications, or size.
Figure 7.
BAD domains in LUC7L3 and RBM25 are necessary for reciprocal interactions with U1-70K and nuclear granule localization. A, immunocytochemistry (ICC) was performed to assess the co-localization of native U1-70K (green) with RBM25 (red) or LUC7L3 (red). DAPI-stained nuclei are shown in blue. Scale bar equates to 10 μm. B, full-length (WT) recombinant GST-fused, Myc-tagged LUC7L3 (rLUC7L3), and variants lacking the BAD domain or the BAD domain alone (upper panel) were overexpressed in HEK293 cells and immunoprecipitated with anti-Myc antibodies. IP with a nonspecific IgG was also performed from mock-transfected cells as a negative control. Western blotting (WB) for recombinant Myc-tagged proteins (green) and native LUC7L3 (red) are shown for both the inputs and co-IPs (bottom panels). Membranes were also re-probed for native U1-70K (red) or RBM25 (red). C, full-length (WT) recombinant GST-fused and Myc-tagged RBM25 (rRBM25) and deletion variants lacking the BAD domain or the BAD domain alone (upper panel) were overexpressed in HEK293 cells and immunoprecipitated with anti-Myc antibodies. A nonspecific IgG was also used on mock-transfected cells as a negative control. Western blotting for recombinant Myc-tagged proteins (green) and native U1-70K (red) or LUC7L3 (red) are shown for both the inputs and co-IPs (bottom panels). D, immunocytochemistry for full-length rLUC7L3, a variant lacking the BAD domain (LUC7L3-ΔBAD), and the BAD domain alone (LUC7L3-BAD) were expressed in HEK293 cells and visualized by confocal microscopy. E, immunocytochemistry for full-length rRBM25, a variant lacking the BAD domain (RBM25-ΔBAD), and the BAD domain alone (RBM25-BAD) were expressed in HEK293 cells and visualized by confocal microscopy. DAPI was used to visualize nuclei (blue). Scale bars are 10 μm for both D and E.
Given the role of the U1-70K LC1/BAD domain in nuclear granule localization, we sought to determine whether the BAD domains of LUC7L3 and RBM25 influence nuclear localization of these proteins to granules (Fig. 7, D and E). Both full-length rLUC7L3 and rRBM25 localize to nuclear granules by immunocytochemistry, consistent with the endogenous LUC7L3 and RBM25 localization pattern in cells (Fig. 7A). However, rLUC7L3-ΔBAD diffusely localizes to the cytoplasm, whereas rRBM25-ΔBAD is primarily nuclear but not localized to nuclear granules. Consistent with U1-70K, the BAD domain of rLUC7L3 was sufficient to localize to nuclear granules, likely due to interactions with U1-70K and other BAD RBPs. However, the BAD domain of rRBM25, which does not interact by co-IP with BAD RBPs, does not localize to nuclear granules in cells. Taken together, our findings suggest a shared functional role for BAD domains in stabilizing protein–protein interactions that likely play a role in nuclear granule assembly.
RNA-binding proteins with BAD domains have elevated insolubility in AD brain
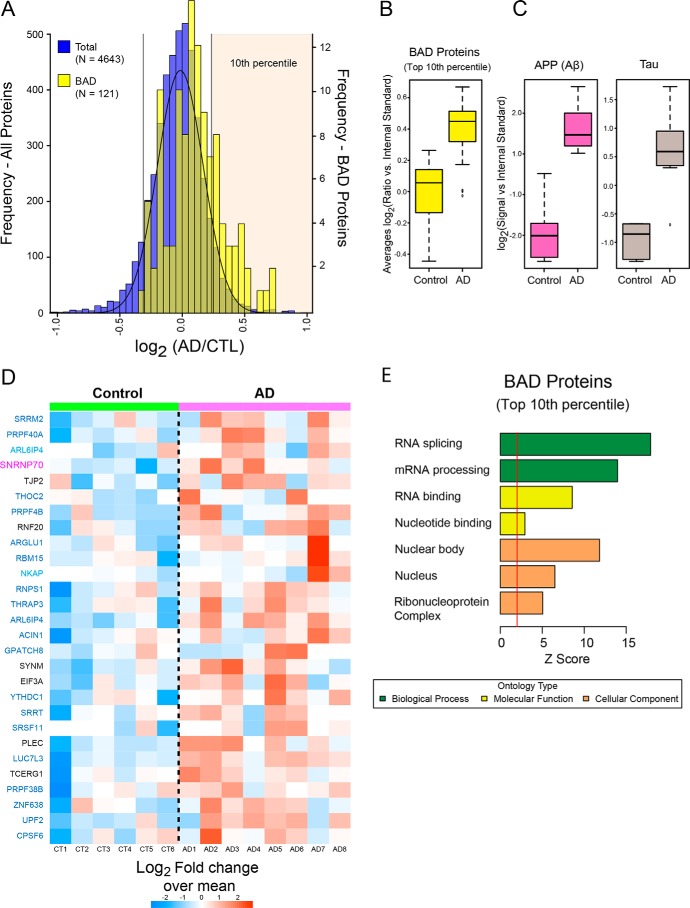
Based on the ability of U1-70K to aggregate in AD brain homogenate, and the key role of the BAD domain in U1-70K oligomerization in vitro, we hypothesized that proteins harboring similar BAD domains would preferentially aggregate in AD brain. To test this hypothesis, we assessed the distribution of insoluble proteins with BAD domains in a recently published comprehensive analysis of the Sarkosyl-insoluble proteome (n = 4,643 proteins quantified) from individual human control and AD cases (47). Protein ratios for all pairwise comparisons (i.e. control versus AD) were converted into log2 values and the resulting histogram fit to a normal Gaussian distribution (Fig. 8A). Compared with the normal distribution of all proteins in the AD insoluble proteome (blue histogram), quantified BAD proteins (yellow histogram) show a global shift toward insolubility in AD (Fig. 8A). This increase is significant using a one-tailed Fisher exact test (p value = 2.028868e-09). Consistently, BAD proteins within the top 10th percentile (n = 28) are significantly elevated in AD cases compared with controls, similar to Aβ and Tau levels (Fig. 8, B and C). Strikingly, 68% of the AD-enriched BAD proteins within the top 10th percentile, including LUC7L3, are members of the blue module identified in the rU1-70K interactome studies (Fig. 4), with shared functions in RNA binding, splicing, and processing (Fig. 8, D and E). However, RBM25 is not significantly elevated in AD brain, despite containing a BAD domain. Thus, although RBPs with BAD domains clearly have a higher likelihood of insolubility and aggregation in the AD brain, the presence of a BAD domain alone is not sufficient for aggregation.
Figure 8.
RNA-binding proteins with BAD domains have increased insolubility in AD brain. A, histogram of average log2 ratios (AD/control) for proteins measured in control (n = 6) and AD (n = 8) brain detergent-insoluble fractions. Protein ratios for all pairwise comparisons (i.e. control versus AD) were converted into log2 values, and the resulting histogram was fit to a normal Gaussian distribution. Compared with the normal distribution of all proteins in the AD insoluble proteome (blue histogram), quantified BAD proteins (n = 112 yellow histogram) showed a global shift toward insolubility in AD. BAD proteins in the top 10th percentile (n = 28) are significantly over-represented in the AD insoluble proteome (Fisher exact p value 2.0e-09). B, cumulative levels of the BAD proteins that fell into the top tenth percentile in control and AD samples. C, amyloid precursor protein (Aβ) and MAPT (Tau) protein levels in control and AD samples. The central bar depicts mean, and box edges indicate 25th and 75th percentiles, with whiskers extending to the 5th and 95th percentiles, excluding outlier measurements. D, heat map representing the fold-change over the mean of BAD proteins in the top 10th percentile across the control and AD cases. Gene symbols are displayed in text colored by their respective module color in the U1-70K interactome (black, not in a module). E, GO analysis of the 28 enriched BAD domain proteins highlights functions in RNA binding and processing. Significant over-representation of the ontology term is reflected with Z score greater than 1.96, which is equivalent to p < 0.05 (above red line).
BAD domain of U1-70K interacts with pathological Tau specifically from AD brain
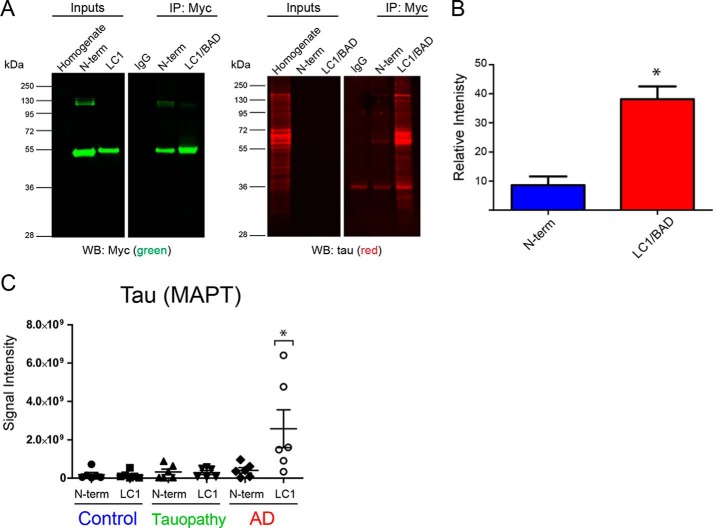
We have previously reported an association of aggregated U1-70K with Tau neurofibrillary tangles in both sporadic and familial cases of AD but not in other tauopathies (5–7, 48). However, mechanisms underlying the specificity of Tau-U1-70K co-aggregation in AD are poorly understood. Similar to the biophysical properties of RBPs, recent evidence indicates that Tau undergoes LLPS in vitro (49). This process is enhanced by polyanions, such as heparin (49) and RNA (50), as well as phosphorylation on Tau (49). Based on these observations, we sought to assess whether the LC1/BAD domain of U1-70K interacts with pathological Tau from the human AD brain. Equivalent amounts of GST-purified, Myc-tagged LC1/BAD domain or, as a control, the N-terminal domain of rU1-70K were added to AD brain homogenates and immunoprecipitated with anti-Myc antibodies followed by a Western blotting for Tau (Fig. 9A). Compared with the N-terminal domain, the LC1/BAD domain of rU1-70K co-immunoprecipitated significantly more Tau, including modified Tau species of altered molecular weights (Fig. 9, A and B).
Figure 9.
LC1/BAD domain of U1-70K interacts with Tau specifically in AD brain. A, GST-purified N-terminal domain or the LC1/BAD domain (4 μg) of rU1-70K was added separately to AD brain homogenates and immunoprecipitated with anti-Myc antibodies. IP with a nonspecific IgG was also performed as a negative control. Inputs and IPs were analyzed by Western blotting using anti-Tau antibodies (red) and Myc antibodies (green). B, LC1/BAD domain interacted with significantly higher levels of Tau from AD brain than the N-terminal domain (t test one-tail p value = 0.0156). The experiment was done in biological triplicate (n = 3) from independent AD cases with the mean and standard deviation shown. C, GST-purified N-terminal fragment or the LC1 domain (4 μg) of rU1-70K was added separately to brain homogenates from control (n = 6), AD (n = 6), or non-AD tauopathy (n = 6) brain tissue and immunoprecipitated with anti-Myc antibodies followed by mass spectrometry (MS) analysis. Label-free quantification was used to determine the signal intensities (y axis) of Tau across co-IP conditions (LC1 or N-term) and brain tissue homogenates. The mean and standard deviations for each condition are shown. Statistical significance for Tau interactions with the LC1 and N-terminal domain was determined by ANOVA. Significantly more Tau interacted with the LC1 domain in AD brain homogenates compared with all other experimental conditions (*, p value <0.05).
To test whether the interaction between the BAD domain of rU1-70K and Tau was specific to the AD brain, purified LC1/BAD or the N-terminal rU1-70K proteins were added to homogenates generated from control (n = 6), AD (n = 6), or non-AD tauopathy (n = 6) brain tissue. The latter group included progressive supranuclear palsy (n = 1) and corticobasal degeneration (n = 5) cases (51, 52). Following IP with anti-Myc antibodies, samples were analyzed by MS to identify and quantify Tau. Consistent with Western blotting results, significantly more Tau in AD brain homogenates interacted with the LC1/BAD domain compared with the N-terminal domain (Fig. 9, A–C). Furthermore, MS analysis revealed that the LC1/BAD domain interacts with pathological Tau from AD brain homogenates but not other tauopathies (Fig. 9C). This suggests that BAD domains in U1-70K and other RBPs mediate interactions with pathological Tau isoforms specific to AD.
Discussion
In this study, we show that the BAD domain of rU1-70K can directly self-interact in vitro to form high molecular weight oligomers and that this domain is also necessary and sufficient for U1-70K self-association in cells. Using quantitative proteomics, functional classes of U1-70K-interacting proteins were identified that favored interactions with the BAD domain. This analysis revealed a class of structurally similar RBPs that also contained analogous BAD LC domains. We show that for at least two other RBPs, LUC7L3 and RBM25, their respective BAD domains are required for reciprocal interactions with U1-70K and for proper localization to nuclear granules. Comprehensive analysis of the detergent-insoluble proteome in human brain revealed elevated levels of BAD RBPs in AD. Finally, we show that the LC1/BAD domain of U1-70K can interact with Tau from AD brain but not other tauopathies. This supports a hypothesis that BAD domains in U1-70K and related RBPs could mediate cooperative protein–protein interactions with Tau isoforms specifically in AD.
We propose that BAD proteins be considered a class of proteins due to their related biological function and shared primary structure. For example, we provide evidence to support the function of the LC1/BAD domain in U1-70K and more broadly other BAD domains in nuclear granule assembly. Thus, under physiological conditions the reciprocal interactions of BAD domains could form the “glue” that drives granule assembly. BAD domains, such as those found in U1-70K, RBM25, and LUC7L3, have been proposed to self-assemble through the formation of polar zippers (27). Furthermore, the promiscuous nature of BAD domain interactions could be crucial to facilitating the complicated and multifaceted cooperative function of RNA-processing proteins (16, 53). The presence of distinct LC domains like the LC2 (317–407 residues) in U1-70K may further refine the dynamics of this process. For example, U1-70K interacts with both the spliceosome and members of the polyadenylation complex, including FIP1L1. Analogous to U1-70K, FIP1L1 harbors a BAD domain and is the top hub protein in the blue module (54). Thus, the presence of BAD domains in U1-70K enable physical, if not also functional, cross-talk between the role of U1-70K in 5′-splice site recognition and the polyadenylation complex in mRNA processing.
Nuclear U1-70K is found mislocalized to cytoplasmic Tau-immunoreactive neurofibrillary aggregates in AD neurons (6), which may contribute to a loss of spliceosome function, given the recently identified RNA splicing deficits in the disease (8). Mislocalization of other RBPs also contributes to neurodegenerative disease (55, 56). Here, we show that the LC1/BAD domain is important for nuclear localization of U1-70K, supporting a link between aberrant BAD domain interactions and mislocalization of U1-70K. Our findings also shed light on previous studies in which the C-terminal domain (residues 161–437) of U1-70K was found to be sufficient for nuclear localization (30, 57). Moreover, our ΔLC1 + 2 deletion mutant mimics findings for the 1–199 U1-70K C-terminal truncation (57), wherein this N-terminal fragment localized to the nucleus but not to granules. Perhaps, the nuclear localization sequence within the LC1/BAD domain was missed by earlier studies due to the selected sites of truncation, as the LC1/BAD domain was never expressed in its entirety (57).
We show that U1-70K and other BAD proteins share many of the same properties as the Gln/Asn-rich prion-like RBPs, despite their difference in primary sequences. These properties include the homotypic selectivity to self-assemble into high molecular weight oligomers, localize to nuclear granules in cells, and promote aggregation (58). The formation of RNA granules has been viewed as an intermediary step toward protein aggregation (1, 9, 59), and our observations place U1-70K and other BAD RBPs among prion-like RBPs, such as TDP-43, FUS, hnRNPA1, and TIA-1 that associate with granules and aggregate in neurodegenerative disease (4, 21, 26, 60). Furthermore, repeat-associated non-ATG (RAN) translation of C9orf72 generates dipeptide repeat proteins (DPRs) that form pathological aggregates in ALS and frontotemporal dementia (61). Similar to our findings for the LC1/BAD domain of U1-70K, MS analysis of C9orf72 DPR-interacting proteins showed that arginine-containing DPRs, poly-Gly–Arg and poly-Pro–Arg, interact with RBPs with LC domains that mediate the assembly of RNA granules (62). Thus, we hypothesize that the ability of the LC1/BAD domain within U1-70K and other RBPs to self-interact poises them for pathological aggregation in neurodegenerative diseases, which is consistent with their increased insolubility in AD.
Both U1-70K and Tau co-localize to neurofibrillary tangles in late-onset sporadic and familial cases of AD but not in other tauopathies (5–7, 48, 63). U1-70K also aggregates in preclinical or asymptomatic AD cases (63), defined by significant Aβ deposition in the absence of significant cortical Tau deposition and cognitive impairment. Although mechanisms underlying the relationship between Aβ, Tau, and U1-70K aggregation are incompletely understood, our findings suggest that Aβ precedes and influences U1-70K interactions with Tau in brain. For example, many RBPs shuttle between the nucleus and cytoplasm (3), potentially bringing them into contact with Tau and/or under the influence of downstream intracellular signaling events triggered by extracellular Aβ (63). These signaling events could influence Tau post-translational modification and tertiary structures that favor interactions with U1-70K and BAD RBPs in AD. For example, Tau undergoes LLPS via electrostatic interactions in vitro, referred to as coacervation (50, 64, 65), mediated by the Tau microtubule-binding repeats (residues 244–369) (66). This is notable, as a recent study examining the physical structure of Tau filaments in AD brain revealed an exposed BAD motif within the tertiary structure of Tau, composed of residues 338–358 in the microtubule-binding repeat domain (67). Thus, it is tempting to speculate that pathological Tau may behave like other BAD RBPs and sequester U1-70K to neurofibrillary tangles or vice versa (Fig. 10). Specifically, we propose that this BAD surface in pathological Tau in part mediates physical interactions between Tau and the U1-70K LC1/BAD domain. These structural conformations of Tau are likely specific to AD brain, compared with other tauopathies, which may in part underlie its selective co-aggregation with U1-70K and other BAD RBPs in AD. Furthermore, Tau–U1-70K hetero-oligomers may have a unique aggregation propensity, although additional determinants of aggregation may reside in the cytoplasm, including RNA (48).
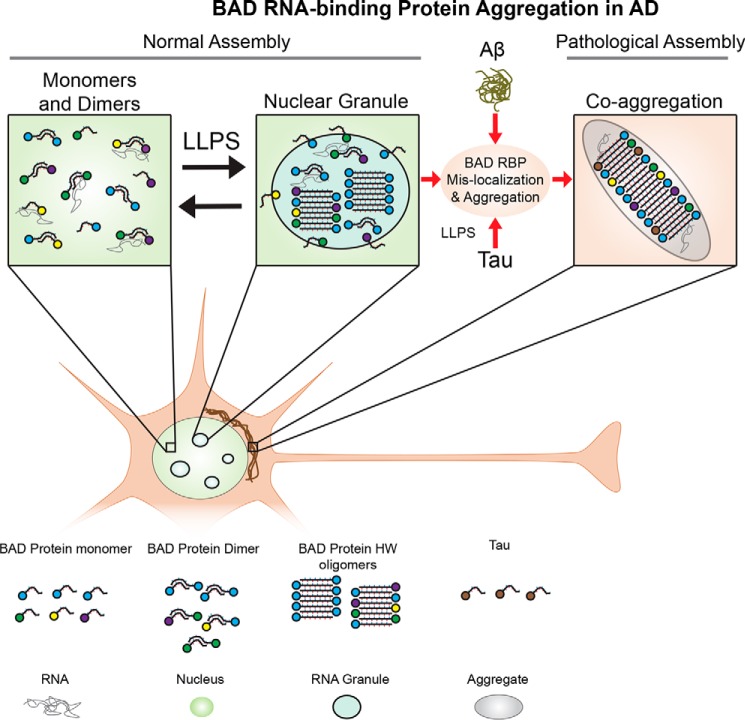
Figure 10.
Model for the assembly and pathological aggregation of U1-70K and other RNA-binding proteins with BAD domains in AD. RNA-binding proteins with BAD domains reciprocally interact to form dimers, oligomers, and RNA granules (via LLPS) under normal endogenous conditions. In AD, RBPs with BAD domains and Tau aberrantly interact in the cytoplasm to form aggregates under pathological conditions, with a mechanism likely influenced by Aβ-directed signaling events.
In summary, we have identified novel functional roles for BAD domains in protein–protein interactions, nuclear localization, granule assembly, and pathological aggregation. We show similarities between BAD domains and Gln/Asn-rich domains found in RBPs and DPRs that aggregate in neurodegenerative diseases. We also demonstrate how a weighted protein–protein interaction network analysis can be used to resolve biologically and structurally distinct complexes. Notably, RBPs with BAD domains showed elevated insolubility in AD brain, and the BAD domain of U1-70K is sufficient to interact with pathological Tau in AD brain but not other tauopathies. These findings support a hypothesis where BAD domains in U1-70K and related RBPs could mediate cooperative interactions with Tau isoforms specific to AD.
Experimental procedures
Materials
Primary antibodies used in these studies include the following: an in-house rabbit polyclonal antibody raised against a synthetic keyhole limpet hemocyanin-conjugated peptide corresponding to a C-terminal epitope of U1-70K (EM439) (7); an anti-Myc tag (clone 9B11, Cell Signaling); an anti-GST (catalog no. ab6613, Abcam) antibody; an anti-LUC7L3 (catalog no. HPA018484-100UL, Sigma); an anti-RBM25 (ab72237, Abcam); an anti-histone H3 (catalog no. ab1791, Abcam); an anti-U1-70K monoclonal (catalog no. 05-1588, Millipore); an anti-Tau (catalog no. ab54193, Abcam); and IgG mouse control (catalog no. 550339, Pharmingen). Secondary antibodies were conjugated to either Alexa Fluor 680 (Invitrogen) or IRDye800 (Rockland) fluorophores.
Plasmids and cloning
The original cDNA of U1-70K containing C-terminal Myc and DDK tags was cloned from pCMV6-Entry vector (Origene) and inserted into the HindIII/BamHI sites in the pcDNA3.1 vector (5). Full-length and U1-70K deletions sequences were subsequently cloned into the EcoRV/XhoI sites in the pLEXM-GST vector for the expression of N- and C-terminal GST-tagged proteins. Similar cloning strategies were performed using LUC7L3 (catalog no. RG214406, Origene) and RBM25 (catalog no. RC212256, Origene) plasmids. All cloning was performed by the Emory Custom Cloning Core Facility, and plasmids were confirmed by DNA sequencing.
Immunoprecipitation
Human embryonic kidney (HEK) 293T cells (CRL-3216, ATCC) were cultured in Dulbecco's modified Eagle's medium (high glucose (Gibco)) supplemented with 10% (v/v) fetal bovine serum (Gibco) and penicillin/streptomycin (Gibco) and maintained at 37 °C under a humidified atmosphere of 5% (v/v) CO2 in air. For transient transfection, the cells were grown to 80–90% confluency in 10-cm2 culture dishes and transfected with 10 μg of expression plasmid and 30 μg of linear polyethyleneimine. Cells were homogenized in ice-cold immunoprecipitation (IP) buffer containing 50 mm HEPES, pH 7.4, 150 mm NaCl, 5% glycerol, 1 mm EDTA, 0.5% (v/v) Nonidet P-40, 0.5% (v/v) CHAPS, Halt phosphatase inhibitor mixture (1:100, ThermoFisher Scientific). Samples were sonicated for 5 s on 5 s off at 30% amplitude for a total of 1.5 min (13 cycles). The samples were cleared (14,000 × g for 10 min), and protein concentrations were determined using a standard bicinchoninic acid (BCA) assay (Pierce). Protein A–Sepharose 4B beads (catalog no. 101042, Invitrogen; 20 μl per IP), were washed twice in IP buffer and then blocked with 0.1 mg/ml BSA (catalog no. 23209, ThermoFisher Scientific) and washed three additional times in IP buffer. Anti-Myc (4 μg) mouse mAb (catalog no. 2276, Cell Signaling) or 4 μg of IgG control (catalog no. 550339, Pharmingen) was allowed to incubate rotating with the bead slurry in IP buffer (500 μl) for a minimum of 90 min to allow antibody conjugation to beads. Beads were washed three times in IP buffer. The pre-cleared protein lysates were added to beads (1.5 mg per IP) and incubated by rotating overnight at 4 °C. The beads were washed three times in IP wash buffer (IP buffer without glycerol or CHAPS) by centrifugation at 500 × g for 5 min at 4 °C and then resuspended in IP wash buffer. Following the last wash, the bead suspension was transferred to a new Eppendorf tube to minimize contamination. The bound protein was eluted with 8 m urea buffered in 10 mm Tris, pH 8.0. For proteomics assays, four independent biological replicates were performed for each condition. For protein digestion, 50% of the eluted protein samples was reduced with 1 mm dithiothreitol (DTT) at 25 °C for 30 min, followed by 5 mm iodoacetamide at 25 °C for 30 min in the dark. Protein was digested with 1:100 (w/w) lysyl endopeptidase (Wako) at 25 °C for 2 h and diluted with 50 mm NH4HCO3 to a final concentration of less than 2 m urea. Samples were further digested overnight with 1:50 (w/w) trypsin (Promega) at 25 °C. Resulting peptides were desalted with in-house stage tips and dried under vacuum.
RNase A treatment
Cells were lysed in IP buffer with the addition of 5 mm MgCl2. Following sonication and centrifugation as described above, the lysates were split and treated with RNase A (Sigma) or buffer alone (control) to a final concentration of 50 μg/ml of RNase A. The RNase A treated and control samples were incubated for 30 min at room temperature followed by centrifugation at 10,000 × g for 10 min at 4 °C. The supernatant was added to beads, and the IP was completed as detailed above.
Blue native gel electrophoresis
Recombinant N-terminal (residues 1–99) and LC1/BAD U1-70K fragments (residues 231–308) were purified, and their concentrations were determined as described previously (5). The purified GST used as control was kindly gifted by the Dr. Richard Kahn (Department of Biochemistry, Emory University). Prior to analysis, purified rU1-70K fragments were cleared by centrifugation at 20,000 × g for 15 min at 4 °C to remove any insoluble precipitates. Each protein (0.8 μg) was added to blue native gel loading buffer (5% glycerol, 50 mm tris(2-carboxyethyl)phosphine, 0.02% (w/v) G-250 Coomassie, 1× Native Page Running Buffer (catalog no. BN2001, Invitrogen)) and allowed to incubate at room temperature for 30 min. Samples were loaded onto a 3–12% native-PAGE BisTris gel (catalog no. BN2011BX10, Invitrogen) in addition to a native gel molecular weight marker (catalog no. LC0725, ThermoFisher Scientific). Samples were resolved by electrophoresis at 150 V for 1.5 h in anode native-PAGE Running Buffer (catalog no. BN2001, Invitrogen) and cathode buffer with additive (catalog no. BN2002, Invitrogen). Gels were de-stained overnight in a solution of 15% (v/v) methanol and 5% (v/v) acetic acid and protein visualized on the Odyssey IR Imaging System (Li-Cor Biosciences). For Western blot analysis, native gels were prepped with a 30-min incubation at room temperature in 1% (v/v) SDS and then transferred using the semidry iblot transfer system (Invitrogen) onto nitrocellulose (catalog no. IB23001, Invitrogen).
Immunocytochemistry
HEK293 cells were plated on Matrigel (catalog no. 356234, Corning)-coated coverslips and prepared for transfection using Lipofectamine (ThermoFisher Scientific) according to the manufacturer's protocol. Immunocytochemistry was performed 48–72 h after transfection essentially as described (68). After the blocking step, slides were dabbed to remove excess liquid and incubated in primary antibody overnight at 4 °C. Primary antibodies included the following: rabbit anti-U1-70K (EM439); mouse anti-Myc; mouse anti-LUC7L3; mouse anti-RBM25; mouse anti-U1-70K. The slides were washed three times with PBS with 0.05% (v/v) saponin and then incubated with secondary antibody (Dylight 549, Alexa 488) for 1 h of shaking at room temperature. Again, the slides were washed three times with PBS with 0.05% saponin. DAPI diluted in PBS was added to each slide and incubated for at least 30 min while rotating at room temperature. Following additional rinses in PBS, cells were mounted in Vectashield (Vector Laboratories, Burlingame, CA) and sealed with nail polish. Images were captured on an Fluoview FV1000 confocal laser-scanning microscope (Olympus).
Liquid chromatography coupled to tandem MS (LC-MS/MS)
Tryptic peptides were analyzed by LC-MS/MS essentially as described (69). Peptides were resuspended in loading buffer (0.1% formic acid, 0.03% TFA, 1% acetonitrile) and separated on a self-packed C18 (1.9 μm Dr. Maisch, Germany) fused silica column (20 cm × 75 μm internal diameter; New Objective, Woburn, MA) by a NanoAcquity UHPLC (Waters) and monitored on a Q-Exactive Plus mass spectrometer (ThermoFisher Scientific, San Jose, CA). Elution was performed over a 140-min gradient at a rate of 300 nl/min with buffer B ranging from 3 to 80% (buffer A: 0.1% formic acid and 5% DMSO in water; buffer B: 0.1% formic and 5% DMSO in acetonitrile). The mass spectrometer cycle was programmed to collect one full MS scan followed by 10 data-dependent MS/MS scans. The MS scans (300–1,800 m/z range, 1,000,000 AGC, 100-ms maximum ion time) were collected at a resolution of 70,000 at m/z 200 in profile mode, and the MS/MS spectra (2 m/z isolation width, 28 normalized collision energy, 50,000 AGC target, 50 ms maximum ion time) were acquired at a resolution of 17,500 at m/z 200. Dynamic exclusion was set to exclude previous sequenced precursor ions for 30 s. Precursor ions with +1 and +6 or higher charge states were excluded from sequencing. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD008260.
Database search
Raw data files were analyzed using MaxQuant version 1.5.2.8 with Thermo Foundation 2.0 for RAW file reading capability (70). The search engine Andromeda was used to build and search a concatenated target-decoy UniProt Knowledgebase (UniProtKB) containing both Swiss-Prot and TrEMBL human reference protein sequences (90,411 target sequences downloaded April 21, 2015), plus 245 contaminant proteins included as a parameter for Andromeda search within MaxQuant (71). Methionine oxidation (+15.9949 Da), asparagine and glutamine deamidation (+0.9840 Da), and protein N-terminal acetylation (+42.0106 Da) were variable modifications (up to five allowed per peptide); cysteine was assigned a fixed carbamidomethyl modification (+57.0215 Da). Only fully tryptic peptides were considered with up to two miscleavages in the database search. A precursor mass tolerance of ±20 ppm was applied prior to mass accuracy calibration and ±4.5 ppm after internal MaxQuant calibration. Other search settings included a maximum peptide mass of 6,000 Da, a minimum peptide length of six residues, and 0.05-Da tolerance for high resolution MS/MS scans. The false discovery rate for peptide spectral matches, proteins, and site decoy fraction were all set to 1%. The LFQ algorithm in MaxQuant (33, 72) was used for protein quantitation. One limitation of data-dependent LFQ proteomics methods is the inherent missing data (i.e. missing protein identifications or abundance values), especially for low abundance proteins (73). Thus, proteins with 10 or more missing values across the 20 individual samples were not included in the subsequent bioinformatic analysis.
Protein–protein interaction network analysis
The R package WGCNA was used to sort proteins into functional groups by examining relative levels of co-enrichment (74). In WGCNA, correlation coefficients between each protein pair in the dataset are first calculated and transformed continuously with the power adjacency function to generate an adjacency matrix that defines the connection strength between protein pairs. This adjacency matrix is then used to calculate a topological matrix (TO), which measures the interconnectedness or correlation between two proteins and all other proteins in the matrix. All proteins are then hierarchically clustered (e.g. average linkage) using 1-TO as a distance measure, and module assignments are subsequently determined by dynamic tree cutting (74). Threshold power Beta for reduction of false-positive correlations (i.e. the beneficial effect of enforcing scale-free topology) was sampled in increments of 0.5, and as the target scale free topology R2 was approached, 0.1. The power selected was the lowest power at which scale-free topology R2 was ∼0.80 or, in the case of not reaching 0.80, the power at which a horizontal asymptote (plateau) was nearly approached before further increasing the power, had a negative effect on scale-free topology R2. Other parameters were selected as optimized previously for protein abundance networks (69). Thus, for the signed network built on protein LFQ abundances obtained from IP-LC-MS/MS, parameters were input into the WGCNA:: blockwiseModules() function as follows: Beta 10.9; mergeCutHeight 0.07; pamStage TRUE; pamRespectsDendro TRUE; reassignThreshold p < 0.05; deepSplit 4; minModuleSize 15; corType bicor; and maxBlockSize greater than the total number of proteins. T-Distributed Stochastic Neighbor Embedding (tSNE) analysis was performed as described (75). Proteins with WGCNA intramodular kME> = 0.50 were retained, and all duplicated values were removed, as well as proteins with any missing values for the 16 non-IgG measurements. Then Rtsne R package Barnes-Hut-Stochastic Neighbor Embedding (SNE) Rtsne function was run on the LFQ expression matrix to reduce dimensionality from 16 to 2. The remaining points or proteins (n = 375) were colored according to WGCNA module membership. GO Elite analysis on each module was performed as described previously (69).
Bioinformatic analysis of BAD proteins in the detergent-insoluble proteome in AD brain
Quantitative proteomic analysis using isobaric tagging of Sarkosyl-insoluble fractions (frontal cortex) from eight AD and six control cases was previously performed as described (47). Supplementary proteomic data were downloaded, and R was used to generate histograms, Fisher exact p values, box plots, and the clustered heat map with the Nonnegative Matrix Factorization (NMF) package.
Nuclear and cytoplasmic fractionation
The fractionation protocol was performed as essentially described in Ref. 76, with slight modifications. Briefly, after transfections with full-length rU1-70K plasmids or respective variants, HEK293 cells were harvested by scraping and washed with PBS, including 1× Protease Inhibitor Mixture (Sigma). Cells were then spun down at 1000 × g at 4 °C for 5 min and carefully resuspended in hypotonic buffer (10 mm HEPES, pH 7.9, 20 mm KCl, 0.1 mm EDTA, 1 mm DTT, 5% glycerol, 0.5 mm phenylmethylsulfonyl fluoride, and Halt phosphatase inhibitor mixture (1:100, ThermoFisher Scientific)) and incubated on ice for 15 min. The detergent Nonidet P-40 was then added to a 0.1% (v/v) final concentration, and cells were briefly agitated by vortex and left on ice for 5 additional min followed by centrifugation for 10 min at 4 °C at 15,600 × g, affording the supernatant (S1) as the cytoplasmic fraction and the pellet (P1) as the nuclear fraction. To determine whether differences in nuclear and cytoplasmic distributions were significant across conditions, repeated measures ANOVA with post-hoc Tukey was performed in GraphPad Prism.
Immunoprecipitation of rU1-70K fragments from human brain homogenates
Post-mortem frontal cortex tissue from pathologically confirmed AD cases were provided by the Emory Alzheimer's Disease Research Center (ADRC) brain bank (Table S2). Neuropathological evaluation of amyloid plaque distribution was performed according to the Consortium to Establish a Registry for AD (CERAD) semi-quantitative scoring criteria (77), whereas neurofibrillary tangle pathology was assessed in accordance with the Braak staging system (78). The corticobasal degeneration and progressive supranuclear palsy cases included in this study also underwent extensive neuropathological characterization required for diagnosis. Tissues were homogenized in Nonidet P-40 lysis buffer (25 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 5% glycerol + protease + phosphatase inhibitors) using a bullet blender (69) followed by centrifugation at 10,000 × g for 10 min at 4 °C to clear tissue debris. Immunoprecipitation was performed from 1 mg of brain homogenate from three independent AD cases. Homogenates were first pre-cleared using 30 μl of protein A-Sepharose–conjugated beads (Invitrogen catalog no. 101041) rotating at 4 °C for 1 h. GST-purified rU1-70K fragments (4 μg) were added independently to the pre-cleared homogenates. IP was performed using anti-Myc tag (clone 9B11, Cell Signaling) in samples containing the rU1-70K proteins. An IgG control antibody (catalog no. 550339, Pharmingen) was used as negative control. Immunocomplexes were captured using the Dynabeads protein G magnetic beads (catalog no. 1003D, Invitrogen), which were washed three times using wash buffer (50 mm Tris-HCl, pH 8, 150 mm NaCl, and 1% Nonidet P-40) followed by 5 min boiling in Laemmli sample buffer to elute bound proteins prior to Western blot analysis. Samples were prepared for MS as described above and analyzed on the Orbitrap Fusion mass spectrometer (ThermoFisher Scientific) (79). A total of 25 Tau (MAPT) were also added to an inclusion list to increase the likelihood of identification and quantification following database searching and quantification by MaxQuant as described above. The intensities of Tau were compared across conditions. ANOVA was used to determine significance with Tau levels across samples in GraphPad Prism.
Western blotting
Western blotting was performed according to standard procedures as reported previously (5, 10, 68). Samples in Laemmli sample buffer (8% glycerol, 2% SDS, 50 mm Tris, pH 6.8, 3.25% β-mercaptoethanol) were resolved by SDS-PAGE before an overnight wet transfer to 0.2-μm nitrocellulose membranes (Bio-Rad) or a semi-dry transfer using the iBlot2 system. Membranes were blocked with casein blocking buffer (catalog no. B6429, Sigma) and probed with primary antibodies (see under “Materials”) at a 1:1,000 dilution overnight at 4 °C. Membranes were incubated with secondary antibodies conjugated to Alexa Fluor 680 (Invitrogen) or IRDye800 (Rockland) fluorophores for 1 h at room temperature. Images were captured using an Odyssey IR Imaging System (Li-Cor Biosciences), and band intensities were quantified using Odyssey imaging software.
Assessment of protein similarity to LC1/BAD domain of U1-70K
The U1-70K LC1/BAD domain protein similarity list was created using the Uniprot pBLAST feature (http://www.uniprot.org/blast/)5 using the following parameters: Target Database Human containing 160,363 entries (updated March 2015), E-threshold: 10, Matrix: Auto, Filtering: None, Gapped: Yes, Hits: 1000. The input blast entry was the LC1/BAD domain of U1-70K (residues 231–308). The resulting list of proteins was subsequently filtered to remove unreviewed entries, producing 255 proteins with E-values less than 0.005 and similarity to the LC1/BAD domain of higher than 20% (Table S3). The “biotin-isoxazole” list originates from a previous study (43) using the biotinylated isoxazole compound to precipitate proteins from HEK293 nuclear extracts. The “prion-like” list of RNA-binding proteins originates from a previous work study using in silico methods to identify Gln/Asn-rich prion-like proteins (42). Finally, protein alignment was done using Clustal Omega multiple sequence alignment (https://www.ebi.ac.uk/Tools/msa/clustalo/)5 (80).
Author contributions
I. B., A. I. L., and N. T. S. conceptualization; I. B., E. B. D., D. M. D., and N. T. S. formal analysis; I. B., A. I. L., and N. T. S. funding acquisition; I. B., E. B. D., and N. T. S. methodology; I. B. and N. T. S. writing-original draft; I. B., E. B. D., D. M. D., S. R. K., J. J. L., A. I. L., and N. T. S. writing-review and editing; E. B. D., D. M. D., and S. R. K. investigation; M. G., A. I. L., and N. T. S. resources; A. I. L. and N. T. S. supervision.
Supplementary Material
Acknowledgments
We acknowledge Drs. Anita Corbett (Emory Department of Biology) and Daniel Reines (Emory Department of Biochemistry) for providing helpful comments to the manuscript. We also thank Dr. Measho Abreha for technical advice on brain homogenization and immunoprecipitation conditions. Finally, we acknowledge David S. Sanders who on Twitter created the Basic-Acidic Dipeptide (BAD) domain acronym after reading our paper on bioRxiv (https://www.biorxiv.org/). Research reported in this publication was supported in part by the Emory Neuroscience NINDS Core Facilities (Grant P30NS055077) from the National Institutes of Health, NINDS.
This work was supported in part by National Institutes of Health Grants 5R01AG053960 and R21AG054206 (to N. T. S.), Accelerating Medicine Partnership for AD Grant U01AG046161, and Emory Alzheimer's Disease Research Center Grant P50 AG025688. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1–S3.
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier accession no. PXD008260.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- RBP
- RNA-binding protein
- BAD
- basic-acidic dipeptide
- AD
- Alzheimer's disease
- GO
- Gene Ontology
- snRNP
- small nuclear ribonucleoprotein
- snRNA
- small nuclear RNA
- LLPS
- liquid–liquid phase separation
- co-IP
- co-immunoprecipitation
- LFQ
- label-free quantification
- WGCNA
- weighted co-expression network analysis
- PPI
- protein–protein interaction
- tSNE
- T-distributed stochastic neighbor embedding
- DPR
- dipeptide repeat protein
- IP
- immunoprecipitation
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- LFQ
- label-free quantification
- ANOVA
- analysis of variance
- DAPI
- 4,6-diamidino-2-phenylindole
- Aβ
- β-amyloid
- LC
- low complexity
- ALS
- amyotrophic lateral sclerosis
- BN-PAGE
- blue native-gel PAGE
- GST
- glutathione S-transferase
- FUS
- fused in sarcoma
- N-term
- N-terminal.
References
- 1. Ramaswami M., Taylor J. P., and Parker R. (2013) Altered ribostasis: RNA-protein granules in degenerative disorders. Cell 154, 727–736 10.1016/j.cell.2013.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wolozin B., and Apicco D. (2015) RNA-binding proteins and the genesis of neurodegenerative diseases. Adv. Exp. Med. Biol. 822, 11–15 10.1007/978-3-319-08927-0_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maziuk B., Ballance H. I., and Wolozin B. (2017) Dysregulation of RNA binding protein aggregation in neurodegenerative disorders. Front. Mol. Neurosci. 10, 89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vanderweyde T., Yu H., Varnum M., Liu-Yesucevitz L., Citro A., Ikezu T., Duff K., and Wolozin B. (2012) Contrasting pathology of the stress granule proteins TIA-1 and G3BP in tauopathies. J. Neurosci. 32, 8270–8283 10.1523/JNEUROSCI.1592-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Diner I., Hales C. M., Rabenold L., Bishof I., Duong D. M., Yi H., Laur O., Gearing M., Troncoso J., Thambisetty M., Lah J. J., Levey A. I., and Seyfried N. T. (2014) Aggregation properties of the small nuclear ribonucleoprotein U1-70K in Alzheimer disease. J. Biol. Chem. 289, 35296–35313 10.1074/jbc.M114.562959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hales C. M., Seyfried N. T., Dammer E. B., Duong D., Yi H., Gearing M., Troncoso J. C., Mufson E. J., Thambisetty M., Levey A. I., and Lah J. J. (2014) U1 small nuclear ribonucleoproteins (snRNPs) aggregate in Alzheimer's disease due to autosomal dominant genetic mutations and trisomy 21. Mol. Neurodegener. 9, 15 10.1186/1750-1326-9-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bai B., Hales C. M., Chen P. C., Gozal Y., Dammer E. B., Fritz J. J., Wang X., Xia Q., Duong D. M., Street C., Cantero G., Cheng D., Jones D. R., Wu Z., Li Y., et al. (2013) U1 small nuclear ribonucleoprotein complex and RNA splicing alterations in Alzheimer's disease. Proc. Natl. Acad. Sci. U.S.A. 110, 16562–16567 10.1073/pnas.1310249110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Raj T., Li Y., Wong G., Ramdhani S., Wang Y.-C., Ng B, Wang M., Gupta I., Haroutunian V., Zhang B., Schadt E. E., Young-Pearse T., Mostafavi S., Sklar P., Bennett D., and De Jager P. L. (2017) Integrative analyses of splicing in the aging brain: role in susceptibility to Alzheimer's disease. bioRxiv 10.1101/174565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kato M., Han T. W., Xie S., Shi K., Du X., Wu L. C., Mirzaei H., Goldsmith E. J., Longgood J., Pei J., Grishin N. V., Frantz D. E., Schneider J. W., Chen S., Li L., et al. (2012) Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Seyfried N. T., Gozal Y. M., Donovan L. E., Herskowitz J. H., Dammer E. B., Xia Q., Ku L., Chang J., Duong D. M., Rees H. D., Cooper D. S., Glass J. D., Gearing M., Tansey M. G., Lah J. J., et al. (2012) Quantitative analysis of the detergent-insoluble brain proteome in frontotemporal lobar degeneration using SILAC internal standards. J. Proteome Res. 11, 2721–2738 10.1021/pr2010814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Anderson P., and Kedersha N. (2006) RNA granules. J. Cell Biol. 172, 803–808 10.1083/jcb.200512082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Banani S. F., Rice A. M., Peeples W. B., Lin Y., Jain S., Parker R., and Rosen M. K. (2016) Compositional control of phase-separated cellular bodies. Cell 166, 651–663 10.1016/j.cell.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nott T. J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., Craggs T. D., Bazett-Jones D. P., Pawson T., Forman-Kay J. D., and Baldwin A. J. (2015) Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57, 936–947 10.1016/j.molcel.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Allen M., Wang X., Burgess J. D., Watzlawik J., Serie D. J., Younkin C. S., Nguyen T., Malphrus K. G., Lincoln S., Carrasquillo M. M., Ho C., Chakrabarty P., Strickland S., Murray M. E., Swarup V., et al. (2018) Conserved brain myelination networks are altered in Alzheimer's and other neurodegenerative diseases. Alzheimers Dement. 14, 352–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Falkenberg C. V., Carson J. H., and Blinov M. L. (2017) Multivalent molecules as modulators of RNA granule size and composition. Biophys. J. 113, 235–245 10.1016/j.bpj.2017.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Coletta A., Pinney J. W., Solís D. Y., Marsh J., Pettifer S. R., and Attwood T. K. (2010) Low-complexity regions within protein sequences have position-dependent roles. BMC Syst. Biol. 4, 43 10.1186/1752-0509-4-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li P., Banjade S., Cheng H. C., Kim S., Chen B., Guo L., Llaguno M., Hollingsworth J. V., King D. S., Banani S. F., Russo P. S., Jiang Q. X., Nixon B. T., and Rosen M. K. (2012) Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 10.1038/nature10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A. P., Kim H. J., Mittag T., and Taylor J. P. (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 10.1016/j.cell.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cohen T. J., Hwang A. W., Unger T., Trojanowski J. Q., and Lee V. M. (2012) Redox signalling directly regulates TDP-43 via cysteine oxidation and disulphide cross-linking. EMBO J. 31, 1241–1252 10.1038/emboj.2011.471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Neumann M., Rademakers R., Roeber S., Baker M., Kretzschmar H. A., and Mackenzie I. R. (2009) A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain 132, 2922–2931 10.1093/brain/awp214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kwong L. K., Neumann M., Sampathu D. M., Lee V. M., and Trojanowski J. Q. (2007) TDP-43 proteinopathy: the neuropathology underlying major forms of sporadic and familial frontotemporal lobar degeneration and motor neuron disease. Acta Neuropathol. 114, 63–70 10.1007/s00401-007-0226-5 [DOI] [PubMed] [Google Scholar]
- 22. Shaw D. J., Morse R., Todd A. G., Eggleton P., Lorson C. L., and Young P. J. (2010) Identification of a self-association domain in the Ewing's sarcoma protein: a novel function for arginine-glycine-glycine rich motifs? J. Biochem. 147, 885–893 10.1093/jb/mvq025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang I. F., Chang H. Y., Hou S. C., Liou G. G., Way T. D., and James Shen C. K. (2012) The self-interaction of native TDP-43 C terminus inhibits its degradation and contributes to early proteinopathies. Nat. Commun. 3, 766 10.1038/ncomms1766 [DOI] [PubMed] [Google Scholar]
- 24. Sreedharan J., Blair I. P., Tripathi V. B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J. C., Williams K. L., Buratti E., Baralle F., de Belleroche J., Mitchell J. D., Leigh P. N., Al-Chalabi A., et al. (2008) TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319, 1668–1672 10.1126/science.1154584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu-Yesucevitz L., Lin A. Y., Ebata A., Boon J. Y., Reid W., Xu Y. F., Kobrin K., Murphy G. J., Petrucelli L., and Wolozin B. (2014) ALS-linked mutations enlarge TDP-43-enriched neuronal RNA granules in the dendritic arbor. J. Neurosci. 34, 4167–4174 10.1523/JNEUROSCI.2350-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vance C., Rogelj B., Hortobágyi T., De Vos K. J., Nishimura A. L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P., Ganesalingam J., Williams K. L., Tripathi V., Al-Saraj S., Al-Chalabi A., et al. (2009) Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323, 1208–1211 10.1126/science.1165942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Perutz M. (1994) Polar zippers: their role in human disease. Protein Sci. 3, 1629–1637 10.1002/pro.5560031002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Apicco D. J., Ash P. E. A., Maziuk B., LeBlang C., Medalla M., Al Abdullatif A., Ferragud A., Botelho E., Ballance H. I., Dhawan U., Boudeau S., Cruz A. L., Kashy D., Wong A., Goldberg L. R., et al. (2018) Reducing the RNA-binding protein TIA1 protects against tau-mediated neurodegeneration in vivo. Nat. Neurosci. 21, 72–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wittig I., Braun H.-P., and Schägger H. (2006) Blue native PAGE. Nat. Protoc. 1, 418–428 10.1038/nprot.2006.62 [DOI] [PubMed] [Google Scholar]
- 30. Romac J. M., and Keene J. D. (1995) Overexpression of the arginine-rich carboxy-terminal region of U1 snRNP 70K inhibits both splicing and nucleocytoplasmic transport of mRNA. Genes Dev. 9, 1400–1410 10.1101/gad.9.11.1400 [DOI] [PubMed] [Google Scholar]
- 31. Stejskalová E., and Staněk D. (2014) The splicing factor U1-70K interacts with the SMN complex and is required for nuclear gem integrity. J. Cell Sci. 127, 3909–3915 10.1242/jcs.155838 [DOI] [PubMed] [Google Scholar]
- 32. Howell V. M., Jones J. M., Bergren S. K., Li L., Billi A. C., Avenarius M. R., and Meisler M. H. (2007) Evidence for a direct role of the disease modifier SCNM1 in splicing. Hum. Mol. Genet. 16, 2506–2516 10.1093/hmg/ddm206 [DOI] [PubMed] [Google Scholar]
- 33. Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., and Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Raj T., Shulman J. M., Keenan B. T., Chibnik L. B., Evans D. A., Bennett D. A., Stranger B. E., and De Jager P. L. (2012) Alzheimer disease susceptibility loci: evidence for a protein network under natural selection. Am. J. Hum. Genet. 90, 720–726 10.1016/j.ajhg.2012.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Langfelder P., Zhang B., and Horvath S. (2008) Defining clusters from a hierarchical cluster tree: the dynamic tree cut package for R. Bioinformatics 24, 719–720 10.1093/bioinformatics/btm563 [DOI] [PubMed] [Google Scholar]
- 36. Zambon A. C., Gaj S., Ho I., Hanspers K., Vranizan K., Evelo C. T., Conklin B. R., Pico A. R., and Salomonis N. (2012) GO-Elite: a flexible solution for pathway and ontology over-representation. Bioinformatics 28, 2209–2210 10.1093/bioinformatics/bts366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Egloff S., Vitali P., Tellier M., Raffel R., Murphy S., and Kiss T. (2017) The 7SK snRNP associates with the little elongation complex to promote snRNA gene expression. EMBO J. 36, 934–948 10.15252/embj.201695740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morris G. E. (2008) The Cajal body. Biochim. Biophys. Acta 1783, 2108–2115 10.1016/j.bbamcr.2008.07.016 [DOI] [PubMed] [Google Scholar]
- 39. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Seraphin B., and Rosbash M. (1989) Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell 59, 349–358 10.1016/0092-8674(89)90296-1 [DOI] [PubMed] [Google Scholar]
- 41. Kondo Y., Oubridge C., van Roon A. M., and Nagai K. (2015) Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5′ splice site recognition. Elife 4, 04986 10.7554/eLife.04986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. King O. D., Gitler A. D., and Shorter J. (2012) The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 1462, 61–80 10.1016/j.brainres.2012.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kwon I., Kato M., Xiang S., Wu L., Theodoropoulos P., Mirzaei H., Han T., Xie S., Corden J. L., and McKnight S. L. (2013) Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low complexity domains. Cell 155, 1049–1060 10.1016/j.cell.2013.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jaiswal M., Sandoval H., Zhang K., Bayat V., and Bellen H. J. (2012) Probing mechanisms that underlie human neurodegenerative diseases in Drosophila. Annu. Rev. Genet. 46, 371–396 10.1146/annurev-genet-110711-155456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhou A., Ou A. C., Cho A., Benz E. J. Jr., and Huang S. C. (2008) Novel splicing factor RBM25 modulates Bcl-x pre-mRNA 5′ splice site selection. Mol. Cell. Biol. 28, 5924–5936 10.1128/MCB.00560-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li Y., Ito M., Sun S., Chida T., Nakashima K., and Suzuki T. (2016) LUC7L3/CROP inhibits replication of hepatitis B virus via suppressing enhancer II/basal core promoter activity. Sci. Rep. 6, 36741 10.1038/srep36741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cherry J. D., Zeineddin A., Dammer E. B., Webster J. A., Duong D., Seyfried N. T., Levey A. I., Alvarez V. E., Huber B. R., Stein T. D., Kiernan P. T., McKee A. C., Lah J. J., and Hales C. M. (2018) Characterization of detergent insoluble proteome in chronic traumatic Encephalopathy. J. Neuropathol. Exp. Neurol. 77, 40–49 10.1093/jnen/nlx100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hales C. M., Dammer E. B., Diner I., Yi H., Seyfried N. T., Gearing M., Glass J. D., Montine T. J., Levey A. I., and Lah J. J. (2014) Aggregates of small nuclear ribonucleic acids (snRNAs) in Alzheimer's disease. Brain Pathol. 24, 344–351 10.1111/bpa.12133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ambadipudi S., Biernat J., Riedel D., Mandelkow E., and Zweckstetter M. (2017) Liquid–liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein Tau. Nat. Commun. 8, 275 10.1038/s41467-017-00480-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang X., Lin Y., Eschmann N. A., Zhou H., Rauch J. N., Hernandez I., Guzman E., Kosik K. S., and Han S. (2017) RNA stores tau reversibly in complex coacervates. PLoS Biol. 15, e2002183 10.1371/journal.pbio.2002183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Flament S., Delacourte A., Verny M., Hauw J.-J., and Javoy-Agid F. (1991) Abnormal Tau proteins in progressive supranuclear palsy. Acta Neuropathol. 81, 591–596 10.1007/BF00296367 [DOI] [PubMed] [Google Scholar]
- 52. Ksiezak-Reding H., Morgan K., Mattiace L. A., Davies P., Liu W. K., Yen S. H., Weidenheim K., and Dickson D. W. (1994) Ultrastructure and biochemical composition of paired helical filaments in corticobasal degeneration. Am. J. Pathol. 145, 1496–1508 [PMC free article] [PubMed] [Google Scholar]
- 53. Patil A., Kinoshita K., and Nakamura H. (2010) Domain distribution and intrinsic disorder in hubs in the human protein–protein interaction network. Protein Sci. 19, 1461–1468 10.1002/pro.425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Elkon R., Ugalde A. P., and Agami R. (2013) Alternative cleavage and polyadenylation: extent, regulation and function. Nat. Rev. Genet. 14, 496–506 10.1038/nrg3482 [DOI] [PubMed] [Google Scholar]
- 55. Scotti M. M., and Swanson M. S. (2016) RNA mis-splicing in disease. Nat. Rev. Genet. 17, 19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vance C., Scotter E. L., Nishimura A. L., Troakes C., Mitchell J. C., Kathe C., Urwin H., Manser C., Miller C. C., Hortobágyi T., Dragunow M., Rogelj B., and Shaw C. E. (2013) ALS mutant FUS disrupts nuclear localization and sequesters wild-type FUS within cytoplasmic stress granules. Hum. Mol. Genet. 22, 2676–2688 10.1093/hmg/ddt117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Romac J. M., Graff D. H., and Keene J. D. (1994) The U1 small nuclear ribonucleoprotein (snRNP) 70K protein is transported independently of U1 snRNP particles via a nuclear localization signal in the RNA-binding domain. Mol. Cell. Biol. 14, 4662–4670 10.1128/MCB.14.7.4662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Harrison A. F., and Shorter J. (2017) RNA-binding proteins with prion-like domains in health and disease. Biochem. J. 474, 1417–1438 10.1042/BCJ20160499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vanderweyde T., Apicco D. J., Youmans-Kidder K., Ash P. E. A., Cook C., Lummertz da Rocha E., Jansen-West K., Frame A. A., Citro A., Leszyk J. D., Ivanov P., Abisambra J. F., Steffen M., Li H., Petrucelli L., and Wolozin B. (2016) Interaction of tau with the RNA-binding protein TIA1 regulates tau pathophysiology and toxicity. Cell Rep. 15, 1455–1466 10.1016/j.celrep.2016.04.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kim H. J., Kim N. C., Wang Y. D., Scarborough E. A., Moore J., Diaz Z., MacLea K. S., Freibaum B., Li S., Molliex A., Kanagaraj A. P., Carter R., Boylan K. B., Wojtas A. M., Rademakers R., et al. (2013) Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495, 467–473 10.1038/nature11922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Freibaum B. D., and Taylor J. P. (2017) The role of dipeptide repeats in C9ORF72-related ALS-FTD. Front. Mol. Neurosci. 10, 35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lee K.-H., Zhang P., Kim H. J., Mitrea D. M., Sarkar M., Freibaum B. D., Cika J., Coughlin M., Messing J., Molliex A., Maxwell B. A., Kim N. C., Temirov J., Moore J., Kolaitis R.-M., Shaw T., et al. (2016) C9orf72 dipeptide repeats impair the assembly, dynamics and function of membrane-less organelles. Cell 167, 774–788.e17 10.1016/j.cell.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hales C. M., Dammer E. B., Deng Q., Duong D. M., Gearing M., Troncoso J. C., Thambisetty M., Lah J. J., Shulman J. M., Levey A. I., and Seyfried N. T. (2016) Changes in the detergent-insoluble brain proteome linked to amyloid and tau in Alzheimer's disease progression. Proteomics 16, 3042–3053 10.1002/pmic.201600057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hernández-Vega A., Braun M., Scharrel L., Jahnel M., Wegmann S., Hyman B. T., Alberti S., Diez S., and Hyman A. A. (2017) Local nucleation of microtubule bundles through tubulin concentration into a condensed tau phase. Cell Rep. 20, 2304–2312 10.1016/j.celrep.2017.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wegmann S., Eftekharzadeh B., Tepper K., Zoltowska K. M., Bennett R. E., Dujardin S., Laskowski P. R., MacKenzie D., Kamath T., Commins C., Vanderburg C., Roe A. D., Fan Z., Molliex A. M., Hernandez-Vega A., et al. (2018) Tau protein liquid–liquid phase separation can initiate tau aggregation. EMBO J. 37, e98049 10.15252/embj.201798049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ambadipudi S., Biernat J., Riedel D., Mandelkow E., and Zweckstetter M. (2017) Liquid–liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein Tau. Nat. Commun. 8, 275 10.1038/s41467-017-00480-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fitzpatrick A. W. P., Falcon B., He S., Murzin A. G., Murshudov G., Garringer H. J., Crowther R. A., Ghetti B., Goedert M., and Scheres S. H. W. (2017) Cryo-EM structures of tau filaments from Alzheimer's disease. Nature 547, 185–190 10.1038/nature23002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dammer E. B., Fallini C., Gozal Y. M., Duong D. M., Rossoll W., Xu P., Lah J. J., Levey A. I., Peng J., Bassell G. J., and Seyfried N. T. (2012) Coaggregation of RNA-binding proteins in a model of TDP-43 proteinopathy with selective RGG motif methylation and a role for RRM1 ubiquitination. PLoS One 7, e38658 10.1371/journal.pone.0038658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Seyfried N. T., Dammer E. B., Swarup V., Nandakumar D., Duong D. M., Yin L., Deng Q., Nguyen T., Hales C. M., Wingo T., Glass J., Gearing M., Thambisetty M., Troncoso J. C., Geschwind D. H., et al. (2017) A multi-network approach identifies protein-specific co-expression in asymptomatic and symptomatic Alzheimer's disease. Cell Syst. 4, 60–72.e4 10.1016/j.cels.2016.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- 71. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- 72. Luber C. A., Cox J., Lauterbach H., Fancke B., Selbach M., Tschopp J., Akira S., Wiegand M., Hochrein H., O'Keeffe M., and Mann M. (2010) Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 32, 279–289 10.1016/j.immuni.2010.01.013 [DOI] [PubMed] [Google Scholar]
- 73. Karpievitch Y. V., Dabney A. R., and Smith R. D. (2012) Normalization and missing value imputation for label-free LC-MS analysis. BMC Bioinformatics 2012 13, S5 10.1186/1471-2105-13-S16-S5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Langfelder P., and Horvath S. (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. van der Maaten L, and Hinton G. (2008) Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 [Google Scholar]
- 76. Webb C. H., and Hertel K. J. (2014) Preparation of splicing competent nuclear extracts. Methods Mol. Biol. 1126, 117–121 10.1007/978-1-62703-980-2_8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mirra S. S., Heyman A., McKeel D., Sumi S. M., Crain B. J., Brownlee L. M., Vogel F. S., Hughes J. P., van Belle G., and Berg L. (1991) The consortium to establish a registry for Alzheimer's disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer's disease. Neurology 41, 479–486 10.1212/WNL.41.4.479 [DOI] [PubMed] [Google Scholar]
- 78. Braak H., and Braak E. (1991) Neuropathological staging of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 10.1007/BF00308809 [DOI] [PubMed] [Google Scholar]
- 79. Umoh M. E., Dammer E. B., Dai J., Duong D. M., Lah J. J., Levey A. I., Gearing M., Glass J. D., and Seyfried N. T. (2018) A proteomic network approach across the ALS-FTD disease spectrum resolves clinical phenotypes and genetic vulnerability in human brain. EMBO Mol. Med. 10, 48–62 10.15252/emmm.201708202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Li W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S., Park Y. M., Buso N., and Lopez R. (2015) The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 43, W58–W584 10.1093/nar/gkv279 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.