Abstract
The BRAIN Initiative arose from a grand challenge to “accelerate the development and application of new technologies that will enable researchers to produce dynamic pictures of the brain that show how individual brain cells and complex neural circuits interact at the speed of thought.” The BRAIN Initiative is a public-private effort focused on the development and use of powerful tools for acquiring fundamental insights about how information processing occurs in the central nervous system (CNS). As the Initiative enters its fifth year, NIH has supported >500 principal investigators, who have answered the Initiative's challenge via hundreds of publications describing novel tools, methods, and discoveries that address the Initiative's seven scientific priorities. We describe scientific advances produced by individual laboratories, multi-investigator teams, and entire consortia that, over the coming decades, will produce more comprehensive and dynamic maps of the brain, deepen our understanding of how circuit activity can produce a rich tapestry of behaviors, and lay the foundation for understanding how its circuitry is disrupted in brain disorders. Much more work remains to bring this vision to fruition, and the National Institutes of Health continues to look to the diverse scientific community, from mathematics, to physics, chemistry, engineering, neuroethics, and neuroscience, to ensure that the greatest scientific benefit arises from this unique research Initiative.
Introduction
The U.S. Brain Research through Advancing Innovative Neurotechnologies (BRAIN) Initiative is the most ambitious neuroscience project to date, focusing on transforming our understanding of how the nervous system processes massive amounts of information, in real time, to generate our experience of the world and our actions in it. Truly a public-private endeavor, the National Institutes of Health (NIH) is one of myriad federal and nonfederal organizations supporting the researchers attacking the questions of the CNS (Alivisatos et al., 2012). Although the success of the BRAIN Initiative will undoubtedly rest on the shoulders of all those involved, this article highlights the NIH contribution over the past 5 years. Guided by broad input from the scientific community, BRAIN 2025: A Scientific Vision, presented to the Advisory Committee to the NIH Director (ACD) in 2013, serves as the strategic plan for the NIH BRAIN Initiative (https://braininitiative.nih.gov/2025/index.htm). Bolstered by the scientific input of the BRAIN Multi-Council Working Group (MCWG) of nongovernmental experts, BRAIN progress has accelerated with each succeeding year. With the Initiative now in its fifth year, the NIH recently made >100 new BRAIN Initiative awards, totaling $169 million (Fig. 1). This acceleration has been powered by increases in funds appropriated by Congress specifically for the Initiative, not by diversion of funds from other important neuroscience research. Despite its high profile, it is important to note that the BRAIN Initiative remains <10% of the total NIH investment in Neuroscience (https://report.nih.gov/categorical_spending.aspx). As a result of this investment, NIH has been able to support a suite of projects with a breadth and depth that previously could only be imagined. Indeed, in colloquial circles, the BRAIN Initiative has been compared with the space program and called the “moonshot between our ears.” A stipulated goal of the Initiative is to develop and disseminate tools that will advance a wide spectrum of neuroscience discovery, synergizing with the overall neuroscience investments at NIH to bolster scientific advancement. Looking ahead, these increases in funding and the need for synergy warrant the creation of a new ACD Working Group focused on the second half of the Initiative (https://acd.od.nih.gov/working-groups/brain2.0.html). BRAIN 2025 notes that the NIH BRAIN Initiative must adapt in response to the evolving scientific landscape, and this new Working Group will review Initiative progress to suggest changes to the specific goals of BRAIN 2025 and identify new and unique opportunities for research and technology development, incorporating neuroethics, and the training and empowerment of the broader neuroscience community.
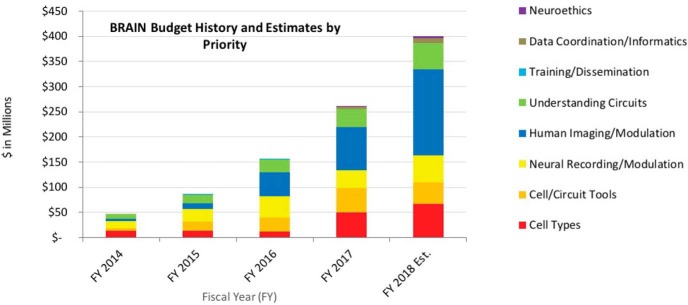
Figure 1.
Thanks to strong bipartisan support in Congress and the Innovation Funds of the 21st Century Cures Act, the budget for the BRAIN Initiative at NIH has increased each fiscal year (FY) since 2014, bringing the total investment to: $46M in FY2014, $85M in FY2015, $155M in FY2016, and $262M in FY2017. With the guidance of the BRAIN 2025 strategic plan and the expertise of BRAIN Institutes and Centers' advisory councils, scientific program staff at NIH have distributed funds across the 7 scientific goals and overarching principles of the Initiative. NIH grants are usually funded incrementally (i.e., one award for each year of the project period). Some BRAIN awards in FY2017 and FY2018 are multi-year funded awards, where funding for the entire project period (more than one year) is provided in the first fiscal year and comparing investment with previous years can become complex. The FY2018 bar represents a very preliminary estimate.
Structure of the BRAIN Initiative
Within NIH, the NIH BRAIN Initiative is managed by staff from the 10 Institutes and Centers (ICs) whose missions and current research portfolios complement the goals of the Initiative: National Center for Complementary and Integrative Health, National Eye Institute, National Institute on Aging, National Institute on Alcohol Abuse and Alcoholism, National Institute of Biomedical Imaging and Bioengineering, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institute on Drug Abuse, National Institute of Deafness and Communication Disorders, National Institute of Mental Health, and National Institute of Neurological Disorders and Stroke. To ensure that NIH keeps the rapid pace outlined in BRAIN 2025, the 10 ICs coordinate at multiple levels, integrating strategic planning, management, and support of BRAIN research across NIH. In addition, the BRAIN MCWG, comprised of scientific members from the Advisory Councils of the 10 ICs that contribute to the NIH BRAIN Initiative, plus additional ad-hoc members, provides regular input and scientific guidance.
The NIH BRAIN Initiative is part of a larger effort with federal and private partners and, indeed, has taken on global proportions. In the United States, the BRAIN Initiative Alliance seeks to coordinate and facilitate communications on BRAIN-related efforts via a centralized website and includes the Kavli Foundation, the Simons Foundation, the National Science Foundation, the Intelligence Advanced Research Projects Activity, the Food and Drug Administration, the Allen Institute for Brain Science, and NIH (www.braininitiative.org). These organizations coordinate at a number of venues, including topic-specific workshops and the annual BRAIN Principal Investigators Meeting. At the international level, NIH has memoranda of understanding with Brain Canada, the Lundbeck Foundation of Denmark, and the Australian National Health and Medical Research Council to fund investigators from these countries working on NIH-funded BRAIN Initiative projects. There is complementarity between the NIH BRAIN Initiative and the European Commission's Human Brain Project, with collaborations encouraged by both funding agencies. Further, national brain projects in Korea, Japan, China, Australia, and Israel, as well as work in other countries developing neurotechnologies, could synergize with the Initiative to enable a deeper understanding of brain circuit structure and function. Recently, a group of countries participating in an International Brain Initiative, including the United States, issued a statement emphasizing the value of international communication and collaboration to accelerate scientific progress (https://www.nsf.gov/news/news_summ.jsp?cntn_id=244044).
Strong interest has surfaced for addressing neuroethical implications of burgeoning neuroscience research efforts around the world, as captured by the annual Global Neuroethics Summit (https://globalneuroethicssummit.com/). Notably, BRAIN 2025 details the importance of integrating neuroethics throughout the research supported by BRAIN. The plan acknowledges that brain research entails ethical issues that are common to other areas of biomedical science while raising special ethical considerations as well. Given that consciousness, our innermost thoughts, and our most basic human needs arise from the brain, mechanistic studies of the brain regularly produce important social and ethical questions. Indeed, neuroscience research in general and the BRAIN Initiative specifically, with its focus on unraveling the mysteries of the human brain, generate many important ethical questions about how these new tools could be responsibly incorporated into medical research and clinical practice. In recognition of this, NIH established a Neuroethics Division of the BRAIN MCWG, which is composed of neuroethics experts and neuroscientists. This division serves to help ensure neuroethical considerations are fully integrated into the NIH BRAIN Initiative, provides input to the MCWG about ethical issues in current and proposed areas of BRAIN investment, holds consultations on individual BRAIN projects, and conveys information to investigators on identifying and addressing ethical issues. The Division organizes topical workshops that explore neuroethical implications of BRAIN Initiative research; topics to date have included research with human neural tissue and human neuroscience research using invasive and noninvasive neural devices, and privacy of brain activity data. Grounded in BRAIN research and informed by the scientific priorities outlined in BRAIN 2025, the Division developed a set of Guiding Principles to help frame and navigate the neuroethical questions that BRAIN-funded research will prompt. Additionally, in FY 2017, NIH began funding neuroethics research as part of the BRAIN Initiative. NIH relies on the perspectives and expertise of these MCWG working group and Division members, as well as the other federal and nonfederal organizations participating in the BRAIN Initiative, to form the best strategies to address the goals of the Initiative.
Circuit-focused tools and maps for a new era of neuroscience
The BRAIN Initiative is inspired by the drive to understand how our brains make us uniquely human and distinct individuals. From the discovery of electrical transmission of nerve impulses by Galvani, later explained by Hodgkin and Huxley (1952), to the complex connectivity of Cajal's Golgi-stained neurons (Cajal, 1888) and Katz's explanation of synaptic signaling (Fatt and Katz, 1952; del Castillo and Katz, 1954), neuroscience developed a more sophisticated, even molecular, understanding of the building blocks upon which information processing occurs in neural tissue. Typically, individual neuroscientists have chosen to work at specific spatial scales, ranging from the nanometer or even atomic scale of ion channels and transmitter receptors, to the intracellular level of molecular pathways, to the intercellular level of synaptic activity and its modulation, or to the systems level of transmission across the brain. Moreover, a range of temporal scales is equally daunting, from the millisecond of synaptic firing to the entire lifespan. Indeed, brain circuit activity is molded during development and by experiences throughout life. More complete descriptions are often feasible in organisms with simpler neural networks, or in specific nuclei in mammals. The breadth and extent of these scales make it daunting to attempt to advance theories that span them, yet spanning these scales is crucial to explain how the human brain actually works.
Within this diverse set of scales, the circuit is a key point of focus for two primary reasons: (1) neural circuits perform the calculations necessary to produce behavior; and (2) dysfunction at the level of the circuit is the basis of disability in many neurological and psychiatric disorders. In most psychiatric and substance use disorders, there is no pathology to be seen on brain examination. Yet, in these conditions as well as in neurological disorders, the psychiatric or neurologic signs and symptoms, which can be profoundly incapacitating, are the result of brain circuit activity gone awry. Unfortunately, the relevant brain circuit abnormalities remain invisible, relegating accurate diagnoses and rational therapy developments in many conditions to a “best guess.” The promise of the BRAIN Initiative is to facilitate an understanding of how neural circuits produce behavior, thereby enabling us to understand the nature of circuit dysfunction in brain disorders. In the service of developing a neural circuit-based understanding of the brain, and building toward a more comprehensive understanding that bridges across scales, the BRAIN Initiative consolidates its mission into three goals: (1) to build tools to monitor and modulate circuit activity; (2) to construct detailed maps of the brain that start with circuits and extend across multiple scales; and (3) to employ these tools and maps to expand knowledge of how circuits function to enable perception, emotion, cognition, and action. Guided by BRAIN 2025, these efforts are organized into seven pillars of investigation, each with an aspirational goal.
The BRAIN Initiative and the Seven Pillars
1. Discovering diversity. Identify and provide experimental access to the different brain cell types to determine their roles in health and disease.
Aspirational goal: Recent high-throughput molecular profiling of individual brain cells by RNA-sequencing and chromatin methylation analysis puts a comprehensive regional cell census of the human brain within reach of the BRAIN Initiative.
The brain is an intricate cellular matrix of neurons, glia, immune, and vascular cells that are highly ordered in networks that subserve brain-specific functions. A bottom-up approach to understanding these networks requires the ability to functionally classify and categorize cell types. Importantly the -omic-based classification of brain cell types leads not simply to a library of cells but also provides the “-omic keys” that open the door for investigators to specifically label cells for connectomic tracing, or to record and modulate their activity to define their roles in the network.
Progress to date has been remarkable. In 2014, ten 3-year pilot grants were awarded via the BRAIN Initiative to develop classification strategies and gather data/metadata for a comprehensive brain cell census. These projects focused on multiple brain regions from several vertebrates (e.g., zebrafish, mouse, rat, and human) using a variety of advanced technologies. Awardees collaborated to define collection processes and standards to describe experiments and datasets, as well as combine analyses. These BRAIN-funded projects led to technological advances in high-throughput single-cell transcriptomics (Macosko et al., 2015; Habib et al., 2016; McKenna et al., 2016; Shekhar et al., 2016; Tasic et al., 2016), epigenomics (Mo et al., 2015; Mo et al., 2016), new tools for targeting brain cell types (Deverman et al., 2016; He et al., 2016), as well as tools to establish their developmental lineage. As a sign of their potential clinical value, initial projects have described novel cell types and provided insights into mechanisms of human brain development (Pollen et al., 2015; Liu et al., 2016; Nowakowski et al., 2016a; Silbereis et al., 2016; Won et al., 2016), as well as into developmental abnormalities associated with Down syndrome (Olmos-Serrano et al., 2016) and Zika virus infection (Nowakowski et al., 2016b; Onorati et al., 2016; Retallack et al., 2016).
The high-throughput single-cell transcriptomics component has grown at such a pace that last year NIH launched a BRAIN Initiative Cell Census Network (BICCN) team science project to build a comprehensive mouse brain cell atlas and to advance scalable technologies to integrate morphological, connectivity, and physiologic data (Ecker et al., 2017). The BICCN will collaborate with a broad community to define and build the required informatics backbone for this project, allowing the universe of scientists to explore it with their own classification algorithms. Technology developments, such as classification of postmortem human brain cells by single-cell analysis of nuclei (i.e., nuclear RNA-seq) (Krishnaswami et al., 2016) or chromatin structure (Luo et al., 2017), suggest that a reference brain cell census of at least parts of the human brain may be within reach by 2025.
2. Maps at multiple scales. Generate circuit diagrams that vary in resolution from synapses to the whole brain.
Aspirational goal: Electron microscopy (EM) level connectomes of the Drosophila and zebrafish brain are under development. Major advances in tract-tracing through 3D EM datasets make EM level maps of the mouse brain and multiple human nuclei feasible. In addition, mesoscale connectome maps of the human brain at the level of a few microns might be possible.
The various cell types of the brain are in turn organized anatomically into circuits of formidable complexity. A given neuron, for example, receives myriad synapses on its intricate array of dendrites, processes those inputs, and sends outputs throughout large expanses of the brain, making multiple, ordered connections. Defining these anatomical substrates of brain processing is a challenge met by various BRAIN projects. EM-based reconstructions of brains of zebrafish (Hildebrand et al., 2017) and Drosophila (Takemura et al., 2017), as well as pieces of mouse cortex (Kasthuri et al., 2015) have demonstrated the feasibility of mapping aspects of this complexity at a fine scale, enabled by advances in computational techniques that facilitate automated segmentation and tract tracing (Fig. 2). Methods to clear and expand tissue can enable super-resolution imaging (Murray et al., 2015; Treweek et al., 2015; F. Chen et al., 2016; Ku et al., 2016; Tillberg et al., 2016; Zhang et al., 2016) on an even finer scale. Still, the scaling problem is enormous, and additional high-throughput approaches will be needed to take full advantage of these methods.
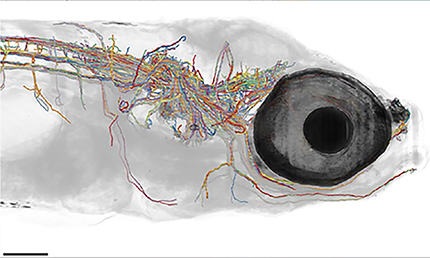
Figure 2.
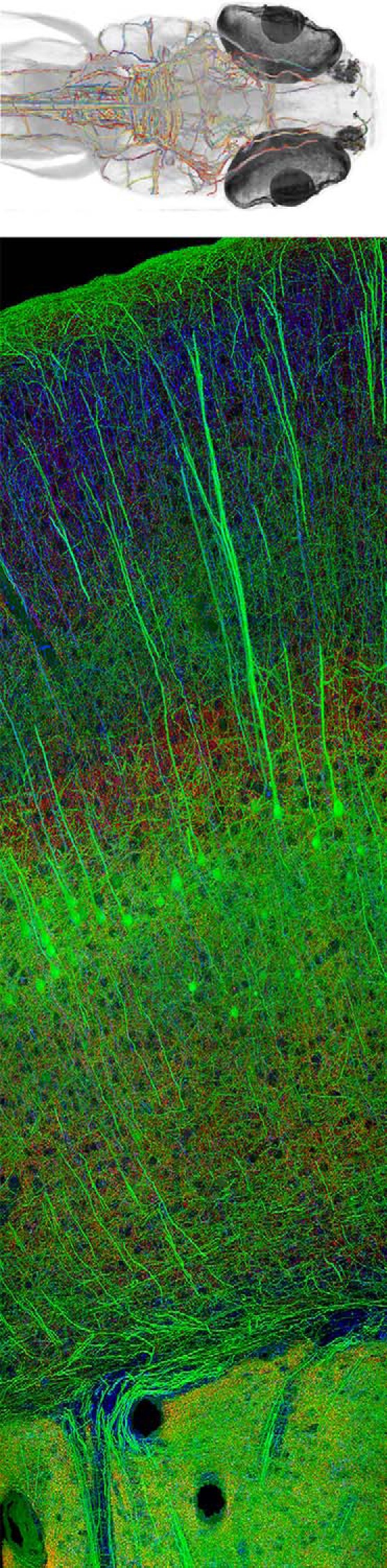
A, BRAIN Initiative scientists are using whole-brain activity imaging and connectomics to chart a detailed diagram of zebrafish brain circuits. Here is a reconstruction of myelinated axons found in one brain (see also Movie 1). Courtesy of the F. Engert laboratory (Harvard University). Reproduced with permission of Springer. B, New high-throughput imaging methods developed by BRAIN researchers offer unprecedented capabilities for imaging tissue molecular architectures. This is an example of array tomography of mouse cortex. Courtesy of the S. Smith laboratory (Stanford University).
Video of Figure 2A. 3D rotation of projections throughout zebrafish brain. Courtesy of the Engert laboratory (Harvard University).
Novel technologies promise to open up new horizons in connectivity mapping. Cell-specific reagents, such as anterograde transported viral tracers allow for dissection of connections to and from genetically identified cells, promise the possibility of cellular connectivity maps (Wall et al., 2016). Progress in “barcode” technologies that insert cell-type specific RNA labels at the synapse may better expose the brain's wiring for examination by high-throughput sequencing technologies (Peikon et al., 2017). Whether these microscale techniques can ever be scaled up to mapping of the whole human brain remains aspirational. Meanwhile, researchers are growing increasingly complex cerebral organoids in vitro derived from human pluripotent stem cells that may inform our understanding of the development of brain circuits in humans, as well as some level of their functional properties (Di Lullo and Kriegstein, 2017). This represents an area where integrated neuroethics considerations will be crucial because, as the models improve (i.e., the more accurately they approximate aspects of human brain function), the greater the level of ethical concern.
Mesoscale maps in people are possible with advanced MRI techniques. The NIH Blueprint for Neuroscience Research's Human Connectome Project acquires and shares data from structural and functional neuroimaging studies in thousands of living individuals across the lifespan (https://www.neuroscienceblueprint.nih.gov/connectome/index.htm). New techniques, such as optical computed tomography (Men et al., 2016; Tong et al., 2017) and high-density x-ray computed tomography (Dyer et al., 2017), offer the promise of high resolution, postmortem, whole-brain connectomics as gold standards for in vivo mesoscale maps.
3. Brain in action. Produce a dynamic picture of the functioning brain by developing and applying improved methods for large-scale monitoring of neural activity.
Aspirational goal: Technological advances from the BRAIN Initiative will achieve the goal of recording from ensembles of hundreds of thousands of neurons to understand how network activity underlies complex behaviors in mammals.
Integral to the BRAIN Initiative is the development of tools that enable scientists to capture and characterize circuit activity. Genetic approaches allowing cell-specific readouts of optical signals tied to brain activity, as well as advances in electrode design, have ushered in a new age of neurophysiology. Combining multiple technologies enables exploration of circuits at a new level of detail (Yamawaki et al., 2016). Additionally, recruiting scientists from engineering, physics, material science, chemistry, and other disciplines to provide solutions to the problem of capturing neural signaling with high fidelity, minimal distortion, and at multiple spatial scales simultaneously is crucial for the Initiative. Indeed, the BRAIN Initiative funded an equal number of investigators trained in engineering relative to those trained in neuroscience in 2016. Still, discovering technical solutions to these complex biological problems will require finding new, better ways to tap the tremendous skill and knowledge in a diverse set of disciplines outside of neuroscience.
In the first 2 years of the Initiative, NIH issued Requests for Applications aimed at recording and modulating neural activity with cellular resolution. The Requests for Applications targeted distinct stages in the technology cycle, including early concept development, proof-of-principle experiments, and optimization/iteration with end-users. Early published results include sophisticated electronics and novel electrodes enabling massively parallel, more stable electrophysiological recordings from neurons (Bennet et al., 2016; Liberti et al., 2016; Patel et al., 2016; Trumpis et al., 2017). Advanced optical methods are similarly enabling high-speed imaging of wide expanses of cortex in the mouse (Bouchard et al., 2015; Lauterbach et al., 2015; Kong et al., 2016; Prevedel et al., 2016), in other tissues (Horstmeyer et al., 2015; Ruan et al., 2015), or with noninvasive methods (Ouzounov et al., 2017). To achieve even deeper imaging, lightweight endoscopes have been developed both through Small Business Innovation Research funding (https://projectreporter.nih.gov/project_info_description.cfm?icde=0&aid=9255696) and a noncommercial open source alternative: www.miniscope.org/ (Cai et al., 2016). Optical voltage sensors promise even more rapid imaging of neurons and subcellular compartments (Gong et al., 2015; Abdelfattah et al., 2016; Marshall et al., 2016; Yang et al., 2016). These and other fluorophore probes are being optimized for brightness, photostability, and for red-shifted spectral characteristics for deeper brain imaging (Bajar et al., 2016; Chu et al., 2016a; Rodriguez et al., 2016). Even more sophisticated probes for subcellular calcium (Berlin et al., 2015) and neurotransmitters, such as serotonin, are showing promise (Liang et al., 2015). Finally, entirely new ways of imaging cellular-level activity, such as photoacoustic methods (Yao et al., 2015, 2016a, b; Wan et al., 2016; D. Wang et al., 2016; L. V. Wang and Yao, 2016; Y. Wang et al., 2016; Deán-Ben et al., 2017), offer the promise of expanding our ability to reach deep brain regions currently inaccessible to optical signals that become scattered as they pass through brain tissue.
Throughout the BRAIN Initiative, there is an attempt to coordinate technology development for animal experimentation that may someday be useful in humans. In that vein, multiple projects are underway to advance current technologies for invasive as well as noninvasive monitoring of human neural activity. Techniques, such as functional magnetic resonance imaging (fMRI), moved us toward being able to understand higher-order cognitive functions, but the BRAIN Initiative holds promise of delivering tools that give us unprecedented precision of measuring brain activity and accessibility for widespread use in humans. Comparing simultaneously obtained intracranial neural recordings with noninvasive signals can greatly advance the power of the latter to reveal the meaning of neural activity. For example, researchers and practicing physicians might one day use lightweight brain-scanning helmets that could precisely, rapidly, and noninvasively assess brain circuit function to facilitate diagnosis of mental illnesses. Human EEG was developed in the 1920s, and more innovative brain circuit-monitoring technology is critically needed to improve current abilities to read brain activity. If successful, there would be a host of attendant issues for societies to navigate, including the ethical implications of identifying preclinical and prodromal states for various neurodevelopmental or neurodegenerative disorders. This has been discussed at length with respect to Alzheimer's disease (see, e.g., Klein and Karlawish, 2013; Peters et al., 2013). Additionally, as we develop better tools to monitor neural activity, we will need thoughtful discussion of issues around the privacy, ownership, and use of these brain data, and consideration of how brain data are unique in a way that clinical data about other organ systems are not. The ethical implications here extend beyond the medical domain, as our improved ability to measure the neural substrate for cognition, emotion, and behavior will likely shift our conceptions of privacy, agency, and personal responsibility (Farah and Wolpe, 2004).
4. Demonstrating causality. Link brain activity to behavior with precise interventional tools that change neural circuit dynamics underlying complex behavior.
Aspirational goal: Investigators will be able to create patterns of activity in animal and human brains that cause complex behaviors.
Recording complex patterns of activity of many neurons during behavior will yield correlative information about the relationship between neuronal circuit activity and behavior. Demonstrating that these patterns are causally related to behavior requires the development of tools that can modify neuronal activity in equally complex spatiotemporal patterns. Commonly, investigators now use tools that rely on application of light or chemicals to change the firing rate of classes of neurons specifically engineered to contain responsive proteins capable of modulating neural activity or biochemistry. While the toolbox of such proteins and their actuators is expanding, the available tools have a dynamic range of effects that poorly approximate cells' natural behavior. New tools are needed to simulate the precise, complicated firing patterns that neurons use to transmit information if we are to achieve the ambitious goal of modeling the complex network activity of entire ensembles of neurons.
Accordingly, NIH is funding the development of a suite of invasive and noninvasive tools for interrogating and modulating circuits. Early published results include new silicon-based electrodes with integrated micro- Light emitting diode (LEDs), facilitating experiments that combine directed optogenetic stimulation and electrical recording (Buzsáki et al., 2015; Wu et al., 2015). New wavefront-shaping techniques permit complex 3D holography for precisely patterned optogenetic stimulation (Emiliani et al., 2015; Paluch-Siegler et al., 2015; Chaigneau et al., 2016; Conti et al., 2016; Hernandez et al., 2016). Novel channel opsins are being optimized for targeted, single-cell stimulation (Govorunova et al., 2016), as are photoswitchable receptors for precise interventions (Lin et al., 2015; Reiner et al., 2015; Berlin et al., 2016), and the toolbox for chemogenetic neuromodulation now has a novel designer ligand-receptor pair (English and Roth, 2015; Vardy et al., 2015). New strategies for noninvasive imaging and modulation include MR contrast agents for imaging calcium dynamics (Bartelle et al., 2016), which would revolutionize fMRI. Other methods under development harness radio frequency (R. Chen et al., 2015; Stanley et al., 2016), or ultrasound energy (Ibsen et al., 2015) to noninvasively modulate specific neurons.
In tandem, neuroethics research is needed to inform ethical questions that arise when considering using these tools in people: How best can we manipulate neural activity in a way that is respectful of individual agency, autonomy, and personal identity? How should we conceptualize risk in human intracranial recording and stimulation studies, and how should investigators convey such risks to research participants as part of the informed consent process? How should the informed consent process be designed to respect consent capacity fluctuations due to brain disorder, treatment, or both? These are important questions to address as our ability to manipulate neural activity increases in sophistication.
5. Identifying fundamental principles. Produce conceptual foundations for understanding the biological basis of mental processes through development of new theoretical and data analysis tools.
Aspirational goal: Teams of scientists working with high-density structural and functional maps of specific neural circuits will develop mathematical models that describe the context-dependent information processing in key brain circuits in a variety of animal models.
One might consider the discovery of a novel alphabet and its application toward a new language as a metaphor of taking a census of brain cell types and building new maps of the functioning brain. As a deluge of data on the firing patterns of ensembles of neurons (or circuits) in the context of behavior becomes available, there grows a need for new analytic methods and infrastructure established by tested theories. Expanding the metaphor, this research that links firing patterns and behaviors will tease out the words, phrases, sentences, and paragraphs in these data. The BRAIN Initiative provides a unique opportunity to assess and integrate existing theories in neuroscience. The Theories, Models, and Methods program promotes the development of new theories that can be widely used by the broader community. These theories and their resulting instantiation in mathematic applications and computational models facilitate model-driven experimental designs to help test the model's predictive power. Indeed, a complete understanding of a circuit can depend on the ability to construct robust computational models that not only describe the linked repertoire of behaviors, but also how behaviors may change by altering circuit activity (Zhou et al., 2018). Standardized protocols for data management and acquisition can be linked to metadata that describe the details of the experiments so that others can replicate, refute, or offer different theories that better fit the data. Ultimately, the goals are to reveal the building blocks of the neural code, and how the brain processes information. This aspect of the BRAIN Initiative calls for multidisciplinary research teams of scientists with sophisticated information processing skills to explore neural data, develop and apply new analytical tools, and construct models that predict behavior from the firing patterns, predict changes in behavior that result from precise circuit manipulations, and even predict firing patterns for novel behaviors. In contemplating the overall success of the Initiative, incorporating such theories and models will speed the development of transformative projects with the potential to create or overturn fundamental paradigms in neuroscience (Antic et al., 2018). In the end, only when we have complete models for systems as complex as the circuits underlying behavior will we achieve a complete understanding of the brain.
To apply newly emerging technologies toward understanding brain circuits, NIH is funding team projects to develop wide-ranging studies of circuit-level anatomy and function in the context of specific behaviors. Initial data from these studies yielded insights into circuit processing in a variety of animals and neural systems. These include small organisms, such as the larval zebrafish (Bianco and Engert, 2015; Dunn et al., 2016; Naumann et al., 2016) and fruit fly (Weir and Dickinson, 2015; Suver et al., 2016; Tuthill and Wilson, 2016; Weir et al., 2016), which allow comprehensive assays of entire circuits. In songbirds, learned behaviors can be studied on millisecond time scales aligned with neural activity (Liberti et al., 2016), and nonhuman primates offer neocortical brain regions that are direct orthologues of human counterparts (Sunkara et al., 2016). Using rodents, projects have elucidated circuit mechanisms in the retina (Baden et al., 2016; Greene et al., 2016), olfactory bulb (Chu et al., 2016b), brainstem and spinal cord (Deschênes et al., 2016), midbrain and hypothalamus (Kunwar et al., 2015; N. Chen et al., 2016; Hou et al., 2016; Xiao et al., 2016), hippocampus (Buzsáki, 2015; Danielson et al., 2016a,b; Kaifosh and Losonczy, 2016; Senzai and Buzsáki, 2017), amygdala (Namburi et al., 2015), striatum (Hunnicutt et al., 2016), thalamus (Moore et al., 2015; Morgan et al., 2016; Whitmire et al., 2016), and cortex (N. Chen et al., 2015; Rikhye and Sur, 2015; Scott et al., 2015; J. L. Chen et al., 2016; Makino et al., 2016; Sugihara et al., 2016; Watson et al., 2016; Khoshkhoo et al., 2017). In addition to the insights from these experiments, the BRAIN Initiative is supporting investigators who review and assess theories and data analysis methods to uncover fundamental principles of neural coding (Lisman, 2015; Aljadeff et al., 2016; Rajan et al., 2016; Panzeri et al., 2017) and, together with the National Science Foundation, the Initiative funds the Collaborative Research in Computational Neuroscience program.
6. Advancing human neuroscience: Develop innovative technologies to understand the human brain and treat its disorders and create and support integrated human brain research.
Aspirational goals: BRAIN technologies for recording and modulating circuit activity in patients will enable the diagnosis and treatment of neurological, psychiatric, and substance use disorders with accuracy and effectiveness that far exceed current methods and at earlier stages of illness than is currently possible.
Comparative species studies will identify structural and functional features that are shared among species as well as those that underlie unique human neural capabilities.
Advancing understanding of what gives rise to human characteristics and experiences (language, thoughts, actions) is a critical component of the BRAIN Initiative. Discovering the diversity of cell types, connections, and neural activity patterns, and demonstrating the causal relationships between these structural and functional characteristics and behavior, are the fundamental building blocks of a scientific approach toward understanding the brain. Applying this approach to the human brain is crucial if we are to attempt to measure and alter circuit activity to treat neurological, psychiatric, and substance use disorders. The Initiative's Research Opportunities in Humans program provides resources for understanding human neuroscience in unprecedented mechanistic detail by bringing together neurosurgeons, neurologists, neuroscientists, neuroengineers, and theoretical modelers with direct access to brain circuits. These interdisciplinary teams will explore underlying important human capacities. Additionally, the project teams will build our knowledge base of capacities shared across species, such as volition, emotion, decision-making and action-regulation, memory, and complex navigation, with a focus on circuitry in the human brain. This program encourages the preservation of acquired human data so that it can be shared with other researchers, maximizing the usefulness of the extremely limited data collected during clinical procedures.
To develop such comprehensive understanding, circuit technologies, such as chemogenetics, optogenetics, and optical neuroimaging, must be developed for use in humans. To this end, BRAIN investigators are developing viral vectors capable of delivering genes into specific brain regions via injections; such vectors are already being tested in human clinical trials. Their application for circuit modulation with increased precision above what is currently possible with electrical deep brain stimulation may not be far behind. Noninvasive targeting of neural circuits in animals and humans presents another area of investigation in BRAIN, including new methods to monitor neural activity, such as high-resolution fMRI measurement of cortical column activation (Feinberg et al., 2018; Vu et al., 2018), and rigorous characterization of the biological bases of fMRI signals (Bartelle et al., 2016). As noted, NIH is funding grants for the development of next-generation imaging technologies for application to humans, with promising innovations based on improved coil designs, new materials, and new contrast agents, as well as projects for novel noninvasive neuromodulation technologies (Fig. 3) (Chang et al., 2015; Bauer et al., 2016; Lu et al., 2016; Feinberg et al., 2018; Setsompop et al., 2018).
Figure 3.
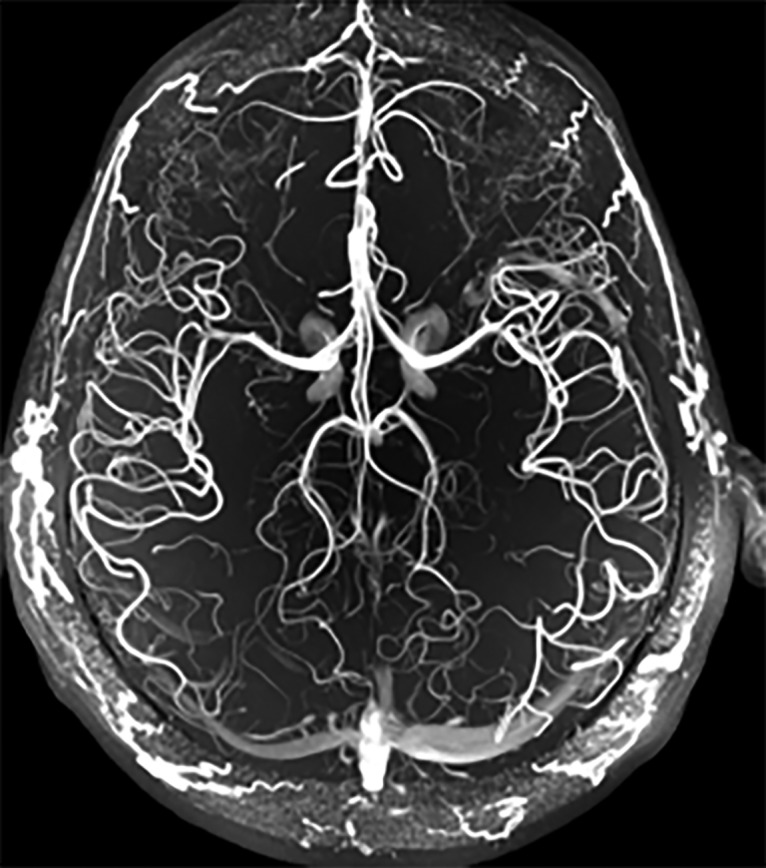
The BRAIN Initiative supports innovations in the next generation of human imaging technologies, as well as projects for novel noninvasive neuromodulation technologies. This magnetic resonance angiography highlights the vasculature throughout the human brain in high resolution, without the use of any contrast agent, on a 7 tesla MRI scanner. Courtesy of J.R. Plimeni and L.L. Wald (Massachusetts General Hospital).
While these technologies in development are the promise of the future, there are clear opportunities to improve upon and use implantable devices for novel treatments in the near term. The BRAIN Initiative supports innovative research to improve upon current electrical deep brain stimulation for neurological and psychiatric disorders. A Public-Private Partnership Program (https://braininitiative.nih.gov/resources/BRAIN_PPP/) engages commercial device manufacturers, and the Initiative funds translational projects for therapeutic benefit in a variety of disorders (with additional funded projects expected in 2018), including Parkinson's disease, essential tremor, epilepsy, obsessive-compulsive disorder, major depressive disorder, traumatic brain injury, stroke, tetraplegia, and blindness (Swann et al., 2018). These projects aim to produce early feasibility clinical studies to serve as a bridge to larger-scale efforts for market approval for a sustained therapeutic treatment.
Because of the inherent uncertainties in developing neuromodulation therapies, NIH staff monitor these projects carefully, with strict milestones for device testing and patient recruitment. This is also an area ripe for neuroethics inquiry and integration, as advances in human neuroscience carry unique ethical, legal, and social implications. In the words of the French philosopher Bruno Latour, “Science not only provides solutions, it also generates deeper and more expansive questions.” Indeed, our quest to understand the human brain better will raise profound questions about who we are, how we relate to each other, and what that might mean for the societies in which we live. The NIH BRAIN Initiative's Neuroethics Division is developing a set of points to consider for investigators in this space, regarding issues related to invasiveness, risk assessment, informed consent, and long-term obligations to implanted patients. Relatedly, the NIH BRAIN Initiative launched a neuroethics research program, with grants supporting inquiry on the neuroethical issues associated with BRAIN Initiative research, including the use of novel neurotechnologies for clinical indications.
7. From BRAIN Initiative to the brain. Integration of the new technological and conceptual approaches that are produced to discover how dynamic patterns of activity transform into cognition, emotion, perception, and action in health and disease.
Aspirational goal: Data infrastructure built for BRAIN will serve as a platform for data from across the larger neuroscience community and catalyze an even more complete understanding of brain function.
The BRAIN Initiative presents exciting, unique opportunities to explore a variety of paths to understand brain function. Countless possibilities arise by engaging teams of scientists to work in a coordinated fashion to build knowledge to answer important, often complex, questions. The integrated neuroscience projects in the BRAIN Circuit Programs (BCP) offer a new way to organize teams of investigators to bring together approaches that span methodological domains, spatiotemporal scales, and multiple species in a quantitative and model-driven manner. The aim is to understand well-defined behaviors and neural systems at real-time resolution in specifically identified circuits: that is, with in vivo mechanistic understanding of subsecond activity patterns for whole ensembles of cells, as well as the emergent neural information embedded in those patterns. The Initiative BCP teams are coordinating the data management and data science efforts of these awards to maximize the reuse of these data for any interested scientists, for years to come. Additionally, the Initiative allows the scientific community to pursue complex projects that were previously considered too labor-intensive to be undertaken in single laboratories. The BICCN comprehensive center on establishing a mouse brain atlas represents such a project. Both of these examples (BCP and BICCN) require sophisticated means for acquiring data in a standardized manner to enable data sharing among the participating laboratories to assemble the final product. Such data archives should prove valuable not only to the project scientist, but to the wider community of neuroscientists and computational scientists as well. These researchers need to enable data analyses to occur across the globe as opposed to the confines of individual laboratories. As the BRAIN Initiative builds knowledge bases along the range of spatial and temporal scales using multiple techniques, and with the application of a variety of analytic tools, the challenge becomes integrating information across the spectrum of studies to inform a complete, synthetic model of brain function. The success of the Initiative rests, in large part, on whether it can spur a cultural change in how neuroscience research is performed and its findings are shared.
The Initiative attempts to integrate technologies and conceptual approaches on all fronts, ranging from mapping cell types and monitoring circuits for understanding their function (El-Boustani and Sur, 2014; Randlett et al., 2015; Dunn et al., 2016; Morgan et al., 2016; Naumann et al., 2016; Rajan et al., 2016; Panzeri et al., 2017) to technology dissemination and therapy development (Krook-Magnuson et al., 2015; Thung et al., 2016). To tackle the challenges associated with aggregating and maintaining the diverse data types emerging from BRAIN Initiative studies, NIH supports a multipronged approach to address the unique needs associated with different subdomains of neuroscience data. In 2017, NIH funded a Brain Cell Data Center, which will integrate data collected from BRAIN-funded cell census production sites, and from collaborating investigators, and NIH released solicitations for developing standardized data descriptions, archives for cloud-based storage and community access, and software tools for data processing and analysis (Ai-Awami et al., 2016). In addition to disseminating data, NIH must determine how best to circulate the complex tools generated by the Initiative, and how to train investigators to use these new tools optimally. This requires generosity and cooperation of the tool creators, as well as an infrastructure to support manufacturing, distribution, and training with the tools. The NIH BRAIN Initiative small business grants support the commercialization of BRAIN tools, but there is concern that this may not be sufficient for some technologies. The effort, time, and expense of the process of establishing safety before use in humans could slow the translation of neurotechnologies funded by the Initiative from the preclinical phase to benefitting patient populations. Unique types of public-private partnerships could address these challenges that lie outside the scope of NIH, as established in legislation.
Conclusions and looking ahead
Informed by the BRAIN 2025 strategies and regular discussions with the scientific experts on the BRAIN MCWG, the NIH BRAIN Initiative is a dynamic and multifaceted science program focused on understanding the dynamics of information flow occurring in neural circuits. In charting the second phase of the Initiative, the NIH is embarking on a process of obtaining input from the wider scientific community. With BRAIN 2025 as a guide, the ACD BRAIN Initiative Working Group will seek to identify new specific scientific questions from high priority research areas that now can be interrogated given the emerging set of tools and neurotechnologies. This planning effort will include a focused exploration of the neuroethical implications that may arise as the science produces new neurotechnologies, and those technologies are applied toward advancing the goals of the NIH BRAIN Initiative. The excitement of the current progress will enable our partners in science to expand the vision of what the BRAIN Initiative can accomplish in the years ahead.
We are at a new threshold of discovery; already, an explosion in techniques and methods has advanced science in particular areas. More than 350 years after Descartes famously noted “I think, therefore I am,” we are finally crafting the tools to understand how we think—and how that makes us who we are. However, technological challenges remain, especially in domains requiring large-scale efforts to achieve a comprehensive analysis of circuit activity in human brains, and in recording ensemble activity deep in the brain. The BRAIN Initiative demonstrates that our inherent inquisitiveness about how the brain functions can be a powerful draw for scientists from myriad disciplines. Looking ahead, we anticipate an increasing need for innovative computational scientists and theorists to unravel the neural information code, and for scientists from physics, chemistry, material science, and bioengineering to present breakthrough technologies that enable experiments on a scale not currently possible. The experimental neuroscience community must make primary data available for analysis by the most powerful methods and strive for rich collaborations with scientists from other disciplines. Moreover, the necessity of theoretical insights to transform how we understand the brain and achieve novel paradigms should not be understated. Theorists can find areas to contribute across the entirety of the Initiative but only if quality data and metadata are available for anlysis. Together, we can develop the tools that break through our current limitations to monitor and modulate neural circuits. With diverse scientists jointly working in novel team structures, often in partnership with industry, and sharing unprecedented types and quantities of data, the BRAIN Initiative offers a unique opportunity to open the door to a golden age in brain science and improved brain health for all.
Footnotes
We thank Dr. Nancy Desmond, Dr. Christopher Thomas, Dr. Jacqueline Ward, Alissa Gallagher, and Margo Warren for their contributions to the BRAIN Initiative and the editing of this manuscript.
The authors, all federal employees of the National Institutes of Health, declare no competing financial interests.
References
- Abdelfattah AS, Farhi SL, Zhao Y, Brinks D, Zou P, Ruangkittisakul A, Platisa J, Pieribone VA, Ballanyi K, Cohen AE, Campbell RE (2016) A bright and fast red fluorescent protein voltage indicator that reports neuronal activity in organotypic brain slices. J Neurosci 36:2458–2472. 10.1523/JNEUROSCI.3484-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai-Awami AK, Beyer J, Haehn D, Kasthuri N, Lichtman JW, Pfister H, Hadwiger M (2016) NeuroBlocks: visual tracking of segmentation and proofreading for large connectomics projects. IEEE Trans Vis Comput Graph 22:738–746. 10.1109/TVCG.2015.2467441 [DOI] [PubMed] [Google Scholar]
- Alivisatos AP, Chun M, Church GM, Greenspan RJ, Roukes ML, Yuste R (2012) The brain activity map project and the challenge of functional connectomics. Neuron 74:970–974. 10.1016/j.neuron.2012.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aljadeff J, Lansdell BJ, Fairhall AL, Kleinfeld D (2016) Analysis of neuronal spike trains, deconstructed. Neuron 91:221–259. 10.1016/j.neuron.2016.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antic SD, Hines M, Lytton WW (2018) Embedded ensemble encoding hypothesis: The role of the “Prepared” cell. J Neurosci Res. Advance online publication. Retrieved Apr. 6, 2018. doi: 10.1002/jnr.24240. 10.1002/jnr.24240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden T, Berens P, Franke K, Román Rosón M, Bethge M, Euler T (2016) The functional diversity of retinal ganglion cells in the mouse. Nature 529:345–350. 10.1038/nature16468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajar BT, Wang ES, Lam AJ, Kim BB, Jacobs CL, Howe ES, Davidson MW, Lin MZ, Chu J (2016) Improving brightness and photostability of green and red fluorescent proteins for live cell imaging and FRET reporting. Sci Rep 6:20889. 10.1038/srep20889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartelle BB, Barandov A, Jasanoff A (2016) Molecular fMRI. J Neurosci 36:4139–4148. 10.1523/JNEUROSCI.4050-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer CE, Brefczynski-Lewis J, Marano G, Mandich MB, Stolin A, Martone P, Lewis JW, Jaliparthi G, Raylman RR, Majewski S (2016) Concept of an upright wearable positron emission tomography imager in humans. Brain Behav 6:e00530. 10.1002/brb3.530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennet KE, Tomshine JR, Min HK, Manciu FS, Marsh MP, Paek SB, Settell ML, Nicolai EN, Blaha CD, Kouzani AZ, Chang SY, Lee KH (2016) A diamond-based electrode for detection of neurochemicals in the human brain. Front Hum Neurosci 10:102. 10.3389/fnhum.2016.00102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin S, Carroll EC, Newman ZL, Okada HO, Quinn CM, Kallman B, Rockwell NC, Martin SS, Lagarias JC, Isacoff EY (2015) Photoactivatable genetically encoded calcium indicators for targeted neuronal imaging. Nat Methods 12:852–858. 10.1038/nmeth.3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin S, Szobota S, Reiner A, Carroll EC, Kienzler MA, Guyon A, Xiao T, Trauner D, Isacoff EY (2016) A family of photoswitchable NMDA receptors. Elife 5:1–29. 10.7554/eLife.12040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco IH, Engert F (2015) Visuomotor transformations underlying hunting behavior in zebrafish. Curr Biol 25:831–846. 10.1016/j.cub.2015.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard MB, Voleti V, Mendes CS, Lacefield C, Grueber WB, Mann RS, Bruno RM, Hillman EM (2015) Swept confocally-aligned planar excitation (SCAPE) microscopy for high speed volumetric imaging of behaving organisms. Nat Photonics 9:113–119. 10.1038/nphoton.2014.323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsáki G. (2015) Hippocampal sharp wave-ripple: a cognitive biomarker for episodic memory and planning. Hippocampus 25:1073–1188. 10.1002/hipo.22488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsáki G, Stark E, Berényi A, Khodagholy D, Kipke DR, Yoon E, Wise KD (2015) Tools for probing local circuits: high-density silicon probes combined with optogenetics. Neuron 86:92–105. 10.1016/j.neuron.2015.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai DJ, Aharoni D, Shuman T, Shobe J, Biane J, Song W, Wei B, Veshkini M, La-Vu M, Lou J, Flores SE, Kim I, Sano Y, Zhou M, Baumgaertel K, Lavi A, Kamata M, Tuszynski M, Mayford M, Golshani P, et al. (2016) A shared neural ensemble links distinct contextual memories encoded close in time. Nature 534:115–118. 10.1038/nature17955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajal SR. (1888) Estrcuctura de los centros nerviosos de las aves. Rev Trimest Histol Norm Patol 1:1–10. [Google Scholar]
- Chaigneau E, Ronzitti E, Gajowa MA, Soler-Llavina GJ, Tanese D, Brureau AY, Papagiakoumou E, Zeng H, Emiliani V (2016) Two-photon holographic stimulation of ReaChR. Front Cell Neurosci 10:234. 10.3389/fncel.2016.00234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HC, Sundman M, Petit L, Guhaniyogi S, Chu ML, Petty C, Song AW, Chen NK (2015) Human brain diffusion tensor imaging at submillimeter isotropic resolution on a 3Tesla clinical MRI scanner. Neuroimage 118:667–675. 10.1016/j.neuroimage.2015.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Wassie AT, Cote AJ, Sinha A, Alon S, Asano S, Daugharthy ER, Chang JB, Marblestone A, Church GM, Raj A, Boyden ES (2016) Nanoscale imaging of RNA with expansion microscopy. Nat Methods 13:679–684. 10.1038/nmeth.3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Voigt FF, Javadzadeh M, Krueppel R, Helmchen F (2016) Long-range population dynamics of anatomically defined neocortical networks. Elife 5:1–26. 10.7554/eLife.14679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Sugihara H, Sur M (2015) An acetylcholine-activated microcircuit drives temporal dynamics of cortical activity. Nat Neurosci 18:892–902. 10.1038/nn.4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Sugihara H, Kim J, Fu Z, Barak B, Sur M, Feng G, Han W (2016) Direct modulation of GFAP-expressing glia in the arcuate nucleus bi-directionally regulates feeding. Elife 5:1–21. 10.7554/eLife.18716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Romero G, Christiansen MG, Mohr A, Anikeeva P (2015) Wireless magnetothermal deep brain stimulation. Science 347:1477–1480. 10.1126/science.1261821 [DOI] [PubMed] [Google Scholar]
- Chu J, Oh Y, Sens A, Ataie N, Dana H, Macklin JJ, Laviv T, Welf ES, Dean KM, Zhang F, Kim BB, Tang CT, Hu M, Baird MA, Davidson MW, Kay MA, Fiolka R, Yasuda R, Kim DS, Ng HL, et al. (2016a) A bright cyan-excitable orange fluorescent protein facilitates dual-emission microscopy and enhances bioluminescence imaging in vivo. Nat Biotechnol 34:760–767. 10.1038/nbt.3550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu MW, Li WL, Komiyama T (2016b) Balancing the robustness and efficiency of odor representations during learning. Neuron 92:174–186. 10.1016/j.neuron.2016.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti R, Assayag O, de Sars V, Guillon M, Emiliani V (2016) Computer generated holography with intensity-graded patterns. Front Cell Neurosci 10:236. 10.3389/fncel.2016.00236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson NB, Zaremba JD, Kaifosh P, Bowler J, Ladow M, Losonczy A (2016a) Sublayer-specific coding dynamics during spatial navigation and learning in hippocampal area CA1. Neuron 91:652–665. 10.1016/j.neuron.2016.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson NB, Kaifosh P, Zaremba JD, Lovett-Barron M, Tsai J, Denny CA, Balough EM, Goldberg AR, Drew LJ, Hen R, Losonczy A, Kheirbek MA (2016b) Distinct contribution of adult-born hippocampal granule cells to context encoding. Neuron 90:101–112. 10.1016/j.neuron.2016.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deán-Ben XL, Gottschalk S, McLarney B, Shoham S, Razansky D (2017) Advanced optoacoustic methods for multiscale imaging of in vivo dynamics. Chem Soc Rev 46:2158–2198. 10.1039/C6CS00765A [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Castillo J, Katz B (1954) Quantal components of the end-plate potential. J Physiol 124:560–573. 10.1113/jphysiol.1954.sp005129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschênes M, Takatoh J, Kurnikova A, Moore JD, Demers M, Elbaz M, Furuta T, Wang F, Kleinfeld D (2016) Inhibition, not excitation, drives rhythmic whisking. Neuron 90:374–387. 10.1016/j.neuron.2016.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deverman BE, Pravdo PL, Simpson BP, Kumar SR, Chan KY, Banerjee A, Wu WL, Yang B, Huber N, Pasca SP, Gradinaru V (2016) Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat Biotechnol 34:204–209. 10.1038/nbt.3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lullo E, Kriegstein AR (2017) The use of brain organoids to investigate neural development and disease. Nat Rev Neurosci 18:573–584. 10.1038/nrn.2017.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn TW, Gebhardt C, Naumann EA, Riegler C, Ahrens MB, Engert F, Del Bene F (2016) Neural circuits underlying visually evoked escapes in larval zebrafish. Neuron 89:613–628. 10.1016/j.neuron.2015.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer EL, Gray Roncal W, Prasad JA, Fernandes HL, Gürsoy D, De Andrade V, Fezzaa K, Xiao X, Vogelstein JT, Jacobsen C, Körding KP, Kasthuri N (2017) Quantifying mesoscale neuroanatomy using X-ray microtomography. eNeuro 4:ENEURO.195–17.2017. 10.1523/ENEURO.0195-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker JR, Geschwind DH, Kriegstein AR, Ngai J, Osten P, Polioudakis D, Regev A, Sestan N, Wickersham IR, Zeng H (2017) The BRAIN initiative cell census consortium: lessons learned toward generating a comprehensive brain cell atlas. Neuron 96:542–557. 10.1016/j.neuron.2017.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Boustani S, Sur M (2014) Response-dependent dynamics of cell-specific inhibition in cortical networks in vivo. Nat Commun 5:5689. 10.1038/ncomms6689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emiliani V, Cohen AE, Deisseroth K, Häusser M (2015) All-optical interrogation of neural circuits. J Neurosci 35:13917–13926. 10.1523/JNEUROSCI.2916-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English JG, Roth BL (2015) Chemogenetics: a transformational and translational platform. JAMA Neurol 72:1361–1366. 10.1001/jamaneurol.2015.1921 [DOI] [PubMed] [Google Scholar]
- Farah MJ, Wolpe PR (2004) Monitoring and manipulating brain function: new neuroscience technologies and their ethical implications. Hastings Cent Rep 34:35–45. 10.2307/3528418 [DOI] [PubMed] [Google Scholar]
- Fatt P, Katz B (1952) Spontaneous subthreshold activity at motor nerve endings. J Physiol 117:109–128. 10.1113/jphysiol.1952.sp004735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg DA, Vu AT, Beckett A (2018) Pushing the limits of ultra-high resolution human brain imaging with SMS-EPI demonstrated for columnar level fMRI. Neuroimage 164:155–163. 10.1016/j.neuroimage.2017.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Huang C, Li JZ, Grewe BF, Zhang Y, Eismann S, Schnitzer MJ (2015) High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science 350:1361–1366. 10.1126/science.aab0810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorunova EG, Sineshchekov OA, Spudich JL (2016) Structurally distinct cation channelrhodopsins from cryptophyte algae. Biophys J 110:2302–2304. 10.1016/j.bpj.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene MJ, Kim JS, Seung HS, EyeWirers (2016) Analogous convergence of sustained and transient inputs in parallel on and off pathways for retinal motion computation. Cell Rep 14:1892–1900. 10.1016/j.celrep.2016.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib N, Li Y, Heidenreich M, Swiech L, Avraham-Davidi I, Trombetta JJ, Hession C, Zhang F, Regev A (2016) Div-seq: single-nucleus RNA-seq reveals dynamics of rare adult newborn neurons. Science 353:925–928. 10.1126/science.aad7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Tucciarone J, Lee S, Nigro MJ, Kim Y, Levine JM, Kelly SM, Krugikov I, Wu P, Chen Y, Gong L, Hou Y, Osten P, Rudy B, Huang ZJ (2016) Strategies and tools for combinatorial targeting of GABAergic neurons in mouse cerebral cortex. Neuron 91:1228–1243. 10.1016/j.neuron.2016.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez O, Papagiakoumou E, Tanese D, Fidelin K, Wyart C, Emiliani V (2016) Three-dimensional spatiotemporal focusing of holographic patterns. Nat Commun 7:11928. 10.1038/ncomms11928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand DG, Cicconet M, Torres RM, Choi W, Quan TM, Moon J, Wetzel AW, Scott Champion A, Graham BJ, Randlett O, Plummer GS, Portugues R, Bianco IH, Saalfeld S, Baden AD, Lillaney K, Burns R, Vogelstein JT, Schier AF, Lee WA, et al. (2017) Whole-brain serial-section electron microscopy in larval zebrafish. Nature 545:345–349. 10.1038/nature22356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin AL, Huxley AF (1952) A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol 117:500–544. 10.1113/jphysiol.1952.sp004764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstmeyer R, Ruan H, Yang C (2015) Guidestar-assisted wavefront-shaping methods for focusing light into biological tissue. Nat Photonics 9:563–571. 10.1038/nphoton.2015.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou XH, Hyun M, Taranda J, Huang KW, Todd E, Feng D, Atwater E, Croney D, Zeidel ML, Osten P, Sabatini BL (2016) Central control circuit for context-dependent micturition. Cell 167:73–86.e12. 10.1016/j.cell.2016.08.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunnicutt BJ, Jongbloets BC, Birdsong WT, Gertz KJ, Zhong H, Mao T (2016) A comprehensive excitatory input map of the striatum reveals novel functional organization. Elife 5:1–32. 10.7554/eLife.19103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibsen S, Tong A, Schutt C, Esener S, Chalasani SH (2015) Sonogenetics is a non-invasive approach to activating neurons in Caenorhabditis elegans. Nat Commun 6:8264. 10.1038/ncomms9264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaifosh P, Losonczy A (2016) Mnemonic functions for nonlinear dendritic integration in hippocampal pyramidal circuits. Neuron 90:622–634. 10.1016/j.neuron.2016.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasthuri N, Hayworth KJ, Berger DR, Schalek RL, Conchello JA, Knowles-Barley S, Lee D, Vázquez-Reina A, Kaynig V, Jones TR, Roberts M, Morgan JL, Tapia JC, Seung HS, Roncal WG, Vogelstein JT, Burns R, Sussman DL, Priebe CE, Pfister H, et al. (2015) Saturated reconstruction of a volume of neocortex. Cell 162:648–661. 10.1016/j.cell.2015.06.054 [DOI] [PubMed] [Google Scholar]
- Khoshkhoo S, Vogt D, Sohal VS (2017) Dynamic, cell-type-specific roles for GABAergic interneurons in a mouse model of optogenetically inducible seizures. Neuron 93:291–298. 10.1016/j.neuron.2016.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein E, Karlawish J (2013) Ethical issues in the neurology of aging and cognitive decline. Handb Clin Neurol 118:233–242. 10.1016/B978-0-444-53501-6.00020-2 [DOI] [PubMed] [Google Scholar]
- Kong L, Tang J, Cui M (2016) In vivo volumetric imaging of biological dynamics in deep tissue via wavefront engineering. Opt Express 24:1214–1221. 10.1364/OE.24.001214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswami SR, Grindberg RV, Novotny M, Venepally P, Lacar B, Bhutani K, Linker SB, Pham S, Erwin JA, Miller JA, Hodge R, McCarthy JK, Kelder M, McCorrison J, Aevermann BD, Fuertes FD, Scheuermann RH, Lee J, Lein ES, Schork N, et al. (2016) Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat Protoc 11:499–524. 10.1038/nprot.2016.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krook-Magnuson E, Gelinas JN, Soltesz I, Buzsáki G (2015) Neuroelectronics and biooptics: closed-loop technologies in neurological disorders. JAMA Neurol 72:823–829. 10.1001/jamaneurol.2015.0608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku T, Swaney J, Park JY, Albanese A, Murray E, Cho JH, Park YG, Mangena V, Chen J, Chung K (2016) Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat Biotechnol 34:973–981. 10.1038/nbt.3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunwar PS, Zelikowsky M, Remedios R, Cai H, Yilmaz M, Meister M, Anderson DJ (2015) Ventromedial hypothalamic neurons control a defensive emotion state. Elife 4:1–30. 10.7554/eLife.06633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauterbach MA, Ronzitti E, Sternberg JR, Wyart C, Emiliani V (2015) Fast calcium imaging with optical sectioning via HiLo microscopy. PLoS One 10:1–13. 10.1371/journal.pone.0143681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang R, Broussard GJ, Tian L (2015) Imaging chemical neurotransmission with genetically encoded fluorescent sensors. ACS Chem Neurosci 6:84–93. 10.1021/cn500280k [DOI] [PubMed] [Google Scholar]
- Liberti WA 3rd, Markowitz JE, Perkins LN, Liberti DC, Leman DP, Guitchounts G, Velho T, Kotton DN, Lois C, Gardner TJ (2016) Unstable neurons underlie a stable learned behavior. Nat Neurosci 19:1665–1671. 10.1038/nn.4405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WC, Tsai MC, Davenport CM, Smith CM, Veit J, Wilson NM, Adesnik H, Kramer RH (2015) A comprehensive optogenetic pharmacology toolkit for in vivo control of GABA(A) receptors and synaptic inhibition. Neuron 88:879–891. 10.1016/j.neuron.2015.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman J. (2015) The challenge of understanding the brain: where we stand in 2015. Neuron 86:864–882. 10.1016/j.neuron.2015.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SJ, Nowakowski TJ, Pollen AA, Lui JH, Horlbeck MA, Attenello FJ, He D, Weissman JS, Kriegstein AR, Diaz AA, Lim DA (2016) Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol 17:67. 10.1186/s13059-016-0932-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Zhu XH, Chen W (2016) In vivo P-31 MRS assessment of intracellular NAD metabolites and NAD(+)/NADH redox state in human brain at 4 T. NMR Biomed 29:1010–1017. 10.1002/nbm.3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Keown CL, Kurihara L, Zhou J, He Y, Li J, Castanon R, Lucero J, Nery JR, Sandoval JP, Bui B, Sejnowski TJ, Harkins TT, Mukamel EA, Behrens MM, Ecker JR (2017) Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science 357:600–604. 10.1126/science.aan3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA (2015) Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161:1202–1214. 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino H, Hwang EJ, Hedrick NG, Komiyama T (2016) Circuit mechanisms of sensorimotor learning. Neuron 92:705–721. 10.1016/j.neuron.2016.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall JD, Li JZ, Zhang Y, Gong Y, St-Pierre F, Lin MZ, Schnitzer MJ (2016) Cell-type-specific optical recording of membrane voltage dynamics in freely moving mice. Cell 167:1650–1662.e15. 10.1016/j.cell.2016.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Findlay GM, Gagnon JA, Horwitz MS, Schier AF, Shendure J (2016) Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353:1–17. 10.1126/science.aaf7907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men J, Huang Y, Solanki J, Zeng X, Alex A, Jerwick J, Zhang Z, Tanzi RE, Li A, Zhou C (2016) Optical coherence tomography for brain imaging and developmental biology. IEEE J Sel Top Quantum Electron 22:6803213. 10.1109/JSTQE.2015.2513667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Picard S, Urich MA, Nery JR, Sejnowski TJ, Lister R, Eddy SR, Ecker JR, Nathans J (2015) Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86:1369–1384. 10.1016/j.neuron.2015.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo A, Luo C, Davis FP, Mukamel EA, Henry GL, Nery JR, Urich MA, Picard S, Lister R, Eddy SR, Beer MA, Ecker JR, Nathans J (2016) Epigenomic landscapes of retinal rods and cones. Elife 5:1–29. 10.7554/eLife.11613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JD, Mercer Lindsay N, Deschênes M, Kleinfeld D (2015) Vibrissa self-motion and touch are reliably encoded along the same somatosensory pathway from brainstem through thalamus. PLoS Biol 13:e1002253. 10.1371/journal.pbio.1002253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JL, Berger DR, Wetzel AW, Lichtman JW (2016) The fuzzy logic of network connectivity in mouse visual thalamus. Cell 165:192–206. 10.1016/j.cell.2016.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray E, Cho JH, Goodwin D, Ku T, Swaney J, Kim SY, Choi H, Park YG, Park JY, Hubbert A, McCue M, Vassallo S, Bakh N, Frosch MP, Wedeen VJ, Seung HS, Chung K (2015) Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. Cell 163:1500–1514. 10.1016/j.cell.2015.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namburi P, Beyeler A, Yorozu S, Calhoon GG, Halbert SA, Wichmann R, Holden SS, Mertens KL, Anahtar M, Felix-Ortiz AC, Wickersham IR, Gray JM, Tye KM (2015) A circuit mechanism for differentiating positive and negative associations. Nature 520:675–678. 10.1038/nature14366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann EA, Fitzgerald JE, Dunn TW, Rihel J, Sompolinsky H, Engert F (2016) From whole-brain data to functional circuit models: the zebrafish optomotor response. Cell 167:947–960.e20. 10.1016/j.cell.2016.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ, Pollen AA, Sandoval-Espinosa C, Kriegstein AR (2016a) Transformation of the radial glia scaffold demarcates two stages of human cerebral cortex development. Neuron 91:1219–1227. 10.1016/j.neuron.2016.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ, Pollen AA, Di Lullo E, Sandoval-Espinosa C, Bershteyn M, Kriegstein AR (2016b) Expression analysis highlights AXL as a candidate Zika virus entry receptor in neural stem cells. Cell Stem Cell 18:591–596. 10.1016/j.stem.2016.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos-Serrano JL, Kang HJ, Tyler WA, Silbereis JC, Cheng F, Zhu Y, Pletikos M, Jankovic-Rapan L, Cramer NP, Galdzicki Z, Goodliffe J, Peters A, Sethares C, Delalle I, Golden JA, Haydar TF, Sestan N (2016) Down syndrome developmental brain transcriptome reveals defective oligodendrocyte differentiation and myelination. Neuron 89:1208–1222. 10.1016/j.neuron.2016.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onorati M, Li Z, Liu F, Sousa AM, Nakagawa N, Li M, Dell'Anno MT, Gulden FO, Pochareddy S, Tebbenkamp AT, Han W, Pletikos M, Gao T, Zhu Y, Bichsel C, Varela L, Szigeti-Buck K, Lisgo S, Zhang Y, Testen A, et al. (2016) Zika virus disrupts phospho-TBK1 localization and mitosis in human neuroepithelial stem cells and radial glia. Cell Rep 16:2576–2592. 10.1016/j.celrep.2016.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouzounov DG, Wang T, Wang M, Feng DD, Horton NG, Cruz-Hernández JC, Cheng YT, Reimer J, Tolias AS, Nishimura N, Xu C (2017) In vivo three-photon imaging of activity of GCaMP6-labeled neurons deep in intact mouse brain. Nat Methods 14:388–390. 10.1038/nmeth.4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluch-Siegler S, Mayblum T, Dana H, Brosh I, Gefen I, Shoham S (2015) All-optical bidirectional neural interfacing using hybrid multiphoton holographic optogenetic stimulation. Neurophotonics 2:031208. 10.1117/1.NPh.2.3.031208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzeri S, Harvey CD, Piasini E, Latham PE, Fellin T (2017) Cracking the neural code for sensory perception by combining statistics, intervention, and behavior. Neuron 93:491–507. 10.1016/j.neuron.2016.12.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PR, Zhang H, Robbins MT, Nofar JB, Marshall SP, Kobylarek MJ, Kozai TD, Kotov NA, Chestek CA (2016) Chronic in vivo stability assessment of carbon fiber microelectrode arrays. J Neural Eng 13:066002. 10.1088/1741-2560/13/6/066002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peikon ID, Kebschull JM, Vagin VV, Ravens DI, Sun YC, Brouzes E, Corrêa IR Jr, Bressan D, Zador AM (2017) Using high-throughput barcode sequencing to efficiently map connectomes. Nucleic Acids Res 45:e115. 10.1093/nar/gkx292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters KR, Lynn Beattie B, Feldman HH, Illes J (2013) A conceptual framework and ethics analysis for prevention trials of alzheimer disease. Prog Neurobiol 110:114–123. 10.1016/j.pneurobio.2012.12.001 [DOI] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Chen J, Retallack H, Sandoval-Espinosa C, Nicholas CR, Shuga J, Liu SJ, Oldham MC, Diaz A, Lim DA, Leyrat AA, West JA, Kriegstein AR (2015) Molecular identity of human outer radial glia during cortical development. Cell 163:55–67. 10.1016/j.cell.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevedel R, Verhoef AJ, Pernía-Andrade AJ, Weisenburger S, Huang BS, Nöbauer T, Fernández A, Delcour JE, Golshani P, Baltuska A, Vaziri A (2016) Fast volumetric calcium imaging across multiple cortical layers using sculpted light. Nat Methods 13:1021–1028. 10.1038/nmeth.4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan K, Harvey CD, Tank DW (2016) Recurrent network models of sequence generation and memory. Neuron 90:128–142. 10.1016/j.neuron.2016.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randlett O, Wee CL, Naumann EA, Nnaemeka O, Schoppik D, Fitzgerald JE, Portugues R, Lacoste AM, Riegler C, Engert F, Schier AF (2015) Whole-brain activity mapping onto a zebrafish brain atlas. Nat Methods 12:1039–1046. 10.1038/nmeth.3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A, Levitz J, Isacoff EY (2015) Controlling ionotropic and metabotropic glutamate receptors with light: principles and potential. Curr Opin Pharmacol 20:135–143. 10.1016/j.coph.2014.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retallack H, Di Lullo E, Arias C, Knopp KA, Laurie MT, Sandoval-Espinosa C, Mancia Leon WR, Krencik R, Ullian EM, Spatazza J, Pollen AA, Mandel-Brehm C, Nowakowski TJ, Kriegstein AR, DeRisi JL (2016) Zika virus cell tropism in the developing human brain and inhibition by azithromycin. Proc Natl Acad Sci U S A 113:14408–14413. 10.1073/pnas.1618029113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikhye RV, Sur M (2015) Spatial correlations in natural scenes modulate response reliability in mouse visual cortex. J Neurosci 35:14661–14680. 10.1523/JNEUROSCI.1660-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez EA, Tran GN, Gross LA, Crisp JL, Shu X, Lin JY, Tsien RY (2016) A far-red fluorescent protein evolved from a cyanobacterial phycobiliprotein. Nat Methods 13:763–769. 10.1038/nmeth.3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan H, Jang M, Yang C (2015) Optical focusing inside scattering media with time-reversed ultrasound microbubble encoded light. Nat Commun 6:8968. 10.1038/ncomms9968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott BB, Constantinople CM, Erlich JC, Tank DW, Brody CD (2015) Sources of noise during accumulation of evidence in unrestrained and voluntarily head-restrained rats. Elife 4:1–23. 10.7554/eLife.11308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senzai Y, Buzsáki G (2017) Physiological properties and behavioral correlates of hippocampal granule cells and mossy cells. Neuron 93:691–704.e5. 10.1016/j.neuron.2016.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setsompop K, Fan Q, Stockmann J, Bilgic B, Huang S, Cauley SF, Nummenmaa A, Wang F, Rathi Y, Witzel T, Wald LL (2018) High-resolution in vivo diffusion imaging of the human brain with generalized slice dithered enhanced resolution: Simultaneous multislice (gSlider-SMS). Magn Reson Med 79:141–151. 10.1002/mrm.26653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, McCarroll SA, Cepko CL, Regev A, Sanes JR (2016) Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell 166:1308–1323.e30. 10.1016/j.cell.2016.07.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silbereis JC, Pochareddy S, Zhu Y, Li M, Sestan N (2016) The cellular and molecular landscapes of the developing human central nervous system. Neuron 89:248–268. 10.1016/j.neuron.2015.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SA, Kelly L, Latcha KN, Schmidt SF, Yu X, Nectow AR, Sauer J, Dyke JP, Dordick JS, Friedman JM (2016) Bidirectional electromagnetic control of the hypothalamus regulates feeding and metabolism. Nature 531:647–650. 10.1038/nature17183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara H, Chen N, Sur M (2016) Cell-specific modulation of plasticity and cortical state by cholinergic inputs to the visual cortex. J Physiol Paris 110:37–43. 10.1016/j.jphysparis.2016.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkara A, DeAngelis GC, Angelaki DE (2016) Joint representation of translational and rotational components of optic flow in parietal cortex. Proc Natl Acad Sci U S A 113:5077–5082. 10.1073/pnas.1604818113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suver MP, Huda A, Iwasaki N, Safarik S, Dickinson MH (2016) An array of descending visual interneurons encoding self-motion in Drosophila. J Neurosci 36:11768–11780. 10.1523/JNEUROSCI.2277-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swann NC, de Hemptinne C, Thompson MC, Miocinovic S, Miller AM, Gilron R, Ostrem JL, Chizeck HJ, Starr PA (2018) Adaptive deep brain stimulation for Parkinson's disease using motor cortex sensing. J Neural Eng 15:046006. 10.1088/1741-2552/aabc9b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura SY, Nern A, Chklovskii DB, Scheffer LK, Rubin GM, Meinertzhagen IA (2017) The comprehensive connectome of a neural substrate for ‘ON’ motion detection in Drosophila. Elife 6:1–16. 10.7554/eLife.24394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, Bertagnolli D, Goldy J, Shapovalova N, Parry S, Lee C, Smith K, Bernard A, Madisen L, Sunkin SM, Hawrylycz M, et al. (2016) Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci 19:335–346. 10.1038/nn.4216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thung KH, Adeli E, Yap PT, Shen D (2016) Stability-weighted matrix completion of incomplete multi-modal data for disease diagnosis. Med Image Comput Comput Assist Interv 9901:88–96. 10.1007/978-3-319-46723-8_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillberg PW, Chen F, Piatkevich KD, Zhao Y, Yu CC, English BP, Gao L, Martorell A, Suk HJ, Yoshida F, DeGennaro EM, Roossien DH, Gong G, Seneviratne U, Tannenbaum SR, Desimone R, Cai D, Boyden ES (2016) Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat Biotechnol 34:987–992. 10.1038/nbt.3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong MQ, Hasan MM, Lee SS, Haque MR, Kim DH, Islam MS, Adams ME, Park BH (2017) OCT intensity and phase fluctuations correlated with activity-dependent neuronal calcium dynamics in the Drosophila CNS [Invited]. Biomed Opt Express 8:726–735. 10.1364/BOE.8.000726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treweek JB, Chan KY, Flytzanis NC, Yang B, Deverman BE, Greenbaum A, Lignell A, Xiao C, Cai L, Ladinsky MS, Bjorkman PJ, Fowlkes CC, Gradinaru V (2015) Whole-body tissue stabilization and selective extractions via tissue-hydrogel hybrids for high-resolution intact circuit mapping and phenotyping. Nat Protoc 10:1860–1896. 10.1038/nprot.2015.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpis M, Insanally M, Zou J, Elsharif A, Ghomashchi A, Sertac Artan N, Froemke RC, Viventi J (2017) A low-cost, scalable, current-sensing digital headstage for high channel count muECoG. J Neural Eng 14:026009. 10.1088/1741-2552/aa5a82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuthill JC, Wilson RI (2016) Parallel transformation of tactile signals in central circuits of Drosophila. Cell 164:1046–1059. 10.1016/j.cell.2016.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardy E, Robinson JE, Li C, Olsen RH, DiBerto JF, Giguere PM, Sassano FM, Huang XP, Zhu H, Urban DJ, White KL, Rittiner JE, Crowley NA, Pleil KE, Mazzone CM, Mosier PD, Song J, Kash TL, Malanga CJ, Krashes MJ, et al. (2015) A new DREADD facilitates the multiplexed chemogenetic interrogation of behavior. Neuron 86:936–946. 10.1016/j.neuron.2015.03.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu AT, Beckett A, Setsompop K, Feinberg DA (2018) Evaluation of SLIce Dithered Enhanced Resolution Simultaneous MultiSlice (SLIDER-SMS) for human fMRI. Neuroimage 164:164–171. 10.1016/j.neuroimage.2017.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall NR, De La Parra M, Sorokin JM, Taniguchi H, Huang ZJ, Callaway EM (2016) Brain-wide maps of synaptic input to cortical interneurons. J Neurosci 36:4000–4009. 10.1523/JNEUROSCI.3967-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Zhou Y, Ying L, Meng J, Song L, Xia J (2016) Enabling high-speed wide-field dynamic imaging in multifocal photoacoustic computed microscopy: a simulation study. Appl Opt 55:3724–3729. 10.1364/AO.55.003724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Wang Y, Zhou Y, Lovell JF, Xia J (2016) Coherent-weighted three-dimensional image reconstruction in linear-array-based photoacoustic tomography. Biomed Opt Express 7:1957–1965. 10.1364/BOE.7.001957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LV, Yao J (2016) A practical guide to photoacoustic tomography in the life sciences. Nat Methods 13:627–638. 10.1038/nmeth.3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang D, Zhang Y, Geng J, Lovell JF, Xia J (2016) Slit-enabled linear-array photoacoustic tomography with near isotropic spatial resolution in three dimensions. Opt Lett 41:127–130. 10.1364/OL.41.000127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson BO, Levenstein D, Greene JP, Gelinas JN, Buzsáki G (2016) Network homeostasis and state dynamics of neocortical sleep. Neuron 90:839–852. 10.1016/j.neuron.2016.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir PT, Dickinson MH (2015) Functional divisions for visual processing in the central brain of flying Drosophila. Proc Natl Acad Sci U S A 112:E5523–E5532. 10.1073/pnas.1514415112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir PT, Henze MJ, Bleul C, Baumann-Klausener F, Labhart T, Dickinson MH (2016) Anatomical reconstruction and functional imaging reveal an ordered array of skylight polarization detectors in Drosophila. J Neurosci 36:5397–5404. 10.1523/JNEUROSCI.0310-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmire CJ, Waiblinger C, Schwarz C, Stanley GB (2016) Information coding through adaptive gating of synchronized thalamic bursting. Cell Rep 14:795–807. 10.1016/j.celrep.2015.12.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H, de la Torre-Ubieta L, Stein JL, Parikshak NN, Huang J, Opland CK, Gandal MJ, Sutton GJ, Hormozdiari F, Lu D, Lee C, Eskin E, Voineagu I, Ernst J, Geschwind DH (2016) Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538:523–527. 10.1038/nature19847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Stark E, Ku PC, Wise KD, Buzsáki G, Yoon E (2015) Monolithically integrated mu LEDs on silicon neural probes for high-resolution optogenetic studies in behaving animals. Neuron 88:1136–1148. 10.1016/j.neuron.2015.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Cho JR, Zhou C, Treweek JB, Chan K, McKinney SL, Yang B, Gradinaru V (2016) Cholinergic mesopontine signals govern locomotion and reward through dissociable midbrain pathways. Neuron 90:333–347. 10.1016/j.neuron.2016.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamawaki N, Suter BA, Wickersham IR, Shepherd GM (2016) Combining optogenetics and electrophysiology to analyze projection neuron circuits. Cold Spring Harb Protoc 2016:840–846. 10.1101/pdb.prot090084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HH, St-Pierre F, Sun X, Ding X, Lin MZ, Clandinin TR (2016) Subcellular imaging of voltage and calcium signals reveals neural processing in vivo. Cell 166:245–257. 10.1016/j.cell.2016.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Song L, Wang LV (2015) Photoacoustic microscopy: superdepth, superresolution, and superb contrast. IEEE Pulse 6:34–37. 10.1109/MPUL.2015.2409100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Xia J, Wang LV (2016a) Multiscale functional and molecular photoacoustic tomography. Ultrason Imaging 38:44–62. 10.1177/0161734615584312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Kaberniuk AA, Li L, Shcherbakova DM, Zhang R, Wang LD, Li G, Verkhusha VV, Wang LV (2016b) Multiscale photoacoustic tomography using reversibly switchable bacterial phytochrome as a near-infrared photochromic probe. Nat Methods 13:67–73. 10.1038/nmeth.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YS, Chang JB, Alvarez MM, Trujillo-de Santiago G, Aleman J, Batzaya B, Krishnadoss V, Ramanujam AA, Kazemzadeh-Narbat M, Chen F, Tillberg PW, Dokmeci MR, Boyden ES, Khademhosseini A (2016) Hybrid microscopy: enabling inexpensive high-performance imaging through combined physical and optical magnifications. Sci Rep 6:1–10. 10.1038/srep22691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Qiao L, Li W, Zhang L, Shen D (2018) Simultaneous estimation of low- and high-order functional connectivity for identifying mild cognitive impairment. Front Neuroinform 12:1–8. 10.3389/fninf.2018.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video of Figure 2A. 3D rotation of projections throughout zebrafish brain. Courtesy of the Engert laboratory (Harvard University).