Abstract.
Extracting, harvesting, and building large-scale annotated radiological image datasets is a greatly important yet challenging problem. Meanwhile, vast amounts of clinical annotations have been collected and stored in hospitals’ picture archiving and communication systems (PACS). These types of annotations, also known as bookmarks in PACS, are usually marked by radiologists during their daily workflow to highlight significant image findings that may serve as reference for later studies. We propose to mine and harvest these abundant retrospective medical data to build a large-scale lesion image dataset. Our process is scalable and requires minimum manual annotation effort. We mine bookmarks in our institute to develop DeepLesion, a dataset with 32,735 lesions in 32,120 CT slices from 10,594 studies of 4,427 unique patients. There are a variety of lesion types in this dataset, such as lung nodules, liver tumors, enlarged lymph nodes, and so on. It has the potential to be used in various medical image applications. Using DeepLesion, we train a universal lesion detector that can find all types of lesions with one unified framework. In this challenging task, the proposed lesion detector achieves a sensitivity of 81.1% with five false positives per image.
Keywords: medical image dataset, lesion detection, convolutional neural network, deep learning, picture archiving and communication system, bookmark
1. Introduction
Computer-aided detection/diagnosis (CADe/CADx) has been a highly prosperous and successful research field in medical image processing. Recent advances have attracted much interest to the application of deep learning approaches.1,2 Convolutional neural network (CNN) based deep learning algorithms perform significantly better than conventional statistical learning approaches combined with handcrafted image features. However, these performance gains are often achieved at the cost of requiring tremendous amounts of labeled training data. Unlike general computer vision tasks, medical image analysis currently lacks a large-scale annotated image dataset (comparable to ImageNet3 and MS COCO4), which is mainly because the conventional methods for collecting image labels via Google search + crowd-sourcing from average users cannot be applied in the medical image domain, as medical image annotation requires extensive clinical expertise.
Detection and characterization of lesions are important topics in CADe/CADx. Existing detection/characterization algorithms generally target one particular lesion type, such as skin lesions,5 lung nodules,6,7 liver lesions,8 sclerotic lesions, and colonic polyps.9 While some common types of lesions receive much attention, vast infrequent types are ignored by most CADe programs. Besides, studying one lesion type at a time differs from the method radiologists routinely apply to read medical images and compile radiological reports. In practice, multiple findings can be observed and are often correlated. For instance, metastases can spread to regional lymph nodes or other body parts. By obtaining and maintaining a holistic picture of relevant clinical findings, a radiologist will be able to make a more accurate diagnosis. However, it remains challenging to develop a universal or multicategory CADe framework, capable of detecting multiple lesion types in a seamless fashion, partially due to the lack of a multicategory lesion dataset. Such a framework is crucial to building an automatic radiological diagnosis and reasoning system.
In this paper, we attempt to address these challenges. First, we introduce a paradigm to harvest lesion annotations from bookmarks in a picture archiving and communication system (PACS) with minimum manual effort. Bookmarks are metadata10 marked by radiologists during their daily work to highlight target image findings. Using this paradigm, we collected a large-scale dataset of lesions from multiple categories (Fig. 1). Our dataset, named DeepLesion, is composed of 32,735 lesions in 32,120 bookmarked CT slices from 10,594 studies of 4427 unique patients. Different from existing datasets, it contains a variety of lesions including lung nodules, liver lesions, enlarged lymph nodes, kidney lesions, bone lesions, and so on. DeepLesion is publicly released and may be downloaded from Ref. 11.
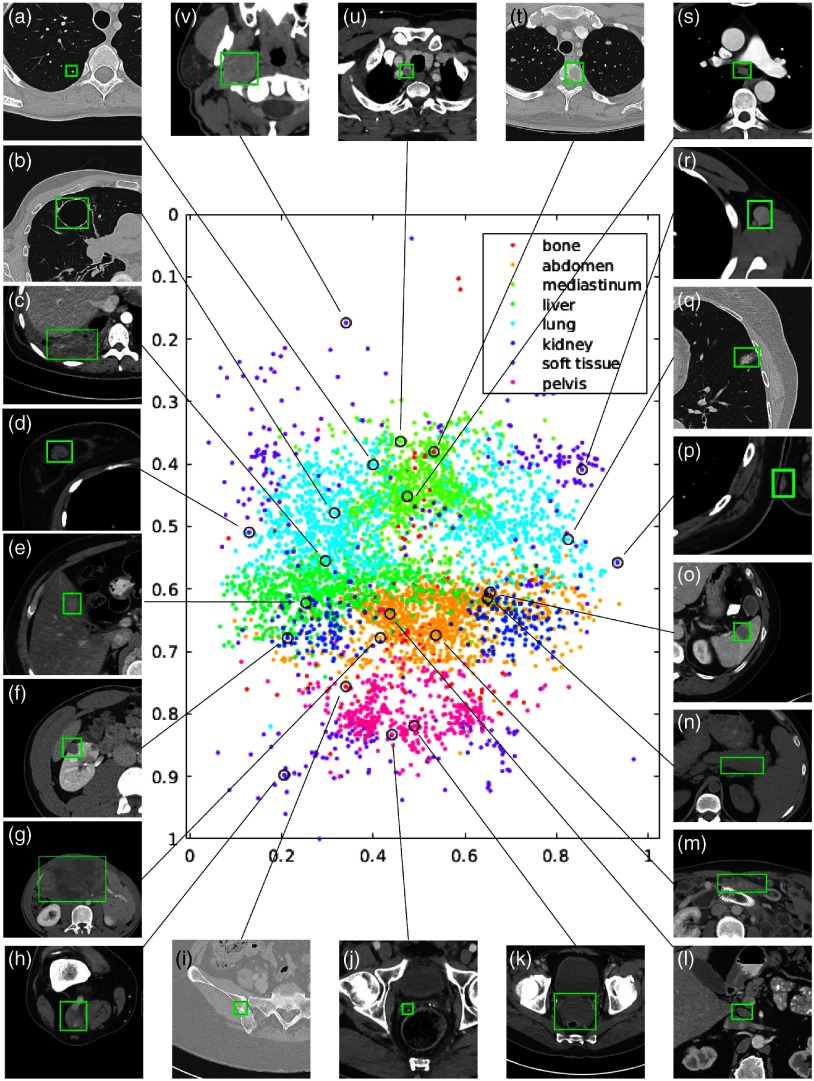
Fig. 1.
Visualization of a subset (15%) of the DeepLesion dataset. The - and -axes of the scatter map correspond to the - and -coordinates of the relative body location of each lesion, respectively. Therefore, this map is similar to a frontal view of the human body. Colors indicate the manually labeled lesion types. Sample lesions are exhibited to show the great diversity of DeepLesion, including: (a) lung nodule; (b) lung cyst; (c) costophrenic sulcus (lung) mass/fluid; (d) breast mass; (e) liver lesion; (f) renal mass; (g) large abdominal mass; (h) posterior thigh mass; (i) iliac sclerotic lesion; (j) perirectal lymph node (LN); (k) pelvic mass; (l) periportal LN; (m) omental mass; (n) peripancreatic lesion; (o) splenic lesion; (p) subcutaneous/skin nodule; (q) ground glass opacity; (r) axillary LN; (s) subcarinal LN; (t) vertebral body metastasis; (u) thyroid nodule; and (v) neck mass. Reproduced from the supplementary material of Ref. 12.
Using this dataset, we develop an automatic lesion detection algorithm to find all types of lesions with one unified framework. Our algorithm is based on a regional convolutional neural network (faster RCNN13). It achieves a sensitivity of 77.31% with three false positives (FPs) per image and 81.1% with five FPs. Note that the clinical bookmarks are not complete annotations of all significant lesions on a radiology image. Radiologists typically only annotate lesions of focus to facilitate follow-up studies of lesion matching and growth tracking. There are often several other lesions left without annotation. We empirically find that a large portion of the so-called FPs is actually true lesions, as demonstrated later. To harvest and distinguish those clinician unannotated lesions from “true” FPs will be an important future work.
2. Materials and Methods
In this section, we will first introduce bookmarks as radiology annotation tools. Then, we will describe the setup procedure and data statistics of the DeepLesion dataset. The proposed universal lesion detector will be presented afterward.
2.1. Bookmarks
Radiologists routinely annotate and measure hundreds of clinically meaningful findings in medical images, which have been collected for two decades in our institute’s PACS. Figure 2 shows a sample of a bookmarked image. Many of the bookmarks are either tumors or lymph nodes measured according to the response evaluation criteria in solid tumors (RECIST) guidelines.14 According to RECIST, assessment of the change in tumor burden is an important feature of the clinical evaluation of cancer therapeutics. Therefore, bookmarks usually indicate critical lesion findings. It will be extremely useful if we can collect them into a dataset and develop CADe/CADx algorithms to detect and characterize them.
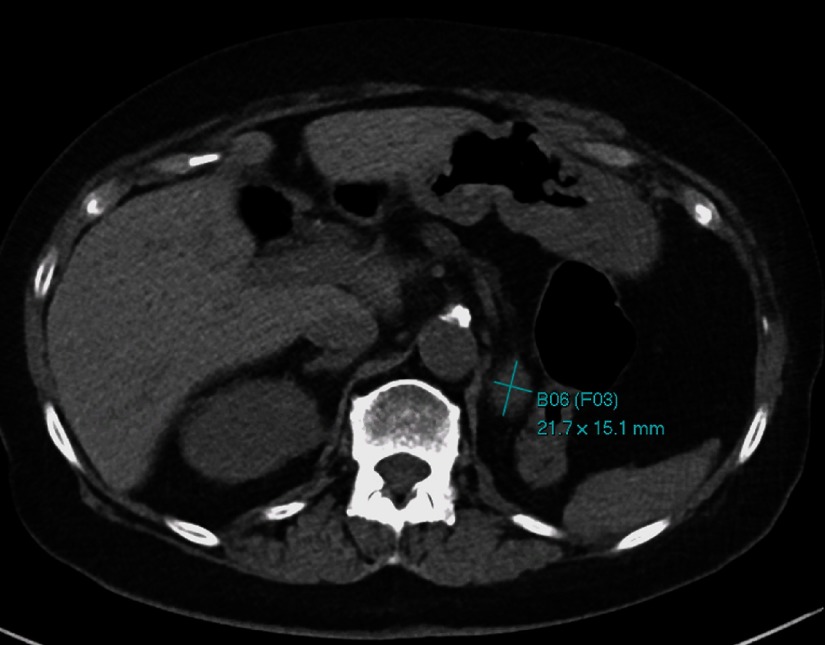
Fig. 2.
An example of a bookmarked image. A mass in or adjacent to the left nephrectomy bed is bookmarked using the RECIST-diameter tool. The bookmark identifiers indicate that this is bookmark number 6 (B06) and that this bookmark is part of follow-up set number 3 of bookmarks on the same lesion (F03).
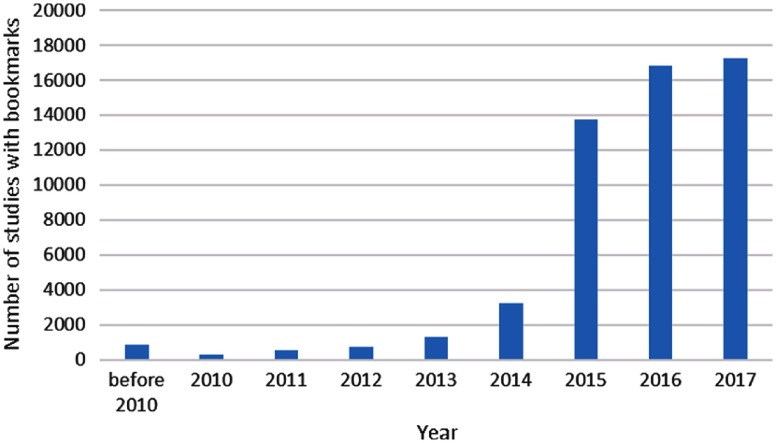
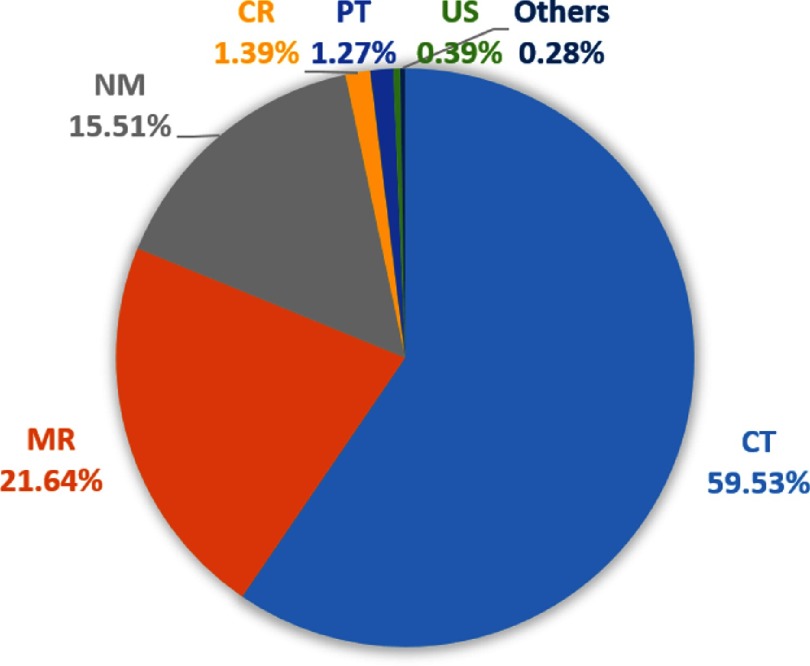
To get an overview of the bookmarks, we analyze them by year, image modality, and annotation tool. From Fig. 3, we can see that the number of studies with bookmarks increases each year with a boost in 2015. This indicates that bookmarks are becoming more and more popular as radiologists discover that it is a helpful tool.15 By collecting these bookmarks every year, we can easily obtain a large-scale lesion dataset. The image modalities of the bookmarks are shown in Fig. 4. CT images make up the largest percentage, followed by MR and nuclear medicine.
Fig. 3.
Number of studies with bookmarks in the PACS of our institute in each year (all image modalities included).
Fig. 4.
Proportion of different image modalities of the bookmarks in our institute. CT, computed tomography; MR, magnetic resonance; NM, nuclear medicine; CR, computed radiography; PT, positron emission tomography (PET); and US, ultrasound.
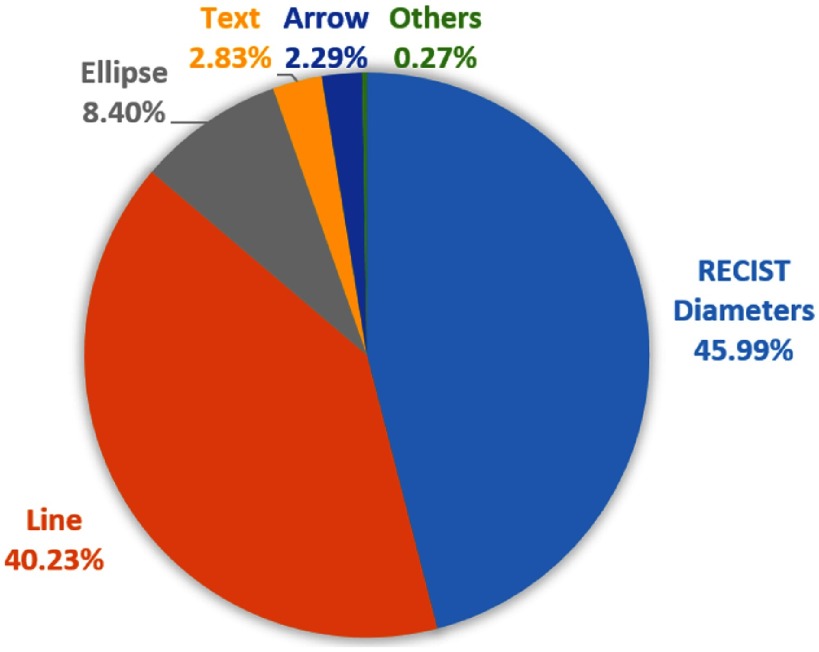
Radiologists can use various annotation tools to annotate the bookmarks, including arrows, lines, ellipses, bidimensional RECIST diameters, segmentations, and text. We downloaded all the bookmarks in CT studies and counted the usage of the tools (Fig. 5). RECIST diameters were applied most frequently. Each RECIST-diameter bookmark consists of two lines: one measuring the longest diameter of the lesion and the second measuring its longest perpendicular diameter in the plane of measurement. Examples can be found in Fig. 2. The RECIST-diameter bookmarks can tell us the exact location and size of a lesion. A line bookmark contains only one length measurement, which may be the longest or shortest diameter of a lesion, or even a measurement of a nonlesion. For line, ellipse, text, or arrow bookmarks, while we can infer the approximate location of a lesion, the exact location and/or size is not available.
Fig. 5.
Proportion of different annotation tools of the CT bookmarks in our institute.
2.2. DeepLesion Dataset
Because bookmarks can be viewed as annotations of critical lesions, we collected them to build a lesion dataset for CADe/CADx algorithms. This research has been approved by our Institutional Research Board. Without loss of generality, currently, we only focus on CT bookmarks, which are the most abundant. As for the annotation tools, now, we only consider RECIST diameters. Until January 2017, we have collected 33,418 bookmarks of this type. After filtering some noisy bookmarks (detailed in Sec. 2.2.1), we obtained the DeepLesion dataset with 32,120 axial slices from 10,594 CT studies of 4427 unique patients. There are one to three bookmarks in each slice, for a total of 32,735 bookmarks. The dataset will be introduced in detail from the following aspects: setup procedure, data statistics, advantages, limitations, and potential applications.
2.2.1. Setup procedure
First, we acquired the accession numbers of the CT studies with bookmarks by querying the PACS (Carestream Vue V12.1.6.0117). Then, the bookmarks were downloaded according to them using a Perl script provided by the PACS manufacturer. We selected only the RECIST-diameter ones, which are represented by four vertices. Most of them were annotated on the axial plane. We filtered the nonaxial ones, and then converted the vertices to image coordinates. The conversion was done by first subtracting the “ImagePositionPatient” (extracted from the DICOM file) from each vertex and then dividing the coordinates of each vertex with the pixel spacing.
The CT volumes that contain these bookmarks were also downloaded. We used MATLAB to convert each image slice from DICOM files to 16-bit portable network graphics (PNG) files for lossless compression and anonymization. Real patient IDs, accession numbers, and series numbers were replaced by self-defined indices of patient, study, and series (starting from 1) for anonymization. We named each volume with the format “{patient index}_{study index}_{series index}.” Note that one patient often underwent multiple CT examinations (studies) for different purposes or follow-up. Each study contains multiple volumes (series) that are scanned at the same time point but differ in image filters, contrast phases, etc. Every series is a three-dimensional (3-D) volume composed of tens to hundreds of axial image slices. Metadata,10 such as pixel spacing, slice interval, intensity window, and patient gender and age, were also recorded. The slice intervals were computed by differentiating the “ImagePositionPatient” (extracted from DICOM) of neighboring slices. We made sure that the slice indices increased from head to feet.
To facilitate applications such as computer-aided lesion detection, we converted the RECIST diameters into bounding-boxes. Denote the four vertices as , , , and . The coordinates are omitted since the vertices are on the same axial plane. A bounding box (left, top, right, and bottom) was computed to enclose the lesion measurement with 5-pixel padding in each direction, i.e., , where , , and similarly for and . The 5-pixel padding was applied to cover the lesion’s full spatial extent.
There are a limited number of incorrect bookmarks. For example, some bookmarks are outside the body, which is possibly caused by annotation error by the user. To remove these label noises, we computed the area and width-height-ratio of each bounding-box, as well as the mean and standard deviation of the pixels inside the box. Boxes that are too small/large/flat/dark or small in intensity range were manually checked. Another minor issue is duplicate annotations. A small number of lesions were bookmarked more than once possibly by different radiologists. We merged bounding-boxes that have more than 60% overlap by averaging their coordinates.16
2.2.2. Data statistics
The slice intervals of the CT studies in the dataset range between 0.25 and 22.5 mm. About 48.3% of them are 1 mm and 48.9% are 5 mm. The pixel spacings range between 0.18 and with a median of . Most of the images are and 0.12% of them are or . Figure 6 displays the distribution of the sizes of the bounding-boxes. The median values of the width and height are 22.9 and 22.3 mm, respectively. The diameter range of the lesions is 0.42 to 342.5 mm for long diameter and 0.21 to 212.4 mm for short diameter.
Fig. 6.
Distribution of the sizes of the bounding-boxes in DeepLesion. The bounding-boxes were computed from the RECIST diameters after dilation by 5 pixels. The width and height are the size of the - and -axes of the boxes, respectively.
To explore the lesion types in DeepLesion, we randomly selected 9816 lesions and manually labeled them into eight types: lung (2426), abdomen (2166), mediastinum (1638), liver (1318), pelvis (869), soft tissue (677), kidney (490), and bone (232). These are coarse-scale attributes of the lesions. The mediastinum type mainly consists of lymph nodes in the chest. Abdomen lesions are miscellaneous ones that are not in liver or kidney. The soft tissue type contains lesions in the muscle, skin, and fat. Examples of the lesions in the eight types can be found in Fig. 1, where a subset of the lesions is drawn on a scatter map to show their types and relative body coordinates. The map is similar to a frontal view of the human body. To obtain the approximate -coordinate of each lesion, we adopted the unsupervised body part regressor17 to predict the slice score of each image slice. From Fig. 1, we can find that the dataset is clinically diversified.
2.3. Universal Lesion Detection
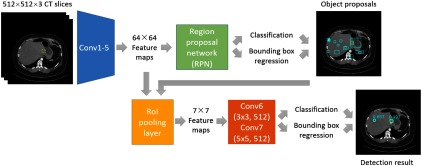
In this section, we will introduce our universal lesion detector in detail. It is trained on DeepLesion, thus can detect all types of lesions that radiologists are interested in measuring with one unified framework. The algorithm is adapted from the faster RCNN method.13 Its flowchart is illustrated in Fig. 7.
Fig. 7.
Flowchart of the lesion detection algorithm. Yellow dashed and cyan solid boxes in each image indicate the ground-truth and the predicted bounding-boxes, respectively.
2.3.1. Image preprocessing
The 12-bit CT intensity range was rescaled to floating-point numbers in [0,255] using a single windowing ( to 3071 HU) that covers the intensity ranges of lung, soft tissue, and bone. Every image slice was resized to . To encode 3-D information, we used three axial slices to compose a three-channel image and input it to the network. The slices were the center slice that contains the bookmark and its neighboring slices interpolated at 2-mm slice intervals. No data augmentation was used since our dataset is large enough to train a deep neural network.
2.3.2. Network architecture
The VGG-1618 model was adopted as the backbone of the network. We also compared deeper architectures including ResNet-5019 and DenseNet-12120 and the shallower AlexNet21 on the validation set and observed that VGG-16 had the highest accuracy. As shown in Fig. 7, an input image was first processed by the convolutional blocks in VGG-16 (Conv1–Conv5) to produce feature maps. We removed the last two pooling layers (pool4 and pool5) to enhance the resolution of the feature map and to increase the sampling ratio of positive samples (candidate regions that contain lesions), since lesions are often small and sparse in an image.
Next, a region proposal network13 parsed the feature maps and proposes candidate lesion regions. It estimated the probability of “lesion/nonlesion” on a fixed set of anchors on each position of the feature maps. At the same time, the location and size of each anchor were fine-tuned via bounding box regression. After investigating the sizes of the bounding-boxes in DeepLesion, we used five anchor scales (16, 24, 32, 48, and 96) and three anchor ratios (1:2, 1:1, and 2:1) in this paper.
Afterward, the lesion proposals and the feature maps were sent to a region of interest (RoI) pooling layer, which resampled the feature maps inside each proposal to a fixed size ( in this paper). These feature maps were then fed into two convolutional layers, Conv6 and Conv7. Here, we replaced the original 4096D fully-connected (FC) layers in VGG-16 so that the model size was cut to 1/4 while the accuracy was comparable. Conv6 consisted of 512 filters with zero padding and stride 1. Conv7 consisted of 512 filters with zero padding and stride 1. Rectified linear units were inserted after the two convolutional layers. The 512D feature vector after Conv7 then underwent two FC layers to predict the confidence scores for each lesion proposal and ran another bounding box regression for further fine-tuning. Nonmaximum suppression (NMS)13 was then applied to the fine-tuned boxes to generate the final predictions. The intersection-over-union (IoU) thresholds for NMS were 0.7 and 0.3 in training and testing, respectively.
2.3.3. Implementation details
The proposed algorithm was implemented using MXNet.22 The weights in Conv1 to Conv5 were initialized with the ImageNet pretrained VGG-16 model, whereas all the other layers were randomly initialized. During training, we fixed the weights in Conv1 and Conv2. The two classification and two regression losses were jointly optimized. This end-to-end training strategy is more efficient than the four-step strategy in the original faster RCNN implementation.13 Each mini-batch had eight images. The number of region proposals per image for training was 32. We adopted the stochastic gradient descent optimizer and set the base learning rate to 0.002, and then reduced it by a factor of 10 after six epochs. The network converged within eight epochs.
3. Results
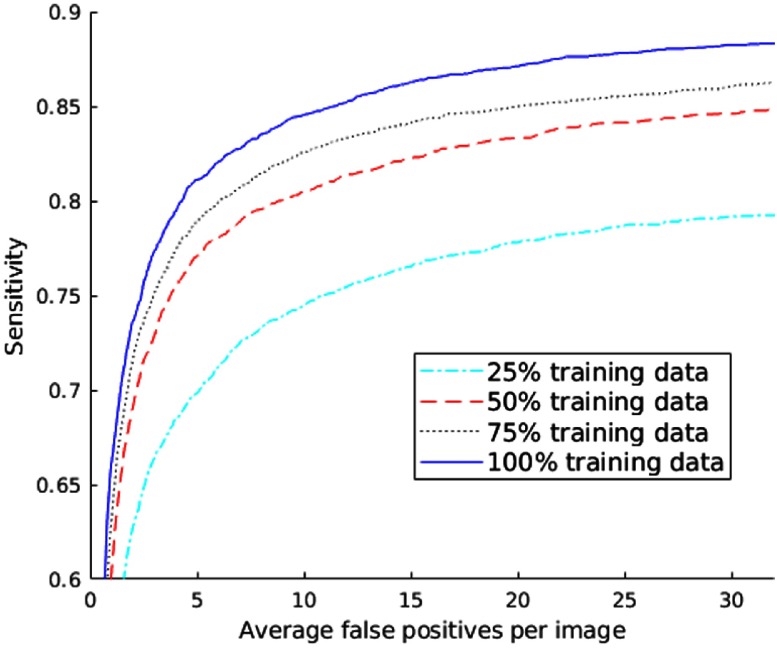
To evaluate the proposed algorithm, we divided DeepLesion into training (70%), validation (15%), and test (15%) sets by randomly splitting the dataset at the patient level. The proposed algorithm only took 34 ms to process a test image on a Titan X Pascal GPU. Here, we report the free receiver operating characteristic (FROC) curves on the test set in Fig. 8. The sensitivity reaches 81.1% when there are five FPs on average on each image. In addition, the performance steadily improves as more training samples are used. As a result, the accuracy is expected to be better as we harvest more data in the future.
Fig. 8.
FROC curves of lesion detection on the test set of DeepLesion when different proportions of training data are used.
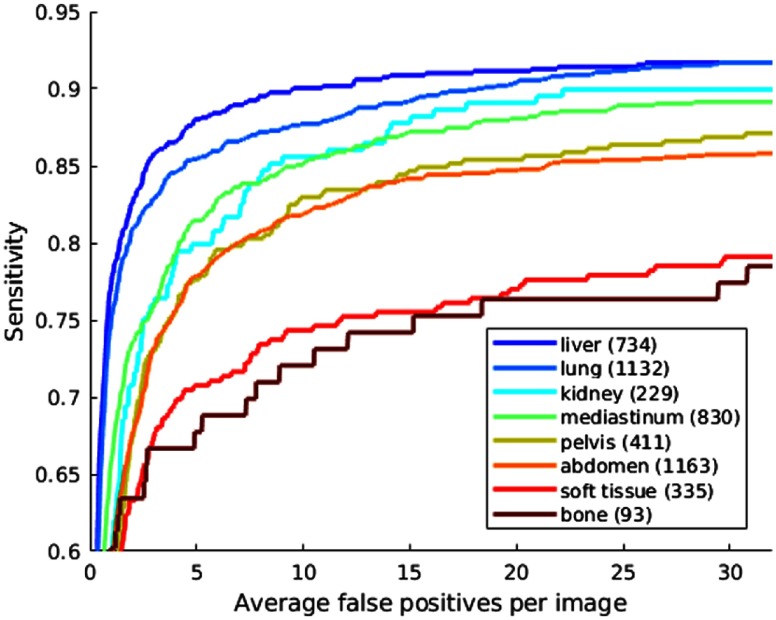
The FROC curves of different lesion types are shown in Fig. 9. Note that our network does not predict the type of each detected lesion, so the -axis in Fig. 9 is the average number of FPs of all lesion types per image. Thus, the curves could not be directly compared with the literature.7–9 Instead, they reflect the relative performance of different types and sizes. From Fig. 9, we can find that liver, lung, kidney, and mediastinum lesions are among the easiest ones to detect. This is probably because their intensity and appearance is relatively distinctive from the background. It is more difficult to detect abdominal and pelvic lesions, where normal and abnormal structures including bowel and mesentery clutter the image and may have similar appearances (Figs. 18–21). Soft tissue and bone lesions have fewer training samples and small contrast with normal structures, thus have the lowest sensitivity.
Fig. 9.
FROC curves of lesion detection on the test set of DeepLesion with respect to different lesion types. The -axis is the average number of FPs of all lesion types per image. The numbers in the legend are the numbers of lesions of a specific type in the test set.
Fig. 18.
The ground-truth and another liver lesion are detected. A small liver lesion is missed. Two small lymph nodes are FPs.
Fig. 21.
The ground-truth inguinal lymph node is detected with high confidence, although its appearance is similar to the surrounding muscles and vessels. FP is on a normal bladder.
The FROC curves of different lesion sizes are shown in Fig. 10. The size is computed by averaging the long and short diameters. In Fig. 10, it is not surprising that small lesions () are harder to detect. It is also easy to find very large () lesions. However, when lesion size is between 10 and 50 mm, the sensitivity is not proportional with lesion size, which is possibly because detection accuracy can be affected by multiple factors, such as lesion size, lesion type, number of training samples, etc. The algorithm performs the best when the lesion size is 15 to 20 mm.
Fig. 10.
FROC curves of lesion detection on the test set of DeepLesion with respect to different lesion sizes. The -axis is the average number of FPs of all sizes per image. The numbers in the legend are the numbers of lesions of a specific size in the test set. Accuracy can be affected by multiple factors, such as lesion size, lesion type, number of training samples, etc. Thus, its order does not strictly follow the order of lesion size.
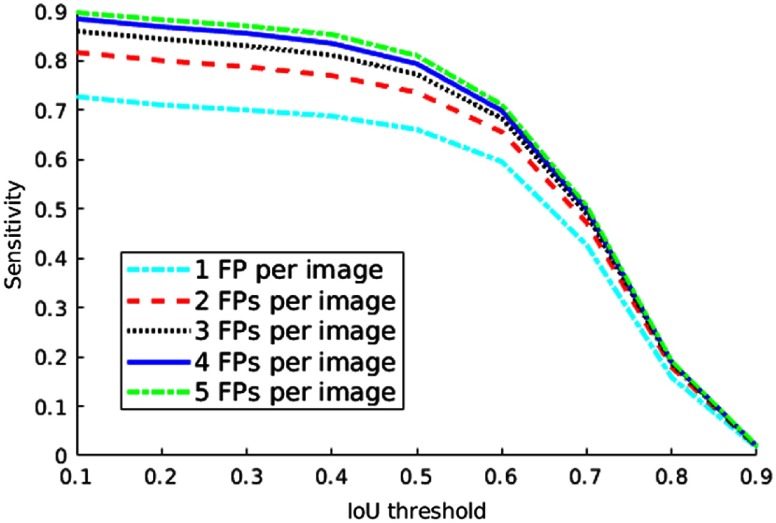
The detection accuracy also depends on the selected IoU threshold. From Fig. 11, we can find that the sensitivity decreases if the threshold is set higher.
Fig. 11.
Sensitivity of lesion detection on the test set of DeepLesion with respect to different IoU thresholds and the numbers of average FPs per image.
Some qualitative results are randomly chosen from the test set and are shown in Figs. 12–21. The figure shows examples of true positives, FPs, and false negatives (FNs).
Fig. 12.
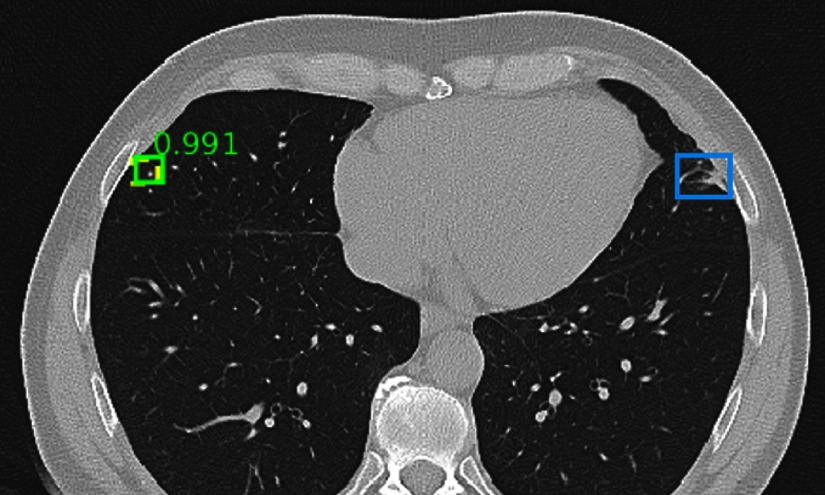
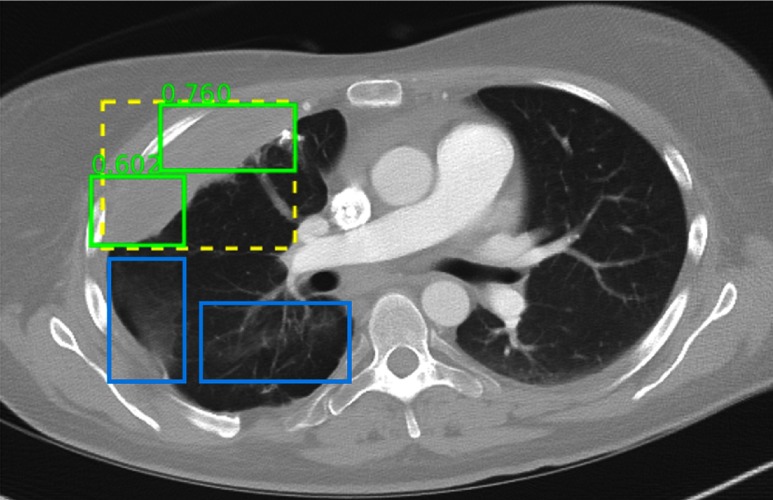
Detection results randomly chosen from the test set. The ground-truth and correct predictions are marked with yellow dashed boxes and green solid boxes, respectively. FPs and FNs are marked with red and blue solid boxes, respectively. The numbers beside the predictions are confidence scores. Predictions with scores are shown. The same explanation applies to Figs. 13–21. In the figure, a tiny lung nodule is detected with high confidence. An area of scarring in the lingula is not detected, which is possibly because there are few bookmarks of scars in the dataset.
Fig. 13.
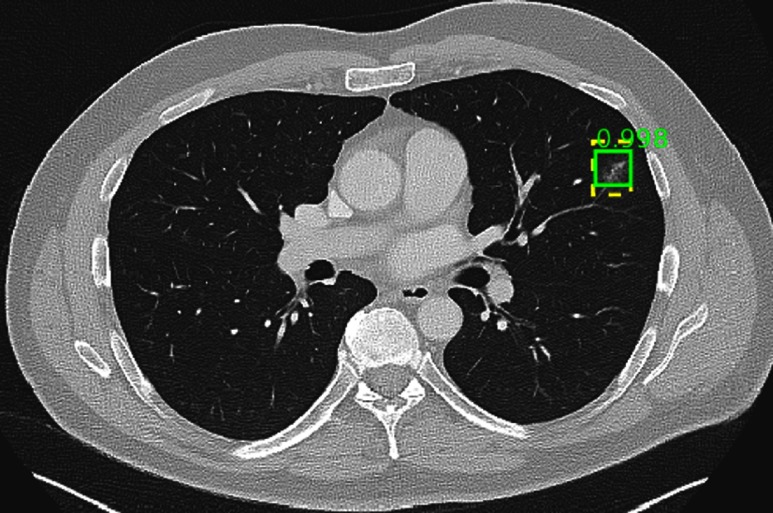
The ground-truth is detected but split into two parts. Some minor areas of scarring are not marked.
Fig. 14.
A correct detection with high confidence.
Fig. 15.
An enlarged lymph node is correctly detected, but two unenlarged ones are also marked (red boxes). This is probably because the universal lesion detector is robust to small scale changes. Therefore, small and large lymph nodes are sometimes both detected.
Fig. 16.
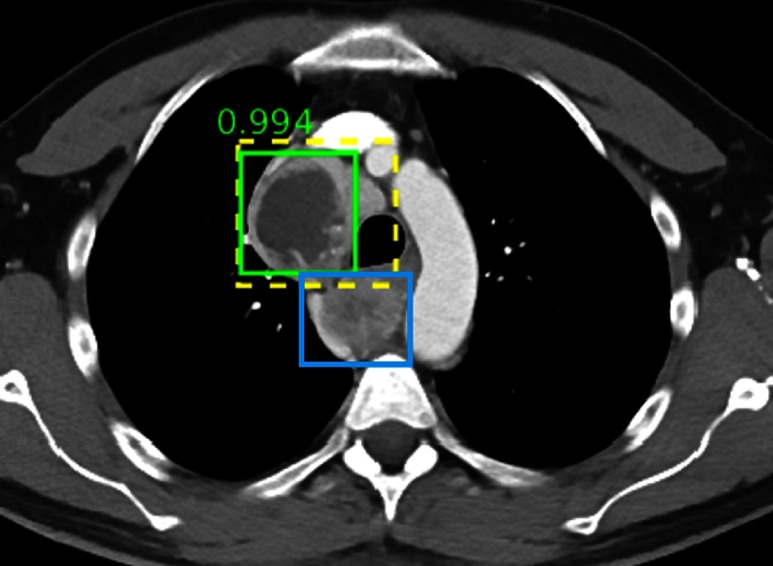
A mass is correctly detected with high confidence, but another one posterior to the trachea is missed.
Fig. 17.
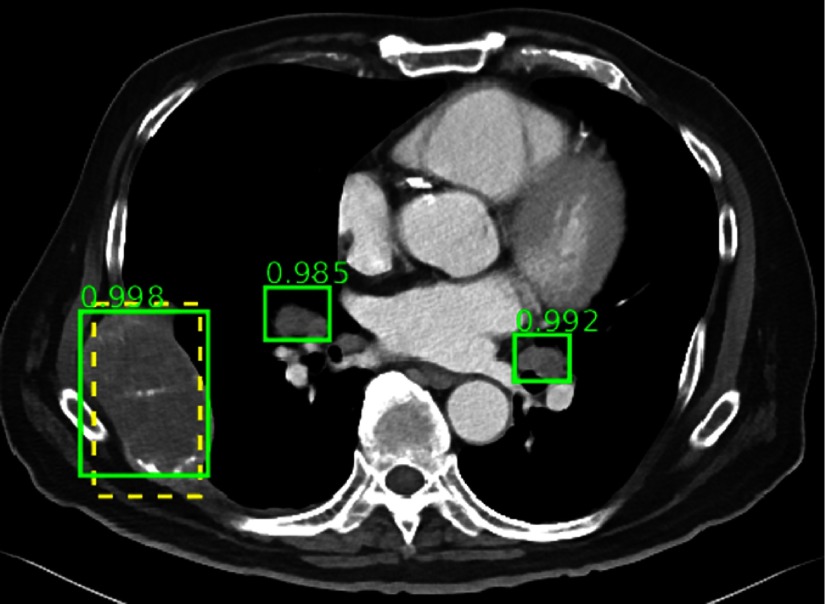
The ground-truth and two enlarged lymph nodes are correctly detected, even though the lymph nodes are not annotated in the dataset.
Fig. 19.
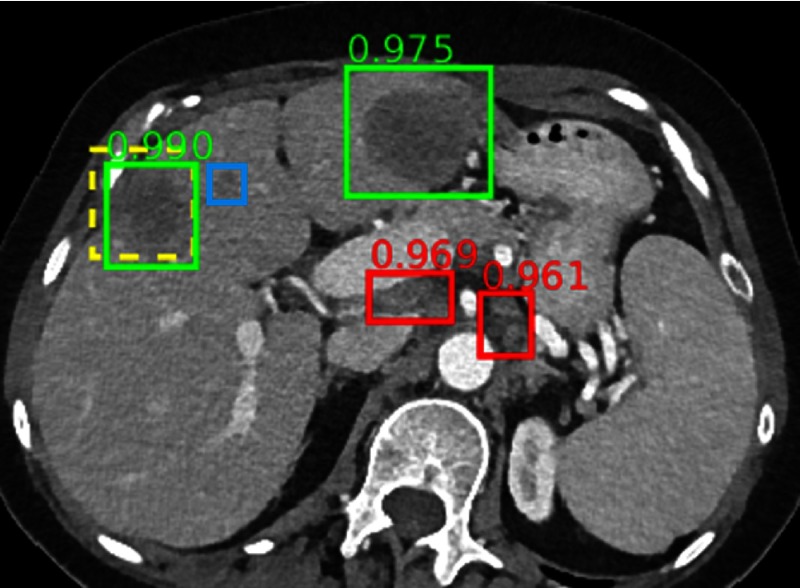
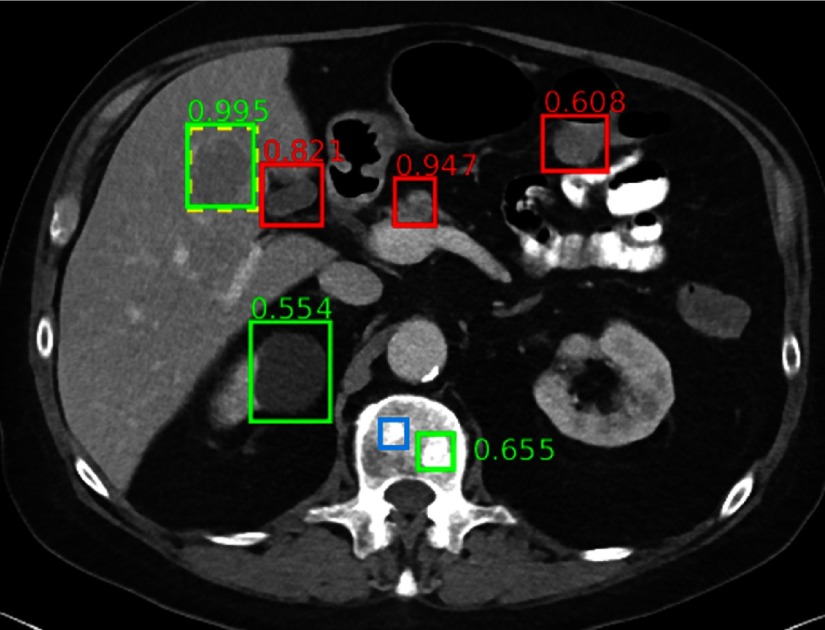
The ground-truth liver lesion is detected with high confidence. A renal cyst and a bone metastasis are also detected correctly. FPs include normal pancreas (0.947), gallbladder (0.821), and bowel (0.608). A subtle bone metastasis (blue box) is missed. Note the complexity and clutter of the appearance of abdominal structures.
Fig. 20.
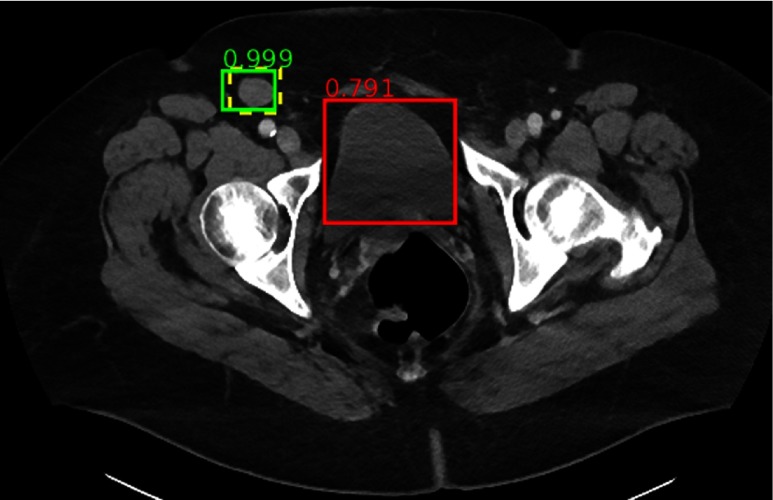
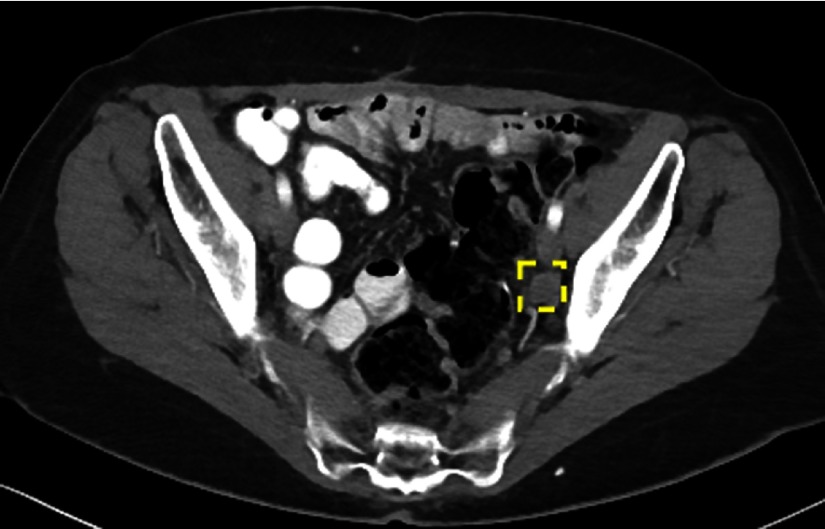
The ground-truth iliac lymph node is missed. Note the complexity and clutter of the appearance of pelvic structures.
4. Discussion
4.1. DeepLesion Dataset
4.1.1. Advantages
Compared to most other lesion medical image datasets23–28 that consist of only certain types of lesions, one major feature of our DeepLesion database is that it contains all kinds of critical radiology findings, ranging from widely studied lung nodules, liver lesions, and so on, to less common ones, such as bone and soft tissue lesions. Thus, it allows researchers to:
-
•
Develop a universal lesion detector. The detector can help radiologists find all types of lesions within one unified computing framework. It may open the possibility to serve as an initial screening tool and send its detection results to other specialist systems trained on certain types of lesions.
-
•
Mine and study the relationship between different types of lesions.12 In DeepLesion, multiple findings are often marked in one study. Researchers are able to analyze their relationship to make discoveries and improve CADe/CADx accuracy, which is not possible with other datasets.
Another advantage of DeepLesion is its large size and small annotation effort. ImageNet3 is an important dataset in computer vision, which are composed of millions of images from thousands of classes. In contrast, most publicly available medical image datasets have tens or hundreds of cases, and datasets with more than 5000 well-annotated cases are rare.10,29 DeepLesion is a large-scale dataset with over 32K annotated lesions from over 10K studies. It is still growing every year, see Fig. 3. In the future, we can further extend it to other image modalities, such as MR, and combine data from multiple hospitals. Most importantly, these annotations can be harvested with minimum manual effort. We hope the dataset will benefit the medical imaging area just as ImageNet benefitted the computer vision area.
4.1.2. Potential applications
-
•
Lesion detection: This is the direct application of DeepLesion. Lesion detection is a key part of diagnosis and is one of the most labor-intensive tasks for radiologists.2 An automated lesion detection algorithm is highly useful because it can help human experts to improve the detection accuracy and decrease the reading time.
-
•
Lesion classification: Although the type of each lesion was not annotated along with the bookmarks, we can extract the lesion types from radiology reports coupled with each study. Nowadays, radiologists often put hyperlinks in reports to link bookmarks with lesion descriptions.15 Consequently, we can use natural language processing algorithms to automatically extract lesion types and other information cues.30,31
-
•
Lesion segmentation: With the RECIST diameters and bounding-boxes provided in the dataset, weakly supervised segmentation algorithms32 can be developed to automatically segment or measure lesions. One can also select lesions of interest and manually annotate them for training and testing. During the annotation process, active learning may be employed to alleviate human burden.
-
•
Lesion retrieval: Considering its diversity, DeepLesion is a good data source for the study of content-based or text-based lesion retrieval algorithms.33,34 The goal is to find the most relevant lesions given a query text or image.
-
•
Lesion growth analysis: In the dataset, lesions (e.g., tumors and lymph nodes) are often measured multiple times for follow-up study.14 With these sequential data, one may be able to analyze or predict the change of lesions based on their appearance and other relative information.35
4.1.3. Limitations
Since DeepLesion was mined from PACS, it has a few limitations:
-
•
Lack of complete labels: DeepLesion contains only two-dimensional diameter measurements and bounding-boxes of lesions. It has no lesion segmentations, 3-D bounding-boxes, or fine-grained lesion types. We are now working on extracting lesion types from radiology reports.
-
•
Missing annotations: Radiologists typically mark only representative lesions in each study.14 Therefore, some lesions remain unannotated. The unannotated lesions may harm or misrepresent the performance of the trained lesion detector because the negative samples (nonlesions) are not purely true. To solve this problem, one can leverage machine learning strategies, such as learning with noisy labels.36 It is also feasible to select negative samples from another dataset of healthy subjects. Furthermore, to more accurately evaluate the trained detector, it is better to have a fully labeled test set with all lesions annotated. The newly annotated lesions should also be similar to those already in DeepLesion, so lesions that do not exist in DeepLesion should not be annotated.
-
•
Noise in lesion annotations: According to manual examination, although most bookmarks represent abnormal findings or lesions, a small proportion of the bookmarks is actually measurement of normal structures, such as lymph nodes of normal size. We can design algorithms to either filter them (e.g., by using extracted lesion types from reports) or ignore them (e.g., by using machine learning models that are robust to noise).
4.2. Universal Lesion Detection
Because radiologists typically mark only representative lesions in each study,14 there are missing annotations in the test set. Therefore, the actual FP rates should be lower. We would argue that the current result is still a nonperfect but reasonable surrogate of the actual accuracy. From the qualitative detection results in Figs. 12–21, we can find that the universal lesion detector is able to detect various types of lesions in the test set of DeepLesion, including the annotated ones (ground-truth) as well as some unannotated ones, although a few FPs and FNs still present.
-
•
Lung, mediastinum, and liver lesions can be detected more accurately, as their intensity and appearance patterns are relatively distinctive from the background.
-
•
Lung scarring is not always detected, which is possibly because it is not commonly measured by radiologists, thus DeepLesion contains very few training samples.
-
•
Unenlarged lymph nodes are sometimes detected as FNs. This is probably because the design of faster RCNN (e.g., the RoI pooling layer) allows it to be robust to small scale changes. We can amend this issue by training a special lymph node detector and a lesion size regressor.
-
•
There are more FPs and FNs in the abdominal and pelvic area, as normal and abnormal structures bowel and mesentery clutter inside the image and may have similar appearances (Figs. 18–21). This problem may be mitigated by applying ensemble of models and enhancing the model with 3-D context.6,7,9
It is not proper to directly compare our results with others’ since most existing work7–9 can only detect one type of lesion. However, we can use them as references. Roth et al.9 proposed CNNs with random view aggregation to detect sclerotic bone lesions, lymph nodes, and colonic polyps. Their detection results are 70%, 77%, and 75% at three FPs per patient for the three types of lesions, respectively. Ben-Cohen et al.8 applied fully convolutional network and sparsity-based dictionary learning for liver lesion detection in CT. Their result is 94.6% at 2.9 FPs per case. Multilevel contextual 3-D CNNs were used7 to detect lung nodules with a sensitivity of 87.9 at two FPs per scan. The main reason that our result (77.31% at three FPs per image) is still inferior than those in Refs. 7–9 is that our task is considerably harder, which tries to detect all kinds of lesions including lung nodules, liver lesions, bone lesions, lymph nodes, and so on. Besides, our dataset is much larger (32,735 lesions with about 25% lung lesions and 13% liver ones, versus 123 liver lesions8 and 1186 lung nodules7) with lesion sizes ranging widely from 0.21 to 342.5 mm. Furthermore, we did not use a fully annotated dataset of a specific lesion to train a sophisticated detection model such as those in Refs. 7–9. Improving the detection accuracy is one of our future works.
5. Conclusion
In this paper, we introduced a paradigm to collect lesion annotations and build large-scale lesion datasets with minimal manual effort. We made use of bookmarks in PACS, which are annotations marked by radiologists during their routine work to highlight significant clinical image findings that would serve as references for longitudinal studies. After analyzing their characteristics, we harvested and sorted them to create DeepLesion, a dataset with over 32K lesion bounding-boxes and measurements. DeepLesion is composed of a variety of lesions and has many potential applications. As a direct application, we developed a universal lesion detector that can find all types of lesions with one unified framework. Qualitative and quantitative results proved its effectiveness.
In the future, we will keep on improving the DeepLesion dataset by collecting more data and extracting lesion types from radiology reports. We also plan to improve the universal lesion detector by leveraging 3-D and lesion type information.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institutes of Health, Clinical Center. We thank Sue Powell and Douglas Milewski in our department’s PACS/RIS section for help downloading the bookmarks.
Biographies
Ke Yan received his BS and PhD degrees both from the Department of Electronic Engineering, Tsinghua University, Being, China. He was the winner of the 2016 Tsinghua University Excellent Doctoral Dissertation Award. Currently, he is a postdoctoral researcher at the National Institutes of Health, USA. His research interests include medical image analysis, deep learning, computer vision, and machine learning. He has published 16 journal and conference papers (including CVPR, MICCAI).
Xiaosong Wang received his PhD in computer vision from the University of Bristol, UK, 2011. From 2011 to 2015, he was an algorithm engineer, manager in CAD Department and then product manager of medical image postprocessing workstations in the Software Business Unit at Shanghai United Imaging Healthcare. Currently, he is a visiting fellow at National Institutes of Health Clinical Center, focusing on machine learning, deep learning, and their applications in medical imaging.
Le Lu is a director of Ping An Technology US Research Labs, and was a senior research manager of medical imaging and clinical informatics at NVIDIA. He was a staff scientist at NIH Clinical Center during 2013–2017 and a senior staff scientist at Siemens since 2006. He has been named on 23 patents and 32 inventions. He has authored 120 peer-reviewed publications. He received his Ph.D. in computer science from Johns Hopkins University in 2007. He won the NIH Mentor of the Year award in 2015 and NIH-CC CEO award in 2017. He serves the Area chair for MICCAI 2018,2016,2015; CVPR 2019,2017; ICIP 2017; and Demo chair of CVPR 2017.
Ronald M. Summers received his BA degree in physics and his MD and PhD degrees in medicine/anatomy and cell biology from the University of Pennsylvania. He is a tenured senior investigator and staff radiologist in the Radiology and Imaging Sciences Department at the NIH Clinical Center in Bethesda, Maryland. His awards include being named a fellow of the Society of Abdominal Radiologists, the Presidential Early Career Award for scientists and engineers, the NIH Director’s award, and the NIH Clinical Center director’s award. He is a member of the editorial boards of the Journal of Medical Imaging, Radiology: Artificial Intelligence and Academic Radiology and a past member of the editorial board of radiology. He has coauthored over 400 articles and is a coinventor on 14 patents.
Disclosures
Author R.M.S. has pending and/or awarded patents for automated image analyses, and received royalty income from iCAD, Zebra Medical, Imbio, ScanMed and Koninklijke Philips. His lab received research support from Ping An Technology Company Ltd. and NVIDIA. The other authors declare no disclosures.
References
- 1.Greenspan H., van Ginneken B., Summers R. M., “Deep learning in medical imaging: overview and future promise of an exciting new technique,” IEEE Trans. Med. Imaging 35(5), 1153–1159 (2016). 10.1109/TMI.2016.2553401 [DOI] [Google Scholar]
- 2.Litjens G., et al. , “A survey on deep learning in medical image analysis,” Med. Image Anal. 42(1995), 60–88 (2017). 10.1016/j.media.2017.07.005 [DOI] [PubMed] [Google Scholar]
- 3.Deng J., et al. , “ImageNet: a large-scale hierarchical image database,” in IEEE Conf. Computer Vision Pattern Recognition, pp. 248–255 (2009). 10.1109/CVPR.2009.5206848 [DOI] [Google Scholar]
- 4.Lin T.-Y., et al. , “Microsoft COCO: common objects in context,” in European Conf. on Computer Vision, pp. 740–755 (2014). 10.1007/978-3-319-10602-1_48 [DOI] [Google Scholar]
- 5.Esteva A., et al. , “Dermatologist-level classification of skin cancer with deep neural networks,” Nature 542(7639), 115–118 (2017). 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teramoto A., et al. , “Automated detection of pulmonary nodules in PET/CT images: ensemble false-positive reduction using a convolutional neural network technique,” Med. Phys. 43(6), 2821–2827 (2016). 10.1118/1.4948498 [DOI] [PubMed] [Google Scholar]
- 7.Dou Q., et al. , “Multilevel contextual 3-D CNNs for false positive reduction in pulmonary nodule detection,” IEEE Trans. Biomed. Eng. 64(7), 1558–1567 (2017). 10.1109/TBME.2016.2613502 [DOI] [PubMed] [Google Scholar]
- 8.Ben-Cohen A., et al. , “Fully convolutional network and sparsity-based dictionary learning for liver lesion detection in CT examinations,” Neurocomputing 275, 1585–1594 (2018). 10.1016/j.neucom.2017.10.001 [DOI] [Google Scholar]
- 9.Roth H. R., et al. , “Improving computer-aided detection using convolutional neural networks and random view aggregation,” IEEE Trans. Med. Imaging 35(5), 1170–1181 (2016). 10.1109/TMI.2015.2482920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohli M. D., Summers R. M., Geis J. R., “Medical image data and datasets in the era of machine learning—whitepaper from the 2016 C-MIMI meeting dataset session,” J. Digit. Imaging 30(4), 392–399 (2017). 10.1007/s10278-017-9976-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.https://nihcc.box.com/v/DeepLesion.
- 12.Yan K., et al. , “Deep lesion graphs in the wild: relationship learning and organization of significant radiology image findings in a diverse large-scale lesion database,” in IEEE 2018 Conf. Computer Vision Pattern Recognition (2018). [Google Scholar]
- 13.Ren S., et al. , “Faster R-CNN: towards real-time object detection with region proposal networks,” in Proc. of the 28th Int. Conf. on Neural Information Processing Systems, pp. 91–99 (2015). [Google Scholar]
- 14.Eisenhauer E. A., et al. , “New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1),” Eur. J. Cancer 45(2), 228–247 (2009). 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 15.Machado L. B., et al. , “Radiology reports with hyperlinks improve target lesion selection and measurement concordance in cancer trials,” Am. J. Roentgenol. 208(2), W31–W37 (2017). 10.2214/AJR.16.16845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Setio A. A. A., et al. , “Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: the LUNA16 challenge,” Med. Image Anal. 42, 1–13 (2017). 10.1016/j.media.2017.06.015 [DOI] [PubMed] [Google Scholar]
- 17.Yan K., Le L., Summers R. M., “Unsupervised body part regression via spatially self-ordering convolutional neural networks,” in IEEE Int. Conf. on Biomedical Imaging (ISBI 2018), pp. 1022–1025 (2018). [Google Scholar]
- 18.Simonyan K., Zisserman A., “Very deep convolutional networks for large-scale image recognition,” in Int. Conf. on Learning Representations, pp. 1–14 (2015). [Google Scholar]
- 19.He K., et al. , “Deep residual learning for image recognition,” in IEEE Conf. on Computer Vision and Pattern Recognition (CVPR), pp. 770–778 (2016). 10.1109/CVPR.2016.90 [DOI] [Google Scholar]
- 20.Huang G., et al. , “Densely connected convolutional networks,” in IEEE Conf. on Computer Vision and Pattern Recognition (CVPR) (2017). 10.1109/CVPR.2017.243 [DOI] [Google Scholar]
- 21.Krizhevsky A., Sutskever I., Hinton G., “ImageNet classification with deep convolutional neural networks,” in Proc. of the 25th Int. Conf. on Neural Information Processing Systems, pp. 1097–1105 (2012). [Google Scholar]
- 22.Chen T., et al. , “MXNet: a flexible and efficient machine learning library for heterogeneous distributed systems,” in Neural Information Processing Systems (NIPS), Workshop on Machine Learning Systems (2016). [Google Scholar]
- 23.Clark K., et al. , “The cancer imaging archive (TCIA): maintaining and operating a public information repository,” J. Digit. Imaging 26(6), 1045–1057 (2013). 10.1007/s10278-013-9622-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Open-Access Medical Image Repositories, “aylward.org,” http://www.aylward.org/notes/open-access-medical-image-repositories (10 January 2018).
- 25.LIDC-IDRI, “The cancer imaging archive (TCIA) public access,” https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI (13 January 2018).
- 26.CT Colonography, “The cancer imaging archive (TCIA) public access,” 2011, https://wiki.cancerimagingarchive.net/display/Public/CT+COLONOGRAPHY (13 January 2018).
- 27.SPIE-AAPM-NCI PROSTATEx Challenges, “The cancer imaging archive (TCIA) public access—cancer imaging archive wiki,” https://wiki.cancerimagingarchive.net/display/Public/SPIE-AAPM-NCI+PROSTATEx+Challenges#521f3ddfc6a94cea8a9178ca8f35009c (13 January 2018).
- 28.CT Lymph Nodes dataset, “The cancer imaging archive (TCIA) public access,” 2016, https://wiki.cancerimagingarchive.net/display/Public/CT+Lymph+Nodes (13 January 2018).
- 29.The Cancer Imaging Archive (TCIA), “A growing archive of medical images of cancer,” http://www.cancerimagingarchive.net/ (10 January 2018).
- 30.Wang X., et al. , “ChestX-ray8: hospital-scale chest X-ray database and benchmarks on Weakly-supervised classification and localization of common thorax diseases,” in IEEE Conf. on Computer Vision and Pattern Recognition (CVPR), pp. 2097–2106 (2017). [Google Scholar]
- 31.Depeursinge A., et al. , “From radiological image data: preliminary results with liver lesions in CT,” IEEE Trans. Med. Imaging 33(8), 1669–1676 (2014). 10.1109/TMI.2014.2321347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai J., He K., Sun J., “BoxSup: exploiting bounding boxes to supervise convolutional networks for semantic segmentation,” in Proc. of the IEEE Int. Conf. on Computer Vision, pp. 1635–1643 (2015). 10.1109/ICCV.2015.191 [DOI] [Google Scholar]
- 33.Li Z., et al. , “Large-scale retrieval for medical image analytics: a comprehensive review,” Med. Image Anal. 43, 66–84 (2018). 10.1016/j.media.2017.09.007 [DOI] [PubMed] [Google Scholar]
- 34.Wei G., Qiu M., “Similarity measurement of lung masses for medical image retrieval using kernel based semisupervised distance metric,” Med. Phys. 43(12), 6259–6269 (2016). 10.1118/1.4966030 [DOI] [PubMed] [Google Scholar]
- 35.Zhang L., et al. , “Convolutional invasion and expansion networks for tumor growth prediction,” IEEE Trans. Med. Imaging 37, 638–648 (2018). 10.1109/TMI.2017.2774044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Natarajan N., et al. , “Learning with noisy labels,” in Proc. of the 26th Int. Conf. on Neural Information Processing Systems, pp. 1196–1204 (2013). [Google Scholar]