Unlike the benzene ring, the uneven distribution of the electron density of the pyridine ring influences the positional reactivity and interaction with enzymes; e.g., the ortho and para oxidations are more difficult than the meta oxidations. Hydroxylation is an important oxidation process for the pyridine derivative metabolism. In previous reports, the ortho hydroxylations of pyridine derivatives were catalyzed by multicomponent molybdenum-containing monooxygenases, while the meta hydroxylations were catalyzed by monocomponent FAD-dependent monooxygenases. This study identified the new monocomponent FAD-dependent monooxygenase HpaM that catalyzed the ortho decarboxylative hydroxylation of 5HPA. In addition, we found that the maiA gene coding for maleic acid cis-trans isomerase was pivotal for the metabolism of 5HPA, nicotinic acid, and picolinic acid in A. faecalis JQ135. This study provides novel insights into the microbial metabolism of pyridine derivatives.
KEYWORDS: Alcaligenes faecalis JQ135, 5-hydroxypicolinic acid, biodegradation, hpa operon, 5-hydroxypicolinic acid 2-monooxygenase, decarboxylative hydroxylation
ABSTRACT
5-Hydroxypicolinic acid (5HPA), a natural pyridine derivative, is microbially degraded in the environment. However, the physiological, biochemical, and genetic foundations of 5HPA metabolism remain unknown. In this study, an operon (hpa), responsible for 5HPA degradation, was cloned from Alcaligenes faecalis JQ135. HpaM was a monocomponent flavin adenine dinucleotide (FAD)-dependent monooxygenase and shared low identity (only 28 to 31%) with reported monooxygenases. HpaM catalyzed the ortho decarboxylative hydroxylation of 5HPA, generating 2,5-dihydroxypyridine (2,5DHP). The monooxygenase activity of HpaM was FAD and NADH dependent. The apparent Km values of HpaM for 5HPA and NADH were 45.4 μM and 37.8 μM, respectively. The genes hpaX, hpaD, and hpaF were found to encode 2,5DHP dioxygenase, N-formylmaleamic acid deformylase, and maleamate amidohydrolase, respectively; however, the three genes were not essential for 5HPA degradation in A. faecalis JQ135. Furthermore, the gene maiA, which encodes a maleic acid cis-trans isomerase, was essential for the metabolism of 5HPA, nicotinic acid, and picolinic acid in A. faecalis JQ135, indicating that it might be a key gene in the metabolism of pyridine derivatives. The genes and proteins identified in this study showed a novel degradation mechanism of pyridine derivatives.
IMPORTANCE Unlike the benzene ring, the uneven distribution of the electron density of the pyridine ring influences the positional reactivity and interaction with enzymes; e.g., the ortho and para oxidations are more difficult than the meta oxidations. Hydroxylation is an important oxidation process for the pyridine derivative metabolism. In previous reports, the ortho hydroxylations of pyridine derivatives were catalyzed by multicomponent molybdenum-containing monooxygenases, while the meta hydroxylations were catalyzed by monocomponent FAD-dependent monooxygenases. This study identified the new monocomponent FAD-dependent monooxygenase HpaM that catalyzed the ortho decarboxylative hydroxylation of 5HPA. In addition, we found that the maiA gene coding for maleic acid cis-trans isomerase was pivotal for the metabolism of 5HPA, nicotinic acid, and picolinic acid in A. faecalis JQ135. This study provides novel insights into the microbial metabolism of pyridine derivatives.
INTRODUCTION
Pyridine and its derivatives (such as nicotine, nicotinic acid [NA], picolinic acid [PA], vitamin B6, etc.) are among the most abundant natural N-heterocyclic compounds. Furthermore, artificial pyridine derivatives are widely used in agriculture and pharmaceutical and chemical industries as solvents, dyes, pharmaceuticals, herbicides, and pesticides (1–3). In addition, many of the pyridine derivatives are also toxic and carcinogenic (4, 5). Therefore, the biodegradation, detoxification, and transformation of pyridine derivatives are of significant interest. Unlike the benzene ring, the pyridine ring contains an N atom with stronger electronegativity than that of the C atom. The electron cloud shifted to the N atom, and the cloud density of the C atom on the pyridine ring is thus significantly reduced, especially at the ortho and para positions. In turn, the electrophilic substitution reaction (oxidation reaction) is more difficult to occur in the ortho and para positions than in the meta position.
Hydroxylation increases the hydrophilicity and polarity of the substrate and is thus typically the initial and key degradation step for pyridine derivatives. The hydroxylation could occur in ortho, meta, and para positions of the pyridine ring. Generally, the position and substituted group affect the ring hydroxylation of pyridine derivatives. As previously shown, hydroxylations that occur at the ortho position of the pyridine ring are catalyzed via multicomponent molybdenum-containing monooxygenases, e.g., nicotinate hydroxylase, 3-succinoyl-pyridine monooxygenase, and nicotine hydroxylase (6–8). On the other hand, the hydroxylations at the meta position are catalyzed via monocomponent flavin adenine dinucleotide (FAD)-dependent monooxygenases, e.g., 6-hydroxynicotinate 3-monooxygenase (NicC), 2,6-dihydroxypyridine 3-monooxygenase (2,6-DHPH), and 6-hydroxy-3-succinoyl-pyridine 3-monooxygenase (HspB) (6, 9, 10). However, a monocomponent FAD-dependent monooxygenase that catalyzes the ortho hydroxylation of the pyridine ring and its coding gene have not been identified thus far.
5-Hydroxypicolinic acid (5HPA) is a typical natural pyridine derivative with an ortho-substituted carboxyl group and a meta hydroxy group in the pyridine. 5HPA is produced either by the marine bacterium Nocardia sp. (11) or by microalgae (12). Recently, 5HPA was also found as a biologically active compound in several traditional Chinese medicines, including Hericium erinaceus, Gynura divaricata, and purslane (13–15). The microbial degradation or transformation of 5HPA has not been extensively studied to date. Previously, Karvelis et al. isolated a 5HPA-degrading strain, Pusillimonas sp. strain 5HP, from soil (16). In this strain, the degradation of 5HPA was found to be started by ortho decarboxylative hydroxylation, producing 2,5-dihydroxypyridine (2,5DHP). A 5-hydroxypicolinic acid 2-monooxygenase was partially purified from Pusillimonas sp. strain 5HP. However, both the amino acid sequence of the 5-hydroxypicolinic acid 2-monooxygenase and its coding genes remain unknown. In this study, we cloned an operon (hpa) responsible for 5HPA degradation in Alcaligenes faecalis JQ135, a picolinic acid-degrading bacterium reported previously (17). HpaM (5-hydroxypicolinic acid 2-monooxygenase) was identified as a monocomponent monooxygenase catalyzing the ortho decarboxylative hydroxylation of 5HPA (Fig. 1). In addition, the functions of the hpaDXF and maiA genes in the metabolism of 5HPA were investigated.
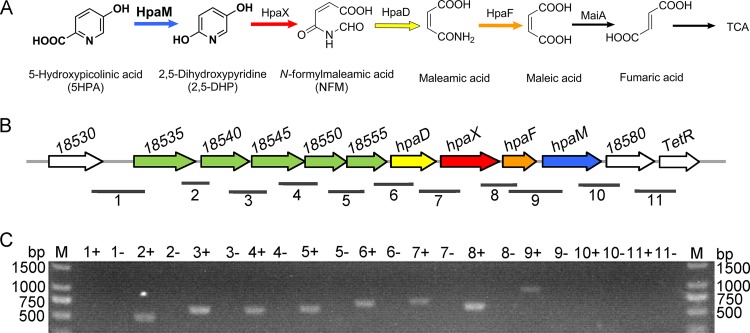
FIG 1.
Metabolism of 5HPA in A. faecalis JQ135. (A) Proposed 5HPA degradation pathway in A. faecalis JQ135. HpaM, 5-hydroxypicolinic acid 2-monooxygenase; HpaX, 2,5-dihydroxypyridine 5,6-dioxygenase; HpaD, N-formylmaleamic acid deformylase; HpaF, maleamic acid amidohydrolase; MaiA, maleic acid cis-trans isomerase; TCA, tricarboxylic acid cycle. (B) Organization of the hpa operon. Numbers above each gene represent the locus tags for each gene. The numbered horizontal lines below the hpa operon show the location and size of the PCR fragments in panel C. (C) Agarose gel electrophoresis of RT-PCR products. Plus and minus indicate that the cells were cultured with 5HPA and glycerol, respectively.
RESULTS
Degradation of 5HPA, NA, and nicotine by A. faecalis JQ135.
A. faecalis JQ135 was tested for its ability to degrade 5HPA, NA, and nicotine in mineral salts medium (MSM) supplemented with each substrate as sole carbon and nitrogen sources. The results showed that A. faecalis JQ135 completely degraded 1.0 mM 5HPA and NA but not nicotine within 8 h; correspondingly, the optical density at 600 nm (OD600) of the culture increased from approximately 0.3 to 0.5 (see Fig. S1 in the supplemental material). These results indicated that A. faecalis JQ135 could degrade and utilize 5HPA and NA, but not nicotine, as sole carbon and nitrogen sources for growth. Moreover, attempts to detect metabolic intermediates of 5HPA within the culture failed, which might be due to little or no intermediate excretion from strain JQ135 cells during the degradation of 5HPA.
Transposon mutagenesis and cloning of an operon involved in 5HPA degradation.
To clone the genes involved in 5HPA degradation, a transposon mutant library of A. faecalis JQ135 was constructed. One mutant (Z10) that could not grow in MSM agar containing 1.0 mM 5HPA was identified from approximately 5,000 mutants (Fig. S2). The genome of A. faecalis JQ135 was determined via PacBio sequencing. The complete genome of the strain contained one circular chromosome (4,078,346 bp), and no plasmid was found. A total of 3,723 open reading frames (ORFs) were predicted. The insertion position of the transposon, determined via DNA walking (18), was located in gene AFA_18575 (genome position 4070825). AFA_18575 (named hpaM) is 1,218 bp in length and has a G+C content of 56.08%. The predicted protein was used to search the NCBI database (http://blast.ncbi.nlm.nih.gov/) with BLASTP (Table 1). HpaM shared low identities with reported flavin-containing monooxygenases, e.g., 28% identity with salicylate 1-monooxygenase (SalM) from Pseudomonas putida KF715 (19) and 31% identity with NicC from P. putida KT2440 (6) (Fig. S3 and S4). NicC and SalM catalyze the decarboxylative hydroxylation of 6-hydroxynicotinic acid (6HNA) and salicylate, respectively. Downstream of hpaM, a gene (AFA_18580) coding for the DUF2236 family enzyme and a TetR-type regulator gene (AFA_18585) were predicted (Fig. 1; Table 1). Upstream of hpaM, eight genes were identified. AFA_18535, AFA_18540, and AFA_18545 were predicted to be branched-chain amino acid ABC transporter substrate-binding proteins, while AFA_18550 and AFA_18555 were predicted to be branched-chain amino acid ABC transporter ATP-binding proteins. Based on the above analysis, the five consecutive genes putatively encoded an ABC-type transporter. Genes hpaD, hpaX, and hpaF showed 57%, 53%, and 41% identities, respectively, with NicD (N-formylmaleamic acid deformylase), NicX (2,5DHP dioxygenase), and NicF (maleamate amidohydrolase) from P. putida KT2440 at the amino acid level (Fig. S3). Interestingly, NicC, NicX, NicD, and NicF formed a complete metabolic pathway for the transformation of 6HNA into maleic acid (6). Considering the structural similarity between 5HPA and 6HNA, we presumed that these contiguous genes were organized in an operon and were involved in the degradation of 5HPA in A. faecalis JQ135. In this study, this operon was designated hpa (5-hydroxypicolinic acid).
TABLE 1.
Sequence comparison of the hpa operon of A. faecalis JQ135 with database entries
| Gene locus | Product sizea | Databaseb | Homologous proteinc | Accession no. | Identity (%) | Proposed function |
|---|---|---|---|---|---|---|
| AFA_18535 | 369 | NR | Branched-chain amino acid ABC transporter, substrate-binding protein LivK | CUI69837 | 99 | ATP binding and 5-hydroxypicolinic acid transportation |
| Swiss-Prot | Leu/Ile/Val/Thr/Ala-binding protein | P21175 | 30 | |||
| AFA_18540 | 291 | NR | Branched-chain amino acid ABC transporter, substrate-binding protein LivH | WP_035269511 | 100 | |
| Swiss-Prot | High-affinity branched-chain amino acid transport system permease protein LivH | P0AEX8 | 35 | |||
| AFA_18545 | 319 | NR | Branched-chain amino acid ABC transporter, substrate-binding protein LivM | WP_083055103 | 99 | |
| Swiss-Prot | High-affinity branched-chain amino acid transport system permease protein LivM | P22729 | 30 | |||
| AFA_18550 | 252 | NR | Branched-chain amino acid ABC transporter, ATP-binding protein LivG | WP_042485129 | 100 | |
| Swiss-Prot | High-affinity branched-chain amino acid transport system permease protein LivG | P0A9S8 | 41 | |||
| AFA_18555 | 242 | NR | Branched-chain amino acid ABC transporter, ATP-binding protein LivF | WP_035269507 | 100 | |
| Swiss-Prot | High-affinity branched-chain amino acid transport system permease protein LivF | P0A191 | 46 | |||
| AFA_18560 (hpaD) | 275 | NR | Alpha/beta hydrolase | WP_042485331 | 98 | N-Formylmaleamic acid deformylase HpaD |
| Swiss-Prot | N-Formylmaleamate deformylase NicD | Q88FY3 | 57 | |||
| AFA_18565 (hpaX) | 346 | NR | 2,5-Dihydroxypyridine 5,6-dioxygenase | WP_083055106 | 98 | 2,5-Dihydroxypyridine 5,6-dioxygenase HpaX |
| Swiss-Prot | 2,5-Dihydroxypyridine 5,6-dioxygenase NicX | Q88FY1 | 53 | |||
| AFA_18570 (hpaF) | 205 | NR | N-Carbamoylsarcosine amidase | WP_060186798 | 100 | Maleamic acid amidohydrolase HpaF |
| Swiss-Prot | Maleamate amidohydrolase NicF | Q88FY5 | 41 | |||
| AFA_18575 (hpaM) | 405 | NR | Salicylate 1-monooxygenase | ARP55611 | 100 | 5-Hydroxypicolinic acid 2-monooxygenase HpaM |
| Swiss-Prot | 6-Hydroxynicotinate 3-monooxygenase NicC | Q88FY2 | 35 | |||
| AFA_18580 | 291 | NR | DUF2236 domain-containing protein | WP_086060754 | 99 | Unknown function |
| Swiss-Prot | Ubiquitin-activating enzyme E1C | Q99MI7 | 25 | |||
| AFA_18585 | 235 | NR | TetR family transcriptional regulator | WP_035269502 | 100 | Transcriptional regulator |
| Swiss-Prot | HTH type transcriptional regulator BetI | Q13NG5 | 41 |
Number of amino acids.
NR, NCBI nonredundant protein sequence database.
The top BLASTP hit was selected.
Heterologous expression and purification of HpaM and function determination.
The gene hpaM was cloned into pET29a(+) and expressed in Escherichia coli BL21(DE3). The N-terminal 6×His-tagged HpaM was purified using nickel affinity chromatography. The purified enzyme migrated as a single band, which was in agreement with its theoretical value (45.2 kDa) (Fig. S5). The result of gel filtration chromatography indicated that HpaM was a dimer (Fig. S6).
HpaM was predicted to contain FAD binding domains (such as GXGXXG) (Fig. S4). However, purified HpaM was colorless, suggesting that no flavin was associated with the enzyme after purification. In addition, the UV-visible spectrum of HpaM also did not show the typical absorption of a FAD cofactor at 380 nm to 480 nm (Fig. S7). Purified HpaM showed enzymatic activity only after the addition of external FAD (but not flavin mononucleotide [FMN]). NADH (but not NADPH) was used as an electron donor, and no activity was detected under anaerobic conditions. These results indicate that HpaM was FAD, NADH, and O2 dependent, which resembles 5-hydroxypicolinic acid 2-monooxygenase purified from Pusillimonas sp. strain 5HP (16). High-performance liquid chromatography (HPLC) results showed that 5HPA (with a retention time of 5.52 min) decreased and a product (with a retention time of 5.22 min) accumulated (Fig. S8). The retention time of the product was equal to that of the authentic 2,5DHP standard, and liquid chromatography-mass spectrometry (LC-MS) analysis showed that the product had a molecular ion at m/z 112.0400 [M+H]+, which was identical to that of 2,5DHP (16, 20, 21). Thus, the product was identified as 2,5DHP. Based on the above analysis, HpaM was identified as a monocomponent FAD-dependent monooxygenase catalyzing the ortho decarboxylative hydroxylation of 5HPA producing 2,5DHP.
Analysis of kinetic constants and biochemical properties.
The Km and kcat values of HpaM for 5HPA were 45.4 ± 4.2 μM and 10.2 ± 0.3 s−1, respectively (Fig. S9). The apparent catalytic efficiency (kcat/Km) of HpaM was 225.5 s−1 mM−1, and the apparent Km value of HpaM for NADH was 37.8 ± 3.4 μM. The optimal pH of HpaM was 7.0; when the pH exceeded 8.0, the enzyme activity decreased dramatically (Fig. S10).
The following structural analogs of 5HPA were tested as the substrates in a standard activity test (Table S1): PA, NA, 5-aminopicolinic acid, 5-methylpicolinic acid, 5-chloropicolinic acid, 5-bromoopicolinic acid, 3-hydroxypicolinic acid, 6-hydroxypicolinic acid, 2,3-pyridinedicarboxylic acid, 2,5-pyridinedicarboxylic acid, 2,6-pyridinedicarboxylic acid, 2-hydroxynicotinic acid, 4-hydroxynicotinic acid, 5-hydroxynicotinic acid, 6-hydroxynicotinic acid, 3-hydroxyisonicotinic, and 6-hydroxy-3-succinoylpyridine (HSP). HpaM showed no activity against any of these compounds, indicating that the hydroxylase activity was 5HPA specific.
Functional study of the genes hpaD, hpaX, and hpaF.
As mentioned above, HpaD, HpaX, and HpaF shared the highest homology with NicD, NicX, and NicF from P. putida KT2440, respectively, suggesting that HpaX, HpaD, and HpaF convert 2,5DHP into maleic acid. To study the function of hpaX, the gene was cloned into pET29a(+) and expressed in E. coli BL21(DE3) (Fig. S11). HPLC results showed that 2,5DHP was degraded by purified HpaX (Fig. S12). When Fe2+ was absent or under anaerobic conditions, no 2,5DHP dioxygenase activity was detected. These characteristics were similar to those of previously reported Fe2+- and O2-dependent 2,5DHP dioxygenases (6, 16, 20, 22). HpaX degraded 2,5DHP with Km and kcat values of 77.9 ± 6.2 μM and 11.6 ± 0.3 s−1, respectively (Fig. S13).
The activities of HpaD and HpaF were more difficult to determine. To investigate the function of hpaD and hpaF, a DNA fragment containing the hpaDXFM genes plus a maleic acid cis-trans isomerase gene (maiA) was cloned into the 5HPA-nondegrading bacterium Sphingomonas wittichii DC-6 (23). The reason for inclusion of the maiA gene was that the MaiA converts maleic acid to fumaric acid (6, 22) and the hpa operon lacks the maiA gene. Bioinformatic analysis showed that the gene AFA_16520, which was physically distant from the hpa operon, encoded a putative protein with 99% identity to a previously reported MaiA from A. faecalis IFO13111 (24). Thus, AFA_16520 (maiA) was predicted to convert maleic acid to fumaric acid in A. faecalis JQ135. The whole-cell transformation experiments showed that recombinant DC-6/pBBR-hpaDXFM-maiA acquired the ability to degrade and utilize 5HPA for growth while DC-6/pBBR-hpaDXFM and DC-6/pBBR-maiA did not (Fig. S14). These results suggest that hpaD, hpaX, and hpaF were responsible for the conversion of 2,5DHP to maleic acid.
Deletion of the hpa genes and maiA.
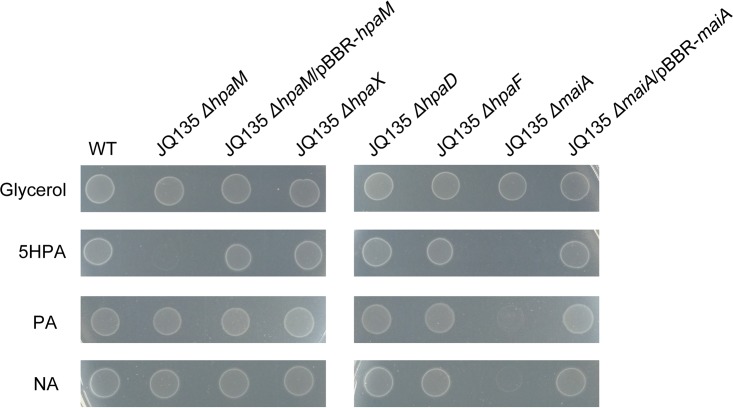
To determine the physiological functions of hpaD, hpaX, hpaF, and hpaM in vivo, the in-frame deletion mutants JQ135 ΔhpaD, JQ135 ΔhpaX, JQ135 ΔhpaF, and JQ135 ΔhpaM, respectively, were constructed. These four mutants could still grow on MSM plates supplemented with glycerol as the carbon source, indicating that these four genes did not affect the basic metabolism (Fig. 2). However, mutant JQ135 ΔhpaM lost the ability to utilize 5HPA for growth, while the complemented strain JQ135 ΔhpaM/pBBR-hpaM recovered the ability to grow on 5HPA. The result clearly demonstrated that hpaM was essential for the catabolism of 5HPA in A. faecalis JQ135. Furthermore, mutant JQ135 ΔhpaM could grow on NA and PA, indicating that hpaM was not involved in the degradation of NA or PA in A. faecalis JQ135. Interestingly, the mutants JQ135 ΔhpaD, JQ135 ΔhpaX, and JQ135 ΔhpaF could still grow on 5HPA, which might be because the other isoenzymes of HpaX, HpaD, and HpaF were present in A. faecalis JQ135.
FIG 2.
Growth phenotype of wild-type A. faecalis JQ135 and mutant derivatives on MSM plates with 1.0 mM glycerol, 5HPA, PA, and NA as sole carbon sources.
To investigate the function of maiA, this gene was also deleted from A. faecalis JQ135. The mutant JQ135 ΔmaiA could still degrade 5HPA, NA, and PA but lost the ability to utilize the three substrates for growth (Fig. 2). These results indicated maiA to be essential for 5HPA, NA, and PA metabolism in A. faecalis JQ135.
Transcription of hpa genes in response to 5HPA, NA and PA.
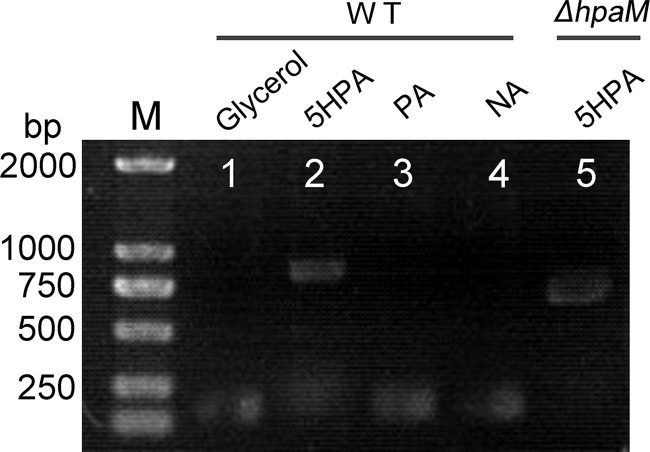
Reverse transcription-PCR (RT-PCR) was conducted to investigate whether the transcription of hpa genes was induced by 5HPA. The results showed that the five putative transporter genes (AFA_18535 to AFA_18555), hpaD, hpaX, hpaF, and hpaM comprised a transcriptional operon and were transcribed in cells grown on 5HPA but not on glycerol (Fig. 1C). RT-PCR results showed that hpaM was not transcribed in cells grown on NA or PA (Fig. 3). Moreover, in mutant JQ135 ΔhpaM, the transcription of the hpa operon was still induced by 5HPA, suggesting that 5HPA but not its metabolites was the inducer.
FIG 3.
Agarose gel electrophoresis of RT-PCR products generated using RNA of A. faecalis JQ135 or JQ135 ΔhpaM. Lane M, ladder DNA marker; lanes 1, 2, 3, and 4, RT-PCR products of A. faecalis JQ135 grown in MSM supplemented with 1.0 mM glycerol, 5HPA, PA, and NA, respectively; lane 5, RT-PCR products of JQ135 ΔhpaM incubated in MSM supplemented with 1.0 mM 5HPA.
DISCUSSION
5HPA is a natural pyridine derivative produced by many marine bacteria, microalgae, and plants. The microbial metabolism of 5HPA has been studied only preliminarily in Pusillimonas sp. 5HP (16). In this study, we cloned the operon hpa from strain A. faecalis JQ135, which was involved in the metabolism of 5HPA. 5HPA was initially decarboxylatively hydroxylated at the ortho position of the pyridine ring by the monocomponent FAD-dependent monooxygenase HpaM, yielding 2,5DHP. Furthermore, 2,5DHP was further decomposed by HpaX, HpaD, HpaF, and MaiA. Thus, this study provided a physiological and genetic base of the microbial metabolism of 5HPA.
Flavin-dependent monooxygenases constitute an important class of enzymes that utilize a reduced flavin to oxidize their substrates. Flavin-dependent monooxygenases catalyze a variety of reactions, such as hydroxylation, Baeyer-Villiger oxidation, sulfoxidation, halogenation, and epoxidation reactions, and play an important role in the metabolism of natural and xenobiotic compounds (25). So far, a number of flavin-dependent monooxygenases catalyzing the hydroxylation of aromatic compounds have been identified. Most of them catalyzed the hydroxylation of the substituted/unsubstituted carbon atom in the benzene ring (26–28). Only three of them (NicC, 2,6-DHPH, and HspB) catalyzed the hydroxylation of the pyridine ring, and their hydroxylation happened at the meta position (6, 10, 29). To the best of our knowledge, HpaM is the only reported monocomponent FAD-dependent monooxygenase that catalyzes the ortho decarboxylative hydroxylation of the pyridine ring. Based on their structural features, protein sequence motifs, electron donor, and type of oxygenation reaction, flavin-dependent monooxygenases can be divided into eight groups (A to H) (25). Group A flavin monooxygenases are monocomponent, contain a glutathione reductase (GR-2) type Rossmann fold for FAD binding, and use NAD(P)H as the electron donor. HpaM was monocomponent and FAD dependent and used NADH as the electron donor. Sequence alignment and structure prediction results suggested that HpaM has a classical GR-2 type Rossmann fold (GXGXXG) (see Fig. S4 in the supplemental material). A phylogenetic tree of related monocomponent flavin-dependent monooxygenases showed that HpaM was clustered with group A enzymes and formed a subclade with NicC (Fig. 4) (25, 30). These indications suggest that HpaM belongs to group A flavin-dependent monooxygenases. However, HpaM shared very low identities with reported group A flavin-dependent monooxygenases, showed strict substrate specificity for 5HPA, and did not hydroxylate PA, NA, 6HNA, or HSP. Strict substrate specificity appears to be a common feature for flavin monooxygenases in group A, which may be due to the diversity of substrate binding sites among these enzymes; e.g., the 6HNA binding sites His47, Cys202, Met213, and His302 (NicC numbering) in NicC of P. putida KT2440 were not found in HpaM (Fig. S4). The diverse binding sites of group A flavin monooxygenases have led to their adaptation to different substrates. Based on the above analysis, HpaM was a novel group A flavin-dependent monooxygenase.
FIG 4.
Phylogenetic analysis of HpaM. (A) The phylogenetic tree was constructed based on alignment of HpaM with related monocomponent flavin-dependent monooxygenases. Multiple-alignment analysis was performed using ClustalX v2.0. The phylogenetic tree was constructed via the neighbor-joining algorithm using MEGA 6.0, and bootstrap values (based on 1,000 replications) are indicated at branch nodes. Bar, 0.20 substitutions per amino acid position. Each item was arranged in the following order: protein name, UniProtKB/Swiss-Prot accession number, and EC number. (B) Comparison of reactions catalyzed via HpaM (red, ortho hydroxylation) and three monooxygenases (blue, meta hydroxylation), NicC, HspB, and 2,6-DHPH. (C) Five C—C bond cleavage hydroxylation reactions of the benzene ring. SalM, salicylate 1-monooxygenase; pHB1M, p-hydroxybenzoate 1-monooxygenase; 4AB1M, 4-aminobenzoate 1-monooxygenase; HipH, 4-hydroxyisophthalate 3-monooxygenase; PhzS, 5-methylphenazine-1-carboxylate 1-monooxygenase. Proteins in panels B and C are indicated in bold in the phylogenetic tree.
2,5DHP is a common catabolic intermediate of several pyridine derivatives, such as 6HNA (from NA) in P. putida KT2440, HSP (from nicotine) in P. putida S16, 2-hydroxypyridine in Arthrobacter sp., and 3-methyl-pyridine in Pseudomonas sp. KM-3 (31). This study showed that 2,5DHP was also an intermediate of 5HPA degradation in A. faecalis JQ135. The metabolism of 2,5DHP involves four steps: 2,5DHP is initially ring cleaved into N-formylmaleamic acid by a 2,5DHP dioxygenase; N-formylmaleamic acid is subsequently transformed to maleamic acid by N-formylmaleamate deformylase, which is then converted to maleic acid by maleamate amidohydrolase; and maleic acid is finally transformed into fumaric acid, a metabolite of citric acid cycle, by maleic acid cis-trans isomerase (6, 20, 22). Our study demonstrated that the metabolism of 2,5DHP produced from 5HPA degradation also followed the same pathway (Fig. 1A). The degradation of 6HNA, HSP, and 5HPA differed only at the initial steps, which started with three different flavin monooxygenases, NicC, HspB, and HpaM (Fig. S3). However, it is interesting that the JQ135 ΔhpaD, JQ135 ΔhpaX, and JQ135 ΔhpaF mutants could still grow on 5HPA. Bioinformatic analysis revealed that there were two other gene clusters (AFA_14960 to AFA_14970 and AFA_15130 to AFA_15140) that contained homologous genes, sharing 54 to 69% identities with hpaD, hpaX, and hpaF predicted in the genome of A. faecalis JQ135. It is possible that these homologous genes were involved in the metabolism of other pyridine derivatives (such as NA or PA) with 2,5DHP as an intermediate. It is interesting that only one copy of the maleic acid cis-trans isomerase gene maiA (AFA_16520) was found in the genome of A. faecalis JQ135 and that maiA was not located in the two clusters and the hpa operon. In addition, gene deletion results demonstrated that the maiA gene was essential for the degradation of 5HPA, PA, and NA in A. faecalis JQ135. These analyses suggest that MaiA was a pivotal enzyme of the metabolism of the pyridine derivatives in A. faecalis JQ135, i.e., each pyridine derivative was converted to maleic acid, and then MaiA isomerized maleic acid to fumaric acid, which is a metabolite of citric acid cycle. The possible reason for this arrangement is that it is more advantageous for the flexible regulation of the metabolism of various pyridine derivatives and saves genetic resources. Moreover, maleic acid can also be hydrated to d-malic acid by some microorganisms (32–34). However, the inability to utilize 5HPA, NA, and PA of mutant JQ135 ΔmaiA suggests that this d-malic acid shunt pathway does not exist in A. faecalis JQ135.
MATERIALS AND METHODS
Chemicals and media.
5HPA (99%) and its structural analogs were purchased from J&K Scientific Ltd. (Shanghai, China). 2,5DHP (98%) was purchased from SynChem OHG (Altenburg, Germany). All other chemicals and solvents used for this experiment were commercially available. Enzymes used in this study were purchased from Vazyme Biotech Co., Ltd. (Nanjing, China). Mineral salts medium (MSM) and Luria-Bertani medium (LB) have been described previously (17). To prepare nitrogen-free MSM, NH4NO3 was omitted.
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 2. A. faecalis JQ135 has previously been identified as a picolinic acid-degrading bacterium (17). E. coli strains were grown at 37°C, while other strains were grown at 30°C. Media were supplemented with chloramphenicol (Cm; 34 μg/ml), kanamycin (Km; 50 μg/ml), gentamicin (Gm; 50 μg/ml), or streptomycin (Str; 50 μg/ml) as required.
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| Alcaligenes faecalis | ||
| JQ135 | Strr; wild type; 5HPA-degrading strain; G− | 17 |
| Z10 | Strr Kmr; transposon mutant of JQ135; 5HPA growth deficient | This study |
| JQ135 ΔhpaM | Strr; hpaM deletion mutant of JQ135 | This study |
| JQ135 ΔhpaM/pBBR-hpaM | Strr Gmr; JQ135 ΔhpaM containing pBBR-hpaM | This study |
| JQ135 ΔhpaD | Strr; hpaD deletion mutant of JQ135 | This study |
| JQ135 ΔhpaX | Strr; hpaX deletion mutant of JQ135 | This study |
| JQ135 ΔhpaF | Strr; hpaF deletion mutant of JQ135 | This study |
| JQ135 ΔmaiA | Strr; maiA deletion mutant of JQ135 | This study |
| JQ135 ΔmaiA/pBBR-maiA | Strr Gmr; JQ135 ΔmaiA containing pBBR-maiA | This study |
| Sphingomonas wittichii | ||
| DC-6 | Chloroacetanilide herbicide degrader; cannot degrade 5HPA | 21 |
| DC-6/pBBR-hpaDXFM-maiA | DC-6 containing pBBR-hpaDXFM-maiA | This study |
| DC-6/pBBR-hpaDXFM | DC-6 containing pBBR-hpaDXFM | This study |
| DC-6/pBBR-maiA | DC-6 containing pBBR-maiA | This study |
| Escherichia coli | ||
| DH5α | F− recA1 endA1 thi-1 hsdR17 supE44 relA1 deoRΔ(lacZYA-argF) U169 ϕ80lacZΔM15 | TaKaRa |
| SM10λpir | Donor strain for biparental mating | 40 |
| BL21(DE3) | F− ompT hsdS(rB− mB−) gal dcm lacY1 (DE3) | Novagen |
| Plasmids | ||
| pET29a(+) | Kmr; expression plasmid | Novagen |
| pSC123 | Cmr Kmr; suicide plasmid, mariner transposon | 40 |
| pJQ200SK | Gmr; mob+ orip15A lacZα+ sacB; suicide plasmid | 41 |
| pBBR1-MCS5 | Gmr; broad-host-range cloning plasmid | 42 |
| pJQΔhpaM | Gmr; hpaM gene deletion plasmid; the upstream and downstream regions of hpaM gene fused into SacI/PstI-digested pJQ200SK | This study |
| pJQΔhpaD | Gmr; hpaD gene deletion plasmid; the upstream and downstream regions of hpaD gene fused into SacI/PstI-digested pJQ200SK | This study |
| pJQΔhpaX | Gmr; hpaX gene deletion plasmid; the upstream and downstream regions of hpaX gene fused into SacI/PstI-digested pJQ200SK | This study |
| pJQΔhpaF | Gmr; hpaF gene deletion plasmid; the upstream and downstream regions of hpaF gene fused into SacI/PstI-digested pJQ200SK | This study |
| pJQΔmaiA | Gmr; maiA gene deletion plasmid; the upstream and downstream regions of maiA gene fused into SacI/PstI-digested pJQ200SK | This study |
| pBBR-hpaM | Gmr; hpaM gene complementation plasmid; the hpaM gene fused into XhoI/HindIII-digested pBBR1-MCS5 | This study |
| pBBR-maiA | Gmr; hpaM gene complementation plasmid; the maiA gene fused into XhoI/HindIII-digested pBBR1-MCS5 | This study |
| pBBR-hpaDXFM | Gmr; hpaDXFM genes fused into XhoI/HindIII-digested pBBR1-MCS5 | This study |
| pBBR-hpaDXFM-maiA | Gmr; hpaDXFM and maiA genes fused into XhoI/HindIII-digested pBBR1-MCS5 | This study |
| pET-hpaM | Kmr; NdeI-XhoI fragment containing hpaM inserted into pET29a(+) | This study |
| pET-hpaX | Kmr; NdeI-XhoI fragment containing hpaX inserted into pET29a(+) | This study |
Isolation of 5HPA growth-deficient mutants and determination of the transposon insertion site.
Both generation and selection of 5HPA growth-deficient mutants of A. faecalis JQ135 were performed according to a selection method previously used to identify picolinic acid (PA) growth-deficient mutants (17). The plasmid pSC123 (carrying the mariner transposon) was used as a transposon delivery plasmid. 5HPA was substituted for PA. The insertion site of the transposon was determined via self-formed adaptor PCR, as previously described (17, 18).
Gene deletion and complementation.
Standard DNA manipulation techniques were performed as previously described (35). All primers used in this study are listed in Table 3. Deletion of the hpaM, hpaD, hpaX, hpaF, and maiA genes in A. faecalis JQ135 was performed via the two-step homogenetic recombination method using the suicide plasmid pJQ200SK. Using the deletion of hpaM as an example, two primer pairs (kohpaM-UF/kohpaM-UR and kohpaM-DF/kohpaM-DR) were used to amplify the homologous recombination-directing sequences. Both PCR fragments were then cloned into SacI/PstI-digested pJQ200SK using the ClonExpress MultiS one-step cloning kit (Vazyme Biotech Co., Ltd., Nanjing, China). The resulting plasmid, pJQ-ΔhpaM, was then introduced into A. faecalis JQ135 cells. Single-crossover mutants were screened on LB plates containing Str and Gm. A double-crossover mutant (JQ135 ΔhpaM) was selected on LB plates containing Str and 10% (wt/vol) sucrose.
TABLE 3.
Primers used in the study
| Primer | Sequence (5′–3′) | Description |
|---|---|---|
| kohpaM-UF | AGCTTGATATCGAATTCCTGCAGCCATCATCATGACGGTCGAGAACG | To construct plasmid pJQΔhpaM |
| kohpaM-UR | TCTAGAACTAGTGGATCCGACCGCATTCGGAGTCATTTGAATC | |
| kohpaM-DF | GGATCCACTAGTTCTAGAGGCATCAAGACGGACTGGGTTTA | |
| kohpaM-DR | CTAAAGGGAACAAAAGCTGGAGCTCCAATGTGGTACCGGCATAGCCAG | |
| kohpaD-UF | AGCTTGATATCGAATTCCTGCAGAAACCCAGATTGTTGATGCTGG | To construct plasmid pJQΔhpaD |
| kohpaD-UR | TCTAGAACTAGTGGATCCGCCATAGCTAAGTCGCGATAGG | |
| kohpaD-DF | GGATCCACTAGTTCTAGATCTTACGTGTACCAGACGCTGG | |
| kohpaD-DR | CTAAAGGGAACAAAAGCTGGAGCTACTTCATTATTGGGGCCAAAGG | |
| kohpaX-UF | AGCTTGATATCGAATTCCTGCAGATGGCGTGTCCATTCTGGTGG | To construct plasmid pJQΔhpaX |
| kohpaX-UR | TCTAGAACTAGTGGATCCAGACTCAGCACCTCATTCCAGG | |
| kohpaX-DF | GGATCCACTAGTTCTAGAAAAGAGCAAACCCTGGGCATGG | |
| kohpaX-DR | CTAAAGGGAACAAAAGCTGGAGCTCTCGGGCAAGAAACCAACCTTGG | |
| kohpaF-UF | AGCTTGATATCGAATTCCTGCAGTCGGGCACCAAAATTCTCCTGG | To construct plasmid pJQΔhpaF |
| kohpaF-UR | TCTAGAACTAGTGGATCCCTGTCGCTCGTAAACACTCTGG | |
| kohpaF-DF | GGATCCACTAGTTCTAGATTTGTCCTGGCAGATTGCGTGG | |
| kohpaF-DR | CTAAAGGGAACAAAAGCTGGAGCTCGGTTCTTCCTCCCCAAAGATGG | |
| komaiA-UF | AGCTTGATATCGAATTCCTGCAGGCGAAACGGGTAGGCTTCTATG | To construct plasmid pJQΔmaiA |
| komaiA-UR | TCTAGAACTAGTGGATCCAGTGGGTCTGTTGAACCCGGAAAAC | |
| komaiA-DF | GGATCCACTAGTTCTAGAGCAACCAGGCAAGCGTAGGCCATCACG | |
| komaiA-DR | TAAAGGGAACAAAAGCTGGAGCTCGACATGATTTTTCAGCAACAC | |
| hpaM-F | GGTACCGGGCCCCCCCTCGAGGTACGTGCTTCCGTCGTGGA | To construct plasmid pBBR-hpaM |
| hpaM-R | CAGGAATTCGATATCAAGCTTGCAAGCACAATGCAGAAATCC | |
| maiA-F | GGTACCGGGCCCCCCCTCGAGGTACGTGCTTCCGTCGTGGA | To construct plasmid pBBR-maiA |
| maiA-R | CAGGAATTCGATATCAAGCTTGCAAGCACAATGCAGAAATCC | |
| hpaDXFM-F | CGAGGTCGACGGTATCGATAAGCTTAGCATGGCGTGTCCATTCTGG | To construct plasmid pBBR-hpaDXFM-maiA |
| hpaDXFM-R | GGCCGATGCGGTAGGTTTTCATTTAAACCTGTAATGCTTTCGGCTG | |
| maiA-F2 | CAGCCGAAAGCATTACAGGTTTAAATCTCTGCCACTCCTGCCATCG | |
| maiA-R2 | ACTCACTATAGGGCGAATTGGAGCTCTTAGGCTTGTGGCTTGGCACCCGA | |
| exphpaMF | TAAGAAGGAGATATACATATGAAAAAAACGTTGAAAGTAGGTGTGAT | To construct plasmid pET-hpaM |
| exphpaMR | GTGGTGGTGGTGGTGCTCGAGAACCTGTAATGCTTTCGGCTGA | |
| exphpaXF | TAAGAAGGAGATATACATATGGCTGTATCTGATCATCAAATGGTTCA | To construct plasmid pET-hpaX |
| exphpaXR | GTGGTGGTGGTGGTGCTCGAGTGCCAACTCCTCTACCACTTTGC | |
| RT1F | GAGATGGTCAAACAGTTCCAGG | To amplify fragment 1 in Fig. 1 |
| RT1R | TACGCTCTTTGACAGCCAACTTGG | |
| RT2F | GATCCGGACAAGATTCGTCTGG | To amplify fragment 2 in Fig. 1 |
| RT2R | TTGTCCCGGATACGAGCAATCAGG | |
| RT3F | AGCTCCATTTATGTCGCGCAGG | To amplify fragment 3 in Fig. 1 |
| RT3R | CCAGCGTCACAATGACAAAGAAGG | |
| RT4F | GCTACTCGCAGTTTGTCTTTGTGG | To amplify fragment 4 in Fig. 1 |
| RT4R | AAACACTCAGAGCCCGCAGCAC | |
| RT5F | TGGCCAGCTCCTTGTCTTGTGG | To amplify fragment 5 in Fig. 1 |
| RT5R | TGTCTTCAACGGTCATTTCAGG | |
| RT6F | TGAAGTGGAGCGGCGTAAAGG | To amplify fragment 6 in Fig. 1 |
| RT6R | ATGGAGTGCCCCACATAAATGG | |
| RT7F | TATCCGGCGAATATCGCCTGG | To amplify fragment 7 in Fig. 1 |
| RT7R | TGTTCGGGAGAGAACAGCAAGG | |
| RT8F | GGCTTACGCCATGTCCCACATAGG | To amplify fragment 8 in Fig. 1 |
| RT8R | CAGTACCAAAGAAGGCCGATGG | |
| RT9F | TATTCGGTCTGAAAGTCCCCGG | To amplify fragment 9 in Fig. 1 |
| RT9R | TACTTCCGAACGCACTCCATCGG | |
| RT10F | GGTGAATGAAACAGCCAGCTGG | To amplify fragment 10 in Fig. 1 |
| RT10R | CGGGTCATTGGCATCGTAAGG | |
| RT11F | TCTGCAATCCCATATGCGCTGG | To amplify fragment 11 in Fig. 1 |
| RT11R | GTAGACGCTCATCGGAAATGG | |
| RT-ΔhpaMR | TAAACCTGTAATGCTTTCGGCTG | To amplify fragment 5 with RT9F in Fig. 3 |
The plasmid pBBR-hpaM was constructed for gene complementation. hpaM was amplified using the primers hpaM-F and hpaM-R and then ligated to XhoI/HindIII-digested pBBR1-MCS5, thus generating pBBR-hpaM. The pBBR-hpaM plasmid was then transferred into the JQ135 ΔhpaM mutant via biparental mating to generate the complemented strain JQ135 ΔhpaM/pBBR-hpaM. The plasmid pBBR-maiA and complementation strain JQ135 ΔmaiA/pBBR-maiA were constructed in a similar manner.
A 3.9-kbp DNA fragment containing hpaDXFM was amplified and fused into XhoI/HindIII-digested pBBR1-MCS5, thus yielding the plasmid pBBR-hpaDXFM. The 3.9-kbp DNA fragment containing hpaDXFM and a 0.9-kbp DNA fragment containing maiA were amplified and fused into XhoI/HindIII-digested pBBR1-MCS5, thus yielding plasmid pBBR-hpaDXFM-maiA. These plasmids were transferred into Sphingomonas wittichii DC-6 by biparental mating (36), generating several recombinant strains (Table 2).
RNA extraction and RT-PCR.
The A. faecalis JQ135 wild-type and JQ135 ΔhpaM mutant strains were cultured in glycerol-containing medium and harvested at mid-exponential phase, washed twice with MSM, and resuspended in MSM. The cell suspension (OD600 of 0.6) was transferred into a 50-ml flask containing 20 ml MSM supplemented with 1 mM substrate (glycerol, 5HPA, NA, or PA). After incubation at 30°C for 6 h, the cells were harvested. Total RNA was isolated using an RNA isolation kit (TaKaRa). Reverse transcription (RT)-PCR was conducted with a PrimeScript RT reagent kit (TaKaRa). All RT-PCR primers are listed in Table 3. All samples were run in triplicate.
Cloning and expression of hpaM and hpaX genes.
The hpaM gene (excluding the stop codon) was PCR amplified with primers exphpaMF and exphpaMR and fused into NdeI/XhoI-digested pET29a(+), thus producing pET-hpaM. The hpaX gene was PCR amplified with primers exphpaXF and exphpaXR and fused into NdeI/XhoI-digested pET29a(+), thus producing pET-hpaX. The recombinant plasmids pET-hpaM and pET-hpaX were transformed into E. coli BL21(DE3), and the respective genes were overexpressed. Cells were cultured in LB at 37°C to an OD600 of 0.5 and then induced for 16 h via the addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 16°C. His-tagged proteins were purified via Ni2+-nitrilotriacetic acid agarose chromatography (Novagen), eluted using 100 mM imidazole, dialyzed against phosphate-buffered saline (PBS) (50 mM, pH 7.0) at 4°C for 24 h, and analyzed via 12.5% SDS-PAGE. The oligomeric state of the native protein was determined via gel filtration chromatography (37). HpaM was loaded onto a column of Superdex 2000 (GE Healthcare AKTA Prime liquid system) that was equilibrated with 10 mM Tris-HCl buffer (pH 7.5, with 0.1 M NaCl and 5% glycerol) at a flow rate of 1 ml min−1. The standard proteins myosin (200 kDa), β-galactosidase (116 kDa), phosphorylase b (97 kDa), bovine serum albumin (66 kDa), and ovalbumin (45 kDa) were used.
Enzyme assays.
All enzyme activities were measured at 25°C in 50 mM PBS (pH 7.0), in a total volume of 1 ml using a UV2450 spectrophotometer (Shimadzu). HpaM activity was analyzed by measuring NADH oxidation at 370 nm (ε = 2,470 M−1 cm−1) (38) instead of at its λmax of 340 nm to avoid interference with the absorption spectra of 5HPA and 2,5DHP. The standard reaction mixture contained 0.2 mM FAD, 0.5 mM NADH, 0.2 mM 5HPA, and 1 μg purified HpaM. The reference cuvette contained all of these components except 5HPA. The assay was initiated via the addition of the substrate 5HPA. One unit of HpaM activity was defined as the amount of enzyme required for the oxidation of 1 μmol of NADH per min. HpaM activity toward 5HPA analogs was determined by replacing 5HPA with the equimolar analogs based on the standard reaction. The reactions were monitored for 10 min. To determine the Km for 5HPA, a range of 5HPA concentrations (3.6 to 360 μM) were employed at a fixed concentration of the other components in the standard reaction mixture. Similarly, when the Km for NADH was determined, a range of NADH concentrations (5 to 500 μM) were employed with fixed concentrations of the other components in the standard reaction mixture. The values were calculated via nonlinear regression fitting to the Michaelis-Menten equation using ORIGIN software version 8.5. Buffers at pH values from 4.0 to 6.0 (citric acid-sodium phosphate), 6.0 to 8.0 (KH2PO4-K2HPO4 buffer), and 8.5 to 10.0 (glycine-NaOH) were used in pH dependency experiments (39). To determine enzyme activity under anaerobic conditions, PBS was purged with high-purity nitrogen (99.99% N2) for 20 min to remove oxygen before the substrate was added.
HpaX (2,5DHP dioxygenase) activity was measured by assaying for 2,5DHP degradation at 320 nm (ε = 5,200 M−1 cm−1), according to several previously published reports (6, 16, 20, 22). The reaction mixture contained 1 mM 2,5DHP, 1 mM Fe2+, and 10 ng purified HpaX protein in 1 ml of 50 mM PBS buffer (pH 7.0). One unit of activity was defined as the amount of enzyme that catalyzed the degradation of 1 μmol of 2,5DHP in 1 min. For determination of kinetic constants, 2,5DHP was appropriately diluted at several concentrations (18 to 315 μM).
Analytical methods.
Determinations of 5HPA and 2,5DHP were performed via HPLC analysis on a Shimadzu AD20 system equipped with a Phecda C18 reversed-phase column (250 mm by 4.60 mm, 5 μm) with array detection at 310 nm. The mobile phase consisted of methanol-water-formic acid (12.5:87.5:0.2, vol/vol/vol) at a flow rate of 0.6 ml/min at 30°C. All assays in this study were independently performed three times, and the means and standard errors of measurements were calculated. LC-MS analysis was performed in a Thermo (America) DECA-60000 XLCQ Deca XP Plus instrument as previously described (21).
Accession number(s).
The hpa operon sequence and the complete genome sequence of A. faecalis JQ135 have been deposited in the GenBank database under accession numbers KY230187 and CP021641, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the anonymous reviewers for their valuable comments, which greatly improved the manuscript.
This work was supported by the State's Key Project of Research and Development Plan (grant 2016YFD0801102), the National Natural Science Foundation of China (grants 31500082 and 31770117), the China Postdoctoral Science Foundation (grant 2016M601826), and the Postdoctoral Foundation of Jiangsu Province (grant 1601035A).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00910-18.
REFERENCES
- 1.Guan A-Y, Liu C-L, Sun X-F, Xie Y, Wang M-A. 2016. Discovery of pyridine-based agrochemicals by using intermediate derivatization methods. Bioorg Med Chem 24:342–353. doi: 10.1016/j.bmc.2015.09.031. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um KS, Wilson T, Collins D, De Lisle G, Jacobs WR Jr. 1994. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263:227–229. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 3.Hamaker JW, Johnston H, Martin RT, Redemann CT. 1963. A picolinic acid derivative: a plant growth regulator. Science 141:363–363. doi: 10.1126/science.141.3578.363. [DOI] [PubMed] [Google Scholar]
- 4.Tekle-Röttering A, Reisz E, Jewell KS, Lutze HV, Ternes TA, Schmidt W, Schmidt TC. 2016. Ozonation of pyridine and other N-heterocyclic aromatic compounds: kinetics, stoichiometry, identification of products and elucidation of pathways. Water Res 102:582–593. doi: 10.1016/j.watres.2016.06.021. [DOI] [PubMed] [Google Scholar]
- 5.Brinkmann M, Maletz S, Krauss M, Bluhm K, Schiwy S, Kuckelkorn J, Tiehm A, Brack W, Hollert H. 2014. Heterocyclic aromatic hydrocarbons show estrogenic activity upon metabolization in a recombinant transactivation assay. Environ Sci Technol 48:5892–5901. doi: 10.1021/es405731j. [DOI] [PubMed] [Google Scholar]
- 6.Jiménez JI, Canales A, Jiménez-Barbero J, Ginalski K, Rychlewski L, García JL, Díaz E. 2008. Deciphering the genetic determinants for aerobic nicotinic acid degradation: the nic cluster from Pseudomonas putida KT2440. Proc Natl Acad Sci U S A 105:11329–11334. doi: 10.1073/pnas.0802273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Xie K, Yu W, Hu L, Huang H, Xie H, Wang S. 2016. Nicotine dehydrogenase complexed with 6-hydroxypseudooxynicotine oxidase involved in the hybrid nicotine-degrading pathway in Agrobacterium tumefaciens S33. Appl Environ Microbiol 82:1745–1755. doi: 10.1128/AEM.03909-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu H, Tang H, Li Y, Xu P. 2015. Molybdenum-containing nicotine hydroxylase genes in a nicotine degradation pathway that is a variant of the pyridine and pyrrolidine pathways. Appl Environ Microbiol 81:8330–8338. doi: 10.1128/AEM.02253-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandsch R. 2006. Microbiology and biochemistry of nicotine degradation. Appl Microbiol Biotechnol 69:493–498. doi: 10.1007/s00253-005-0226-0. [DOI] [PubMed] [Google Scholar]
- 10.Yu H, Hausinger RP, Tang H-Z, Xu P. 2014. Mechanism of the 6-hydroxy-3-succinoyl-pyridine 3-monooxygenase flavoprotein from Pseudomonas putida S16. J Biol Chem 289:29158–29170. doi: 10.1074/jbc.M114.558049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makar'eva T, Kalinovskii A, Stonik V, Vakhrusheva E. 1989. Identification of 5-hydroxypicolinic acid among the products biosynthesized by Nocardia sp. Chem Nat Compd 25:125–126. doi: 10.1007/BF00596721. [DOI] [Google Scholar]
- 12.Yoshikuni Y, Wargacki AJ, Cooper SR, Raisner R, Gill A, Tripathi SA, Enquist-Newman MK. 2012. A method of producing 5-hydroxypyridine-2-carboxylic acid from alginate. U.S. patent WO 2012/142326 A1.
- 13.Chen L, Song Z, Wang J, Song H, Zhang G, Wang J. 2010. Studies on the chemical constituents from aerial parts of Gynura divaricata. Zhong Yao Cai 33:373–376. (In Chinese.) [PubMed] [Google Scholar]
- 14.Li G, Yu K, Li F, Xu K, Li J, He S, Cao S, Tan G. 2014. Anticancer potential of Hericium erinaceus extracts against human gastrointestinal cancers. J Ethnopharmacol 153:521–530. doi: 10.1016/j.jep.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Tian JL, Liang X, Gao PY, Li LZ, Song SJ. 2015. Chemical constituents of Portulaca oleracea. Chem Nat Compd 51:760–761. doi: 10.1007/s10600-015-1403-8. [DOI] [Google Scholar]
- 16.Karvelis L, Gasparavičiūtė R, Klimavičius A, Jančienė R, Stankevičiūtė J, Meškys R. 2014. Pusillimonas sp. 5HP degrading 5-hydroxypicolinic acid. Biodegradation 25:11–19. doi: 10.1007/s10532-013-9636-3. [DOI] [PubMed] [Google Scholar]
- 17.Qiu J, Zhang J, Zhang Y, Wang Y, Tong L, Hong Q, He J. 2017. Biodegradation of picolinic acid by a newly isolated bacterium Alcaligenes faecalis strain JQ135. Curr Microbiol 74:508–514. doi: 10.1007/s00284-017-1205-2. [DOI] [PubMed] [Google Scholar]
- 18.Wang SM, He J, Cui ZL, Li SP. 2007. Self-formed adaptor PCR: a simple and efficient method for chromosome walking. Appl Environ Microbiol 73:5048–5051. doi: 10.1128/AEM.02973-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J, Oh J, Min KR, Kim Y. 1996. Nucleotide sequence of salicylate hydroxylase gene and its 5′-flanking region of Pseudomonas putida KF715. Biochem Biophys Res Commun 218:544–548. doi: 10.1006/bbrc.1996.0097. [DOI] [PubMed] [Google Scholar]
- 20.Yu H, Tang H, Zhu X, Li Y, Xu P. 2015. Molecular mechanism of nicotine degradation by a newly isolated strain, Ochrobactrum sp. strain SJY1. Appl Environ Microbiol 81:272–281. doi: 10.1128/AEM.02265-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma Y, Wei Y, Qiu J, Wen R, Hong J, Liu W. 2014. Isolation, transposon mutagenesis, and characterization of the novel nicotine-degrading strain Shinella sp. HZN7. Appl Microbiol Biotechnol 98:2625–2636. doi: 10.1007/s00253-013-5207-0. [DOI] [PubMed] [Google Scholar]
- 22.Tang H, Yao Y, Wang L, Yu H, Ren Y, Wu G, Xu P. 2012. Genomic analysis of Pseudomonas putida: genes in a genome island are crucial for nicotine degradation. Sci Rep 2:377. doi: 10.1038/srep00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Q, Wang C-H, Deng S-K, Wu Y-D, Li Y, Yao L, Jiang J-D, Yan X, He J, Li S-P. 2014. Novel three-component Rieske non-heme iron oxygenase system catalyzing the N-dealkylation of chloroacetanilide herbicides in sphingomonads DC-6 and DC-2. Appl Environ Microbiol 80:5078–5085. doi: 10.1128/AEM.00659-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatakeyama K, Asai Y, Uchida Y, Kobayashi M, Terasawa M, Yukawa H. 1997. Gene cloning and characterization of maleate cis-trans isomerase from Alcaligenes faecalis. Biochem Biophys Res Commun 239:74–79. doi: 10.1006/bbrc.1997.7430. [DOI] [PubMed] [Google Scholar]
- 25.Huijbers MM, Montersino S, Westphal AH, Tischler D, van Berkel WJ. 2014. Flavin dependent monooxygenases. Arch Biochem Biophys 544:2–17. doi: 10.1016/j.abb.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Tsuji H, Ogawa T, Bando N, Sasaoka K. 1986. Purification and properties of 4-aminobenzoate hydroxylase, a new monooxygenase from Agaricus bisporus. J Biol Chem 261:13203. [PubMed] [Google Scholar]
- 27.Chao H-J, Chen Y-F, Fang T, Xu Y, Huang WE, Zhou N-Y. 2016. HipH catalyzes the hydroxylation of 4-hydroxyisophthalate to protocatechuate in 2,4-xylenol catabolism by Pseudomonas putida NCIMB. 9866. Appl Environ Microbiol 82:724–731. doi: 10.1128/AEM.03105-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uemura T, Kita A, Watanabe Y, Adachi M, Kuroki R, Morimoto Y. 2016. The catalytic mechanism of decarboxylative hydroxylation of salicylate hydroxylase revealed by crystal structure analysis at 2.5 Å resolution. Biochem Biophys Res Commun 469:158–163. doi: 10.1016/j.bbrc.2015.11.087. [DOI] [PubMed] [Google Scholar]
- 29.Hicks KA, Yuen ME, Zhen WF, Gerwig TJ, Story RW, Kopp MC, Snider MJ. 2016. Structural and biochemical characterization of 6-hydroxynicotinic acid 3-monooxygenase, a novel decarboxylative hydroxylase involved in aerobic nicotinate degradation. Biochemistry 55:3432–3446. doi: 10.1021/acs.biochem.6b00105. [DOI] [PubMed] [Google Scholar]
- 30.van Berkel W, Kamerbeek N, Fraaije M. 2006. Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J Biotechnol 124:670–689. doi: 10.1016/j.jbiotec.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 31.Kaiser J-P, Feng Y, Bollag J-M. 1996. Microbial metabolism of pyridine, quinoline, acridine, and their derivatives under aerobic and anaerobic conditions. Microbiol Rev 60:483–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu K, Xu Y, Zhou N-Y. 2015. Identification of a specific maleate hydratase in the direct hydrolysis route of the gentisate pathway. Appl Environ Microbiol 81:5753–5760. doi: 10.1128/AEM.00975-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asano Y, Ueda M, Yamada H. 1993. Microbial production of d-malate from maleate. Appl Environ Microbiol 59:1110–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hopper D, Chapman P, Dagley S. 1968. Enzymic formation of d-malate. Biochem J 110:798. doi: 10.1042/bj1100798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 36.Qiu J, Ma Y, Wen Y, Chen L, Wu L, Liu W. 2012. Functional identification of two novel genes from Pseudomonas sp. strain HZN6 involved in the catabolism of nicotine. Appl Environ Microbiol 78:2154–2160. doi: 10.1128/AEM.07025-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J-P, Zhang W-M, Chao H-J, Zhou N-Y. 2017. PnpM, a LysR-type transcriptional regulator activates the hydroquinone pathway in para-nitrophenol degradation in Pseudomonas sp. strain WBC-3. Front Microbiol 8:1714. doi: 10.3389/fmicb.2017.01714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luanloet T, Sucharitakul J, Chaiyen P. 2015. Selectivity of substrate binding and ionization of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase. FEBS J 282:3107–3125. doi: 10.1111/febs.13220. [DOI] [PubMed] [Google Scholar]
- 39.Yun H, Liang B, Qiu J, Zhang L, Zhao Y, Jiang J, Wang A. 2017. Functional characterization of a novel amidase involved in biotransformation of triclocarban and its dehalogenated congeners in Ochrobactrum sp. TCC-2. Environ Sci Technol 51:291–300. doi: 10.1021/acs.est.6b04885. [DOI] [PubMed] [Google Scholar]
- 40.Rietsch A, Wolfgang MC, Mekalanos JJ. 2004. Effect of metabolic imbalance on expression of type III secretion genes in Pseudomonas aeruginosa. Infect Immun 72:1383–1390. doi: 10.1128/IAI.72.3.1383-1390.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quandt J, Hynes MF. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 42.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.