Algal biofuels are one of the more promising forms of renewable energy. In our study, we investigate whether ecological interactions between species of microalgae regulate two important factors in cultivation—the biomass of the crop produced and the quality of the biomass that is produced. We found that species interactions often improved production yields, especially the fatty acid content of the algal biomass, and that differentially expressed genes involved in fatty acid metabolism are predictive of improved quality metrics of bio-oil. Other studies have found that diversity often improves productivity and stability in agricultural and natural ecosystems. Our results provide further evidence that growing multispecies crops of microalgae may improve the production of high-quality biomass for bio-oil.
KEYWORDS: biofuels, diversity-function, transcriptomics
ABSTRACT
Algal biofuels have the potential to curb the emissions of greenhouse gases from fossil fuels, but current growing methods fail to produce fuels that meet the multiple standards necessary for economical industrial use. For example, algae grown as monocultures for biofuel production have not simultaneously and economically achieved high yields of the high-quality lipid-rich biomass desired for the industrial-scale production of bio-oil. Decades of study in the field of ecology have demonstrated that simultaneous increases in multiple functions, such as the quantity and quality of biomass, can occur in natural ecosystems by increasing biological diversity. Here, we show that species consortia of algae can improve the production of bio-oil, which benefits from both a high biomass yield and a high quality of biomass rich in fatty acids. We explain the underlying causes of increased quantity and quality of algal biomass among species consortia by showing that, relative to monocultures, species consortia can differentially regulate lipid metabolism genes while growing to higher levels of biomass, in part due to a greater utilization of nutrient resources. We identify multiple genes involved in lipid biosynthesis that are frequently upregulated in bicultures and further show that these elevated levels of gene expression are highly predictive of the elevated levels in biculture relative to that in monoculture of multiple quality metrics of algal biomass. These results show that interactions between species can alter the expression of lipid metabolism genes and further demonstrate that our understanding of diversity-function relationships from natural ecosystems can be harnessed to improve the production of bio-oil.
IMPORTANCE Algal biofuels are one of the more promising forms of renewable energy. In our study, we investigate whether ecological interactions between species of microalgae regulate two important factors in cultivation—the biomass of the crop produced and the quality of the biomass that is produced. We found that species interactions often improved production yields, especially the fatty acid content of the algal biomass, and that differentially expressed genes involved in fatty acid metabolism are predictive of improved quality metrics of bio-oil. Other studies have found that diversity often improves productivity and stability in agricultural and natural ecosystems. Our results provide further evidence that growing multispecies crops of microalgae may improve the production of high-quality biomass for bio-oil.
INTRODUCTION
Just as early humans were hunter-gatherers with respect to food, modern humans remain hunter-gatherers with respect to energy. We drill, mine, or extract wherever we can find resources. To become a sustainable society, we must transition from this hunter-gatherer mode to one that provides a continuous, stable, and abundant supply of energy. Bio-oil derived from microalgae is one of the more promising forms of renewable energy, as algae can produce more oil per acre than land plants and can be grown nearly anywhere, including in degraded habitats and nonarable lands (1, 2). Fatty acid methyl esters (FAMEs) are the main constituent of biodiesel produced by the extraction and transesterification of fatty acids from algal biomass. Thus, the key metrics of the quality of algal biomass for bio-oil production are the proportion of the biomass composed of fatty acids and the composition of different types of fatty acids. Also, high FAME yields lead to improved bio-oil yield and quality from newer more promising technologies such as hydrothermal liquefaction that transform the entire wet algal biomass into biocrude oil (3).
A key bottleneck that limits the economic viability of energy cultivation by algae is our inability to maximize biomass quality, based on fatty acid content, while at the same time maximizing biomass quantity. The physiological reasoning behind this limitation is that organisms with finite resources choose either to use energy to grow or to store energy in the form of lipids for future growth (4–6). One approach that has proven moderately successful has been to genetically engineer microalgal strains (7), but these strains are expensive to produce and unlikely to thrive under the variable field conditions inherent in outdoor commercial-production facilities. However, there is an alternative approach that uses principles in ecology to engineer communities of multiple species (1, 4, 8, 9). This approach is based on ecological research that has shown that multispecies ecosystems regularly outperform monocultures in many functions, including biomass production (10–14).
These positive causal relationships between biodiversity and function are especially strong when considering multiple functions at once (15). While simultaneously improving multiple functions would have clear benefits for the extraction of sustainable resources, our knowledge of biodiversity-ecosystem function relationships in natural systems has yet to be applied to commercial applications such as biofuels and bioproducts. For example, a biorefinery, in which plant biomass is grown and refined to yield multiple products such as biofuel and biochemicals, would benefit from simultaneous improvements of multiple functions (16). Specific for biofuels, studies suggest that algal species richness may influence the quantity or quality of the biomass (13, 17). However, the simultaneous effects of species richness on the biomass yield and biomass quality needed for improved bio-oil production, along with the genomic drivers underlying such a phenomenon, remain to be rigorously investigated.
Here, we incorporated new data and analyses into an experiment that was previously analyzed to test the effect of evolutionary relatedness and gene expression on community stability and coexistence (18–20). In this experiment, we grew monocultures of eight species of green freshwater microalgae, Chlorella sorokiniana, Closteriopsis acicularis, Cosmarium turpinii, Pandorina charkowiensis, Scenedesmus acuminatus, Selenastrum capricornutum, Staurastrum punctulatum, and Tetraedron minutum, and all 28 of the possible species pairs under controlled laboratory conditions for 48 days until the majority of the communities had reached steady-state biomass. These species were specifically selected to span a large gradient in phylogenetic distances on our molecular phylogeny (21). The charophytes included the two desmids Cosmarium turpinii and Staurastrum punctulatum, while the remaining species belong to the chlorophytes. At the end of the experiment (i.e., day 48), for each species, we quantified the total cell density and RNA transcription levels. For each culture, we measured the biomass quality as total fatty acid methyl ester (FAME) content, which consisted of a mixture of 28 different fatty acids. The proportions of these different fatty acids were used to calculate two important qualities of extracted total FAME, cetane number and higher heating value. Previously, these data were used to show that bicultures consisting of more closely related species generally exhibited more similar gene expression, weaker competitive interactions, and greater temporal stability of biomass (19, 20). In our current analyses, we show that multispecies communities of freshwater microalgae outperform monocultures in producing a high quantity and quality of biomass representative of algal feedstock for bio-oil production. To assess the causes underlying our observation that species richness simultaneously improved the algal biomass yield and quality, we also measured gene expression in each algal population in each community. We used these data to contrast lipid metabolism, nutrient assimilation, and photosynthetic gene expression of each algal species alone in monoculture versus those in each biculture growing condition, and related genome-wide expression patterns to the biomass attained by each algal population and our metric for biomass quality, FAME.
RESULTS
We first tested whether algal bicultures outperformed monocultures in the production of bio-oil, considering the metrics of biomass quantity and quality. We considered these measures, on average, in bicultures versus monocultures, but also tested whether bicultures overyield with respect to monocultures. Overyielding refers to a biculture that significantly exceeds expectations on the basis of monoculture yields. Second, we evaluated differential expression in bicultures relative to that in their monocultures, focusing specifically on gene functions involved in lipid metabolism that we hypothesized would be important for the production of bio-oil. We first considered these genes categorized into broad Gene Ontology functional groups and then identified specific sets of orthologous genes. Lastly, we linked differential expression of genes involved in lipid metabolism with the overyielding and relative composition of fatty acids, which are key indicators of fuel quality.
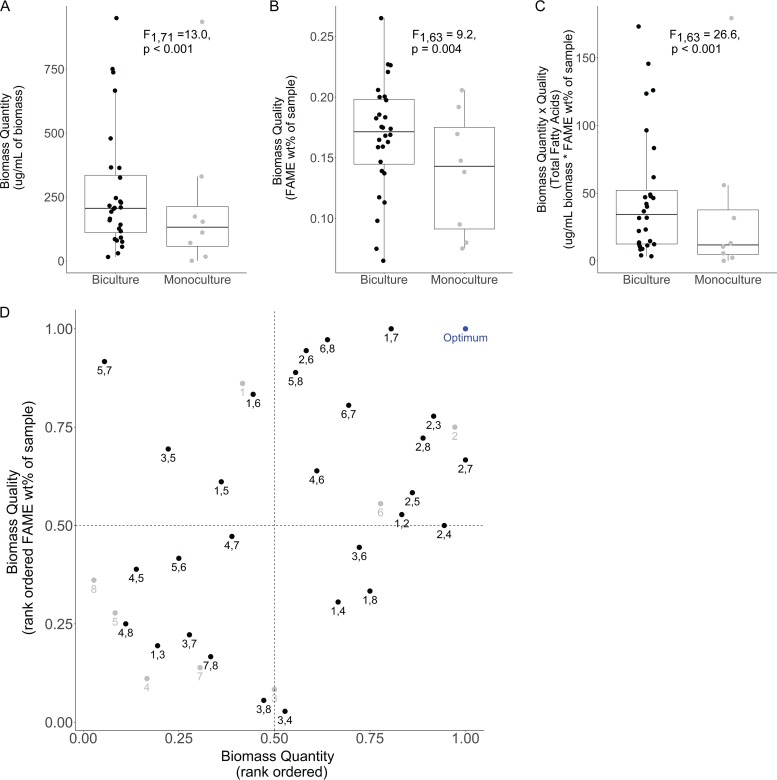
We found that bicultures, on average, produced significantly higher levels of algal biomass and of FAMEs, which we measured in two ways: FAME yield as a weight percentage of the biomass, and the total yield of FAMEs calculated by multiplying FAME yield and algal biomass (Fig. 1A to C). We found that bicultures frequently overyielded with respect to their comprising monocultures in the individual function of either biomass yield or quality. Nearly half of all bicultures produced a biomass of significantly higher quality (i.e., FAME yields) relative to the quality of the biomass produced by their constituent monocultures (FAME overyielding occurred in 13 of 28 combinations). Additionally, five bicultures significantly overyielded in biomass. Further, bicultures generated greater biomass than monocultures in part through a greater utilization of nutrient pools from the growth medium. This trend was significant throughout the experiment (NO3: F1,34 = 6.42, P = 0.012; PO4: F1,34 = 15.32, P < 0.001), but was greater in magnitude toward the end of the experiment, when PO4 levels were 51% lower and NO3 levels were 35% lower in bicultures than in monocultures.
FIG 1.
Bicultures outperform monocultures in three measures of bio-oil production. Bicultures, on average, yield greater algal biomass (A), fatty acid methyl ester as a weight percentage of the algal biomass (B), and total estimated mass of fatty acids (C). (D) Bicultures outperform monocultures when simultaneously considering quantity and quality of algal biomass (nested linear model on Euclidean distance to optimum: F1,34 = 2.9, P < 0.01). The “optimum” is the top rank of each metric observed in the study. As illustrated with the multifunctionality threshold approach, 43% of bicultures versus 25% of monocultures fall within the upper quadrant, indicating higher than average biomass yield and quality, via FAME percentage of biomass. Numbers indicate species identities: 1, Chlorella sorokiniana; 2, Closteriopsis acicularis; 3, Cosmarium turpinii; 4, Pandorina charkowiensis; 5, Scenedesmus acuminatus; 6, Selenastrum capricornutum; 7, Staurastrum punctulatum; 8, Tetraedron minutum.
Furthermore, we found that bicultures significantly outperformed monocultures when simultaneously considering biomass yield and quality, as they were, on average, closer to the optimum than monocultures (nested linear model on the Euclidean distances from the 1:1 “optimum” point; F1,34 = 2.9, P < 0.01) (Fig. 1D). The optimum was defined as the combination of the highest biomass observed across all experiments (i.e., biomass of the Closteriopsis, Staurastrum biculture) with the highest FAME yield observed (i.e., FAME yield of the Chlorella, Staurastrum biculture). This conclusion that bicultures outperformed monocultures was robust using either the “threshold” or “averaging” method for calculating multifunctionality (see Fig. S1 in the supplemental material). While bicultures, on average, significantly outperformed monocultures when considering multifunctionality, and the best biculture exceeded the function of the best monoculture when considering each function separately, the best biculture was not significantly better than the best monoculture when simultaneously considering both functions (Euclidean distance to optimum of the best monoculture fell within 0.27 standard deviations of a bootstrapped distribution of the top three bicultures).
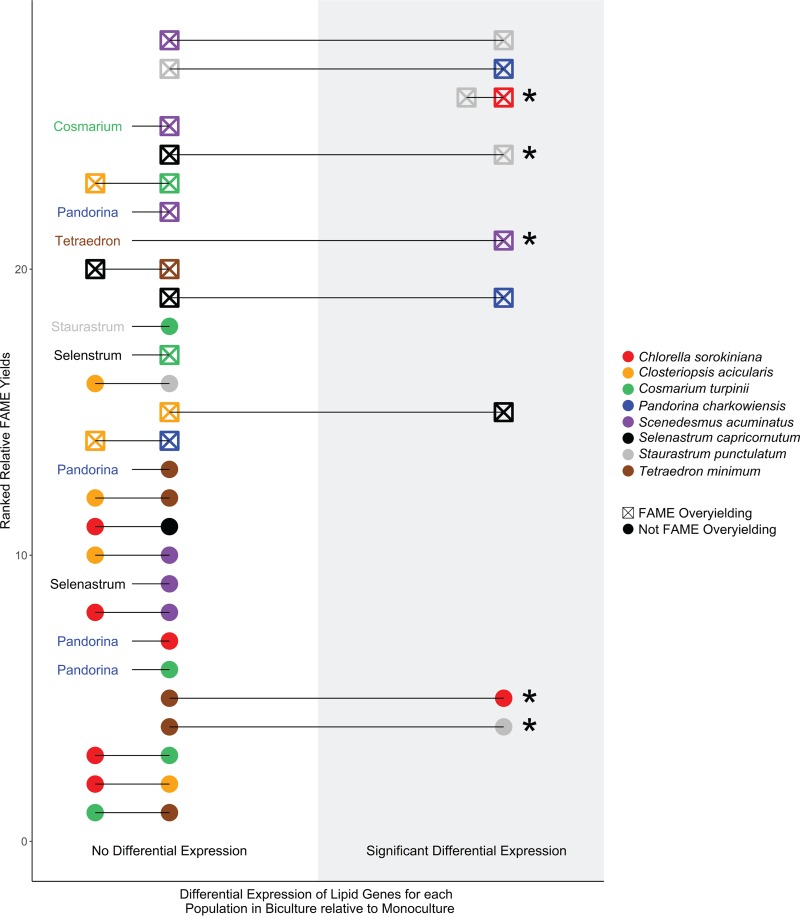
We also found that biomass quality can be predicted by gene expression. Results from our genetic analyses in part explain how species richness promotes multifunctionality: 64% of all bicultures that overyielded in FAME also contained at least one algal species that was differentially expressing genes involved in lipid metabolism (Fig. 2). The relative FAME yield was calculated as the mean FAME yield of A plus B biculture/(mean [mean yield of species A in monoculture and mean yield of species B in monoculture]). We indicate an overyielding in FAME only where the biculture yield was significantly higher than the average of the two monocultures. Differential expression indicates that the algal species expressed significantly greater or lower levels of the gene in the biculture growing condition relative to that in the monoculture growing condition. We identified candidate genes that we can infer are involved in lipid metabolism using a keyword search of lipid-relevant terms in the Gene Ontology annotation database. For conciseness, we refer to these genes as “lipid metabolism genes.” Importantly, one-half of the algal populations that were differentially expressing their lipid genes were also growing at biomass levels greater than those produced in monoculture (Fig. 2). The differential expression of genes involved in lipid metabolism was predictive of FAME overyielding in bicultures relative to the expectations based on monoculture yields (logistic regression: relative FAME, χ2 = 6.9, P < 0.01). Furthermore, the differential expression of these genes was predictive of the total FAME yield in bicultures (logistic regression: total FAME, χ2 = 4.8, P = 0.027). We directly assessed the effect of species richness on lipid metabolism by measuring the gene expression in each algal population in each biculture compared to the expression by that species in monoculture. For example, a community can contain either two species that do not differentially express lipid genes (i.e., Chlorella and Cosmarium), one species that does differentially express lipid genes and one that does not (i.e., Chlorella and Tetraedron), or two species that both differentially express lipid genes (i.e., Chlorella and Staurastrum). With this approach, we identified 10 occasions in which at least one of the two species in biculture differentially expressed genes with known lipid functions compared to all other genes (Kolmogorov-Smirnov tests, all false-discovery-rate-corrected P values were <0.05) (Fig. 2) and found that overyielding in FAME and/or biomass occurred in all 10 of these occasions. Furthermore, among the top 10 bicultures with the highest relative FAME yields, six bicultures had one or both species differentially regulating their lipid genes (Fig. 2). Lastly, we confirmed the reproducible effects of these treatment conditions on algal gene expression by calculating Euclidean distances between biological replicates that were visualized with multidimensional scaling. We show that Euclidean distances among biological replicates within the same species combination were significantly less than the distances among biological replicates of different combinations (see Table S2).
FIG 2.
Significant differential expression of lipid metabolism genes was observed only in communities overyielding in biomass quantity and/or quality. Solid lines connect populations growing together in biculture, symbol color indicates species identities in each biculture, and symbol fill indicates whether that biculture is overyielding in biomass quality (i.e., wt % FAMEs). Location on the x axis indicates whether those populations significantly differentially express their lipid genes via Kolmogorov-Smirnov tests on the distributions of log2-fold change values of lipid versus nonlipid genes. Populations differentially expressing their lipid genes and maintaining biomass at levels greater than those attained in monoculture are marked with asterisks. Bicultures are ranked from high (top) to low (bottom) on the y axis on the basis of their FAME yield in biculture relative to that in monocultures. Overyielding in FAME is designated where biculture yield was significantly higher than the average of the two monocultures. Missing symbol pairs are for those populations with insufficient gene-library sizes for inclusion in transcriptome analyses.
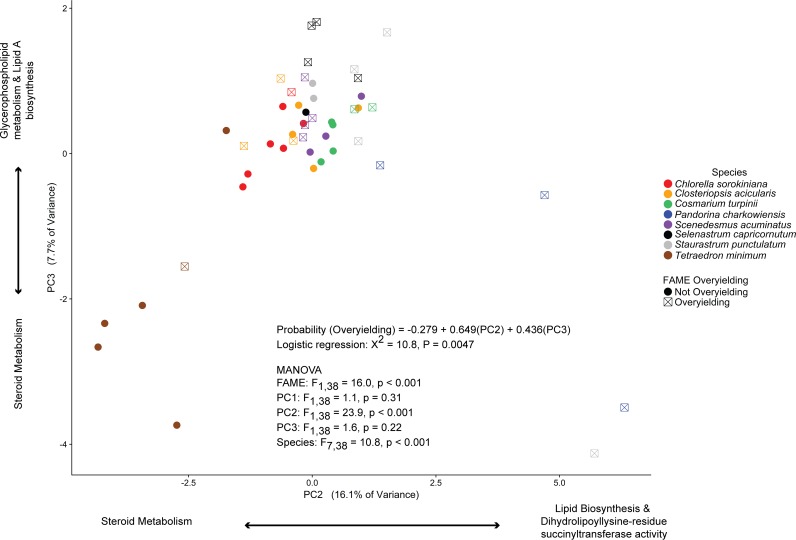
We also found that the expression of genes known to regulate specific lipid functions was a significant predictor of overyielding in FAME (Fig. 3). Specifically, overyielding bicultures diverged from nonoveryielding bicultures via the overexpression of genes regulating the saponifiable lipids, including glycerophospholipids and lipid A, as well as the regulation of dihydrolipoyllysine-residue succinyltransferase activity, which is essential for the production of coenzyme A and fatty acid synthesis (22). Overyielding bicultures also diverged from nonoveryielding bicultures via the underexpression of genes regulating the metabolism of steroids, which are nonsaponifiable lipids (Fig. 3, PC2 and PC3). This trend was common across multiple species, with overyielding bicultures diverging significantly from nonoveryielding bicultures in lipid gene expression, as summarized using the mean change in gene expression of 28 Gene Ontology terms involved in lipid metabolism (see principal-component analysis [PCA] in Fig. 3) (logistic regression: χ2 = 10.7, P = 0.0047; multivariate analysis of variance: F1,38 = 16.0, P < 0.001) (see Table S3 for variable loadings and Fig. S2 for a PCA plot of PC1 versus PC2).
FIG 3.
Bicultures overyielding versus nonoveryielding in FAME production exhibit notably distinct expression patterns of genes regulating synthesis and oxidation of fatty acids and other lipids. These patterns are conserved across multiple algal species. We illustrate these patterns via principal-component analysis incorporating mean log2-fold change values in the monoculture versus biculture conditions for each algal species of 28 different Gene Ontology groups involved in lipid regulatory functions.
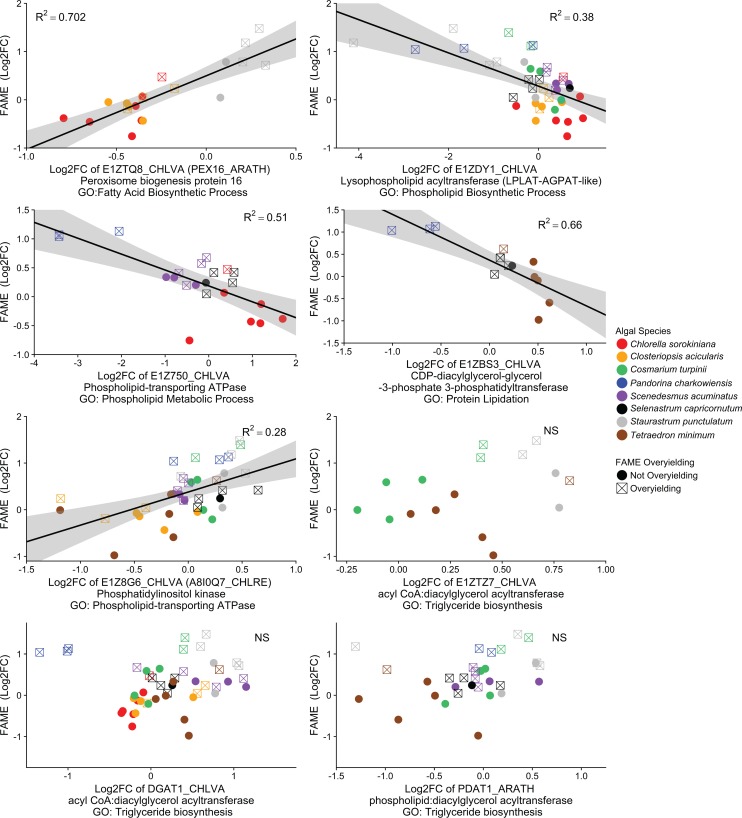
We then identified orthologous sets of genes across all eight species of microalgae by searching for best matches in Chlorella variabilis, a more-well-studied species of algae. With this approach, we found that differential expression of orthologous sets of genes in the monoculture versus biculture growing conditions was predictive of the elevated FAME yields observed in bicultures relative to that in monocultures. Specifically, the upregulation of a set of orthologous genes involved in peroxisome biogenesis, where fatty acid oxidation occurs, was predictive of elevated FAME yields (Fig. 4). Furthermore, the downregulation of a set of orthologous genes involved in the modification of phospholipids was predictive of an elevated FAME yield among bicultures (Fig. 4). In addition to searching for orthologs in Chlorella variabilis, we searched Arabidopsis thaliana for two enzymes that were poorly described in algae but are especially notable for fatty acid biosynthesis (diacylglycerol acyltransferase [DGAT] and phospholipid diacylglycerol acyltransferase [PDAT]). Differential expression levels of DGAT and PDAT genes were not predictive of an elevated FAME yield (Fig. 4). However, we found that the differential expression of several other orthologous sets of genes was predictive of the fold change in other important metrics of biomass quality measured in the monoculture versus biculture conditions. The differential expression among four orthologous sets of genes was predictive of the fold change in cetane number, a measure of fuel ignition delay (see Fig. S3), and differential expression among five orthologous sets of genes was predictive of the fold change in higher heating value, a measure of the quantity of heat released during fuel combustion (see Fig. S4). Cetane number and higher heating value are largely determined by the relative composition of fatty acids that differ in carbon chain length and saturation.
FIG 4.
Across multiple species of algae, the relative change in gene expression between monoculture and biculture growing conditions significantly predicts the relative change in FAMEs. Correlations between log2-fold changes of gene expression and FAME in biculture relative to those in monoculture were evaluated via simple linear regression models with false discovery rate significance corrections. Orthologs in Chlorella variabilis were used for gene annotation when possible. Occasionally, we note orthologs from other model organisms from which we needed to infer gene annotation. For genes especially notable for fatty acid biosynthesis (DGATs and PDATs inferred from Arabidopsis thaliana), we show a lack of correlation.
Overall, our differential expression analyses contribute to understanding the underlying mechanism by identifying which algal species within the community contributes to total fatty acid generation. Linking lipid gene expression to specific community members enabled us to merge these data with population densities to determine that lipid overyielding does not necessitate a reduction in growth. To further elucidate mechanistically how bicultures may be outperforming monocultures in fatty acid production, we evaluated whether differential expression among genes regulating photosynthesis and nutrient assimilation was associated with increased FAME production or total fatty acid production, which accounts for both increased biomass production and increased FAMEs as a weight percentage of the biomass. The genes we tested for their role in regulating photosynthesis included those encoding carbonic anhydrases, glutamate semialdehyde aminotransferases, and light harvesting chlorophyll a-b complexes. We tested iron permeases, nitrite transporters, nitrite reducers, nitrate transporters, nitrate reducers, and phosphate transporters for their roles in regulating nutrient assimilation. Although we found that 39% of bicultures contained at least one algal species upregulating photosynthesis genes (see Fig. S7), and 25% of bicultures contained at least one algal species upregulating nutrient assimilation genes (see Fig. S8), we found no systematic association between genes regulating photosynthesis or nutrient assimilation with the magnitude of FAME overyielding or production of total fatty acids (see Fig. S9).
DISCUSSION
Our results illustrate that in addition to improving each aspect of bio-oil production individually, species richness can simultaneously improve biomass yield and quality. Our findings are consistent with the majority of studies that have found that species richness enhances biomass yield (10, 13, 14), though there are exceptions (23). Our study is also consistent with a prior study that suggests richness may help improve biomass quality (17). However, our study shows that species richness can help us accomplish both of these at once. Furthermore, our results demonstrate that key metrics of bio-oil quality have significant transcriptomic correlates that are common across the multiple species of algae studied here. We show that species richness directly affects the regulation of fatty acid metabolic processes that are known to be key for the production of high-quality bio-oil based on both a fundamental understanding of fatty acid biosynthesis (22) and the validation here from finding correlations of gene expression with our measurements of bio-oil quality. By tracing lipid gene expression to individual populations of algae, we found that species richness can simultaneously improve both the biomass yield and the fatty acid content of algal biomass.
Our results add to several other studies that document numerous advantages of incorporating diversity. For example, compared to monocultures, multispecies crops are more stable, require less fertilizer, generate less nutrient pollution, and are more resistant to pest invasions (23–26). While recent studies have used gene engineering approaches to improve fatty acid production under controlled laboratory conditions (7), few monocultures thrive when exposed to the abiotic and biotic stressors of outdoor conditions that are essential for commercially scaled production. Our results show that ecological engineering is a promising approach that harnesses multispecies communities to improve the quantity, quality, and environmental sustainability of bio-oil production. However, while diversity improves the production of bio-oil on average, we did not find evidence that the best diverse crop significantly outperformed the best monoculture crop. An important next step is to apply our findings toward identifying whether diversity would improve yields among our best-performing monoculture crops, for example by using a more diverse group of algal species beyond green algae.
Lipid content is arguably the most important measure of quality for an algal feedstock, regardless of whether the biofuel industry relies on direct lipid extraction or adopts newer methods, such as hydrothermal liquefaction, that use the entire algal biomass. By experimentally introducing interactions among species, we show that algae frequently respond to interspecific interactions by differentially expressing genes that regulate lipid biosynthesis. Although it is challenging to definitively identify the functions of genes in nonmodel organisms, we show that the degree of differential expression of our genes that were purportedly involved in lipid metabolism was indeed significantly predictive of the degree of overyielding in FAME in bicultures relative to that in monocultures. We used a cautious approach for these transcriptomics analyses by focusing only on a subset of transcripts that were most taxonomically relevant and functionally informative. By retaining only those transcripts mapping to taxa in the Diaphoretickes (i.e., the Plants+Hacrobia+SAR [stramenopiles, alveolates, and Rhizaria] megagroup of photosynthetic organisms) that have Gene Ontology annotations, we avoided the possible inclusion of transcripts with unknown lipid functions in our “nonlipid” category of genes and minimized the possible misinterpretations from functions inferred from taxa outside the Diaphoretickes megagroup, as the algal cultures we used have associated bacteria and fungi. However, this approach could exclude genes that may be poorly annotated but biologically relevant, which could explain why certain bicultures overyielded in FAMEs but showed no underlying genomic signature (i.e., the Cosmarium-Closteriopsis biculture). We note that our study did not exclude bacteria and fungi in the algal “phycosphere”: those microbes residing in the mucilaginous layer immediately surrounding the algal cell that are known to be facilitating or even essential for algal growth (27–29). Further efforts to improve the cultivation of outdoor algal biofuels should test the effects of these bacteria and fungi, which will inevitable colonize ponds, on the outcomes of biofuel production.
We found that overyielding bicultures could be differentiated from nonoveryielding bicultures on the basis of the differential expression of genes that regulated the synthesis of different types of lipids. Genes regulating the saponifiable lipids, including glycerophospholipids and lipid A, tended to be upregulated among overyielding bicultures. While lipid A is best known from bacterial systems, studies have confirmed that plant mitochondrial membranes contain the full biosynthetic pathway for lipid A (30, 31). Also notable is that overyielding and nonoveryielding bicultures diverged significantly in the expression of the enzyme dihydrolipoyllysine-residue succinyltransferase. This enzyme catalyzes the reaction generating coenzyme A, which is a key player in the synthesis of fatty acids (22). Specifically, the amounts of coenzyme A and its close derivatives, such as malonyl-CoA, that have accumulated in plant plastids are directly predictive of the rate of fatty acid biosynthesis (32). In contrast, overyielding bicultures also diverged from nonoveryielding bicultures via the underexpression of genes regulating the metabolism of steroids, which are nonsaponifiable lipids that are not convertible to FAMEs (33).
Our results also identified several sets of orthologous genes for which differential expression caused by the diversity treatment was strongly predictive of improved biofuel quality metrics among bicultures. Interestingly, we found that the upregulation of peroxisome biogenesis protein 16 (PEX16) was positively associated with the overyielding of FAMEs. This is notable because the peroxisome and PEX genes are best described for their role in the β-oxidation or degradation of fatty acids, which is often induced in plants under stress from pathogens or herbivores (34). However, PEX16 has been implicated in the synthesis of fatty acids in Arabidopsis (35). Lin et al. hypothesized that peroxisomes may therefore be involved in both the degradation and the synthesis of fatty acids, perhaps with some degradation necessary to sustain net synthesis (35). We also identified genes involved in phospholipid metabolism that were predictive of fuel metrics; however, the directional effects were mixed. Phospholipids themselves, with only two fatty acid tails, yield less bio-oil than triglycerides. Resources dedicated toward modifications among the diverse forms of glycerophospholipids or the synthesis of additional phospholipids at the expense of triglycerides may decrease bio-oil yields. For example, triglycerides yield more than 99% bio-oil, while phospholipids typically yield less than 70% bio-oil (36, 37). However, the genes involved in increasing glycerophospholipid transformations into free fatty acids, such as those encoding phospholipase A2s (PLA2s), may improve the bio-oil yield (38). This may explain why some genes, such as those involved in phospholipid translocation, were positively associated with bio-oil production, while others were negatively associated with production. Lysophospholipid acyltransferase and CDP-diacylglycerol-glycerol-3-phosphate 3-phosphatidyltransferase, which are involved in modifications within phospholipids, were both negatively associated with bio-oil production. Such modifications to the phosphate head of the phospholipid that leave the fatty acid tails unchanged may still affect bio-oil yield by changing the proportional mass of the molecule comprising the fatty acids tails.
Lastly, we found that the differential expression of several orthologous sets of genes was associated with changes in cetane number and higher heating value. A main predictor of cetane number is the relative composition of saturated, monounsaturated, and polyunsaturated fatty acids. Our algal diversity treatment drove the differential expression of fatty acid desaturases, which significantly increased cetane number in bicultures. The upregulation of PEX16 also significantly predicted greater cetane number among bicultures. Several findings further suggested that algal diversity contributed to the long-term storage of lipids. Diversity caused an upregulation in lipid-droplet-associated hydrolases, which have been shown with experimental work in cell lines to increase triacylglycerol levels (39). Diversity also upregulated very-long-chain 3-ketoacyl-coenzyme A synthases, which are involved in elongating lipids prior to storage (40). The upregulation of these genes involved in lipid storage corresponded to increased cetane number and higher heating value metrics in bicultures relative to those in monocultures.
Combined, our ecological and transcriptomics analyses show that algal populations growing within overyielding communities are differentially regulating lipid metabolism while frequently maintaining growth at biomass levels above those attained in monoculture. This finding provides an underlying explanation for our observation of a beneficial effect of species richness on both biomass yield and quality, which is somewhat contrary to expectations from within-species tradeoffs. Indeed, attempts to improve multiple functions within monocultures have been met with failure due to an inability to overcome tradeoffs that are inherent to biological populations. One such tradeoff that has been proposed is between the biomass that a population of organisms produces versus the quality of this biomass for bio-oil production. Organisms with access to excess resources tend to grow, while those under nutrient stress instead tend to accumulate lipids that improve the quality of algal biomass (5, 41, 42). It has therefore been argued that populations can increase either biomass quantity or quality or produce suboptimal levels of both. In contrast, our findings suggest that species consortia can better utilize nutrient resources for an improved production of biomass. While our data indeed showed that there was a greater utilization of available nutrients in bicultures, this was unlikely to be causing nutrient stress, because nutrient levels remained orders of magnitude higher than the levels typically reported as the lowest at which green algal species can maintain a stable population (resource ratio [R*] values) (43, 44).
Thus, interspecific interactions evidently trigger the accumulation of fatty acids without necessarily compromising biomass yield. While tradeoffs may be an inherent aspect of biology at the species level, our results begin to show that we can harness what decades of research is seeming to prove as an inherent aspect of ecology: diversity improves functions, and quite often, improves functions simultaneously. By rooting our findings in the genomic foundation of species interactions, our results contribute to our understanding of the drivers of elevated function in nature and may increase our ability to both predict when and where to expect such patterns to occur and apply these phenomena to agricultural and biotechnological uses.
MATERIALS AND METHODS
Culture growth and sampling.
Species cultures were supplied either from the University of Texas Culture Collection of Algae (UTEX; Austin, Texas, USA) or from the Sammlung von Algenkulturen Göttingen (SAG; Göttingen, Germany) culture collections. For each species, we grew batch cultures from laboratory stocks in 125-ml Erlenmeyer flasks filled with 100 ml of COMBO growth medium, which contained 1 mM NaNO3, 0.05 mM K2HPO4, 0.1 mM KCl, and 30 μM NH4Cl at the beginning of the experiment (45). We then measured declines in nitrate and phosphate by repeated measures over eight time points throughout the experiment, but levels remained orders of magnitude above what is considered the lowest at which green algae can maintain a stable population (i.e., N* and P*). We also verified each species grew well in COMBO by running a smaller scale study of each species inoculated into multiple types of growth media (see Fig. S6 in the supplemental material). To minimize contamination, we transferred algae from stock to batch cultures with a flame-sterilized loop or an autoclave-sterilized pipette in a laminar flow cabinet and capped batch cultures with a breathable 0.22-μm vented lid. We filled 108 1-liter media bottles (Wheaton, borosilicate glass) with 1 liter of the modified COMBO growth medium and inoculated bottles with one of the 8 monocultures or 28 biculture combinations at a total initial cell density of 200 cells · ml−1. Bicultures were prepared in a 1:1 ratio substitutive design with each species in biculture inoculated at 100 cells · ml−1. All species combinations were grown in triplicates and placed on Wheaton roller racks (349000-A; Millville, NJ, USA) at 0.75 rotations per min, on a 16:8 h light/dark cycle at 20°C under a light intensity of 81 μE. We completed 10% medium exchanges every second day with 100 ml of sterile COMBO beginning 4 days after the initial inoculation. When cultures reached steady-state densities as indicated by chlorophyll a fluorescence measures, we examined each replicate via compound microscopy to confirm species identification and the absence of visible contaminants.
We measured biomass at the community level every second day by counting cell density, where we estimated total biomass (μg/ml) as the population cell density counts (cells/ml) × volume of algal cells based on the equivalent circle diameter (μm3/cell) × specific gravity of water (1 g/1012 μm3) × (1 × 106 μg/g). For this calculation, we used mean volume estimates from our culture stocks (see Table S1). To measure the cell densities of each population, samples of 1 ml of algae were preserved with 250 μl of sugared buffered Formalin every second day until day 30, and then every fourth day until day 46. Algal densities (either cells or colonies depending on the natural unit for each algal species) were measured via a FlowCam (Fluid Imaging Technologies Inc., Scarborough, ME, USA). Phosphate concentrations were measured using the automated ascorbic acid-molybdenum blue EPA method 365.1, and nitrate was assayed using the automated Cd reduction EPA method 353.2 on a SEAL AutoAnalyzer 3 segmented flow nutrient analyzer at the University of Michigan Biological Station (46).
RNA extraction, transcriptome sequencing, de novo transcriptome assembly, and transcript quantification.
We obtained a pellet of algal cells on day 48 from 100 to 900 ml of the culture medium via serial centrifugation. The growth curve data indicated that the majority of algal monocultures and bicultures were at steady state during this collection period (see Fig. S5). We extracted mRNA from the algal pellet using an Ambion RNAqueous kit (Thermo Fisher Scientific, Waltham, MA, USA). RNA was poly(A)-selected, and we constructed libraries using an Illumina TruSeq RNA sample preparation kit, v2. The sequencing of 91-base pair paired-end reads was performed on an Illumina HiSeq 2000 platform at Beijing Genome Institute (BGI; Shenzhen, Guangdong, China). We assessed the quality of sequence libraries using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). We removed Illumina primers and adapters from raw sequencing reads using cutadapt v1.1, trimmed low quality bases below a Phred score of 20 with the SeqTrim Python module (https://github.com/bastodian/Dimensions/blob/master/QC-and-Assembly/IpythonNotebooks/QTrim.ipynb), and retained reads greater than or equal to 45 base pairs for further analysis. To assemble reference transcriptomes for all eight species, we combined all of the short reads from our monoculture replicates and assembled references using the de Bruijn graph assembler, Trinity, using default settings (47). We then mapped short read sets back to the assembled transcriptomes using Bowtie and estimated the abundance of reads for each species in monoculture and biculture using an expectation-maximization algorithm, RSEM (48, 49). Previous measures of the phylogenetic distances between algal species have shown that the number of shared 25-bp sequences between species in their metatranscriptomes is negligible (50). Therefore, our mapping methods of reads of at least 45 bp in length minimize the potential for misassigning reads between species in biculture. We then identified genes by only retaining sequences with an open reading frame that contained known domains by using hidden Markov model searches against the Pfam A database with Transdecoder (47) and HMMER (51). See supplements in reference 20 and https://github.com/bastodian/Dimensions for further details.
Lipid quantification.
Biocrude and biodiesel are the two renewable liquid fuels that can be produced directly from microalgal biomass. Biocrude can be produced via hydrothermal liquefaction. This thermochemical process uses the entire wet biomass; however, a biomass rich in lipid content produces higher biocrude yield and quality (52, 53). Biodiesel uses the lipid fraction of the algal biomass. Lipid content is therefore a strong proxy for both biocrude and biodiesel yield.
To determine total lipid content, we simultaneously extracted and catalytically (trans)esterified all classes of lipids in algal biomass. We use this analytical-scale acid-catalyzed in situ transesterification method, because this procedure recovers more fatty acids than the traditional two-step extraction and transesterification procedure (54–56). We weighed 20 to 100 mg of dry solids into 16 mm by 100 mm glass tubes sealed with Teflon-lined screw caps. We vigorously stirred solids for 90 min with 2 ml of 99% methanol (MeOH) containing 5% acetyl chloride at 100°C. We stopped the reaction with 1 ml of water and extracted FAMEs into 4 ml total of n-heptane spiked with an internal standard of 100 to 200 mg · liter−1 tricosanoic methyl ester (C23:0 FAME; Supelco, Sigma). We injected 1-μl aliquots onto an HP-InnoWax column (30 m by 0.32 mm by 0.25 μm; J&W 1909BD-113) at an initial oven temperature of 150°C, 10 to 100:1 pulsed split ratio, and 260°C inlet temperature. The temperature was held constant for 3 min and then increased at 6°C · min−1 to 260°C. The flame ionization detector (FID) temperature was 300°C. We used helium as the carrier gas, with a constant flow rate of 1.0 ml · min−1, and N2 as the make-up gas. FAME retention times were identified against a Supelco 37 component FAME mix analytical standard (Sigma). We converted the peak area to mass quantity using the following formula: mg esterx = (mg/liter ISTD in solvent/area ISTD)(liter workup solvent)(area esterx)(RRFISTD,x), where esterx refers to each of the fatty acid groups identified in the algal lipids, ISTD refers to the internal standard used (tricosanoic methyl ester), and RRFISTD,x is the theoretical response factor for each esterx relative to the appropriate internal standard (57).
Total FAME content consisted of 28 different fatty acids. The proportions of these different fatty acids were used to calculate two important qualities of extracted total FAME, cetane number and higher heating value. These measures of fuel ignition delay and the quantity of heat released during fuel combustion can be predicted by the molecular weight of the fatty acid methyl ester and the number of double bonds using the equations detailed by Ramírez-Verduzco et al. (58).
Overyielding and multifunctionality statistical analyses.
To determine which bicultures overyielded in biomass and/or FAME yields, we calculated the expected yields for biculture production by summing the 0.5× monoculture yield produced for each species. As we had multiple biological replicates for each monoculture and biculture combination, we calculated all possible pairwise combinations to obtain a range of expected values. The observed versus expected yields were compared via two-sample t tests. We infer “overyielding” when the biculture yield was significantly higher than the average of the two monocultures, in contrast to “underyielding,” when the biculture yield was significantly lower than the average of the two monocultures. Given the rarity at which we observed significant underyielding, we refer to both nonsignificant differences and underyielding as a combined category of “nonoveryielding” bicultures.
High bio-oil production requires both high biomass yield and quality, and so we have now applied newly developed methods for simultaneously assessing multiple functions. We use two of these “multifunctionality” metrics, including a threshold approach that considers whether each function for a particular community exceeds a predefined threshold of functionality and an averaging approach that reduces multiple functions to a single standardized measure per community (15, 59). Our threshold multifunctionality metric was calculated by standardizing the scales for each function by ranking each culture and dividing each rank by the total number of cultures (n = 99 rather than 108 due to incomplete sampling). The culture with the highest performance for a function has rank 1, and the culture with the lowest performance has a rank of 1/99. We calculated the “distance from optimum” for each culture, defined as the Euclidean distance between the performance ranks for a given culture and the point where both ranks are equal to 1 (i.e., the maximum possible multifunction performance). Smaller distances indicate more optimal performance considering both functions. We also calculated the “maximum performance threshold,” defined as the highest performance rank at which a culture can perform both functions simultaneously. We calculated the percentage of monocultures versus bicultures that performed both functions greater than each threshold (between 0 and 1 at 0.01 increments). To calculate multifunctionality via the averaging index, we converted FAME (wt % of sample) and biomass (i.e., log-transformed values of population cell density multiplied by estimates of cell biovolume) values to standardized Z-scores. Standard scores were averaged together to yield a multifunctionality index. For both multifunctionality methods, we assessed the effect of species richness on multifunctionality via a nested analysis of variance (ANOVA) in which species combination was nested within our monoculture versus biculture variable of interest. To compare the best performing monoculture against the top performing bicultures, we subsampled the six biculture replicates across the top three performing bicultures in groups of two with replacement and iterated 10,000 times. We calculated the Z-score for the mean monoculture value (i.e., Euclidean distance to optimum in our multifunctionality metric) within the distribution of mean biculture values.
Transcriptome statistical analyses.
Assembled transcripts were aligned via BLAST to a UniProt custom database that contains sprot and trembl for fungi and plants (obtained 30 August 2013). We discarded all transcripts with UniProt identifiers (IDs) matching outside the green plant taxa (Diaphoretickes or Plant+SAR+HA megagroup) from further analysis. All remaining transcripts were assigned Gene Ontology terms via their UniProt IDs. The transcripts assigned to UniProt IDs with no known Gene Ontology assignments were excluded from further analyses. After this Gene Ontology filtering step, we excluded nine populations from our bicultures from further analyses, because their sequence libraries averaged fewer than 500,000 total read counts (i.e., sequence depth). The transcript data were analyzed via the EdgeR package in R using standard procedures (60). The transcript counts were normalized using the calcNormFactors function, and read counts for each gene were fit with negative binomial generalized log-linear models using the glmFit function in EdgeR. Model contrasts of the monoculture condition against each biculture condition were used to generate log-fold change values of each transcript in monoculture versus biculture. The standard EdgeR modeling procedure for the calculation of differential expression automatically accounts for variation in sequencing depth or library size between the monoculture and biculture growing conditions. Therefore, the variation in the relative abundance of each algal species due to the treatment condition does not interfere with the evaluation of differential expression.
A primary aim of our study was to determine the effect of growing condition on fatty acid metabolism and the genes involved in this function. We chose to use Gene Ontology terms as a method for identifying genes likely playing important roles in fatty acid metabolism in nonmodel organisms. We note that any interpretation of gene function from nonmodel organisms should always be interpreted with caution. The Gene Ontology database categorizes functions using a hierarchy of descriptive terms. These categorizations are supported by scientific literature either from direct experimental evidence, phylogenetically inferred annotations, or computationally inferred annotations (61). These annotations identify the most specific function for each gene that can be inferred using currently available data. Indeed, for many algal species, more than 40% of their genes could not be reliably assigned to any Gene Ontology (GO) term. We used GO term categorizations as the currently best available means of cautiously predicting function in nonmodel organisms. We divided all Gene Ontology terms into two categories. We designated GO terms as “lipid terms” if the term names contained any of the following keywords or partial words: “fat,” “stero,” “lip,” “steryl,” and “wax.” These key terms were chosen to encompass different classes of fats and their enzymes, such as lipid, lipase, lipoate, steroid, and sterol.
To detect shifts in lipid gene expression compared to that for all other gene functions, we categorized genes and their log-fold change (logFC) values into either the lipid or nonlipid group based on Gene Ontology assignments. For each algal species in each biculture condition, the distributions of logFC values between the lipid and nonlipid groups were compared with a Kolmogorov-Smirnov test. We tested whether significant differential expression of lipid genes was positively associated with relative and total FAME levels using logistic regression.
We measured gene expression similarity among algal populations within and among species combinations using multidimensional scaling plots for each algal species independently (plotMDS function, R package “Limma”). The Euclidean distances between points within versus among treatments were compared via two-sample t tests. To visualize broader patterns across all algal species simultaneously, we calculated mean logFC values of transcripts within each lipid GO term and then used these 28 GO terms as variables in a principal-component analysis. We tested whether clusters of FAME overyielding versus nonoveryielding bicultures were significantly divergent in principal-component (PC) space via multiple analysis of variance on principal-component scores as well as logistic regression.
Since we found these lipid GO terms significantly separated FAME overyielding bicultures from nonoveryielding cultures, we then tested whether sets of orthologous genes within these GO terms were significant predictors of FAME yield via linear regression. To test the effects of specific gene functions across multiple algal species, we first needed to identify gene orthologs. We completed a more restricted BLASTp search of transcripts in the lipid Gene Ontology group to the single algal species that most frequently appeared in our unrestricted blast matches (Chlorella variabilis) to identify orthologs across species. We then completed linear regression on each gene, now restricted to a CHLVA UniProt ID, that was present in two or more of our eight algal species. We were especially interested in two genes involved in fatty acid production, diacylglycerol acyltransferase and phospholipid diacylglycerol acyltransferase, yet we did not find orthologs for each species to Chlorella variabilis. We therefore completed a second BLASTp search using genes annotated as DGATs and PDATs from the Arabidopsis thaliana genome and queried these genes against our reference transcriptome. Lastly, to elucidate whether other physiological processes may explain why certain bicultures overyielded in FAMEs while others did not, we determined whether differential expression among genes regulating photosynthesis and nutrient assimilation were predictive of FAME yields in biculture relative to that in monoculture. We identified genes annotated as photosynthesis regulatory genes (including those encoding carbonic anhydrase, glutamate semialdehyde aminotransferase, and light harvesting chlorophyll a-b complexes), and nutrient assimilation genes (including those encoding iron permease, nitrite transporters, nitrite reducers, nitrate transporters, nitrate reducers, and phosphate transporters) in the Chlamydomonas reinhardtii genome. We identified these genes in our study species using BLASTp searches of C. reinhardtii queries against our reference transcriptomes. To detect shifts in photosynthesis gene expression compared to those for all other gene functions, and nutrient assimilation gene expression compared to that for all other gene functions, we compared the distributions of gene logFC values between groups using Kolmogorov-Smirnov tests as described above for our lipid gene differential expression analysis.
We calculated relative changes in FAMEs, higher heating values, and cetane numbers as log-fold changes to correspond to the standard log-fold change measure for reporting gene differential expression. For all figures referring to relative FAME yield, we calculated a combined relative FAME yield per biculture. These combined relative FAME values were calculated as log2FC {[mean biculture yield]/[mean (mean yield of species A in monoculture, mean yield of species B in monoculture)]}. In addition to testing the effects of gene logFC on FAME yields, we also tested the effects of gene logFC on the bio-oil quality metrics cetane number and higher heating value. Log ratios of these two metrics were calculated in the same way as described for FAME levels. All significance values reported in the above analyses account for multiple comparisons via false discovery rate corrections (62). All analyses were completed in R Studio (version 0.99.903).
Data availability.
Data and analyses can be found at https://github.com/sjackrel/Biofuel-genomics.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the U.S. National Science Foundation DIMENSIONS of Biodiversity program (DEB-1046121) to B.J.C. and T.H.O. and Emerging Frontiers in Research and Innovation in Photosynthetic Biorefineries (EFRI-PSBR 1332343) to B.J.C., V.J.D., P.E.S., and T.H.O.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
The authors declare no competing interests.
B.J.C. and T.H.O. conceived the ideas and developed them with P.E.S. and V.J.D. A.N. designed and carried out algal growth experiments. R.B.L. and D.C.H. measured algal fatty acid content. For transcriptomics analyses, B.B. completed quality control, library construction, and read mapping. S.L.J. completed gene annotation and differential expression analyses, analyzed data sets, and generated figures with advisement from V.J.D. and B.J.C. S.L.J. drafted the paper with conceptual and editorial input from all authors.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00953-18.
REFERENCES
- 1.Smith VH, Sturm BSM, deNoyelles FJ, Billings SA. 2010. The ecology of algal biodiesel production. Trends Ecol Evol 25:301–309. doi: 10.1016/j.tree.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Schenk PM, Thomas-Hall SR, Stephens E, Marx UC, Mussgnug JH, Posten C, Kruse O, Hankamer B. 2008. Second generation biofuels: high-efficiency microalgae for biodiesel production. Bioenergy Res 1:20–43. doi: 10.1007/s12155-008-9008-8. [DOI] [Google Scholar]
- 3.Brown TM, Duan P, Savage PE. 2010. Hydrothermal liquefaction and gasification of Nannochloropsis sp. Energy Fuels 24:3639–3646. doi: 10.1021/ef100203u. [DOI] [Google Scholar]
- 4.Shurin JB, Abbott RL, Deal MS, Kwan GT, Litchman E, McBride RC, Mandal S, Smith VH. 2013. Industrial-strength ecology: trade-offs and opportunities in algal biofuel production. Ecol Lett 16:1393–1404. doi: 10.1111/ele.12176. [DOI] [PubMed] [Google Scholar]
- 5.Ho S-H, Chen C-Y, Chang J-S. 2012. Effect of light intensity and nitrogen starvation on CO2 fixation and lipid/carbohydrate production of an indigenous microalga Scenedesmus obliquus CNW-N. Bioresour Technol 113:244–252. doi: 10.1016/j.biortech.2011.11.133. [DOI] [PubMed] [Google Scholar]
- 6.Rodolfi L, Chini Zittelli G, Bassi N, Padovani G, Biondi N, Bonini G, Tredici M. 2009. Microalgae for oil: strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. Biotechnol Bioeng 102(1):100–112. doi: 10.1002/bit.22033. [DOI] [PubMed] [Google Scholar]
- 7.Ajjawi I, Verruto J, Aqui M, Soriaga LB, Coppersmith J, Kwok K, Peach L, Orchard E, Kalb R, Xu W, Carolson TJ, Francis K, Konigsfeld K, Bartalis J, Schultz A, Lambert W, Schwartz AS, Brown R, Moellering ER. 2017. Lipid production in Nannochloropsis gaditana is doubled by decreasing expression of a single transcriptional regulator. Nat Biotechnol 35:647–652. doi: 10.1038/nbt.3865. [DOI] [PubMed] [Google Scholar]
- 8.Kazamia E, Aldridge DC, Smith AG. 2012. Synthetic ecology—a way forward for sustainable algal biofuel production? J Biotechnol 162:163–169. doi: 10.1016/j.jbiotec.2012.03.022. [DOI] [Google Scholar]
- 9.Kazamia E, Riseley AS, Howe CJ, Smith AG. 2014. An engineered community approach for industrial cultivation of microalgae. Ind Biotechnol (New Rochelle N Y) 10:184–190. doi: 10.1089/ind.2013.0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tilman D, Wedin D, Knops J. 1996. Productivity and sustainability influenced by biodiversity in grassland ecosystems. Nature 379:718–720. doi: 10.1038/379718a0. [DOI] [Google Scholar]
- 11.Ives AR, Carpenter SR. 2007. Stability and diversity of ecosystems. Science 317:58–62. doi: 10.1126/science.1133258. [DOI] [PubMed] [Google Scholar]
- 12.Cardinale BJ, Duffy JE, Gonzalez A, Hooper DU, Perrings C, Venail P, Narwani A, Mace GM, Tilman D, Wardle DA, Kinzip AP, Daily GC, Loreau M, Grace JB, Larigauderie A, Srivastava DS, Naeem S. 2012. Biodiversity loss and its impact on humanity. Nature 486:59–67. doi: 10.1038/nature11148. [DOI] [PubMed] [Google Scholar]
- 13.Shurin JB, Mandal S, Abbott RL. 2014. Trait diversity enhances yield in algal biofuel assemblages. J Appl Ecol 51:603–611. doi: 10.1111/1365-2664.12242. [DOI] [Google Scholar]
- 14.Duffy JE, Godwin CM, Cardinale BJ. 2017. Biodiversity effects in the wild are common and as strong as key drivers of productivity. Nature 549:261–264. doi: 10.1038/nature23886. [DOI] [PubMed] [Google Scholar]
- 15.Lefcheck JS, Byrnes JEK, Isbell F, Gamfeldt L, Griffin JN, Eisenhauer N, Hensel MJS, Hector A, Cardinale BJ, Duffy JE. 2015. Biodiversity enhances ecosystem multifunctionality across trophic levels and habitats. Nat Commun 6:6936. doi: 10.1038/ncomms7936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cherubini F. 2010. The biorefinery concept: using biomass instead of oil for producing energy and chemicals. Energy Convers Manag 51:1412–1421. doi: 10.1016/j.enconman.2010.01.015. [DOI] [Google Scholar]
- 17.Stockenreiter M, Graber A-K, Haupt F, Stibor H. 2012. The effect of species diversity on lipid production by micro-algal communities. J Appl Phycol 24:45–54. doi: 10.1007/s10811-010-9644-1. [DOI] [Google Scholar]
- 18.Narwani A, Alexandrou MA, Oakley TH, Carroll IT, Cardinale BJ. 2013. Experimental evidence that evolutionary relatedness does not affect the ecological mechanisms of coexistence in freshwater green algae. Ecol Lett 16:1373–1381. doi: 10.1111/ele.12182. [DOI] [PubMed] [Google Scholar]
- 19.Venail PA, Alexandrou MA, Oakley TH, Cardinale BJ. 2013. Shared ancestry influences community stability by altering competitive interactions: evidence from a laboratory microcosm experiment using freshwater green algae. Proc Biol Sci 280:20131548. doi: 10.1098/rspb.2013.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narwani A, Bentlage B, Alexandrou MA, Fritschie KJ, Delwiche C, Oakley TH, Cardinale BJ. 2017. Ecological interactions and coexistence are predicted by gene expression similarity in freshwater green algae. J Ecol 105:580–591. doi: 10.1111/1365-2745.12759. [DOI] [Google Scholar]
- 21.Alexandrou MA, Cardinale BJ, Hall JD, Delwiche CF, Fritschie K, Narwani A, Venail PA, Bentlage B, Pankey S, Oakley TH. 2015. Evolutionary relatedness does not predict competition and co-occurrence in natural or experimental communities of green algae. Proc Biol Sci 282:20141745. doi: 10.1098/rspb.2014.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berg JM, Tymoczko JL, Stryer L. 2002. Biochemistry, 5th ed WH Freeman, New York, NY. [Google Scholar]
- 23.Narwani A, Lashaway AR, Hietala DC, Savage PE, Cardinale BJ. 2016. Power of plankton: effects of algal biodiversity on biocrude production and stability. Environ Sci Technol 50:13142–13150. doi: 10.1021/acs.est.6b03256. [DOI] [PubMed] [Google Scholar]
- 24.Godwin CM, Hietala DC, Lashaway AR, Narwani A, Savage PE, Cardinale BJ. 2017. Ecological stoichiometry meets ecological engineering: using polycultures to enhance the multifunctionality of algal biocrude systems. Environ Sci Technol 51:11450–11458. doi: 10.1021/acs.est.7b02137. [DOI] [PubMed] [Google Scholar]
- 25.Godwin CM, Hietala DC, Lashaway AR, Narwani A, Savage PE, Cardinale BJ. 2017. Algal polycultures enhance coproduct recycling from hydrothermal liquefaction. Bioresour Technol 224:630–638. doi: 10.1016/j.biortech.2016.11.105. [DOI] [PubMed] [Google Scholar]
- 26.Nolan M. 2015. Diversity of algae slows growth of Microcystis. MSc thesis University of Michigan, Ann Arbor, MI. [Google Scholar]
- 27.Fukami K, Nishijima T, Ishida Y. 1997. Stimulative and inhibitory effects of bacteria on the growth of microalgae, p 185–191. In Hagiwara A, Snell T, Lubzens E, Tamaru CS (ed), Live food in aquaculture. Springer Netherlands, Dordrecht, Netherlands. [Google Scholar]
- 28.Amin SA, Green DH, Hart MC, Küpper FC, Sunda WG, Carrano CJ. 2009. Photolysis of iron-siderophore chelates promotes bacterial-algal mutualism. Proc Natl Acad Sci U S A 106:17071–17076. doi: 10.1073/pnas.0905512106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. 2005. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438:90–93. doi: 10.1038/nature04056. [DOI] [PubMed] [Google Scholar]
- 30.Li C, Guan Z, Liu D, Raetz CRH. 2011. Pathway for lipid A biosynthesis in Arabidopsis thaliana resembling that of Escherichia coli. Proc Natl Acad Sci U S A 108:11387–11392. doi: 10.1073/pnas.1108840108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duncan O, van der Merwe MJ, Daley DO, Whelan J. 2013. The outer mitochondrial membrane in higher plants. Trends Plant Sci 18:207–217. doi: 10.1016/j.tplants.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 32.Post-Beittenmiller D, Roughan G, Ohlrogge JB. 1992. Regulation of plant fatty acid biosynthesis. Plant Physiology 100:923. doi: 10.1104/pp.100.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Callahan DL, Martin GJO, Hill DRA, Olmstead ILD, Dias DA. 2015. Analytical approaches for the detailed characterization of microalgal lipid extracts for the production of biodiesel, p 331–46. In Kim S-K, Chojnacka K (ed), Marine algae extracts. Wiley-VCH, Weinheim, Germany. [Google Scholar]
- 34.Lopez-Huertas E, Charlton WL, Johnson B, Graham IA, Baker A. 2000. Stress induces peroxisome biogenesis genes. EMBO J 19:6770–6777. doi: 10.1093/emboj/19.24.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin Y, Cluette-Brown JE, Goodman HM. 2004. The peroxisome deficient Arabidopsis mutant sse1 exhibits impaired fatty acid synthesis. Plant Physiol 135:814–827. doi: 10.1104/pp.103.036772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagle N, Lemke P. 1990. Production of methyl ester fuel from microalgae. Appl Biochem Biotechnol 24:355–361. doi: 10.1007/BF02920259. [DOI] [Google Scholar]
- 37.Williams PJB, Laurens LML. 2010. Microalgae as biodiesel and biomass feedstocks: review and analysis of the biochemistry, energetics and economics. Energy Environ Sci 3:554–590. doi: 10.1039/b924978h. [DOI] [Google Scholar]
- 38.Shindou H, Hishikawa D, Harayama T, Yuki K, Shimizu T. 2009. Recent progress on acyl CoA: lysophospholipid acyltransferase research. J Lipid Res 50:S46–S51. doi: 10.1194/jlr.R800035-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goo Y-H, Son S-H, Paul A. 2017. Lipid droplet-associated hydrolase promotes lipid droplet fusion and enhances ATGL degradation and triglyceride accumulation. Sci Rep 7:2743. doi: 10.1038/s41598-017-02963-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Todd J, Post-Beittenmiller D, Jaworski JG. 1999. KCS1encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J 17:119–130. doi: 10.1046/j.1365-313X.1999.00352.x. [DOI] [PubMed] [Google Scholar]
- 41.Parrish CC. 1987. Time series of particulate and dissolved lipid classes during spring phytoplankton blooms in Bedford Basin, a marine inlet. Mar Ecol Prog Ser 35:129–139. doi: 10.3354/meps035129. [DOI] [Google Scholar]
- 42.Parrish CC, Wangersky PJ. 1987. Particulate and dissolved lipid classes in cultures of Phaeodactylum tricornutum grown in cage culture turbidostats with a range of nitrogen supply rates. Mar Ecol Prog Ser 35:119–128. doi: 10.3354/meps035119. [DOI] [Google Scholar]
- 43.Narwani A, Alexandrou MA, Herrin J, Vouaux A, Zhou C, Oakley TH, Cardinale BJ. 2015. Common ancestry is a poor predictor of competitive traits in freshwater green algae. PLoS One 10:e0137085. doi: 10.1371/journal.pone.0137085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tilman D. 1981. Tests of resource competition theory using four species of Lake Michigan algae. Ecology 62:802–815. doi: 10.2307/1937747. [DOI] [Google Scholar]
- 45.Kilham SS, Kreeger DA, Lynn SG, Goulden CE, Herrera L. 1998. COMBO: a defined freshwater culture medium for algae and zooplankton. Hydrobiologia 377:147–159. doi: 10.1023/A:1003231628456. [DOI] [Google Scholar]
- 46.United States Environmental Protection Agency. 1983. Methods for chemical analysis of water and wastes. United States Environmental Protection Agency, Washington, DC. [Google Scholar]
- 47.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, LeDuc RD, Friedman N, Regev A. 2013. De novo transcript sequence reconstruction from RNA-Seq: reference generation and analysis with Trinity. Nat Protoc 8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cooper ED, Bentlage B, Gibbons TR, Bachvaroff TR, Delwiche CF. 2014. Metatranscriptome profiling of a harmful algal bloom. Harmful Algae 37:75–83. doi: 10.1016/j.hal.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finn RD, Clements J, Eddy SR. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sheehan JD, Savage PE. 2017. Modeling the effects of microalga biochemical content on the kinetics and biocrude yields from hydrothermal liquefaction. Bioresour Technol 239:144–150. doi: 10.1016/j.biortech.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 53.Leow S, Witter JR, Vardon DR, Sharma BK, Guest JS, Strathmann TJ. 2015. Prediction of microalgae hydrothermal liquefaction products from feedstock biochemical composition. Green Chem 17:3584–3599. doi: 10.1039/C5GC00574D. [DOI] [Google Scholar]
- 54.Lewis T, Nichols PD, McMeekin TA. 2000. Evaluation of extraction methods for recovery of fatty acids from lipid-producing microheterotrophs. J Microbiol Methods 43:107–116. doi: 10.1016/S0167-7012(00)00217-7. [DOI] [PubMed] [Google Scholar]
- 55.Rodríguez-Ruiz J, Belarbi E-H, Sánchez JLG, Alonso DL. 1998. Rapid simultaneous lipid extraction and transesterification for fatty acid analyses. Biotechnol Tech 12:689–691. doi: 10.1023/A:1008812904017. [DOI] [Google Scholar]
- 56.Tran H-L, Hong S-J, Lee C-G. 2009. Evaluation of extraction methods for recovery of fatty acids from Botryococcus braunii LB 572 and Synechocystis sp. PCC 6803. Biotechnol Bioprocess Eng 14:187–192. doi: 10.1007/s12257-008-0171-8. [DOI] [Google Scholar]
- 57.Ackman RG, Sipos JC. 1964. Application of specific response factors in the gas chromatographic analysis of methyl esters of fatty acids with flame ionization detectors. J Am Oil Chem Soc 41:377–378. doi: 10.1007/BF02654818. [DOI] [Google Scholar]
- 58.Ramírez-Verduzco LF, Rodríguez-Rodríguez JE, Jaramillo-Jacob ADR. 2012. Predicting cetane number, kinematic viscosity, density and higher heating value of biodiesel from its fatty acid methyl ester composition. Fuel 91:102–111. doi: 10.1016/j.fuel.2011.06.070. [DOI] [Google Scholar]
- 59.Byrnes JEK, Gamfeldt L, Isbell F, Lefcheck JS, Griffin JN, Hector A, Cardinale BJ, Hooper DU, Dee LE, Duffy JE. 2014. Investigating the relationship between biodiversity and ecosystem multifunctionality: challenges and solutions. Methods Ecol Evol 5:111–124. doi: 10.1111/2041-210X.12143. [DOI] [Google Scholar]
- 60.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinksi K, Dwight SS, Eppig JT, Harris MA. 2000. Gene Ontology: tool for the unification of biology. Nat Genet 25:25. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and analyses can be found at https://github.com/sjackrel/Biofuel-genomics.