Identifying the sources of microbial contamination is important for developing best management practices to protect the water quality of urban streams for recreational uses. This study collected a large number of water samples from an urban stream under both dry and wet weather conditions and provided quantitative information on the relative contributions of various environmental compartments to the overall microbial contamination in the stream under the two weather conditions. The watershed in this study represents urban watersheds where no dominant fecal sources are consistently present. The findings highlight the importance of reducing the direct contribution of microbes from street surfaces in the watershed to urban streams under wet weather conditions. The methods and findings from this study are expected to be useful to stormwater managers and regulatory agencies.
KEYWORDS: dry weather condition, microbial source tracking, urban stream, wet weather condition
ABSTRACT
Investigating sources of microbial contamination in urban streams, especially when there are no contributions from combined sewer overflows or sewage effluent discharges, can be challenging. The objectives of this study were to identify the sources of microbes in an urban stream and quantify their relative contributions to the microbial community in the stream under dry and wet weather conditions. A microbial source tracking method relying on the 16S rRNA gene was used to investigate the microbial communities in water samples of an urban stream (i.e., from 11 dry and 6 wet weather events), as well as in streambed sediment, soils, street sweepings, sanitary sewage, an upstream lake, and feces of animals and birds collected between 2013 and 2015. The results showed that the Escherichia coli levels in the stream were significantly higher in wet weather flow than in dry weather flow. The upstream lake contributed approximately 93% of the microbes in dry weather flows. Water discharged from storm drain outfalls was the biggest source of microbes in wet weather flows, with a median contribution of approximately 90% in the rising limb and peak flow and about 75% in the declining limb of storms. Furthermore, about 70 to 75% of the microbes in the storm drain outfall water came from materials washed off from the street surfaces in the watershed. Fecal samples did not appear to contribute substantially to the microbes in environmental samples. The results highlight the significance of street surfaces in contributing microbial loads to urban streams under wet weather conditions.
IMPORTANCE Identifying the sources of microbial contamination is important for developing best management practices to protect the water quality of urban streams for recreational uses. This study collected a large number of water samples from an urban stream under both dry and wet weather conditions and provided quantitative information on the relative contributions of various environmental compartments to the overall microbial contamination in the stream under the two weather conditions. The watershed in this study represents urban watersheds where no dominant fecal sources are consistently present. The findings highlight the importance of reducing the direct contribution of microbes from street surfaces in the watershed to urban streams under wet weather conditions. The methods and findings from this study are expected to be useful to stormwater managers and regulatory agencies.
INTRODUCTION
Fecal contamination can make urban streams unsuitable for recreational use (1, 2) and create public health concerns (3, 4). Fecal bacteria in urban streams under dry and wet weather conditions may originate from several different environmental sources. In urban watersheds, fecal bacteria may originate from sources like sanitary sewage and feces of wild animals, birds, and domestic pets (5–7). Embankment soil and streambed sediment may act as a repository of these fecal bacteria (8–10) and serve as environmental sources of fecal bacteria to stream water during wet weather events. Storm drainage from subwatersheds can be a major nonpoint source of microbes in urban streams under wet weather conditions (11–13). Pollutant buildup via processes like wind erosion and deposition during dry periods that precede storms may also contribute to bacterial load (14, 15). Identifying the predominant sources of fecal bacteria in a particular urban stream can be challenging (6, 13, 16).
Levels of fecal indicator bacteria (FIB) in urban streams often rise during storms (3, 17), and the elevated levels can persist from 24 h (18) to up to 5 days after a storm (19). Levels of fecal bacteria in stormwater can vary within a wet weather event, too. Some studies have reported that the earlier phases of a stormwater hydrograph typically have the highest bacterial concentrations compared to later phases (20–22). A better understanding of the temporal variation in bacterial load and composition of stormwater can be helpful for designing stormwater control measures and best management practices.
Culture-based methods of enumerating FIB cannot differentiate fecal bacteria from different sources (23, 24). Quantitative real-time PCR (qPCR)-based methods targeting host-specific markers (25, 26) can detect and quantify fecal contamination from specific hosts; however, they require separate analysis for each host (27) and cannot assess the relative contributions of multiple sources (28). On the other hand, microbial source tracking (MST) tools relying on 16S rRNA gene sequencing libraries have the potential to quantify the relative contributions of multiple sources to the microbial composition of an environment (29, 30). These methods often involve the characterization of microbial communities in relevant samples by targeting the hypervariable region(s) of the 16S rRNA gene, and they have been successfully used to identify the hosts of fecal bacteria in water (24, 26, 31, 32). The use of DNA sequencing data with computational mixing models, such as SourceTracker, enables the employment of Bayesian inference for estimating the relative contributions of microbes from multiple sources to an environment (33). In SourceTracker analyses, the relative contributions from different sources to a sink environment are modeled as a probabilistic mixture of the composition of the sources. The microbial composition of the sink environment is assumed to result from mixing of the given sources and an additional unknown source. The tool assigns each taxon in the sink environment to different sources using Gibbs sampling to generate the posterior distribution. SourceTracker has been used to track the sources of microbial communities in sewage (30), public restrooms (35), indoor air (36), the skin of newborn infants (37), and neonatal intensive care units (38), as well as lake estuaries (39), coastal waters (40), and urban stormwater (41).
The objectives of this study were to identify the sources of microbes in an urban stream and quantify their relative contributions to the microbial community in the stream under dry and wet weather conditions. The study site was a segment of Antelope Creek, a concrete-lined urban stream located in Lincoln, Nebraska. Antelope Creek is impaired due to high levels of Escherichia coli during summer months (42). The microbial communities were characterized for water collected from multiple in-stream locations in 11 dry weather events and multiple stages of six wet weather events, as well as for samples representing storm drain outfall water, streambed sediment, embankment soil along the stream, erodible soil around the watershed, street sweepings from impervious surfaces, an upstream lake reservoir, and sanitary sewage and feces of birds and animals in the watershed. Water quality parameters were also monitored for the in-stream water samples. Bioinformatic and statistical analyses were conducted to elucidate the relationships within the environmental samples and between environmental and fecal samples. Information from the study could be useful in developing best management practices to reduce the levels of fecal contamination in urban streams.
RESULTS
E. coli in Antelope Creek.
The E. coli levels in Antelope Creek water were significantly higher in wet weather flow than in dry weather flow (Fig. 1). The median concentrations of E. coli across the four sampling sites (AC1 to AC4) were 0.36 × 103 to 1.04 × 103 CFU/100 ml in dry weather flow, 0.20 × 105 to 1.05 × 105 CFU/100 ml in rising limb, 4.6 × 104 to 6.5 × 104 CFU/100 ml in peak flow, and 0.91 × 104 to 4.79 × 104 CFU/100 ml in declining limb of wet weather flow (Fig. 1). The E. coli concentrations in rising limb, peak flow, and declining limb were all significantly higher than those in dry weather water (P < 0.001). The average E. coli concentrations among the four sampling sites were not significantly different from each other under dry or wet weather conditions (P > 0.05). The E. coli concentrations in the three stages of the wet weather flows were also not significantly different from each other (P = 0.521).
FIG 1.
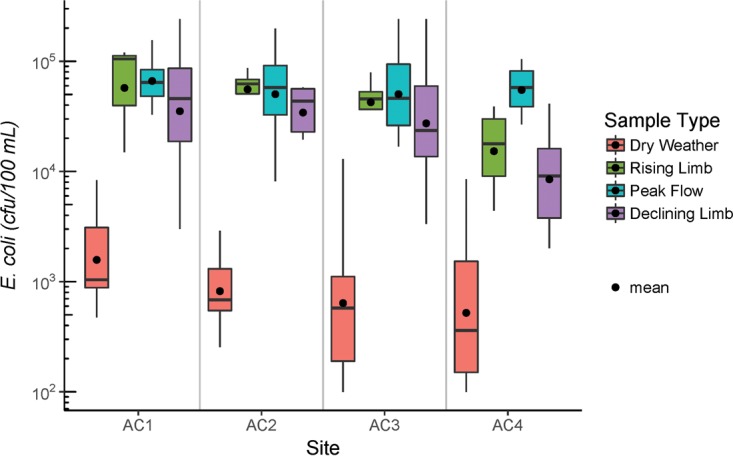
Boxplot showing E. coli concentrations in Antelope Creek water under dry weather conditions and in the three stages under wet weather conditions. The bottom and top of each box represent the 25th and 75th percentiles, respectively. The bold line inside each box represents the median concentration, while the dot represents the average concentration. Whiskers on either side of each box represent data within 1.5 times of the interquartile range (25th percentile to 75th percentile).
General description of DNA sequences.
The average length of DNA sequences was 253 bp. The number of sequences per library ranged from 4,040 to 307,972. The libraries were rarefied to 4,000 reads per sample for downstream analyses, and libraries with <4,000 reads were discarded. The remaining 306 libraries consisted of more than 14 million reads representing 1,759 unique operational taxonomic units (OTUs). Rarefaction curves (see Fig. S2 in the supplemental material) suggest that the sequencing effort covered the dominant taxa at a genetic distance of 3%.
Microbial composition in samples.
Not surprisingly, Proteobacteria, Bacteroidetes, and Actinobacteria were among the major bacterial phyla detected in most environmental and fecal samples (Fig. S3 to S5). In addition to the phylum level, the taxonomic composition of the samples at the family level is summarized in Table S3 and presented in Fig. S6. Some major families in the environmental and fecal samples include Comamonadaceae, Moraxellaceae, Oxalobacteraceae, and Enterobacteriaceae. The α-diversity indices (Chao1, Shannon index, and observed OTUs) of the fecal samples were consistently lower than those of the environmental samples (Table S1). Streambed sediment and wet weather water had higher α-diversity indices than dry weather water. Embankment soil, street sweepings, and erodible soil in the watershed had similar α-diversity indices. Finally, pairwise comparison using permutational analysis of variance (PERMANOVA) revealed that the microbial compositions between samples were, in general, significantly different from one another (P < 0.05, Fig. S7).
Spatial and temporal variation.
The results from PERMANOVA, also sometimes referred to as adonis analysis, showed greater temporal variation than spatial variation for the microbial community present in water samples from Antelope Creek (Table 1). For dry weather water samples, grouping by sampling event explained 80.6% of the variation (P < 0.001), while grouping by sampling site explained only 3.0% of the variation and was not significant (P = 0.997). Similarly, for wet weather water samples, grouping by sampling event better explained the variation than grouping by sampling site (36.7% versus 3.0%; Table 1). The grouping of the three stages of wet weather flows explained 16.9% of the variation in the structure of the microbial community (P < 0.001). For the microbial community in storm drain outfall water, groupings by site and event were both significant.
TABLE 1.
PERMANOVA results showing the significance of spatial and temporal grouping factors for the microbial community in the water samples collected from Antelope Creek and storm drain outfalls along the creek
| Sample type | Grouping factora | No. of groups | R2 | P |
|---|---|---|---|---|
| Dry weather water | Site | 4 | 0.030 | 0.997 |
| Event | 11 | 0.806 | <0.001 | |
| Wet weather water | Site | 4 | 0.030 | 0.918 |
| Event | 6 | 0.367 | <0.001 | |
| Stage (rising limb, peak flow, declining limb) | 3 | 0.169 | <0.001 | |
| Storm drain outfalls | Site | 8 | 0.395 | 0.039 |
| Event | 3 | 0.215 | 0.006 |
Site and event information for each sample type can be found in Table 2.
Correlation with water quality parameters.
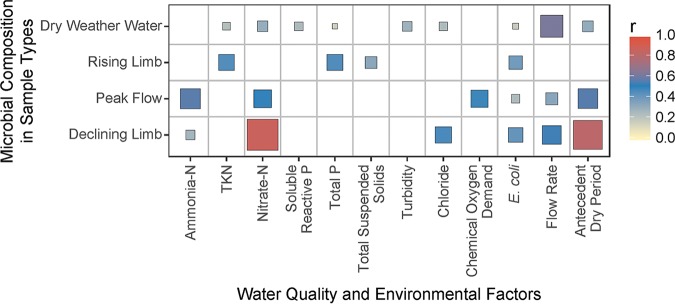
Mantel tests were carried out to test the correlation of the microbial community in water with water quality parameters and environmental factors (Fig. 2). A summary of the water quality parameters is provided in Table S2. The microbial composition in dry weather water samples had a moderate correlation (0.3 < r < 0.7, P < 0.05) with flow rate at the time of sampling. The microbial composition in rising limb samples had moderate correlations with total Kjeldahl nitrogen (TKN), total phosphorus (TP), total suspended solids (TSS), and E. coli. The microbial composition in peak flow samples had moderate correlations with ammonia (NH3-N), nitrate (NO3-N), chemical oxygen demand (COD), flow rate, and antecedent dry period. The microbial composition in declining limb samples had strong correlations (0.7 < r < 1.0, P < 0.05) with NO3-N and antecedent dry period and moderate correlations with chloride, E. coli, and flow rate.
FIG 2.
Mantel test results showing significant correlation (P < 0.05) between weighted UniFrac distance for overall microbial community and Bray-Curtis dissimilarity coefficient for water quality parameters and environmental factors. The sizes of the boxes and the fill color both represent the r value. Boxes are shown only for significant correlations (P < 0.05).
SourceTracker analysis. (i) Conceptual models.
Two conceptual models were established to define the sinks and sources of microbes in the watershed (Fig. 3). For dry weather conditions, water in Antelope Creek (i.e., dry weather water) was considered a sink, while the base flow from Holmes Lake and the resuspension of streambed sediment were considered two sources of microbes (Fig. 3A). For wet weather conditions, water in Antelope Creek (i.e., wet weather water) was considered a sink, while the flow from Holmes Lake, the resuspension of streambed sediment, the erosion of embankment soil, and the storm drain outfall water were considered sources of microbes (Fig. 3B). In the wet weather model, erodible soil, sanitary sewage, and street sweepings were considered the sources of microbes in storm drain outfall water. The storm drains receive runoff from impervious surfaces (i.e., street sweepings from road and parking lots) as well as pervious surfaces (i.e., erodible soils in the subwatershed). Even though the Antelope Creek watershed does not have combined sewer overflows (CSOs) and Antelope Creek does not receive discharge from wastewater treatment plants, sanitary sewage was included as a potential source of microbes to account for sanitary sewage leakage and accidental cross-connections (12).
FIG 3.
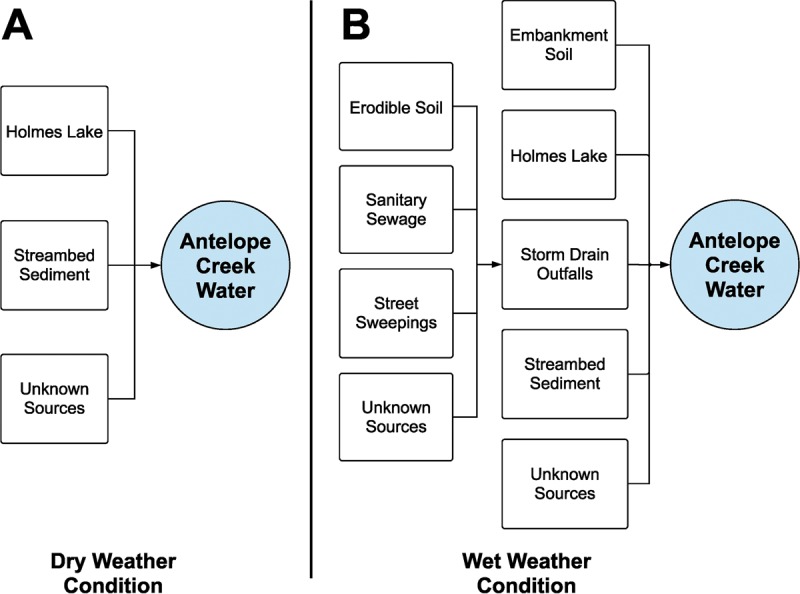
Conceptual model for SourceTracker analysis showing major sources of microbes to Antelope Creek for dry weather conditions (A) and wet weather conditions (B).
(ii) Dry weather conditions.
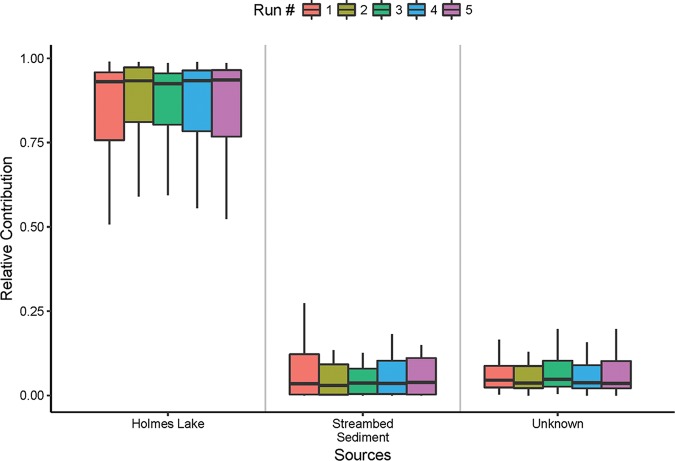
Based on SourceTracker analyses, the base flow from Holmes Lake was predicted to be the largest contributor of microbes to Antelope Creek under dry weather conditions, with the median value of the relative contribution being between 93% and 94% based on five separate runs of SourceTracker analysis (Fig. 4). Streambed sediment and the unknown sources each contributed to less than 5% of the microbes in dry weather water. The families with a relative contribution greater than 0.50% to dry weather water are presented in Table S4. For example, both Holmes Lake and streambed sediment contributed Comamonadaceae to the dry weather water, with relative contributions of 6.42% and 0.51%, respectively.
FIG 4.
Five runs of SourceTracker analysis showing the relative contributions of potential environmental sources to the overall microbial community in dry weather water.
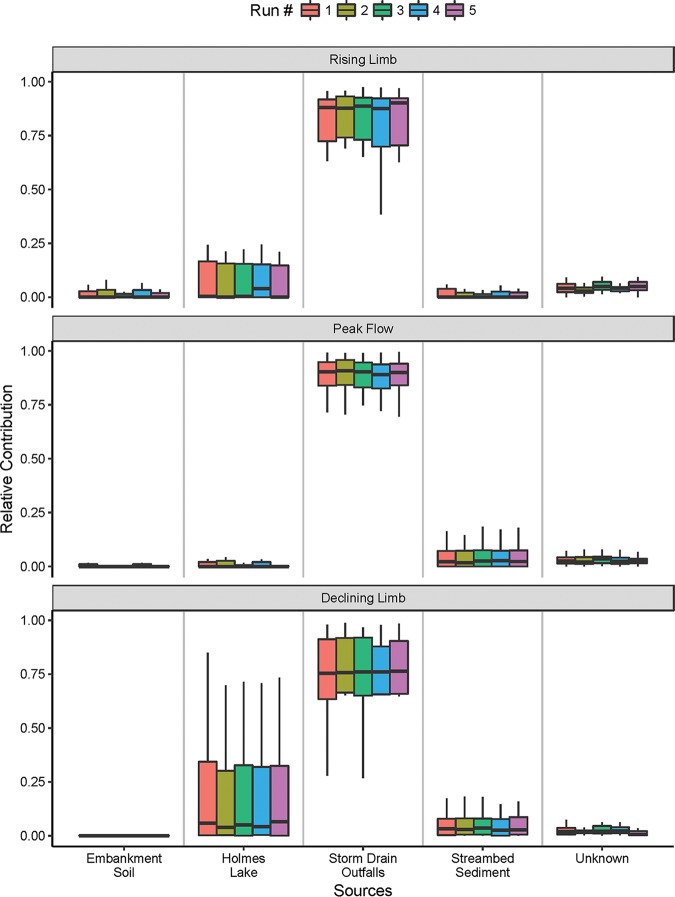
(iii) Wet weather conditions.
For wet weather events, the rising limb, peak flow, and declining limb were separately treated as sinks in SourceTracker analyses. The microbes in runoff water from storm drain outfalls accounted for 88 to 90%, 89 to 91%, and 75 to 76% of the microbes in the rising limb, peak flow, and declining limb, respectively (Fig. 5). A closer look at the SourceTracker data reveals that the relative contribution of Comamonadaceae from the runoff water in storm drain outfalls increased from 5.13%, to 9.55%, and to 18.65% over the three phases (Tables S5 to S7). The relative contributions of microbes from Holmes Lake, embankment soil, and streambed sediment to wet weather flows were generally less than 5% (Fig. 5). The family Comamonadaceae from Holmes Lake and streambed sediment made 1.24% and 0.77%, respectively, relative contributions to the microbial communities in the declining limb of wet weather flow (Table S7).
FIG 5.
Five runs of SourceTracker analysis showing the relative contributions of potential environmental sources to the overall microbial community in the three stages of wet weather flow.
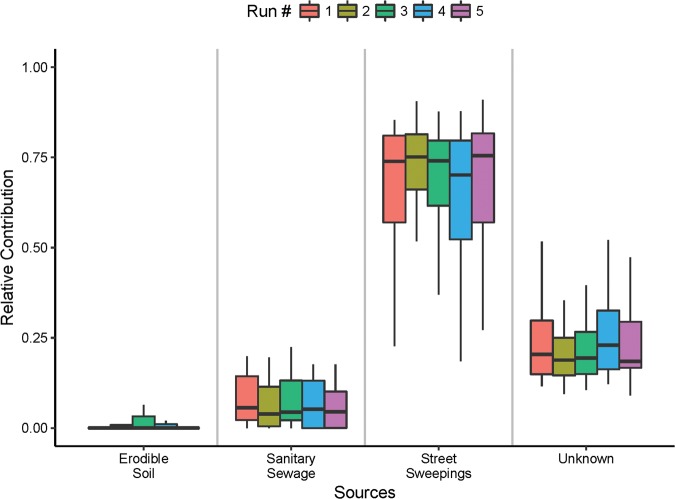
When water from storm drain outfalls was treated as a sink, the median relative contribution from street sweepings ranged from 70% to 75%, while those from the sanitary sewage and unknown sources accounted for 4 to 6% and 18 to 23%, respectively (Fig. 6). The top three families that made the highest relative contributions from street sweepings to the water in storm drain outfalls were Pseudomonadaceae (7.78%), Oxalobacteraceae (6.66%), and Comamonadaceae (5.62%) (Table S8).
FIG 6.
Five runs of SourceTracker analysis showing the relative contributions of potential environmental sources to the overall microbial community in storm drain outfall water.
(iv) Hosts of fecal contamination.
In the SourceTracker analysis of fecal contamination, the feces of animals and birds were treated as sources, and different environmental compartments were treated as sinks (Fig. S8 to S12). Contributions from any fecal samples were predicted to be trivial in embankment soil, erodible soil, streambed sediment, and Holmes Lake samples, while the largest contributions were attributed to unknown sources. Pigeon feces was predicted to account for a median of 14 to 21% of the microbial community in street sweepings.
DISCUSSION
Microbial communities in Antelope Creek water.
Temporal variation in the microbial composition of dry weather and wet weather water samples collected from Antelope Creek was found to be more pronounced than spatial variation (Table 1). This suggests that in Antelope Creek, event-based environmental characteristics can better explain the microbial community in water samples than location-based characteristics can. Since Antelope Creek is mostly composed of concrete-lined straight channels, the retention time for water is fairly short, and the flow is well connected throughout. There are no known hotspots of microbial contamination, as evidenced by similar E. coli levels at different sampling sites along the stream (Fig. 1). In the absence of localized site-specific factors, environmental factors affect microbial composition throughout the stream in a similar manner. The absence of hotspots of microbial contamination also suggests that the microbial contamination in Antelope Creek is likely from nonpoint sources.
Environmental conditions that can affect the temporal variation of microbial composition include parameters, such as antecedent weather, rainfall amount, temperature, and exposure to sunlight UV radiation (43, 44). In this study, an antecedent dry period was found to be correlated with microbial community in dry weather, peak flow, and declining limb water samples, suggesting that an antecedent dry period has an influence on microbial composition of urban water bodies. Nutrient concentrations may also vary seasonally (45, 46). Noticeably, NO3-N had moderate to strong correlations with the water from dry weather flows as well as the peak flow and declining limb of wet weather flows (Fig. 2), indicating that the nitrate and microbial community in Antelope Creek may originate from similar environmental sources. The lack of correlation between NO3-N and the microbial community in rising limb may be due to the contribution of atmospheric NO3-N to Antelope Creek, which was likely to be substantial in the initial stage and negligible in the later stages of the storm (47, 48). Flow rate was found to correlate well with microbial composition, particularly in dry weather and declining limb samples, suggesting that contributing flows brought both water and microbes to Antelope Creek. The E. coli levels in Antelope Creek had significant correlations with microbial composition in dry and wet weather water. This suggests that the same environmental sources could be responsible for both the overall microbial community and the E. coli population in Antelope Creek.
Interestingly, for the microbial composition in storm drain outfall water, spatial variation was more pronounced than temporal variation (Table 1). This highlights the existence of site-specific factors that were distinct among the subwatersheds contributing flow to each storm drain outfall. Some of these factors could be related to storm drainage infrastructure in the neighborhood (30), specific land use within the contributing area (49), and tree cover (50).
Microbial community in street sweepings.
DNA sequence data show that the microbial communities in street sweeping samples had the following major families (Table S3): Rhodobacteraceae (8.1% relative abundance), Comamonadaceae (7.3%), Enterobacteriaceae (7.3%), and Oxalobacteraceae (7.0%). The high relative abundance of Enterobacteriaceae is noticeable, as this is a large family of Gram-negative bacteria that includes many familiar pathogens, such as Salmonella spp., Klebsiella spp., Shigella spp., and E. coli.
SourceTracker analysis. (i) Dry weather conditions.
Holmes Lake was the biggest contributor of microbes in dry weather flow. The lack of substantial contribution from streambed sediments is likely due to the fact that the dry weather flow in Antelope Creek does not contain a large amount of suspended sediments. Only very fine particles are likely to enter the stream from Holmes Lake, as larger particles settle upstream of the dam. The mean particle size in typical dry weather flows was measured to be approximately 1 μm (Fig. S13). On the other hand, the mean particle size in typical streambed sediment was measured to be about 2 mm. This explains why the resuspension of bed sediments into the water column was not substantial during dry weather flow and why dry weather water and streambed sediment formed separate clusters in the principal-coordinate analysis plot shown in Fig. S14 (9, 51).
(ii) Wet weather conditions.
Field observation and model simulation suggest that streambed sediment could be a major source of fecal bacteria in wet weather flows due to resuspension (52–55). Streambed sediment can act as a repository of bacteria because it provides favorable growth conditions (8–10) and protection from UV radiation (56). However, the streambed sediment in Antelope Creek was not a major contributor of the microbes in wet weather flows, likely because Antelope Creek is concrete-lined and does not contain a large amount of erodible streambed sediment.
A large portion of the stormwater in urban streams comes from the impervious areas in the watershed. This partially explains why the flow rate in Antelope Creek was up to 300 times higher during storms than during dry periods (Fig. S1). Indeed, SourceTracker analyses showed that the runoff discharged at the storm drain outfalls accounted for a large proportion of the microbial community in wet weather water in Antelope Creek, with the highest contribution to the peak flow, followed by the rising limb and the declining limb. Conversely, the contribution from Holmes Lake was lowest in the peak flow and highest in the declining limb, suggesting that contributions from Holmes Lake become increasingly important during the later parts of the flow hydrograph.
According to SourceTracker analyses, street sweeping samples, which represent materials deposited on impervious surfaces, were the largest contributors to the microbes in the storm drain outfall water. Connected imperious surfaces can directly convey water to a stream through a network of pipes and lined channels (57). In addition to affecting chemical parameters, such as electrical conductivity, total phosphorus, and filterable reactive phosphorus (57, 58), impervious areas in residential areas were found to contribute 78% of the fecal coliform load in an urban watershed in Wisconsin (59). A simulation study suggested that a combination of structural management practices and regular street sweepings can reduce fecal coliform loads by 7.5% to 17% in urban stormwater (60). The findings of this study also suggest that connected impervious surfaces can serve as an immediate source of microbes to urban streams and should be targeted for implementing best management practices.
Conclusions.
The high E. coli levels in Antelope Creek likely resulted from nonpoint sources. Holmes Lake, the lake located upstream of the study area, contributed >90% of the microbes in Antelope Creek during dry weather flows, while the contribution of streambed sediment was less than 5%. Stormwater runoff discharged into Antelope Creek from the storm drain outfalls contributed more than 75% of the microbes in Antelope Creek during wet weather flows, while the runoff mostly consisted of microbes washed off from streets in the watershed. Feces of pets, animals, and wild birds were not found to be major contributors of microbial contamination in Antelope Creek.
MATERIALS AND METHODS
Study area.
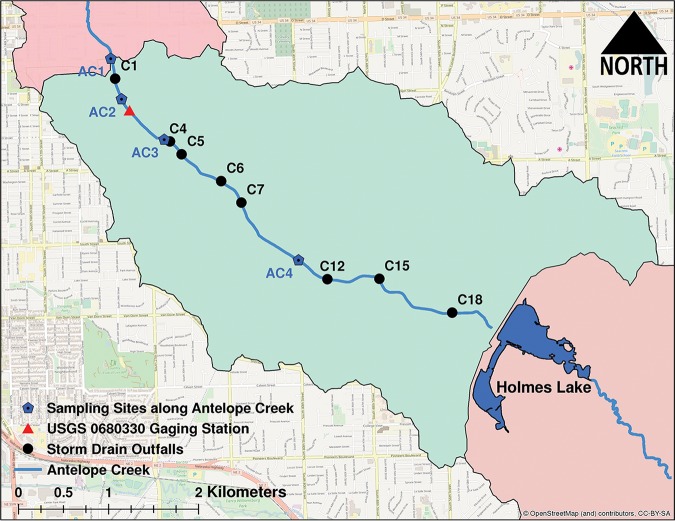
The study area is an approximately 5.8 km (3.6 mile) long reach within Antelope Creek located in Lincoln, NE. The reach is bounded at the upstream end by Holmes Lake, a 45-ha (112-acre) flood control reservoir, and by a weir at the downstream end (Fig. 7). The stream channel in the study area is fairly straight, and nearly 80% of the channel is concrete lined. The E. coli levels in summer months routinely exceed the bacteriological water quality standard of 126 CFU/100 ml (61).
FIG 7.
Map of the study area (in green) marked with in-stream sampling sites, storm drain outfalls, and a USGS gaging station. Direction of flow in Antelope Creek is from Holmes Lake toward AC1. OpenStreetMap was used to create this figure (https://www.openstreetmap.org/).
The Antelope Creek watershed is primarily a residential watershed, with more than 75% of the land use classified as residential area and roads. Under dry weather conditions, the reach in the study area receives base flow from Holmes Lake and intermittent flows, such as landscape irrigation. There are no combined sewers or known industrial and municipal discharges into Antelope Creek. With the exception of the main channel, the storm drainage system in the area consists of subsurface pipes and culverts. In the study area, the sediment layer on the bed of Antelope Creek is generally thin, patchy, and composed of mostly sand and gravel.
Sampling.
Sampling sites along Antelope Creek consisted of, from downstream to upstream, AC1, AC2, AC3, and AC4. Storm drain outfalls sampled consisted of, from downstream to upstream, C1, C4, C5, C6, C7, C12, C15, and C18 (Fig. 7). Time series data for precipitation, flow, and stage were collected from the U.S. Geological Service (USGS) no. 0680330 gaging station in Antelope Creek.
All sampling events are summarized in Table 2. Specifically, grab samples of dry weather water and streambed sediment were collected from four sites (AC1 to AC4) in 11 sampling events between 2013 and 2015. Dry weather water samples were collected in 1-liter Nalgene bottles from about 3 cm below the water surface. Streambed sediment samples were collected in sterile 50-ml centrifuge tubes from the top 3 cm of sediment in the streambed at the same location and time as the dry weather water samples. In the four dry weather sampling events of 2015, embankment soil samples were also collected from the stream embankment above the concrete lining of the channel, 100 m and 200 m upstream of each sampling site (AC1 to AC4), totaling 64 soil samples (4 sites × 4 events × 2 banks × 2 distances). The embankment soils were collected because they may be transported to the stream through overland runoff during storms.
TABLE 2.
List of the environmental samples collected
| Sample type | Site(s) | Yr | No. of sampling events by mo |
||||
|---|---|---|---|---|---|---|---|
| June | July | August | September | October | |||
| Dry weather water | AC1, AC2, AC3, AC4 | 2013 | 1 | 1 | 1 | ||
| 2014 | 1 | 1 | 2 | ||||
| 2015 | 2 | 1 | 1 | ||||
| Dry weather sediment | AC1, AC2, AC3, AC4 | 2013 | 1 | 1 | 1 | ||
| 2014 | 1 | 1 | 2 | ||||
| 2015 | 2 | 1 | 1 | ||||
| Embankment soil | Upstream of AC1, AC2, AC3, AC4 | 2013 | |||||
| 2014 | |||||||
| 2015 | 2 | 1 | 1 | ||||
| Rising limb water | AC1, AC2, AC3, AC4 | 2013 | |||||
| 2014 | 1 | ||||||
| 2015 | 1 | 1 | 1 | ||||
| Peak flow water | AC1, AC2, AC3, AC4 | 2013 | 2 | ||||
| 2014 | 2 | 1 | |||||
| 2015 | 2 | 1 | 1 | ||||
| Declining limb water | AC1, AC2, AC3, AC4 | 2013 | 1 | ||||
| 2014 | 1 | 1 | |||||
| 2015 | 1 | 1 | |||||
| Storm drain outfalls | C1, C4, C5, C6, C7, C12, C15, C18 | 2013 | |||||
| 2014 | 1 | ||||||
| 2015 | 2 | ||||||
| Holmes Lake water | Holmes Lake | 2013 | |||||
| 2014 | 1 | 4 | 4 | 2 | |||
| 2015 | 4 | 2 | 1 | 3 | |||
Wet weather water samples were collected from four sites (AC1 to AC4) in six storms. The wet weather water samples were collected in 1-liter Nalgene bottles using rope-suspended samplers from bridges over the sampling sites. The water samples were classified as rising limb, peak flow, or declining limb based on the timing of the sample collection in relation to the flow hydrograph generated at the USGS gaging station (Fig. S1). Out of the 72 wet weather water samples, 16 samples were from rising limb (4 events × 4 sites), 36 samples were from peak flow (9 events × 4 sites), and 20 samples were from declining limb (5 events × 4 sites).
Grab samples of water (n = 21) were collected from Holmes Lake under dry weather conditions. Grab samples of water (n = 24) were collected at eight storm drain outfalls (from downstream to upstream, C1, C4, C5, C6, C7, C12, C15, and C18) during storm one time in the summer of 2014 and two times in the summer of 2015 (3 events × 8 outfalls). Samples were collected at the storm drain outfalls as early as possible after the start of rain events in 1-liter Nalgene bottles using either rope-suspended samplers or sampling poles.
Other samples included street sweepings, sanitary sewage, and erodible soil in the watershed. Street sweepings refer to the residuals collected by motorized sweeping equipment from sweeping the streets (62) and represent materials that have deposited on streets. Street sweepings (n = 3) were collected in collaboration with the City of Lincoln street sweeping crew from stockpiles of freshly collected sweepings. Twenty-four-hour composite samples of untreated sanitary sewage (n = 7) were collected from a local wastewater treatment plant. Erodible soil samples (n = 20) were collected from exposed (i.e., without grass cover) surfaces in the watershed.
Fecal samples were collected from the Antelope Creek watershed and its vicinity. Samples of horse feces were obtained from a nearby horse riding stable. Fecal samples from other hosts were identified at the time of collection based on reconnaissance surveys, animal sightings, surrounding evidence, and field guides (63–65). A total of 30 fecal samples were collected from pigeons (n = 3), swallows (n = 6), ducks (n = 4), geese (n = 5), dogs (n = 4), horses (n = 4), and small mammals (e.g., opossums, raccoons, and mice; n = 4).
All the samples were transported to the lab on ice within 2 h after sampling. The water samples were filtered through 0.45-μm nitrocellulose filters (Millipore, Billerica, MA) to retain the microbial cells on the filters (66). The filter papers, along with other samples, were frozen at −20°C until DNA extraction.
Water quality analysis.
For water samples collected from Antelope Creek, ammonia (NH3-N), nitrate (NO3-N), total Kjeldahl nitrogen (TKN), soluble reactive phosphorus (SRP), total phosphorus (TP), total suspended solids (TSS), turbidity, surfactants, chloride, conductivity, and chemical oxygen demand (COD) were measured according to the standard methods (67). E. coli enumeration was performed using the Quanti-Tray/2000 method (Idexx Laboratories, ME). Particle size analysis for selected dry weather water samples was carried out using a Zeta potential analyzer (ZetaPALS; Brookhaven Instruments Corporation, NY). Grain size analysis for selected streambed sediment samples was carried out using the ASTM D422-63(2007)e2 method (68).
DNA extraction and sequencing.
DNA was extracted from the filters and source samples using the Mo Bio PowerSoil DNA isolation kit (Carlsbad, CA) and purified using the Zymo OneStep PCR inhibitor removal kit (Irvine, CA). To ensure that the PCR inhibitors were successfully removed, PCR tests were carried out on the purified DNA extracts using the 27F-1492R primer set (69). Purified DNA extracts were sent to Argonne National Laboratory to sequence the V4 region of the 16S rRNA gene on an Illumina MiSeq platform using the 515F/806R primer set reported in the Earth Microbiome project (70).
Bioinformatic and statistical analyses.
Sequence data were analyzed using QIIME (71), an open-source bioinformatic pipeline. Demultiplexing and quality filtering were carried out in QIIME using the default settings. Chimeric sequences were identified and removed from the demultiplexed reads using Usearch6.1 (72). Taxonomy was assigned to sequences using the RDP Classifier based on the Greengenes database v13_8 with 97% sequence similarity (72). Sequences were further filtered to remove operational taxonomic units (OTUs) that accounted for less than 0.005% of the total sequence counts (73).
The relative contributions of microbes from potential sources to sink environments were estimated using SourceTracker 1.0.1, a computational tool that performs Bayesian inference using the Markov chain Monte Carlo (MCMC) method (33). For each SourceTracker analysis, five independent runs were carried out to reduce the effect of false predictions (40).
Statistical analysis and data visualization were carried out using R version 3.41 (74). E. coli concentrations for the Antelope Creek water and the results of SourceTracker analysis were visualized as box plots using the package ggplot2 (version 2.2.1) in R (75). Permutational analysis of variance (PERMANOVA) (76) and a mantel test (77) were carried out using the package vegan version 2.4.3 (78). The package APE version 4.1 was used to carry out principal-coordinate analysis based on weighted UniFrac distance (79).
Accession number(s).
The DNA sequences have been deposited in the NCBI repository with the accession number SRP073625.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the City of Lincoln.
We thank Rock Krzycki and Ben Higgins of the City of Lincoln for providing watershed data and facilitating the collection of samples. We thank Cody Kimball, Bret Shald, Paige Schneider, Lauren Klesenmeyer, Ally Pietrok, Phillip Weibe, David Hansen, Adarsh Jnawali, and Sara Mollamohammada for help with sampling and analysis. We also thank Aina Kekilova, Mohammed Jalloh, Scott Speicher, Steven Speicher, Vincent Kuppig, Shivam Aashirbadam, Zhe Du, Rachel Levine, and Shaobin Li for help with sampling.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00896-18.
REFERENCES
- 1.Santo Domingo JW, Bambic DG, Edge TA, Wuertz S. 2007. Quo vadis source tracking? Towards a strategic framework for environmental monitoring of fecal pollution. Water Res 41:3539–3552. [DOI] [PubMed] [Google Scholar]
- 2.Rabinovici SJ, Bernknopf RL, Wein AM, Coursey DL, Whitman RL. 2004. Economic and health risk trade-offs of swim closures at a Lake Michigan beach. Environ Sci Technol 38:2737–2745. doi: 10.1021/es034905z. [DOI] [PubMed] [Google Scholar]
- 3.Cho KH, Cha SM, Kang J, Lee SW, Park Y, Kim J, Kim JH. 2010. Meteorological effects on the levels of fecal indicator bacteria in an urban stream: a modeling approach. Water Res 44:2189–2202. doi: 10.1016/j.watres.2009.12.051. [DOI] [PubMed] [Google Scholar]
- 4.Pintar K, Fazil A, Pollari F, Charron D, Waltner-Toews D, McEwen S. 2010. A risk assessment model to evaluate the role of fecal contamination in recreational water on the incidence of cryptosporidiosis at the community level in Ontario. Risk Anal 30:49–64. doi: 10.1111/j.1539-6924.2009.01321.x. [DOI] [PubMed] [Google Scholar]
- 5.Bushon RN, Brady AM, Christensen ED, Stelzer EA. 2017. Multi-year microbial source tracking study characterizing fecal contamination in an urban watershed. Water Environ Res 89:127–143. doi: 10.2175/106143016X14798353399412. [DOI] [PubMed] [Google Scholar]
- 6.Sidhu JPS, Ahmed W, Gernjak W, Aryal R, McCarthy D, Palmer A, Kolotelo P, Toze S. 2013. Sewage pollution in urban stormwater runoff as evident from the widespread presence of multiple microbial and chemical source tracking markers. Sci Total Environ 463–464: 488–496. doi: 10.1016/j.scitotenv.2013.06.020. [DOI] [PubMed] [Google Scholar]
- 7.Propst CW, Harwood VJ, Morrison G. 2011. Case studies of urban and suburban watersheds, p 433–450. In Hagedorn C, Blanch A, Harwood V (ed), Microbial source tracking: methods, applications, and case studies. Springer, New York, NY. [Google Scholar]
- 8.Cho KH, Pachepsky Y, Kim JH, Guber A, Shelton D, Rowland R. 2010. Release of Escherichia coli from the bottom sediment in a first-order creek: experiment and reach-specific modeling. J Hydrol 391:322–332. doi: 10.1016/j.jhydrol.2010.07.033. [DOI] [Google Scholar]
- 9.Garzio-Hadzick A, Shelton D, Hill R, Pachepsky Y, Guber A, Rowland R. 2010. Survival of manure-borne E. coli in streambed sediment: effects of temperature and sediment properties. Water Res 44:2753–2762. doi: 10.1016/j.watres.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 10.Burton GA Jr, Gunnison D, Lanza GR. 1987. Survival of pathogenic bacteria in various freshwater sediments. Appl Environ Microbiol 53:633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sauvé S, Aboulfadl K, Dorner S, Payment P, Deschamps G, Prévost M. 2012. Fecal coliforms, caffeine and carbamazepine in stormwater collection systems in a large urban area. Chemosphere 86:118–123. doi: 10.1016/j.chemosphere.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 12.Sauer EP, VandeWalle JL, Bootsma MJ, McLellan SL. 2011. Detection of the human specific Bacteroides genetic marker provides evidence of widespread sewage contamination of stormwater in the urban environment. Water Res 45:4081–4091. doi: 10.1016/j.watres.2011.04.049. [DOI] [PubMed] [Google Scholar]
- 13.Parker JK, McIntyre D, Noble RT. 2010. Characterizing fecal contamination in stormwater runoff in coastal North Carolina, USA. Water Res 44:4186–4194. doi: 10.1016/j.watres.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 14.Torno HC, Marsalek J, Desbordes M. 2013. Urban runoff pollution. Springer Verlag, Heidelberg, Germany. [Google Scholar]
- 15.Aryal R, Kandasamy J, Vigneswaran S, Naidu R, Lee S. 2009. Review of stormwater quality, quantity and treatment methods part 1: stormwater quantity modelling. Environ Eng Res 14:71–78. doi: 10.4491/eer.2009.14.2.071. [DOI] [Google Scholar]
- 16.Petersen TM, Rifai HS, Suarez MP, Stein AR. 2005. Bacteria loads from point and nonpoint sources in an urban watershed. J Environ Eng 131:1414–1425. doi: 10.1061/(ASCE)0733-9372(2005)131:10(1414). [DOI] [Google Scholar]
- 17.Lee D, Lee H, Trevors JT, Weir SC, Thomas JL, Habash M. 2014. Characterization of sources and loadings of fecal pollutants using microbial source tracking assays in urban and rural areas of the Grand River Watershed, Southwestern Ontario. Water Res 53:123–131. doi: 10.1016/j.watres.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Kleinheinz GT, McDermott CM, Hughes S, Brown A. 2009. Effects of rainfall on E. coli concentrations at Door County, Wisconsin beaches. Int J Microbiol 2009:876050. doi: 10.1155/2009/876050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ackerman D, Weisberg SB. 2003. Relationship between rainfall and beach bacterial concentrations on Santa Monica Bay beaches. J Water Health 1:85–89. doi: 10.2166/wh.2003.0010. [DOI] [PubMed] [Google Scholar]
- 20.Galfi H, Haapala J, Nordqvist K, Westerlund C, Blecken G, Marsalek J, Viklander M. 2016. Inter-event and intra-event variations of indicator bacteria concentrations in the storm sewer system of the city of Östersund, Sweden. J Environ Eng 142:06016003. doi: 10.1061/(ASCE)EE.1943-7870.0001067. [DOI] [Google Scholar]
- 21.Tiefenthaler L, Stein ED, Schiff KC. 2011. Levels and patterns of fecal indicator bacteria in stormwater runoff from homogenous land use sites and urban watersheds. J Water Health 9:279–290. doi: 10.2166/wh.2010.056. [DOI] [PubMed] [Google Scholar]
- 22.Krometis LH, Characklis GW, Simmons OD III, Dilts MJ, Likirdopulos CA, Sobsey MD. 2007. Intra-storm variability in microbial partitioning and microbial loading rates. Water Res 41:506–516. doi: 10.1016/j.watres.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 23.Staley ZR, Grabuski J, Sverko E, Edge TA. 2016. Comparison of microbial and chemical source tracking markers to identify fecal contamination sources in the Humber River (Toronto, Ontario, Canada) and associated storm water outfalls. Appl Environ Microbiol 82:6357–6366. doi: 10.1128/AEM.01675-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newton RJ, Bootsma MJ, Morrison HG, Sogin ML, McLellan SL. 2013. A microbial signature approach to identify fecal pollution in the waters off an urbanized coast of Lake Michigan. Microb Ecol 65:1011–1023. doi: 10.1007/s00248-013-0200-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diston D, Sinreich M, Zimmermann S, Baumgartner A, Felleisen R. 2015. Evaluation of molecular-and culture-dependent MST markers to detect fecal contamination and indicate viral presence in good quality groundwater. Environ Sci Technol 49:7142–7151. doi: 10.1021/acs.est.5b00515. [DOI] [PubMed] [Google Scholar]
- 26.Cao Y, Van De Werfhorst Laurie C, Dubinsky EA, Badgley BD, Sadowsky MJ, Andersen GL, Griffith JF, Holden PA. 2013. Evaluation of molecular community analysis methods for discerning fecal sources and human waste. Water Res 47:6862–6872. doi: 10.1016/j.watres.2013.02.061. [DOI] [PubMed] [Google Scholar]
- 27.Harwood VJ, Staley C, Badgley BD, Borges K, Korajkic A. 2014. Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS Microbiol Rev 38:1–40. doi: 10.1111/1574-6976.12031. [DOI] [PubMed] [Google Scholar]
- 28.Wang D, Silkie SS, Nelson KL, Wuertz S. 2010. Estimating true human and animal host source contribution in quantitative microbial source tracking using the Monte Carlo method. Water Res 44:4760–4775. doi: 10.1016/j.watres.2010.07.076. [DOI] [PubMed] [Google Scholar]
- 29.McLellan SL, Eren AM. 2014. Discovering new indicators of fecal pollution. Trends Microbiol 22:697–706. doi: 10.1016/j.tim.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shanks OC, Newton RJ, Kelty CA, Huse SM, Sogin ML, McLellan SL. 2013. Comparison of the microbial community structures of untreated wastewaters from different geographic locales. Appl Environ Microbiol 79:2906–2913. doi: 10.1128/AEM.03448-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahmed W, Staley C, Sadowsky MJ, Gyawali P, Sidhu JP, Palmer A, Beale DJ, Toze S. 2015. Toolbox approaches using molecular markers and 16S rRNA gene amplicon data sets for identification of fecal pollution in surface water. Appl Environ Microbiol 81:7067–7077. doi: 10.1128/AEM.02032-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Unno T, Jang J, Han D, Kim JH, Sadowsky MJ, Kim OS, Chun J, Hur HG. 2010. Use of barcoded pyrosequencing and shared OTUs to determine sources of fecal bacteria in watersheds. Environ Sci Technol 44:7777–7782. doi: 10.1021/es101500z. [DOI] [PubMed] [Google Scholar]
- 33.Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, Bushman FD, Knight R, Kelley ST. 2011. Bayesian community-wide culture-independent microbial source tracking. Nat Methods 8:761–763. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reference deleted.
- 35.Flores GE, Bates ST, Knights D, Lauber CL, Stombaugh J, Knight R, Fierer N. 2011. Microbial biogeography of public restroom surfaces. PLoS One 6:e28132. doi: 10.1371/journal.pone.0028132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prussin AJ II, Marr LC. 2015. Sources of airborne microorganisms in the built environment. Microbiome 3:78. doi: 10.1186/s40168-015-0144-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shin H, Pei Z, Martinez KA II, Rivera-Vinas JI, Mendez K, Cavallin H, Dominguez-Bello MG. 2015. The first microbial environment of infants born by C-section: the operating room microbes. Microbiome 3:59. doi: 10.1186/s40168-015-0126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hewitt KM, Mannino FL, Gonzalez A, Chase JH, Caporaso JG, Knight R, Kelley ST. 2013. Bacterial diversity in two neonatal intensive care units (NICUs). PLoS One 8:e54703. doi: 10.1371/journal.pone.0054703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown CM, Staley C, Wang P, Dalzell B, Chun CL, Sadowsky MJ. 2017. A high-throughput DNA-sequencing approach for determining sources of fecal bacteria in a Lake Superior estuary. Environ Sci Technol 51:8263–8271. doi: 10.1021/acs.est.7b01353. [DOI] [PubMed] [Google Scholar]
- 40.Henry R, Schang C, Coutts S, Kolotelo P, Prosser T, Crosbie N, Grant T, Cottam D, O'Brien P, Deletic A, McCarthy D. 2016. Into the deep: evaluation of SourceTracker for assessment of faecal contamination of coastal waters. Water Res 93:242–253. doi: 10.1016/j.watres.2016.02.029. [DOI] [PubMed] [Google Scholar]
- 41.McCarthy DT, Jovanovic D, Lintern A, Teakle I, Barnes M, Deletic A, Coleman R, Rooney G, Prosser T, Coutts S, Hipsey MR, Bruce LC, Henry R. 2017. Source tracking using microbial community fingerprints: method comparison with hydrodynamic modelling. Water Res 109:253–265. doi: 10.1016/j.watres.2016.11.043. [DOI] [PubMed] [Google Scholar]
- 42.NDEQ. 2006. 2006 surface water quality integrated report. Water Quality Division, Nebraska Department of Environmental Quality, Lincoln, NE. [Google Scholar]
- 43.McCarthy D, Hathaway J, Hunt W, Deletic A. 2012. Intra-event variability of Escherichia coli and total suspended solids in urban stormwater runoff. Water Res 46:6661–6670. doi: 10.1016/j.watres.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 44.Hathaway J, Hunt W, Simmons O III. 2010. Statistical evaluation of factors affecting indicator bacteria in urban storm-water runoff. J Environ Eng 136:1360–1368. doi: 10.1061/(ASCE)EE.1943-7870.0000278. [DOI] [Google Scholar]
- 45.Lampman G, Caraco N, Cole J. 1999. Spatial and temporal patterns of nutrient concentration and export in the tidal Hudson River. Estuaries 22:285–296. doi: 10.2307/1352984. [DOI] [Google Scholar]
- 46.Jaworski NA. 1981. Sources of nutrients and the scale of eutrophication problems in estuaries, p 83–110. In Neilson BJ, Cronin LE (ed), Estuaries and nutrients. Contemporary issues in science and society. Humana Press, New York, NY. [Google Scholar]
- 47.Baral D, Fisher JR, Florek MJ, Dvorak BI, Snow DD, Admiraal DM. 2017. Atmospheric contributions of nitrate to stormwater runoff from two urban watersheds. J Environ Eng 144:05017009. doi: 10.1061/(ASCE)EE.1943-7870.0001323. [DOI] [Google Scholar]
- 48.Lim B, Jickells T, Davies T. 1991. Sequential sampling of particles, major ions and total trace metals in wet deposition. Atmos Environ Part A 25:745–762. doi: 10.1016/0960-1686(91)90073-G. [DOI] [Google Scholar]
- 49.Mhuireach G, Johnson BR, Altrichter AE, Ladau J, Meadow JF, Pollard KS, Green JL. 2016. Urban greenness influences airborne bacterial community composition. Sci Total Environ 571:680–687. doi: 10.1016/j.scitotenv.2016.07.037. [DOI] [PubMed] [Google Scholar]
- 50.Lymperopoulou DS, Adams RI, Lindow SE. 2016. Contribution of vegetation to the microbial composition of nearby outdoor air. Appl Environ Microbiol 82:3822–3833. doi: 10.1128/AEM.00610-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bott TL, Kaplan LA. 1985. Bacterial biomass, metabolic state, and activity in stream sediments: relation to environmental variables and multiple assay comparisons. Appl Environ Microbiol 50:508–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jamieson R, Joy DM, Lee H, Kostaschuk R, Gordon R. 2005. Transport and deposition of sediment-associated Escherichia coli in natural streams. Water Res 39:2665–2675. doi: 10.1016/j.watres.2005.04.040. [DOI] [PubMed] [Google Scholar]
- 53.Pandey PK, Soupir ML. 2014. Assessing linkages between E. coli levels in streambed sediment and overlying water in an agricultural watershed in Iowa during the first heavy rain event of the season. Trans ASABE 57:1571–1581. doi: 10.13031/trans.57.10371. [DOI] [Google Scholar]
- 54.Gao G, Falconer RA, Lin B. 2013. Modelling importance of sediment effects on fate and transport of enterococci in the Severn Estuary, UK. Mar Pollut Bull 67:45–54. doi: 10.1016/j.marpolbul.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 55.Rehmann CR, Soupir ML. 2009. Importance of interactions between the water column and the sediment for microbial concentrations in streams. Water Res 43:4579–4589. doi: 10.1016/j.watres.2009.06.049. [DOI] [PubMed] [Google Scholar]
- 56.Koirala SR, Gentry RW, Perfect E, Schwartz JS, Sayler GS. 2008. Temporal variation and persistence of bacteria in streams. J Environ Qual 37:1559–1566. doi: 10.2134/jeq2007.0310. [DOI] [PubMed] [Google Scholar]
- 57.Hatt BE, Fletcher TD, Walsh CJ, Taylor SL. 2004. The influence of urban density and drainage infrastructure on the concentrations and loads of pollutants in small streams. Environ Manage 34:112–124. [DOI] [PubMed] [Google Scholar]
- 58.Fisher J. 2011. Water quality models for stormwater runoff in two Lincoln, Nebraska urban watersheds. Masters thesis University of Nebraska-Lincoln, Lincoln, NE. [Google Scholar]
- 59.Bannerman RT, Owens DW, Dodds RB, Hornewer NJ. 1993. Sources of pollutants in Wisconsin stormwater. Water Sci Technol 28:241–259. doi: 10.2166/wst.1993.0426. [DOI] [Google Scholar]
- 60.Zarriello PJ, Breault RF, Weiskel PK. 2002. Potential effects of structural controls and street sweeping on stormwater loads to the lower Charles River, Massachusetts. Water-Resources Investigations Report 02-4220. U.S. Geological Survey, Richmond, VA; https://pubs.usgs.gov/wri/wri024220/. [Google Scholar]
- 61.NDEQ. 2007. Total maximum daily loads for Antelope Creek: LP2-20900. Parameters of concern: total ammonia and E. coli. Water Quality Division, Nebraska Department of Environmental Quality, Lincoln, NE. [Google Scholar]
- 62.Jang Y, Jain P, Tolaymat T, Dubey B, Townsend T. 2009. Characterization of pollutants in Florida street sweepings for management and reuse. J Environ Manage 91:320–327. doi: 10.1016/j.jenvman.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 63.Elbroch M, Kresky M, Evans JW. 2012. Field guide to animal tracks and scat of California. University of California Press, Los Angeles, CA. [Google Scholar]
- 64.Elbroch M. 2003. Mammal tracks & sign: a guide to North American species. Stackpole Books, Mechanicsburg, PA. [Google Scholar]
- 65.Elbroch M, Marks EM, Boretos CD. 2001. Bird tracks & sign: a guide to North American species. Stackpole Books, Mechanicsburg, PA. [Google Scholar]
- 66.Eichmiller JJ, Hicks RE, Sadowsky MJ. 2013. Distribution of genetic markers of fecal pollution on a freshwater sandy shoreline in proximity to wastewater effluent. Environ Sci Technol 47:3395–3402. doi: 10.1021/es305116c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.APHA. 2012. Standard methods for the examination of water and wastewater. American Public Health Association, Washington, DC. [Google Scholar]
- 68.ASTM. 2007. ASTM D422-63(2007)e2 Standard test method for particle-size analysis of soils (withdrawn 2016). ASTM International, West Conshohocken, PA. doi: 10.1520/D0422-63R07E02. [DOI] [Google Scholar]
- 69.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 73.Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. 2013. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.R Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 75.Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer, New York, NY. [Google Scholar]
- 76.Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x. [DOI] [Google Scholar]
- 77.Legendre P, Legendre LF. 2012. Numerical ecology, 3rd ed Elsevier, Oxford, United Kingdom. [Google Scholar]
- 78.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H. 2011. The vegan package: community ecology package. R package version 2.0-2. The vegan package: community ecology Package.R package version 2.0-2. [Google Scholar]
- 79.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.